Abstract
The topological similarity of voltage-gated proton channels (HV1s) to the voltage-sensing domain (VSD) of other voltage-gated ion channels raises the central question of whether HV1s have a similar structure. We present the construction and validation of a homology model of the human HV1 (hHV1). Multiple structural alignment was used to construct structural models of the open (proton-conducting) state of hHV1 by exploiting the homology of hHV1 with VSDs of K+ and Na+ channels of known three-dimensional structure. The comparative assessment of structural stability of the homology models and their VSD templates was performed using massively repeated molecular dynamics simulations in which the proteins were allowed to relax from their initial conformation in an explicit membrane mimetic. The analysis of structural deviations from the initial conformation based on up to 125 repeats of 100-ns simulations for each system reveals structural features consistently retained in the homology models and leads to a consensus structural model for hHV1 in which well-defined external and internal salt-bridge networks stabilize the open state. The structural and electrostatic properties of this open-state model are compatible with proton translocation and offer an explanation for the reversal of charge selectivity in neutral mutants of Asp112. Furthermore, these structural properties are consistent with experimental accessibility data, providing a valuable basis for further structural and functional studies of hHV1. Each Arg residue in the S4 helix of hHV1 was replaced by His to test accessibility using Zn2+ as a probe. The two outermost Arg residues in S4 were accessible to external solution, whereas the innermost one was accessible only to the internal solution. Both modeling and experimental data indicate that in the open state, Arg211, the third Arg residue in the S4 helix in hHV1, remains accessible to the internal solution and is located near the charge transfer center, Phe150.
INTRODUCTION
Voltage-gated proton channels (HV1s) enable phagocytes to kill pathogens (Henderson et al., 1988; DeCoursey, 2010; Demaurex and El Chemaly, 2010), basophils to secrete histamine (Musset et al., 2008), airway epithelia to control surface pH (Fischer, 2012), sperm to capacitate and fertilize eggs (Lishko et al., 2010), and B lymphocyte signaling (Capasso et al., 2010), and may exacerbate breast cancer metastasis (Wang et al., 2012) and ischemic brain damage (Wu et al., 2012). When the gene was discovered in 2006, however, the most remarkable feature of the human HV1 (hHV1) protein was its resemblance to the voltage-sensing domain (VSD) of other voltage-gated ion channels (Ramsey et al., 2006; Sasaki et al., 2006).
The VSD is a protein module that confers the ability to respond to potential changes across a membrane (Jiang et al., 2003; Bezanilla, 2008; Swartz, 2008). Classes of proteins with VSDs include many voltage-gated cation channels, HV1s, voltage-sensing phosphatases (VSPs), and C15orf27 proteins of unknown function. The VSD contains four transmembrane (TM) segments, S1–S4, and intervening intracellular and extracellular loops. Voltage-sensitive cation channels contain one to several repeats of the fundamental unit comprising a VSD and a TM pore segment consisting of two TM regions; the fundamental unit may have additional N- and C-terminal domains that confer, for example, the ability to respond to cyclic nucleotides. Cation channels arrange themselves in the membrane so that four VSDs surround an ion pore that assembles from four pairs of TM helices (Swartz, 2008). The conduction pathway in hHV1 is contained within S1–S4 and does not require accessory proteins (Lee et al., 2009). Our aim is to define the structure of the conducting (open) conformation of hHV1.
Several structural features characterize VSDs. The S4 helix contains two to seven positively charged residues (mostly arginine and less frequently lysine), each separated by two hydrophobic residues (Bezanilla, 2008). S1–S3 contain negatively charged residues thought to form both intracellular and extracellular charge clusters together with the cationic charges in S4 (Papazian et al., 1995; Long et al., 2007; Swartz, 2008; Jensen et al., 2012). Finally, VSDs contain a “gating charge transfer center” (Tao et al., 2010) or “hydrophobic center” (Yarov-Yarovoy et al., 2012) characterized by a highly conserved phenylalanine residue on S2, thought to delimit internal from external access. A wealth of evidence, including crystal structures of potassium and, more recently, sodium channels, indicates that the S4 helix of the VSD moves in response to the membrane potential, and that the mechanical transduction of this motion opens or closes the pore (Bezanilla, 2008; Swartz, 2008; Gonzalez et al., 2013). As S4 moves, its arginines participate in salt bridges with intracellular and extracellular charge clusters, which are separated by the constriction at the charge transfer center (Papazian et al., 1995; Tiwari-Woodruff et al., 2000; Long et al., 2007; Tao et al., 2010; Lin et al., 2011).
We previously undertook a phylogenetic analysis (Musset et al., 2011) that included VSDs not only from eukaryotic voltage-gated potassium (KV), sodium (NaV), and calcium channels (CaV), but also from VSD homologues that lack an ion pore (HV1, VSP, and C15orf27). This analysis showed that the VSDs that lack an ion pore comprise a subfamily distinct from the VSDs of eukaryotic cation channels. Despite this subfamily occupying a separate branch of the phylogenetic tree, several lines of evidence indicate that its S4 moves qualitatively like the S4 of other VSDs (Murata et al., 2005; Okamura et al., 2009; Gonzalez et al., 2010; 2013). In VSPs, this motion presumably controls the activity of the phosphatase; in hHV1s, this motion is thought to regulate proton conduction. The VSDs of CiVSP and of human C15orf27 do not appear to conduct protons or other ions (Murata et al., 2005; Okamura et al., 2009; Musset et al., 2011). VSDs of voltage-gated potassium and sodium channels do not conduct ions but can be converted into voltage-gated proton channels by point mutations on the S4 segment (Starace et al., 1997; Starace and Bezanilla, 2001, 2004; Struyk and Cannon, 2007; Gosselin-Badaroudine et al., 2012). The hHV1s are VSD homologues that lack a separate ion pore (Ramsey et al., 2006; Sasaki et al., 2006) but include a proton-specific conduction pathway within the VSD (Lee et al., 2009; Musset et al., 2011).
Recently, Musset et al. (2011) discovered that Asp112 is a crucial component of the proton selectivity filter in hHV1. Neutralizing mutants at position 112 not only exhibited impaired proton selectivity but were anion selective. Only the conservative Asp→Glu mutant remained proton selective. The presence of a carboxyl side chain is not per se sufficient to provide selectivity, as Asp185 is also in the pore but can be neutralized without affecting selectivity (Musset et al., 2011). These findings raise important questions regarding a mechanistic understanding of proton-specific conduction. How does Asp112 modulate ionic selectivity? What other residues form the selectivity filter? Do titratable groups of the protein participate directly in proton relay, or does proton translocation occur via a water-mediated mechanism as in simple, water-filled pores such as the gramicidin channel (Levitt et al., 1978; Pomès and Roux, 1996). Explaining the molecular mechanism of proton conduction of hHV1 requires a deeper exploration of molecular structure.
Given the absence of an experimentally determined structure of hHV1, we used homology modeling to generate a structural model of hHV1 from its primary amino acid sequence using sequence alignments and structural templates of experimentally determined homologous VSDs. On the one hand, this task is made possible by the availability of high resolution atomic structures of homologous VSDs (Jiang et al., 2003; Lee et al., 2005; Long et al., 2007; Payandeh et al., 2011). On the other hand, below-average sequence identity between hHV1 and its homologues makes homology modeling a difficult task. A minimum 30% sequence identity is typically pursued in generating homology models (Forrest et al., 2006). Homology models based on low sequence identity have been regarded as likely to be inaccurate (Jacobson and Sali, 2004). Therefore, sequence alignment for hHV1 is nontrivial.
Here, we focus on the structure of the open conformation of hHV1 as a starting point for future studies of closed states, gating, and other properties. We used a three-pronged approach to gain insight into the structural basis for the function of hHV1. First, a more comprehensive phylogenetic analysis of VSD sequences was undertaken, which provided a more reliable understanding of the relationships among VSDs and a more structurally informed alignment of the arginines that control S4 movement. Second, we used alternative alignments and multiple structural templates to develop several homology models of the VSD of hHV1. We performed structure-stability tests of two select models, one based on our phylogenetically informed alignment and the other based on another widely accepted alignment (Ramsey et al., 2010; Wood et al., 2012). To perform these tests, the computational efficiency afforded by a lipid membrane mimetic was combined with an ensemble approach consisting of many molecular dynamics (MD) simulation repeats to obtain meaningful statistics. The analysis of these simulations allowed us to choose a preferred homology model that was further validated by simulations in a phospholipid bilayer, and led to a testable hypothesis regarding the solvent accessibility of the S4 arginines of hHV1 in the open state. Finally, we used His scanning of the S4 Arg residues to evaluate accessibility by Zn2+ effects on currents under voltage clamp. Previous studies have evaluated accessibility by mutating specific residues to Cys (Cys scanning) and then using MTS reagents as probes, detected either by Western blotting (“PEGylation” assay) (Sakata et al., 2010) or by altered channel gating (Gonzalez et al., 2010). The Zn2+ accessibility results support the hypothesis generated by the computational study and provide a new basis for further mechanistic investigations into hHV1.
MATERIALS AND METHODS
Sampling of taxa and phylogenetic methods
Sequences of VSD homologues were identified by similarity using BLAST (Altschul et al., 1997) and HMMer (Finn et al., 2011) searching, and an effort was made to include prokaryotic and fungal sequences as well as a diversity of metazoan sequences (e.g., plants, invertebrates, and vertebrates). VSD sequences were aligned using PromalS3D (Pei et al., 2008), constraining the alignment to two VSD crystal structures, 2R9R (paddle chimera; Long et al., 2007) and 3RVY (NaVAb; Payandeh et al., 2011). Two other crystal structures of KV channels were deliberately omitted to avoid biasing the structural alignment toward a KV-type structure. The resulting alignment was inspected visually and analyzed with Maxalign (Gouveia-Oliveira et al., 2007). TM helices were identified by homology, and intervening loops were removed. Sequences were submitted to Prottest, which predicted the LG amino acid substitution model (Le and Gascuel, 2008) with I (proportion of invariant sites) and G model (gamma shape) parameters. A maximum likelihood phylogenetic tree was generated from this alignment using PhyML (Guindon and Gascuel, 2003) at the ATGC site (http://www.atgc-montpellier.fr) using the LG model, four substitution rate categories, estimated proportion of variable sites, estimated gamma shape parameter, and nearest neighbor plus subtree pruning and regrafting tree improvements. The sequence order in the TM-only alignment was randomized, and parsimony trees were generated from subsets of the randomized full alignment using Protpars (Phylogeny Inference Package [PHYLIP] version 3.5c) at the Mobyle site (Néron et al., 2009); maximum likelihood trees from the same subsets were also generated using PhyML. A different alignment was constructed from the full set of sequences using the default TCoffee program (Notredame et al., 2000) in which there is no structural constraint, and a maximum likelihood tree was generated using PhyML. Trees were visualized at ITOL (Letunic and Bork, 2007) and in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Gene expression
Site-directed mutants were created using the Stratagene QuikChange (Agilent Technologies) procedure according to the manufacturer’s instructions. Clones were sequenced commercially to confirm the mutations. COS-7 cells were maintained in 5% CO2 and 95% air in a humidified incubator at 37°C in Dulbecco’s modified Eagle’s medium containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 IU/ml penicillin, and 100 µg/ml streptomycin. The coding sequence of hHV1 (Hvcn1) was cloned into pQBI25-fC3 (to make GFP-HV1) vectors as described previously (Ramsey et al., 2006). COS-7 cells were grown to ∼80% confluency in 35-mm cultures dishes. Cells were transfected with ∼0.5 µg of the appropriate cDNA using Lipofectamine 2000 (Invitrogen) or polyethylenimine (Sigma-Aldrich). After a 6-h incubation at 37°C in 5% CO2, cells were trypsinized and replated onto glass coverslips at low density in medium containing fetal bovine serum for patch-clamp recording.
Electrophysiology
GFP-tagged proton channels were studied. Fluorescent cells were identified using inverted microscopes with fluorescence capability (Nikon). Micropipettes were pulled using an automatic pipette puller (Flaming Brown; Sutter Instrument) from glass (PG52165-4; World Precision Instruments, Inc.) coated with Sylgard 184 (Dow Corning Corp.) and heat polished to a tip resistance ranging typically from 5 to 15 MΩ with the pipette solutions used. Electrical contact with the pipette solution was achieved by a thin sintered Ag-AgCl pellet (In Vivo Metric Systems) attached to a Teflon-encased silver wire, or simply a chlorided silver wire. A reference electrode made from a Ag-AgCl pellet was connected to the bath through an agar bridge made with Ringer’s solution (mM: 160 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, and 5 HEPES, pH 7.4). The current signal from the patch clamp (EPC-9; HEKA, or Axopatch 200B; Axon Instruments) was recorded and analyzed using Laboratory View SCB-68 (National Instruments), Pulse and PulseFit (HEKA), or P-CLAMP (Molecular Devices) software supplemented by Microsoft Excel, Origin 7.5, and Sigmaplot (SPSS Inc.). Seals were formed with Ringer’s solution in the bath, and the potential zeroed after the pipette was in contact with the cell. Where stated, reversal potentials were corrected for measured liquid junction potentials.
Inside-out patches were formed by forming a seal and then briefly lifting the pipette into the air or solution. For whole-cell and inside-out patch recording, bath and pipette solutions contained 60–100 mM buffer, 1–2 mM CaCl2 or MgCl2 (intracellular solutions were Ca2+ free), 1–2 mM EGTA, and TMAMeSO3 to adjust the osmolality to roughly 300 mOsm, titrated with TMAOH. Buffers used were Mes, pH 5.5–6.0, and PIPES, pH 7.0. We omitted EGTA from solutions for Zn2+ measurements. No leak correction has been applied to any current records. Measurements were done at 21°C or at room temperature (20–25°C). The bath temperature was controlled by a system from Brooks Industries or by an in-house–built system using Peltier devices in a feedback arrangement, monitored by a resistance temperature detector element (Omega Scientific) immersed in the bath.
