Abstract
Temporally and spatially controlled activation of the Aurora A kinase (AURKA) regulates centrosome maturation, entry into mitosis, formation and function of the bipolar spindle, and cytokinesis. Genetic amplification and mRNA and protein overexpression of Aurora A are common in many types of solid tumor, and associated with aneuploidy, supernumerary centrosomes, defective mitotic spindles, and resistance to apoptosis. These properties have led Aurora A to be considered a high-value target for development of cancer therapeutics, with multiple agents currently in early-phase clinical trials. More recently, identification of additional, non-mitotic functions and means of activation of Aurora A during interphase neurite elongation and ciliary resorption have significantly expanded our understanding of its function, and may offer insights into the clinical performance of Aurora A inhibitors. Here we review the mitotic and non-mitotic functions of Aurora A, discuss Aurora A regulation in the context of protein structural information, and evaluate progress in understanding and inhibiting Aurora A in cancer.
Keywords: Aurora A, AURKA, Cancer, Mitosis, Cell cycle, Kinase, Centrosome, Cilia
Introduction
Aurora A kinase (official gene symbol AURKA) has many aliases, including, most commonly, Aurora, and also Aurora-2, serine/threonine kinase 15 (STK15), serine/threonine kinase 6 (STK6), breast tumor amplified kinase (BTAK), Aurora-related kinase 1 (ARK1), Homo sapiens Aurora/IPL1-related kinase (HsAirk1), Eg2, and Ipl- and Aurora-related kinase 1 (IAK1). As these names indicate, this protein is a member of the Aurora/IPL1-related kinase family of serine/threonine kinases. The founding member of the family, Ipl (increase-in-ploidy) 1, was first identified in 1993 in a screen for mitotic mutants that failed to undergo normal chromosome segregation in Saccharomyces cerevisiae [1]. Ipl1-like kinases were independently identified in cell cycle studies in Xenopus laevis and Drosophila melanogaster [1–5]. The Xenopus laevis Eg2 transcript emerged in a screen for Xenopus egg mRNAs that became deadenylated after fertilization. The Eg2 gene encodes a kinase that was subsequently defined as a regulator of the G2/meiosis I transition in Xenopus oocytes and of mitotic spindle function in Xenopus laevis eggs [3, 4]. Severe mutations at the Drosophila aurora locus result in pupal lethality and a mitotic arrest characterized by the presence of monopolar spindles; less severe mutations include defects in centrosome separation, formation of astral microtubules, chromosome segregation, and spindle positioning [5–9]. All of the early studies in model organisms indicated a requirement for this protein in mitotic progression.
Subsequent studies determined that Ipl1 is the unique S. cerevisiae representative of a family that diverges into two Ipl1-like kinases (Aurora A and Aurora B) in Drosophila, C. elegans, and X. laevis, and three Ipl1-like kinases (Aurora A, Aurora B, and Aurora C) in mammals. All of these kinases have been found to have essential functions in mitosis and/or meiosis. However, among the three mammalian kinases, Aurora A has attracted very significant attention in the past decade, based on the recognition that it is overexpressed in many tumors arising from breast, colon, ovary, skin, and other tissues, and because it has been shown to function as an oncogene when exogenously expressed in numerous cell line models [10–14]. Aurora A overexpression, whether in naturally occurring tumors or following deliberate overexpression, is associated with increased numbers of centrosomes and multipolar spindles, which arise as a consequence of failed cytokinesis. For these reasons, Aurora A has been a popular target for development of targeted therapeutic agents for cancer, with multiple Aurora-A-specific or pan-Aurora kinase inhibitors undergoing clinical assessment. At the same time, continuing investigation of Aurora A has unexpectedly suggested that the activity of this protein is not confined to regulation of mitosis, with new functions observed in interphase and post-mitotic cells. Given the recent nature of these discoveries, they are not yet broadly appreciated, although they may both impact the use of Aurora-A-targeted drugs in the clinic and also expand the general understanding of the biological role of this protein in cell physiology.
In this review, we describe and discuss signaling by human Aurora A in normal cell division and the deregulation of Aurora A signaling in cancer. We also review the current status of drugs targeting Aurora A for cancer and summarize recent insights into Aurora A function emerging from structural biology and from systems biology resources. Because of space constraints, we do not discuss functions of the other Aurora kinases in depth. For useful recent reviews of the function of Aurora B, see [15, 16]; at present, Aurora C function is much less well understood.
Aurora A actions relevant to mitosis
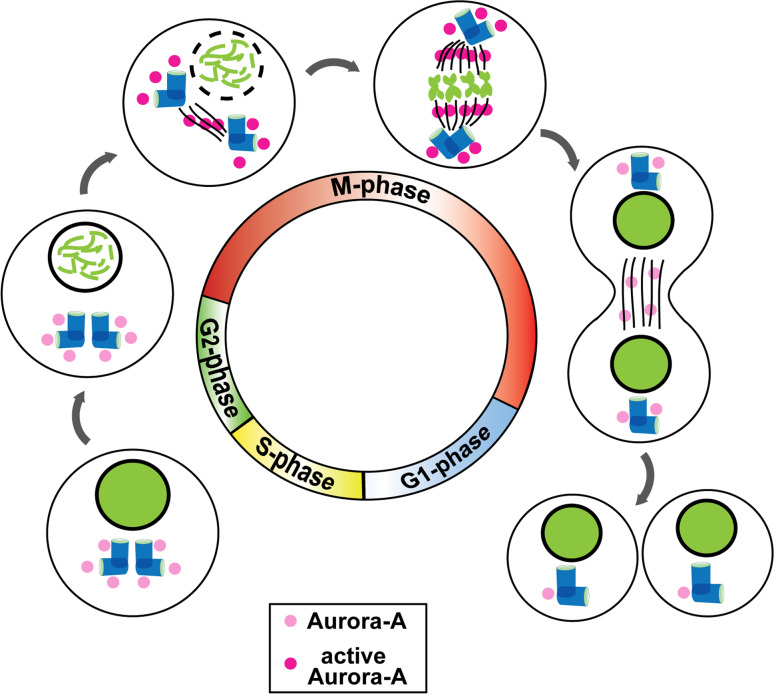
Aurora A expression, localization, and activities as cells enter and exit mitosis are summarized in Fig. 1. Initial genetic studies of Aurora A mutants all identified defects in the formation and control of the bipolar spindle in mitosis. A more detailed analysis of Aurora A expression, activation, and direct phosphorylation substrates parses Aurora A’s contribution to a series of steps that extend earlier in the cell cycle, establishing conditions for appropriate progress into and through mitosis. Aurora-A-regulated processes include centrosome maturation and separation, followed by assembly of a bipolar spindle, trigger of mitotic entry, alignment of chromosomes in the metaphase, and cytokinesis/abscission: proteolytic degradation of Aurora A is necessary before cells progress to G1. Aurora-A-interacting proteins relevant to these activities are listed in Table 1, and the functions of some of the most important are summarized below.
Fig. 1.
Aurora A in the cell cycle. Aurora A begins to accumulate significantly at centrosomes in the S phase and is activated at the boundary between the G2 and M phases. Active Aurora A propagates along the mitotic spindle to the midzone, with most of the protein being inactivated and degraded before cytokinesis with only low levels detectable in early G1 cells
Table 1.
| Gene | Aurora A phosphorylates | Phosphorylation on Aurora A | Localization | Function | Reference | |
|---|---|---|---|---|---|---|
| Activator | TPX2 | ND | T288 | Centrosome | Activation of kinase activity, targeting to microtubules | [78, 146] |
| Ajuba | ND | T288 | Centrosome | [86] | ||
| NEDD9 (HEF1) | S296 | T288 | Centrosome | Activation of kinase activity | [21] | |
| BORA | T288 | Cytoplasm | Activation | [20] | ||
| Ca2+/calmodulin | S51/S53/54/S66/67/S98 | Centrosome | Rapid activation | [105] | ||
| Nucleophosmin B23 (NPM) | S89 | Centrosome | Activation | [104] | ||
| Phosphatase inhibitor (I-2) | T288 independent | Centrosome | Activation | [101] | ||
| Arpc1b | T21 | T288 | Centrosome | Activation | [99] | |
| Calmodulin | T288 | Centrosome | Activation | [143] | ||
| PAK1 | T288, S342 | Centrosome | Activation | [74] | ||
| Inhibitor | PP1 | Dephosphorylate T288 | ? | [102] | ||
| P53 | Nucleus | [172] | ||||
| Gadd45a | [241] | |||||
| PP6 | Spindle pole | [102] | ||||
| Substrate | BRCA1 | S308 | – | Centrosome | G2/M transition of cell cycle | [41] |
| PLK1 | T210 | Cytoplasm | [31] | |||
| CDC25B | Cytoplasm | [33] | ||||
| Centrin | S122/S170 | Centrosome | [242] | |||
| LATS2 | S83 | – | Centrosome | [23] | ||
| GEF-H1 | S885 | – | Centrosome | [72] | ||
| TACC3 (human) | S34/S552/S558 (human) | – | Centrosome | [7, 49, 52] | ||
| NDEL1 | S251 | – | Centrosome | [26] | ||
| HDAC6 | ND | – | Basal body of primary cilium | Activation of kinase activity | [130] | |
| Ski | ND | – | Centrosome |
? Mitosis ? Oncogenic |
[243] | |
| Hepatoma upregulated protein (HURP) | S627/S725/S757/S830 | – | Protein stabilization | [61] | ||
| p53 | S215/S315 | – | Protein degradation | [244] | ||
| PP1 | [102] | |||||
| TPX2 | S48/S90/S94 | T288 | Nucleus and mitotic spindle | [79] | ||
| Eg5 | – | Mitotic spindle | Centrosome maturation and separation | [57] | ||
| Histone H3 | S10 | – | Chromosomes | [245] | ||
| CENP-A | S7 | – | Centrosome | [65] | ||
| CENP-E | T422 | – | [64] | |||
| CEP192 | T295 (equivalent to T288 in humans) | Centrosome | Activation, targeting to centrosomes | [19] | ||
| CPEB | Cytoplasm | [246] | ||||
| LIMK1 | S307/T508 | ND (not at T288) | Centrosome | Activation | [56] | |
| LIMK2 | S283/T494/T505 | Cytoplasm, nucleus | [247] | |||
| SRC | ND | ND | Cytoplasm | Kinase activity enhancement | [196] | |
| RalA | S194 | ND | Cytoplasm | RalA translocation; increased motility, anchorage-independent growth | [182, 183] | |
| AKT | S473 | – | Cytoplasm | Kinase activity enhancement | [181] | |
| IκBα | S32/S36 | – | Cytoplasm | Degradation | [180] | |
| Polycystin-2 (PC2) | S829 | ? ER, primary cilium ? | Negative regulation of protein function | [131] | ||
| Binding partners | TACC1 | Centrosome, spindle pole | [25] | |||
| Centrosomin (CNN) | Spindle pole | [248] | ||||
| N-Myc | Spindle | Protein stabilization | [187] | |||
| Destruction | Chfr | Spindle pole | Promotes Ubq-dependent proteosomal degradation | [112] | ||
| Protein phosphatase 2A (PP2A) | – | – | Spindle pole | Promotes Ubq-dependent proteosomal degradation | [117] | |
| Cdh1 | Spindle pole | Promotes Ubq-dependent proteosomal degradation | [111] | |||
| Cdc20 | Spindle pole | Promotes Ubq-dependent proteosomal degradation | [111] | |||
| AURKAIP1 (AIP) | – | – | Spindle pole | Ubq-independent proteasome dependent degradation | [122, 123] | |
| Antizyme 1 (Az1) | – | – | Spindle pole | Ubq-independent proteasome dependent degradation | [121] | |
| GSK-3β | S245/S387 | S290/S291/S349 | Spindle pole | Promotes Ubq-independent proteasome dependent degradation | [106] | |
| FBXW7 | T217/E221 binding sites | – | Spindle pole | Promotes Ubq-independent proteasome dependent degradation | [124] | |
| Fas-associated factor 1 (FAF1) | S289/S291 | ND |
Promotes Ubq-independent proteasome dependent degradation |
[249] | ||
| PHLDA1 | S98 | Cytoplasm | Promotes Ubq- dependent degradation | [250] | ||
| PUM2 | – | – | Centrosome | Protective against degradation | [107] | |
| Ubiquitin-specific cysteine protease 2a (USP2a) | – | – | Centrosome | Inhibits degradation, deubiquitination | [119] |
Centrosome maturation
During the S phase, following centrosomal duplication, Aurora A starts to accumulate at the centrosomes. As cells progress toward the point of mitotic initiation, centrosomes undergo a maturation process that renders them capable of nucleating the many microtubules that form the mitotic spindle and allows them to act as a signaling platform for mitotic regulators [17, 18]. During this maturation process, which is most noticeable in the late G2 phase, centrosomes expand in diameter owing to the accretion of a pericentriolar mass (PCM) composed of γ-tubulin, the γ-tubulin ring complex (γ-TURC), and a number of additional regulatory proteins [18]. PCM protein recruitment during centrosome maturation is controlled by centrosome protein of 192 kDa [Cep192/spindle defective 2 (Spd-2)], which helps target Aurora A to the centrosome as well as activate the protein in mitosis [19]. Defective expression of Aurora A or some of its activators, such as Bora and NEDD9, similarly leads to defects in centrosome maturation [20, 21].
At the centrosome, Aurora A helps in recruiting γ-tubulin, centrosomin, LATS2, TACC, and NDEL1 to the PCM [22–26]. Comparison of studies performed in Drosophila, C. elegans, and mammals indicates some variance in Aurora-A-dependent PCM growth depending on the organismal and cellular context, with Aurora A being required for γ-tubulin recruitment in C. elegans and in Drosophila sensory organ precursor cells [22, 27], but not in Drosophila neuroblasts or S2 cells [7]. Aurora A phosphorylation of the LATS2 kinase promotes its recruitment to the centrosome [23]. At least in some cell systems, LATS2 is required for recruitment of γ-tubulin, working in concert with the Aurora A partner Ajuba [24]. Another target of Aurora A in centrosome maturation is nuclear distribution element-like 1 (NDEL1), an evolutionarily conserved coiled-coil-containing protein. Aurora A phosphorylation of NDEL1 on S251 in the late G2 phase is required for NDEL1 localization to the centrosome and also subsequently triggers the ubiquitin-mediated degradation of NDEL1; expression of a phosphomimetic mutant of NDEL1 fully compensates for the centrosome maturation defect seen with depletion of Aurora A [26]. Downstream, one important role of NDEL1 in the G2 phase is targeting of TACC3 (also known as maskin, and discussed extensively below in relation to control of spindle function) to the centrosome [26].
Timing of mitotic entry
Concurrent with centrosomal maturation, Aurora A also supports the activation of the CDK1/cyclin B complex to allow nuclear entry. CDK1/cyclin B activation occurs initially at the centrosome before propagating throughout the cell [28, 29], with this initial activation being dependent on positive-reinforcement cycles involving Aurora A. Aurora A in association with its partner Bora phosphorylates T210 in the T-loop of the PLK1 phosphatase during the G2 phase; Bora is later degraded in a PLK1-dependent manner after entry into mitosis [30–32]. PLK1 promotes the recruitment of Aurora A to the centrosome in the late G2 phase, where Aurora A phosphorylates the CDK-activating phosphatase CDC25B (cell division cycle 25B) on S353, promoting mitotic entry [33]. Activated PLK1 also promotes the activation of CDK1/cyclin B by inducing the degradation of the CDK-inhibitory kinase WEE1, and by activating phosphatase CDC25C [34, 35].
Studies in Xenopus oocytes have identified an additional role for Aurora A in regulating the accumulation of cyclin B1 by enhancing its mRNA translation [36–40]. In cycling extracts from non-mitotic oocytes, a complex containing TACC3/maskin, the translational control factor CPEB, eIF4E, and PUM2 binds the maternal cyclin B1 mRNA, precluding the formation of an active translation initiation complex. Upon cell entry into the M phase, Eg2 (Aurora A) phosphorylates CPEB on S174, causing the complex to dissociate and inducing cyclin B1 mRNA polyadenylation and translation [36–40]. Although well documented in Xenopus, this particular activity of Aurora A has not been demonstrated in mammalian somatic cells, and may be species-specific.
Aurora A also controls the G2/M transition via interactions with the C-terminal domain of centrosomally localized BRCA1 (breast cancer associated gene 1) [41] that help localize BRCA1 to the centrosome [42]. Further, Aurora A phosphorylation of S308 of BRCA1 is required for M phase entry; exposure of cells to the DNA damage induced by ionizing radiation triggers a cell cycle checkpoint in part through elimination of this phosphorylation. At present, downstream targets of BRCA1 in this pathway are not defined.
Some Aurora A activities that are critical for mitotic entry do not impact centrosomally localized partners and are also coordinated with CDK1/Cyclin B effector function. For example, mitochondria must undergo fission, allowing equal post-mitotic segregation between daughter cells. Aurora A phosphorylation of the Ras family GTPase RALA on S194 drives RALA to the mitochondria, where it associates with its effector RALBP1 and with the dynamin-related GTPase DRP1. DRP1 is separately directed to the mitochondria by phosphorylation by activated CDK1/cyclin B. Together, the RALA–RALBP1–DRP1 complex effectively regulates mitochondrial fission [43].
