Summary
Factor-induced reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) is inefficient, complicating mechanistic studies. Here, we studied defined intermediate cell populations poised to becoming iPSCs by genome-wide analyses. We show that induced pluripotency elicits two transcriptional waves, which are driven by c-Myc/Klf4 (first wave) and Oct4/Sox2/Klf4 (second wave). Cells that become refractory to reprogramming activate the first but fail to initiate the second transcriptional wave and can be rescued by elevated expression of all four factors. The establishment of bivalent domains occurs gradually after the first wave, while changes in DNA methylation take place after the second wave when cells acquire stable pluripotency. This integrative analysis allowed us to identify genes that act as roadblocks during reprogramming and surface markers that further enrich for cells prone to forming iPSCs. Collectively, our data offer new mechanistic insights into the nature and sequence of molecular events inherent to cellular reprogramming.
Introduction
Induced pluripotent stem cells (iPSCs) have been generated from a number of mouse and human cell types upon enforced expression of transcription factors such as Oct4, Klf4, Sox2, and c-Myc (OKSM)(Takahashi et al., 2007; Takahashi and Yamanaka, 2006). iPSCs provide a valuable source of patient-specific cells for the study and potential treatment of human diseases (Wu and Hochedlinger, 2011). In addition, iPSC technology offers a unique tool to dissect the principles of cell fate determination during normal development and its dysregulation in disease (Stadtfeld and Hochedlinger, 2010).
In general, less than 3% of somatic cells expressing OKSM give rise to iPSC colonies, complicating efforts to dissect the mechanisms of reprogramming. Owing to this limitation, most previous studies focused on the immediate response of somatic cells to factor expression. For example, fibroblasts were shown to go through a process that was reminiscent of a mesenchymal-to-epithelial transition (MET) within a few days of OKSM expression (Li et al., 2010; Samavarchi-Tehrani et al., 2010). At the epigenetic level, widespread remodeling of certain histone modifications, but not of DNA methylation patterns, was seen within the first few cell divisions of iPSC induction (Koche et al., 2011). However, intermediate and late stages of reprogramming have remained inaccessible for more detailed molecular analyses.
We and others have documented that fibroblasts undergoing reprogramming pass through a number of defined intermediates (Brambrink et al., 2008; Stadtfeld et al., 2008). Briefly, cells expressing OKSM from doxycycline (dox)-inducible lentiviral vectors initially downregulate the fibroblast-associated marker Thy1 (day 1–2), then activate the SSEA1 antigen (day 3–5) and eventually upregulate an Oct4-GFP reporter (day 8–10) before forming stable iPSC colonies at approximately 1.5 weeks. Importantly, isolation of these rare cell populations with the aforementioned markers allowed us to significantly enrich for cells that are poised to becoming iPSCs. Here, we have utilized this approach, in combination with a transgenic system that enables homogeneous dox-inducible OKSM expression in somatic cells (Stadtfeld et al., 2010), to purify intermediate stages of iPSC formation with the goal to elucidate the nature and sequence of molecular changes specific to cellular reprogramming.
Results
Experimental approach to studying rare reprogramming intermediates
We first determined whether the reprogramming of fibroblasts with a recently reported dox-inducible transgenic system (“reprogrammable system”)(Stadtfeld et al., 2010) generates the same subpopulations of cells that we have previously described using direct lentiviral infection (Stadtfeld et al., 2008). As shown in Figure 1A, murine embryonic fibroblasts (MEFs) carrying the Col1a1-tetO-OKSM transgene, the ROSA26-M2-rtTA allele and an Oct4-GFP knock-in reporter gave rise to Thy1− cells, SSEA1+ cells and Oct4-GFP+ cells with the expected kinetics. To verify that these intermediate populations were indeed enriched for cells that would form iPSCs, we sorted cells on feeders based on Thy1, SSEA1 and GFP expression and treated them with dox for an equal number of days (see Supplementary Experimental Procedures). Consistent with our previous report, intermediate cells with the potential to give rise to iPSCs were initially present within both, the Thy1− and SSEA1+ populations, then progressed to SSEA1+ cells and ultimately transited to the SSEA1+, Oct4-GFP+ population (Figure 1B, C). Importantly, sorting of Thy1+ cells after day 3 and of Thy1− cells after day 6 consistently failed to yield iPSC colonies, indicating that these cell populations had become refractory to reprogramming.
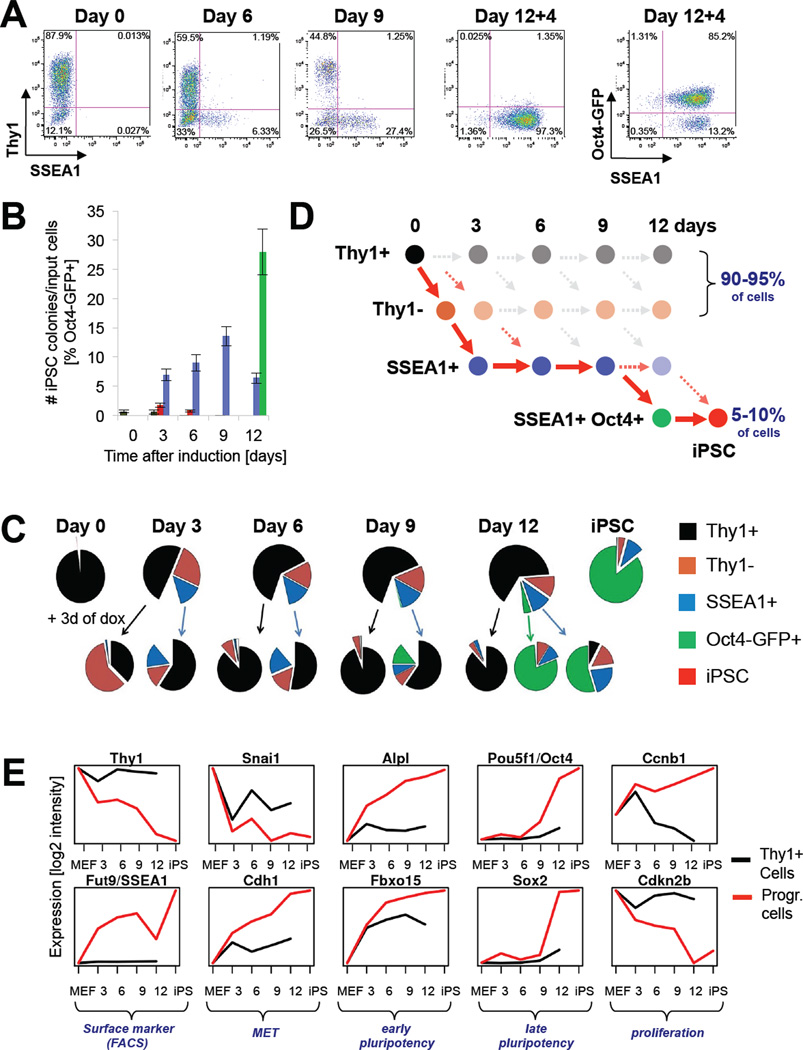
Figure 1. Strategy for isolating reprogramming intermediates.
(A) FACS analysis of reprogrammable MEFs at indicated time points. 12+4 denotes transgene-independent growth for 4 days. (B) Comparison of reprogramming efficiencies of intermediates purified at indicated time points. Note that established iPSCs have a colony formation efficiency of ~30% (Stadtfeld et al., 2008). Data are represented as mean +/− S.E.M. (n=3). (C) Pie charts summarizing FACS analysis of reprogrammable cells at indicated time points (top row). Bottom row shows FACS analysis for Thy1, SSEA1 and Oct4-GFP 3 days after sorting and plating of the above cell populations in the presence of doxycycline. (D) Scheme illustrating the different subpopulations throughout reprogramming. Solid red arrows connect cell populations progressing towards iPSCs as inferred from data in (B). (E) Expression analyses of indicated genes at day 0, 3, 6, 9, 12 of reprogramming and in established iPSCs (black lines depict Thy1+ populations; red lines depict cells undergoing successful reprogramming as defined by red arrows in Figure 1D).
