Abstract
The nutrient germinant receptors (nGRs) of spores of Bacillus species are clusters of three proteins that play a critical role in triggering the germination of dormant spores in response to specific nutrient molecules. Here we report the crystal structure of the C protein of the GerB germinant receptor, so called GerBC, of Bacillus subtilis spores at 2.3 Å resolution. The GerBC protein adopts a previously uncharacterized type of protein fold consisting of three distinct domains, each of which is centered by a β sheet surrounded by multiple α helices. Secondary structure prediction and structure-based sequence alignment suggest that the GerBC structure represents the prototype for C subunits of nGRs from spores of all Bacillales and Clostridiales species and defines two highly conserved structural regions in this family of proteins. GerBC forms an interlocked dimer in the crystalline state but is predominantly monomeric in solution, pointing to the possibility that GerBC oligomerizes as a result of either high local protein concentrations or interaction with other nGR proteins in spores. Our findings provide the first structural view of the nGR subunits and a molecular framework for understanding the architecture, conservation and function of nGRs.
Keywords: Bacillus subtilis, spores, spore germination, germinant receptors, protein structure
One of the most remarkable features of Bacillus species is their ability to form spores in sporulation, a process triggered by starvation for one or more nutrients.1,2 These spores are metabolically dormant and extremely resistant to a large variety of environmental stresses, properties that allow spores to survive for years, with some reports suggesting that spores may survive for millions of years.3–5 However, during their long period of dormancy, spores are constantly sensing the environment, and when nutrients return, spores can rapidly come back to life in the process of spore germination followed by outgrowth.6–8 Since spores of a number of species are major agents of food spoilage and food borne disease, as well as bioterrorism (Bacillus anthracis), there is much interest in methods to efficiently kill the extremely resistant dormant spores.1 Consequently, a detailed understanding of the spore germination process may have significant applied importance, because spores lose their resistance properties upon germination and are thus relatively easy to kill.1,2,7,8
The sensors of nutrients that trigger spore germination are termed nutrient germinant receptors (nGRs). Spores of Bacillus species have multiple individual nGRs, proteins of which share obvious amino acid sequence homology, although different nGRs respond to different nutrient germinants. The nGRs in Bacillus species are most commonly encoded by tricistronic operons encoding proteins A, B and C, and these operons are expressed only late in sporulation and only within the developing spore.6–8 In Bacillus subtilis spores there are three important nGRs, GerA, GerB and GerK. The GerA nGR triggers spore germination in response to L-alanine or L-valine, while the GerB and GerK nGRs are required together for spore germination with a mixture of L-asparagine, D-glucose, D-fructose and K+ ions (AGFK). Loss of any subunit of a particular nGR generally abolishes the function of the whole nGR, but has minimal effects on the function of other nGRs. Indeed, there generally appears to be little if any exchange of subunits between different nGRs in B. subtilis spores with normal nGR levels.9 In addition to the nGRs, the GerD protein of Bacillus species is also required for efficient spore germination in response to all nutrient germinants.10,11 While GerD is likely located in the spores’ inner membrane, where the nGRs are located, the exact function of GerD in spore germination remains unknown.
Despite extensive study of spore germination over the past decade, the mechanism of nGR action remains to be determined. What is known about nGR function includes the following.6–8; 12–14 1) Metabolism of nutrient germinants is not involved in nGR function, as the appropriate nutrient germinant(s) appears only to bind to the nGRs, with this binding then somehow triggering spore germination. 2) The nGRs appear to contain all three proteins encoded by a particular nGR operon, although the evidence for this is largely genetic. 3) The nGRs are located in the spore’s inner membrane that surrounds the spore’s central region, and two of the nGR subunits, the A and B proteins, are almost certainly integral membrane proteins based on their primary sequence, with multiple predicted membrane spanning regions. 4) The third subunit of nGRs, the C protein, is not an integral membrane protein, but is most likely located on the membrane periphery and probably held there by a diacylglycerol anchor attached to an amino-terminal cysteine residue (Fig. S6). This diacylglycerol anchor often is essential for nGR function. 5) The A proteins of different nGRs exhibit significant sequence identity and this is also the case for the B and C proteins. In B. subtilis the amino acid sequence identities are between 19–42% for the A, B or C proteins from the three functional nGRs. 6) The sequences of the B subunits of the nGRs show weak homology with a subfamily of single component bacterial membrane transporters; however, the A and C proteins of nGRs exhibit no sequence homology to other proteins. 7) Average levels of the various nGRs in spores are likely very low, probably < 100 molecules/spore.8
Despite what is known about nGRs as noted above, there are some major unknowns including: i) what the nGRs actually do to trigger germination; ii) the oligomerization states of the subunit proteins and the subunit stoichiometry in the nGRs; iii) how individual subunit proteins function in the nGRs; and iv) how germinant binding triggers nGR function. In order to obtain insight into the function and regulation of nGRs, we have initiated attempts to obtain high-resolution structures of nGR proteins by X-ray crystallography. In this report we present the first crystal structure of an nGR subunit, that of the B. subtilis GerBC protein. The crystal structure reveals a novel protein fold characterized by three distinct domains. Both secondary structure prediction and structure-based alignment suggest that this fold represents the prototype of the nGR C proteins and is likely shared by the C subunits of all nGRs. Biochemical and biophysical studies indicate that GerBC exists predominately as a monomer in solution, although it appears to be a dimer in crystals. Our results open an avenue for understanding the structural organization of nGRs and the molecular basis of their functions in the germination process.