Construction and control of a bipolar spindle
A critical mitotic role of Aurora A is in supporting the appropriate functioning of the centrosome as a microtubule-organizing center (MTOC) in mitosis. In all metazoans assessed to date, mutation or depletion of Aurora A causes formation of spindles with abnormally organized poles, including characteristic monopolar structures, and weak, sparse, or short astral microtubules [5, 7, 22, 44–47]. Aurora A control of the mitotic spindle has been the target of intense study, and a number of critical effectors have been identified.
Transforming acidic coiled-coil (TACC) family proteins are evolutionarily conserved, lack known catalytic activity, and feature a 200 amino acid coiled-coil motif at their C-terminus to promote protein interactions; their role in association with Aurora A in the formation and action of a bipolar spindle is exhaustively discussed elsewhere [48]. Briefly, Aurora A expression is correlated with that of TACC1, TACC2, and TACC3, and Aurora A forms complexes with TACCs 1 and 3 [49–51]. TACC proteins interact with proteins of the highly conserved ch-TOG/XMAP215 family to stabilize microtubules at centrosomes. This stabilization involves the complex binding to the minus ends of microtubules and thus opposing the activity of a microtubule-destabilizing kinesin, mitotic centromere associated kinesin (MCAK/XKCM1) [49, 52]. In Drosophila cells with inactivating aurora mutations, or aurora depleted by RNAi, the abnormal organization of the spindle poles and abnormally short arrays of astral microtubules correlate with the loss of the Drosophila D-TACC from centrosomes [7]. Aurora A also influences formation of bipolar spindles by directly phosphorylating MCAK on S196 to regulate its activity on mitotic asters, and on S719 to promote its association with spindle poles [53]. Together, these events induce conversion of mitotic asters to a fully developed bipolar spindle.
Besides its role in regulated M phase entry, Aurora A inhibits an additional activity of the BRCA1 ubiquitin ligase in negative regulation of centrosomal microtubule nucleation, working in tandem with protein phosphatase 1α (PP1α) [42, 54]. At present, the essential targets of BRCA1 relevant to mitotic MTOC activity for the spindle pole are not known. Centrosomal LIMK1 has been proposed to support spindle formation through modulating a dialog with the actin filament system [55, 56]. Aurora A phosphorylates LIMK1 on S307, with this phosphorylation being important for maintaining mitotic co-localization of the two proteins, and hence directs LIMK1 activity.
The earliest defined mitotic target of Aurora A phosphorylation was the kinesin-related, evolutionarily conserved protein XlEg5 of Xenopus and this phosphorylation was demonstrated to be important for spindle assembly and stability [57]. Subsequent studies suggested a model in which Aurora A control of Eg5 influences centrosomal separation by regulating Eg5 specification of microtubule sliding, i.e., by directly forcing apart centrosomes [48]. A more recent study focused on the interaction between Aurora A/Eg5 and a kinetochore-based spindle assembly pathway involving the protein Mcm21R, which uses poleward microtubule flux to help push centrosomes apart [58].
Spindle nucleation arises from the centrosome, from chromatin, or in the absence of either given an appropriate initiating cue [59]. While most attention focuses on the role of Aurora A at centrosomes in the formation of the spindle aster, Aurora A is capable of inducing formation of spindle-like asters in the absence of the nucleating function of the centrosome or chromatin [60]. This activity involves interaction between Aurora A and TPX2 (target protein for X enopus kinesin-like protein 2), and a group of proteins including HURP [61], XMAP215, and Eg5 [62]. Under physiological conditions, these interactions and Aurora A activation are stimulated by Ran-GTP, which is enriched in the vicinity of the kinetochore and chromatin (a pathway reviewed extensively elsewhere [63]). Aurora A phosphorylates a single conserved residue close to the motor domain of another kinesin, centromere protein A (CENP-E/kinesin-7), which is a major mitotic regulator of bipolar spindle dynamics. A binding site for the PP1 phosphotase overlaps this phosphorylation site; Aurora A phosphorylation disrupts PP1 binding to CENP-E. This Aurora A/PP1 switch is required not only for congression of polar chromosomes through modulation of CENP-E motor activity, but also for CENP-E-dependent delivery of PP1 to the kinetochore, which is important for stable biorientation of chromosomes [64]. CENP-A is a variant of histone H3 that is a component of the nucleosome core of centromeric chromatin at the kinetochore. Aurora A phosphorylates S7 of CENP-A, an essential step for the attachment of microtubules to the kinetochore, and consequently for chromosome alignment and segregation [65, 66].
Another Aurora A partner, RASSF1A, is a tumor suppressor that binds microtubules to arrest cell growth in the M phase [67]. Aurora A phosphorylation of RASSF1A interrupts its microtubule binding activity, and also relieves RASSF1A-dependent inhibition of the APC/Cdc20 protein degradation complex, permitting M phase progression [67, 68]. Aurora A also coordinates chromosome segregation and anaphase microtubule dynamics later in mitosis. Aurora A propagates from the centrosome to the spindle and then to the midzone, before much of the protein is degraded at the midbody [69]. Aurora A has recently been found to be mitotically SUMOylated, which may contribute to its localization control [70]. After moving onto the spindle, Aurora A contributes actively to the ability of the APC/Cdh1 complex to form a robust spindle midzone in late mitosis [69]. SAF-A (scaffold-attachment factor A) is required for chromosome segregation, in part through association with the Aurora A–TPX2 complex. An essential function of SAF-A is to recruit Aurora A to the spindle microtubules [71]; together, SAF-A, Aurora A, and TPX-2 influence chromosome congression, kinetochore–microtubule attachments, and stability of kinetochore microtubules. Finally, together with other mitotic kinases, Aurora A also helps time mitotic progression by phosphorylating and thus restraining the activity of the RhoA activator GEF-H1; loss of Aurora A and GEF-H1 activation during the latter stages of mitosis facilitates RhoA activation at the cleavage furrow and during abscission [72].
Control of Aurora A activation and degradation in mitosis
Activation of Aurora A
Activation of Aurora A as cells enter mitosis involves spatially and temporally constrained interactions with multiple partner proteins. Proteins regulating this process are indicated in Table 1 and Fig. 2. Littlepage et al. [73] first defined mitotic phosphorylation of Aurora A incubated with metaphase-arrested frog oocytes, with phosphorylation observed on residues S53, T295, and S349 (equivalent to S51, T288, and S342 of human Aurora A). Activating interactions that induce Aurora A autophosphorylation at T288 (in human Aurora A; T295 in Xenopus Aurora A) in the kinase activation loop have been most studied. Relevant partner proteins for this phosphorylation include TPX2, Ajuba/JUB, NEDD9 (also known as HEF1 and CasL), and BORA. In addition, PAK1 (p21-activating kinase 1) has been reported to activate Aurora A via transphosphorylation of T288 [74].
Fig. 2.
Aurora A interaction with partners regulating activation and destruction. Proteins interacting with Aurora to promote activation (green line, star), promote destruction (blue line, arrow), or protect from destruction (orange line, shield) are indicated in context of phases of cell cycle in which they associate with Aurora A. When interaction involves specific phosphorylations on Aurora A, these are indicated in a circle at the left of drawing
The structural basis of the TPX2 [75] interaction has been most intensively studied. Activation of the GTPase Ran at nuclear envelope breakdown GTP stimulates spindle assembly by releasing the spindle assembly factor TPX2 from an importin-α/β inhibitory complex. The liberated TPX2 binds to Aurora A kinase, promoting a conformational change that activates Aurora A autophosphorylation, and moves the activation loop to a central position from which it is protected from dephosphorylation by the negative regulatory factor PP1, providing access for Aurora A substrates [76–82]. This autophosphorylation is a dominant factor in initiating and maintaining Aurora A activation in mitosis, and is lost when cells are treated with Aurora A kinase inhibitors [83, 84]. Phosphorylation of TPX2 by Plx1 increases its ability to activate Aurora A [85]. The TPX2 interaction also helps target Aurora A to mitotic spindles, proximal to substrates [51, 79].
Ajuba (JUB) is a scaffolding protein, containing multiple LIM domains, that was described as an interactor and activator of Aurora A in 2003 [86]. Both Aurora A and Ajuba are phosphorylated by Aurora A during their interaction; this interaction was shown to be important for activation of the cyclin-B/CDK1 complex and for committment of cells to mitosis [86]. No studies of this activating interaction have subsequently been reported in humans. However, one recent study in Drosophila sheds potential light on the Aurora A/Ajuba interaction [87]. Drosophila jub (ajuba) mutants die in the larval–pupal transition. Jub localizes to the centrosomes of neural stem cells; mutation of jub led to centrosome separation defects and abnormal mitotic spindles. Surprisingly, in jub mutants Aurora A activity is not perturbed, but Aurora A recruitment and maintenance at the centrosome is defective. The authors of this study proposed that a major function of Jub in Drosophila is to restrict active Aurora A to the centrosome during mitosis, but not to activate Aurora A. This failure of Jub to regulate Aurora A might also reflect a difference in the way Aurora A is regulated between vertebrates and invertebrates [87]: more investigation is required.
Aurora A directly interacts with another scaffolding protein, NEDD9, also known as CAS-L and HEF1 [88–90]. Identification of NEDD9 as an Aurora A activator was initially surprising, given a predominant role for this protein at focal adhesions in the regulation of cell migration and invasion [91–94]. However, NEDD9 was subsequently found to accumulate at the centrosome as cells move into the G2 and M phases [21, 95]. Depletion of NEDD9 does not affect Aurora A accumulation at the centrosome, but blocks the T288 phosphorylation and activation of Aurora A at mitotic entry and leads to accumulation of cells with monopolar spindles and cleavage furrow regression. Overexpression of NEDD9 induces Aurora A hyperactivation and produces cells with both multipolar spindles and supernumerary centrosomes and failure of cytokinesis [21, 96]. Interestingly, a protein closely related to NEDD9, p130Cas/BCAR1, has been shown to bind directly to Ajuba [97]: potentially, NEDD9 and Ajuba similarly interact during Aurora A activation.
The serine/threonine kinase Pak1 has well-known functions in proliferation and cell migration signaling, but moonlights with additional roles in the cell cycle, including the control of centrosome number and mitotic progression. Zhao et al. defined interactions between Pak1 and a centrosomal adaptor protein, PIX/GIT, that lead to centrosomal activation of Pak1: PAK1, or two related PAK kinases, PAK2 and PAK3, each bind Aurora A and can phosphorylate Aurora A on the T288 activation loop site or an alternative site, S342, in vitro [74]. Like Pak1, LIM kinase 1 (LIMK1) is better known as a regulator of the actin cytoskeleton through phosphorylation of substrates such as cofilin. However, a pool of LIMK1 colocalizes with γ-tubulin and Aurora A at the centrosome between early prophase through anaphase and phosphorylates Aurora A. The target of LIMK1 is not the well-defined T288 motif, but has otherwise not been determined [56].
Mutations in the Drosophila borealis (bora) gene phenocopy Aurora A mutations. One study has shown that Cdc2-dependent nuclear exclusion of Bora releases the protein to the cytoplasm, where it binds and activates Aurora A [20]. Part of the function of Bora in promoting Aurora A activity may involve binding to substrates as a cofactor with Aurora A, increasing phosphorylation site availability [30]. However, in contrast, two independent studies have suggested that Plk1 phosphorylation of Bora leads to β-TrCP-dependent degradation of Bora, with one study indicating that human Bora competed with TPX2 for Aurora A binding, limiting Aurora A activation [32, 98]. The kinase Plk1 is known to be required for Aurora A association with centrosomes, important during centrosome maturation [42, 61]. Overexpressed cytoplasmic hBora, found during Plk1 inhibition, was conjectured to titrate away Aurora A from relevant substrates necessary for spindle assembly, providing a separate means of controlling activity and linking the function of Aurora A with a second important mitotic kinase. Clearly the function of Bora will require more work to elucidate.
Arpc1b is another upstream activator of Aurora A kinase, better known as a component of the Arp2/3 actin regulatory complex. Overexpression of Arpc1b leads to abnormal centrosomal amplification, whereas depletion of Arpc1b drastically reduces the ability of cells to enter the cell cycle; this is accompanied by failure to accumulate active Aurora A at the centrosome at the G2/M transition. Coupled in vitro experiments have demonstrated that Arpc1b interaction with the N-terminal domain of Aurora A activate Aurora A, suggesting the interaction sterically influences the Aurora A active site [99].
The centrosomal protein CEP192 is also important for bipolar spindle assembly. Direct interactions between CEP192 and Aurora A were shown to help concentrate Aurora A at the centrosome, allowing the protein to form homodimers and homo-oligomers. This interaction supports Aurora A activation and microtubule assembly via a different mechanism from that described for TPX2 and the other activators [19].
As early as 1994, the activity of protein type 1 phosphatase was shown to oppose that of IPL1 kinase in mitosis [100], and IPL1 mutations were found to be suppressed by overexpression of Glc8, an ortholog of phosphatase inhibitor 2 (I-2, Inh-2) [101]. In humans, protein phosphatase 1 (PP1) dephosphorylation of T288 limits Aurora A activity [102], whereas I-2/Inh-2 opposes PP1 in this function, thus enhancing Aurora A activity [101]. The opposition involves direct binding of I-2/Inh-2 to Aurora A, is not additive with TPX2 activation, and interestingly, does not involve increased Aurora A activation–loop phosphorylation [101]. A subsequent evolutionary analysis of I-2/Inh-2 proteins demonstrated that vertebrate (human and Xenopus), but not Drosophila, C. elegans, or yeast orthologs of the group, possessed this activation activity [60]. More recently, the protein phosphatase 6 (PP6) holoenzyme has been described as the major negative regulator of Aurora A activation–loop phosphorylation, inhibiting the stability of the Aurora A/TPX2 complex [103].
Aurora A also interacts with nucleophosmin/B23 (NPM). Although NPM is best known as a nucleolar protein, it also accumulates with Aurora A at the centrosome in G2 cells, and co-immunoprecipitates with the protein. Interestingly, although NPM strongly induces Aurora A activity in in vitro kinase assays with canonical substrates, it neither induces T288 phosphorylation on Aurora A nor protects T288-phosphorylated Aurora A from dephosphorylation by PP1 [104]. Instead, this activation was associated with and depends on S89 phosphorylation of Aurora A. These data suggest a third, independent means of activating Aurora A, together with the mechanisms associated with TPX2 and Cep192 (also see [105], and the discussion of Ca2+/calmodulin (CaM), below, for a fourth mechanism that has been described in interphase cells); we note that recent work by Dodson and Bayliss [81] also supports the idea of Aurora A activation without obligate T288 phosphorylation.
Littlepage et al. found that S349A-mutated Xenopus Aurora A had modestly reduced kinase activity, but that an S349D phosphomimic completely blocked kinase activation [73]. Sarkissian et al. [106] extended this work, finding that glycogen synthase kinase 3 (GSK3) phosphorylation of Aurora A on S290/S291 subsequently induces Aurora A autophosphorylation on S349, which is inhibitory. S290 and 291 are just prior to T295 of the activation loop in Xenopus Aurora A, whereas S349 is on the αG helix of the C-terminal domain (see below). The equivalent residues in human Aurora A are S283, S284, T288, and S342. They also found that mutation of these sites to non-phosphorylatable alanines (S349A, S290A/S291A) results in a constitutively active form of the kinase. Interestingly, this group inspected immature versus mature Xenopus oocytes and found that activity of Aurora A correlated with loss of S349 phosphorylation rather than changes in the level of T295 phosphorylation (equivalent to T288 in humans) [106].
Another Aurora A binding protein, PUM2, has been identified as promoting in vitro activation of Aurora A; to date, the mechanism of action involved is unknown [107]. Similarly, the Aurora A partner and substrate RASSF1A activates Aurora A through an undefined mechanism [108]. It is likely that there are additional regulators of Aurora A activity that have not yet been defined. For example, recently Aurora A was identified as a proximal target of a mitotic checkpoint associated with Golgi fragmentation, which inhibited both Aurora A recruitment to centrosomes and Aurora A activation [109]. The molecular basis of this checkpoint requires further study.
Aurora A degradation in late mitosis
Destruction of Aurora A is also regulated via interactions with partner proteins (Table 1; Fig. 2). Aurora A is degraded by the proteasome at the end of mitosis [110–112], with inhibition of the proteasome in cycling cells leading to the accumulation of ubiquitinated forms of Aurora A [110–112]. Aurora A is targeted for proteasomal degradation via interaction with the anaphase-promoting complex/cyclosome (APC/C [113]) and particularly with phosphorylation- and protein partner-controlled association with the APC/C co-activator subunits, Cdc20 and Cdh1 [114]. Aurora A kinase contains two different sequences, an A-box and a D-box, that must both be present for Cdh1-dependent protein destruction. The N-terminal A-box (residues 47–59, in Xenopus) encompasses a short sequence, Q47RILGPSNVPQRV, which is highly conserved in vertebrate forms of Aurora A. The A-box is contained in the N-terminal disordered region of Xenopus Aurora A (residues 1–128). The E3 ubiquitin ligase Chfr (checkpoint protein with forkhead and ring domain) directly targets Aurora A for ubiquitination and degradation, interacting with an N-terminal region (res 1–61) containing the A-box [115]. This region also includes the Xenopus S53 (human S51), phosphorylated during mitosis. Although S53A mutation does not block destruction, an S53D phosphomimic mutation completely blocks Cdh1-induced destruction, results interpreted as suggesting that S53 phosphorylation might negatively regulate Aurora A destruction until the last stages of mitotic exit [116]. In support of this idea, protein phosphatase 2A (PP2A) associates with Aurora A at the spindle poles in mitosis, and dephosphorylates S51; inhibition of PP2A reduces the destruction of Aurora A at the end of mitosis [78, 117].