To examine the phenotypic progression of reprogramming intermediates, we sorted Thy1+, SSEA1+ and Oct4-GFP+ cells after 3, 6, 9 and 12 days of dox induction, followed by culture in dox for another 3 days before re-assessing their surface phenotype (Figure 1C). This analysis, combined with the abovementioned reprogramming results (Figure 1B), documents that (i) cells undergoing successful reprogramming with the Col1a1-tetO-OKSM transgenic system transit in a linear fashion from a Thy1+ to a Thy1− to a SSEA1+ state in the first 6 days and eventually to a SSEA1+, Oct4-GFP+ state by days 9–12 (see Figure 1D for graphic summary; red arrows connect intermediates progressing towards iPSCs); (ii) SSEA1+ cells are phenotypically still plastic until days 9–12 when they undergo commitment to a stable pluripotent cell fate; and (iii) Thy1+ cells lose their ability to progress towards a Thy1− and SSEA1+ state as early as day 3. Note that progressing intermediates account for only 5–10% of cells in regular reprogramming cultures compared with 90–95% Thy1+/Thy1− cells.
We generated gene expression profiles for these intermediate cell populations as well as for non-induced Thy1+ MEFs (day 0) and dox-independent iPSCs. Examination of candidate genes confirmed that Thy1 was downregulated and SSEA1 (synthesized by the gene product of Fut9 (Kudo et al., 2004)) was upregulated in the Thy1− and SSEA1+ populations, respectively (Figure 1E; note that red line depicts progressive intermediates as defined in Figure 1D by red solid arrows while black line shows refractory Thy1+ cells). In addition, we noticed that Snai1 became downregulated while E-Cadherin was upregulated at day 3, consistent with the occurrence of a MET. Alkaline phosphatase (Alpl) and Fbxo15, early markers of pluripotent cells, gradually increased their expression whereas endogenous Oct4 and Sox2 transcripts were detectable only late during iPSC generation. Lastly, Cyclin B1 became upregulated and the CDK inhibitor Cdkn2b (encoding for p15) was downregulated early in reprogramming. It is worth mentioning that Thy1+ cells mirrored the gene expression changes of progressing cells until day 3 but then failed to sustain this trend at later time points, which correlated with their inability to produce iPSCs after day 3 (Figure 1B). We conclude that our sorting strategy allows us to analyze gene expression patterns of progressive intermediate cell populations transitioning towards iPSCs (Figure 1C), and to distinguish these from patterns in the bulk population of cells that are refractory to reprogramming.
Major gene expression changes occur in two discernible phases during iPSC formation
Principle component analysis (PCA) of the cell populations revealed a molecular connectivity reflecting their progression from the initial Thy1+ cells towards Thy1− cells and ultimately SSEA1+ cells, Oct4-GFP+ cells and iPSCs as depicted by the dashed red line (Figure 2A). PCA analysis further showed that all intermediates at day 3 clustered together, indicating that cells responded homogeneously to OKSM activation within the first few days. After day 3, however, SSEA1+ cells progressed towards Oct4-GFP cells, which were most closely related to established iPSCs, demonstrating that the SSEA1+ population gradually evolved towards a bona fide pluripotent state with time. Unsupervised clustering confirmed the similarities of SSEA1+ cells at days 3–9 and of Oct4-GFP+ cells at day 12 and iPSCs (Figure 2B). An examination of the number of differentially expressed genes between progressing (SSEA1+) and refractory (Thy1+) cell populations at each time point showed a gradual increase, which culminated at ~1,500 genes by day 12 (Figure 2C). These observations document that the isolation of subpopulations with experimentally proven distinct reprogramming potentials are distinguishable by global gene expression patterns.
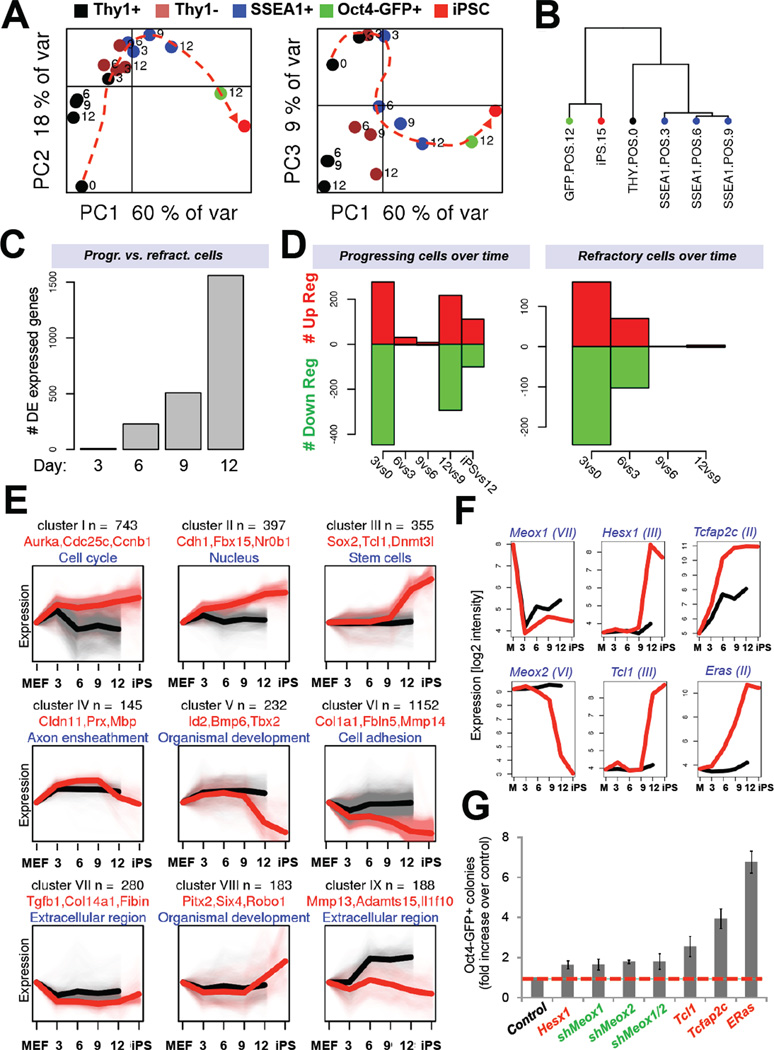
Figure 2. Gene expression dynamics during iPSC formation (see also Figure S1).
(A) Principal component analyses (PCA) of global gene expression data of FACS-sorted subpopulations at indicated time points. (B) Unsupervised hierarchical clustering of gene expression profiles of indicated cell populations. (C) Number of differentially expressed genes between Thy1+ and SSEA1+ cells at indicated time points. (D) Number of differentially expressed (DE) genes in progressing SSEA1+ cells at successive time points. Right panel shows gene expression changes in refractory Thy1+ cells. (E) Gene expression categories (I to IX) clustered by common expression changes during reprogramming (black trendlines depict gene expression patterns in Thy1+ population; red trendlines depict gene expression patterns in cells undergoing successful reprogramming as defined in Figure 1D). Each gene is only represented once per category. (F) Expression analysis of candidate genes selected from (E) for overexpression or knock-down experiments shown in (G). (G) Reprogramming potential of OKSM transgenic MEFs infected with dox-inducible lentiviral vectors expressing the indicated candidate genes or hairpins. Data are represented as mean +/− S.E.M. (n=3).
Remarkably, a comparison of relative gene expression changes among pairs of progressing cell populations at successive time points revealed two distinct waves of major gene activity (Figure 2D, left panel). The first wave occurred between days 0 and 3, while a second wave was detectable towards the end of reprogramming, after day 9. Refractory Thy1+ cells initiated the first wave but failed to undergo the second wave (Figure 2D, right panel). Gene ontology (GO) analysis showed that expression changes within the first phase involved activation of processes related to cell proliferation, metabolism, cytoskeleton organization and downregulation of genes associated with development. Genes upregulated during the second phase were associated with embryonic development and stem cell maintenance. A parallel study, which applied proteomics to study the same intermediates of reprogramming, concurs with our findings and further shows that molecular changes are highly coordinated during both phases (Hansson et al., 2012). Together, these data demonstrate that cells undergoing reprogramming into iPSCs, as defined by populations upregulating SSEA1+ at early time points and SSEA1+/Oct4-GFP at late time points, undergo a biphasic process at the transcriptional level that is separated by a period of less pronounced transcriptional change.