Overall structure of the GerBC protein
Crystals of the GerBC protein were obtained by using a truncated B. subtilis GerBC (residues 25–374) lacking the N-terminal signal and lipobox sequences in 24 mostly hydrophobic residues. The GerBC crystals contain one protein molecule per asymmetric unit. The GerBC structure was determined and refined at a resolution of 2.3 Å by the single-wavelength anomalous dispersion (SAD) method using data from a selenomethionine-substituted crystal (Table 1). The calculated electron density map allowed unambiguous tracing of most of the protein except three disordered loops (residues 57–63, 147–168 and 287–291). The final structure was refined to an R factor of 21.6% and an Rfree of 23.4% with good stereochemistry (Table 1).
Table 1.
Summary of crystallographic analysis
| Native | SeMet (peak) | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.075 | 0.9791 |
| Space group | R32 | R32 |
| Cell dimensions (Å) | ||
| a, b, c (Å) | 142.8, 142.8, 187.8 | 142.4, 142.4, 187.7 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 50.0-2.30 (2.34-2.30) | 50.0-2.54 (2.59-2.54) |
| Rsym (%) | 6.8 (84.0) | 10.2 (87.4) |
| I/σ̣I | 45.1 (4.8) | 28.8 (4.1) |
| Completeness (%) | 99.9 (100.0) | 100.0 (100.0) |
| Redundancy | 21.8 (21.3) | 22.3 (22.4) |
| SAD Analysis | ||
| Rms FH/ε | 0.85 | |
| Mean FOM before DM | 0.43 | |
| Mean FOM after DM | 0.64 | |
| Refinement | ||
| Resolution (Å) | 50.0–2.30 | |
| No. of reflections (|F|>0σ) | 31,175 | |
| R-factor/Rfree | 21.6/23.4 | |
| Total protein atoms | 2,510 | |
| Water molecules | 178 | |
| B-factors (Å2) | ||
| Protein | 31.64 | |
| Water | 39.15 | |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 1.161 | |
| Ramachandran (%) | ||
| Within favored | 98.0 | |
| Within allowed | 99.8 | |
| Outliers | 2 (Gln285 and Lys294) | |
The detailed experimental procedures are provided in SI Materials and Methods. In brief, recombinant B. subtilis GerBC (residues 25–374) was expressed as a His6 fusion protein in Escherichia coli. The GerBC protein was crystallized from a solution consisting of 1.5–1.7 M ammonium sulfate and 0.1 M sodium acetate (pH 4.6) by the hanging-drop vapor diffusion method at 4°C. The structure of GerBC was determined by the single-wavelength anomalous dispersion (SAD) method using data from a selenomethionine-substituted crystal.
Values in parentheses are for the highest-resolution shell. Rsym = ΣhΣj |Ih,j−Ih|/ΣhΣj Ih,j for the intensity (I) of i observations of reflection h. Rms FH/ε = (1/nΣFH2)1/2/(1/nΣε2)1/2, where FH is the structure factor amplitude for anomalous scatterers and ε is the residual lack of closure error. Figure of Merit (FOM) = <ΣP(α) exp(iα)/ΣP(α)>, where P(α) is the probability distribution for the phase. FOM before DM indicates the figure of merit before density modification. R-factor = Σ‖Fobs|−|Fcalc‖/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively. Rfree = R-factor calculated using 5% of the reflection data chosen randomly and omitted from the start of refinement.
