Abstract
Nuclear microenvironments are architecturally organized subnuclear sites where the regulatory machinery for gene expression, replication, and repair resides. This compartmentalization is necessary to attain required stoichiometry for organization and assembly of regulatory complexes for combinatorial control. Combined and methodical application of molecular, cellular, biochemical, and in vivo genetic approaches is required to fully understand complexities of biological control. Here we provide methodologies to characterize nuclear organization of regulatory machinery by in situ immunofluorescence microscopy.
Keywords: Nuclear organization, Runx, confocal microscopy, immunofluorescence microscopy, FRAP, live cell microscopy, nuclear matrix
1. Introduction
The focal distribution of regulatory macromolecules within the nucleus can effectively support the integration of regulatory networks and establish threshold levels of factors for positive and negative control in a broad spectrum of biological contexts that include development and tissue remodeling. Equally important, changes in the composition and organization of regulatory machinery in nuclear microenvironments provides insight into perturbed mechanisms that relate to human disease which is strikingly illustrated by, but not restricted to, skeletal disorders and tumorigenesis (1-5). Examples are modifications in the size, number, and composition of intranuclear sites that support transcription, replication, repair and altered regulatory domains that are causally associated with cleidocranial dysplasia and competency for metastatic breast cancer cells to form osteolytic lesions in bone.
Our understanding of the location of regulatory machinery for gene expression, replication, and repair and its role in functional outcome of various biological processes is increasingly evident. However, it is important to define and develop techniques that provide both specific and quantitative insight into various parameters of nuclear architecture. Development and deployment of such approaches is essential for establishing the biological relevance of subnuclear organization as well as necessary for diagnosing disease or providing a platform for development of targeted therapies.
Traditionally, compartmentalization of regulatory machinery has been identified and characterized by subnuclear fractionation followed by biochemical and molecular analyses. These are informative approaches, but with limitations. During the past several years, advances in microscopy, together with the development of highly specific antibodies and epitope tags have allowed to examine the assembly and activities of regulatory machinery at single cell level in both the fixed as well as live cell preparations. Thus, the combined use of high resolution cellular, biochemical and molecular approaches maximizes the extent to which regulatory mechanisms can be defined.
We will focus on visualization of nuclear microenvironment using Runx transcription factors as an example for compartmentalization of regulatory machinery within nuclei of osteoblastic cells. We will present approaches for imaging of focally localized regulatory complexes in interphase nuclei as well as throughout mitosis. Specificity and quantitation of regulatory complexes that are visualized by microscopy are required to informatively relate cell morphology with regulatory mechanisms. In addition, we will describe a recently developed approach in our laboratory, designated “Intranuclear Informatics”, that quantitatively assimilates multiple parameters of regulatory protein localization within the nucleus into contributions towards skeletal gene expression from a temporal/spatial perspective (6).
1. Materials
1.1. Preparation of metaphase chromosome spreads from suspension and adherent cell cultures
Karyomax Colcemid (10 μg/ml)
0.075 M Potassium chloride (KCl) solution. Pre-warmed to 37°C in a water bath.
Fixative: Methanol/Glacial acetic acid at 3:1, made fresh each time. The fixative should be ice cold prior to use.
Microscopy glass slides (pre-chilled at 4°C).
1.2. Nuclear Matrix Intermediate Filament Preparation
Sterile Glass coverslips, 22mm round, coated with 0.5% gelatin.
Cytoskeleton (CSK) Buffer: 10× Stock Solution: 1M NaCl, 100mM PIPES pH 6.8, 30mM MgCl2, 10mM EGTA. 1× Working Solution: Freshly prepare 100 ml of 1× CSK buffer by dissolving 10.27 g sucrose in 77.6 ml of double distilled water. Add 10 ml of 10× stock CSK buffer, 0.5 ml of Triton X-100, 0.8 ml of ribonucleoside-vanadyl complex (RVC, New England Biolabs, Ipswich, MA) and 0.8 ml of 150 mM AEBSF [4-(2-aminoethyl) benzenesulfonyl fluoride] (Sigma).
Digestion Buffer (DB): 10× Stock Solution: 0.5 M NaCl, 100 mM PIPES pH 6.8, 30 mM MgCl2, 10 mM EGTA. Freshly prepare 1× DB as described above for 1× CSK buffer except use 10× DB instead of 10× CSK buffer.
Phosphate buffered saline (PBS): 9.1 mM dibasic sodium phosphate (Na2HPO4), 1.7 mM monobasic sodium phosphate (NaH2PO4) and 150 mM NaCl. Adjust pH to 7.4 with NaOH.
Fixatives: 3.7% formaldehyde in PBS (WC fixative), or in 1× CSK buffer (CSK fixative), or in 1× DB (NMIF fixative). All fixatives should be freshly prepared.
Stop solution: 250 mM ammonium sulfate in 1× DB. (Add 1 volume of 2M ammonium sulfate to 8 volumes of 1× DB).
Permeabilizing solution: 0.25% Triton X-100 in PBS.
RNAse-free DNAse
PBSA: 0.5% bovine serum albumin (BSA) in PBS.
Prolong Gold (Invitrogen, Carlsbad, CA).