Defining reprogramming-specific gene expression patterns
To gain further insights into the mechanisms of iPSC induction, we next determined categories of genes that changed their expression in characteristic patterns (Figure 2E). A large number of genes became abruptly upregulated (cluster I; ~750 genes) or downregulated (cluster VI; ~1,200 genes) early in reprogramming and then remained largely unchanged until the iPSC state. Genes in these two categories were mainly involved in controlling DNA replication and cell division processes (upregulated genes) as well as cell adhesion and cell-cell contacts (downregulated genes) and account for the first transcriptional wave during reprogramming. Another category was comprised of ~400 genes that were gradually upregulated, such as the pluripotency-associated genes Alpl, Fbx15, Nr0b1, Tcfcp2l1 and Sall1 (cluster II) while roughly 350 genes were induced late during reprogramming and contained genes enriched for the categories stem cells and DNA binding (cluster III). The latter group, which contained well-known core pluripotency factors such as Nanog, Oct4 and Sox2 as well as Esrrb, Dnmt3L, Tcl1 and Nr5a2, is in part responsible for the second transcriptional wave and marks the acquisition of a stable pluripotent state.
Intriguingly, we also identified categories of genes that were either transiently up- or downregulated during iPSC formation (cluster IV and VIII, respectively) or upregulated early and downregulated late (cluster V). Genes within those categories included a number of developmental and cell type-specific regulators such as Bcl11a, Prx and Tbx21 among the transiently upregulated genes and Spp1, Pitx2 and Six4 among the transiently downregulated genes (Figure 2E).
Lastly, we hypothesized that the manipulation of dynamically regulated genes from these categories might enhance reprogramming. We selected the Akt coactivator Tcl1, the transcription factors Tcfap2c and Hesx1 and the ESC-specific Ras isoform ERas for overexpression experiments and the fibroblast-enriched genes Meox1 and Meox2 for knock-down experiments (Figure 2F). Accordingly, upregulation or downregulation of theses genes gave rise to up to seven times more Oct4-GFP+ colonies compared with control cells (Figure 2G). Collectively, these experiments prove that our gene expression categories facilitate the identification of molecules that positively or negatively influence the reprogramming process.
Gene expression patterns of refractory cells
Another category of genes (cluster IX) contained about 200 genes that were aberrantly activated in refractory Thy1+ cells (Figure 2E). Genes within this class were related to extracellular space/matrix, plasma membrane, retinoic acid binding and immune response processes (e.g., Mmp13, Rarres2, Fgf18, Fndc1, Aqp1 and 4, Il1f10, Hsd11b1 and Supplementary Figure 1A) and likely contributed to the failure of Thy1+ cells to reprogram.
To further understand the molecular reasons for the inability of Thy1+ intermediates to reprogram, we analyzed other genes that were differentially expressed between SSEA1+ and Thy1+ cells. This analysis revealed that mesenchymal genes were not properly downregulated while epithelial genes failed to be upregulated in Thy1+ cells compared to SSEA1+ cells after day 3 (Supplementary Figure 1B). We also searched for differentially expressed genes between Thy1+ and SSEA1+ cells at day 3, when overall gene expression patterns were still highly similar among all populations (Figure 2A). This analysis yielded a small number of significantly up and downregulated genes (e.g., Il6, Nup210, Bex1) that might serve as valuable early discriminators between cells that succeed or fail in reprogramming (Supplementary Figure 1C). We conclude that Thy1+ cells become refractory to reprogramming for a variety of reasons that include (i) an inability to undergo a MET, (ii) aberrant activation of differentiation-associated genes, and (iii) a failure to maintain global gene expression trends beyond day 3.
Impact of cellular heterogeneity on molecular dissection of reprogramming
We employed different strategies to determine the degree of heterogeneity among SSEA1+ cells. First, we used Fluidigm technology to perform single cell expression analysis for 26 genes (see Supplementary Figure 2A) in FACS-purified SSEA1+ intermediates at days 3, 6 and 9 as well as in day 0 Thy1+ MEFs and established iPSCs. Correspondence Analysis (COA) of all 26 genes across the different cellular groups showed that the three intermediate populations formed separate clusters that partially overlapped and gradually progressed from MEFs to iPSCs (Figure 3A). COA confirmed the early and late transcriptional waves and illustrated the variation of gene expression within each group (Figure 3B); MEFs and iPSCs showed the least variation while SSEA1+ intermediates exhibited increased variation that was, however, comparable among the three time points. Biplot analysis suggested that activation of Nr5a2, Eras, Zfp42, Esrrb, Dnmt3l and PECAM was most informative for predicting the iPSC state, followed by activation of Nanog, Lin28 and EpCAM (Figure 3C). A comparison of the expression dynamics of individual genes between SSEA1+ bulk populations and single SSEA1+ cells revealed a similar overall kinetics and allowed us to differentiate between MEF and ESC-associated genes that were either downregulated or upregulated immediately, gradually or late upon reprogramming factor expression (Figure 3D).
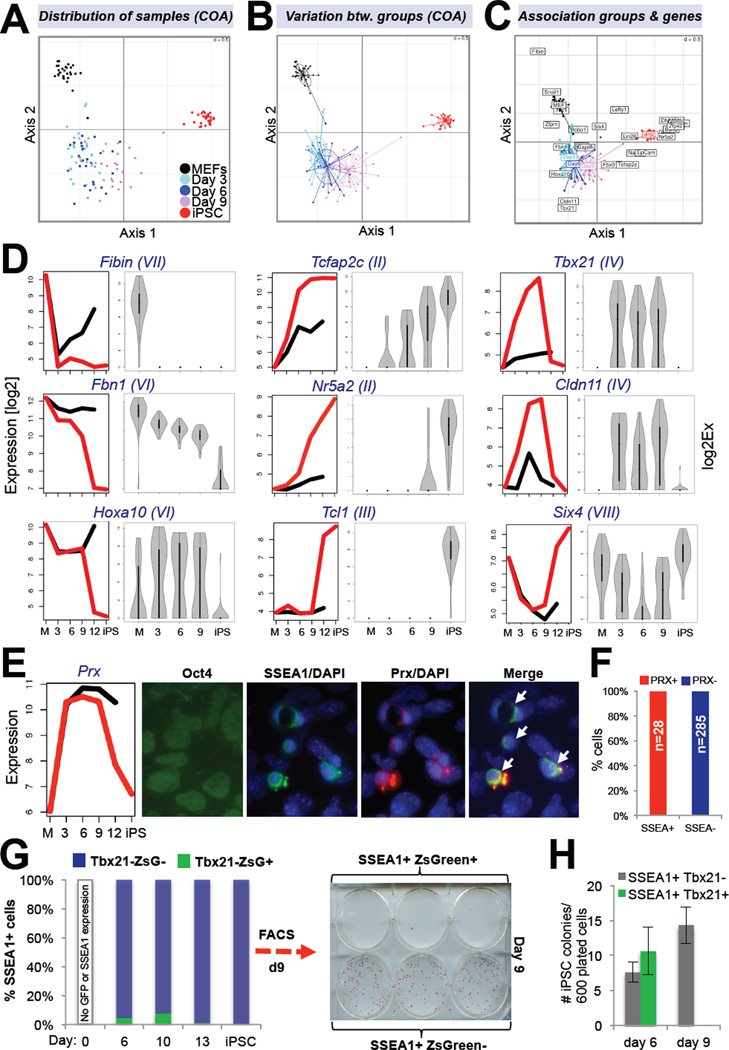
Figure 3. Characterization of cellular heterogeneity within SSEA1+ cells (see also Figure S2).
(A) Correspondence analysis (COA) of single cell expression data obtained with Fluidigm technology for 26 genes in indicated cell populations. (B) COA of same groups as shown in (A) illustrates variation in gene expression. Size of ovals indicates degree of variation. (C) Biplot displaying overlay of COA with genes associated with individual groups. (D) Comparison of Affymetrix (left) and single cell (right) expression data for 12 selected genes. Gene expression categories, as defined in Figure 2E, of selected candidates are shown in brackets. (E) Immunofluorescence for Oct4, SSEA1 and Prx on reprogrammable MEFs treated with dox for nine days. (F) Quantification of data shown in (E). (G) FACS analysis of Tbx21-ZsGreen tail fibroblasts infected with dox-inducible lentivirus expressing OKSM at indicated time points. (H) Reprogramming potentials of indicated cell populations at days 6 and 9. Data are represented as mean +/− S.E.M. (n=3).