The C-terminus of Aurora A contains a functional destruction box (D-box, residues 378–381) [114]. The translational regulator PUM2 binds to the D-box of Aurora A, preventing its ubiquitination and enhancing protein stability [107]. Aurora A also contains a KEN box (residues 6–9), which is required for the degradation of other mitotic proteins (Nek2 and B99); however, this is not crucial for Aurora A degradation [116, 118]. The USP2a (ubiquitin-specific cysteine protease 2a) stabilizes mitotic Aurora A through the direct protein interaction of deubiquitination; it appears that multiple interaction motifs mediate the association of the two proteins [119].
Other studies have indicated an additional, ubiquitin-independent pathway of Aurora A degradation. Aurora A kinase interacting protein 1 (AURKAIP1 or AIP) promotes degradation of Aurora A in a proteasome-dependent but ubiquitin-independent manner [120]. AURKAIP1 acts upstream of antizyme 1 (Az1), a well-studied mediator of ubiquitin-independent protein degradation, and has been proposed to enhance the binding affinity of Az1 to Aurora A to promote proteasomal targeting [121, 122]. AURKAIP1, Aurora A, and GSK-3β colocalize at the spindle poles in the metaphase. Further, depletion of AURKAIP1 stabilizes and activates Aurora A in the early mitotic phase, causing mitotic cell arrest. AURKAIP1 phosphorylation by GSK-3β decreases its ability to downregulate Aurora A, suggesting that GSK-3β positively regulates Aurora A expression in early mitosis [123]. This opposes the effect of GSK-3β phosphorylation on Aurora A discussed above, in which phosphorylation on S290/291 negatively regulates kinase activity [106], suggesting either a delicate and potentially dynamic balance between Aurora A, AURKAIP1, and GSK-3β at different phases of mitosis (or alternatively, differences between assay systems). GSK-3β also regulates the F-box protein FBXW7 to induce degradation of Aurora A; this process is impaired in cancer, after loss of the tumor suppressor PTEN causes inhibition of AKT and GSK-3β [124].
Non-mitotic activation and signaling of Aurora A
While the main focus on Aurora A has been its actions in mitosis or cancer, it is increasingly apparent that the protein has important functions in non-transformed, non-mitotic cells. Better-defined examples include influence on microtubule dynamics, cell migration, and polarity, particularly in the context of neurite extension [22, 26, 27, 125–129], induction of disassembly of cilia [130], and regulation of intracellular calcium signaling [131]. Given the elaborate spatial and temporal assembly of the Aurora A activation machinery described above to support the high level and sustained activation of Aurora A in mitosis, an important question linked to these observed non-mitotic activities relates to activity control in the absence of the machinery. Although this field is much less mature, the emerging answers appear to indicate the existence of alternative means of Aurora A activation [104, 105], and also to indicate that some Aurora A activities may not depend on protein activity, as opposed to scaffolding function provided by inactive Aurora A [132]. We note that there is very clear evidence for multiple additional activities of Aurora A during the interphase in cancer cell lines, generally in cases in which the protein is overexpressed. As it is not yet clear whether these activities occur in untransformed cells, albeit at lower levels, or are specific to the transformed environment, they are discussed in the following section on Aurora A in cancer.
Aurora A in microtubule dynamics and cell polarity control: emphasis on neurite extension
In interphase human cells, the majority of Aurora A is localized to the centrosome, although a smaller pool of the protein is also found in the cytoplasm and nucleus [127]. The centrosome nucleates microtubular networks throughout all phases of the cell cycle, helping organize cell structure, polarity, and migration [95]. Chemical inhibition of Aurora A in interphase mammalian cells significantly disrupts interphase microtubule dynamics, suggesting that at least some of the protein is in an active form and in a non-centrosomal compartment [125]. It is also possible that the accumulating pool of inactive Aurora A serves some functional role in regulating interphase microtubular dynamics; a kinase-independent stabilizing role for Aurora A in stabilizing spindle microtubules was very recently reported for Aurora A in C. elegans [132], but a possible similar function has not been investigated in interphase cells.
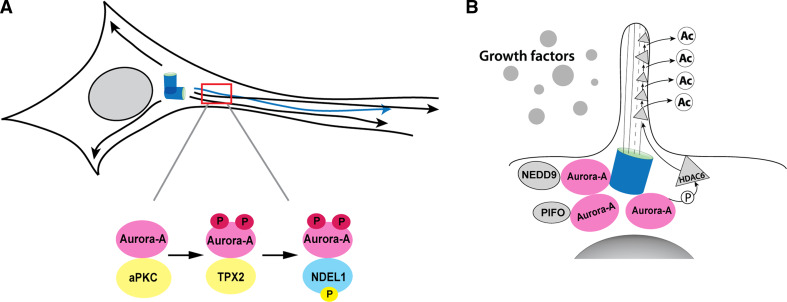
As noted above, studies of mitotic functions of Aurora A demonstrated that Aurora A bound and phosphorylated NDEL1 and TACC3 as part of the completion of centrosomal maturation, and that expression of an NDEL1 phosphorylation-mimetic can compensate for loss of Aurora A activity in allowing mitotic entry [26]. In a fascinating extension of their initial work, Mori and co-workers pursued the observation that NDEL1 is an important binding partner of LIS1, the first gene identified as a target of mutation in the severe neuronal developmental disorder lissencephaly investigating whether Aurora A might contribute to NDEL1 function in this second context [129]. This study found that Aurora A was abundantly expressed in post-mitotic neurons and was colocalized with and phosphorylated by atypical PKC (aPKC) on T287 of the activation loop, with this activating phosphorylation essential for the microtubule-dependent process of neurite extension (Fig. 3a). Although the TPX2–Aurora A interaction leading to T288 phosphorylation was observed in these cells, and this interaction supported the aPKC-Aurora A–NDEL1 pathway, the latter was particularly important for the microtubule reorganization that supported neurite elongation [129].
Fig. 3.
Active Aurora A in non-cycling cells. a aPKC activates Aurora A, which interacts with NDEL1 to induce microtubule-based extension of post-mitotic neurons. b NEDD9 and PIFO activate Aurora A at the basal body of cilia in quiescent cells, leading to Aurora-A-dependent activation of HDAC6, and ciliary resorption
aPKC interacts with two partner proteins, Par-3 and Par-6, in an evolutionarily conserved complex that governs asymmetric cell division at mitosis and regulates cell polarity [126, 133]. Interestingly, Aurora A had previously been defined as promoting aPKC activation in mitosis, with Aurora A phosphorylation of Par-6 derepressing aPKC activity [6]. Activity of the PP2A protein phosphatase opposes this activation of aPKC in neuroblasts [133]. Studies of Aurora A in mitotic cells demonstrated that PP2A dephosphorylation of Aurora A residue S51 promotes Aurora A degradation [117]. To date, the extent to which Aurora A mitotic interactions with PP2A, Par-3, and Par-6 are maintained in post-mitotic functions such as neurite extension remains relatively unexplored. However, given recent identification of roles for Par-3 at the centrosome in the polarization of other post-mitotic populations, such as intestinal epithelial cells [134], it seems likely that this is a fertile area for investigation.
Aurora A in ciliary disassembly, calcium regulation, and PKD
One of the more surprising activities to emerge recently for Aurora A was based on genetic studies of a very distant Aurora A ortholog, the CALK protein of the green algae Chlamydomonas reinhardtii [135, 136]. Chlamydomonas utilize organelles termed flagella for environmental sensing, mating responses, and movement, but resorb or shed flagella during mating response to pheromone and transient ionic shock. CALK is activated during and required for flagellar loss [136]. In mammals, most cells have a single, non-motile cilium—a structure paralogous to a flagellum—protruding as an antenna from the cell surface, acting as a receiver for mechanical and chemical cues from fluid flow or the extracellular matrix (Fig. 3b). The cilia extend from a perimembrane basal body: this basal body differentiates from the centrosome in quiescent cells but redifferentiates to a centrosome as cells return to cycle, paralleled by a cell-cycle-related protrusion and resorption cycle for cilia [137]. Defects in cilia are associated with numerous clinically important syndromes, with “ciliopathies” including polycystic kidney disease (PKD), nephronopthisis (NPHP), Joubert syndrome (JS), Bardet–Biedl syndrome (BBS), and others [29]. Signaling systems dependent on or influenced by intact cilia include Hedgehog, Notch, Wnt, Par3-aPKC, and PDGFa. Predictably, links are increasing between ciliary signaling defects and both developmental disorders and cancer.
In 2007, in studies modeled on those in Chlamydomonas, it was demonstrated that serum growth factors induce Aurora A activation at the basal body of the cell cilium in non-cycling G0/G1 mammalian cells, with this activation necessary and sufficient for ciliary resorption [130]. Activation of Aurora A in ciliary resorption was preceded by and dependent on upregulation of its mitotic activating partner NEDD9. Both cilia and flagella are organized around a cytoskeletal core (the axoneme) composed of nine microtubule doublets arranged in a ring; acetylation of the tubulin subunits stabilizes the axonemal structure [29]. During ciliary disassembly, Aurora A specifically phosphorylated the tubulin deacetylase HDAC6, increasing HDAC6 deacetylase activity. Inhibition of HDAC6 activity by the small molecule inhibitor tubacin or depletion of HDAC6 by siRNA in each case resulted in cilium stabilization. Interestingly, HDAC6 has been reported to associate with PP1 [138], which binds microtubules and dephosphorylates and inactivates Aurora A in pre-mitotic cells [102]. Similar feedback may limit Aurora A activation in cilia. An additional factor, Pitchfork (PIFO), also localizes to the base of the cilia and was recently described as required for Aurora A activation during the disassembly process; as yet, the mechanism is not known [139].
Further work in Chlamydomonas has suggested increasing intraflagellar Ca2+ concentrations during the mating response [140], shortly before the activation of the CALK kinase, whereas a separate study indicated rapid spatiotemporal patterning of Ca2+ distribution as a critical signal for flagellar excision [141]. An indirect association between Aurora A and Ca2+ in vertebrates was provided by an analysis of oocyte maturation in Xenopus, which indicated that inhibition of Ca2+ signaling led to eventual failure to accumulate and activate Aurora A [142]. In a direct investigation of a Ca2+–Aurora A interaction, Plotnikova et al. demonstrated that numerous stimuli that transiently increase cytoplasmic Ca2+ [including arginine vasopressin (AVP), histamine, and thapsigargin] induce Aurora A activation with extremely rapid kinetics. Ca2+-induced Aurora A activity peaks within 1 min of stimulation, returns to baseline within 2–5 min, and depends on a direct interaction between the N-terminal unstructured domain of Aurora A with Ca2+/CaM [105]. In vitro, incubation of Ca2+/CaM with Aurora A results in robust autophosphorylation of Aurora A on S51 or S53, as well as additional phosphorylations on S66 or S67, and S98. The S51/S53 phosphorylation suggested that this method of Aurora A activation may also be relevant to mitotic activation of the kinase [105]. A very recent study has provided the first evidence that Ca2+/CaM governs activation of Aurora A in ciliary resorption and in mitosis, and involves potentiation of the Aurora A/NEDD9 interaction [143]. It is possible that these activators also regulate Aurora A in post-mitotic neurons, but this has not yet been investigated. This group of studies had been performed in renal cells because of the previously defined critical interaction between cilia, Aurora A, and the function of the heterodimeric complex between polycystins 1 and 2, encoded by the PKD1 and PKD2 genes, which serves as a cilia-localized transmembrane Ca2+ channel: mutations in these genes are responsible for the most clinically prevalent of the ciliopathies, PKD [144]. Plotnikova et al. [105] determined that Aurora A is abundant and active in a subset of normal, quiescent renal cells and is overexpressed and hyperactivated in human PKD specimens. Moreover, in reciprocal action, Aurora A binds and phosphorylates the polycystin 2/PKD2 protein, limiting PKD2 activity during rapid Ca2+ signaling responses [131]. These findings suggest that Aurora A activity changes may be relevant to the pathology of PKD.
Aurora A protein structure
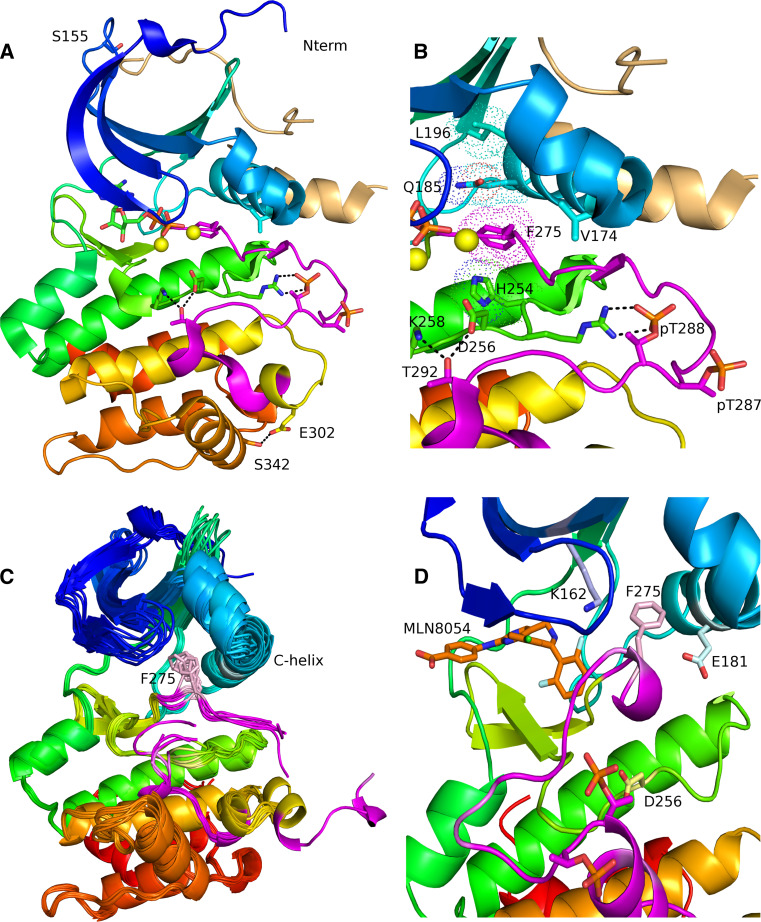
Human Aurora A is a protein of 403 amino acids, with a canonical kinase domain comprising residues 133–383. The first experimental structure of Aurora A was determined in 2002 (PDB entry 1MQ4) [145] and remains the highest-resolution crystal structure of the human kinase. There are now 57 crystal structures of human Aurora A in complex with ATP, ADP, ANP (an analog of ATP), and various inhibitors. Three of these structures are in complex with the Aurora A binding activator protein TPX2 [146–148]. In addition, there are eight structures of mouse Aurora A and five structures of Xenopus laevis Aurora B. The majority of human Aurora A structures contain coordinates only for residues 127–388, regardless of whether the proteins crystallized were longer on either or both ends. The first 123 amino acids of Aurora A are predicted to be intrinsically disordered, as are the C-terminal 16 amino acids [149]. The experimental structures confirm the disorder predictions, because even in those structures of protein constructs consisting of residues 100–403, the first ordered residue in any structure is S123 and the last ordered residue is Q394.
The Aurora A kinase domain shares the common structural features of other protein kinases. The N-terminal part of this domain consists of a five-stranded β-sheet and an α-helix termed the “C-helix”; Aurora A also has a short α-helix called the “B-helix”, seen in some kinases, that is just prior and perpendicular to the C-helix. The C-terminal part of the kinase domain consists of seven α-helices and a two-stranded β-sheet, and contains the catalytic aspartic acid of the HRD motif (sequence His-Arg-Asp at positions 254–256) and the mobile activation loop, the position and conformation of which determine whether the kinase is active or inactive. The Aurora A activation loop spans residues 274–299, beginning with a DFG sequence motif and ending with an APE motif (sequence PPE in Aurora A). Active forms of Aurora A include species that are autophosphorylated on T288, following interaction of Aurora A with partners such as TPX2 (discussed elsewhere [81]), phosphorylated on T287 and T288 by PAK [74] and PKA [112], and phosphorylated on T287 by atypical protein kinase C [129].
A given kinase may have many different conformations depending on the binding of activators or inhibitors and phosphorylation of various sites. These conformations vary primarily in the position of the activation loop as well as the position of the C-helix or the entire N-terminal domain relative to the C-terminal domain. Kornev and Taylor [150] recently provided an analysis of protein kinases in terms of two “spines” of interacting residues that span the two domains. These spine residues are not contiguous in sequence but form stacks of contacts in active kinases that are disrupted in inactive or inhibited kinases. We have identified the spine residues in Aurora A structures using sequence and structure alignments of Aurora A to PKA, based on a detailed analysis of PKA by Kornev and Taylor [150]. The so-called regulatory spine consists of Q185, L196, H254 (of the HRD motif), and F275 (of the DFG motif). These residues correspond to PKA residues L95, L95, Y164, and F185. The catalytic spine consists of V147, A160, V218, L262, L263, L264, L318, and F322 (corresponding to PKA residues V57, A70, M128, L172, L173, L174, L227, and M231, respectively).