Homology model
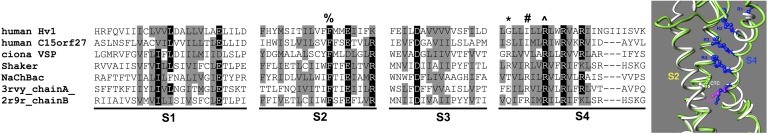
The VSD of potassium channel KVAP (Protein Data Bank accession no. 1ORS; Jiang et al., 2003), the paddle chimera (Protein Data Bank accession no. 2R9R; Long et al., 2007), and the VSD of chain A of the NaVAb sodium channel (Protein Data Bank accession no. 3RVY; Payandeh et al., 2011) were used as templates. These structures are presumed to be in the open state because they experience no potential during crystallization, so that their VSDs are presumed to be “activated.” The three template structures were structurally aligned using Modeller 9.7 (Sali and Blundell, 1993) to obtain a multiple-template, structure-based sequence alignment. The query sequence of hHV1 was then aligned to the template alignment using the large multiple sequence alignment of VSDs (abstracted in Fig. 1) as a guide; two alignments, “R3D” and “R2D” (differing in the register of the arginines in S4) were generated. Each alignment was then used to create a set of 100 homology models, and the best five models were selected by the smallest value of the normalized discrete optimized molecule energy (DOPE) (Shen and Sali, 2006) score available in Modeller 9.7.
Figure 1.
A subset of a larger multiple sequence alignment of the four TM regions of VSDs informed by alignment of the open state produced by structural superposition of crystal structures of the paddle chimera (green ribbon) and NaVAB (white ribbon). The arginines of S4 (blue sticks) are labeled according to their position on the paddle chimera. Note that the most extracellular R of NaVAB corresponds to R2 of the paddle chimera, and the most intracellular R of NaVAB corresponds to K5 of the paddle chimera. The completely conserved phenylalanine on S2 is shown and labeled as “CTC.” The following residues are indicated with symbols over the alignment: %, CTC; *, R1 of Shaker; #, R2 of Shaker; ^, R3 of Shaker.
The S1–S2 loop is of variable length in the four sequences considered: 7, 8, 32, and 15 residues, respectively, in KVAP, NaVAb, the paddle chimera, and hHV1. In the modeling of hHV1, the S1–S2 loop was by far the least reliable part of the homology model, as quantified by a DOPE score above −0.03. For this reason, in each of the selected models (five from each alignment), the S1–S2 loop (HV1 sequence LDLKIIQPDKNNYAA) was reconstructed using SuperLooper (Hildebrand et al., 2009). The starting structures were then energy minimized to correct the splice points. This procedure resulted in an acceptable DOPE score.
MD simulations
Octane simulations.
Each template and five starting configurations of each model were then embedded in a hydrated octane slab with an approximate thickness of 35 Å. Depending on the size of the protein, large enough box sizes were used to avoid boundary condition artifacts. KVAP and the paddle chimera were in a 6 × 6 × 7.5–nm3 box size, whereas NaVAb was in a 5 × 5 × 7.5–nm3 box size, with 349 and 227 octane molecules, respectively. Both homology models were in a 6 × 6 × 7.5–nm3 cubic box with 381 octane molecules. For a given box size, the number of water molecules varies with the length and conformation of the loops. The KVAP, paddle chimera, and NaVAb systems contained 4,700, 4,509, and 3,085 water molecules, respectively. R2D and R3D systems had a mean water content of 4,420 and 4,418 water molecules, respectively. Enough Na+ and Cl− ions were added to neutralize the systems and maintain an ionic concentration at 500 mM. The systems were first energy minimized in 1,000 steps, followed by an equilibration phase of 100 ps with backbone position restraints on protein and water oxygen atoms. This procedure allowed octane molecules to relax around the protein and at the water–octane interface until the octane density stabilized. After this equilibration step, 25 independent replicas for each system were set up by randomizing the starting velocities to initiate the equilibration with position restraints on backbone protein for another 100 ps. During an extra equilibration phase of 2 ns, proteins were treated as rigid bodies by restraining the distance between Cα atoms with a force constant of 1,000 kJ mol−1.
The production runs consisted of unrestrained simulations of 100 ns for each system, with subsequent extension to 200 ns for the R2D model. All simulations were generated using Gromacs (v4.0; Hess et al., 2008) using the OPLS-AA force field (Jorgensen et al., 1996) for protein and octane and the TIP3P model for water (Jorgensen et al., 1983). The integration time step was 2 fs. Twin-range cutoffs of 10 Å for the van der Waals interactions and 10 Å for direct electrostatic interactions calculated by particle-mesh Ewald (Essmann et al., 1995) were used together with 10-Å neighbor lists updated every 10 steps. Constant NPT conditions were applied using Parrinello–Rahman (Parrinello and Rahman, 1980) semi-isotropic pressure coupling in XY directions, with a constant pressure of one bar applied via a coupling constant of τp = 2.0 ps and zero compressibility in the z direction. The aqueous solution, octane, and the protein were coupled separately to a temperature bath at 300 K with a coupling constant of τT = 0.1 ps using the Nosé–Hoover algorithm (Nosé, 1984). The LINCS algorithm was used to constrain bond lengths (Hess et al., 1997).
Lipid bilayer simulations.
The x-ray structure of KVAP and the R2D model were simulated in a phospholipid bilayer. A configuration containing a 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) bilayer with 64 lipids per leaflet was obtained from Tieleman et al. (1999) and equilibrated at 300 K for 50 ns in the absence of any solute. The OPLS-AA protein force field was mixed with the Berger lipid parameters by applying the half-ε double-pairlist method (Chakrabarti et al., 2010). InflateGRO (Kandt et al., 2007) was used to embed the proteins in the bilayer. The bilayer was then hydrated, and 500 mM NaCl was added as in the octane simulations. The final KVAP system contained 126 POPC and 5,862 water molecules in a 6.6 × 6.9 × 7.8–nm3 box. The system was first energy minimized for 2,500 steps, followed by an equilibration phase of 25 ns with backbone position restraints on protein and an unrestrained production run of 430 ns. All parameters were the same as those of the octane simulation except for pressure coupling. A reference pressure of one bar was imposed by semi-isotropic pressure coupling separately in the directions of the bilayer plane and the z direction using a coupling time constant of 1.0 ps and equal compressibility.
Four representative conformations of R2D in its hydrated state (see Results), including pore water, were selected from octane simulations as the starting conformations for unrestrained 100-ns simulations in POPC after a 25-ns equilibration in the presence of position restraints on protein and pore water oxygen atoms. The 7.2 × 7.5 × 7.5–nm3 simulation cells contained 126 POPC and 5,877 water molecules on average. Simulations of the same four protein conformations were extended in octane to 100 ns. Molecular graphics were generated by VMD 1.8.7 (Humphrey et al., 1996), and all trajectories were analyzed using Gromacs tools and in-house–built codes.
Continuum electrostatic calculations
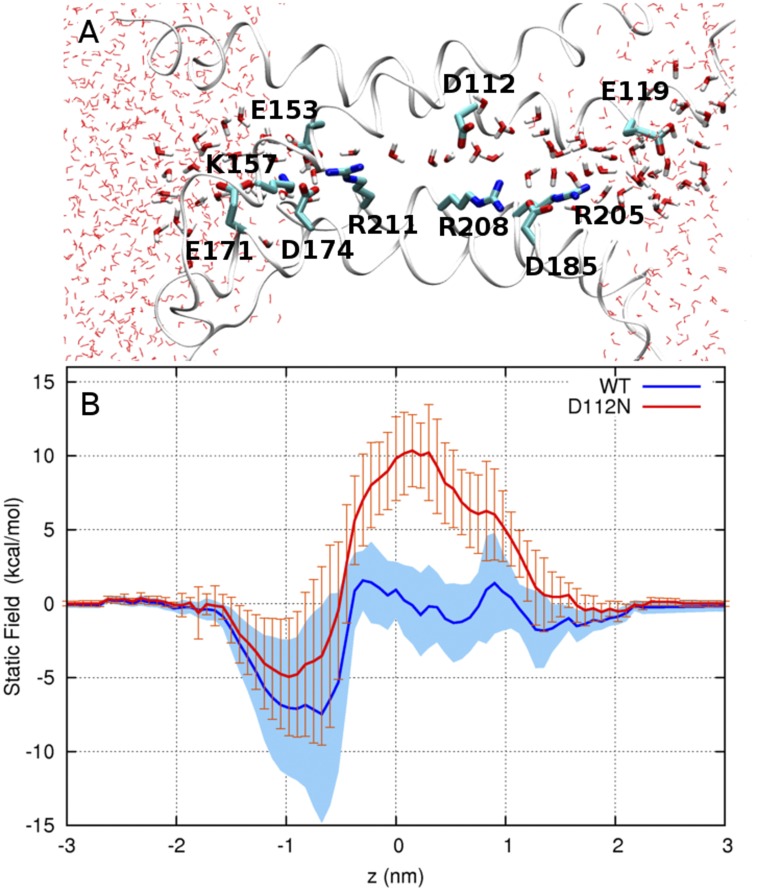
We calculated the static field for the passage of a positive point charge through the hHV1 pore by solving the Poisson–Boltzmann (PB) equation using the PBEQ module of CHARMM (version 31; Brooks et al., 1983; MacKerell et al., 1998), together with a set of optimized atomic radii for amino acids (Nina et al., 1997). The regions occupied by n-octane, protein, and water were assigned dielectric constants of 2, 4, and 80, respectively. 150 conformations were selected by picking every 50th snapshot from the combined trajectory segments in the fully hydrated conformation of the channel (III state in Fig. 7 E), in which a water pathway is present throughout the pore. For each of these snapshots, the coordinates of the oxygen atoms of water molecules along the pore were stored before deleting the water molecules along with ions and octane molecules. The probe point charge was placed at these O atom coordinates to calculate the static field for each protein conformation. The PB equation was solved on a grid of 115 × 115 × 141 points with an initial cell size of 1.0 Å, followed by focusing on a finer grid of 0.25 Å. The implicit membrane was assigned a thickness of 34 Å, and no membrane potential was applied. The calculation was repeated for the same set of snapshots after neutralizing the charge of D112 to mimic the effect of neutral mutants of that residue on charge transfer through the pore of hHV1.
Figure 7.
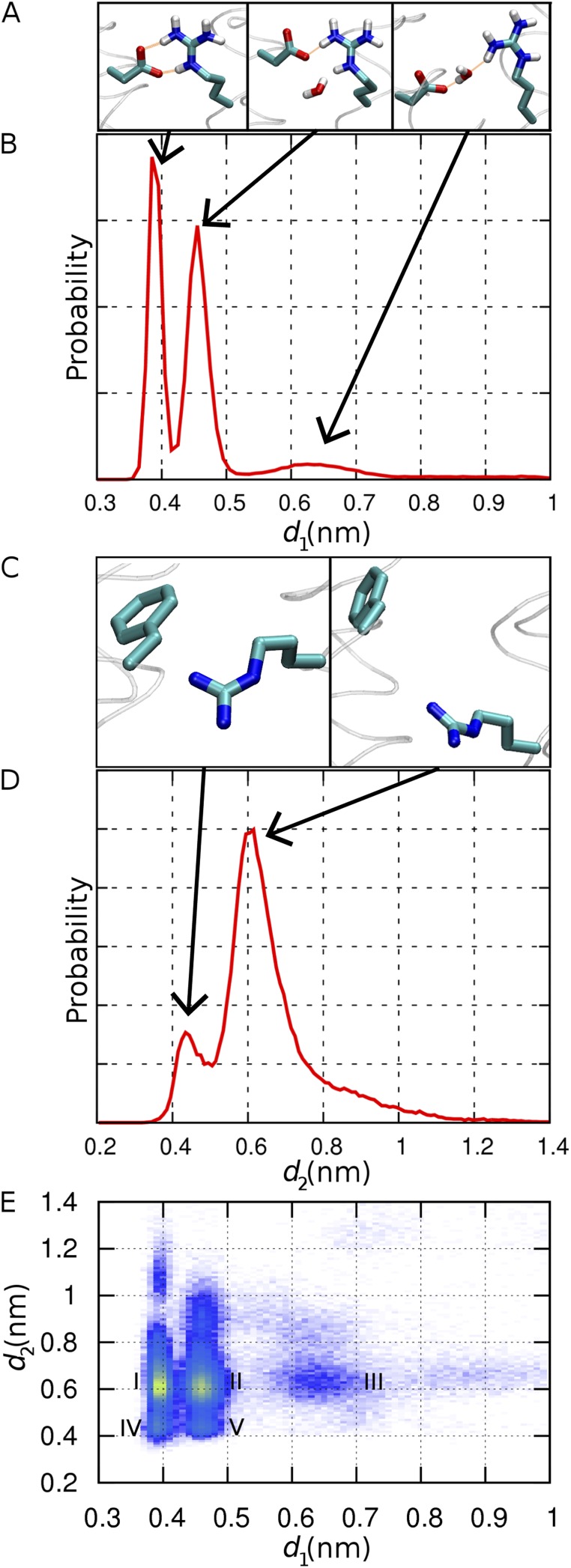
Structural fluctuations of the two channel constrictions in model R2D. (A) Configurations of the D112–R208 salt bridge (from left to right: bidentate, monodentate, and open). Hydrogen bonds are shown as orange lines. (B) Probability distribution of d1, the distance between atom Cγ of D112 and atom Cz of R208. (C) Closed (left) and open (right) conformations of F150 and R211. (D) Probability distribution of d2, the distance between the center of the benzyl ring of F150 and atom Cz of R211. (E) Two-dimensional probability distribution of d1 and d2, with increasing probability from purple to yellow.