We next examined the shape of the violin plots (Figure 3D) in order to deduce whether gene expression changes took place in a minority or majority of SSEA1+ intermediates; while unimodal plots are consistent with uniform gene expression in a majority of cells, bimodal plots are indicative of distinct expression patterns and thus heterogeneous cell populations. We could distinguish between three characteristic patterns of gene expression change (Figure 3D and Supplementary Figure 2B): (i) Exclusively unimodal expression patterns. This group was mostly characterized by genes that changed in one of the two transcriptional waves and included MEF genes that were silenced early (Fibin, Snai1) or gradually (Fbn1) as well as all examined pluripotency genes that were activated late (Zfp42, Esrrb, Nr5a2, Eras, Lin28, PECAM, Tcl1, Dnmt3l); (ii) Unimodal expression early and late with bimodal expression at intermediate stages of reprogramming. This category contained the MEF gene Zfpm and the early iPSC marker EpCAM, Nanog and Tcfap2c; (iii) Bimodal expression patterns at all time points. Examples included the MEF gene Hoxa10 and the intermediate-specific genes Cldn11, Tbx21 and Six4. Coexpression analysis of representative genes from each category showed that they were indeed activated within the same cells (Supplementary Figure 2C).
To determine whether intermediate-specific genes were always expressed heterogeneously within SSEA1+ cells (Figure 3D), we performed immunohistochemistry (IHC) for Prx (Figure 3E) in reprogrammable MEFs induced with dox for 9 days. Co-staining with antibodies recognizing SSEA1 and Prx revealed that all SSEA1+ cells also expressed Prx (28/28 examined cells)(Figure 3F). This result suggested that Prx and probably other intermediate-specific genes are expressed homogeneously among SSEA1+ cells and thus mark cells poised to becoming iPSCs. In contrast, other intermediate-specific genes such as Tbx21 are expressed in more rare subsets of SSEA1+ cells (Figure 3D) whose fate remains unclear. To test whether activation of the latter group of genes correlated with their ability to form iPSCs, we infected tail fibroblasts isolated from Tbx21-ZsGreen mice with a polycistronic viral vector expressing OKSM. Flow cytometric analysis of SSEA1+ intermediates at days 6 and 9 confirmed the heterogeneous expression pattern (Figure 3G). Plating of equal numbers of SSEA1+ Tbx21-ZsGreen+ and of SSEA1+ Tbx21-ZsGreen− cells from day 6 on feeders gave rise to roughly equal numbers of iPSC colonies, indicating that Tbx21 upregulation at this time point was neither necessary nor inhibitory for reprogramming (Figure 3H). However, SSEA1+ Tbx21-ZsGreen+ cells isolated at day 9 of reprogramming almost entirely lost their ability to form iPSC colonies, suggesting that a failure to downregulate this marker at later stages of reprogramming prohibited iPSC formation (Figure 3G, H). We conclude that our analysis of SSEA1+ bulk populations correlates well with expression patterns in individual SSEA1+ cells, thus validating our approach to study FACS-enriched intermediates of reprogramming. However, our observation that SSEA1+ cells exhibited some degree of heterogeneity warrants a search for markers that allow for further purification of reprogramming intermediates destined to form iPSCs (see last section).
Comparison with piPSCs and bulk populations expressing OKSM
Partially reprogrammed iPSCs (piPSCs) are assumed to represent intermediate stages of reprogramming (Mikkelsen et al., 2008; Sridharan et al., 2009). piPSC lines are stable cell lines that have silenced the somatic program but failed to activate the pluripotency program and depend on continuous expression of viral transgenes. To assess whether their overall gene expression signature closely resembled any of our profiled cell populations, we performed PCA analysis between our datasets and published results from six different piPSC lines. Consistent with previous observations, piPSC lines derived from distinct cell types and produced in different laboratories clustered together, suggesting a similar molecular makeup (Supplementary Figure 1D, grey symbols). Unexpectedly, these cell lines were quite distinct from any of our profiled intermediate populations along the depicted PC axes, showing essentially no overlap with one notable exception; piPSC line BIV1 (Mikkelsen et al., 2008) that was generated with dox-inducible lentiviruses clustered together with retrovirally induced piPSCs in the presence of dox (“dox+” marked triangle) but grouped with our late SSEA1+ intermediates after ~10 days of dox withdrawal (“dox−” triangle). We conclude that piPSCs originate from cells that have exited the normal reprogramming route at an early time point and became immortalized, hence showing little overlap with progressing intermediates.
A comparison of gene expression profiles from SSEA1+ intermediates with those obtained from reprogrammable “secondary” cells exposed as bulk populations to dox for 0, 4, 8, 12 or 16 days (Mikkelsen et al., 2008) further showed that the latter samples clustered most closely with Thy1+ and Thy1− cells around day 3 but not with SSEA1+ intermediates at comparable later time points (Supplementary Figure 1D, turquois triangles). This finding indicated that this previous study of bulk populations predominantly captured expression changes of cells that failed to reprogram after day 3 and underscores the importance of enriching for the rare subsets of cells that are prone to generating iPSCs, particularly at later stages of reprogramming.
Transcription dynamics predicts distinct reprogramming factor activities
We next wondered whether the biphasic transcriptional pattern could be explained by the activity of any individual or combinations of transcription factors. To this end, we compared our gene expression data with published genome-wide occupancy studies for Oct4, Sox2, Klf4, c-Myc and Nanog in pluripotent stem cells (see Supplementary Experimental Procedures). While a similar number of targets of Oct4, Sox2 and Klf4 were up- and downregulated during both transcriptional waves, targets of c-Myc were mostly upregulated (~80%) with a bias for the first wave (Supplementary Figure 3A).
Given that most pluripotency-associated genes are targets of Oct4, Sox2, Klf4 and c-Myc in ESCs/iPSCs, we next distinguished between expression changes of individual and combinatorial OKSM targets during both waves. This analysis confirmed that c-Myc alone or in combination with other factors is a dominant force behind early gene induction (shown for c-Myc and Klf4 targets in Supplementary Figure 3B). To assess the contributions from individual factors to reprogramming, we applied a mathematical approach that models and predicts transcription factor activities using network component analysis (Chang et al., 2008). As expected, c-Myc targets showed a striking upregulation during the first few days of OKSM expression, with no major changes detectable until the end of reprogramming (Figure 4A, left panel).
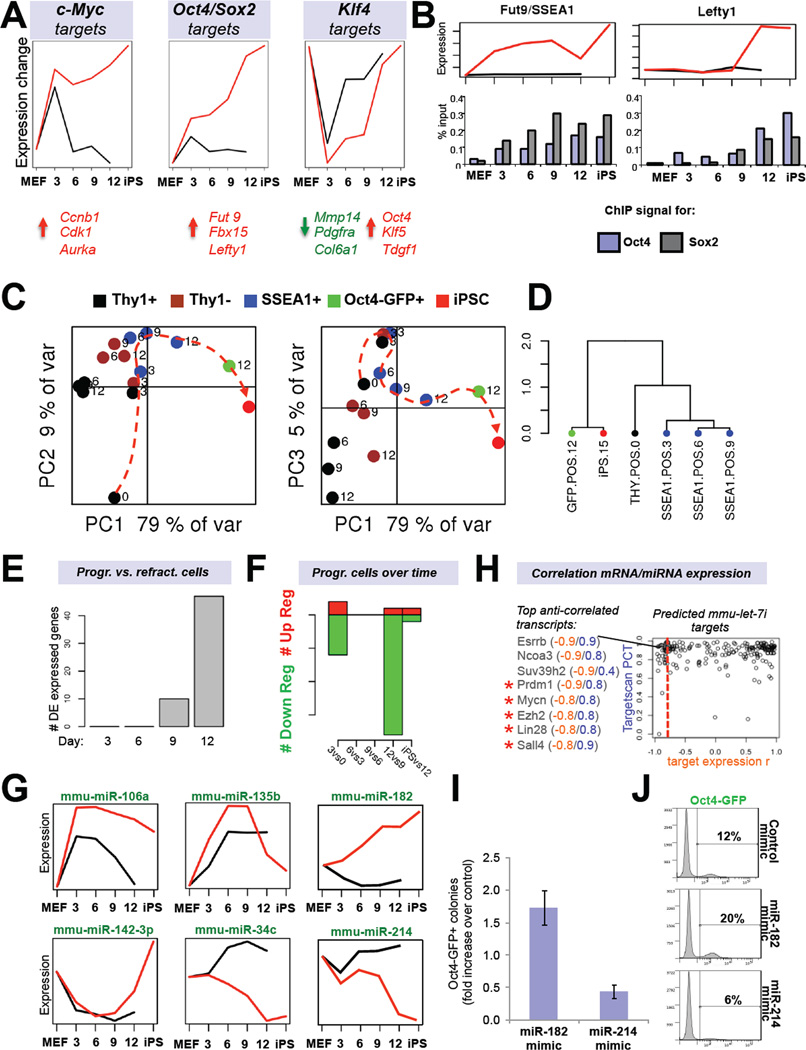
Figure 4. Predicted reprogramming factor activities and microRNA expression dynamics (see also Figure S3).