The integrity of the regulatory spine is dependent on the position of the DFG motif, such that in an active kinase, the F275 residue of DFG points into the kinase domain and sits under the C-helix. Twenty-nine Aurora A structures contain this motif in this so-called DFGin position. As discussed below, Aurora A is activated by phosphorylation and by binding of the mitotic spindle protein TPX2 [79]. There are three structures of Aurora A with TPX2 bound (PDB entries 1OL5, 3E5A, and 3HA6), and only one of these has ADP bound (1OL5) [146], whereas the other two contain small molecule inhibitors. An image of active Aurora A kinase with ADP and TPX2 is shown in Fig. 4a. A close-up of the active site residues is shown in Fig. 4b. This structure is phosphorylated on T287 and T288. Binding of TPX2 alters the conformation of the Aurora A activation loop [146], bringing phosphorylated T288 into a position where it forms a salt bridge with R255 of the HRD motif, forming an active kinase. In the absence of TPX2, pT288 faces out and the activation loop is more extended than it is in the TPX2-bound state. In addition, in the active conformation with TPX2 bound, the side chain of T292 forms a hydrogen bond with the active site D254 of the HRD motif. In other DFGin structures without TPX2, neither of these interactions is formed. As discussed above, the regulatory spine of Aurora A kinase consists of H252, F275, Q185, and L196. In an active kinase, these residues form a column with each residue in the spine in contact with its neighbors. The van der Waals surfaces of these residues are shown in Fig. 4b.
Fig. 4.
a Ribbon diagram of the ADP-bound structure of human Aurora A from PDB entry 1OL5. The chain is colored from blue (N-terminus) to red (C-terminus). The activation loop is shown in magenta and the activator TPX2 is colored beige. Aurora A in this structure is phosphorylated on T287 and T288, and the DFG motif at the beginning of the activation loop is in the “DFG-in” position, such that the F275 of the DFG motif (magenta sticks) is located under the C-helix (cyan). b Close-up of the active site of Aurora A from PDB entry 1OL5. Hydrogen bonds are shown in dotted black lines between pT288 and R255 of the HRD motif, T292 of the activation loop and D256 of the HRD motif and K258. The yellow spheres are active site magnesium ions. The regulatory spine of Aurora A is shown in dots (H254, F275, Q195, and L196). c Several structures in the “DFGup” conformation are shown. F275 points upwards between the C-helix and the N-terminal domain β sheet. d Compound MLN8054 bound to human Aurora A from PDB entry 2WTV. F275 of the DFG motif is in a “DFGup” position, disrupting the salt-bridge interaction between E181 and K162. The activation loop is in an inactive position, in contrast to the active position in a and b
Many protein kinases exhibit an inactive conformation referred to as “DFGout.” Looking at the active site as shown in Fig. 4a, b, the Phe of DFG in DFGout structures sits underneath the ADP rather than under the C-helix. This conformation is present in one Aurora A kinase structure (PDB entry 2C6E) [151]. Besides the DFGin conformations, many Aurora A crystal structures contain an unusual conformation of the DFG loop, referred to by Dodson et al. as “DFGup” [145]. In the DFGup conformation, the F275 points upwards into the N-terminal domain and is wedged between the C-helix and the N-terminal domain β-sheet. A superposition of several DFGup structures of Aurora A kinase is shown in Fig. 4c. This position disrupts a salt bridge between K162 of the β-sheet and E181 of the C-helix that is formed in active Aurora A, as are the equivalent residues in other kinases. Aurora A kinase with the Aurora-specific inhibitor MLN8054 bound and a DFGup conformation of the activation loop is shown in Fig. 4d.
Bibby et al. [152] observed three somatic mutations of Aurora A associated with cancer. One of these, S155R, occurs in the interface between Aurora A and TPX2 and disrupts Aurora A–TPX2 interaction. S155 is the fourth residue in a type I β turn (residues 152–155, sequence EKQS) between strands 2 and 3 of the N-terminal kinase domain. S155 forms a hydrogen bond across the β turn to the side chain of E142. The likely rotamers of R155 all clash with residues in TPX2 and cannot easily form a hydrogen bond with E152 without significant rearrangement of the β turn. This residue is shown in Fig. 4a. This mutation results in a form of Aurora A with greatly reduced cellular activity [152]; this is a noteworthy physiological demonstration that abnormally low, as well as abnormally high, Aurora A activity is potentially transforming based on ability to induce aneuploidy. A second somatic mutation is V174M occurs at the junction of the B-helix and the C-helix in the N-terminal domain. This mutation leads to constitutive activation of Aurora A, probably by altering the interaction of the N- and C-terminal domains. This residue is also shown in Fig. 4b. The third somatic mutation, S361Stop, deletes the last 30 or so residues of the C-terminal domain of the kinase, likely leading to instability of the folded protein as well as deletion of the C-terminal disordered residues.
As noted above, PAK1 phosphorylated S342 of human Aurora A, whereas mutation of the equivalent residue, S349, in Xenopus Aurora A to Ala and Asp leads to less active and inactive kinase, respectively. The serine residue S342 is shown in Fig. 4a in an active kinase structure. In this conformation, S342 forms a hydrogen bond to E302, which is in a segment just after the activation loop of Aurora A (residues 274–299). Phosphorylation of S342 or mutation of S342 to Asp would be expected to break this interaction, potentially altering the conformation or dynamics of the activation loop. Phosphorylation may destabilize the conformation of the activation loop such that it may extend away from the kinase domain in a manner suitable for trans-autophosphorylation, thus leading to activation. Subsequent dephosphorylation of S342 may be required for an active monomeric kinase. Mutation to D242 would lead to permanent inactivation due to the repulsive interaction with E302. One recent report has identified a SUMOylation event on residue K249 of mouse Aurora A (K258 in human Aurora A). Mutation of this residue to a non-SUMOylatable Arg results in defective and multipolar spindles, abnormal localization to the mitotic spindle, but normal kinase activity [70]. This residue (K258), shown in Fig. 4b, is located two residues C-terminal of D256 of the HRD motif and forms hydrogen bonds with the side chains of D256 and T292 of the activation loop.
Aurora A and cancer
Phenotype and mechanism
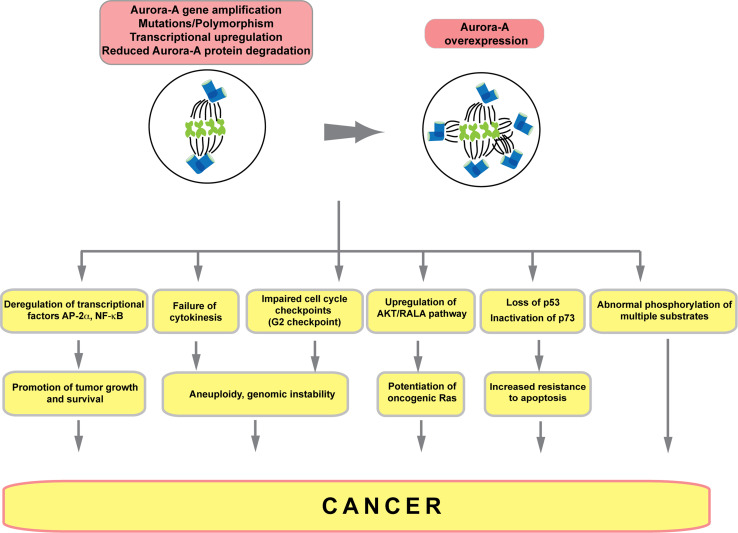
In 1998, two independent studies for the first time identified significant upregulation of Aurora A as a common feature of multiple classes of common solid cancers, including colorectal, breast, ovarian, prostate, neuroblastoma, and cervical, in both primary tumor tissue and cell lines [153, 154]. Aurora A is situated on chromosome 20q13.2, a locus that is frequently amplified in solid tumors, accounting for some of the elevated expression. However, in some cases Aurora A protein was increased in the absence of DNA rearrangements, based on changes of gene expression or protein stabilization. Aurora A transcription is regulated by the ERK-responsive Ets pathway, by STAT5a, by estrogen/GATA3 [155, 156], by HIF1 [157], and by additional pathways that are frequently elevated in cancer (also see review [158]). A number of Aurora A interacting and/or activating proteins are themselves elevated in cancer, and through their enhanced interactions stabilize Aurora A from protein degradation. Examples of these include NEDD9/HEF1 [21, 90], IQGAP1 [159], and TPX2 [77, 160]; as with Aurora A, overexpression of each of these interactors is oncogenic. Conversely, some proteins known to participate in degradation of Aurora A during the cell cycle, such as Chfr, are commonly downregulated via promoter hypermethylation or other means in cancer [161].
The pathological function of Aurora A has been frequently reviewed, and the protein has attracted much interest as a therapeutic target [10, 162–167]. There are a number of phenotypes consistently associated with Aurora A overexpression. These include the presence of supernumerary centrosomes associated with multipolar spindles, aneuploidy, increased resistance to apoptosis, and deficient cell cycle checkpoint functions (Fig. 5). A number of these phenotypes arise from the failure of cytokinesis in cells with overexpressed Aurora A, resulting in initial accumulation of centrosomes [11, 154]. How Aurora A mediates these effects involves altered interaction with numerous partners and substrates, although surprisingly, even overexpression of a kinase-dead form of Aurora A can induce centrosomal amplification [11], indicating both that non-catalytic activities of the protein may be particularly important, and also that cancer treatment strategies based on inhibiting Aurora A kinase activity may be problematic. Upon overexpression, Aurora A is readily detected in cells in all stages of the cell cycle and in both cytoplasmic and nuclear compartments. Hence, a major and open question has been whether the predominant oncogenic activities of Aurora A arise from newly acquired interactions with partners with which it does not normally associate, or from constitutive, enforced interactions replacing normally transient or cyclical functions.
Fig. 5.
Aurora A activities in cancer. Aurora A expression is elevated based on changes in DNA copy number, transcription, and/or changes in protein stability. Overexpressed Aurora A induces multiple categories of growth defect that promote cancer, outlined here schematically; see text for details
One early focus of attention has been the dialog between Aurora A and loss of p53 in tumorigenesis. The aneuploidy associated with overexpression of Aurora A (or of Aurora-A-stabilizing proteins such as NEDD9/HEF1) results in triggering of mitotic checkpoints and rapid clearance of a large part of the cell population due to subsequent apoptosis [11, 21, 95]. Loss of p53 eliminates these checkpoints, alleviating the clearance associated with Aurora A overexpression [11]. In a model for mouse mammary tumors, Aurora A was unable to induce tumorigenesis except in a p53-deficient background, because of the induction of cellular senescence and p16 expression [168]. Suggestively, genomic instability and supernumerary centrosomes also characterize the phenotype of cells or primary tumors that lack p53 [169, 170]. Subsequent work demonstrated that Aurora A and p53 are involved in a tightly regulated negative feedback loop that is lost in cancer. Aurora A phosphorylation of S315 on p53 increases MDM2-dependent degradation of p53 [171]. Reciprocally, interaction of the p53 with the Aurora A A-box inhibits Aurora A kinase activity and potential for transformation [172]. This homeostatic relationship is deregulated in cancer. p53+/− mice develop tumors with elevated Aurora A levels, but tumors derived from p53−/− mice often have Aurora A elevation in normal tissue, but gene deletion in lymphomas [173]. A genome-wide expression array screen of human breast tumors similarly indicated correlation of loss of p53 expression and Aurora A mRNA levels [173]. As one explanation for this functional interaction, loss of p53 in tumors is also associated with loss of heterozygosity (LOH) or mutation of the FBXW7/Cdc4 gene, encoding a ubiquitin ligase that degrades Aurora A [173, 174]. Latent infection with Kaposi’s sarcoma herpesvirus (KSHV) is predisposing for cancer. KSHV encodes a latency-associated nuclear antigen (LANA) that induces Aurora A transcription; the elevated Aurora A causes phosphorylation of p53 on S315 and a second residue, S215, creating a binding site for a cellular ubitiquitin ligase complex that degrades p53 [175, 176]. Elevated Aurora A in tumors also targets the inactivation of the p53-related protein p73, contributing to the loss of mitotic checkpoint by disrupting the CDC20–MAD2 complex and promoting apoptosis resistance [177].
Overexpressed Aurora A also influences the activity of numerous transcription factors relevant for cancer progression, binding and promoting degradation of the AP-2α tumor suppressor [178], and phosphorylating S32 and S36 of IκBα, leading to degradation of this protein [179, 180]. Loss of IκBα activates its partner, the transcription factor NF-κB, which has wide action in promoting tumor growth and survival. Aurora A phosphorylation also activates the prosurvival kinase AKT [181] and potentiates the signaling of oncogenic Ras by activating its interphase effector RALA [182, 183]. Figure 6 is a current summary of proteins known to interact with Aurora A either directly (physically) or functionally; there are almost certainly more that have not yet been determined. In addition, two recent extensive peptide scanning and phosphoproteomics studies have both refined our understanding of previously defined Aurora A targets and identified still more candidates for Aurora A regulation [89, 94]. For each, increased Aurora A action in cancer has the potential to alter functionality.
Fig. 6.
Extended Aurora A interaction network. Schematic indicates proteins that known or highly likely to interact physically and/or functionally (indirect interaction) with Aurora A/AURKA. All proteins are indicated by official gene symbol. Interaction data were collected using online databases String [251] (using medium confidence score 0.4 and excluding text-mining-only results) and Ingenuity [http://www.ingenuity.com/] (extracting only experimental and high confidence predicted interactions). Data were imported, merged, and visualized in Cytoscape [252]. Blue rim around protein indicates direct protein–protein interaction; lack of rim indicates indirect interaction. Green circles represent highly validated interactions generally reported in publications. Blue circles represent high confidence hits from larger screens, but are less well studied. Green lines indicate interactions with Aurora A/AURKA; blue lines indicate interactions among proteins described as interacting with Aurora A/AURKA
Interactions between Aurora A and Myc family members (c-Myc and N-Myc) frequently contribute to carcinogenesis. c-Myc overexpression has been reported to be accompanied by overexpression and/or amplification of Aurora A in multiple types of cancer [184–186]. High expression of Aurora A coupled with amplified MYCN (N-Myc) was observed in human neuroblastoma cell lines and neuroendocrine prostate cancer [187, 188]. In human ovarian and breast epithelial cell lines, Aurora A overexpression stimulated human telomerase reverse transcriptase (hTERT) and telomerase activity in part by inducing c-Myc, which binds the telomerase promoter and induces c-Myc transcription [189].
Several studies have demonstrated that overexpression of Aurora A results in elevated levels of C-myc, whereas inhibition of c-Myc with RNA interference attenuates Aurora-A-induced oncogenic effects [189, 190]. In a study of gastric cancer, Aurora A induction of c-Myc was shown to depend on Aurora A induction of the GSK-3β and β-catenin/TCF transcriptional complex, which positively regulates transcription of c-MYC and additional cancer-relevant genes, including cyclin D1, VEGF, FGF18, and others [191]. Otto and colleagues found that Aurora A overexpression results in enhanced stability of N-Myc protein, thus contributing to proliferation of neuroblastoma cells regardless of the presence of growth factors. This action is due to direct interaction between Aurora A, N-Myc, and the SCFFbxw7 ubiquitin ligase complex and does not depend on the kinase activity of Aurora A [187].
Reciprocally, c-Myc can upregulate the expression of the Aurora A kinase, in part through inducing its transcription [192]. However, although c-Myc binds to some of the E-boxes present in the promoter region of the mouse AurkA gene, it binds none of these in human AURKA gene, implying that c-Myc can regulate Aurora A expression in both a direct and indirect manner [192]. A very recent study on Ba/F3 cells expressing JAK2 V617F mutant also demonstrated c-Myc-dependent induction of Aurora A expression [193]. These findings suggest a positive feedback loop in which Aurora A and c-Myc induce each other. Interestingly, a recent study employing a mouse model with induced expression of an oncogenic gain-of-function mutation in p53, p53R172H, showed that resulting squamous cell tumors were characterized by elevated genomic instability, preferential Myc amplification, and elevated Aurora A expression [194]. These intriguing results suggest close coordination of p53, Myc, and Aurora A activities in the development of solid tumors.
A discussion of all known or likely cancer-relevant Aurora A targets would exceed the length limits of this review. Many of the targets of Aurora A listed in Table 1 have both cell cycle and cancer-relevant roles. Clearly, the potential for Aurora-A-dependent altered function of some, such as RalA, Brca1, and Src, is immediately relevant to cancer, and some studies have begun to address the consequences of these interactions for tumor prognosis [42, 54, 183, 195–197]. For others, much work remains to be done.