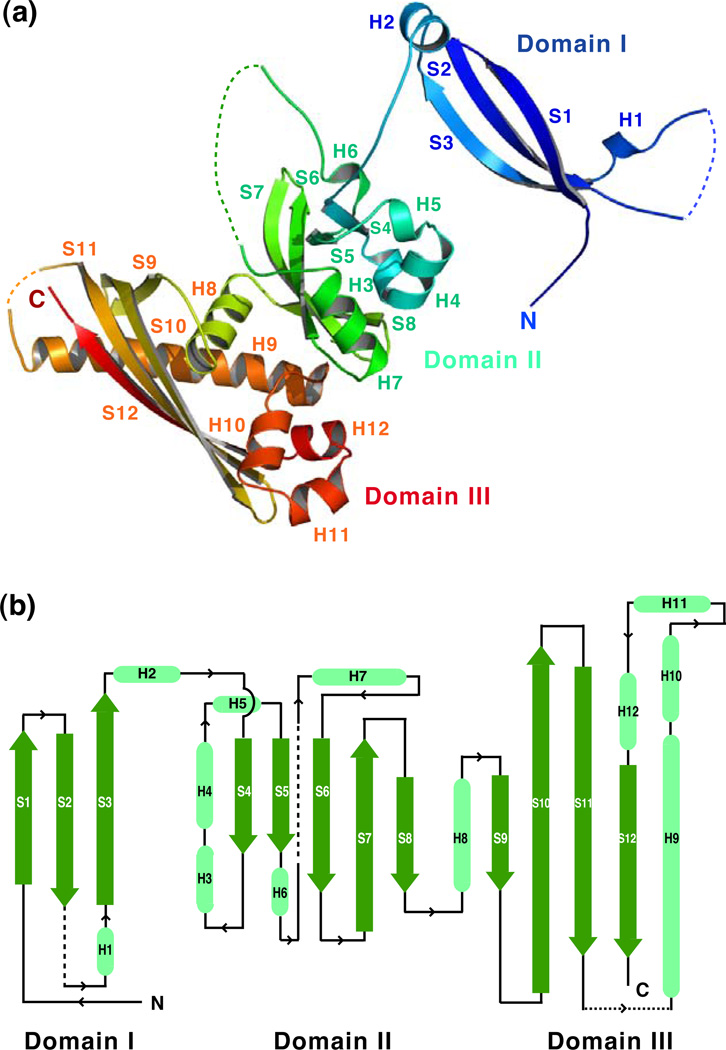
GerBC has an elongated structure resembling a twisted letter M with linear dimensions of approximately 99 Å × 59 Å × 24 Å (Fig. 1a). The overall architecture of the protein is organized into three distinct domains, each of which is centered by a β sheet (Fig. 1a and b). The N-terminal domain (Domain I) is dominated by a β structure composed of three β strands forming an antiparallel β sheet that is edged by two short α helices (strands S1–S3 and helices H1–H2). The middle domain (Domain II) adopts a compact α/β fold composed of a central five-stranded β sheet surrounded by five short α helices (strands S4–S8 and helices H3–H7). The C-terminal domain (Domain III) exhibits an intertwined α/β sandwich structure featured by a curved four-stranded β sheet (strands S9–S12 and helices H8–H11). The electronic potential surface of the GerBC protein reveals two highly positively charged regions in the β sheets of Domains II and III, and two negatively charged patches on the back of these sheets (Fig. S1).
Figure 1. a,b. Overall structure of the GerBC protein.
(a) Ribbon diagram of the GerBC protein colored in rainbow from blue to red.
(b) The topology diagram of the GerBC protein. α helices and β strands are shown in light green and green, respectively. Dotted lines represent disordered regions in the crystal structure.
Interestingly, Domain I of GerBC is positioned well away from Domain II of the protein by an extended 10 amino-acid residue loop between helix H2 and strand S4 (Fig. 1a and b). We should point out that in the crystal structure two GerBC molecules assemble into an interlocked dimer created by a crystallographic two-fold axis (Fig. S2). In the dimer, the entire Domain I of one monomer is placed between Domains I and II of the other molecule, thus allowing the formation of an extended intermolecular 16-stranded β sheet connecting Domains I and II from both protomers (Fig. S2). The GerBC dimer interface buries a large amount of the surface area in two monomers, i.e. ~7000 Å2 or ~20% of the total solvent-accessible surface, raising the possibility that formation of the domain-swapped dimer could help stabilize the relative orientation of Domains I and II in the monomer. Nevertheless, the dimeric nature of GerBC was not observed in solution with relatively low concentrations of the protein (see below).
The structure of GerBC is unique
The crystal structure of GerBC reveals a peculiar topology, which does not seem homologous to any other known fold in the Protein Data Bank. An extensive search in the structural database, using the Dali15, CACH16, ProFunc17 and Pfam18 servers, did not produce any significant matches (Z-scores < 5; root-mean-square deviation (rmsd) > 10 Å). In some cases, a particular arrangement of a few elements of secondary structure was recognized in proteins with known structures, but no protein was found to have an overall fold similar to that of GerBC. We have thus performed the Dali search using the individual domains of GerBC.