(A) Transcription factor (TF) activities for c-Myc, Klf4 and the Sox2-Oct4 dimer based on network component analysis. Shown below are examples of activated (red) or repressed (green) targets. (B) Expression dynamics of an early (Fut9; left panel) and late (Lefty1; right panel) Oct4/Sox2 target during reprogramming. Shown below is promoter ChIP analysis for Oct4 and Sox2. (C) Principle component analysis of microRNA expression data of FACS-sorted subpopulations at the indicated time points. (D) Unsupervised hierarchical clustering of indicated microRNA expression profiles. (E) Number of differentially expressed (DE) microRNAs between Thy1+ and SSEA1+ cells at indicated time points. (F) Number of differentially expressed microRNAs between progressing SSEA1+ cell populations at successive time points. (G) Examples of microRNA profiles that change in dynamic patterns. (H) Predicted target genes of let-7c (Targetscan database) are shown based on inverse expression patterns with let-7c. Examples of putative targets with an inverse expression score of −0.8 or higher are shown. Targets marked by red asterisks have previously been validated. (I) iPSC formation efficiencies and (J) Oct4-GFP FACS quantification of reprogrammable MEFs treated with mimics for miR-182 or miR-214. Data are represented as mean +/− S.E.M. (n=3).
Notably, an analysis of transcriptional activity for genes that are bound by pairs of factors in ESCs showed a gradual change, as exemplified for Oct4/Sox2 (OS) targets, suggesting that the combined activity of certain pluripotency factors is more likely to modulate targets than individual factors (Figure 4A, middle panel). To experimentally verify this mathematical prediction, we picked 3 OS targets that became upregulated early (Fut9, day 3) or late (Nanog and Lefty1, days 9–12) during reprogramming and performed chromatin immunoprecipitation (ChIP) on SSEA1+ intermediates with Oct4- and Sox2-specific antibodies, combined with real time PCR. Indeed, we found that Fut9 was occupied by both Oct4 and Sox2 as early as day 3 while Nanog and Lefty1 were occupied by Oct4 alone at early time points and by both Oct4 and Sox2 by day 12, consistent with their robust transcriptional activation (Figure 4B and Supplementary Figure 3C). The different susceptibilities of OS targets to be transcriptionally activated correlated well with an underlying permissive or repressive chromatin structure (Supplementary Figure 3D).
Klf4 was exceptional in that its targets changed their expression early and late in reprogramming with a phase of less activity during intermediate stages (days 3 to 9), supporting a possible dual role of Klf4 in early somatic gene repression and subsequent pluripotency gene activation (Figure 4A, right panel). Accordingly, regulated Klf4 targets were comprised of factors associated with differentiation, such as Tgfb1, Pdgfra and Col6a1, at early time points and of pluripotency-associated genes including Pou5f1 (Oct4), Tdgf1 and Klf5 at late time points of reprogramming. Altogether, these results suggest that the first transcriptional wave is mostly mediated by c-Myc and occurs in both progressing and non-progressing cells while the second wave is the consequence of a gradual upregulation of OS targets, ultimately leading to the activation of other pluripotency genes, including Nanog, to consolidate the pluripotent transcription factor network. Klf4 seems to support both phases by suppressing genes during the first phase and enhancing pluripotency gene expression during the second phase.
MicroRNA expression follows biphasic pattern and inversely correlates with known and predicted target mRNAs
Similar to the expression analysis for coding genes, miRNA expression analysis allowed us to cluster cell populations into different groups based on their phenotype by using PCA and unsupervised clustering (Figure 4C–E). Pairwise comparisons of progressing SSEA1+ populations at successive time points again revealed two transcriptional waves, which both showed an over representation of downregulated versus upregulated miRNAs (Figure 4F). miRNAs changed their expression in similar patterns to mRNAs over the course of reprogramming (see Figure 4G for representative examples). Moreover, miRNAs that have previously been documented to inhibit (e.g., let-7, miR-34c) or promote (e.g., miR-294, miR-106a)(Huo and Zambidis, 2012) iPSC formation, showed the expected downregulation and upregulation, respectively, in progressing intermediates (Figure 4G and Supplementary Figure 3E). We conclude that forced expression of OKSM controls the expression of both coding and non-coding loci in a similar fashion.
A comparison of miRNAs and their known targets indicated an inverse correlation (Supplementary Figure 3E). This is exemplified for miR-294, which targets TgfbR2, and for let-7, which targets Lin28 (Subramanyam and Blelloch, 2011). To extend this analysis beyond well-established miRNA-mRNA pairs, we built a table that links the expression changes during reprogramming of all differentially expressed miRNAs to their putative mRNA targets (Figure 4H, Supplementary Figure 3F and Supplementary Information). Indeed, the previously validated let-7c targets Lin28, N-Myc and Sall4 and the miR-294 targets Lats2, TgfbR2 and Akt1 exhibited high negative correlation scores (Supplementary Figure 3F). This analysis further suggested that the pluripotency factor Esrrb, the histone methyltransferases Suv39h1/2 and the coactivator Ncoa3, all of which are implicated in cellular reprogramming, are likely targets of let-7c while the documented iPSC-inhibitory factor Prrx1 (Yang et al., 2011) is predicted to be targeted by miR-294.
Unexpectedly, miR-302a, whose forced expression was also shown to enhance iPSC generation in the context of the Yamanaka factors (Subramanyam and Blelloch, 2011), exhibited transient activation specifically in Oct4-GFP+ cells at day 12 but remained otherwise unchanged (Supplementary Figure 3G, H). Mir-302a is normally expressed in mouse epiblast stem cells (Huo and Zambidis, 2012) but barely detectable in mouse ESCs, suggesting that iPSC induction might entail a transient passage through an epiblast-like state before reaching naiive pluripotency. In agreement with this idea, we detected transient downregulation of a number of putative mir-302a targets in Oct4-GFP+ cells such as Rbl2, Rab11fip5, Rbbp6 and transient, albeit modest, upregulation of epiblast stem cell-associated markers including Brachyury (T), Cer1, Foxa2, Eomes and Fgf5 (Supplementary Figure 3G, I). It remains to be tested whether activation of miR-302a and associated transcripts takes place in all or only a subset of Oct4-expressing intermediates.
We finally wondered whether our miRNA expression data would allow us to identify novel modulators of reprogramming. Indeed, gain-of-function of miR-183 by mimics increased while that of miR-214 decreased iPSC formation, consistent with their transcriptional changes during reprogramming (Figure 4G, I, J).