1.3. Microscopy
40 mm glass coverslips (Bioptechs, Butler, PA)
60 mm Corning culture dishes
Microwave oven
McCoy’s 5A complete media: McCoy’s 5A media with L-glutamine, supplemented with 10% FBS, 1% Penicillin-Streptomycin and 1% L-glutamine (200 mM)
Live Cell Stage-Closed System Chamber (Bioptechs)
Aqueduct slide, gaskets (Bioptechs)
Perfusion fluid: McCoy’s 5A complete medium with 20mM Hepes Buffer Solution (1 M), heated to 37°
Bioptechs objective heater and slide heater
2. Methods
2.1. Preparation of metaphase chromosome spreads from suspension and adherent cell cultures
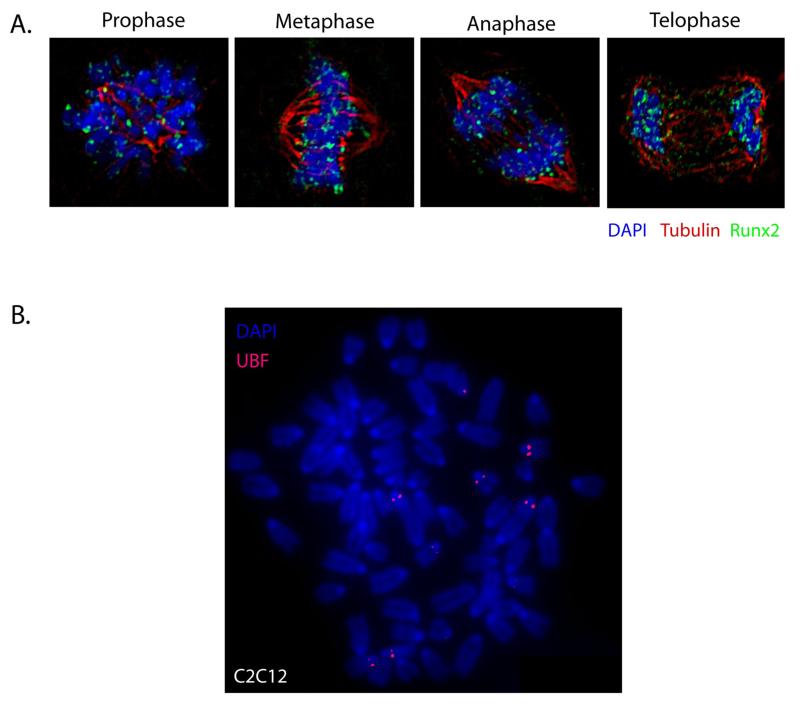
Metaphase chromosome spreads are traditionally used for identification of chromosomal abnormalities (translocation, deletions, and insertions) in patients. It has been recently observed that some lineage-specific proteins, such as Runx transcription factors, retain association with chromosomes during mitosis (Figure 1A). In such cases, metaphase chromosome spreads provide powerful means for the identification of chromosomes where Runx transcription factors reside during mitosis. For example, Runx proteins associate with nucleolar organizing regions (NORs) on metaphase chromosomes. NORs can be visualized in situ by immunolabeling metaphase chromosome spread preparations for Upstream Binding Factor (UBF), a known regulator of ribosomal RNA transcription (Figure 1B). Below is a protocol used in our laboratory for preparation of metaphase chromosome spreads (7-9).
Figure 1. Metaphase Chromosome Spreads.
A. Runx2 is stable throughout mitosis. Synchronously growing Saos-2 cells were fixed and stained for DNA by using DAPI and for Runx2 by using a rabbit polyclonal antibody. Mitotic cells were identified by chromosome morphology. High-resolution images obtained by three-dimensional deconvolution algorithms reveal that Runx2 (green) is localized in mitotic chromosomes. A subset of Runx2 colocalizes with the microtubules, labeled by α-tubulin staining (red). B. A metaphase chromosome spread of mouse pre-myoblast C2C12 cells, immuno-labeled for Upstream Binding Factor (UBF; red) to identify nucleolar organizing regions (NORs).
2.1.1. Suspension Cells
Passage 1×106 cells in regular growth medium, 1 to 2 days prior to performing the chromosome spreads.
Feed cells with fresh media 12-14 hours prior to harvesting and add Colcemid to a final concentration of 0.05 μg/ml; incubate at 37°C for 3-4 hours.
Transfer the cells into a centrifuge tube and pellet at 750 g for 5 min. (see Note 1)
Aspirate the supernatant completely.
Add 10 ml of 0.075M KCl solution drop by drop i.e., hypotonic treatment. Resuspend the pellet by pipetting up and down gently. Incubate at 37°C for 30 min. (see Note 2)
Add 1ml of ice cold fixative (Methanol/Glacial acetic acid 3:1) to the cell suspension and keep at room temperature for 15 min.
Spin the cells at 400 g for 5 min.
Aspirate the supernatant completely. Add 2 ml of fresh fixative (methanol/Glacial acetic acid in a 3:1 ratio) to the cells and keep at 4°C for slide preparation. (At this stage the cell suspension can be stored at 4°C for few days or at −20°C long term.)