Predictive value of Aurora A amplification and overexpression
For effective clinical management of cancer, one goal has been to better understand how the presence of Aurora A amplification and/or overexpression impacts the likely course of disease: in particular, whether it predicts greater or lesser aggression or response to specific classes of drugs. At present, the scientific literature tends to associate elevated Aurora A expression with a poorer outcome in tumors, although some studies contradict the trend. Particularly in cases in which Aurora A levels are increased based on genomic amplification, it is difficult to conclusively assign outcomes to contributions of Aurora A, in part because the Aurora A amplicon on chromosome 20 frequently causes the enhanced expression of multiple additional genes. We here summarize a number of studies regarding Aurora A and prognosis in several common cancers.
Increased copy number of the Aurora A gene, including aneuploidy involving the region of chromosome 20 encompassing Aurora A, is common in ovarian cancer [198–200]. However, among six published clinical studies of Aurora A and disease prognosis, various conclusions were reached [186, 201–205]. For instance, Lassmann et al. [186] showed that high Aurora A expression is associated with improved overall survival in patients with stage III ovarian cancer receiving taxol/carboplatin therapy, but significantly worse survival in patients who received the carboplatin-based treatment without taxol. A second group found increased overall and progression-free survival (PFS) in patients with Aurora A positive tumors in comparison with the patients whose tumors did not express Aurora A [205]. In contrast, Kulkarni et al. found that expression of Aurora A strongly predicted shorter disease-free survival in early-stage ovarian carcinomas, but not in advanced-stage tumors [201]; three other groups also correlated high expression with aggressive disease and poor outcome [202–204]. Interestingly, some recent work has identified a negative correlation between expression of BRCA2 and Aurora A that more strongly predicts prognosis in patients with ovarian cancer. In these studies, the nuclear accumulation of BRCA2 was significantly associated with good overall survival and disease-free survival in patients with high-grade ovarian carcinoma, whereas strong expression of Aurora A was significantly associated with poor outcome [206, 207].
Increased copy number of Aurora A is often associated with progression from a colonic polyp to an invasive malignancy in colon cancer [208] and is one of the most common copy number alterations in cancer. A number of genes involved in cell cycle regulation are part of an Aurora A amplicon, including TPX2 [209]: overexpression of the TPX2 and Aurora A proteins in tumors has been correlated by immunohistochemical analysis. In a multivariate analysis, Aurora A protein overexpression was associated with chromosomal instability (identified as loss of heterozygosity in 2p, 5q, 17q, and 18q) but did not correlate with clinical outcomes [210]. In a second study, higher Aurora A expression was associated with recurrence in stage II and III in a large, homogenous cohort of colon cancer patients [211]. In a third study, Aurora A was most commonly detected in well or moderately differentiated, versus poorly differentiated tumors [212]. In another study, elevated Aurora A copy number correlated with increased PFS and overall survival, but only in the context of a wild-type Ras allele [213]. An Aurora A F31I polymorphism is associated with increased aneuploidy in colon tumors and has been described as a low penetrance cancer susceptibility allele affecting multiple cancer types [214, 215]. A rare S155R somatic mutation associated with colon cancer has been suggested to promote aneuploidy by reducing the interactions between Aurora A and TPX2 [152].
High expression of Aurora A overexpression is very strongly linked to decreased survival in primary breast tumors and is associated with high expression of HER2 and progesterone receptor [216]. Correlated expression of Aurora A, progesterone receptor (PR), and estrogen receptor (ER) was found in an independent study [203]. In investigations of the codon 31 polymorphism, the F/I and I/I genotypes were associated with increased risk of breast cancer, particularly in overweight women; in analysis of a second polymorphism, V57I, the 57V allele was associated with an increased risk of invasive breast cancer [217–219]. Overexpression of Aurora A localized to the perimembrane compartment was associated with decreased 5-year survival in non-small cell lung cancer (NSCLC) and was also associated with poor survival prognosis [220]. Aurora A mRNA and protein are overexpressed in poorly or moderately differentiated lung cancer [221]. In lung cancer, a large case control study of Aurora A codon 31 polymorphisms found that the I/I genotype reduced odds for lung cancer, but only in men [222]. A novel protein, SLAN (suppressed in lung cancer), was recently identified as binding and inactivating Aurora A [223]. Although SLAN is frequently downregulated in lung cancer tissues overexpressing Aurora A, the prognostic value of SLAN expression is not yet known [223]. Patients with head and neck carcinoma and elevated Aurora A mRNA had a shorter disease-free and overall survival [224].
Aurora A inhibitors in the clinic
On the basis of the information summarized above, Aurora A has been of high interest as a drug target, with drug development and assessment effort much reviewed (e.g., [163, 225]). Numerous candidate drugs have undergone preclinical testing in vitro and in animal models. As of 2012, compounds that had made it through preclinical testing into phase I or II trials include MK-0457 and MK-5108 (Merck), AZD1152 (Astra Zeneca), AT9283 (Astex Therapeutics), PF-03814735 (Pfizer), AS703569 (EMD Serono), PHA-739358 (Nerviano), and MLN8054 and MLN8237 (Millennium). Many of these compounds have activity against multiple structurally related kinases including ABL, SRC, JAK2, VEGFR2, FLT3, and FGFR1, which has influenced the clinical development of these drugs towards certain tumor types with relevance to their off-target activity. For example, one such application has been in chronic myeloid leukemia (CML) where inhibitors of Aurora kinase have been active against ABL kinase with a T315I resistance mutation [226]. Those compounds still in clinical assessment as of June 2012 are summarized in Table 2.
Table 2.
| Aurora kinase inhibitor | Activity | Aurora A IC50 (in vitro) (nM) | Aurora B IC50 (in vitro) | Aurora C IC50 (in vitro) | Clinical trial status | Clinical trial registry number |
|---|---|---|---|---|---|---|
| AMG900 | Pan-Aurora kinase | 5 | 4 nM | 1 nM | Active | NCT01380756 |
| NCT00858377 | ||||||
| AT9283 | Pan-Aurora kinase | 5 | 3 nM | – | Active | NCT01431664 |
| NCT01145989 | ||||||
| NCT00985868 | ||||||
| MLN8237 | Aurora A | 61 | 200-fold higher | – | Active | NCT01045421 |
| NCT01091428 | ||||||
| NCT00962091 | ||||||
| NCT00697346 | ||||||
| NCT01397825 | ||||||
| NCT01034553 | ||||||
| NCT01316692 | ||||||
| NCT01471964 | ||||||
| NCT01094288 | ||||||
| NCT01540682 | ||||||
| NCT01601535 | ||||||
| NCT00739427 | ||||||
| NCT01512758 | ||||||
| NCT01567709 | ||||||
| NCT01154816 | ||||||
| NCT01482962 | ||||||
| NCT01466881 | ||||||
| AZD1152 | Aurora A and Aurora B | 1,368 | 0.37 nM | – | Active | NCT00952588 |
| NCT01354392 | ||||||
| ENMD2076 | Aurora A and Aurora B | 13 | 350 nM | – | Active | NCT00658671 |
| NCT01104675 |
Overall, Aurora kinase inhibitors have been assessed in a broad range of hematological and solid tumors, with efficacy primarily in disease stabilization in a minority of patients. For example, a selective orally administered inhibitor of Aurora A kinase, MLN8054, has completed three phase I studies which identified useful clinical biomarkers coupled with promising early indications of antitumor activity measured by durable partial or minor responses [227–229]. However, a striking somnolence associated with use of MLN8054 was unexpected and remains unexplained. One possibility is that benzodiazepine-like central nervous system effects may be involved, as MLN8054 is structurally related to this compound and is active against the GABAA α1 receptor [228]. It is also interesting to speculate that the recently detected interphase activities of Aurora A in regulation of Ca2+ signaling, cilia, and in the function of post-mitotic neurons may be relevant. At present, trials involving MLN8054 have been stopped (Clinicaltrials.gov).
A number of phase II trials are ongoing using a second-generation related compound, MLN8237/alisertib (ClinicalTrials.gov identifiers NCT00830518, NCT00500903, NCT01091428, and others). Toxicities consisted mainly of reversible neutropenia along with mucositis and somnolence, with neutropenia as the dose-limiting toxicity. The predominant toxicities of MLN8237 reflect the mechanism of action in highly proliferating tissues (bone marrow, GI epithelium, and hair follicles). Several applications of MLN8237 are showing considerable promise. Preclinical studies combining MLN8237 with other drugs showed efficacy of this agent in combination with cytarabine (ara-C) in cell lines, primary cells, and mouse models for acute myeloid leukemia (AML), suggesting that this may be a productive clinical application of this compound [230]. A recent open-label, multicenter phase II trial assessed MLN8237 in adults with advanced AML or intermediate-2/high-risk myelodysplastic syndrome (MDS) in 57 patients. Fourteen patients withdrew from the study because of adverse events, which included neutropenia, thrombocytopenia, anemia, fatigue, and somnolence. Six (13 %) patients achieved complete or partial responses; the predominant result was stable disease, which was observed in 17 (49 %) AML patients and in 2 (20 %) MDS patients (NCT00830518 [231]). Another phase II trial has been performed in patients with aggressive lymphomas [232]. Here, the overall response rate was 32 % (95 % CI 0.181–0.481) and response by histology varied from 20 to 23 % in aggressive B cell lymphomas to 57 % in T cell non-Hodgkin lymphomas. Interestingly, the activity of this Aurora A inhibitor did not correlate with Aurora A protein expression level on tissue sections, in contrast to some other target-directed therapies such as EGFR [233]. A phase III trial was recently initiated (NCT01482962) in patients with relapsed or treatment-refractory peripheral T cell lymphoma (PTCL, a rare, aggressive form of non-Hodgkin lymphoma) [234]. A discussion of the use of MLN8237 in combination with paclitaxel in a phase I trial for recurrent epithelial ovarian, fallopian, or primary peritoneal cancer or breast cancer indicated that the drug combination is well tolerated and that antitumor activity is observed in ovarian and breast cancer: dose escalation is in progress [230].
In the case of the solid tumors, the reasons for the overall modest clinical effect are also unclear and contrast sharply with the results obtained in cell lines and xenografts discussed above. One possibility for such discrepancy might be the existence of redundant signaling pathways in tumor cells allowing for bypass signaling (i.e., drug-selected activation of Aurora A partners or effectors) to drive cellular proliferation despite the blockade of Aurora A kinase. In a large-scale screen in breast cancer cell lines, high Aurora A copy number conferred resistance to GSK1070916, an inhibitor of Aurora B/C [235], thus suggesting a bypass mechanism between functionally related Aurora A paralogs. Practical considerations in regard to allowable dose and schedule of drug administration may also play a role in limiting efficacy; significant neutropenia is the main dose-limiting toxicity of Aurora inhibition, thus preventing prolonged Aurora A signaling inhibition. Dose-escalation studies [236] demonstrated a trend toward higher antitumor activity when neutropenia was prevented by administration of growth factors.
In spite of all the biological properties discussed above, it remains unresolved if Aurora kinase A is a true oncogenic driver in human cancers. As noted above, the Aurora A locus at 20q13 is frequently amplified as part of large chromosomal amplicons, or even the entire arm or chromosome copy number is increased. The relationship of the Aurora copy number and its kinase activity to the sensitivity to its specific inhibitors is presently unknown. The Aurora gene copy number may have differential interaction with the treatment drugs. Suggestively, in some studies, a higher level of AURKA expression was noted in low-grade, less aggressive tumors [212, 226]. Alternatively, increased AURKA gene copy number may be an oncogenic driver, but simultaneously render cells more susceptible to chemotherapy owing to enhancement of a “mutator phenotype” [237] associated with abnormal cell divisions and genomic instability in cells with hyperactive Aurora A kinase.
Abundant preclinical data provide support for combining Aurora A inhibitors with a wide variety of existing agents targeting the Aurora A partners and effectors discussed above, or with less specific antimitotic agents such as microtubular poisons or ionizing radiation [238]. For the last possibility, one rationale is based on the known activity of Aurora A in enhancing tumor radioresistance [239] through p53 phosphorylation and increased Akt activity [171, 181]. In practice, inhibition of Aurora A sensitizes cancer cells to radiation therapy (RT), even in tumors such as head and neck carcinomas that contain p53-inactivating mutations [163]. Moving further afield, we have previously identified synergy in combining inhibitors targeting Aurora A and members of core proliferation and survival pathways such as EGFR [240] and SRC [196]; EGFR and SRC inhibition each independently enhance RT. Future strategies may combine Aurora A inhibition with EGFR inhibitors such as cetuximab or SRC inhibitors such as dasatinib to improve clinical efficacy. If there is one consistent lesson emerging from the field of systems biology, it is that inhibition of a single target, no matter how promising, is likely to be insufficient for cancer therapy except in the most unusual cases.
Acknowledgments
The authors were supported by R01s CA63366 and CA113342 (to EAG); by NIH R21 CA-164205 and 1K22CA-160725 (to IA); NIH R01 GM84453 (to RLD); and NIH core grant CA-06927, the Commonwealth of Pennsylvania, and the Pew Charitable Fund (to Fox Chase Cancer Center). The authors apologize for not being able to include discussion of additional relevant Aurora-A-related studies, because of the length limitations of this article.