Online supplemental material
The online supplemental material includes VSD sequence information (Table S1), a plot of the root mean square deviation (RMSD) versus time for KVAP in a phospholipid bilayer and in octane (Fig. S1), representative conformations of the two homology models (Fig. S2), and an analysis of salt-bridge populations in the two homology models (Fig. S3). The supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201210856/DC1.
RESULTS
Multiple sequence alignment and phylogenetic analysis
A previous multiple sequence alignment and phylogenetic analysis including 123 genes showed that the VSDs of HV1s, VSPs, and C15orf27s comprise a subfamily distinct from other, mainly eukaryotic VSDs (Musset et al., 2011). To get a more comprehensive view of the relationships among VSD sequences, a multiple sequence alignment of almost 300 diverse VSD sequences was created using PromalS3D (Pei et al., 2008), an algorithm that incorporates both structural and sequence information. The sequences were chosen to include preferentially VSDs known to respond directly to membrane potential and were drawn from a wide spectrum of organisms. The parameters were chosen so that the algorithm used the VSDs from the NaVAb crystal structure (3RVY) and the paddle-chimera crystal structure (2R9R) to constrain the alignment structurally. A representative subset of the TM-only alignment is shown in Fig. 1.
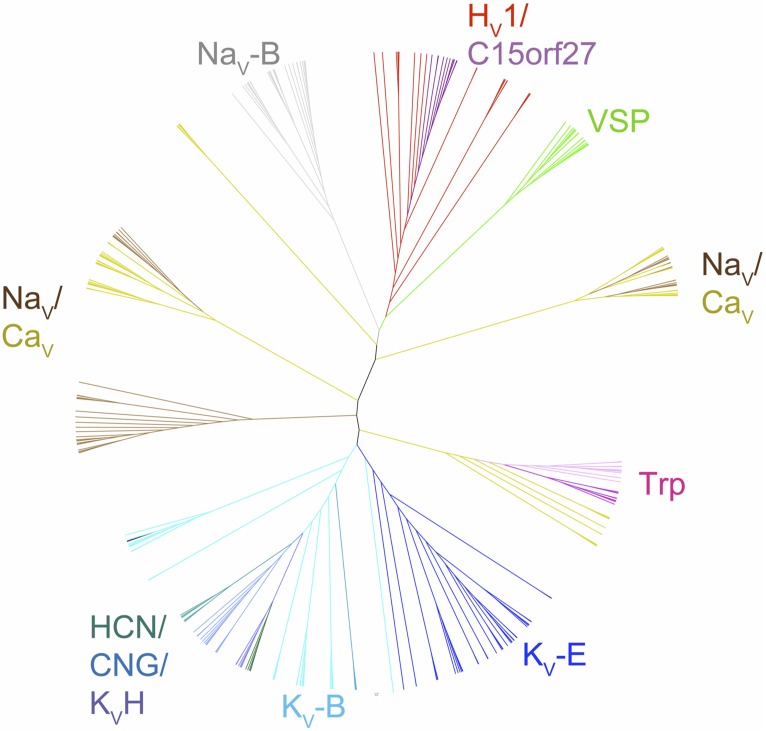
The tree produced by maximum likelihood phylogenetic analysis of this multiple sequence alignment (Fig. 2) has very similar topology to trees previously constructed both from full-length ion channel sequences and from the sequences of the ion pores only (Nelson et al., 1999; Yu and Catterall, 2004). As was concluded in the previous full-length and pore-only alignments, it seems clear from this tree that KV- and NaV/CaV-type channels separated in bacteria, at the latest approximately one billion years ago. The tree in Fig. 2 also agrees with topologies of previous trees obtained from pore alignments in that it shows VSDs from CNG-type channels as more closely related to VSDs from KV channels, and VSDs from transient receptor potential channels as more closely related to VSDs of NaV/CaV channels. Interestingly, phylogenies created from various multiple sequence alignments (see below) of VSD sequences contain no indication of NaV/CaV-type VSDs being associated with KV pores, or vice versa, indicating that swapping VSDs and pore domains among the ion channel families is extremely rare to nonexistent. The similarity of the phylogeny based on pore-only and the VSD-only (pore-excluded) sequences reinforces the validity of both.
Figure 2.
Phylogenetic tree constructed from the multiple sequence alignment of VSDs exemplified in Fig. 1. Branches and names of subfamilies of VSDs are color coded. KV-E, eukaryotic potassium channel; KV-B, prokaryotic potassium channel; KV-H, H family potassium channel; HCN, eukaryotic hyperpolarization–activated, cyclic nucleotide–gated; CNG, cyclic nucleotide–gated channel; Trp, transient receptor potential channel; NaV-E, eukaryotic sodium channel; NaV-B, prokaryotic sodium channel; CaV-E, eukaryotic calcium channel; HV1, voltage-gated proton channel; VSP, voltage-sensitive phosphatase; C15orf27, homologues of human C15orf27, a protein of unknown function.
The phylogenetic analysis of VSDs shows that the HV1/C15orf27/VSP subfamily lies on the same branch that contains the bacterial sodium channel VSDs (Fig. 2). This relationship is robust: it is preserved in trees constructed with different subsets of taxa, in trees constructed using parsimony as a criterion, and in trees derived from different underlying alignments (Table S1). In addition, BLAST and HMMer searches of databases using probes from the HV1/C15orf27/VSP subfamily return sodium channels as the top scoring hits outside of the HV1/C15orf27/VSP subfamily. These results strongly indicate that the VSD subfamily exemplified by HV1/C15orf27/VSP, i.e., VSDs not associated with a separate ion pore, became distinct from the KV-type VSD at or before the split between NaV/CaV and KV channels, which occurred in bacteria.
We now focus on the charged residues in the S4 region that are considered to sense membrane potential. The full multiple sequence alignment (abstracted in Fig. 1) shows an alignment in S4 in which R4 (the fourth charged residue counting from the N terminus) of NaVAb occupies a position equivalent to K5 of Shaker, in agreement with the structural alignment obtained by superposition of the crystal structures of NaVAb and the paddle chimera (Fig. 1). This same alignment also predicts that R3 of HV1 is equivalent to R4 of NaVAb and K5 of the paddle chimera. Similar to alignments produced by HHpred (Yarov-Yarovoy et al., 2012), PromalS3D alignments that included just NaV/CaV and KV VSDs and that were guided by the NaVAb and paddle-chimera crystal structures also confirmed the homology of R4 of NaV/CaV with K5 of KV (not depicted). The consensus from the phylogenetic analysis and the structurally guided multiple sequence alignments leads to the conclusion that R3 of HV1 is equivalent to K5 of Shaker.
Homology modeling
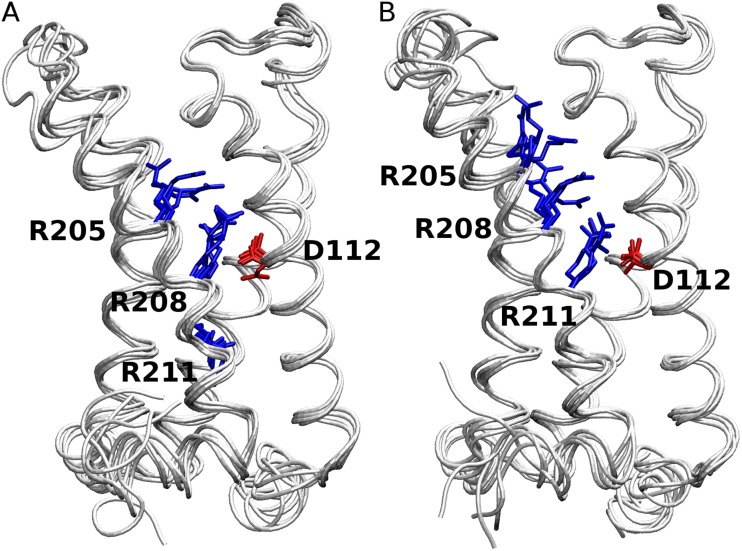
These results, along with the availability of the NaVAb crystal structure (Payandeh et al., 2011), suggested a need for the generation of a new homology model of hHV1. The model generated is presumed to be in the activated state (see Materials and methods). The two alternative sequence alignments of hHV1 to the template structural alignment, R2D and R3D (see Materials and methods), differed significantly only in the Arg registry in the fourth TM helix. The Arg registry of R2D obtained from the analysis in Fig. 1 has been suggested in only two previous studies (Alabi et al., 2007; Smith et al., 2011). The R3D Arg registry closely resembles that in most previous studies (Freites et al., 2006; Ramsey et al., 2010; Sakata et al., 2010; Wood et al., 2012). As expected from the underlying alignment, the subsequent models from these two alignments also differed in the positioning of the Arg residues of S4. From this point, we shall refer to models in which D112 is positioned opposite the second and third Arg residue (i.e., R208 and R211) as derived from the R2D and R3D alignments, respectively (Fig. 3).
Figure 3.
Energy-minimized starting structures of the five models of R2D (A) and R3D (B) are superimposed. D112 in S1 and the three Arg residues in S4 are shown in red and blue, respectively. The channels are viewed from the side (membrane), and the external end is at the top.
Evaluation of alternative models
To evaluate the quality of the two structural models, R2D and R3D, we performed MD simulations of the protein embedded in a hydrated membrane-mimetic n-octane slab. The principal advantage of using an octane slab rather than an explicit lipid bilayer is that the shorter relaxation time scale of the octane phase speeds up the conformational relaxation of the protein and the statistical convergence of the simulations. This computationally efficient feature of the n-octane slab allows the use of many independent samples for a statistically meaningful study (see Materials and methods for more details).
To validate the use of octane as a membrane mimetic, we performed a 430-ns-long control simulation of the highest resolution VSD template, KVAP, in a lipid bilayer and compared the results with those of octane/water simulations. The results, which are illustrated in Fig. S1, show that the average structure of the octane-solvated VSD is closer to that of the lipid-solvated VSD (RMSD of 0.12 nm) than either of these structures are to the initial crystallographic model (0.18 and 0.24 nm, respectively). These results demonstrate the validity of using this membrane mimetic for VSD-like domains.
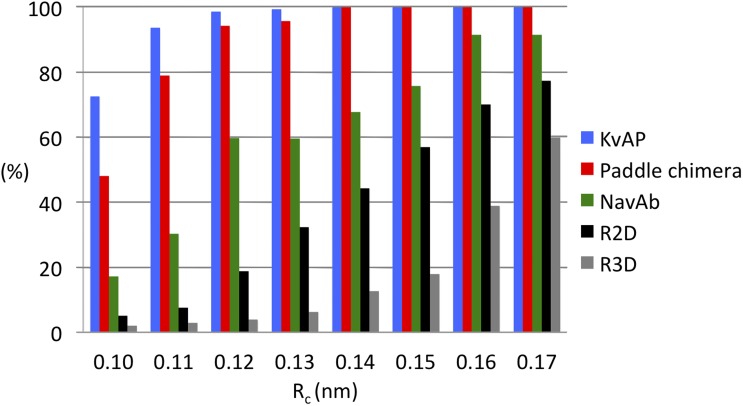
25 simulations were performed on each of the top five models from each alignment (see Materials and methods). In each of these simulations, the system was allowed to evolve in the octane slab at 300 K for 100 ns without any structural restraint. A clustering analysis was performed to characterize the conformational heterogeneity within an ensemble of structures generated by pooling together snapshots taken at 2-ns intervals during the last 20 ns of each of the 25 or 125 simulations for the templates and the homology models, respectively. The criterion for the clustering analysis was the RMSD of TM Cα atoms from the starting conformation of the protein, a measure of overall structural stability. The single-linkage clustering method of GROMACS clustering tool g_cluster was used to add a particular snapshot to a cluster when its RMSD value is less than a predefined cutoff value (Rc (nm)). The clustering of 1,250 snapshots from each homology model was repeated with increasing cutoff values ranging from 0.1 to 0.17 nm in 0.01-nm increments. The partition of the individual snapshots into clusters is a measure of structural similarity within the structural ensemble. The smaller the cutoff, the larger the number of clusters and the smaller the number of structures in each cluster. Fig. 4 illustrates the number of clusters and number of structures in the most highly populated cluster as a percentage of the total population at each cutoff value of the templates, R2D and R3D. Because model variation within R2D and R3D was found to be trivial, the structures from the top five models are combined for each R2D and R3D.
Figure 4.
Fraction of structures in the most populated cluster, shown as a percentage of the total population at each clustering cutoff value, Rc, for each of the three VSD templates and the two homology models.
Analysis of structural divergence
The statistics of the clustering itself highlights structural divergence. As the cluster cutoff value increases, the number of clusters decreases, indicating that the boundary for grouping the related structures together becomes gradually less stringent. The number of clusters at any cutoff value is always higher in the homology models than in the templates, indicating that structural divergence is greater in the homology models than in the templates. In KVAP and the paddle chimera, the population of the most highly populated cluster (NC) at every cutoff value is a large percentage (from 50% at 0.1 nm to 100% at a 0.17-nm cutoff) of its total population, i.e., the total number of snapshots (Fig. 4). In contrast, NC for NaVAb is always less populated, further indicating that its structure is more divergent than those of KVAP and the paddle chimera. The same interpretation applies to the homology models, with a significantly larger structural divergence in R3D than in R2D models.
