Abstract
To analyze HIV-1 genotypes in Lithuania and the transmission of drug-resistant viruses, HIV-1 sequences were obtained from 138 individuals, who were diagnosed as HIV-1 infected in 1990–2008 and represented all major risk groups. Subtype A strains, dominating in the former Soviet Union (90% of cases), were found in 60% of individuals, followed by subtype B (22%) and CRF03_AB (12%) strains. The remaining 7% of the strains included variants belonging to subtype C, CRF01_AE, CRF02_AG, more complex recombinant forms, and strains that could not be reliably genotyped. Analysis of virus genotypes per risk group revealed the circulation of distinct HIV-1 strains in different risk groups: subtype A viruses were present in 82% of injecting drug users (IDUs), but less than a half of heterosexually infected individuals and cases with unknown transmission route, and none of men having sex with men (MSM). We observed no mutations causing drug resistance among 27 newly diagnosed HIV-1 cases.
The new independent states (NIS) of the former Soviet Union (FSU) have experienced an explosive HIV-1 epidemic since the mid-1990s, when the increase in the incidence of HIV-1 in this region was the largest in the world. This epidemic primarily affects injecting drug users (IDUs) and their sexual partners and started after the introduction of two HIV-1 strains into populations of IDUs in the South Ukraine in 1994: subtype A (designated IDU-A) and subtype B (IDU-B) viruses.1 Molecular epidemiological studies demonstrated that of these two viruses, the IDU-A strains have spread throughout the whole FSU territory and account for around 90% of over a million of HIV-1-infected individuals in the region.2 These strains are dominating in Azerbaijan,3 Belarus,4,5 Georgia,6 Kazakhstan,7,8 Latvia,9,10 Moldova,11 Russia,2,12 Tajikistan,13 Ukraine,1,14,15 and Uzbekistan16,17 and are the second (after CRF06_cpx strains) major cause of infections in Estonia.18,19
In this explosive HIV-1 epidemic in the FSU, Lithuania remains among the least affected countries with 1,900 registered HIV-1 infections (as of January 1, 2012, 0.06% of the population, our own data), with the UNAIDS estimation of the prevalence of HIV-1 in the adult population being 0.1% (all UNAIDS estimations are from www.unaids.org/en/dataanalysis/tools/aidsinfo/). This includes a large single outbreak of HIV-1 infection, when >300 prisoners were infected by contaminated drugs and injecting equipment in the Alytus prison within a few days to weeks. The low number of HIV-1 infections in Lithuania is especially remarkable considering the fact that three other Baltic territories of the FSU—the countries of Estonia and Latvia and the Russian enclave of the Kaliningrad region—are among the most affected regions of the HIV-1 epidemic in the FSU. In fact, as estimated by UNAIDS, Estonia has the highest prevalence of HIV-1 in adults among all NIS (9,900 cases, 1.2% of the population) and Latvia is above the average (8,600 cases, 0.7%). The registered HIV-1 prevalence in the Kaliningrad region of Russia (7,563 cases, 1.02%) is more than twice as high as it is in all of Russia (650,100 cases, 0.46%; the data are from www.hivrussia.ru/stat/2011.shtml, as of January 1, 2012), and the UNAIDS estimation for the prevalence of HIV-1 in adults in Russia is 1.0%. Unlike all other Russian regions, where the IDU-A strains are dominant, the HIV-1 outbreak in the Kaliningrad region is caused by a circulating recombinant form, CRF03_AB, that resulted from a recombination of the original IDU-A and IDU-B strains.20,21
While molecular epidemiological data are available for a number of NIS (listed above), no such studies have been carried out in Lithuania. In this study we analyzed HIV-1 genotypes in Lithuania and transmission of drug-resistant viruses.