For GerBC Domain I, its antiparallel β sheet arrangement was found to have a fair similarity with a part of the Pseudomonas aeruginosa lectin that has an overall β sandwich fold19 (Z-score = 3.0; rmsd = 4.3 Å) (Fig. S3). Domain II of GerBC consists of a highly curved five-stranded β sheet in which two of the strands (S4 and S5) are almost perpendicular to the other three strands (S6–S8) (Fig. 1a). The sheet is decorated on two sides by five short α helices (H3–H7). The Dali search found that the Domain II of GerBC has a somewhat similar fold to several PAS domain family proteins; PAS domain-containing proteins participate in a wide variety of biological and biochemical processes and frequently act as environmental sensors.20 The common PAS fold comprises a five-stranded β sheet with 3–5 α helices located on both sides. Still, GerBC Domain II can only be superimposed partially on the N-terminal PAS domain of the HERG voltage-dependent potassium channel21 with a low Z-score (2.4) and a high rmsd (3.3 Å) (Fig. S4). Domain III of GerBC adopts a cylinder-like fold with the highly curved four-stranded β sheet (S9–S12) as well as a 27-residue α helix (H9) that diagonally crosses the β sheet forming the tube, and four short helices (H8, H10–H12) at the base (Fig. 1a). The Dali search found that Domain III of GerBC shows some structural resemblance with a hypothetical osmotically-inducible protein MPN625 from Mycoplasma pneumoniae22 (Z-score = 4.3; rmsd = 3.4 Å) (Fig. S5). MPN625 has a similar overall topology to GerBC Domain III but with a three-stranded β sheet and two long α helices. Taken together, we conclude that GerBC possesses three uniquely folded domains and represents a previously uncharacterized type of protein fold.
The structure of GerBC is conserved among its homologs
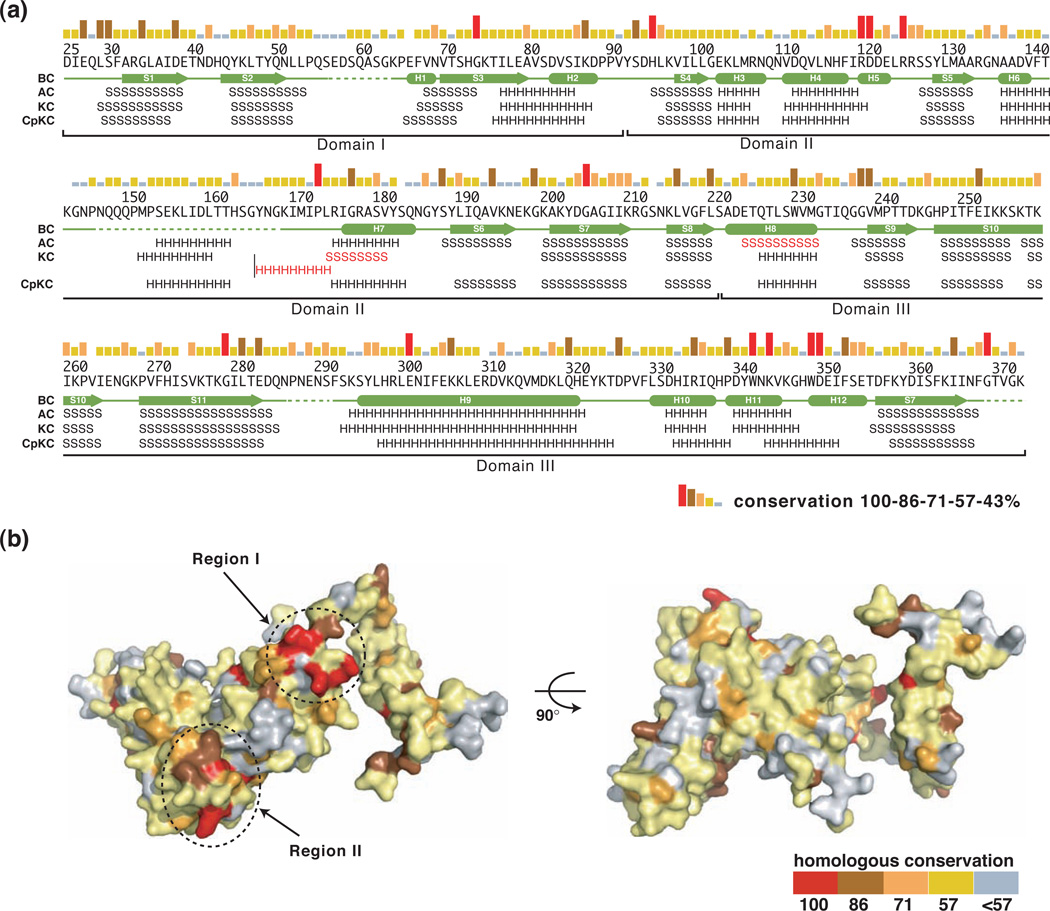
A broad BLAST search using the B. subtilis GerBC protein as the query sequence has identified more than 100 homologs of GerBC in 21 completed spore-forming Bacillacae genomes, with 2–15 C genes per species and an average of five genes per species (data not shown). In addition, a search using a characterized GerBC homolog from Clostridium perfringens (termed GerKC)23 has identified more than 50 GerBC homologs in 23 annotated Clostridiales genomes. Sequence alignments of these GerBC homologs show that they share ~17–66% pairwise sequence identities; in particular, the four likely GerBC proteins from the closely related species Bacillus amyloliquefaciens, Bacillus pumilus, Bacillus clausii, and Bacillus cereus, have sequence identities ranging from 29% to 66% (Figs. 2a and S6). In addition, the C subunits of the two other important B. subtilis nGRs, GerAC and GerKC, share 33% and 24% pairwise sequence identities with GerBC, respectively. As shown in Figure 2a, however, these highly conserved residues are located throughout the entire sequences, and it was not immediately clear whether they play any role in mediating C protein function.