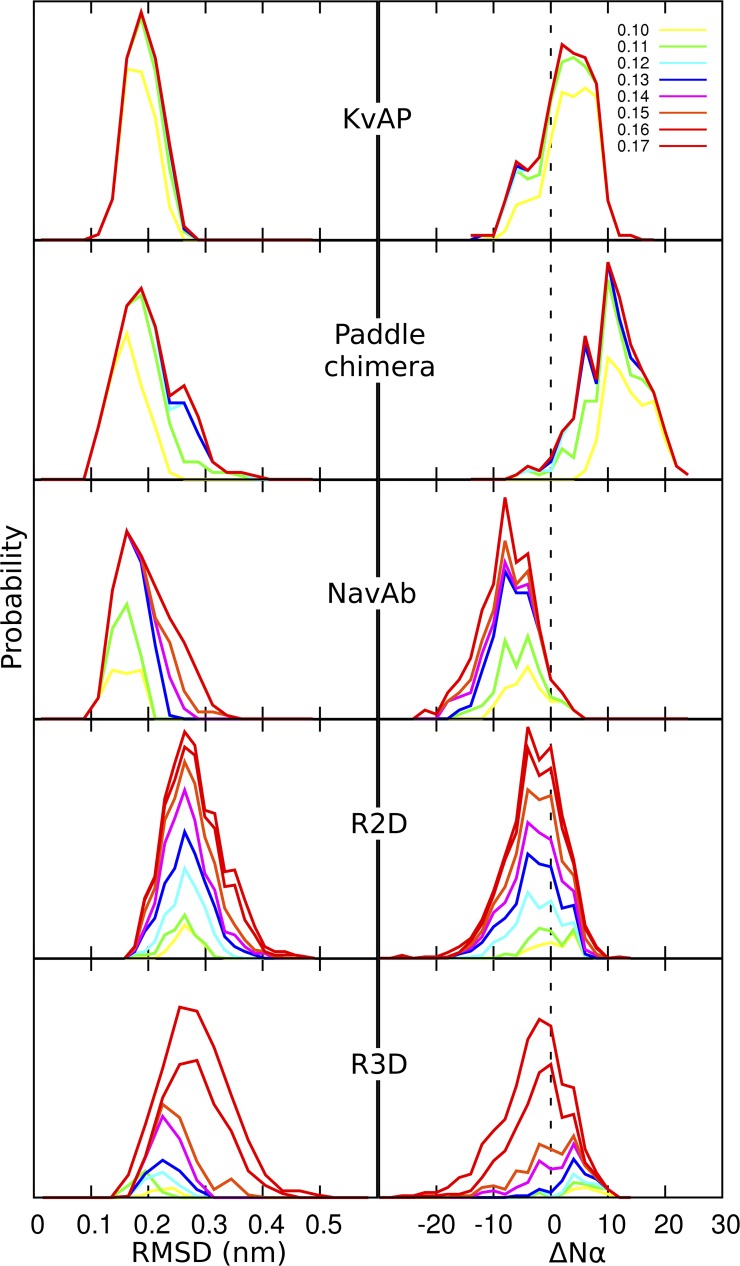
Two measures of structural stability of the models compared to the templates are the RMSD itself and the change in secondary structure. At each cutoff value, the probability distribution of the RMSD values of individual conformations in the most highly populated cluster was plotted for both templates and homology models (Fig. 5, left). This analysis reveals several key features. First, the structures of the three templates, although different from the starting structure, converge to a common single point independent of the cutoff value. Second, there is a trend of increasing deviation among these structures as more and more structures are added onto a cluster (in other words, the cluster grows with increasing cutoff value). The deviation is minimal in KVAP and maximal in NaVAb. This property is correlated to the decreasing atomic resolution of the structural models from KVAP to the paddle chimera and to NaVAb (1.9, 2.4, and 2.7 Å, respectively). For the homology models, deviation was greater for R3D than for R2D. Moreover, the R2D models converge to a single point, whereas R3D models lack such a feature. Finally, the distribution of RMSD values as the cluster grows retains its overall shape better in R2D than in R3D, indicating greater structural divergence in the latter model.
Figure 5.
Analysis of structural relaxation from massively repeated sampling. Relative probability distributions of (left) RMSD values (see Results) and (right) change in the number of α-helical residues, ΔNα, in the most populated cluster at each cutoff value (in nanometers, top right), relative to the starting structure of the simulations. The data are normalized to the total population.
The change in secondary structure, as measured by the loss or gain of α-helical residues in the TM region, is shown at different cutoff values in Fig. 5 (right). Qualitatively, KVAP undergoes both loss and gain of α-helical structure, whereas the paddle chimera and NaVAb undergo mainly a gain and a loss of α-helical structure, respectively. Discrepancies in α-helical content between the crystallographic models and the simulations of the VSD templates could be caused in part by inaccuracies in the OPLS-AA force field (Lindorff-Larsen et al., 2012). However, secondary structure is largely preserved in the structural ensembles of the two homology models, with distributions centered near zero (the greater discrepancy as a function of cluster cutoffs again reflecting the greater conformational diversity of R3D). As such, the main difference between the two homology models lies in the consistency of the relative arrangement of the TM helices, with significantly looser helix packing in R3D relative to R2D (Fig. S2).
Collectively, these data suggest that the R2D model is more stable and less divergent than R3D. Structural divergence of the templates increased with decreasing resolution of the crystal structures. The fact that the R2D model was only moderately more divergent than the lowest resolution structure (NaVAb) supports its validity as a model.
Structural properties
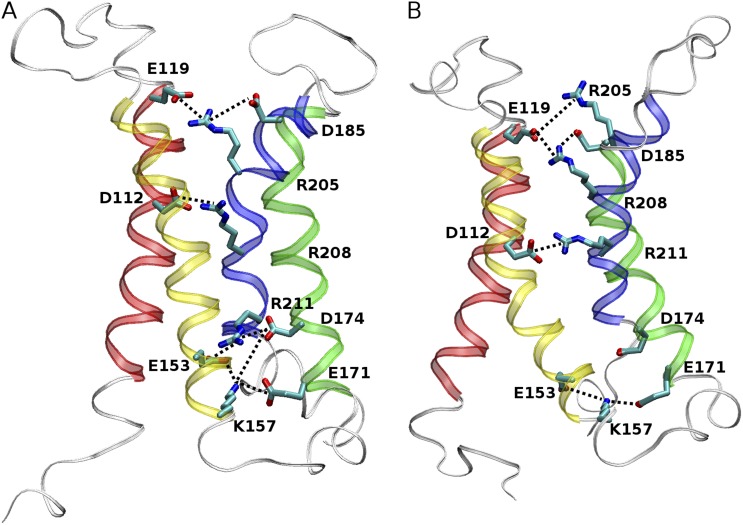
Having compared the overall structural stability and convergence of R2D and R3D, we examined the extent to which these models exhibit previously recognized structural features of VSDs. One “structural signature” of VSDs is two networks of electrostatic interactions (salt bridges), one internal and one external, between basic and acidic residues in the four helices (Papazian et al., 1995; Bezanilla, 2008; Yarov-Yarovoy et al., 2012). We therefore systematically screened R2D and R3D clusters to identify alignment-specific salt bridges adopted during simulations and evaluated the consistency of those signatures as the cluster grew. Fig. S3 illustrates the percentage of structures bearing a particular salt bridge in the highest populated cluster with increasing cutoff value in R2D and R3D. Fig. 6 highlights the salt bridges of both models that were present in >10% of the population of the most highly populated cluster at the 0.l-nm cutoff value.
Figure 6.
Salt-bridge formation in the (A) R2D and (B) R3D model. Acidic and basic residues in the α-helical TM region (red, S1; yellow, S2; green, S3; blue, S4) are shown in licorice representation. The salt bridges present in >10% of the most populated cluster obtained with Rc = 0.1 nm and are highlighted in thick dotted lines.
As shown in Fig. 6 A, R2D exhibits two networks of salt bridges: an external network formed by R205 and R208 in S4, E119 and D112 in S1, and D185 in S3; and an internal network formed by R211 in S4, K157 and E153 in S2, and D174 and E171 in S3. Residue F150, homologous with the conserved phenylalanine of the charge transfer center described by Tao et al. (2010), appears to mark the division between internal and external salt-bridge networks. Note that the population of some prominent salt bridges drops as the cluster cutoff increases, implying that these salt bridges contribute to the observed structural convergence and structural stability. In particular, the salt bridge between D112 and R208 located approximately at the center of the pore, was present in four of the five starting models of R2D and appears to be the most populated salt bridge of the external network during the simulation. The salt bridge of R205 and D185 was not present in any of the starting structures and occurs with a lower probability during the simulation but is still present a large proportion of the time, indicating a probable role in stability. Other salt bridges of the external network do not seem to make a significant contribution to the structural stability and convergence. For example, the R208–D185 salt bridge is absent at low cutoff and increases only to 10% at the highest cutoff examined.
The formation of the internal salt-bridge network in the R2D model is even more striking. The branching interaction of R211 with both E153 and D174 was the only salt bridge present in the starting structures. The consensus conformation after simulation, however, reveals a highly populated five-member salt-bridge network involving E153, K157, E171, D174, and R211.
Comparison of the R3D model with R2D reveals several important differences (Fig. 6). An external salt-bridge network in R3D comprising a chain linking D112–R211–E119–R208 together is present in the initial structures. Structural relaxation leads to stabilization of the R211–D112 ion pair at the center of the helix bundle, whereas the other two Arg residues evolve to form a persistent chain of salt bridges, R205–E119–R208–D185, in the external funnel (Fig. 6 B). These interactions tend to become less populated with increasing cutoff values because of greater structural heterogeneity (Fig. S3). The internal salt-bridge network is missing in R3D. Only K157–E153 is significantly populated, with low occurrence of K157–D174 and K157–E171 ion pairs.
Hydration profile
Water penetration into VSDs via external and internal water-filled crevices has been suggested to focus the membrane electric field and to provide the structural basis for ion translocation in mutant VSDs; therefore, it must be considered a characteristic structural feature of VSDs (Larsson et al., 1996; Yang et al., 1996; Starace et al., 1997). Given that hHV1 is a proton-selective channel that excludes the translocation of other ions, it is reasonable to expect it to contain a narrow pore or a constriction site within the TM region. Having identified R2D as the preferred homology model for hHV1, we next characterize pore hydration from extended simulations of 200 ns for each of 125 replicas.
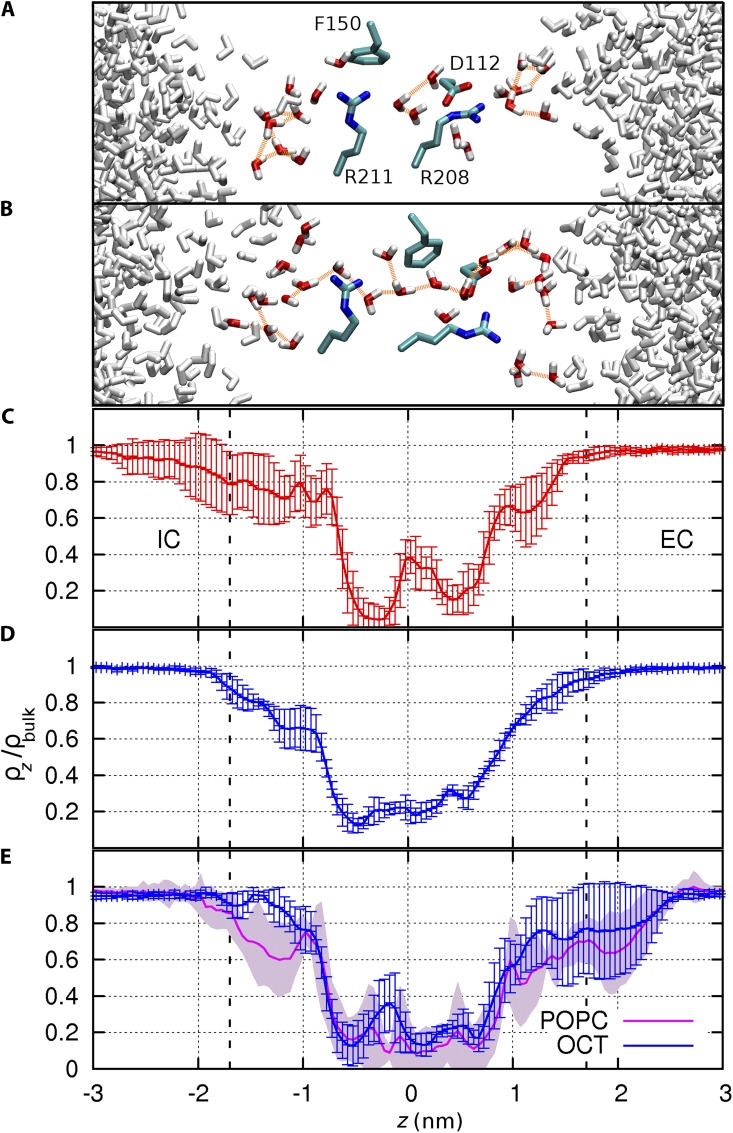
Specifically, we examine the conformational fluctuations of residues located at the narrowest part of the channel (Fig. 7) and the time-averaged probability distribution of water along the mean axis of the helix bundle (Fig. 8) using conformations saved every 10 ps. The hydration profile reveals the presence of external and internal water-filled cones consistent with the structure of other VSDs. Between these two funnels, two bottlenecks, or “constriction sites,” delineate a narrow hydrated region at the pore center (Fig. 8). The external bottleneck corresponds to the location of the salt bridge between D112 and R208, whereas the internal bottleneck is at the interface between R211 and F150. The distances between these two residue pairs were monitored to characterize their spontaneous conformational fluctuations (Fig. 7, A and B). Three distinct populations emerge for the external constriction site (D112–R208). These populations represent the three configurations of the salt bridge: bidentate and monodentate conformations (involving two or one hydrogen bond, respectively), and open, in order of increasing separation. The separation between R211–F150 is essentially bimodal (Fig. 7, C and D). When the distance between the center of mass of the phenyl ring of F150 and the guanidinium Cz atom of R211 is <0.55 nm, the guanidinium group makes a direct contact with the aromatic ring. Rapid conformational fluctuations modulate the size of each of the two constrictions independently of the other. Seven distinct conformational states are resolved in the two-dimensional map, of which five are labeled I–V and illustrated in Fig. 7 E. The most constricted of these states (designated IV in Fig. 7 E) corresponds to a bidentate D112–R208 salt bridge and close nonpolar contact of R211–F150. In contrast, the least constricted state (III) corresponds to disrupted interactions for both D112–R208 and R211–F150 residue pairs. Although the latter residue pair is disrupted 87% of the time, the D112–R208 salt bridge is disrupted (or “open”) only 10% of the time (state III). Here, “open” refers to the conformation of the D112–R208 salt bridge, not that of the channel, which is presumably in the open conformation throughout the simulation.