References
- 1.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-Y. [DOI] [PubMed] [Google Scholar]
- 3.Andresson T, Ruderman JV. The kinase Eg2 is a component of the Xenopus oocyte progesterone-activated signaling pathway. EMBO J. 1998;17:5627–5637. doi: 10.1093/emboj/17.19.5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roghi C, Giet R, Uzbekov R, Morin N, Chartrain I, Le Guellec R, Couturier A, Doree M, Philippe M, Prigent C. The Xenopus protein kinase pEg2 associates with the centrosome in a cell cycle-dependent manner, binds to the spindle microtubules and is involved in bipolar mitotic spindle assembly. J Cell Sci. 1998;111(Pt 5):557–572. doi: 10.1242/jcs.111.5.557. [DOI] [PubMed] [Google Scholar]
- 5.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 6.Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Gene Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Gene Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Hirota T, Marumoto T, Shimizu M, Kunitoku N, Sasayama T, Arima Y, Feng L, Suzuki M, Takeya M, Saya H. Cre-loxP-controlled periodic Aurora-A overexpression induces mitotic abnormalities and hyperplasia in mammary glands of mouse models. Oncogene. 2004;23:8720–8730. doi: 10.1038/sj.onc.1208153. [DOI] [PubMed] [Google Scholar]
- 14.Tatsuka M, Katayama H, Ota T, Tanaka T, Odashima S, Suzuki F, Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–4816. [PubMed] [Google Scholar]
- 15.Waal MS, Hengeveld RC, Horst A, Lens SM. Cell division control by the Chromosomal Passenger Complex. Exp Cell Res. 2012;318(12):1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, Wu X. Centrosome maturation. Curr Top Dev Biol. 2000;49:449–470. doi: 10.1016/S0070-2153(99)49021-0. [DOI] [PubMed] [Google Scholar]
- 18.Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr Opin Cell Biol. 2012;24:14–23. doi: 10.1016/j.ceb.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc Natl Acad Sci U S A. 2010;107:21022–21027. doi: 10.1073/pnas.1014664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutterer A, Berdnik D, Wirtz-Peitz F, Zigman M, Schleiffer A, Knoblich JA. Mitotic activation of the kinase Aurora-A requires its binding partner Bora. Dev Cell. 2006;11:147–157. doi: 10.1016/j.devcel.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7:937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans . J Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- 24.Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- 25.Conte N, Delaval B, Ginestier C, Ferrand A, Isnardon D, Larroque C, Prigent C, Seraphin B, Jacquemier J, Birnbaum D. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene. 2003;22:8102–8116. doi: 10.1038/sj.onc.1206972. [DOI] [PubMed] [Google Scholar]
- 26.Mori D, Yano Y, Toyo-oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, Wynshaw-Boris A, Hirotsune S. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol Cell Biol. 2007;27:352–367. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berdnik D, Knoblich JA. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol. 2002;12:640–647. doi: 10.1016/S0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- 28.De Souza CP, Ellem KA, Gabrielli BG. Centrosomal and cytoplasmic Cdc2/cyclin B1 activation precedes nuclear mitotic events. Exp Cell Res. 2000;257:11–21. doi: 10.1006/excr.2000.4872. [DOI] [PubMed] [Google Scholar]
- 29.Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- 30.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 32.Chan EH, Santamaria A, Sillje HH, Nigg EA. Plk1 regulates mitotic Aurora A function through betaTrCP-dependent degradation of hBora. Chromosoma. 2008;117:457–469. doi: 10.1007/s00412-008-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouche JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, Ducommun B. Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci. 2004;117:2523–2531. doi: 10.1242/jcs.01108. [DOI] [PubMed] [Google Scholar]
- 34.van Vugt MA, Bras A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell. 2004;15:799–811. doi: 10.1016/j.molcel.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 35.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 36.Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6:1253–1259. doi: 10.1016/S1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 37.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/S0092-8674(02)00733-X. [DOI] [PubMed] [Google Scholar]
- 38.Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/S0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 41.Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, Deng C, Sen S, Lee SW, Ouchi T. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- 42.Brodie KM, Henderson BR. Characterization of BRCA1 protein targeting, dynamics, and function at the centrosome: a role for the nuclear export signal, CRM1, and Aurora A kinase. J Biol Chem. 2012;287:7701–7716. doi: 10.1074/jbc.M111.327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Q, Ruderman JV. Aurora A, mitotic entry, and spindle bipolarity. Proc Natl Acad Sci U S A. 2006;103:5811–5816. doi: 10.1073/pnas.0601425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 46.Katayama H, Sasai K, Kloc M, Brinkley BR, Sen S. Aurora kinase-A regulates kinetochore/chromatin associated microtubule assembly in human cells. Cell Cycle. 2008;7:2691–2704. doi: 10.4161/cc.7.17.6460. [DOI] [PubMed] [Google Scholar]
- 47.Bird AW, Hyman AA. Building a spindle of the correct length in human cells requires the interaction between TPX2 and Aurora A. J Cell Biol. 2008;182:289–300. doi: 10.1083/jcb.200802005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 49.Kinoshita K, Noetzel TL, Pelletier L, Mechtler K, Drechsel DN, Schwager A, Lee M, Raff JW, Hyman AA. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeRoy PJ, Hunter JJ, Hoar KM, Burke KE, Shinde V, Ruan J, Bowman D, Galvin K, Ecsedy JA. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 2007;67:5362–5370. doi: 10.1158/0008-5472.CAN-07-0122. [DOI] [PubMed] [Google Scholar]
- 51.Ozlu N, Srayko M, Kinoshita K, Habermann B, O’Toole ET, Muller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Barros TP, Kinoshita K, Hyman AA, Raff JW. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol. 2005;170:1039–1046. doi: 10.1083/jcb.200504097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sankaran S, Crone DE, Palazzo RE, Parvin JD. Aurora-A kinase regulates breast cancer associated gene 1 inhibition of centrosome-dependent microtubule nucleation. Cancer Res. 2007;67:11186–11194. doi: 10.1158/0008-5472.CAN-07-2578. [DOI] [PubMed] [Google Scholar]
- 55.Chakrabarti R, Jones JL, Oelschlager DK, Tapia T, Tousson A, Grizzle WE. Phosphorylated LIM kinases colocalize with gamma-tubulin in centrosomes during early stages of mitosis. Cell Cycle. 2007;6:2944–2952. doi: 10.4161/cc.6.23.4957. [DOI] [PubMed] [Google Scholar]
- 56.Ritchey L, Ottman R, Roumanos M, Chakrabarti R. A functional cooperativity between Aurora A kinase and LIM kinase1: implication in the mitotic process. Cell Cycle. 2012;11:296–309. doi: 10.4161/cc.11.2.18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giet R, Uzbekov R, Cubizolles F, Le Guellec K, Prigent C. The Xenopus laevis aurora-related protein kinase pEg2 associates with and phosphorylates the kinesin-related protein XlEg5. J Biol Chem. 1999;274:15005–15013. doi: 10.1074/jbc.274.21.15005. [DOI] [PubMed] [Google Scholar]
- 58.Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- 60.Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 61.Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol Biol Cell. 2008;19:2083–2091. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 63.Gruss OJ, Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5:853–864. doi: 10.1016/S1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- 66.Hegarat N, Smith E, Nayak G, Takeda S, Eyers PA, Hochegger H. Aurora A and Aurora B jointly coordinate chromosome segregation and anaphase microtubule dynamics. J Cell Biol. 2011;195:1103–1113. doi: 10.1083/jcb.201105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rong R, Jiang LY, Sheikh MS, Huang Y. Mitotic kinase Aurora-A phosphorylates RASSF1A and modulates RASSF1A-mediated microtubule interaction and M-phase cell cycle regulation. Oncogene. 2007;26:7700–7708. doi: 10.1038/sj.onc.1210575. [DOI] [PubMed] [Google Scholar]
- 68.Song SJ, Song MS, Kim SJ, Kim SY, Kwon SH, Kim JG, Calvisi DF, Kang D, Lim DS. Aurora A regulates prometaphase progression by inhibiting the ability of RASSF1A to suppress APC-Cdc20 activity. Cancer Res. 2009;69:2314–2323. doi: 10.1158/0008-5472.CAN-08-3984. [DOI] [PubMed] [Google Scholar]
- 69.Floyd S, Pines J, Lindon C. APC/C Cdh1 targets aurora kinase to control reorganization of the mitotic spindle at anaphase. Curr Biol. 2008;18:1649–1658. doi: 10.1016/j.cub.2008.09.058. [DOI] [PubMed] [Google Scholar]
- 70.Perez de Castro I, Aguirre-Portoles C, Martin B, Fernandez-Miranda G, Klotzbucher A, Kubbutat MHG, Megias D, Arlot-Bonnemains Y, Malumbres M (2011) A SUMOylation motif in Aurora-A: implications fo spindle dynamics and oncogenesis. Front Oncol 1:50. doi:10.3389/fonc.2011.00050 [DOI] [PMC free article] [PubMed]
- 71.Ma N, Matsunaga S, Morimoto A, Sakashita G, Urano T, Uchiyama S, Fukui K. The nuclear scaffold protein SAF-A is required for kinetochore-microtubule attachment and contributes to the targeting of Aurora-A to mitotic spindles. J Cell Sci. 2011;124:394–404. doi: 10.1242/jcs.063347. [DOI] [PubMed] [Google Scholar]
- 72.Birkenfeld J, Nalbant P, Bohl BP, Pertz O, Hahn KM, Bokoch GM. GEF-H1 modulates localized RhoA activation during cytokinesis under the control of mitotic kinases. Dev Cell. 2007;12:699–712. doi: 10.1016/j.devcel.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Littlepage LE, Wu H, Andresson T, Deanehan JK, Amundadottir LT, Ruderman JV. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci U S A. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol Cell. 2005;20:237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 75.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, a novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem. 2004;279:9008–9015. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 77.Giubettini M, Asteriti IA, Scrofani J, De Luca M, Lindon C, Lavia P, Guarguaglini G. Control of Aurora-A stability through interaction with TPX2. J Cell Sci. 2011;124:113–122. doi: 10.1242/jcs.075457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–697. doi: 10.1016/S0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 79.Kufer TA, Sillje HH, Korner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsai MY, Wiese C, Cao K, Martin O, Donovan P, Ruderman J, Prigent C, Zheng Y. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 81.Dodson CA, Bayliss R. Activation of Aurora-A kinase by protein partner binding and phosphorylation are independent and synergistic. J Biol Chem. 2012;287:1150–1157. doi: 10.1074/jbc.M111.312090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu X, Wang X, Xiao Z, Li Y, Wang Y. Two TPX2-dependent switches control the activity of Aurora A. PLoS One. 2011;6:e16757. doi: 10.1371/journal.pone.0016757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scutt PJ, Chu ML, Sloane DA, Cherry M, Bignell CR, Williams DH, Eyers PA. Discovery and exploitation of inhibitor-resistant aurora and polo kinase mutants for the analysis of mitotic networks. J Biol Chem. 2009;284:15880–15893. doi: 10.1074/jbc.M109.005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ, Ray ET, Sells TB, Stringer B, Stroud SG, Vos TJ, Weatherhead GS, Wysong DR, Zhang M, Bolen JB, Claiborne CF. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci U S A. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eckerdt F, Pascreau G, Phistry M, Lewellyn AL, DePaoli-Roach AA, Maller JL. Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle. 2009;8:2413–2419. doi: 10.4161/cc.8.15.9086. [DOI] [PubMed] [Google Scholar]
- 86.Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 2003;114:585–598. doi: 10.1016/S0092-8674(03)00642-1. [DOI] [PubMed] [Google Scholar]
- 87.Sabino D, Brown NH, Basto R. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J Cell Sci. 2011;124:1156–1166. doi: 10.1242/jcs.076711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, Ivins F, Sessa F, Hudecz O, Nigg EA, Fry AM, Musacchio A, Stukenberg PT, Mechtler K, Peters JM, Smerdon SJ, Yaffe MB. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal. 2011;4:ra42. doi: 10.1126/scisignal.2001796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tikhmyanova N, Little JL, Golemis EA. CAS proteins in normal and pathological cell growth control. Cell Mol Life Sci. 2010;67:1025–1048. doi: 10.1007/s00018-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Astier A, Manie SN, Law SF, Canty T, Haghayghi N, Druker BJ, Salgia R, Golemis EA, Freedman AS. Association of the Cas-like molecule HEF1 with CrkL following integrin and antigen receptor signaling in human B-cells: potential relevance to neoplastic lymphohematopoietic cells. Leuk Lymphoma. 1997;28:65–72. doi: 10.3109/10428199709058332. [DOI] [PubMed] [Google Scholar]
- 92.Fashena SJ, Einarson MB, O’Neill GM, Patriotis C, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115:99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- 93.Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, Wang W, Grammer JR, Gladson CL. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–1732. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 94.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4:rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pugacheva EN, Golemis EA. HEF1-aurora A interactions: points of dialog between the cell cycle and cell attachment signaling networks. Cell Cycle. 2006;5:384–391. doi: 10.4161/cc.5.4.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dadke D, Jarnik M, Pugacheva EN, Singh MK, Golemis EA. Deregulation of HEF1 impairs M-phase progression by disrupting the RhoA activation cycle. Mol Biol Cell. 2006;17:1204–1217. doi: 10.1091/mbc.E05-03-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sankaran S, Starita LM, Groen AC, Ko MJ, Parvin JD. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol Cell Biol. 2005;25:8656–8668. doi: 10.1128/MCB.25.19.8656-8668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seki A, Coppinger JA, Du H, Jang CY, Yates JR, 3rd, Fang G. Plk1- and beta-TrCP-dependent degradation of Bora controls mitotic progression. J Cell Biol. 2008;181:65–78. doi: 10.1083/jcb.200712027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molli PR, Li DQ, Bagheri-Yarmand R, Pakala SB, Katayama H, Sen S, Iyer J, Chernoff J, Tsai MY, Nair SS, Kumar R. Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J Cell Biol. 2010;190:101–114. doi: 10.1083/jcb.200908050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Francisco L, Wang W, Chan CS. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol. 1994;14:4731–4740. doi: 10.1128/mcb.14.7.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Satinover DL, Leach CA, Stukenberg PT, Brautigan DL. Activation of Aurora-A kinase by protein phosphatase inhibitor-2, a bifunctional signaling protein. Proc Natl Acad Sci U S A. 2004;101:8625–8630. doi: 10.1073/pnas.0402966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276:46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- 103.Zeng K, Bastos RN, Barr FA, Gruneberg U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol. 2010;191:1315–1332. doi: 10.1083/jcb.201008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Reboutier D, Troadec MB, Cremet JY, Fukasawa K, Prigent C. Nucleophosmin/B23 activates Aurora A at the centrosome through phosphorylation of serine 89. J Cell Biol. 2012;197:19–26. doi: 10.1083/jcb.201107134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Plotnikova OV, Pugacheva EN, Dunbrack RL, Golemis EA. Rapid calcium-dependent activation of Aurora-A kinase. Nat Commun. 2010;1:64. doi: 10.1038/ncomms1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sarkissian M, Mendez R, Richter JD. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Gene Dev. 2004;18:48–61. doi: 10.1101/gad.1136004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang YH, Wu CC, Chou CK, Huang CY. A translational regulator, PUM2, promotes both protein stability and kinase activity of Aurora-A. PLoS One. 2011;6:e19718. doi: 10.1371/journal.pone.0019718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu L, Guo C, Dammann R, Tommasi S, Pfeifer GP. RASSF1A interacts with and activates the mitotic kinase Aurora-A. Oncogene. 2008;27:6175–6186. doi: 10.1038/onc.2008.220. [DOI] [PubMed] [Google Scholar]
- 109.Persico A, Cervigni RI, Barretta ML, Corda D, Colanzi A. Golgi partitioning controls mitotic entry through Aurora-A kinase. Mol Biol Cell. 2010;21:3708–3721. doi: 10.1091/mbc.E10-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukuda T, Mishina Y, Walker MP, DiAugustine RP. Conditional transgenic system for mouse aurora a kinase: degradation by the ubiquitin proteasome pathway controls the level of the transgenic protein. Mol Cell Biol. 2005;25:5270–5281. doi: 10.1128/MCB.25.12.5270-5281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Honda K, Mihara H, Kato Y, Yamaguchi A, Tanaka H, Yasuda H, Furukawa K, Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- 112.Walter AO, Seghezzi W, Korver W, Sheung J, Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene. 2000;19:4906–4916. doi: 10.1038/sj.onc.1203847. [DOI] [PubMed] [Google Scholar]