2.1.2. Adherent Cells
Follow the initial steps 1 and 2 as described above in section 3.2.1 for suspension cells.
During the mitotic block, some adherent cells become rounded and detach from the plate. In this case, media should not be discarded but should be transferred to a centrifuge tube.
Rinse the plate with PBS and detach the cells with 0.5 ml of trypsin. Mix the detached cells with the medium which contains the cells from mitotic shake off (Step 2).
Centrifuge the cells at 750 g for 5 minutes. Discard the supernatant and resuspend pellet in ice-cold PBS and centrifuge again at 750 g for 5 minutes. Repeat the PBS wash.
Continue as in step 5 above for suspension cells and follow each step exactly as described.
2.1.3. Slide Preparation
Cool slides to 4°C before using them for metaphase spreads. Adjust hot plate for medium heat (ideally 50°C to 60°C). (see Note 3)
Take 100-200 μl of the cell suspension and add drop by drop to the slide from a height of about 20 cm. Drain the excess solution by tilting the slide.
Immediately put the slide on hot plate (heat shock) for 1 minute.
Air dry the slide, check for chromosome spreading in the phase contrast microscope.
Keep the slides in a box at 4°C (up to 8 weeks) or at −80°C for a longer period of time.
2.2. Nuclear Matrix Intermediate Filament Preparation
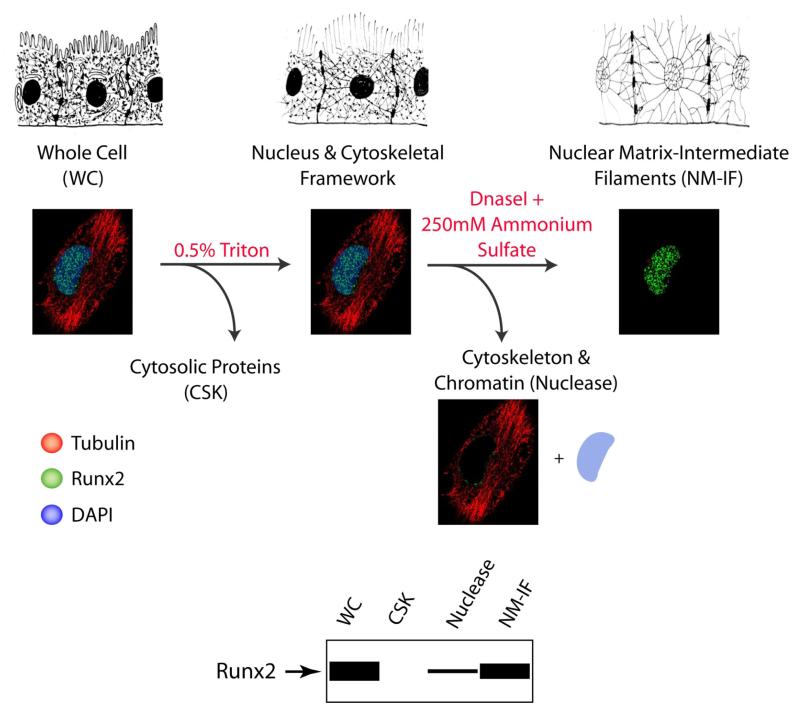
It is becoming increasingly evident that regulatory proteins are organized in highly specialized compartments within the mammalian nucleus. The biological activity of proteins often correlates with their presence or absence in these nuclear microenvironments (1). The subnuclear organization of regulatory proteins can be assessed by the sequential removal of soluble proteins and chromatin from the mammalian cell (Figure 2) followed by either in situ immunofluorescence or western blot analysis. Below is an optimized protocol that we routinely use for in situ assessment of parameters of gene expression.
Figure 2. In Situ Assessment of Nuclear Microenvironments.
Regulatory proteins can be visualized by indirect immunofluorescence in situ. Proteins involved in distinct nuclear processes are localized in specialized nuclear microenvironments. These microenvironments can be further visualized by removing soluble cytosolic and nuclear proteins as well as chromatin. The procedure is schematically outlined. The upper panel shows a cell after sequential extractions that remove soluble cytosolic as well as soluble nuclear proteins. Finally, the chromatin is digested with DNaseI to reveal a network of ribonuclear proteins, designated the nuclear matrix. The middle panel shows corresponding in situ immunofluorescence of an osteoblast co-stained with tubulin (red) and Runx2 (green). DNA is depicted as a blue colored circle. Each fraction can be also resolved by SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) to identify proteins of interest. The bottom panel shows a schematic of western blot analysis of biochemical fractionation for Runx2, which is an architecturally associated protein primarily present in the NM-IF fraction.
2.2.1 Whole Cell (WC) Preparation
-
1
Plate cells at a density of 0.5×106 cells per 6-well plate and incubate in humidified incubator with 5% CO2 at 37°C for 24 hours.
-
2
After 24 hours, wash cells twice with ice cold PBS, fix the WC preparation on ice for 10 minutes (in an experiment, typically two wells of a six well plate are allocated to each of the WC, CSK and NMIF preparations) by adding 2 ml of WC fixative per well.
-
3
Wash cells once with PBS.
-
4
To facilitate antibody staining of WC preparations, permeabilize WC preparations with 1 ml of permeabilizing solution on ice for 20 minutes.