Figure 8.
Pore hydration in the R2D model. Representative snapshots are shown successively for (A) the most constricted conformational state defined as d1 < 0.42 nm and d2 < 0.55 nm and (B) least constricted conformational state defined as d1 > 0.52 nm and d2 > 0.55 nm (see Fig. 7 E). Residues D112, F150, R208, and R211 are shown in licorice representation. Water molecules within 5 Å of these residues are colored in red and white, and hydrogen bonds between these molecules are shown with orange lines. (C) Water density within a 0.7-nm radius of the mean axis of the pore, normalized to the bulk water density in the most constricted and (D) least constricted conformational states, respectively. (E) Comparison of the water density profiles from four control simulations of 100 ns, successively in n-octane (blue) and in a POPC bilayer (magenta). Error bars represent the standard deviation.
Computing the hydration profile separately for the most and least constricted conformations of the bottleneck (respectively, states IV and III in Fig. 7 E) confirms that pore hydration is modulated by the conformation of these two constriction sites (Fig. 8). The two distinct bottlenecks characterizing the constricted state (Fig. 8, A and C) disappear in the relaxed state, where water density becomes nearly homogeneous throughout the narrow, 1-nm-long pore (Fig. 8, B and D). Unlike the constricted state, the dilated state contains a continuous hydrogen-bonded column of water molecules with occasional branching. The formation of a continuous water chain is facilitated by the disruption of the D112–R208 salt bridge, with one third of these opening events lasting longer than 1 ns.
To evaluate the effect of the membrane mimetic on the homology model, four simulations of R2D were extended by another 100 ns, separately in octane and in a phospholipid bilayer. The RMSD between the average backbone structures in the two solvents is 0.15 nm, and the average hydration profile of the pore is similar in the octane and lipid simulations (Fig. 8 E), indicating that the overall structural properties of the model are conserved in a lipid bilayer.
Energetics of ion translocation
As a first step toward understanding the energetics of ion permeation in hHV1, we used continuum electrostatic calculations to compute the static field, i.e., the energetic contribution arising from the distribution of electric charge in the pore, for the transfer of a positive point charge through the water-filled state of the pore (corresponding to conformation III in Fig. 7 E; Fig. 9 A). In the WT channel, the static field profile is compatible with the transfer of a point charge through the narrow part of the pore. Overall, the static field can be approximately divided into three regions. The cytosolic end of the pore (−1.5 < z < −0.5 nm) contains an energy well induced by the net negative charge from the salt-bridge network (E153, K157, E171, D174, and R211) present in the internal water-filled crevice. Importantly, the static field essentially cancels out throughout the narrowest region of the pore (−0.5 < z < 0.8 nm), which contains D112 (0.25 < z < 0.8 nm) and the flexible side chain of R208 (−0.5 < z < 1 nm). The effect of the positive charge of R205 in the extracellular crevice is cancelled out by the proximity of D185.
Figure 9.
Effect of the charge distribution of the channel on the energetics of ion translocation. (A) Representative snapshot of the water-filled open conformation of HV1. Water molecules and charged residues in the pore are shown in licorice representation; the extracellular end is to the right. (B) Static field energy for the transfer of a positive point charge in (blue) WT and (red) neutral-D112 forms of HV1. Two thick lines represent the mean of five running averages, each computed from a dataset of 30 snapshots using a five-point moving window. Blue shading and orange error bars represent the SEM.
Upon neutralization of residue D112, the static field increases by ∼10 kcal/mol in the narrow region of the pore, where the presence of the unpaired positive charge of R208 presents a significant barrier to the movement of cations (Fig. 9 B). Collectively, these findings suggest why D112 is necessary for the movement of protons and why neutral mutants of D112 are selective to anions (see Discussion).
Accessibility studies
The MD results favor the R2D open-state model as more stable than the R3D open-state model. These results suggested that when hHV1 is in the open state, R211 should be accessible only to the cytoplasmic side, and that R205 should be accessible only to the extracellular side. We tested this prediction by replacing each of the three Arg residues in the S4 TM segment with His. We probed the mutants with Zn2+ on the assumption that Zn2+ binding to the introduced His residue should affect the measured current. Because all three Arg residues studied here are located within the putative proton conduction region and face this pathway in existing models (Musset et al., 2010; Ramsey et al., 2010; Wood et al., 2012), it would be surprising if Zn2+ binding to any one of these sites had no detectable effect on current. Because a larger effect does not necessarily indicate better accessibility, we focus only on the qualitative effects of Zn2+ on each introduced His residue. We cannot rule out the possibility that Zn2+ might bind to a different site that is preferentially exposed by a particular mutation.
Strategy for Zn2+ studies.
To minimize Zn2+ binding to other parts of the channel, we replaced both His residues known to bind Zn2+ in WT channels (Ramsey et al., 2006; Musset et al., 2010), namely H140A and H193A. We also truncated the C terminus at position 221 to prevent dimerization (Koch et al., 2008; Tombola et al., 2008; Gonzalez et al., 2010), because Zn2+ binding at the dimer interface appears to interfere with channel opening (Musset et al., 2010). In addition, using monomeric constructs precludes complications caused by interactions thought to occur between protomers during gating (Gonzalez et al., 2010; Tombola et al., 2010) but whose mechanism is unknown. For most measurements of external accessibility, Zn2+ was applied at pHo 7.0, because H+ and Zn2+ compete for binding sites on proton channels (Cherny and DeCoursey, 1999). Whole-cell measurements of mutants that generated small currents were done at pHi 5.5 to increase current amplitude. Measurements in inside-out patches were made at pHi 6.0, also to increase the current amplitude. Effects of Zn2+ were weaker at pHi 6.0 than at pHi 7.0 but not dramatically so; the more profound competition in WT channels is caused by Zn2+ coordination at multiple titratable sites (Cherny and DeCoursey, 1999). We interpret effects of Zn2+ as the result of binding to the His residue introduced by mutation; EGTA application removes Zn2+ and should reverse the effect.
Behavior of the mutants in the absence of Zn2+.
There were no drastic behavioral changes in voltage-clamp studies of the R→H mutants compared with WT channels. All three constructs were nonconducting at negative voltages and opened with time during depolarizing pulses. Specifically, we did not observe evidence for H+ conductance activated at negative voltages, or for proton carrier activity at intermediate voltages, as might have been expected by analogy with the His scanning studies of Starace and Bezanilla in Shaker K+ channels (Starace et al., 1997; Starace and Bezanilla, 2001, 2004). All R→H mutants retained proton selectivity. The change in Vrev for a 1.5-U change in pHo averaged −75 ± 4.5 mV (mean ± SEM; n = 4) for R205H, −83.2 ± 7.3 mV (n = 5) for R208H, and −75.1 ± 2.0 mV (n = 8) for R211H, compared with −80.9 mV in the WT channel (Musset et al., 2011). A report that R211x mutants are permeable to guanidinium (Berger and Isacoff, 2011) evidently does not indicate a general widening of the pore, because smaller cations and anions were impermeant in R211H. The mean change in Vrev (corrected for measured liquid junction potentials) when TMA+ or CH3SO3− was replaced was +0.3 ± 2.0 mV (mean ± SEM; n = 5) for K+, +2.3 ± 0.4 mV (n = 5) for Li+, +3.0 ± 1.4 mV (n = 5) for Na+, and +0.8 ± 1.3 mV (n = 4) for Cl−. We interpret guanidinium permeation as a reflection of its propensity to modify hydrogen bonds and denature proteins (England and Haran, 2011).
External accessibility.
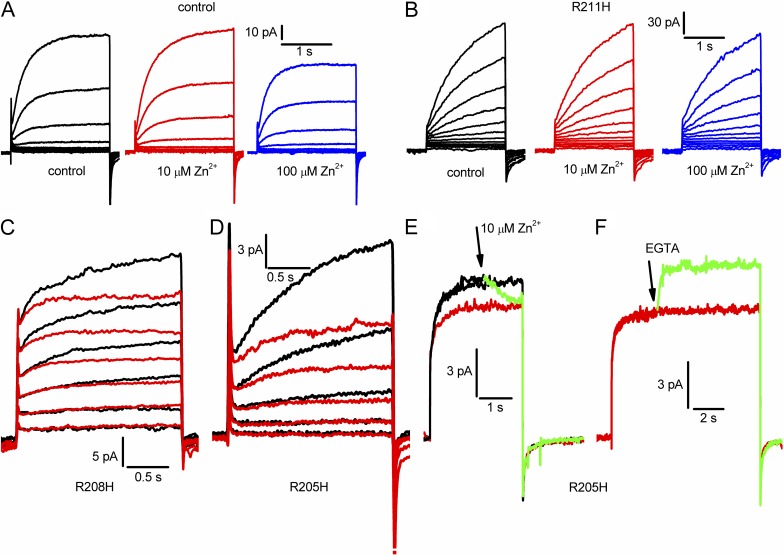
Fig. 10 A shows that the “Zn2+-insensitive” background construct studied in whole-cell configuration was negligibly affected by externally applied Zn2+ at 10 µM but was distinctly inhibited at 100 µM. Similar results were obtained in eight cells. Therefore, we adopted 10 µM Zn2+ as a test concentration. Fig. 10 B, representative of 10 cells, shows that the innermost Arg in S4, R211H, was not sensitive to externally applied Zn2+.
Figure 10.
Evaluation of external accessibility of the three Arg residues in the S4 helix of hHV1. All measurements were made in a construct designed to have low sensitivity to Zn2+, H140A/H193A/K221stop. (A) Currents during identical families of pulses in 10-mV increments up to +60 mV in the background construct in whole-cell configuration, in the presence of 0, 10, or 100 µM Zn2+ in the bath solution. (B) Whole-cell currents in the R211H mutant during identical families of pulses in 10-mV increments up to +80 mV, in the presence of 0, 10, or 100 µM Zn2+. (C) Currents during identical pulse families in 20-mV increments up to +60 mV in the R208H mutant in the absence (black) or presence of 10 µM Zn2+ (red). (D) Currents during identical pulse families in 10-mV increments up to +30 mV in the R205H mutant in the absence (black) or presence of 10 µM Zn2+ (red). (E) Currents during three consecutive pulses to +90 mV in a cell transfected with R205H. At or slightly before the arrow, 10 µM Zn2+ was applied to the bath, reducing the current during the pulse (green). The next pulse (red) shows the steady-state extent of Zn2+ inhibition. (F) Later in the same experiment, EGTA was added during a pulse at or before the arrow, rapidly relieving Zn2+ effects. For all parts, pHo was 7.0; pHi was 7.0 for A, B, E, and F; and pHi was 5.5 for C and D.
The middle Arg, R208, was difficult to evaluate because currents in the R208H mutant were quite small. Most measurements were done with pHi 5.5 to increase the amplitude of the currents. Nevertheless, in 12 experiments, 10 µM Zn2+ applied to the bath reduced currents in 10 cells and had no clear effect in 2 cells. Fig. 10 C shows superimposed currents recorded during an identical family of pulses in the absence (black) and presence (red) of 10 µM Zn2+. The time-dependent component of current was reduced by Zn2+, especially during large depolarizing pulses. A similar picture emerged for the R205H mutant (Fig. 10 D is representative of eight cells). In both R208H and R205H (Fig. 10, C and D), Zn2+ reduced the current progressively during the pulse, consistent with greater accessibility in the open than closed state. Alternatively, these effects can be interpreted as acceleration of activation combined with reduction of maximal current.
To test accessibility of R205 specifically in the open state, Zn2+ was applied during a depolarizing pulse (Fig. 10 E, green trace). The current was reduced rapidly from the control level (Fig. 10 E, black) to approximately the level seen during a subsequent pulse (red). Later in the same experiment, after equilibration with 10 µM Zn2+, EGTA was applied during a long pulse (Fig. 10 F, green). The current was rapidly restored. Similar results were seen in four cells. We imagine that Zn2+ binds and unbinds rapidly to the His residue, and that EGTA simply removes free Zn2+ from the vicinity. In other words, this experiment does not test the accessibility of the site to EGTA but rather the accessibility of the site to Zn2+.
In summary, R205H and R208H were accessible to Zn2+ applied externally, but R211H was not. Although R205H was readily accessible in the open state, and Zn2+ effects on R208H appeared consistent with preferential open-state access, we could not determine whether either mutant was accessible in the closed state. The presumed rapid kinetics of Zn2+ binding/unbinding precludes definitive conclusions on that point.
Internal accessibility.