Figure 2. a,b. Sequence conservation between GerBC and its homologs.
(a) Sequence conservation and secondary structure elements for GerBC and its homologs. Sequence conservation is shown as a bar graph, with red bars indicating identity among the seven GerBC homologs from B. subtilis and the closely related species B. amyloliquefaciens, B. pumilus, B. clausii, and B. cereus (see also Figure S6). Secondary structure assignments of GerBC (BC) from the crystal structure are shown as green cylinders (α helices) and arrows (β strands), while disordered regions are shown as dashed lines. Predicted secondary structure elements (http://www.predictprotein.org) for B. subtilis GerAC and GerKC (AC and KC, respectively), and C. perfringens GerKC (CpKC) are indicated by letters (“H”: α helix; “S”: β strand) and the major differences are highlighted in red.
(b) Molecular surface of GerBC colored according to homologous conservation.
With the structure of GerBC at hand, we can now elaborate on several questions relating to sequence and structure conservations among GerBC homologs. First, other C proteins likely have structures similar to that of B. subtilis GerBC. Most C proteins identified in Bacillacae and Clostridiales have sizes similar to that of B. subtilis GerBC. For example, B. subtilis GerAC has 373 amino acid residues, which is nearly identical to the size of GerBC (374 residues). Secondary structure predictions by the programs PHD24 and Jpred25 suggest that GerAC exhibits essentially identical secondary structure topology to that of GerBC, except for helix H8 of GerBC, which is predicted as a strand in GerAC (Fig. 2a). We note that this site in GerBC was also predicted as a β strand, although all the other predicted secondary elements matched very well with the determined structure. Compared to GerAC and GerBC, B. subtilis GerKC is larger, with 407 amino-acid residues. While the overall topology of GerKC closely resembles that of GerBC, the secondary structure predictions suggest that the sequence insertions in GerKC relative to GerBC occur in two loops joining strand S2 and helix H1 in Domain I as well as the helices H6 and H7 in Domain II (Fig. 2a and S6). Both loops in GerBC are highly flexible and partially disordered in the crystal structure. We then assessed the secondary structure prediction for the evolutionarily more distantly related C protein from C. perfringens SM101 (GenBank accession no. YP_697943 (gerKC)). Again, this C protein shares a stunningly similar predicted secondary structure topology with GerBC, which is significant, given that this C protein has only 19% pairwise sequence identities to GerBC (Fig. 2a). These analyses suggest that the core fold of the GerBC structure is expected to be a prototype fold for the entire C protein family. We posit that differences in the loop regions may be reflective of differences in specific functions of individual C proteins.
Second, the investigation of homologous sequence conservation in the structural context identifies two highly conserved surface areas (regions I and II) in the GerBC homologs (Fig. 2b). To illustrate this feature, we mapped the conservation of five annotated GerBC proteins from the closely related species B. amyloliquefaciens, B. pumilus, B. clausii, and B. cereus, and the B. subtilis GerAC and GerKC onto the surface of the GerBC structure. As shown in Figure 2b, region I is located on the starting edge of the central β sheet in Domain II including five invariant residues, His95 (H2-S4 loop), Arg120 (H5), Asp121 (H5), Arg125 (H5-S5 loop) and Pro173 (H6–H7 loop). Region II is located on the base of the β sheet in Domain III, including four invariant residues, Trp342 (H11), Lys344 (H11), Trp349 (H12) and Asp350 (H12). These two regions have much higher sequence conservation than the rest of the structure. Importantly, these invariant residues in two regions are also highly conserved in more than 100 C proteins identified from other Bacillacae and Clostridiales species (data not shown), suggesting they may play an important role in mediating general functions of the C proteins. Indeed, our preliminary data show that both double (R120A/D121A in Region I) and quadruple (W342A/K344A/W349A/D350A in Region II) GerBC mutants in Regions I and II substantially impaired the B. subtilis germination with the AGFK mixture but not with L-valine (data not shown). Thus, both conserved regions are important for GerBC function in AGFK-mediated spore germination in B. subtilis.