Clinical samples (plasma or serum) and epidemiological information were obtained from 138 HIV-1-infected residents of Lithuania, which is >7% of the total HIV-1-infected population in the country. The individuals were diagnosed as HIV-1 infected in 1990–2008 and represented all major risk groups: IDUs (n=77, 63 males and 14 females), heterosexually infected individuals (n=33, 17 males and 15 females, including one perinatal infection), and men who have sex with men (MSM) (n=21); for seven individuals their transmission route was unknown (five males and two females). Males represented 77% of the total study population and 73% of the non-MSM study participants. Both the proportion of individuals representing each risk group in this study and the proportion of males correspond to their proportions in the total infected population in Lithuania (own data), where of the total of 1,900 HIV-1 cases 70% are among IDUs (88% of whom are males), 15% are among heterosexually infected individuals (55% males), 6% are among MSM, and 9% are among individuals with an unknown transmission route (77% males). Of the study individuals, 27 were new infections diagnosed in 2008 (10 IDUs, nine heterosexually infected, five MSM, and three with unknown transmission routes). The study was approved by the Bioethical Commission of the National AIDS Centre of Lithuania.
For 111 individuals diagnosed in 1990–2007, genetic regions of gag p17/p24 (729 nt in length, corresponding to HIV-1 HXB2 positions 859–1587) and env (270 nt, positions 7032–7307) genes were obtained. For 27 individuals diagnosed in 2008, pol sequences (1,302 nt, positions 2253–3554) were obtained, to assess transmission of drug-resistant HIV-1 strains, next to their subtype characterization. Both approaches make it possible to identify all HIV-1 variants specific for the FSU, including CRF03_AB, as one of the recombination points of this CRF is within the pol gene.
Sequences obtained in this study have been submitted to GenBank with accession numbers JX946435–JX946653.
Phylogenetic analysis was performed with MEGA5 software, www.megasoftware.net/, using the maximum likelihood method based on the general time reversible model with G-distribution (n=5) and invariant sites (ML GTR+G+I). Reference sequences of HIV-1 genetic subtypes and CRFs from the Los Alamos HIV Sequence Database, www.hiv.lanl.gov/content/index, were included in the analysis. The statistical significance of phylogenetic clusters was established by bootstrap analysis, with 1,000 replicates. Recombination analysis was performed by the BootScan method of the SimPlot software, http://sray.med.som.jhmi.edu/SCRoftware/simplot/. Analysis of drug-resistant mutations was performed by the on-line facility of the HIV Drug Resistance Database of Stanford University, http://cpr.stanford.edu.