Control measurements in inside-out patches produced a similar picture for internal application (Fig. 11 A) as seen for external (Fig. 10 A); 10 µM Zn2+ produced negligible effects (n = 5), and 100 µM Zn2+ distinctly attenuated the current. Determining internal accessibility requires inside-out patches, and unfortunately, both R205H and R208H exhibited currents too small to permit clearly interpretable data. In four of five inside-out patches, R208H currents appeared to be inhibited to some extent by 10 µM Zn2+ (not depicted). R211H was amenable to study, and we found that 10 µM Zn2+ applied internally consistently inhibited the current (n = 7), indicating accessibility to the internal solution (Fig. 11 B). The application of 10 µM Zn2+ during long pulses did not produce clear evidence of block during the pulse; instead, currents declined gradually over several pulses (1–2 min). However, as illustrated in Fig. 11 C, the application of 1 mM EGTA during depolarizing pulses did reverse Zn2+ inhibition (n = 3), indicating open-state accessibility of R211H. The apparent contradiction between the effects of Zn2+ and EGTA can be understood by considering that a substantial unstirred volume at the tip of the pipette insulates the inside of the membrane from the bath solution. The concentration of Zn2+ near the membrane will increase gradually after a delay and must approach equilibrium to affect the current markedly. In contrast, the local EGTA concentration need only reach 1% of its final value to remove Zn2+ because of the 100-fold excess of EGTA (1 mM) over Zn2+ (10 µM). Higher concentrations of Zn2+ (100 µM) did sometimes reduce the current when applied during pulses, but this result was ambiguous because this concentration has a distinct effect in the control (Fig. 11 A). In summary, R211H is accessible to the internal solution even in the open state.
Figure 11.
Evaluation of internal accessibility of Arg211 in the S4 helix of hHV1. All measurements are in inside-out patches at pHo 7.0, pHi 6.0. (A) Control families in the absence and presence of 0, 10, or 100 µM Zn2+ with pulses in 10-mV increments up to +60 mV. (B) Currents in an R211H patch during identical families of pulses in 10-mV increments up to +60 mV in the presence of 0 or 10 µM Zn2+. (C) Three consecutive pulses to +60 mV in an inside-out patch with the R211H mutant, first in the presence of 10 µM Zn2+ (red), and then with the addition of EGTA during the pulse (green), and finally in the absence of Zn2+ (black). The reversal of Zn2+ effects shows accessibility of R211H in the open state. For all parts, Vhold = −60 mV.
DISCUSSION
Phylogeny
The results that place the HV1/C15orf27/VSP subfamily of VSDs in the same main branch as the NaV/CaV-type VSDs are robust to different tree construction methods, changes in the included taxa, and even different underlying multiple sequence alignments (Fig. 1 and Table S1). These results indicate that the VSD subfamily that is not associated with a canonical ion pore, typified by HV1, is more closely related to the NaV/CaV family of VSDs and appears to have split from the KV family of VSDs at least a billion years ago. Sequence similarity between the HV1/C15orf27/VSP subfamily and the NaV/CaV subfamily thus most likely represents leftover relationships from “history” rather than positive selection. In this view, the sequence similarity is evidence of the common ancestry of these groups but does not reflect any particular property that evolved in both groups. Despite the presence of Kir-type channels in bacteria, which are homologous to the S5–S6 TM helices of voltage-gated cation channels, the lack of discernible “poreless” VSDs in bacteria makes any assignment of the association of the ancestral VSD with the ancestral ion pore extremely speculative.
The gold standard for a “correct” sequence alignment is one derived from a structural superposition. The sequence alignment in S4 of VSDs is challenging precisely because this gold standard cannot be applied from sequence information alone; a particular arginine within the same VSD is thought to occupy different positions relative to the rest of the VSD, depending on whether the VSD is “activated,” “inactivated,” “resting,” or at some intermediate position. This point has been exemplified dramatically in the presumed inactivated-state crystal structures of NaVAb (Payandeh et al., 2012) and NaVRh (Zhang et al., 2012) that appeared while this paper was in preparation. S4 of the NaVRh crystal is displaced by one full helical turn compared with S4 of NaVAb when using the conserved phenylalanine of the charge transfer center as a reference point (Zhang et al., 2012); this difference is presumed to represent a different position of S4 rather than a different alignment of the four arginines in each S4. The sequence alignment methods that incorporate structural information, as do both HHPred and PromalS3D, provide the crucial comparison that leads to the “correct” representation of the registry of the arginine residues in S4 in the resulting multiple sequence alignments: constraining the alignment to the “activated”-state crystal structures, for example, provides the “activated”-state registry and thus an alignment that will be preserved in the other states of the VSD characterized by S4 motion. The alignment of the S4 arginine in the multiple sequence alignment shown in Fig. 1 is consistent with the structural superposition of the presumed “active” NaVAb and paddle-chimera crystal structures, and also with a recent alignment of NaV/CaV and KV VSDs obtained by using HHpred (Yarov-Yarovoy et al., 2012). The alignment used here also includes the HV1/C15orf/VSP subfamily of VSDs and provides a different alignment in S4 than has been used previously in other credible and useful structural analyses (Freites et al., 2006; Sasaki et al., 2006; Gonzalez et al., 2010; Ramsey et al., 2010; Sakata et al., 2010; Berger and Isacoff, 2011). Only two previous studies aligned R1 in HV1 with R3 in Shaker (Alabi et al., 2007; Smith et al., 2011). These considerations led us to develop a new homology model of hHV1 and to compare it to another model based on a previous, widely accepted alignment. Alignments with significant differences in the placement of charged residues outside of S4 seem implausible on the basis of alignment scores. Only one such alignment (Freites et al., 2006) places a charged residue (HV1 D185) more than one residue away compared with the alignment we use here. The latter alignment was obtained before the sodium channel crystal structures were available and disagrees at that position with the structurally informed alignment obtained by Yarov-Yarovoy et al. (2012).
We used a multi-template approach and different alignments in S4 to produce in the end two homology models of hHV1 using Modeller. We then developed a novel MD method to evaluate the consistency of the two models. This method enabled us to reach consensus-derived structural determinants for both templates and homology models. The VSD in the new inactivated-state crystal structures of NaVAb (Payandeh et al., 2012) is essentially indistinguishable from the VSD in the activated-state structure that we used in our multi-template modeling. The VSD in the NaVRh structure (Zhang et al., 2012) is very similar in S1–S3 to the templates we used, so our model would not be changed greatly by incorporating this structure; implications of significant differences in S4 of this structure are discussed below.
Computational validation of the homology models
MD simulations have been used previously both as assessment and refinement tools for homology models of proteins. For soluble proteins, these simulations typically involve all-atom explicit models in water; gradual model refinement using structural restraints; a simulation time spanning hundreds of nanoseconds to microseconds; and the analysis of ensemble structural properties, such as the conformational drift from the starting conformation, conservation of the secondary structure, and nonbonded residue contacts within the model and its templates (Chen and Brooks, 2007; Raval et al., 2012). Similar efforts for homology models of membrane proteins, which must take into account the anisotropic membrane environment, have until now been limited to comparatively shorter simulation times. These simulations have used either a lipid bilayer or biphasic membrane mimetic n-octane slab (Capener and Sansom, 2002; Arinaminpathy et al., 2003; Law and Sansom, 2004), which have typically been limited to a single simulation. The advantage of using n-octane is that its low viscosity speeds up the conformational fluctuations and relaxation of the embedded protein compared with a lipid bilayer. This property results in a wider conformational ensemble of the protein within a relatively short simulation time. This improved sampling efficiency, together with multiple repeats, was exploited in the current protocol to achieve a broad conformational sampling in a computationally efficient manner. In addition, we used a clustering analysis to identify a consensus conformation for the protein. To our knowledge, this work is the first comparative homology modeling MD simulation based on a consensus-derived three-dimensional model for a membrane protein. The general protocol presented here can readily be applied to evaluate the quality of homology models of other membrane proteins.
Because homology modeling is derived from little or no experimental evidence, and particularly in light of the low sequence identity between crystallized VSDs and hHV1, it is paramount to assess the quality of the model before examining the structural basis of hHV1 function. Although other homology models of hHV1, including another generated by our group, have been proposed recently (Musset et al., 2010; Ramsey et al., 2010; Wood et al., 2012), the present study is the first to offer a comparative assessment of the quality of homology models. In this study, two principal criteria were used to assess and compare the structural stability of the models to its templates: overall structural drift of the TM segments and retention of α-helical structure from the starting conformation. A key aspect of our approach is the evaluation of statistical properties from multiple replicas and subsequent clustering analysis. It is noteworthy that in some cases (Fig. 5), the probability distribution of R2D and R3D models almost overlaps. With clustering, however, this source of ambiguity is resolved. Thus, our results indicate that combining massively repeated simulations with clustering made it possible to identify R2D as the better model for hHV1. This conclusion appears to be corroborated by the subsequent analysis of salt-bridge formation as well as hydration and solvent accessibility (see below).
The validity of using a biphasic membrane mimetic (or simple n-octane slab) for the purpose of estimating the stability of membrane proteins in general, and of VSDs in particular, is demonstrated by the results obtained for the VSD templates and the control simulation in a lipid bilayer, which consistently retained their crystallographic conformation within a mere 0.2 nm RMSD over 100-ns MD trajectories. This value is consistent with what would be expected in lipid bilayers (Sands and Sansom, 2007). Moreover, the analysis reported above shows that the magnitude of structural deviations from the starting point is correlated to the resolution of the crystallographic structure, which demonstrates the sensitivity of the approach to the quality of the structural model. As in any molecular simulation study, it is not possible to predict whether or not conformational changes not observed in a given simulation may occur on longer time scales. As such, we cannot dismiss the possibility that longer simulations of our systems may lead to conformational differences caused by approximations in the membrane representation or by other factors. Nevertheless, the simulations reported in this study represent over 45 µs of sampling, which is, to our knowledge, orders of magnitude longer than any previous validation study of a homology model of a membrane protein.
Although the stability of both homology models is lower than that of the lowest resolution template (the VSD from sodium channel NaVAb) in terms of both the magnitude of deviation from the starting structure and the heterogeneity of the ensemble of structures generated by the simulations, they were clearly differentiated by the analysis. The resulting structural ensemble is much better clustered around a consensus structure for the R2D than for the R3D model, as manifested by the conserved location of the peak in the RMSD distribution, independent of clustering cutoff (Fig. 5). Thus, although the ensemble of structures obtained at 300 K deviates from the initial homology R2D model, these structures retain a high degree of similarity with one another.
These findings indicate that, unlike the R3D model, the R2D model corresponds to a unique conformational basin that is consistently represented by the thermal ensemble obtained at 300 K. Although the quality of the model is evidently inferior to that of the crystallographically derived structural models of the VSD templates from K+ and Na+ channels, the self-similarity of structures making up the R2D conformational ensemble approaches that obtained for the VSD of NaVAb (Figs. 4 and 5). Moreover, the high degree of structural stability of salt bridges in the simulations of R2D is consistent with the proposal that charge pairing contributes to the stability of VSDs (Papazian et al., 1995; Bezanilla, 2008; Swartz, 2008). As shown in Fig. S3, the population of ion pairs making up the salt-bridge networks in the conformational ensemble of R2D is essentially independent of cluster cutoff, whereas alternative salt bridges become populated with increasing cutoff values in the R3D ensemble. These results do not prove that the R2D model is accurate. Nevertheless, the above results, collectively, support the validity of our systematic, ensemble-based computational approach in discriminating between two possible homology models.
Hydration and mechanism of proton conduction
The above structural analysis leads to various considerations regarding the putative functional mechanism of hHV1. Importantly, the overall structural features of our R2D model are a priori consistent with the properties required of a selective ion channel in its open state. Although the helix bundle retains the overall “hourglass” shape of the VSDs of K+ and Na+ channels, the four helices of hHV1 define a narrow, mostly water-filled pore. This narrow pore consists of a 1-nm-long constriction defined by two pairs of highly conserved residues, Arg211–Phe150 and Arg208–Asp112. In the above simulations, the residues lining this putative ion-translocation pathway undergo structural fluctuations resulting in the transient formation of a water chain throughout the pore (Fig. 8 D). Although the presence of a narrow, intermittent chain of water molecules that could serve as a putative proton pathway does not prove that this structure would be selective or even permeable to protons, these features can conceivably play a role in selective proton permeation, as discussed below.
The presence of a salt bridge involving D112 at the narrowest part of the channel suggests possible mechanisms for proton transport and selectivity. Either protons are translocated throughout the entire length of the pore in a water-mediated Grotthuss-like mechanism, as described in simpler, nonselective cation channels such as gramicidin (Pomès and Roux, 1996, 2002), or one or more titratable groups of the protein act as proton-relay groups. Because proton transport in hydrogen-bonded water wires is very efficient (Pomès and Roux, 1996), and because D185N and D185A retained proton selectivity (Musset et al., 2011), it is likely that protein-mediated proton relay, if it occurs, would be restricted to the D112–R208 bottleneck. Even if these residues are directly involved in proton relay, the ion pair would have to dissociate, if only transiently, to enable the changes in protonation states and in the organization of the hydrogen-bonded network inherent in vectorial proton movement. As a result, the fact that the ion pair undergoes reversible opening transitions in our simulations supports proton translocation a priori, although it does not allow us to discriminate between water-only and protein-mediated proton relay mechanisms.
Ultimately, discriminating between these two mechanisms will require detailed examination of the energetics and kinetics for the movement of an excess proton through the pore. In principle, both mechanisms may be invoked in the mechanism of proton selectivity. The rapid positional fluctuations of Arg208 would be expected to lower the pKa of Asp112 when they are close together, resulting in proton ejection. Alternatively, it is conceivable that fluctuations resulting in the formation of a transient water column (∼10% of the time) might allow the rapid translocation of protons but not the slower diffusion of other cations. The gramicidin channel contains a water wire essentially continuously (Levitt et al., 1978; Pomès and Roux, 1996), yet its proton conductance, extrapolated to neutral pH, is an order of magnitude lower than that of hHV1 (Cherny et al., 2003; DeCoursey, 2003). The conductance of hHV1 might be increased, compared with that of gramicidin, by (a) a shorter narrow segment or (b) its negatively charged inner vestibule (Fig. 9). There is a possible parallel between the largely discontinuous water column in hHV1 and suggestions that other ion channels may “close” by hydrophobic collapse (Dzubiella et al., 2004; Beckstein and Sansom, 2006; Roth et al., 2008; Chakrabarti et al., 2010; Jensen et al., 2010). However, our model is exclusively of the open conformation of hHV1; based on existing evidence (Gonzalez et al., 2010; 2013), we imagine that the closed conformation differs significantly in the relative position of S4. In addition, interruption of the water wire by Asp112–Arg208 salt bridge formation occurs on a nanosecond timescale, much faster than macroscopic closing of the channel, which has time constants of milliseconds to seconds. Future simulation studies of hHV1 in a lipid bilayer, including free-energy calculations for the translocation of protons and other ions, will be performed in an effort to relate the microscopic structure of the channel to the mechanism of proton translocation and selectivity.