GerBC is a monomer in solution
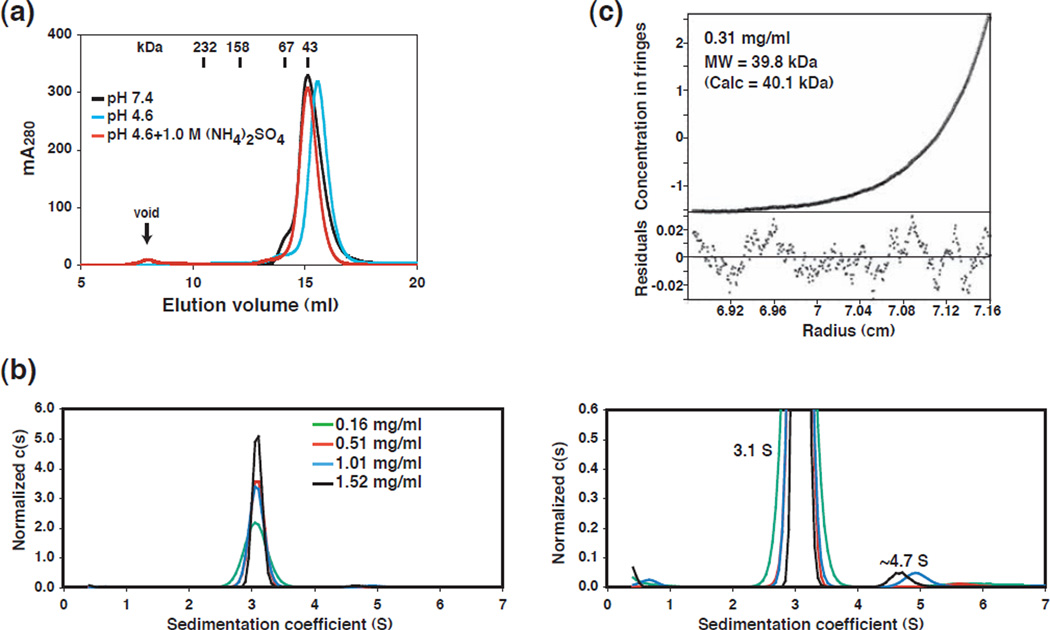
Given that GerBC assembles into a tightly interlocked dimer in the crystal state, we sought to examine whether GerBC also forms a dimer in solution. Gel filtration chromatography showed that GerBC eluted as a single and symmetrical peak in pH 7.4 with an apparent molecular weight of ~43 kDa, comparable to the calculated 40.1 kDa molecular mass of a monomer (Fig. 3a). Moreover, both sedimentation velocity and equilibrium ultracentrifugation experiments confirmed that GerBC forms predominantly a monomer in solution. In particular, the velocity g(s*) analyses showed that the GerBC protein exists mainly as a single species in solution (Fig. S7a). The c(s) analysis indicated that the majority of the GerBC protein (~98%) in solution is present in the main peak at 3.1 S, while a small amount of the protein (~2%) sediments faster than the main peak, and is presumably the GerBC dimer (Fig. 3b). The global fit of the velocity data, using SEDANAL, best fit a model of a mixture of monomer and dimer, with the monomer molecular weight of 40.5 kDa and 2.5% of dimer (by weight) present (data not shown). Moreover, the sedimentation equilibrium data indicated that the system is best modeled by a monomer-dimer self-association along with the presence of a small amount (~7%) of incompetent dimer (Fig. 3c and S7b). The fitted value for the molecular weight (39.8 kDa) agrees quite well with the expected value for the GerBC monomer. The self-association, if real, is very weak (Kd of ~2 mM) and just within the limits of detection for this method.
Figure 3. a–c. Oligomerization state of the GerBC protein.
(a) Overlay of gel filtration chromatography profiles of GerBC under different solution conditions. The retention volumes of proteins of known mass and the void volume of the Superdex 200 column are indicated.
(b) Sedimentation velocity analysis of the GerBC protein loaded at four concentrations as indicated. The sedimentation velocity traces were analyzed using SEDFIT29 to obtain c(s) distributions and were normalized by height (left). The figure on the right shows the c(s) data at a scale of 10× that of the full scale plot, with the peak near s = 3.1 S corresponding to a monomer and the peak near s = 4.7 S to a dimer.
(c) Representative sedimentation equilibrium data for GerBC (0.31 mg/ml and 23 krpm) (see also Figure S7b). The line shows a global nonlinear least squares fit using a monomer-dimer self-association model incorporating an incompetent dimer. The best-fit parameters are as follows: Kd = 2.2 (1.4, 5.5) mM with 7% incompetent dimer and rms = 0.0035 mg/ml.
GerBC was crystallized from a solution consisting of 1.5–1.7 M ammonium sulfate and 0.1 M sodium acetate (pH 4.6). Consequently, we wanted to test the effect of the high ammonium sulfate concentration and low pH on the oligomerization of GerBC. Gel filtration chromatography showed that GerBC also eluted as a monomer in sodium acetate buffer (pH 4.6) alone or supplemented with 1.0 M ammonium sulfate (Fig. 3a). Taken together, these results suggest that GerBC is predominantly monomeric in aqueous solution. It is thus possible that the observed dimeric configuration in the crystalline state is due to the crystal packing at a high protein concentration.