Statistical analysis was performed with Fisher's exact test as implemented in GraphPad Prism 5 software.
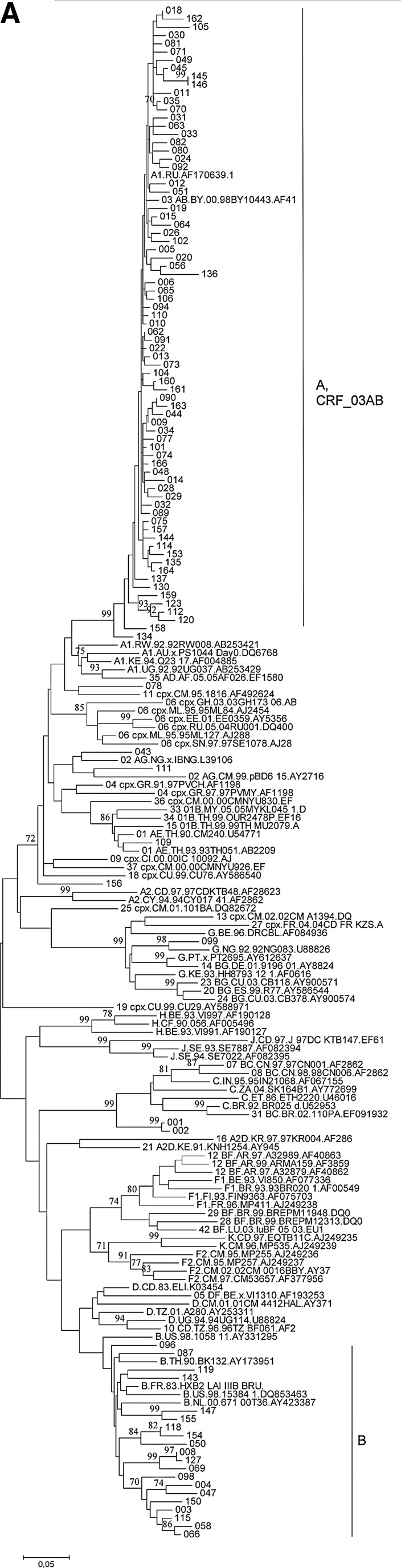
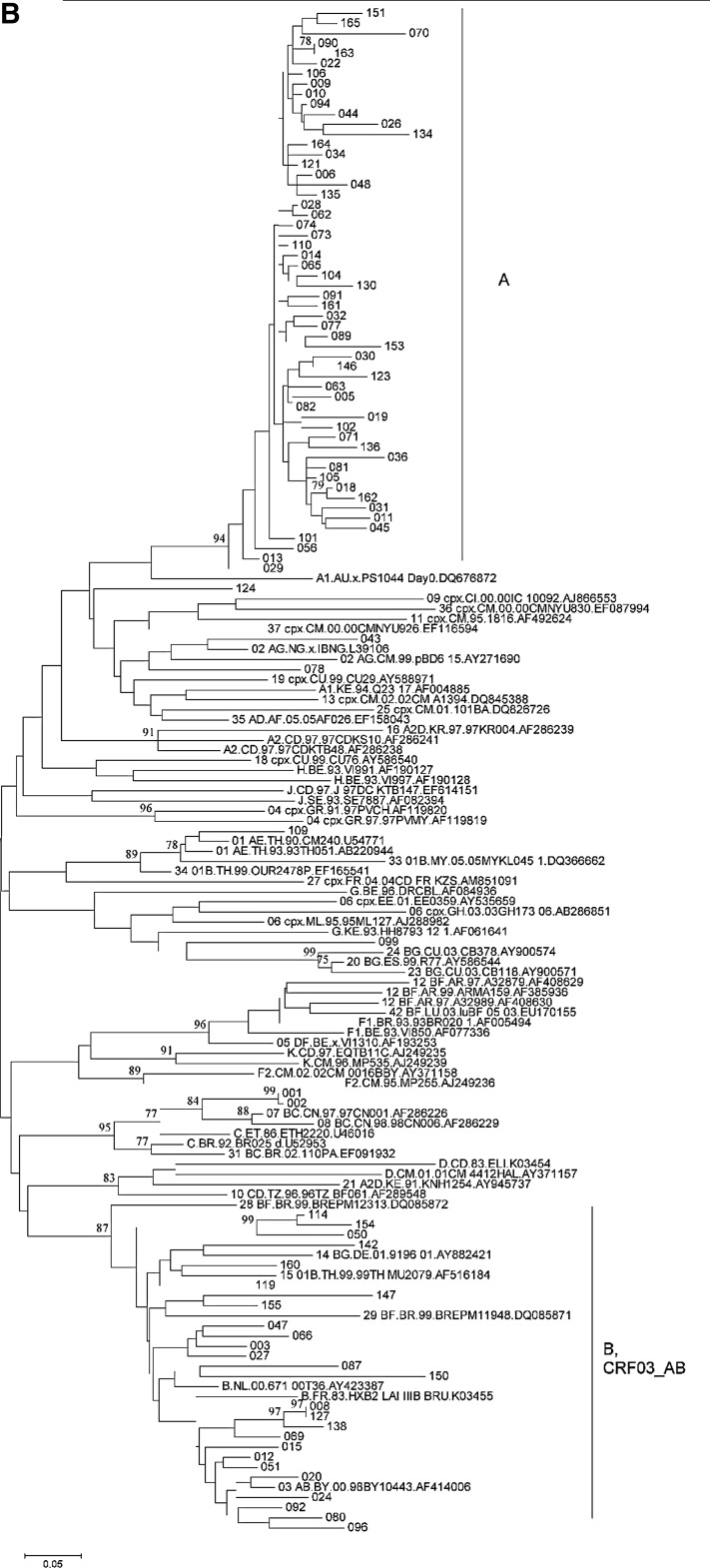
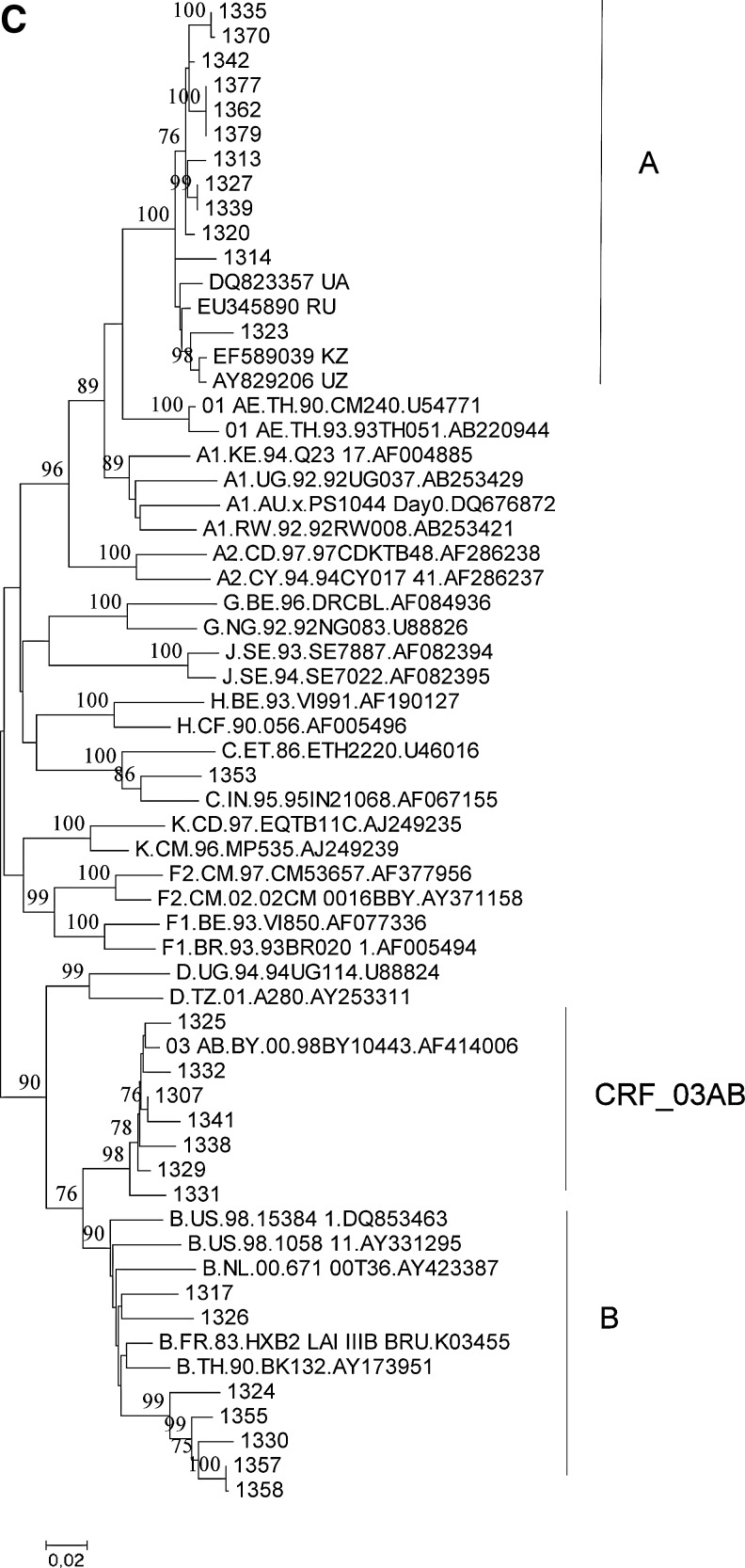
Phylogenetic analysis of HIV-1 strains obtained from 138 infected individuals demonstrated their remarkable variety in Lithuania (Fig. 1). Subtype A strains were found in 82 individuals (60%, including the Alytus prison outbreak), followed by subtype B (30 cases, 22%) and CRF03_AB (16 cases, 12%) strains. Strains of other genotypes (10 cases, 7%) were each present in 1–2% of cases: three cases of infections by subtype C strains (including a mother–child pair), two by CRF02_AG, one by CRF01_AE, and one by a complex recombinant between subtype G and CRF06_cpx; finally, three of the strains obtained could not be reliably genotyped, as their phylogenetic clustering was not significantly supported by bootstrap analysis (one of these strains clustered with CRF11_cpx in the gag, but not in the env region). Recombination analysis of untypeable strains did not provide evidence for their possible recombinant origin (data not shown).