-
5
Aspirate permeabilizing solution and wash twice with PBS.
-
6
Add 1 ml of PBSA to the wells.
2.2.2. Cytoskeleton (CSK) Preparation
-
7
Wash cells twice with ice cold PBS.
-
8
Add 1ml of 1× CSK buffer and incubate plates on ice for 5 minutes while swirling plates once or twice.
-
9
Wash wells allotted for CSK preparation (see section 3.3.1) once with ice-cold PBS and fix cells by adding 2 ml of CSK fixative per well.
-
10
Aspirate CSK fixative after 10 minutes and wash twice with PBS.
-
11
Add 1ml of PBSA to the wells.
2.2.3. Nuclear Matrix Intermediate Filament (NMIF) Preparation
-
12
Wash cells twice with ice cold PBS.
-
13
Add 1 ml of 1× CSK buffer and incubate plates on ice for 5 minutes while swirling plates once or twice.
-
14
Prepare 1 ml of DB by adding 400 units of RNase free DNase I to 1× DB.
-
15
Flatten parafilm on the covers of plates, label and dispense 20 μl drop of DB containing RNase free DNase I on the covers of respective plates. (This step is to conserve the amount of DNase I, otherwise add 1 ml of DB containing RNase free DNase I to each well.)
-
16
To digest the chromatin with DNase I, carefully invert the coverslip, so that cells will face DB containing DNase I.
-
17
Incubate cells for 50 minutes at 30°C. Place coverslips back in their respective labeled wells. Add 1 ml Stop Solution to the wells and incubate plates on ice for 10 minutes to stop the activity of DNase I.
-
18
Wash once with ice-cold PBS and fix NMIF preparations in 2 ml of NMIF fixative on ice for 10 minutes.
-
19
Aspirate fixative and wash twice with PBS.
-
20
Add 1 ml of PBSA.
2.2.4. Immuno-Staining of the Samples
-
21
Dilute antibody in PBSA to an appropriate dilution. As quality and specificity of antibodies vary among suppliers and lots, we recommend testing several dilutions to optimize antibody concentration. When immuno-labeling cells with two proteins, caution must be practiced to assure that the antibodies used are raised in different species (e.g., mouse versus rabbit). If raised in the same species, they must be of different isotypes (e.g., IgG versus IgM).
-
22
On parafilm already flattened on the lids of plates, dispense a 20 μl drop of diluted antibody per well. Carefully invert a coverslip on the drop so that the cells are in direct contact with the antibody. Avoid creating air bubbles by gently placing the coverslip on one edge on the antibody droplet and slowly lowering it. Incubate for 1 hour at 37°C.
-
23
Place coverslips back in respective wells on ice with cells facing upward and wash four times with ice cold PBSA.
-
24
Stain cells with appropriate secondary antibodies conjugated with fluorochromes (e.g., Texas Red or FITC) for 1 hour at 37°C.
-
25
Place coverslips back in their respective wells and wash four times with ice cold PBSA.
-
26
Stain cells with DAPI (0.5 μg DAPI in 0.1% Triton X-100-PBSA) for 5 minutes on ice.
-
27
Wash once with 0.1% Triton X-100-PBSA followed by two washes with PBS. Leave cells in last wash on ice.
-
28
Immediately mount coverslips in an antifade mounting medium (e.g., Prolong Gold) and aspirate excess of mounting medium. After 10-15 minutes, seal coverslips with nail polish and store at −20°C in dark.
2.3. Microscopy
2.3.1. Fluorescence Microscopy
Fluorescence microscopy provides a powerful tool to assess subcellular and subnuclear localization of regulatory proteins as well as nucleic acids. A variety of microscopes are available; each microscope has its own set of unique features. Below are the instructions specific for the Zeiss Axioplan 2 Microscope.
Turn on the mercury lamp, microscope, charged-coupled device (CCD) camera and computer. Clean all lenses with microscopic lens paper.
Wipe your slide with tissue paper and lens cleaner. Place a small drop of lens oil on your slide. It is important that the coverslips are sealed to avoid any mixing of the mounting medium with lens oil.
To find your cells, start at 10× magnification and then proceed to 40× magnification to start your analysis. Once you have identified a cell or field to document, you can then increase the magnification, depending upon your specific requirements.
Once you have selected a cell or field for documentation, you are ready to acquire an image using MetaMorph Software (Molecular Devices, Downingtown, PA).
Before acquiring an image, make sure that the arrow on the knob, which diverts light either towards 35mm camera or towards charged coupled device (commonly called as CCD camera), is pointing towards the CCD camera.
In the main menu, go to ACQUIRE to access the Acquire Dialogue Box.
Enter an exposure time. Set the region of interest by selecting Entire Chip or Central Quadrant option on the Acquire dialogue box.
Acquire the image on all filters (DAPI, Fitc, Texas Red and Phase) if analyzing a dual labelled slide.
The default image depth of the CCD camera is 12 bits. However, these images can not be opened by Adobe Photoshop or Microsoft PowerPoint Software. Copy images to 8 bit by selecting ‘Copy to 8 Bits’ command on the main task bar. Always keep your original image (i.e., raw data) as it contains the most information.