Differential chromatin states provide an epigenetic logic for early, gradual and late gene regulation
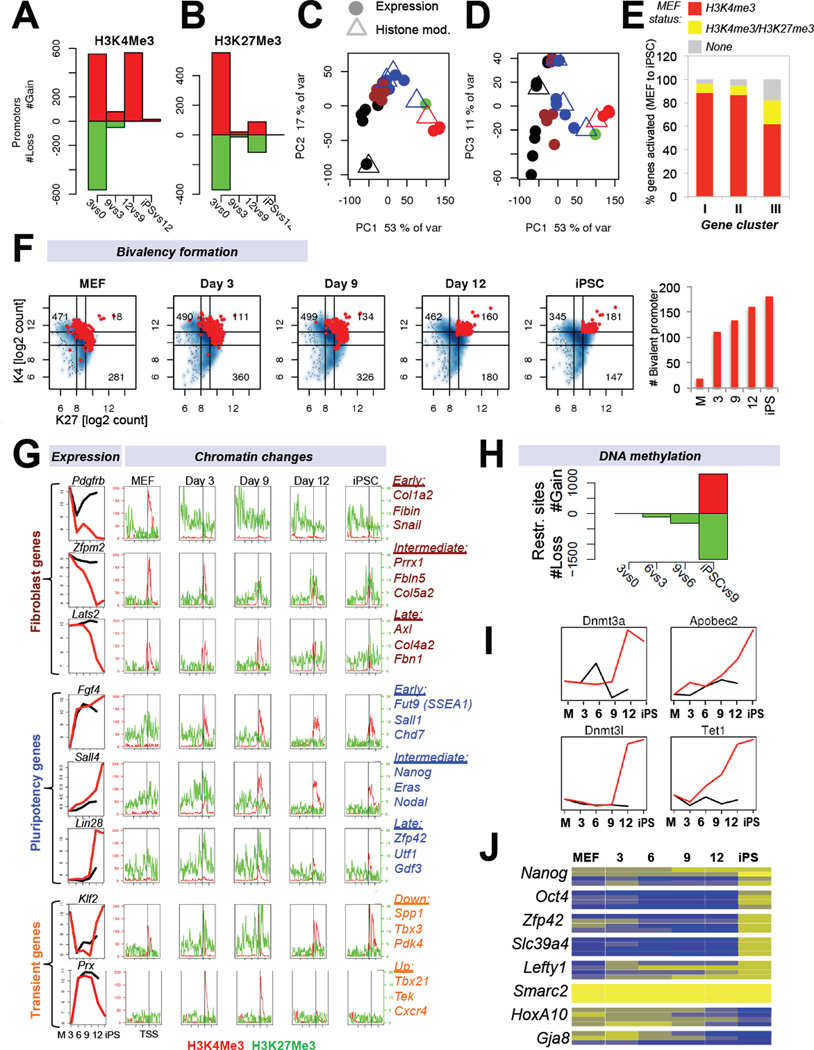
We and others have previously shown that both transcriptional and DNA/histone methylation patterns are reset from a somatic state to a pluripotent state upon reprogramming into iPSCs (Maherali et al., 2007; Mikkelsen et al., 2008). However, it is unknown when these epigenetic changes occur during reprogramming and whether there is a hierarchy in their establishment and erasure, respectively. In an attempt to resolve these questions, we analyzed active and repressive histone methylation marks (histone H3 lysine 4 and lysine 27 trimethylation, H3K4me3/H3K27me3) and DNA methylation patterns at a genome-wide scale in SSEA1+ cells throughout reprogramming. Analysis of H3K4me3 and H3K27me3 ChIP-seq patterns in progressing intermediates showed two waves (Figure 5A, B), which coincided with the observed mRNA and miRNA phases (Figure 5C, D). Kinetic analysis of bivalency formation (H3K4me3/H3K27me3 enriched promoters)(Bernstein et al., 2006) at genes that changed expression during reprogramming showed an initial burst of ~110 targets by day 3, which gradually increased to ~130 at day 9, ~160 by day 12 and ~180 in established iPSCs (Figure 5F, Supplementary Figure 4A). Thus, activating and repressive histone marks individually exhibit a biphasic pattern akin to coding and non-coding genes, whereas the establishment of differentially expressed bivalent promoters is a more gradual process.
Figure 5. Histone and DNA methylation dynamics during cellular reprogramming (see also Figure S4).
(A, B) Enrichment for H3K4me3 or H3K27me3 at promoters of differentially expressed genes in progressing intermediates. (C, D) Superimposition of principal component analyses for genes enriched in H3K4me3 (C) or H3K27me3 (D)(triangles) with gene expression data (circles) of the same cell populations (see Figure 2A for color coding). (E) Display of activated genes from gene expression categories I, II and III (see Figure 2E) in relation to their chromatin status in MEFs. (F) Number of differentially expressed genes that become bivalent (H3K27me3 and H3K4me3 enriched) during reprogramming (red dots = bivalent promoters) and quantification. (G) Integration of gene expression and histone modifications data defines subsets of genes with characteristic expression changes. Shown are examples of fibroblast-associated (top), pluripotency-associated (center) and transiently changing genes (bottom). (H) Number of genes, which change DNA methylation status in progressing cell populations during reprogramming as determined by genome-wide methylation analysis. (I) Expression dynamics of candidate genes associated with DNA methylation and demethylation. (J) Heatmap of DNA methylation analysis of specific CpGs (boxes) in the promoter regions of indicated genes during reprogramming using EpiTYPER DNA methylation analyses. Yellow indicates 0% methylation and blue represents 100% methylation.
An examination of histone marks at genes that changed their transcription in characteristic patterns allowed us to study their underlying chromatin dynamics (Figure 5G). Consistent with the different gene expression categories (Figure 2E), we were able to distinguish between genes that changed their H3K4me3 and H3K27me3 status early (day 3), at intermediate stages (day 6–9) or late (day 12). For example, the fibroblast-associated gene Pdgfrb was downregulated by day 3, which coincided with the early loss of H3K4me3 and subsequent acquisition of H3K27me3 marks, suggesting efficient access of this locus by chromatin silencers. In contrast, the MEF-expressed gene Zfpm2 showed a gradual decrease in H3K4me3 marks and a concomitant increase in H3K27me3 marks, resulting in a bivalent state and transcriptional silencing around day 9, whereas the Lats2 gene became decorated by H3K27me3 and was transcriptionally silenced only by day 12.
Similar to the deposition of H3K27me3 marks and the concomitant silencing of MEF-specific genes, we observed distinct classes of pluripotency genes that gained H3K4me3 and lost H3K27me3 at different time points (e.g., Fgf4, Sall4, Lin28)(Figure 5G). Lastly, genes that changed their expression transiently acquired H3K4me3 or H3K27me3 in a temporal manner (e.g., Prx, Klf2)(Figure 5G). We deduce from these results that the kinetics of silencing of MEF genes and activation of ESC genes is determined by a combination of parameters, including the type and complexity of underlying histone modifications as well as the availability and accessibility of transcription factors to regulate a given target (see Figure 4B). Indeed, the vast majority of genes (~90%) that were activated early or gradually (categories I and II; see Figure 2E) already carried activating H3K4me3 marks in MEFs (Figure 5E and Supplementary Figure 4B), thus complementing and expanding observations made in a previous study (Koche et al., 2011). In contrast, genes that were activated late (category III, e.g., Oct4, Nanog) are often unmarked (~15% of genes) or bivalent (~15%) in MEFs, suggesting that loci associated with these chromatin patterns are more resistant to transcriptional activation.
DNA methylation patterns are reset late in reprogramming
In contrast to gene expression and histone modification patterns, genome-wide promoter DNA methylation changes occurred predominantly late in reprogramming as determined by HELP analysis (Figure 5H). Equal numbers of methylated restriction sites were gained and lost after day 9, indicating that a comparable number of loci became methylated and demethylated, respectively. In agreement, we found that enzymes implicated in DNA methylation and demethylation, such as Dnmt3a/b, Dnmt3L, Apobec2 and Tet1 were transcriptionally upregulated late and specifically in SSEA1+ cells (Figure 5I). To confirm these global changes of DNA methylation at single base-resolution, we investigated promoter methylation levels at a number of candidate loci by mass array EpiTYPEr on genomic DNA isolated from SSEA1+ intermediates. We found that pluripotency-associated genes, such as Nanog, Oct4 and Zfp42 (Rex1) became demethylated very late during reprogramming (~days 9–12)(Figure 5J). Similarly, genes that are normally methylated in pluripotent cells but demethylated in fibroblasts, including HoxA10 and Gja8, became de novo methylated late.