FIG. 1.
Phylogenetic trees of HIV-1 gag (A), env (B), and pol (C) sequences from Lithuania. Sequences from Lithuania are labeled by their numbers. Reference sequences representing HIV-1 genetic subtypes (labeled by the first letter) and circulating recombinant forms (CRFs) (labeled by the two digits and subtypes of parental strains) are included. Clusters of Lithuanian sequences are shown. Bootstrap values of ≥70 are shown.
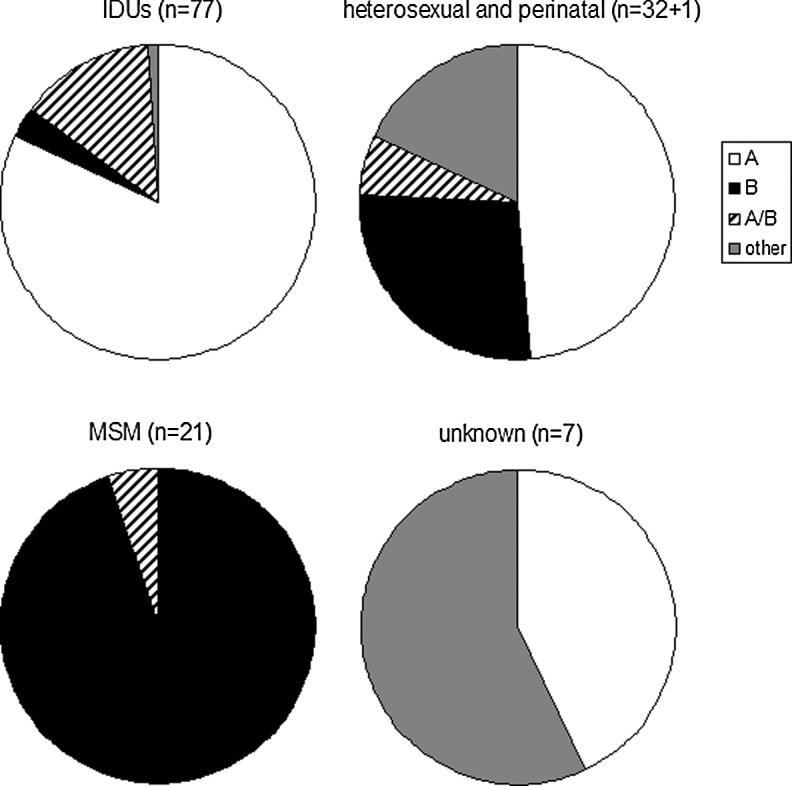
Analysis of virus genotypes per risk group revealed distinct HIV-1 strains circulating in different risk groups (Fig. 2). Virus strains specific for the epidemic in the FSU accounted for 95% of cases among IDUs: 63 (82%) infections by subtype A (IDU-A strains) and 10 (13%) by CRF03_AB strains. Yet almost half (n=14, 42%) of the cases among heterosexually infected individuals and more than half (n=4, 57%) of the cases among individuals with unknown transmission route were infections by viruses unspecific for the epidemic in the FSU (p<0.0001 and p=0.0011, respectively, for the comparisons with IDUs). No statistically significant differences in HIV-1 genotype distribution per gender were observed for these risk groups. All but one MSM (20 cases, 95%) were infected by subtype B virus stains.
FIG. 2.