Once acquired, images can be presented (PowerPoint) or published (Adobe Photoshop, Illustrator) directly or can be further analyzed quantitatively by using MetaMorph Imaging Software or the Intranuclear Informatics mathematical algorithm (see below).
2.3.2. Viewing Live Cells Using the Confocal Microscope
Our lab has characterized the Runx family of transcription factors, describing spatial distribution, subnuclear architectural scaffolding and relationships with coregulatory factors. Much of this work was done with fixed cells on an epifluorescence microscope with verification using a confocal microscope. This naturally led to an interest in documenting the Runx protein dynamics using live cell imaging; looking at mobility, mitotic labeling and protein-protein interactions.
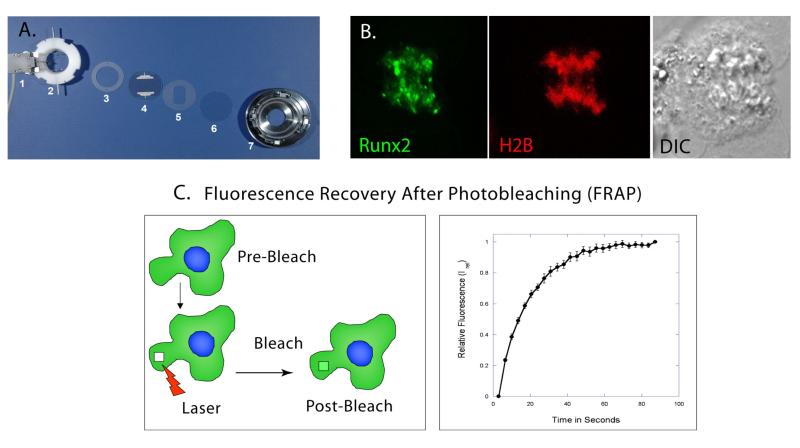
The laser scanning confocal microscope coupled with a Bioptechs micro observation system offers us higher image resolution of live cells with the ability to capture images that are sharply defined optical sections produced by the elimination of out of focus light and background information from which three dimensional renderings can be recreated. This coupled with the live cell stage allow us to verify the location of Runx foci throughout dynamic processes, for example, mitosis and to assess the mobility of Runx foci in interphase and during mitosis (Figure 3B) using Fluorescence Recovery After Photobleaching (FRAP) techniques.
Figure 3. Live Cell Microscopy by Confocal Laser Scanning.
A. The Bioptics Live Cell Stage-Closed System Chamber allows the viewing of living cells by maintaining 37°C temperature; pH and nutrient supply by perfusing media across a cell-laden coverslip. The slide heater (1) warms an Aqueduct Slide (4) to 37°C. The Perfusion Ring Chamber (2) allows media to enter, cross and exit the chamber, and Gaskets (3, 5) sandwich the Aqueduct Slide and cell-laden coverslip (6) to prevent leaks. This assembly sits in the self-locking base (7) that is placed on the microscope stage. B. Live image of U2-OS cell in Anaphase. Runx2-EGFP foci (green) localize to mitotic chromosomes in Anaphase. Histone 2B-RFP and Differential Interference Contrast (DIC) images are used to identify mitotic stage. C. Initially, using high intensity laser power, a Spot or Region of Interest inside the fluorescent cell is bleached. After bleaching, a series of images are taken at predetermined time intervals to measure the recovery of fluorescence in the bleached spot (Left Panel). Y-axis represents the relative fluorescence after photo bleaching in the bleached spot and X-axis represents time in seconds. Post normalization, the relative fluorescence in the bleached spot is zero. This time point is represented as zero time. The relative fluorescence increases with time until it reaches asymptote (Right Panel).
A common problem occurs while live cell imaging GFP transfected cells. Cells become extremely sensitive when exposed to blue filtered UV light (green fluorescence) and die while viewing over long periods of time. For example, in a dynamic process like mitosis, cells permanently stall in Metaphase. Using the scanning laser confocal microscope relieves this situation. Red fluorescence proteins (RFP) are not as sensitive to UV light, thus we transfected a RFP mitotic stage marker, found the stage of mitosis we were interested in, then used the lasers to image the spatial localization of GFP Runx labeled protein through mitosis. Another clear advantage to laser scanning confocal microscopy is the elimination of possible bleed through from double labeling by turning off one of the lasers to confirm specific localization.
Sterilize 40 mm coverslips set in 60 mm dishes in a microwave for 20 minutes.
Plate cells in McCoy’s 5A complete medium at a density of ~0.3 × 106. Allow cells to grow for 40-48 hours to 50-80% confluency.
Transfection. Ascertain and document cell growth. Warm complete and incomplete McCoy’s 5A media. Using Roche FuGENE 6 Transfection Reagent, follow the standard protocol for a 60 mm dish using 5 ml total volume of media, 200-600 ng of DNA and 200 μl total volume of complex per coverslip.