Rescue of refractory cells by increased OKSM expression
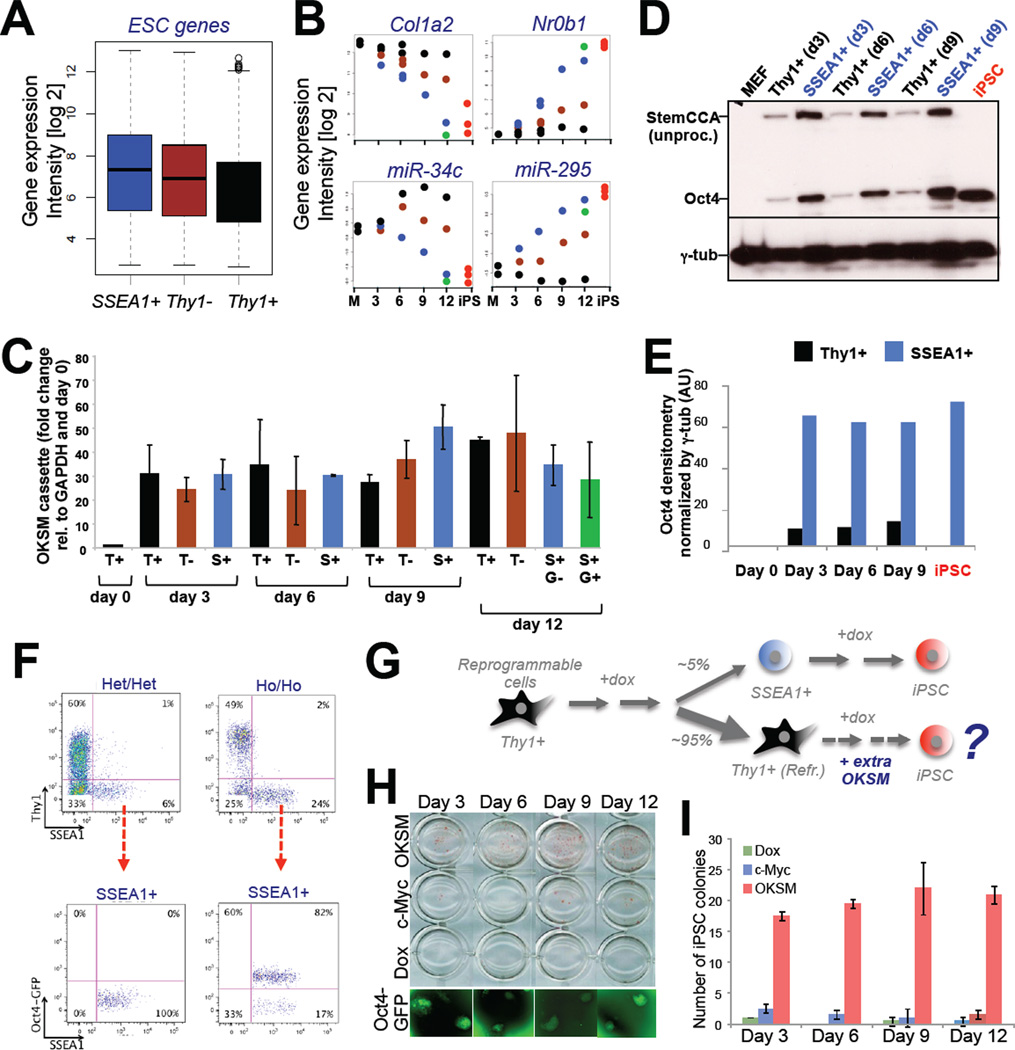
A comparison of gene expression intensities among the different subpopulations showed that Thy1− and Thy1+ cells generally failed to regulate ESC-enriched and MEF-enriched mRNAs and miRNAs to the same extent as SSEA1+ cells (Figure 6A, B). Although this observation cannot be explained by differential transcription of the OKSM transgene in these subpopulations (Figure 6C), we were surprised to detect substantially increased protein levels for Oct4, which represents activity of the entire OKSM polycistronic cassette, in SSEA1+ cells compared with Thy1+ cells (Figure 6D, E). Consistently, we observed a 20–25% reduction in the number of Thy1+ and Thy1− cells and a concomitant 400% increase in the number of SSEA1+ cells when inducing reprogrammable MEFs carrying two copies of the OKSM cassette and Rosa26-M2rtTA allele (Ho/Ho), respectively, with dox compared with MEFs that only contained one copy of each transgene (Het/Het)(Figure 6F).
Figure 6. Rescue of refractory Thy1+ cells by increased OKSM expression.
(A) Box plot depicting average expression levels in Thy1+ cells, Thy1− cells and SSEA1+ cells of genes that are significantly upregulated between MEFs and iPSCs. (B) Expression dynamics of indicated MEF-associated and ESC-associated transcripts in Thy1+, Thy1− and SSEA1+ cells (see Figure 2A for color coding). (C) Exogenous Oct4 expression levels, normalized to GAPDH for the indicated cell populations and time points. (D) Western blot analysis for Oct4 and gamma-tubulin (γ-tub) for the indicated cell populations and time points. Higher molecular weight band for exogenous Oct4 compared with endogenous Oct4 (iPSC) reflects unprocessed protein originating from the polycistronic construct as described previously by Carey, Jaenisch and colleagues (Cell Stem Cell, 2011). (E) Densitometric quantification of Western blot analysis shown in (D). AU, arbitrary units. (F) FACS analysis of reprogrammable fibroblasts carrying one (Het/Het) or two (Ho/Ho) copies each of the OKSM cassette and Rosa26-M2rtTA allele (top row). Bottom row shows FACS analysis for SSEA1 and Oct4-GFP of the same samples. (G) Experimental outline to rescue refractory Thy1+ cells by supplying viral copies of OKSM. (H) Alkaline phosphatase stained colonies obtained after infecting Thy1+ reprogrammable cells at indicated days with a dox-inducible vector expressing OKSM. Controls were uninfected, dox-treated Thy1+ cells (“Dox”) and Thy1+ cells infected with c-Myc virus alone. Representative Oct4-GFP+ colonies are shown at the bottom. (I) Quantification of results in (G). Data are represented as mean +/− S.E.M. (n=3).
To test whether elevated OKSM protein levels could rescue refractory Thy1+ cells at different stages of reprogramming, we infected Thy1+ cells isolated at days 3, 6, 9 and 12 of dox induction from Het/Het reprogrammable MEFs with viral vectors expressing additional copies of OKSM (Figure 6G). Remarkably, Thy1+ cells receiving extra copies of OKSM, but not untreated control cells or cells infected with c-Myc vector alone, gave rise to a substantial number of Oct4-GFP+ iPSC colonies (Figure 6H, I). This result thus documents that the inability to sustain OKSM protein expression in Thy1+ cells on or after day 3 contributes to their failure to form iPSCs.
Identification of molecules to enrich for cells poised to becoming iPSCs
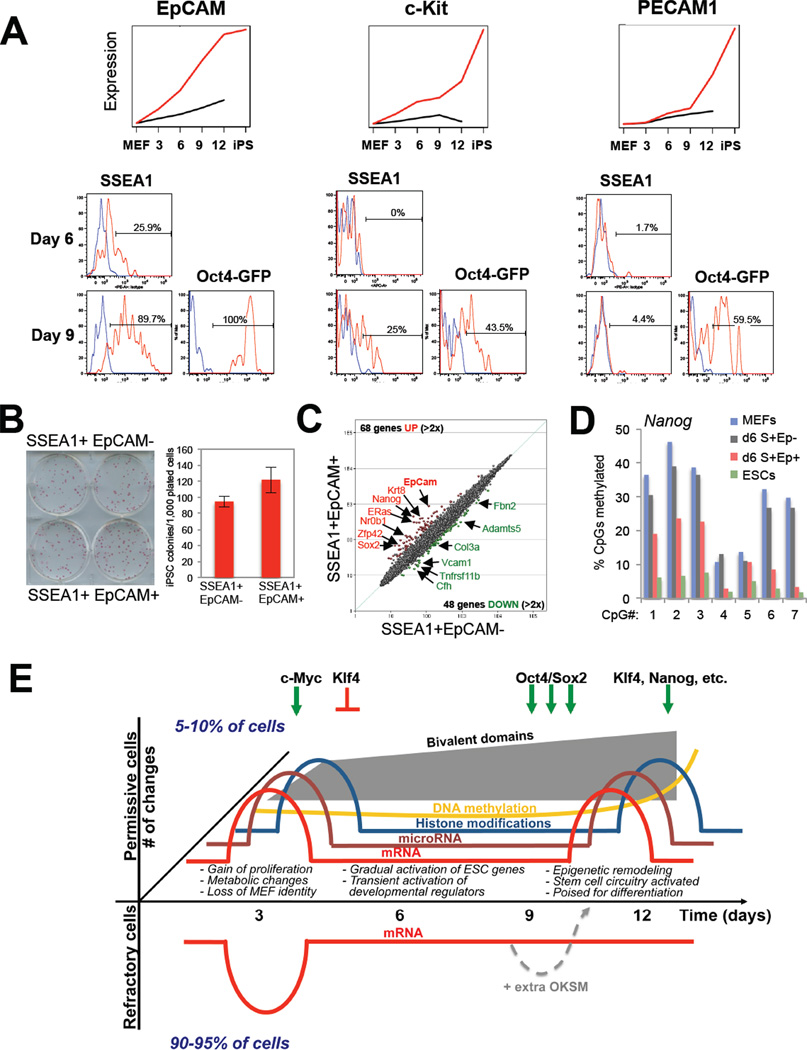
In a last set of experiments, we aimed to identify new surface markers that would allow further enrichment for subpopulations of cells undergoing reprogramming in comparison with Thy1, SSEA1 and Oct4-GFP expression. We focused on the molecules c-Kit, EpCAM and PECAM1 because of their expression patterns specifically in SSEA1+ intermediates (EpCAM, early gene; c-Kit, intermediate gene; PECAM1, late gene)(Figure 7A and Supplementary Figure 5A). Notably, EpCAM+ cells were first detectable at day 6 in a fraction (~25%) of SSEA1+ cells. In contrast, c-Kit became upregulated only by day 9 in ~25% of SSEA1+ cells while PECAM1 was detectable exclusively in SSEA1+, Oct4-GFP+ cells at day 9. Altogether, these results show that EPCAM, c-Kit and PECAM1 become activated at successive time points in subsets of SSEA1+ cells. This experiment further documented that SSEA1+ intermediates that activated the endogenous Oct4(-GFP) locus were generally more homogeneous for these three markers than are SSEA1+ Oct4-GFP− cells at day 6 and day 9.