Distribution of HIV-1 genotypes among risk groups in Lithuania.
Analysis of infections registered in 1990–2007 vs. 2008 demonstrated a trend for increased transmission of viruses specific for the epidemic in the FSU: all 10 infections among IDUs, seven infections (78%) among heterosexually infected individuals, and two infections (67%) among individuals with unknown transmission route were by subtype A or CRF03_AB strains.
Our analysis demonstrated an extremely low prevalence of drug-resistant mutations in 27 newly infected therapy-naive individuals: only one HIV-1 strain (3.7%) had a single mutation, T215S, associated with resistance to nucleoside reverse transcriptase inhibitors (NRTI). In fact, this mutation does not cause phenotypic resistance to NRTI, but can often be detected in virus strains that eventually go on to develop the drug-resistant mutations T215Y or T215F (transitional mutation).
To summarize our findings, four conclusions might be drawn: (1) The HIV-1 epidemic in Lithuania is caused by a variety of HIV-1 genotypes. (2) Distinct HIV-1 strains are circulating in different risk groups. (3) Lithuania appeared to be the only territory outside the Kaliningrad region of Russia in which CRF03_AB strains are epidemiologically significant. (4) No transmission of drug-resistant strains was observed.
Lithuania, Estonia, and Latvia are the three Baltic countries of the FSU that are highly similar to each other in basically all social and economic areas. All three of them were among the Soviet republics with the highest living standards and they were considered to be among the most economically prosperous of the NIS. In spite of their similarity, there is a major contrast in the HIV-1 epidemic rate between these countries: the HIV-1 prevalence in Lithuania is 10-fold lower than in Latvia and Estonia, which are among the most affected by HIV-1 NIS. A possible reason for this contrast might be related to the establishing of the National AIDS Center in Lithuania, which, starting in the late 1980s, has carried out numerous prevention programs,22 as well as the fact that after the declaration of independence Lithuanian citizenship was granted to all inhabitants of the country. That is in contrast to Estonia and Latvia, in which citizenship was not granted to a third of the population, leading to their marginalization.
The marked genetic variety of HIV-1 strains in Lithuania is atypical for the epidemic in the FSU, in which subtype A viruses account for around 90% of all infections, both among IDUs and heterosexually infected individuals. Yet in Lithuania these viruses are present in just 82% of IDUs, less than 50% of heterosexually infected individuals and cases with an unknown transmission route, and none of MSM. Such a variety of circulating strains was characteristic of the nascent stage of the epidemic in the FSU in the 1980s to the early 1990s, when distinct viruses were simultaneously introduced into different risk groups.23
Our study demonstrated that Lithuania is the only country in which CRF03_AB viruses are epidemiologically significant. Before this study, CRF03_AB strains were found only in single cases outside the border with the Lithuania Kaliningrad region of Russia,21 where this CRF dominates.20,21
We demonstrated that IDUs and MSM in Lithuania represent two separate epidemiological networks with different HIV-1 strains in circulation, despite the geographic and likely behavioral overlap of these populations. Such a phenomenon was previously demonstrated in The Netherlands.24,25
Analysis of the pol gene of recently registered infections demonstrated no transmission of drug-resistant viruses: only one of 27 sequences had a single resistant-associated mutation, which does not cause phenotypic resistance. The absence of transmitted drug-resistant strains might be explained by the late onset of the epidemic in Lithuania, when patients are initially treated by modern potent antiretroviral regimens.