2.3.3. Preparation of live cell stage, a closed system chamber
The Focht’s Closed System Chamber (FCS) allows the microscopic observation of living cells by duplicating conditions of an incubator on the microscope. Temperature is controlled by a slide heater and an objective heater. The slide heater works in conjunction with a microaqueduct slide that has a thermally conductive coating which the slide heater arms rest on. The temperature is set by a controller unit. The objective heater’s temperature is also set by a controller unit and has an adjustable loop which surrounds the objective lens. These heaters are designed to eliminate heat-sink loss. Over time, cells under microscopic observation must be fed, pH level maintained and waste eliminated. A micro peristaltic pump working in conjunction with a microaqueduct slide allows medium to perfuse across the coverslip at a precise, very slow rate, feeding the cells, maintaining pH and eliminating waste.
Pre-warm the following items to 37°. One hour prior to using the confocal microscope, warm water bath to 37°, placing a flask with water and a thermometer in the bath to verify bath temperature.
After reaching temperature, place near confocal microscope for perfusion media.
Warm perfusion medium, which is McCoy’s 5A medium with 20mM Hepes Buffered solution (Hepes maintains the media pH throughout the imaging session), to 37°.
Warm the Focht’s Chamber System (FCS) and parts to 37°C as well as an insulated transporter box with 2 × 50 ml centrifuge tubes of 37°C water (on each side of the chamber) that will keep the chamber warm en route from the warm room to the scope.
One half hour prior to viewing, confirm that the objective lens and slide heater controller systems are calibrated to 37°C, then attach the objective heater to objective lens and turn on to warm.
2.3.4. Microscope Preparation
Confirm DIC is aligned, correct condenser (wide field) and rotating condenser prism are in place, and stage height is correct.
Assemble chamber with cells in warm room. All chamber pieces, gaskets and tubing have been cleaned with 70% ethanol and distilled water after the last use and again before current use.
The order of assembly of the Focht’s Chamber System is as follows (Figure 3A): Place upper gasket (part 3) on perfusion ring chamber (part 2) matching 2 holes over 2 pegs, then align the aqueduct slide (part 4) on the pegs. The aqueduct slide allows the perfusion fluid to flow across the coverslip, keeping the cells fed and warm. Place the lower gasket (part 5) on top of the aqueduct slide. Pipet 0.25 ml of medium onto the aqueduct slide from the 60 mm dish containing the coverslip with cells, filling the channels and mid space area. Finally, invert the coverslip (part 6) with cells onto the layered assembly. It is very important to touch one edge of the coverslip to one side of the aqueduct slide and slowly lower the coverslip onto the aqueduct slide in order to avoid air bubbles. Invert this sandwich assembly and secure in the self-locking base (part 7). Wipe the medium off the underside of the apparatus.
Place chamber in the transporter box with the 2 × 50ml, 37° warming tubes to keep it warm while walking from the warm room to the microscope room. Place chamber on stage, attach inlet tubing, and turn on dialysis pump to a very slow rate (ex. 0 and 2) and confirm perfusion fluid is entering, crossing, and exiting the chamber. Then attach exit tubing and drain to receiver vessel. Insert slide heater (part 1, Figure 3A) in the slide warmer receptacle and oil the objective lens.
2.4. Fluorescence Recovery After Photobleaching (FRAP)
A powerful approach for measuring the dynamics of nuclear microenvironments is to track fluorescently tagged molecules in living nuclei. There are always at least two pools of molecules in nuclear microenvironments: free and bound. Molecules will move at diffusion rates when free and at the same rate as the structure when bound. Taking advantage of this difference, photobleaching-recovery techniques can help characterize the binding equilibriums for molecules docking on a simple, stable structure. The analysis becomes complicated if the protein has multiple and heterogeneous interactions. Several studies have examined the dynamics of cellular and nuclear GFP-fusion proteins by Fluorescence Recovery After Photobleaching (FRAP) (13-16).
In FRAP, fluorescence in a Spot of Interest (SOI) or Region Of Interest (ROI) inside the cell is irreversibly bleached with a high intensity focused laser (Figure 3C, left panel). This results in non-fluorescent molecules in the bleached areas surrounded by fluorescent molecules outside the bleached region. Since the binding of these molecules is dynamic, bleached molecules will unbind and diffuse away (10-11). Molecules that are still fluorescent and are bound in the unbleached region will unbind and diffuse into the bleached zone where they can bind. Photobleach recovery rates are determined by unbinding, diffusion, and binding rates (13,14). After bleaching, a series of images of the bleached cell are taken to measure the recovery of fluorescence in the bleached spot (Figure 3C, right panel). Most papers in the biological literature report FRAP recovery rates in terms of half time of recovery or T1/2. Some others report “apparent diffusion coefficients” even though FRAP recovery rates are dependent on binding but not on diffusion (14). Therefore, measurement of binding and unbinding constants is very important in understanding FRAP recovery rates, especially for nuclear proteins (13,14). After FRAP, data from the confocal system is exported into a spreadsheet software package, corrected for the loss of fluorescence in the whole cell and normalized for pre- and immediate post-bleach fluorescence in the bleached zone. Post-normalization, full recovery of the fluorescence in the bleached zone might be expected. If there is no full recovery, then a fraction of the protein is immobile, which means the protein is tightly bound to the subcellular structure and exchanges too slowly to be measured in the post bleach session. FRAP has been valuable in many applications including, but not limited to, measurement of the binding of Histone proteins to DNA (14,17,18), the binding of the estrogen receptor to promoters (19), the binding of the glucocorticoid receptor to promoters (20), and the dynamic binding of Exon Junction Complex proteins to RNA splicing speckled domains (15,16).