Nevertheless, it is pertinent and informative at the present model-validation stage to consider the competence of the model in terms of cation permeation by examining whether the electrostatic environment of the pore is compatible with the movement of a positive point charge (Fig. 9). A previous computational study of aquaporin has shown that the static field contribution of the electrostatic potential for the movement of a positive point charge is similar to the potential of mean force for the translocation of an excess proton in the single-file region of the pore (Chakrabarti et al., 2004). This agreement suggests that the free energy for the movement of H+ in the narrow region is largely determined by the distribution of polar and charged groups of the channel. As such, the static field profile affords a first glimpse into the energetics of ion permeation in hHV1. The cancellation of the static field in the bottleneck region of the channel suggests that there is no intrinsic preference for cations over anions in the WT channel, and that both anions and cations other than H+ could be excluded by the same mechanism. In contrast, the large static field barrier induced by the neutralization of the charge of D112, which results in an excess positive charge (that of R208) in the bottleneck, is consistent with the experimentally observed anion selectivity of neutral mutants of D112 (Musset et al., 2011). On the one hand, reversing the sign of the probe point charge yields the opposite static field profile, turning the broad 10-kcal/mol energy barrier (Fig. 9) into a well of the same depth (not depicted). On the other hand, adding the desolvation penalty for bringing a finite-size ion from bulk water into the narrow part of the pore, a quantity that is always positive in sign, would further disfavor cations while bringing the electrostatic profile of anions closer to zero, consistent with anion selectivity.
How far does S4 move during gating?
In other voltage-gated ion channels, the membrane electrical field is concentrated and much of it drops across a narrow region that is at the focus of an hourglass of water (Larsson et al., 1996; Yang et al., 1996; Starace et al., 1997). The Arg residues are thought to move sequentially past this constriction, altering their accessibility from inside to outside as they do so, and thus transferring gating charge across a large fraction of the membrane field at the expense of a relatively short actual movement. Recent evidence indicates that three (Jensen et al., 2012; Yarov-Yarovoy et al., 2012) or four S4 Arg residues move past the charge transfer center during Na+ or K+ channel opening and inactivation (Lin et al., 2011; Henrion et al., 2012; Zhang et al., 2012). Given the remarkable similarities between hHV1 and the VSD of other voltage-gated ion channels, the movement of S4 might be expected to be similar. However, our preferred homology model (R2D) suggests that in hHV1, R3 (R211) remains internal to F150, the external delimiter of the charge transfer center past which the S4 arginines of the VSD are envisioned to move during gating (Tao et al., 2010). Consistent with this prediction, when the Arg residues in S4 of hHV1 were mutated to His, the two outermost positions (R205H and R208H) were accessible to the external solution in the open state, but the third, R211H, was not. R211H was accessible to the internal solution even in the open state. The accessibility determined here with Zn2+ is somewhat greater than in previous studies using the MTS reagents as probes. PEGylation assays indicated that R3 of mHV1 was fully accessible to the internal solution (Sakata et al., 2010), although the channel was presumably in the closed state. R3 in CiHV1 was accessible to internally applied MTS reagents in closed but not open states (Gonzalez et al., 2010). The latter study reported external accessibility of R1 in open but not closed CiHV1 channels. R2 was inaccessible in both studies. The higher accessibility of the S4 Arg residues observed for Arg→His mutants, together with the higher Zn2+ affinity than that typically observed for binding to imidazole (Gurd and Wilcox, 1956), may reflect favorable contributions of the external and internal charge networks identified here (Figs. 6 and 9) to coordinating Zn2+ at these intimate locations.
The conclusion from our accessibility studies that R211 remains internal to the charge transfer center in the open state is compatible with the finding that proton channel gating and selectivity persisted even when the C terminus was truncated, removing R3 altogether (Sakata et al., 2010). The lowest energy structure of NaChBac predicted by Yarov-Yarovoy et al. (2012) also supports the internal accessibility of its R4 (the equivalent of R3 in our alignment of hHV1) in closed states and at least transiently in open states, although 9 of 10 low energy structures show R4 of NaChBac making contact with the extracellular charged nexus. The recent structure of NaVRh (Zhang et al., 2012) shows that its R4 can ratchet “up” even one more turn in a presumably inactivated state, whereas our accessibility data suggest that the homologous position in hHV1 does not move past the charge transfer center, Phe150. Perhaps the smaller number of charged groups on S4 of hHV1 prevents the full extent of movement predicted in NaChBac and NaVRh, keeping R3 of hHV1 internally accessible. Our results appear to limit the outward movement of S4 and suggest that S4 movement during gating is more restricted in hHV1 than in the K+ channel VSD. A speculative explanation for this difference is that unlike K+, Na+, and Ca2+ channels, hHV1 lacks a separate pore domain. The large displacement of S4 that is needed to open other channels is evidently not needed to produce a proton pathway.
These conclusions depend on the assumption that the mutations introduced here do not drastically affect the position of S4 in open or closed channels. Because in all cases charged residues were mutated, there is a strong possibility that the lowest energy position of the mutated residues will differ at least to some extent from the WT channel. On the other hand, the Arg→His mutation does not necessarily change net charge. Speaking against gross structural changes, the Arg→His mutants displayed relatively normal gating and were all proton selective. Although hHV1 appears to function as a dimer (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008) that exhibits cooperative gating (Gonzalez et al., 2010; Tombola et al., 2010), the present study does not address gating in general or the cooperative gating of the dimer. The main unique features of hHV1 proton-specific permeation and ΔpH-dependent gating occur both in monomeric and in dimeric hHV1 constructs (Koch et al., 2008; Tombola et al., 2008, 2010; Gonzalez et al., 2010; Musset et al., 2010). Nevertheless, we cannot rule out the possibility that dimeric channels might differ in ways that we cannot now predict.
Comparison with Starace and Bezanilla’s His scanning studies.
Our Arg→His mutations mirror those performed in the Shaker K+ channel VSD by Starace and Bezanilla (Starace et al., 1997; Starace and Bezanilla, 2001, 2004), who mutated each of the first four Arg residues of S4 to His. The picture that emerged was of aqueous crevices on either side of a narrow bridge in the VSD that interrupts aqueous access from one side of the membrane to the other. The probes in these studies were effectively water molecules or, more accurately, hydronium ions. The four mutations (numbering the Arg residues starting from the outside) produced a hyperpolarization-activated proton channel (R1→His), a proton carrier (R2→His or R3→His), or a depolarization activation proton channel (R4→His). In the cases of R1→His or R4→His, the appearance is that in closed or open states, respectively, the His is positioned at a constriction where it is accessible to both sides of the membrane simultaneously. Strictly defined, “accessible” means capable of being protonated by H3O+ or donating a proton to H2O to form H3O+. Based on these studies, one might expect the R205H mutant of hHV1 to be a hyperpolarization-activated proton channel. The family of currents in Fig. 10 D shows that this expectation was not met. There is no significant inward current at negative voltages. We might further expect R208H to be a proton carrier, shuttling protons across the membrane at intermediate voltages where open↔closed transitions occur frequently. Even if this prediction were correct, it would be very difficult to distinguish H+ efflux mediated by channel versus carrier modes. At positive voltages, hHV1 functions as a proton channel (in contrast to the K+ channel VSD that does not normally conduct current), and the rate of transport by a shuttle mechanism is unlikely to be as great as the flux in channel mode. Finally, we might predict that R211H would conduct protons at positive voltages. Because hHV1 does this already, we can think of no way to evaluate this possibility. In summary, the only definitive conclusion from the properties of the Arg→His mutants of hHV1 is that R205H does not act as a hyperpolarization-activated proton channel at negative voltages. Several interpretations are possible. R205H may not be located at the constriction of hHV1 in the closed state. Alternatively, R205H may be located at a constriction of hHV1 in the closed state, but, the geometry of the constriction does not allow H3O+ access from both sides. In any case, hHV1 appears to differ significantly from the Shaker VSD.
In summary, our model indicates that the open state of hHV1 is stabilized by an external charge cluster that includes a prominent Asp112–Arg208 salt bridge, and by a large internal charge network. Both model and His scanning indicate that the third S4 Arg, Arg211, remains accessible to the internal solution in the open state. This result may be understood in light of hHV1 having only three Arg residues in S4, in contrast with approximately seven cationic residues in many other voltage-gated ion channels. In K+ channels, after R1–R4 ratchet through the charge transfer center during opening, several cationic residues remain inside to interact with acidic groups. If S4 in hHV1 ratcheted up so that all three Arg residues in S4 were above the constriction at F150, none would remain inside to interact with internal acidic groups. Here, we propose that the middle Arg (R208) is poised at Asp112 in the open state, with R1 outside and R3 inside, both able to interact with external and internal charge clusters, respectively, and stabilize the open state.
Supplementary Material
Acknowledgments
We thank Chris Neale for providing the preequilibrated POPC structure. Computations were performed on the GPC supercomputer at the SciNet, Westgrid, and Clumeq HPC Consortia. SciNet is funded by the Canada Foundation for Innovation under the auspices of Compute Canada, the Government of Ontario, the Ontario Research Fund, and the University of Toronto.
This work was supported by Canadian Institutes of Health Research (operating grant MOP43949 to R. Pomès), National Science Foundation (award MCB-0943362 to T.E. DeCoursey and S.M.E. Smith), and National Institutes of Health (NIH; grant R01-GM087507 to T.E. DeCoursey). The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institute of General Medical Sciences or the NIH.
Kenton J. Swartz served as editor.