Acknowledgments
We thank Margreet Bakker, Laboratory of Experimental Virology, Department of Medical Microbiology, Center for Infection and Immunity Amsterdam (CINIMA), Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands, for processing clinical samples. The study was financially supported by INTAS, grant 05-1000004-7749.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Nabatov AA. Kravchenko ON. Lyulchuk MG. Shcherbinskaya AM. Lukashov VV. Simultaneous introduction of HIV type 1 subtype A and B viruses into injecting drug users in southern Ukraine at the beginning of the epidemic in the former Soviet Union. AIDS Res Hum Retroviruses. 2002;18(12):891–895. doi: 10.1089/08892220260190380. [DOI] [PubMed] [Google Scholar]
- 2.Nabatov AA. Masharsky AE. Verevochkin SV, et al. The rate of epidemiological and virological changes during the transition from nascent to concentrated HIV epidemic stage in the former Soviet Union countries. AIDS Res Hum Retroviruses. 2007;23(2):183–192. doi: 10.1089/aid.2006.0006. [DOI] [PubMed] [Google Scholar]
- 3.Saad MD. Aliev Q. Botros BA, et al. Genetic forms of HIV Type 1 in the former Soviet Union dominate the epidemic in Azerbaijan. AIDS Res Hum Retroviruses. 2006;22(8):796–800. doi: 10.1089/aid.2006.22.796. [DOI] [PubMed] [Google Scholar]
- 4.Lazouskaya NV. Eremin VF. Adema KW, et al. The HIV type 1 epidemic in Belarus: Predominance of Eastern European subtype A strains and circulation of subtype B viruses. AIDS Res Hum Retroviruses. 2005;21(9):830–833. doi: 10.1089/aid.2005.21.830. [DOI] [PubMed] [Google Scholar]
- 5.Lukashov VV. Karamov EV. Eremin VF. Titov LP. Goudsmit J. Extreme founder effect in an HIV type 1 subtype A epidemic among drug users in Svetlogorsk, Belarus. AIDS Res Hum Retroviruses. 1998;14(12):1299–1303. doi: 10.1089/aid.1998.14.1299. [DOI] [PubMed] [Google Scholar]
- 6.Zarandia M. Tsertsvadze T. Carr JK, et al. HIV-1 genetic diversity and genotypic drug susceptibility in the Republic of Georgia. AIDS Res Hum Retroviruses. 2006;22(5):470–476. doi: 10.1089/aid.2006.22.470. [DOI] [PubMed] [Google Scholar]
- 7.Eyzaguirre LM. Erasilova IB. Nadai Y, et al. Genetic characterization of HIV-1 strains circulating in Kazakhstan. J Acquir Immune Defic Syndr. 2007;46(1):19–23. doi: 10.1097/QAI.0b013e318073c620. [DOI] [PubMed] [Google Scholar]
- 8.Bobkov AF. Kazennova EV. Sukhanova AL, et al. An HIV type 1 subtype A outbreak among injecting drug users in Kazakhstan. AIDS Res Hum Retroviruses. 2004;20(10):1134–1136. doi: 10.1089/aid.2004.20.1134. [DOI] [PubMed] [Google Scholar]
- 9.Balode D. Ferdats A. Dievberna I, et al. Rapid epidemic spread of HIV type 1 subtype A1 among intravenous drug users in Latvia and slower spread of subtype B among other risk groups. AIDS Res Hum Retroviruses. 2004;20(2):245–249. doi: 10.1089/088922204773004978. [DOI] [PubMed] [Google Scholar]
- 10.Ferdats A. Konicheva V. Dievberna I. Lilja E. Albert J. An HIV type 1 subtype A outbreak among injecting drug users in Latvia. AIDS Res Hum Retroviruses. 1999;15(16):1487–1490. doi: 10.1089/088922299310007. [DOI] [PubMed] [Google Scholar]
- 11.Pandrea I. Descamps D. Collin G, et al. HIV type 1 genetic diversity and genotypic drug susceptibility in the Republic of Moldova. AIDS Res Hum Retroviruses. 2001;17(13):1297–1304. doi: 10.1089/088922201750461375. [DOI] [PubMed] [Google Scholar]