GerBC does not interact with GerD
Another component in the germination apparatus of spores of Bacillales species is the GerD protein. Mutations in the gerD gene of B. subtilis result in much delayed spore germination with all nutrient germinants, although only germination via nGRs is affected in the gerD mutation.10 Therefore, GerD may be involved in nGR-mediated activation of downstream germination events. Like GerBC, GerD is predicted to be a lipoprotein and is also in the inner membrane of B. subtilis spores.11 In addition, both GerB proteins and GerD have recently been found to colocalize in a small cluster in individual spores (K. K. Griffiths, J. Zhang, A. E. Cowan, J. Yu, and P. Setlow, unpublished data).11 Consequently, it is possible that GerBC and GerD interact directly with each other in spores. We thus assessed the interaction between GerBC and GerD using a Ni2+-NTA affinity pulldown assay. In this experiment, we incubated 14 µM purified His6-GerBC with 14 µM of purified GerD that lacks the latter’s N-terminal signal peptide, precipitated the mixture using Ni2+-NTA resin, eluted proteins from the resin with imidazole, and analyzed the eluates by SDS-PAGE and Coomassie staining. Figure 4 shows that His6-GerBC does not bind untagged GerD (lane 6). This result is confirmed by the reciprocal pulldown experiment using His6-GerD and untagged GerBC (Figure 4, lane12), suggesting the two proteins do not interact with each other directly in vitro.
Figure 4. GerBC does not interact with GerD.
A reciprocal Ni2+-NTA affinity pulldown assay was used to characterize the interaction of the GerBC with GerD. Indicated proteins were incubated and reactions were precipitated with Ni2+-NTA resin. The unbound (U) and eluted bound (B) fractions were analyzed by SDS-PAGE and Coomassie staining.
Implications for GerBC function and regulation
Our structural and biochemical data address three specific questions about our current understanding of the architecture, conservation and function of the GerBC subunit of the B. subtilis GerB germinant receptor. First, the crystal structure of GerBC reveals an unusual configuration in which each of the three distinct domains possesses a unique fold that has not been observed in reported protein structures. Although GerBC forms an interlocked dimer in the crystalline state via the interchange of the Domain I between two monomers, gel filtration and sedimentation equilibrium experiments revealed predominantly a monomeric GerBC in solution. It is possible that the linkage between Domains I and II of GerBC is flexible and thus the protein could have a more globular and compact fold in solution than in the crystals. Because GerBC is anchored to the membrane periphery via an N-terminal cysteine residue immediately upstream of Domain I in the cell, it is also possible that the flexible linker between Domains I and II might allow Domains II and III to interact with other receptor subunits and/or downstream effectors. In addition to crystal packing, the high protein concentration (39 mg/ml) used for crystallization could well contribute to dimer formation, given that the highest protein concentration used in sedimentation experiments was 1.52 mg/ml. It is tempting to speculate that a monomer-dimer equilibrium of GerBC may play a role in GerBC function, however, at present we do not have sufficient evidence to definitively support a dimer configuration for GerBC in spores.
Second, both the sequence alignments and secondary structure prediction analyses suggest that the unique fold of GerBC represents the first structural view of an nGR subunit and is likely conserved among C protein homologs in spores of species ranging from Bacillacae to Clostridiales. Furthermore, two specific sequence-conserved regions in the GerBC crystal structure have been identified and site-directed mutagenesis studies to test the functions of the conserved side chains in vivo are underway. In addition, the predicted structural differences between GerBC and GerKC suggest that two loop regions in the GerBC structure could adapt specific conformations in response to specific protein-protein interactions occurring in different C proteins, which in turn may play important roles in determining the functional specificity of various C proteins.
Third, the affinity pulldown experiments show that GerBC does not interact directly with GerD, a potential mediator for the signal transduction events downstream of nGRs. This negative result emphasizes the major question about nGR proteins: what precisely do individual nGR proteins do, and in particular what does the C protein do and how can the GerBC structure help elucidate this function? In B. subtilis nGRs the C protein as well as the A and B proteins are essential for nGR function.6 Specific nGR functions that can currently be readily tested in vivo, in addition to the ability to trigger spore germination, are the affinity and specificity for various germinants and the cooperation between different nGRs. Mutations have been found that alter the germinant affinities of the B. subtilis GerA nGR and such mutations affect only the GerAB protein.6 Mutations altering B proteins have also been found to affect both the affinity and specificity of Bacillus megaterium nGRs’ for various germinants.26, 27 These results suggest that the B proteins contain the nGRs germinant binding sites. Mutations have also been identified in both the A and B subunits of the B. subtilis GerB nGR that allow this nGR to trigger germination with L-asparagine without the need for the GerK receptor28, suggesting that the A and B proteins may be involved in some fashion in nGR-nGR cooperativity. However, specific amino acid changes in C proteins have not yet been identified that alter nGR function other than to eliminate it. Perhaps site-directed mutagenesis of the B. subtilis gerBC gene guided by the structure of the GerBC protein will lead to some understanding of what C proteins do in nGRs. Alternatively, the determination of the function of C proteins in nGRs may require the elucidation of the structure of a whole nGR.