- 113.Barford D. Structural insights into anaphase-promoting complex function and mechanism. Philos Trans R Soc Lond B Biol Sci. 2011;366:3605–3624. doi: 10.1098/rstb.2011.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Castro A, Arlot-Bonnemains Y, Vigneron S, Labbe JC, Prigent C, Lorca T. APC/Fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, Ward IM, Saya H, Fang G, van Deursen J, Chen J. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 116.Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Gene Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Horn V, Thelu J, Garcia A, Albiges-Rizo C, Block MR, Viallet J. Functional interaction of Aurora-A and PP2A during mitosis. Mol Biol Cell. 2007;18:1233–1241. doi: 10.1091/mbc.E06-12-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Gene Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 119.Shi Y, Solomon LR, Pereda-Lopez A, Giranda VL, Luo Y, Johnson EF, Shoemaker AR, Leverson J, Liu X. Ubiquitin-specific cysteine protease 2a (USP2a) regulates the stability of Aurora-A. J Biol Chem. 2011;286:38960–38968. doi: 10.1074/jbc.M111.231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kiat LS, Hui KM, Gopalan G. Aurora-A kinase interacting protein (AIP), a novel negative regulator of human Aurora-A kinase. J Biol Chem. 2002;277:45558–45565. doi: 10.1074/jbc.M206820200. [DOI] [PubMed] [Google Scholar]
- 121.Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- 122.Lim SK, Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem J. 2007;403:119–127. doi: 10.1042/BJ20061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fumoto K, Lee PC, Saya H, Kikuchi A. AIP regulates stability of Aurora-A at early mitotic phase coordinately with GSK-3beta. Oncogene. 2008;27:4478–4487. doi: 10.1038/onc.2008.92. [DOI] [PubMed] [Google Scholar]
- 124.Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Liu Y, Huang Y, Kang HC, Delrosario R, Jen KY, Perez-Losada J, Wei G, Balmain A, Mao JH. Pten regulates Aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10(6):834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lorenzo C, Liao Q, Hardwicke MA, Ducommun B. Pharmacological inhibition of aurora-A but not aurora-B impairs interphase microtubule dynamics. Cell Cycle. 2009;8:1733–1737. doi: 10.4161/cc.8.11.8617. [DOI] [PubMed] [Google Scholar]
- 126.Yamada M, Hirotsune S, Wynshaw-Boris A. The essential role of LIS1, NDEL1 and Aurora-A in polarity formation and microtubule organization during neurogenesis. Cell Adh Migr. 2010;4:180–184. doi: 10.4161/cam.4.2.10715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rannou Y, Troadec MB, Petretti C, Hans F, Dutertre S, Dimitrov S, Prigent C. Localization of aurora A and aurora B kinases during interphase: role of the N-terminal domain. Cell Cycle. 2008;7:3012–3020. doi: 10.4161/cc.7.19.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/S1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 129.Mori D, Yamada M, Mimori-Kiyosue Y, Shirai Y, Suzuki A, Ohno S, Saya H, Wynshaw-Boris A, Hirotsune S. An essential role of the aPKC-Aurora A-NDEL1 pathway in neurite elongation by modulation of microtubule dynamics. Nat Cell Biol. 2009;11:1057–1068. doi: 10.1038/ncb1919. [DOI] [PubMed] [Google Scholar]
- 130.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Plotnikova OV, Pugacheva EN, Golemis EA. Aurora A kinase activity influences calcium signaling in kidney cells. J Cell Biol. 2011;193:1021–1032. doi: 10.1083/jcb.201012061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Toya M, Terasawa M, Nagata K, Iida Y, Sugimoto A. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat Cell Biol. 2011;13:708–714. doi: 10.1038/ncb2242. [DOI] [PubMed] [Google Scholar]
- 133.Ogawa H, Ohta N, Moon W, Matsuzaki F. Protein phosphatase 2A negatively regulates aPKC signaling by modulating phosphorylation of Par-6 in Drosophila neuroblast asymmetric divisions. J Cell Sci. 2009;122:3242–3249. doi: 10.1242/jcs.050955. [DOI] [PubMed] [Google Scholar]
- 134.Zhao B, Smallwood A, Yang J, Koretke K, Nurse K, Calamari A, Kirkpatrick RB, Lai Z. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008;17:1791–1797. doi: 10.1110/ps.036590.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pan J, Snell WJ. Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p-like protein kinase translocates from the cell body into the flagella during gamete activation in chlamydomonas. J Biol Chem. 2000;275:24106–24114. doi: 10.1074/jbc.M002686200. [DOI] [PubMed] [Google Scholar]
- 136.Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell. 2004;6:445–451. doi: 10.1016/S1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 137.Plotnikova OV, Golemis EA, Pugacheva EN. Cell cycle-dependent ciliogenesis and cancer. Cancer Res. 2008;68:2058–2061. doi: 10.1158/0008-5472.CAN-07-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brush MH, Guardiola A, Connor JH, Yao TP, Shenolikar S. Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J Biol Chem. 2004;279:7685–7691. doi: 10.1074/jbc.M310997200. [DOI] [PubMed] [Google Scholar]
- 139.Kinzel D, Boldt K, Davis EE, Burtscher I, Trumbach D, Diplas B, Attie-Bitach T, Wurst W, Katsanis N, Ueffing M, Lickert H. Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev Cell. 2010;19:66–77. doi: 10.1016/j.devcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol. 2007;179:501–514. doi: 10.1083/jcb.200704069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wheeler GL, Joint I, Brownlee C. Rapid spatiotemporal patterning of cytosolic Ca2+ underlies flagellar excision in Chlamydomonas reinhardtii . Plant J. 2008;53:401–413. doi: 10.1111/j.1365-313X.2007.03349.x. [DOI] [PubMed] [Google Scholar]
- 142.Sun L, Hodeify R, Haun S, Charlesworth A, MacNicol AM, Ponnappan S, Ponnappan U, Prigent C, Machaca K. Ca2+ homeostasis regulates Xenopus oocyte maturation. Biol Reprod. 2008;78:726–735. doi: 10.1095/biolreprod.107.063693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Plotnikova OV, Nikonova AS, Loskutov YV, Kozyulina PY, Pugacheva EN, Golemis EA (2012) Calmodulin activation of Aurora-A (AURKA) is required during ciliary disassembly and in mitosis. Mol Biol Cell. doi:10.1091/mbc.E11-12-1056 [DOI] [PMC free article] [PubMed]
- 144.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nowakowski J, Cronin CN, McRee DE, Knuth MW, Nelson CG, Pavletich NP, Rogers J, Sang BC, Scheibe DN, Swanson RV, Thompson DA. Structures of the cancer-related Aurora-A, FAK, and EphA2 protein kinases from nanovolume crystallography. Structure. 2002;10:1659–1667. doi: 10.1016/S0969-2126(02)00907-3. [DOI] [PubMed] [Google Scholar]
- 146.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/S1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 147.Zhao B, Smallwood A, Yang J, Koretke K, Nurse K, Calamari A, Kirkpatrick RB, Lai Z. Modulation of kinase-inhibitor interactions by auxiliary protein binding: crystallography studies on Aurora A interactions with VX-680 and with TPX2. Protein Sci. 2008;17:1791–1797. doi: 10.1110/ps.036590.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJ, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, O’Keefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B, Morgan BA. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 149.Dodson CA, Kosmopoulou M, Richards MW, Atrash B, Bavetsias V, Blagg J, Bayliss R. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 150.Kornev AP, Taylor SS. Defining the conserved internal architecture of a protein kinase. Biochim Biophys Acta. 2010;1804:440–444. doi: 10.1016/j.bbapap.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Heron NM, Anderson M, Blowers DP, Breed J, Eden JM, Green S, Hill GB, Johnson T, Jung FH, McMiken HH, Mortlock AA, Pannifer AD, Pauptit RA, Pink J, Roberts NJ, Rowsell S. SAR and inhibitor complex structure determination of a novel class of potent and specific Aurora kinase inhibitors. Bioorg Med Chem Lett. 2006;16:1320–1323. doi: 10.1016/j.bmcl.2005.11.053. [DOI] [PubMed] [Google Scholar]
- 152.Bibby RA, Tang C, Faisal A, Drosopoulos K, Lubbe S, Houlston R, Bayliss R, Linardopoulos S. A cancer-associated aurora A mutant is mislocalized and misregulated due to loss of interaction with TPX2. J Biol Chem. 2009;284:33177–33184. doi: 10.1074/jbc.M109.032722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 155.Jiang S, Katayama H, Wang J, Li SA, Hong Y, Radvanyi L, Li JJ, Sen S. Estrogen-induced aurora kinase-A (AURKA) gene expression is activated by GATA-3 in estrogen receptor-positive breast cancer cells. Horm Cancer. 2010;1:11–20. doi: 10.1007/s12672-010-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee HH, Zhu Y, Govindasamy KM, Gopalan G. Downregulation of Aurora-A overrides estrogen-mediated growth and chemoresistance in breast cancer cells. Endocr Relat Cancer. 2008;15:765–775. doi: 10.1677/ERC-07-0213. [DOI] [PubMed] [Google Scholar]
- 157.Xu J, Li H, Wang B, Xu Y, Yang J, Zhang X, Harten SK, Shukla D, Maxwell PH, Pei D, Esteban MA. VHL inactivation induces HEF1 and Aurora kinase A. J Am Soc Nephrol. 2010;21:2041–2046. doi: 10.1681/ASN.2010040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Karthigeyan D, Prasad SB, Shandilya J, Agrawal S, Kundu TK. Biology of Aurora A kinase: implications in cancer manifestation and therapy. Med Res Rev. 2011;31(5):757–793. doi: 10.1002/med.20203. [DOI] [PubMed] [Google Scholar]
- 159.Yin N, Shi J, Wang D, Tong T, Wang M, Fan F, Zhan Q. IQGAP1 interacts with Aurora-A and enhances its stability and its role in cancer. Biochem Biophys Res Commun. 2012;421:64–69. doi: 10.1016/j.bbrc.2012.03.112. [DOI] [PubMed] [Google Scholar]
- 160.Aguirre-Portoles C, Bird AW, Hyman A, Canamero M, Perez de Castro I, Malumbres M. Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 2012;72:1518–1528. doi: 10.1158/0008-5472.CAN-11-1971. [DOI] [PubMed] [Google Scholar]
- 161.Sanbhnani S, Yeong FM. CHFR: a key checkpoint component implicated in a wide range of cancers. Cell Mol Life Sci. 2012;69:1669–1687. doi: 10.1007/s00018-011-0892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Asteriti IA, Rensen WM, Lindon C, Lavia P, Guarguaglini G. The Aurora-A/TPX2 complex: a novel oncogenic holoenzyme? Biochim Biophys Acta. 2010;1806:230–239. doi: 10.1016/j.bbcan.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 163.Dar AA, Goff LW, Majid S, Berlin J, El-Rifai W. Aurora kinase inhibitors—rising stars in cancer therapeutics? Mol Cancer Ther. 2010;9:268–278. doi: 10.1158/1535-7163.MCT-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gautschi O, Heighway J, Mack PC, Purnell PR, Lara PN, Jr, Gandara DR. Aurora kinases as anticancer drug targets. Clin Cancer Res. 2008;14:1639–1648. doi: 10.1158/1078-0432.CCR-07-2179. [DOI] [PubMed] [Google Scholar]
- 165.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/A:1023789416385. [DOI] [PubMed] [Google Scholar]
- 166.Marumoto T, Zhang D, Saya H. Aurora-A—a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 167.Yan A, Wang L, Xu S, Xu J. Aurora-A kinase inhibitor scaffolds and binding modes. Drug Discov Today. 2011;16:260–269. doi: 10.1016/j.drudis.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 168.Zhang D, Shimizu T, Araki N, Hirota T, Yoshie M, Ogawa K, Nakagata N, Takeya M, Saya H. Aurora A overexpression induces cellular senescence in mammary gland hyperplastic tumors developed in p53-deficient mice. Oncogene. 2008;27:4305–4314. doi: 10.1038/onc.2008.76. [DOI] [PubMed] [Google Scholar]
- 169.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 170.Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, Gray J, Bradley A, Medina D, Varmus HE. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- 171.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 172.Chen SS, Chang PC, Cheng YW, Tang FM, Lin YS. Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function. EMBO J. 2002;21:4491–4499. doi: 10.1093/emboj/cdf409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mao JH, Wu D, Perez-Losada J, Jiang T, Li Q, Neve RM, Gray JW, Cai WW, Balmain A. Crosstalk between Aurora-A and p53: frequent deletion or downregulation of Aurora-A in tumors from p53 null mice. Cancer Cell. 2007;11:161–173. doi: 10.1016/j.ccr.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 175.Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2:e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cai Q, Xiao B, Si H, Cervini A, Gao J, Lu J, Upadhyay SK, Verma SC, Robertson ES. Kaposi’s sarcoma herpesvirus upregulates Aurora A expression to promote p53 phosphorylation and ubiquitylation. PLoS Pathog. 2012;8:e1002566. doi: 10.1371/journal.ppat.1002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Katayama H, Wang J, Treekitkarnmongkol W, Kawai H, Sasai K, Zhang H, Wang H, Adams HP, Jiang S, Chakraborty SN, Suzuki F, Arlinghaus RB, Liu J, Mobley JA, Grizzle WE, Sen S. Aurora kinase-A inactivates DNA damage-induced apoptosis and spindle assembly checkpoint response functions of p73. Cancer Cell. 2012;21:196–211. doi: 10.1016/j.ccr.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zou L, Sun Y, Wang M, Zhan Q. Aurora-A interacts with AP-2alpha and down regulates its transcription activity. PloS One. 2011;6:e23110. doi: 10.1371/journal.pone.0023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Briassouli P, Chan F, Savage K, Reis-Filho JS, Linardopoulos S. Aurora-A regulation of nuclear factor-kappaB signaling by phosphorylation of IkappaBalpha. Cancer Res. 2007;67:1689–1695. doi: 10.1158/0008-5472.CAN-06-2272. [DOI] [PubMed] [Google Scholar]
- 180.Chefetz I, Holmberg JC, Alvero AB, Visintin I, Mor G. Inhibition of Aurora-A kinase induces cell cycle arrest in epithelial ovarian cancer stem cells by affecting NFkB pathway. Cell Cycle. 2011;10:2206–2214. doi: 10.4161/cc.10.13.16348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. J Int Cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- 182.Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, Counter CM. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2010;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu JC, Chen TY, Yu CT, Tsai SJ, Hsu JM, Tang MJ, Chou CK, Lin WJ, Yuan CJ, Huang CY. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005;280:9013–9022. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- 184.Neben K, Korshunov A, Benner A, Wrobel G, Hahn M, Kokocinski F, Golanov A, Joos S, Lichter P. Microarray-based screening for molecular markers in medulloblastoma revealed STK15 as independent predictor for survival. Cancer Res. 2004;64:3103–3111. doi: 10.1158/0008-5472.CAN-03-3968. [DOI] [PubMed] [Google Scholar]
- 185.Dimova I, Raitcheva S, Dimitrov R, Doganov N, Toncheva D. Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumours. Eur J Cancer. 2006;42:674–679. doi: 10.1016/j.ejca.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 186.Lassmann S, Shen Y, Jutting U, Wiehle P, Walch A, Gitsch G, Hasenburg A, Werner M. Predictive value of Aurora-A/STK15 expression for late stage epithelial ovarian cancer patients treated by adjuvant chemotherapy. Clin Cancer Res. 2007;13:4083–4091. doi: 10.1158/1078-0432.CCR-06-2775. [DOI] [PubMed] [Google Scholar]
- 187.Otto T, Horn S, Brockmann M, Eilers U, Schuttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, Berwanger B, Eilers M. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 188.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, Macdonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, de la Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang H, Ou CC, Feldman RI, Nicosia SV, Kruk PA, Cheng JQ. Aurora-A kinase regulates telomerase activity through c-Myc in human ovarian and breast epithelial cells. Cancer Res. 2004;64:463–467. doi: 10.1158/0008-5472.CAN-03-2907. [DOI] [PubMed] [Google Scholar]
- 190.Yang S, He S, Zhou X, Liu M, Zhu H, Wang Y, Zhang W, Yan S, Quan L, Bai J, Xu N. Suppression of Aurora-A oncogenic potential by c-Myc downregulation. Exp Mol Med. 2010;42:759–767. doi: 10.3858/emm.2010.42.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dar AA, Belkhiri A, El-Rifai W. The aurora kinase A regulates GSK-3beta in gastric cancer cells. Oncogene. 2009;28:866–875. doi: 10.1038/onc.2008.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.den Hollander J, Rimpi S, Doherty JR, Rudelius M, Buck A, Hoellein A, Kremer M, Graf N, Scheerer M, Hall MA, Goga A, von Bubnoff N, Duyster J, Peschel C, Cleveland JL, Nilsson JA, Keller U. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116:1498–1505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sumi K, Tago K, Kasahara T, Funakoshi-Tago M. Aurora kinase A critically contributes to the resistance to anti-cancer drug cisplatin in JAK2 V617F mutant-induced transformed cells. FEBS Lett. 2011;585:1884–1890. doi: 10.1016/j.febslet.2011.04.068. [DOI] [PubMed] [Google Scholar]