Footnotes
Abbreviations used in this paper:
- hHV1
- human voltage-gated proton channel
- HV1
- voltage-gated proton channel
- MD
- molecular dynamics
- POPC
- 1-palmitoyl-2-oleoylphosphatidylcholine
- RMSD
- root mean square deviation
- TM
- transmembrane
- VSD
- voltage-sensing domain
- VSP
- voltage-sensing phosphatase
References
- Alabi A.A., Bahamonde M.I., Jung H.J., Kim J.I., Swartz K.J. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arinaminpathy Y., Biggin P.C., Shrivastava I.H., Sansom M.S.P. 2003. A prokaryotic glutamate receptor: homology modelling and molecular dynamics simulations of GluR0. FEBS Lett. 553:321–327 10.1016/S0014-5793(03)01036-6 [DOI] [PubMed] [Google Scholar]
- Beckstein O., Sansom M.S.P. 2006. A hydrophobic gate in an ion channel: the closed state of the nicotinic acetylcholine receptor. Phys. Biol. 3:147–159 10.1088/1478-3975/3/2/007 [DOI] [PubMed] [Google Scholar]
- Berger T.K., Isacoff E.Y. 2011. The pore of the voltage-gated proton channel. Neuron. 72:991–1000 10.1016/j.neuron.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla F. 2008. How membrane proteins sense voltage. Nat. Rev. Mol. Cell Biol. 9:323–332 10.1038/nrm2376 [DOI] [PubMed] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. 1983. Charmm—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217 10.1002/jcc.540040211 [DOI] [Google Scholar]
- Capasso M., Bhamrah M.K., Henley T., Boyd R.S., Langlais C., Cain K., Dinsdale D., Pulford K., Khan M., Musset B., et al. 2010. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat. Immunol. 11:265–272 10.1038/ni.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capener C.E., Sansom M.S.P. 2002. Molecular dynamics simulations of a K channel model: Sensitivity to changes in ions, waters, and membrane environment. J. Phys. Chem. B. 106:4543–4551 10.1021/jp0129986 [DOI] [Google Scholar]
- Chakrabarti N., Roux B., Pomès R. 2004. Structural determinants of proton blockage in aquaporins. J. Mol. Biol. 343:493–510 10.1016/j.jmb.2004.08.036 [DOI] [PubMed] [Google Scholar]
- Chakrabarti N., Neale C., Payandeh J., Pai E.F., Pomès R. 2010. An iris-like mechanism of pore dilation in the CorA magnesium transport system. Biophys. J. 98:784–792 10.1016/j.bpj.2009.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Brooks C.L., III 2007. Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins. 67:922–930 10.1002/prot.21345 [DOI] [PubMed] [Google Scholar]
- Cherny V.V., DeCoursey T.E. 1999. pH-dependent inhibition of voltage-gated H+ currents in rat alveolar epithelial cells by Zn2+ and other divalent cations. J. Gen. Physiol. 114:819–838 10.1085/jgp.114.6.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny V.V., Murphy R., Sokolov V., Levis R.A., DeCoursey T.E. 2003. Properties of single voltage-gated proton channels in human eosinophils estimated by noise analysis and by direct measurement. J. Gen. Physiol. 121:615–628 10.1085/jgp.200308813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey T.E. 2003. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83:475–579 10.1152/physrev.00028.2002 [DOI] [PubMed] [Google Scholar]
- DeCoursey T.E. 2010. Voltage-gated proton channels find their dream job managing the respiratory burst in phagocytes. Physiology (Bethesda). 25:27–40 10.1152/physiol.00039.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaurex N., El Chemaly A. 2010. Physiological roles of voltage-gated proton channels in leukocytes. J. Physiol. 588:4659–4665 10.1113/jphysiol.2010.194225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzubiella J., Allen R.J., Hansen J.P. 2004. Electric field-controlled water permeation coupled to ion transport through a nanopore. J. Chem. Phys. 120:5001–5004 10.1063/1.1665656 [DOI] [PubMed] [Google Scholar]
- England J.L., Haran G. 2011. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu. Rev. Phys. Chem. 62:257–277 10.1146/annurev-physchem-032210-103531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593 10.1063/1.470117 [DOI] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer H. 2012. Function of proton channels in lung epithelia. Wiley Interdiscip Rev Membr Transp Signal. 1:247–258 10.1002/wmts.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest L.R., Tang C.L., Honig B. 2006. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys. J. 91:508–517 10.1529/biophysj.106.082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freites J.A., Tobias D.J., White S.H. 2006. A voltage-sensor water pore. Biophys. J. 91:L90–L92 10.1529/biophysj.106.096065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C., Koch H.P., Drum B.M., Larsson H.P. 2010. Strong cooperativity between subunits in voltage-gated proton channels. Nat. Struct. Mol. Biol. 17:51–56 10.1038/nsmb.1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C., Rebolledo S., Perez M.E., Larsson H.P. 2013. Molecular mechanism of voltage sensing in voltage-gated proton channels. J. Gen. Physiol. 141:275–285 10.1085/jgp.201310970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin-Badaroudine P., Keller D.I., Huang H., Pouliot V., Chatelier A., Osswald S., Brink M., Chahine M. 2012. A proton leak current through the cardiac sodium channel is linked to mixed arrhythmia and the dilated cardiomyopathy phenotype. PLoS ONE. 7:e38331 10.1371/journal.pone.0038331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia-Oliveira R., Sackett P.W., Pedersen A.G. 2007. MaxAlign: maximizing usable data in an alignment. BMC Bioinformatics. 8:312 10.1186/1471-2105-8-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Gurd F.R., Wilcox P.E. 1956. Complex formation between metallic cations and proteins, peptides and amino acids. Adv. Protein Chem. 11:311–427 10.1016/S0065-3233(08)60424-6 [DOI] [PubMed] [Google Scholar]
- Henderson L.M., Chappell J.B., Jones O.T.G. 1988. Superoxide generation by the electrogenic NADPH oxidase of human neutrophils is limited by the movement of a compensating charge. Biochem. J. 255:285–290 [PMC free article] [PubMed] [Google Scholar]
- Henrion U., Zumhagen S., Steinke K., Strutz-Seebohm N., Stallmeyer B., Lang F., Schulze-Bahr E., Seebohm G. 2012. Overlapping cardiac phenotype associated with a familial mutation in the voltage sensor of the KCNQ1 channel. Cell. Physiol. Biochem. 29:809–818 [DOI] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J. 1997. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472 [DOI] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. 2008. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4:435–447 10.1021/ct700301q [DOI] [PubMed] [Google Scholar]
- Hildebrand P.W., Goede A., Bauer R.A., Gruening B., Ismer J., Michalsky E., Preissner R. 2009. SuperLooper—a prediction server for the modeling of loops in globular and membrane proteins. Nucleic Acids Res. 37:W571–W574 10.1093/nar/gkp338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jacobson M., Sali A. 2004. Comparative protein structure modeling and its applications to drug discovery. Annu. Rep. Med. Chem. 39:259–276 10.1016/S0065-7743(04)39020-2 [DOI] [Google Scholar]
- Jensen M.O., Borhani D.W., Lindorff-Larsen K., Maragakis P., Jogini V., Eastwood M.P., Dror R.O., Shaw D.E. 2010. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl. Acad. Sci. USA. 107:5833–5838 10.1073/pnas.0911691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.O., Jogini V., Borhani D.W., Leffler A.E., Dror R.O., Shaw D.E. 2012. Mechanism of voltage gating in potassium channels. Science. 336:229–233 10.1126/science.1216533 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B.T., MacKinnon R. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41 10.1038/nature01580 [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935 10.1063/1.445869 [DOI] [Google Scholar]
- Jorgensen W.L., Maxwell D.S., Tirado-Rives J. 1996. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118:11225–11236 10.1021/ja9621760 [DOI] [Google Scholar]
- Kandt C., Ash W.L., Tieleman D.P. 2007. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 41:475–488 10.1016/j.ymeth.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Koch H.P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., Larsson H.P. 2008. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 105:9111–9116 10.1073/pnas.0801553105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson H.P., Baker O.S., Dhillon D.S., Isacoff E.Y. 1996. Transmembrane movement of the shaker K+ channel S4. Neuron. 16:387–397 10.1016/S0896-6273(00)80056-2 [DOI] [PubMed] [Google Scholar]
- Law R.J., Sansom M.S. 2004. Homology modelling and molecular dynamics simulations: comparative studies of human aquaporin-1. Eur. Biophys. J. 33:477–489 10.1007/s00249-004-0398-z [DOI] [PubMed] [Google Scholar]
- Le S.Q., Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307–1320 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Lee A., Chen J., MacKinnon R. 2005. Structure of the KvAP voltage-dependent K+ channel and its dependence on the lipid membrane. Proc. Natl. Acad. Sci. USA. 102:15441–15446 10.1073/pnas.0507651102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Letts J.A., Mackinnon R. 2008. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc. Natl. Acad. Sci. USA. 105:7692–7695 10.1073/pnas.0803277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Letts J.A., MacKinnon R. 2009. Functional reconstitution of purified human Hv1 H+ channels. J. Mol. Biol. 387:1055–1060 10.1016/j.jmb.2009.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23:127–128 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Levitt D.G., Elias S.R., Hautman J.M. 1978. Number of water molecules coupled to the transport of sodium, potassium and hydrogen ions via gramicidin, nonactin or valinomycin. Biochim. Biophys. Acta. 512:436–451 10.1016/0005-2736(78)90266-3 [DOI] [PubMed] [Google Scholar]
- Lin M.C., Hsieh J.Y., Mock A.F., Papazian D.M. 2011. R1 in the Shaker S4 occupies the gating charge transfer center in the resting state. J. Gen. Physiol. 138:155–163 10.1085/jgp.201110642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Maragakis P., Piana S., Eastwood M.P., Dror R.O., Shaw D.E. 2012. Systematic validation of protein force fields against experimental data. PLoS ONE. 7:e32131 10.1371/journal.pone.0032131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko P.V., Botchkina I.L., Fedorenko A., Kirichok Y. 2010. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 140:327–337 10.1016/j.cell.2009.12.053 [DOI] [PubMed] [Google Scholar]
- Long S.B., Tao X., Campbell E.B., MacKinnon R. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 450:376–382 10.1038/nature06265 [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435:1239–1243 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- Musset B., Cherny V.V., Morgan D., Okamura Y., Ramsey I.S., Clapham D.E., DeCoursey T.E. 2008. Detailed comparison of expressed and native voltage-gated proton channel currents. J. Physiol. 586:2477–2486 10.1113/jphysiol.2007.149427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset B., Smith S.M.E., Rajan S., Cherny V.V., Sujai S., Morgan D., DeCoursey T.E. 2010. Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J. Physiol. 588:1435–1449 10.1113/jphysiol.2010.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset B., Smith S.M.E., Rajan S., Morgan D., Cherny V.V., DeCoursey T.E. 2011. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.D., Kuan G., Saier M.H., Jr, Montal M. 1999. Modular assembly of voltage-gated channel proteins: a sequence analysis and phylogenetic study. J. Mol. Microbiol. Biotechnol. 1:281–287 [PubMed] [Google Scholar]
- Néron B., Ménager H., Maufrais C., Joly N., Maupetit J., Letort S., Carrere S., Tuffery P., Letondal C. 2009. Mobyle: a new full web bioinformatics framework. Bioinformatics. 25:3005–3011 10.1093/bioinformatics/btp493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nina M., Beglov D., Roux B. 1997. Atomic radii for continuum electrostatics calculations based on molecular dynamics free energy simulations. J. Phys. Chem. B. 101:5239–5248 10.1021/jp970736r [DOI] [Google Scholar]
- Nosé S. 1984. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81:511 10.1063/1.447334 [DOI] [Google Scholar]
- Notredame C., Higgins D.G., Heringa J. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Okamura Y., Murata Y., Iwasaki H. 2009. Voltage-sensing phosphatase: actions and potentials. J. Physiol. 587:513–520 10.1113/jphysiol.2008.163097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian D.M., Shao X.M., Seoh S.A., Mock A.F., Huang Y., Wainstock D.H. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301 10.1016/0896-6273(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Parrinello M., Rahman A. 1980. Crystal structure and pair potentials: A molecular-dynamics study. Phys. Rev. Lett. 45:1196–1199 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Payandeh J., Scheuer T., Zheng N., Catterall W.A. 2011. The crystal structure of a voltage-gated sodium channel. Nature. 475:353–358 10.1038/nature10238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J., Gamal El-Din T.M., Scheuer T., Zheng N., Catterall W.A. 2012. Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature. 486:135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Kim B.H., Grishin N.V. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36:2295–2300 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomès R., Roux B. 1996. Structure and dynamics of a proton wire: a theoretical study of H+ translocation along the single-file water chain in the gramicidin A channel. Biophys. J. 71:19–39 10.1016/S0006-3495(96)79211-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomès R., Roux B. 2002. Molecular mechanism of H+ conduction in the single-file water chain of the gramicidin channel. Biophys. J. 82:2304–2316 10.1016/S0006-3495(02)75576-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey I.S., Moran M.M., Chong J.A., Clapham D.E. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey I.S., Mokrab Y., Carvacho I., Sands Z.A., Sansom M.S., Clapham D.E. 2010. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat. Struct. Mol. Biol. 17:869–875 10.1038/nsmb.1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval A., Piana S., Eastwood M.P., Dror R.O., Shaw D.E. 2012. Refinement of protein structure homology models via long, all-atom molecular dynamics simulations. Proteins. 80:2071–2079 [DOI] [PubMed] [Google Scholar]
- Roth R., Gillespie D., Nonner W., Eisenberg R.E. 2008. Bubbles, gating, and anesthetics in ion channels. Biophys. J. 94:4282–4298 10.1529/biophysj.107.120493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata S., Kurokawa T., Nørholm M.H., Takagi M., Okochi Y., von Heijne G., Okamura Y. 2010. Functionality of the voltage-gated proton channel truncated in S4. Proc. Natl. Acad. Sci. USA. 107:2313–2318 10.1073/pnas.0911868107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Sands Z.A., Sansom M.S. 2007. How does a voltage sensor interact with a lipid bilayer? Simulations of a potassium channel domain. Structure. 15:235–244 10.1016/j.str.2007.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Takagi M., Okamura Y. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- Shen M.Y., Sali A. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15:2507–2524 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Morgan D., Musset B., Cherny V.V., Place A.R., Hastings J.W., DeCoursey T.E. 2011. Voltage-gated proton channel in a dinoflagellate. Proc. Natl. Acad. Sci. USA. 108:18162–18167 10.1073/pnas.1115405108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace D.M., Bezanilla F. 2001. Histidine scanning mutagenesis of basic residues of the S4 segment of the Shaker K+ channel. J. Gen. Physiol. 117:469–490 10.1085/jgp.117.5.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace D.M., Bezanilla F. 2004. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 427:548–553 10.1038/nature02270 [DOI] [PubMed] [Google Scholar]
- Starace D.M., Stefani E., Bezanilla F. 1997. Voltage-dependent proton transport by the voltage sensor of the Shaker K+ channel. Neuron. 19:1319–1327 10.1016/S0896-6273(00)80422-5 [DOI] [PubMed] [Google Scholar]
- Struyk A.F., Cannon S.C. 2007. A Na+ channel mutation linked to hypokalemic periodic paralysis exposes a proton-selective gating pore. J. Gen. Physiol. 130:11–20 10.1085/jgp.200709755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz K.J. 2008. Sensing voltage across lipid membranes. Nature. 456:891–897 10.1038/nature07620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X., Lee A., Limapichat W., Dougherty D.A., MacKinnon R. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman D.P., Sansom M.S.P., Berendsen H.J.C. 1999. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys. J. 76:40–49 10.1016/S0006-3495(99)77176-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff S.K., Lin M.A., Schulteis C.T., Papazian D.M. 2000. Voltage-dependent structural interactions in the Shaker K+ channel. J. Gen. Physiol. 115:123–138 10.1085/jgp.115.2.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F., Ulbrich M.H., Isacoff E.Y. 2008. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 58:546–556 10.1016/j.neuron.2008.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola F., Ulbrich M.H., Kohout S.C., Isacoff E.Y. 2010. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat. Struct. Mol. Biol. 17:44–50 10.1038/nsmb.1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li S.J., Wu X., Che Y., Li Q. 2012. Clinicopathological and biological significance of human voltage-gated proton channel Hv1 protein overexpression in breast cancer. J. Biol. Chem. 287:13877–13888 10.1074/jbc.M112.345280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.L., Schow E.V., Freites J.A., White S.H., Tombola F., Tobias D.J. 2012. Water wires in atomistic models of the Hv1 proton channel. Biochim. Biophys. Acta. 1818:286–293 10.1016/j.bbamem.2011.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.J., Wu G., Akhavan Sharif M.R., Baker A., Jia Y., Fahey F.H., Luo H.R., Feener E.P., Clapham D.E. 2012. The voltage-gated proton channel Hv1 enhances brain damage from ischemic stroke. Nat. Neurosci. 15:565–573 10.1038/nn.3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., George A.L.J., Jr, Horn R. 1996. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 16:113–122 10.1016/S0896-6273(00)80028-8 [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V., DeCaen P.G., Westenbroek R.E., Pan C.Y., Scheuer T., Baker D., Catterall W.A. 2012. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. USA. 109:E93–E102 10.1073/pnas.1118434109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F.H., Catterall W.A. 2004. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci. STKE. 2004:re15 10.1126/stke.2532004re15 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ren W.L., DeCaen P., Yan C.Y., Tao X., Tang L., Wang J.J., Hasegawa K., Kumasaka T., He J.H., et al. 2012. Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature. 486:130–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.