Once the live cell chamber is set up, fluorescent cells are identified and images of them are collected at low laser power. Optimum gain and offset values for images are determined and the settings are saved under a user profile.
Initially, 5 to 10 images are recorded before bleaching a Region of Interest (ROI) or a Spot of Interest (SOI).
In spot photo bleaching, one or more than one spot inside the fluorescent cell is selected. Bleaching is usually done with maximum laser power from 1 to 3 seconds until about 70% of the fluorescence in the spot is bleached. If more than one spot is selected for bleaching, the confocal system performs bleaching sequentially. Bleaching can be done in larger regions of the cell, for example in half of the nucleus, by zooming up to a ROI within the cell and scanning it 30-50 times at full laser intensity, resulting in the photobleaching of this enlarged region of the cell.
After photobleaching, routinely 30 to 50 images are taken at intervals of 1.7 to 20 seconds (it can be minutes if desired) depending on the dynamics of the fluorescent protein. (If large area bleaching was performed, images are enlarged to the size equal to the prebleach image.)
With the aid of Leica Confocal Software version 2.0, the fluorescence within the bleached spot, the fluorescence in the whole cell or nucleus, and the fluorescence in a region outside the bleach zone are measured for the entire stack of images.
For data analysis, fluorescence intensity values from the Leica software are exported to a spreadsheet software package, for example Microsoft Excel. Normalization is done at two levels. At the first level, the initial postbleach intensity is subtracted for the fluorescence in the ROI so that any fluorescence in the bleached area after bleaching is normalized to zero. At the second level, the prebleach fluorescence intensity, corrected for the fluorescence loss in the whole cell that is caused by the bleach, is normalized to 1.
The relative fluorescence intensity (I rel) in the bleached spot is measured as described by (12): Irel = T0*If / Tt*I0, with T0 being the total cellular intensity before bleach, Tt the total cellular intensity at time t, I0 the intensity in the bleached area before bleach, and If, the intensity in the previously bleached area at time t.
Recovery curves are obtained using Microsoft Excel or Kaleidograph.3.5 (Synergy Development).
The immobile protein fraction is calculated by subtracting the relative intensity at the asymptote of the recovery curve from a relative recovery 1. For example, an asymptote at 0.7 reflects an immobile fraction of 30%.
2.5. Intranuclear Informatics
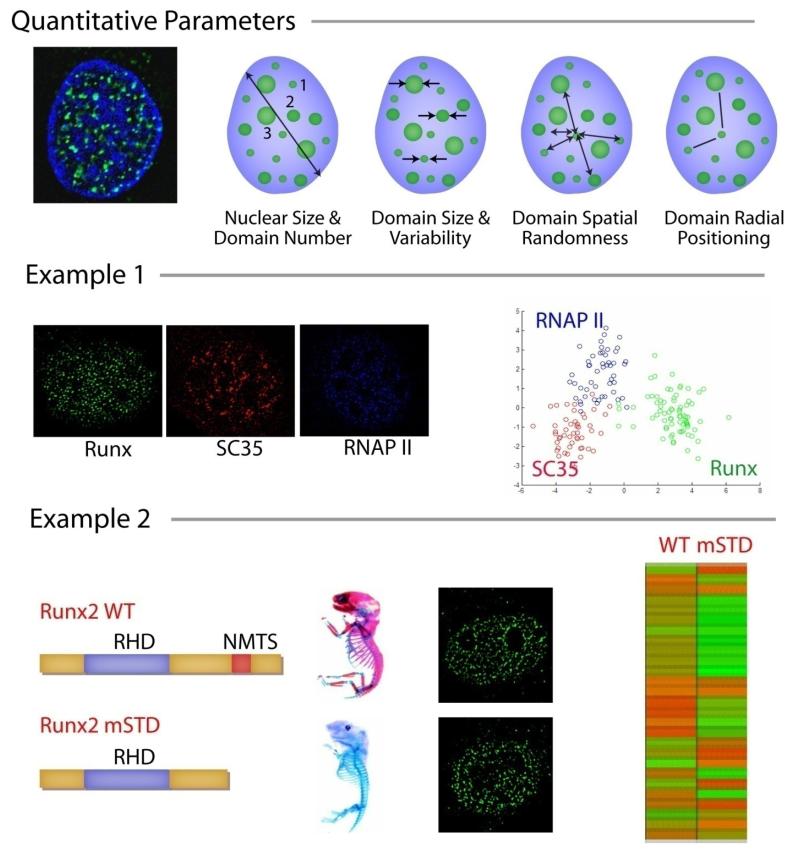
Intranuclear informatics is a mathematical algorithm that is designed to identify and assign unique quantitative signatures that define regulatory protein localization within the nucleus (6); Figure 4). Quantitative parameters that can be assessed include nuclear size and variability in domain number, size, spatial randomness, and radial positioning (Figure 4, top panel).
Figure 4. Intranuclear Informatics.