Supplementary Material
Acknowledgements
We are grateful to Dr. James L. Cole and Jeffrey W. Lary of the National Analytical Ultracentrifugation Facility in University of Connecticut for sedimentation analysis, Dr. Wuxian Shi of the National Synchrotron Light Source X29A beamline for assistance with x-ray data collection and processing, and Dr. Jeff Hoch of the University of Connecticut Health Center for critical reading of the manuscript. This work was supported by a Multidisciplinary University Research Initiative (MURI) award from the Department of Defense to P. Setlow and B. Hao.
Footnotes
Protein Data Bank accession numbers. Coordinates and structure factors have been deposited in the RCSB Protein Data Bank with accession number 3N54.
References
- 1.Setlow P, Johnson EA. Spores and their significance. In: Doyle MP, Beuchat LR, editors. Food Microbiology, Fundamentals and Frontiers. 3rd edition. Washington, DC: ASM Press; 2007. pp. 35–67. [Google Scholar]
- 2.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 3.Cano RJ, Borucki MK. Revival and identification of bacterial spores in 25-to 40-million-year-old Dominican amber. Science. 1995;268:1060–1064. doi: 10.1126/science.7538699. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy MJ, Reader SL, Swierczynski LM. Preservation records of micro-organisms: evidence of the tenacity of life. Microbiology. 1994;140(Pt 10):2513–2529. doi: 10.1099/00221287-140-10-2513. [DOI] [PubMed] [Google Scholar]
- 5.Vreeland RH, Rosenzweig WD, Powers DW. Isolation of a 250 million-year-old halotolerant bacterium from a primary salt crystal. Nature. 2000;407:897–900. doi: 10.1038/35038060. [DOI] [PubMed] [Google Scholar]
- 6.Moir A. How do spores germinate? J Appl Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 7.Paidhungat M, Setlow P. Spore germination and outgrowth. Bacillus subtillis and its Relative: from Genes to Cells. 2002:537–548. [Google Scholar]
- 8.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Igarashi T, Setlow P. Interaction between individual protein components of the GerA and GerB nutrient receptors that trigger germination of Bacillus subtilis spores. J Bacteriol. 2005;187:2513–2518. doi: 10.1128/JB.187.7.2513-2518.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelczar PL, Igarashi T, Setlow B, Setlow P. Role of GerD in germination of Bacillus subtilis spores. J Bacteriol. 2007;189:1090–1098. doi: 10.1128/JB.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelczar PL, Setlow P. Localization of the germination protein GerD to the inner membrane in Bacillus subtilis spores. J Bacteriol. 2008;190:5635–5641. doi: 10.1128/JB.00670-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson KD, Corfe BM, Kemp EH, Feavers IM, Coote PJ, Moir A. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J Bacteriol. 2001;183:4317–4322. doi: 10.1128/JB.183.14.4317-4322.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Igarashi T, Setlow B, Paidhungat M, Setlow P. Effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J Bacteriol. 2004;186:2984–2991. doi: 10.1128/JB.186.10.2984-2991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paidhungat M, Setlow P. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J Bacteriol. 2001;183:3982–3990. doi: 10.1128/JB.183.13.3982-3990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marotte K, Sabin C, Preville C, Moume-Pymbock M, Wimmerova M, Mitchell EP, Imberty A, Roy R. X-ray structures and thermodynamics of the interaction of PA-IIL from Pseudomonas aeruginosa with disaccharide derivatives. Chem Med Chem. 2007;2:1328–1338. doi: 10.1002/cmdc.200700100. [DOI] [PubMed] [Google Scholar]
- 20.Zhong X, Hao B, Chan MK. Structure of the PAS fold and signal transduction mechanisms. In: Crews Stephen T., editor. PAS proteins: regulators and sensors of development and physiology. Kluwer Academic Publishers; 2003. pp. 1–16. [Google Scholar]
- 21.Morais Cabral JH, Lee A, Cohen SL, Chait BT, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 22.Choi IG, Shin DH, Brandsen J, Jancarik J, Busso D, Yokota H, Kim R, Kim SH. Crystal structure of a stress inducible protein from Mycoplasma pneumoniae at 2.85 Å resolution. J Struct Funct Genomics. 2003;4:31–34. doi: 10.1023/a:1024625122089. [DOI] [PubMed] [Google Scholar]
- 23.Paredes-Sabja D, Torres JA, Setlow P, Sarker MR. Clostridium perfringens spore germination: characterization of germinants and their receptors. J Bacteriol. 2008;190:1190–1201. doi: 10.1128/JB.01748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christie G, Lazarevska M, Lowe CR. Functional consequences of amino acid substitutions to GerVB, a component of the Bacillus megaterium spore germinant receptor. J Bacteriol. 2008;190:2014–2022. doi: 10.1128/JB.01687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christie G, Lowe CR. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J Bacteriol. 2008;190:8009–8017. doi: 10.1128/JB.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant D-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.