- 12.Rumyantseva OA. Olkhovskiy IA. Malysheva MA, et al. Epidemiological networks and drug resistance of HIV type 1 in Krasnoyarsk region, Russia. AIDS Res Hum Retroviruses. 2009;25(9):931–936. doi: 10.1089/aid.2009.0075. [DOI] [PubMed] [Google Scholar]
- 13.Beyrer C. Patel Z. Stachowiak JA, et al. Characterization of the emerging HIV type 1 and HCV epidemics among injecting drug users in Dushanbe, Tajikistan. AIDS Res Hum Retroviruses. 2009;25(9):853–860. doi: 10.1089/aid.2008.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Novitsky VA. Montano MA. Essex M. Molecular epidemiology of an HIV-1 subtype A subcluster among injection drug users in the Southern Ukraine. AIDS Res Hum Retroviruses. 1998;14(12):1079–1085. doi: 10.1089/aid.1998.14.1079. [DOI] [PubMed] [Google Scholar]
- 15.Saad MD. Shcherbinskaya AM. Nadai Y, et al. Molecular epidemiology of HIV Type 1 in Ukraine: Birthplace of an epidemic. AIDS Res Hum Retroviruses. 2006;22(8):709–714. doi: 10.1089/aid.2006.22.709. [DOI] [PubMed] [Google Scholar]
- 16.Kurbanov F. Kondo M. Tanaka Y, et al. Human immunodeficiency virus in Uzbekistan: Epidemiological and genetic analyses. AIDS Res Hum Retroviruses. 2003;19(9):731–738. doi: 10.1089/088922203769232520. [DOI] [PubMed] [Google Scholar]
- 17.Carr JK. Nadai Y. Eyzaguirre L, et al. Outbreak of a West African recombinant of HIV-1 in Tashkent, Uzbekistan. J Acquir Immune Defic Syndr. 2005;39(5):570–575. [PubMed] [Google Scholar]
- 18.Adojaan M. Kivisild T. Mannik A, et al. Predominance of a rare type of HIV-1 in Estonia. J Acquir Immune Defic Syndr. 2005;39(5):598–605. [PubMed] [Google Scholar]
- 19.Zetterberg V. Ustina V. Liitsola K, et al. Two viral strains and a possible novel recombinant are responsible for the explosive injecting drug use-associated HIV type 1 epidemic in Estonia. AIDS Res Hum Retroviruses. 2004;20(11):1148–1156. doi: 10.1089/aid.2004.20.1148. [DOI] [PubMed] [Google Scholar]
- 20.Liitsola K. Holm K. Bobkov A, et al. An AB recombinant and its parental HIV type 1 strains in the area of the former Soviet Union: Low requirements for sequence identity in recombination. AIDS Res Hum Retroviruses. 2000;16(11):1047–1053. doi: 10.1089/08892220050075309. [DOI] [PubMed] [Google Scholar]
- 21.Lukashov VV. Huismans R. Rakhmanova AG, et al. Circulation of subtype A and gagA/envB recombinant HIV type 1 strains among injecting drug users in St. Petersburg, Russia, correlates with geographical origin of infections. AIDS Res Hum Retroviruses. 1999;15(17):1577–1583. doi: 10.1089/088922299309874. [DOI] [PubMed] [Google Scholar]
- 22.Caplinskas S. Epidemiology of HIV/AIDS in Lithuania in 1988–2001: Review of present situation and prognosis of HIV transmission trends. Medicina (Kaunas) 2004;40(2):161–168. [PubMed] [Google Scholar]
- 23.Lukashov VV. Cornelissen MTE. Goudsmit J, et al. Simultaneous introduction of distinct HIV-1 subtypes into different risk groups in Russia, Byelorussia and Lithuania. AIDS. 1995;9(5):435–439. [PubMed] [Google Scholar]
- 24.Lukashov VV. Kuiken CL. Vlahov D. Coutinho RA. Goudsmit J. Evidence for HIV type 1 strains of U.S. intravenous drug users as founders of AIDS epidemic among intravenous drug users in Northern Europe. AIDS Res Hum Retroviruses. 1996;12(12):1179–1183. doi: 10.1089/aid.1996.12.1179. [DOI] [PubMed] [Google Scholar]
- 25.Op de Coul EL. Prins M. Cornelissen M, et al. Using phylogenetic analysis to trace HIV-1 migration among western European injecting drug users seroconverting from 1984 to 1997. AIDS. 2001;15(2):257–266. doi: 10.1097/00002030-200101260-00017. [DOI] [PubMed] [Google Scholar]