This figure shows how intranuclear informatics can be used to examine nuclear alterations in cancer cells compared with normal cells. The top panel shows the conceptual framework for the quantitation of subnuclear organization by intranuclear informatics. Four main groups of parameters, selected based on inherent biological variability, can be examined. Example 1. Regulatory proteins with different activities can be subjected to Intranuclear Informatics analysis, which assigns each protein a unique architectural signature. The overlap between the architectural signatures of different proteins is often correlated to their functional overlap. Shown here are Runx transcription factor, SC35 splicing protein, and RNA Pol II. Example 2. The subnuclear organization of Runx domains is linked with subnuclear targeting, biological function, and disease. Biologically active Runx2 and inactive subnuclear targeting defective mutant (mSTD) show distinct architectural signatures, indicating that the biological activity of a protein can be defined and quantified as subnuclear organization.
The significance and implication of Intranuclear Informatics can be shown by two distinct biological examples (Figure 4). Regulatory proteins with different activities can be subjected to Intranuclear Informatics analysis that assigns each protein a unique architectural signature. The overlap between the architectural signatures of different proteins is often correlated to their functional overlap. Alternatively, the subnuclear organization of a protein domain can be linked with subnuclear targeting, biological function, and disease. For example, biologically active Runx2 and its inactive subnuclear targeting defective mutant (mSTD) show distinct architectural signatures, indicating that the biological activity of a protein can be defined and quantified as subnuclear organization.
These architectural signatures have the potential to discriminate between intranuclear localization of proteins in normal and cancer cells. Intranuclear informatics can be combined with proteomics (changes in protein-DNA and protein-protein interactions) and genomics (altered gene expression profiles) to develop a novel platform for identification and targeting of perturbed regulatory pathways in cancer cells.
Acknowledgments
Studies reported in this article were in part supported by grants from NIH (5PO1CA82834-05, 5PO1AR048818-05, 2R01GM32010, 5R01AR049069).
Footnotes
If the metaphase chromosomes are highly condensed, use a lower concentration of Colcemid and decreased time of Colcemid treatment.
Appropriate hypotonic treatment is vital to the quality of metaphase spreads. The concentration of KCl can be changed according to the quality of chromosome spread.
Drop the cell suspension on cold, slightly moist slides. Chromosomes will spread poorly on dry slides. If necessary, breathe on the slide before dropping the suspension.
Reference List
- 1.Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi J-Y, et al. Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol. 2003;13:584–592. doi: 10.1016/j.tcb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, et al. Intranuclear trafficking: organization and assembly of regulatory machinery for combinatorial biological control. J.Biol.Chem. 2004;279:43363–43366. doi: 10.1074/jbc.R400020200. [DOI] [PubMed] [Google Scholar]
- 3.Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, et al. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat.Rev.Cancer. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
- 5.Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat.Rev.Mol.Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- 6.Young DW, Zaidi SK, Furcinitti PS, Javed A, van Wijnen AJ, Stein JL, et al. Quantitative signature for architectural organization of regulatory factors using intranuclear informatics. J.Cell Sci. 2004;117:4889–4896. doi: 10.1242/jcs.01229. [DOI] [PubMed] [Google Scholar]
- 7.Henegariu O, Heerema NA, Lowe WL, Bray-Ward P, Ward DC, Vance GH. Improvements in cytogenetic slide preparation: controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry. 2001;43:101–109. [PubMed] [Google Scholar]
- 8.Claussen U, Michel S, Muhlig P, Westermann M, Grummt UW, Kromeyer-Hauschild K, et al. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet.Genome Res. 2002;98:136–146. doi: 10.1159/000069817. [DOI] [PubMed] [Google Scholar]
- 9.Deng W, Tsao SW, Lucas JN, Leung CS, Cheung AL. A new method for improving metaphase chromosome spreading. Cytometry A. 2003;51:46–51. doi: 10.1002/cyto.a.10004. [DOI] [PubMed] [Google Scholar]
- 10.Berezney R, Basler J, Kaplan SC, Hughes BB. The nuclear matrix of slowly and rapidly proliferating liver cells. Eur.J.Cell Biol. 1979;20:139–142. [PubMed] [Google Scholar]
- 11.Kruhlak MJ, Lever MA, Fischle W, Verdin E, Bazett-Jones DP, Hendzel MJ. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J.Cell Biol. 2000;150:41–51. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 13.Lele T, Oh P, Nickerson JA, Ingber DE. An improved mathematical approach for determination of molecular kinetics in living cells with FRAP. Mech.Chem.Biosyst. 2004;1:181–190. [PubMed] [Google Scholar]
- 14.Lele T, Wagner SR, Nickerson JA, Ingber DE. Methods for measuring rates of protein binding to insoluble scaffolds in living cells: histone H1-chromatin interactions. J.Cell Biochem. 2006;99:1334–1342. doi: 10.1002/jcb.20997. [DOI] [PubMed] [Google Scholar]
- 15.Wagner S, Chiosea S, Ivshina M, Nickerson JA. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J.Cell Biol. 2004;164:843–850. doi: 10.1083/jcb.200307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc.Natl.Acad.Sci.U.S.A. 2003;100:3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- 18.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 19.Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O’Malley BW, et al. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat.Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- 20.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol.Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]