- 194.Torchia EC, Caulin C, Acin S, Terzian T, Kubick BJ, Box NF, Roop DR. Myc, Aurora Kinase A, and mutant p53(R172H) co-operate in a mouse model of metastatic skin carcinoma. Oncogene. 2012;31:2680–2690. doi: 10.1038/onc.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Maxwell CA, Benitez J, Gomez-Baldo L, Osorio A, Bonifaci N, Fernandez-Ramires R, Costes SV, Guino E, Chen H, Evans GJ, Mohan P, Catala I, Petit A, Aguilar H, Villanueva A, Aytes A, Serra-Musach J, Rennert G, Lejbkowicz F, Peterlongo P, Manoukian S, Peissel B, Ripamonti CB, Bonanni B, Viel A, Allavena A, Bernard L, Radice P, Friedman E, Kaufman B, Laitman Y, Dubrovsky M, Milgrom R, Jakubowska A, Cybulski C, Gorski B, Jaworska K, Durda K, Sukiennicki G, Lubinski J, Shugart YY, Domchek SM, Letrero R, Weber BL, Hogervorst FB, Rookus MA, Collee JM, Devilee P, Ligtenberg MJ, Luijt RB, Aalfs CM, Waisfisz Q, Wijnen J, Roozendaal CE, Easton DF, Peock S, Cook M, Oliver C, Frost D, Harrington P, Evans DG, Lalloo F, Eeles R, Izatt L, Chu C, Eccles D, Douglas F, Brewer C, Nevanlinna H, Heikkinen T, Couch FJ, Lindor NM, Wang X, Godwin AK, Caligo MA, Lombardi G, Loman N, Karlsson P, Ehrencrona H, Wachenfeldt A, Barkardottir RB, Hamann U, Rashid MU, Lasa A, Caldes T, Andres R, Schmitt M, Assmann V, Stevens K, Offit K, Curado J, Tilgner H, Guigo R, Aiza G, Brunet J, Castellsague J, Martrat G, Urruticoechea A, Blanco I, Tihomirova L, Goldgar DE, Buys S, John EM, Miron A, Southey M, Daly MB, Schmutzler RK, Wappenschmidt B, Meindl A, Arnold N, Deissler H, Varon-Mateeva R, Sutter C, Niederacher D, Imyamitov E, Sinilnikova OM, Stoppa-Lyonne D, Mazoyer S, Verny-Pierre C, Castera L, de Pauw A, Bignon YJ, Uhrhammer N, Peyrat JP, Vennin P, Fert Ferrer S, Collonge-Rame MA, Mortemousque I, Spurdle AB, Beesley J, Chen X, Healey S, Barcellos-Hoff MH, Vidal M, Gruber SB, Lazaro C, Capella G, McGuffog L, Nathanson KL, Antoniou AC, Chenevix-Trench G, Fleisch MC, Moreno V, Pujana MA. Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 2011;9:e1001199. doi: 10.1371/journal.pbio.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ratushny V, Pathak HB, Beeharry N, Tikhmyanova N, Xiao F, Li T, Litwin S, Connolly DC, Yen TJ, Weiner LM, Godwin AK, Golemis EA. Dual inhibition of SRC and Aurora kinases induces postmitotic attachment defects and cell death. Oncogene. 2011;31(10):1217–1227. doi: 10.1038/onc.2011.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Arai R, Tsuda M, Watanabe T, Ose T, Obuse C, Maenaka K, Minami A, Ohba Y (2012) Simultaneous inhibition of Src and Aurora kinases by SU6656 induces therapeutic synergy in human synovial sarcoma growth, invasion and angiogenesis in vivo. Eur J Cancer. doi:10.1016/j.ejca.2011.12.028 [DOI] [PubMed]
- 198.Gritsko TM, Coppola D, Paciga JE, Yang L, Sun M, Shelley SA, Fiorica JV, Nicosia SV, Cheng JQ. Activation and overexpression of centrosome kinase BTAK/Aurora-A in human ovarian cancer. Clin Cancer Res. 2003;9:1420–1426. [PubMed] [Google Scholar]
- 199.Hu W, Kavanagh JJ, Deaver M, Johnston DA, Freedman RS, Verschraegen CF, Sen S. Frequent overexpression of STK15/Aurora-A/BTAK and chromosomal instability in tumorigenic cell cultures derived from human ovarian cancer. Oncol Res. 2005;15:49–57. doi: 10.3727/096504005775082101. [DOI] [PubMed] [Google Scholar]
- 200.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, Borg A, Isola JJ. Frequent amplification of chromosomal region 20q12–q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–1839. [PubMed] [Google Scholar]
- 201.Kulkarni AA, Loddo M, Leo E, Rashid M, Eward KL, Fanshawe TR, Butcher J, Frost A, Ledermann JA, Williams GH, Stoeber K. DNA replication licensing factors and aurora kinases are linked to aneuploidy and clinical outcome in epithelial ovarian carcinoma. Clin Cancer Res. 2007;13:6153–6161. doi: 10.1158/1078-0432.CCR-07-0671. [DOI] [PubMed] [Google Scholar]
- 202.Landen CN, Jr, Lin YG, Immaneni A, Deavers MT, Merritt WM, Spannuth WA, Bodurka DC, Gershenson DM, Brinkley WR, Sood AK. Overexpression of the centrosomal protein Aurora-A kinase is associated with poor prognosis in epithelial ovarian cancer patients. Clin Cancer Res. 2007;13:4098–4104. doi: 10.1158/1078-0432.CCR-07-0431. [DOI] [PubMed] [Google Scholar]
- 203.Das K, Lorena PD, Ng LK, Shen L, Lim D, Siow WY, Narasimhan K, Teh M, Choolani M, Putti TC, Salto-Tellez M. Aurora-A expression, hormone receptor status and clinical outcome in hormone related cancers. Pathology. 2010;42:540–546. doi: 10.3109/00313025.2010.508789. [DOI] [PubMed] [Google Scholar]
- 204.Lassus H, Staff S, Leminen A, Isola J, Butzow R. Aurora-A overexpression and aneuploidy predict poor outcome in serous ovarian carcinoma. Gynecol Oncol. 2011;120:11–17. doi: 10.1016/j.ygyno.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 205.Mendiola M, Barriuso J, Marino-Enriquez A, Redondo A, Dominguez-Caceres A, Hernandez-Cortes G, Perez-Fernandez E, Sanchez-Navarro I, Vara JA, Suarez A, Espinosa E, Gonzalez-Baron M, Palacios J, Hardisson D. Aurora kinases as prognostic biomarkers in ovarian carcinoma. Hum Pathol. 2009;40:631–638. doi: 10.1016/j.humpath.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 206.Yang G, Chang B, Yang F, Guo X, Cai KQ, Xiao XS, Wang H, Sen S, Hung MC, Mills GB, Chang S, Multani AS, Mercado-Uribe I, Liu J. Aurora kinase A promotes ovarian tumorigenesis through dysregulation of the cell cycle and suppression of BRCA2. Clin Cancer Res. 2010;16:3171–3181. doi: 10.1158/1078-0432.CCR-09-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yang F, Guo X, Yang G, Rosen DG, Liu J. AURKA and BRCA2 expression highly correlate with prognosis of endometrioid ovarian carcinoma. Mod Pathol. 2011;24:836–845. doi: 10.1038/modpathol.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Carvalho B, Postma C, Mongera S, Hopmans E, Diskin S, van de Wiel MA, van Criekinge W, Thas O, Matthai A, Cuesta MA, Terhaar Sive Droste JS, Craanen M, Schrock E, Ylstra B, Meijer GA. Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression. Gut. 2009;58:79–89. doi: 10.1136/gut.2007.143065. [DOI] [PubMed] [Google Scholar]
- 209.Sillars-Hardebol AH, Carvalho B, Tijssen M, Belien JA, de Wit M, Delis-van Diemen PM, Ponten F, van de Wiel MA, Fijneman RJ, Meijer GA (2011) TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. doi:10.1136/gutjnl-2011-301153 [DOI] [PubMed]
- 210.Baba Y, Nosho K, Shima K, Irahara N, Kure S, Toyoda S, Kirkner GJ, Goel A, Fuchs CS, Ogino S. Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia. 2009;11:418–425. doi: 10.1593/neo.09154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Belt EJ, Brosens RP, Delis-van Diemen PM, Bril H, Tijssen M, van Essen DF, Heymans MW, Belien JA, Stockmann HB, Meijer S, Meijer GA (2012) Cell cycle proteins predict recurrence in stage II and III colon cancer. Ann Surg Oncol. doi:10.1245/s10434-012-2216-7 [DOI] [PubMed]
- 212.Lam AK, Ong K, Ho YH. Aurora kinase expression in colorectal adenocarcinoma: correlations with clinicopathological features, p16 expression, and telomerase activity. Hum Pathol. 2008;39:599–604. doi: 10.1016/j.humpath.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 213.Dotan E, Meropol NJ, Zhu F, Zambito F, Bove B, Cai KQ, Godwin AK, Golemis EA, Astsaturov I, Cohen SJ. Relationship of increased aurora kinase A gene copy number, prognosis and response to chemotherapy in patients with metastatic colorectal cancer. Br J Cancer. 2012;106:748–755. doi: 10.1038/bjc.2011.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Yuan J, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J, Ball S, Almeida M, Linardopoulos S, Balmain A. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34:403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 215.Ewart-Toland A, Dai Q, Gao YT, Nagase H, Dunlop MG, Farrington SM, Barnetson RA, Anton-Culver H, Peel D, Ziogas A, Lin D, Miao X, Sun T, Ostrander EA, Stanford JL, Langlois M, Chan JM, Yuan J, Harris CC, Bowman ED, Clayman GL, Lippman SM, Lee JJ, Zheng W, Balmain A. Aurora-A/STK15 T + 91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- 216.Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y. Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res. 2008;14:4455–4462. doi: 10.1158/1078-0432.CCR-07-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dai Q, Cai QY, Shu XO, Ewart-Toland A, Wen WQ, Balmain A, Gao YT, Zheng W. Synergistic effects of STK15 gene polymorphisms and endogenous estrogen exposure in the risk of breast cancer. Cancer Epidemiol Biomark Prev. 2004;13:2065–2070. [PubMed] [Google Scholar]
- 218.Lo YL, Yu JC, Chen ST, Yang HC, Fann CS, Mau YC, Shen CY. Breast cancer risk associated with genotypic polymorphism of the mitosis-regulating gene Aurora-A/STK15/BTAK. J Int Cancer. 2005;115:276–283. doi: 10.1002/ijc.20855. [DOI] [PubMed] [Google Scholar]
- 219.Cox DG, Hankinson SE, Hunter DJ. Polymorphisms of the AURKA (STK15/Aurora Kinase) gene and breast cancer risk (United States) Cancer Causes Control. 2006;17:81–83. doi: 10.1007/s10552-005-0429-9. [DOI] [PubMed] [Google Scholar]
- 220.Ogawa E, Takenaka K, Katakura H, Adachi M, Otake Y, Toda Y, Kotani H, Manabe T, Wada H, Tanaka F. Perimembrane Aurora-A expression is a significant prognostic factor in correlation with proliferative activity in non-small-cell lung cancer (NSCLC) Ann Surg Oncol. 2008;15:547–554. doi: 10.1245/s10434-007-9653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lo Iacono M, Monica V, Saviozzi S, Ceppi P, Bracco E, Papotti M, Scagliotti GV. Aurora kinase A expression is associated with lung cancer histological-subtypes and with tumor de-differentiation. J Transl Med. 2011;9:100. doi: 10.1186/1479-5876-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gu J, Gong Y, Huang M, Lu C, Spitz MR, Wu X. Polymorphisms of STK15 (Aurora-A) gene and lung cancer risk in Caucasians. Carcinogenesis. 2007;28:350–355. doi: 10.1093/carcin/bgl149. [DOI] [PubMed] [Google Scholar]
- 223.Yu CT, Hsia JY, Hseih YC, Su LJ, Lee TC, Ku CF, Chen KS, Chen JM, Wei TY, Lee YC, Huang CY, Wu YC, Yang CY, Hsu SL. The novel protein suppressed in lung cancer down-regulated in lung cancer tissues retards cell proliferation and inhibits the oncokinase Aurora-A. J Thorac Oncol. 2011;6:988–997. doi: 10.1097/JTO.0b013e318212692e. [DOI] [PubMed] [Google Scholar]
- 224.Reiter R, Gais P, Jutting U, Steuer-Vogt MK, Pickhard A, Bink K, Rauser S, Lassmann S, Hofler H, Werner M, Walch A. Aurora kinase A messenger RNA overexpression is correlated with tumor progression and shortened survival in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:5136–5141. doi: 10.1158/1078-0432.CCR-05-1650. [DOI] [PubMed] [Google Scholar]
- 225.Kollareddy M, Zheleva D, Dzubak P, Brahmkshatriya PS, Lepsik M, Hajduch M (2012) Aurora kinase inhibitors: progress towards the clinic. Investig New Drugs. doi:10.1007/s10637-012-9798-6 [DOI] [PMC free article] [PubMed]
- 226.Sonet A, Graux C, Maertens J, Hartog C-M, Duyster J, Gotze K, Greiner J, Hutter M-, Gratwohl A, Heim D, Hess D, Chalandon Y, Gianella-Borradori A, Rejeb N, Ottman O. Phase I, dose-escalation study of 2 dosing regimens of AS703569, an inhibitor of aurora and other kinases, administered orally in patients with advanced hematological malignancies. Blood ASH Ann Meet Abstr. 2008;112:2963. [Google Scholar]
- 227.Chakravarty A, Shinde V, Tabernero J, Cervantes A, Cohen RB, Dees EC, Burris H, Infante JR, Macarulla T, Elez E, Andreu J, Rodriguez-Braun E, Rosello S, Mehren M, Meropol NJ, Langer CJ, ONeil B, Bowman D, Zhang M, Danaee H, Faron-Yowe L, Gray G, Liu H, Pappas J, Silverman L, Simpson C, Stringer B, Tirrell S, Veiby OP, Venkatakrishnan K, Galvin K, Manfredi M, Ecsedy JA. Phase I assessment of new mechanism-based pharmacodynamic biomarkers for MLN8054, a small-molecule inhibitor of Aurora A kinase. Cancer Res. 2011;71:675–685. doi: 10.1158/0008-5472.CAN-10-1030. [DOI] [PubMed] [Google Scholar]
- 228.Dees EC, Infante JR, Cohen RB, O’Neil BH, Jones S, von Mehren M, Danaee H, Lee Y, Ecsedy J, Manfredi M, Galvin K, Stringer B, Liu H, Eton O, Fingert H, Burris H. Phase 1 study of MLN8054, a selective inhibitor of Aurora A kinase in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2011;67:945–954. doi: 10.1007/s00280-010-1377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Macarulla T, Cervantes A, Elez E, Rodriguez-Braun E, Baselga J, Rosello S, Sala G, Blasco I, Danaee H, Lee Y, Ecsedy J, Shinde V, Chakravarty A, Bowman D, Liu H, Eton O, Fingert H, Tabernero J. Phase I study of the selective Aurora A kinase inhibitor MLN8054 in patients with advanced solid tumors: safety, pharmacokinetics, and pharmacodynamics. Mol Cancer Ther. 2010;9:2844–2852. doi: 10.1158/1535-7163.MCT-10-0299. [DOI] [PubMed] [Google Scholar]
- 230.Kelly KR, Nawrocki ST, Espitia CM, Zhang M, Yang JJ, Padmanabhan S, Ecsedy J, Giles FJ, Carew JS (2012) Targeting aurora a kinase activity with the investigational agent alisertib increases the efficacy of cytarabine through a FOXO-dependent mechanism. Int J Cancer. doi:10.1002/ijc.27579 [DOI] [PMC free article] [PubMed]
- 231.Goldberg SL, Fenaux P, Craig MD, Gyan E, Lister J, Kassis J, Pigneux A, Schiller GJ, Jung J, Leonard EJ, Fingert H, Westervelt P. Phase 2 study of MLN8237, an investigational Aurora A kinase (AAK) inhibitor in patients with acute myelogenous leukemia (AML) or myelodysplastic syndromes (MDS) ASH Ann Meet Abstr. 2010;116:3273. [Google Scholar]
- 232.Friedberg J, Mahadevan D, Jung J, Persky DO, Lossos IS, Danaee H, Zhou X, Leonard EJ, Bernstein SH. Phase 2 trial of alisertib (MLN8237), an investigational, potent inhibitor of Aurora A kinase (AAK), in patients (pts) with aggressive B- and T-cell non-hodgkin lymphoma (NHL) ASH Ann Meet Abstr. 2011;118:95. [Google Scholar]
- 233.Mehra R, Serebriiskii IG, Dunbrack RL, Jr, Robinson MK, Burtness B, Golemis EA. Protein-intrinsic and signaling network-based sources of resistance to EGFR- and ErbB family-targeted therapies in head and neck cancer. Drug Resist Updat. 2011;14:260–279. doi: 10.1016/j.drup.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.O’Connor O, Leonard EJ, Benaim E (2012) Phase III study of investigational MLN8237 (alisertib) versus investigator’s choice in patients (pts) with relapsed/refractory (rel/ref) peripheral T-cell lymphoma (PTCL). J Clin Oncol 30(suppl); abstr TPS8110. http://www.asco.org/ASCOv2/Meetings/Abstract?&vmview=abst_detail_view&conflD=114&abstractID=93843
- 235.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, Bayani N, Hu Z, Billig JI, Dueregger A, Lewis S, Jakkula L, Korkola JE, Durinck S, Pepin F, Guan Y, Purdom E, Neuvial P, Bengtsson H, Wood KW, Smith PG, Vassilev LT, Hennessy BT, Greshock J, Bachman KE, Hardwicke MA, Park JW, Marton LJ, Wolf DM, Collisson EA, Neve RM, Mills GB, Speed TP, Feiler HS, Wooster RF, Haussler D, Stuart JM, Gray JW, Spellman PT. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cohen RB, Jones SF, Aggarwal C, von Mehren M, Cheng J, Spigel DR, Greco FA, Mariani M, Rocchetti M, Ceruti R, Comis S, Laffranchi B, Moll J, Burris HA. A phase I dose-escalation study of danusertib (PHA-739358) administered as a 24-hour infusion with and without granulocyte colony-stimulating factor in a 14-day cycle in patients with advanced solid tumors. Clin Cancer Res. 2009;15:6694–6701. doi: 10.1158/1078-0432.CCR-09-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Venkataraman S, Alimova I, Tello T, Harris PS, Knipstein JA, Donson AM, Foreman NK, Liu AK, Vibhakar R. Targeting Aurora Kinase A enhances radiation sensitivity of atypical teratoid rhabdoid tumor cells. J Neurooncol. 2012;107:517–526. doi: 10.1007/s11060-011-0795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guan Z, Wang XR, Zhu XF, Huang XF, Xu J, Wang LH, Wan XB, Long ZJ, Liu JN, Feng GK, Huang W, Zeng YX, Chen FJ, Liu Q. Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res. 2007;67:10436–10444. doi: 10.1158/0008-5472.CAN-07-1379. [DOI] [PubMed] [Google Scholar]
- 240.Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, Devarajan K, Silverman JS, Tikhmyanova N, Skobeleva N, Pecherskaya A, Nasto RE, Sharma C, Jablonski SA, Serebriiskii IG, Weiner LM, Golemis EA. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal. 2010;3:ra67. doi: 10.1126/scisignal.2001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shao S, Wang Y, Jin S, Song Y, Wang X, Fan W, Zhao Z, Fu M, Tong T, Dong L, Fan F, Xu N, Zhan Q. Gadd45a interacts with aurora-A and inhibits its kinase activity. J Biol Chem. 2006;281:28943–28950. doi: 10.1074/jbc.M600235200. [DOI] [PubMed] [Google Scholar]
- 242.Lukasiewicz KB, Greenwood TM, Negron VC, Bruzek AK, Salisbury JL, Lingle WL. Control of centrin stability by Aurora A. PloS One. 2011;6:e21291. doi: 10.1371/journal.pone.0021291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mosquera J, Armisen R, Zhao H, Rojas DA, Maldonado E, Tapia JC, Colombo A, Hayman MJ, Marcelain K. Identification of Ski as a target for Aurora A kinase. Biochem Biophys Res Commun. 2011;409:539–543. doi: 10.1016/j.bbrc.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- 245.Scrittori L, Hans F, Angelov D, Charra M, Prigent C, Dimitrov S. pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J Biol Chem. 2001;276:30002–30010. doi: 10.1074/jbc.M102701200. [DOI] [PubMed] [Google Scholar]
- 246.Sasayama T, Marumoto T, Kunitoku N, Zhang D, Tamaki N, Kohmura E, Saya H, Hirota T. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells. 2005;10:627–638. doi: 10.1111/j.1365-2443.2005.00870.x. [DOI] [PubMed] [Google Scholar]
- 247.Johnson EO, Chang KH, Ghosh S, Venkatesh C, Giger K, Low PS, Shah K. LIMK2 is a crucial regulator and effector of Aurora-A-kinase-mediated malignancy. J Cell Sci. 2012;125:1204–1216. doi: 10.1242/jcs.092304. [DOI] [PubMed] [Google Scholar]
- 248.Terada Y, Uetake Y, Kuriyama R. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol. 2003;162:757–763. doi: 10.1083/jcb.200305048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jang MS, Sul JW, Choi BJ, Lee SJ, Suh JH, Kim NS, Kim WH, Lim DS, Lee CW, Kim E. Negative feedback regulation of Aurora-A via phosphorylation of Fas-associated factor-1. J Biol Chem. 2008;283:32344–32351. doi: 10.1074/jbc.M804199200. [DOI] [PubMed] [Google Scholar]
- 250.Johnson EO, Chang KH, de Pablo Y, Ghosh S, Mehta R, Badve S, Shah K. PHLDA1 is a crucial negative regulator and effector of Aurora A kinase in breast cancer. J Cell Sci. 2011;124:2711–2722. doi: 10.1242/jcs.084970. [DOI] [PubMed] [Google Scholar]
- 251.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics (Oxford, England) 27:431–432 [DOI] [PMC free article] [PubMed]