Figure 7. Identification of surface markers to enrich for reprogramming intermediates, and model (see also Figure S5).
(A) Expression data and histogram plots of FACS analysis for c-Kit, EpCAM and PECAM1 in SSEA1+ and Oct-GFP+ populations at the indicated days of reprogramming. Red lines depict antibody-specific signal, blue lines show signal obtained with isotype control. No expression was seen before day 6. (B) Potential of EpCAM subpopulations at day 6 to form iPSC colonies. (C) Affymetrix expression analysis of EpCAM subpopulations. (D) Methylation analysis of Nanog promoter by bisulfite sequencing of ESCs, MEFs and intermediates shown in (B). S, SSEA1; Ep, EpCAM. (E) Model summarizing the presented data. Permissive cell populations (positive y-axis) show biphasic pattern of mRNA/miRNA expression and individual histone marks. Bivalent domains are generated gradually after an initial burst. DNA methylation changes occur predominantly at the end of reprogramming. Forced expression of OKSM in refractory cells (negative y-axis)can rescue their ability to form iPSCs. c-Myc/Klf4 mostly drive the first phase while Oct4/Sox2/Klf4 drive the second phase.
To assess the functional value of these markers, we sorted SSEA1+ EpCAM− and SSEA1+ EpCAM+ cells 6 days after dox induction and plated equal numbers on feeders for another 8 days in the presence of dox. Counting of dox-independent alkaline phosphatase-positive colonies 5 days later showed a modest but significant increase in reprogramming efficiency among SSEA1+ EpCAM+ intermediates (Figure 7B). In agreement, expression analysis of both subpopulations revealed subtle differences with 68 genes being upregulated and 48 genes being downregulated more than 2-fold (Figure 7C). Upregulated genes included Nanog (11-fold), ERas (7-fold), Sox2, Nr0b1, Sall4, Nr5a2 and Tdgf1 (3–5-fold). Surprisingly, the core transcription factor Oct4 was not differentially expressed, which is consistent with the absence of Oct4-GFP signal at this time point. At the epigenetic level, Nanog promoter methylation levels were reduced by 50% whereas Oct4 promoter methylation levels decreased only mildly (Figure 7D and Supplementary Figure 5B). These findings thus suggest that there may be a hierarchy in the activation of core pluripotency factors within SSEA1+ cells with Nanog being activated before Oct4.
Discussion
Our results constitute the first comprehensive analysis of transcriptional and epigenetic changes in phenotypically defined intermediates of iPSC induction. These data have elucidated the identity and order of molecular changes inherent to transcription factor-induced reprogramming (see Figure 7 for summary of observations) and provide a rich resource of data to further dissect the mechanisms of cell fate determination. Our findings suggest that the reprogramming of somatic cells follows a similar sequence of epigenetic changes as is seen during normal somatic cell differentiation; differentiating cells are thought to undergo transcriptional and histone modification changes before DNA methylation changes (Jones, 2012). It will be interesting to assess whether the rather abrupt loss of methylation after day 9 is solely the consequence of a replication-dependent passive process or also involves active demethylation. Notably, methylation changes coincided with the acquisition of a stably reprogrammed state and are in line with the interpretation that methylation patterns stably lock in the reprogrammed state.
Our molecular analysis allowed us to define nine categories of dynamically expressed genes, which characterize distinct stages of reprogramming and whose overexpression or knockdown enhanced iPSC formation. We surmise that a failure to activate these (Figure 2F) and related genes constitute roadblocks of reprogramming and is part of the reason why iPSC formation is inefficient and takes relatively long. The observed transient activation/repression of developmental regulators may indicate that the OKSM proteins aberrantly activate/repress these targets or, alternatively, that (some) reprogramming intermediates undergo a transient phase of transdifferentiation or dedifferentiation as part of the reprogramming process. Our results might thus explain recent successes in deriving epiblast stem cells, neural progenitors or cardiomyocytes directly from fibroblasts upon brief expression of OKSM and exposure to culture conditions conducive of the respective cell type (Orkin and Hochedlinger, 2011). This finding could be potentially exploited to generate other desired cell fates directly from fibroblasts.
We have developed a mathematical model that faithfully predicts activation of OKSM targets in the course of reprogramming. Our results provide a transcriptional logic for the previously seen early requirement for c-Myc (Sridharan et al., 2009) and the late requirement for Sox2 (Chen et al., 2011) during reprogramming and suggest an unanticipated dual function for Klf4 by predominantly repressing somatic targets early and activating pluripotency targets late in iPSC formation. Our finding that Thy1+ refractory cells produce less OKSM protein and thus fail to properly regulate target gene expression compared with SSEA1+ cells warrants further examination. One plausible molecular explanation is that the OKSM factors are prone to more ubiquitination-mediated degradation in Thy1+ cells (Buckley et al., 2012).
We recognize the fact that SSEA1+ cells, while enriched for cells poised to forming iPSCs, still exhibit some degree of heterogeneity. Single cell analysis of 26 genes as well as FACS analyses of 3 additional genes (EpCAM, c-Kit and PECAM) documented that gene expression changes occur more homogeneously at early (0–3 days) and late time points (day 9 onwards) while they are more heterogeneous at intermediate stages (days 6–9). There is some debate as to whether reprogramming entails a hierarchic/deterministic or probabilistic/stochastic process (Yamanaka, 2009). A previous study identified an early deterministic phase of reprogramming (Smith et al., 2010) while another recent report concluded that reprogramming involves an early stochastic and a late deterministic phase (Buganim et al., 2012). Our data may explain both observations and we therefore suggest that iPSC formation follows an early and late deterministic phase, which is separated by a more probabilistic phase. With the aid of new surface marker, such as EpCAM, PECAM and c-Kit, or novel reporter alleles, it may be possible to identify rare intermediates that progress towards iPSCs in a purely deterministic manner.
Experimental Procedures
Reprogramming experiments
PSCs were derived from murine murine embryonic fibroblasts (MEFs) or tail tip fibroblasts of reprogrammable mice (Stadtfeld et al., 2010) or directly infected with polycistronic lentivirus expressing Oct4, Sox2, Klf4 and c-Myc as described in detail in Supplementary Experimental Procedures.
Flow cytometry and immunofluorescence
Flow cytometry for GFP, Thy1, SSEA1, EpCAM, PECAM and c-Kit was done as reported previously (Stadtfeld et al., 2008) and/or as described in Supplementary Experimental Procedures.
RNA analysis
Gene expression profiling was performed using Affymetrix arrays. MicroRNA profiling was obtained using the Exiqon platform. Details are given in Supplementary Experimental Procedures.
Single cell expression analysis
Single cell expression analysis was performed using Fluidigm technology for 26 genes using Taqman probes as described in Supplementary Experimental Procedures and Supplementary Table 1.
HELP and MassARRAY EpiTyping DNA methylation analysis
These analyses were performed as described before (Polo et al., 2010). Details are given in the Supplementary Experimental Procedures and Supplementary Table 1.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was essentially as described previously (Polo et al., 2010). Please see Supplementary Experimental Procedures and Supplementary Table 1 for details.
ChIP-Seq Analysis
ChIP products from H3K4Me3 and H3K27Me3 pulldowns were subjected to hight throughput sequencing using the GA/IIX Illumina platform. Details are given in the Supplementary Experimental Procedures.
Statistical and Bioinformatic Analyses
Supplementary Material
Highlights.
Transcriptional analysis of iPSC formation reveals biphasic process.
c-Myc/Klf4 drive first phase while Oct4/Sox2/Klf4 drive second phase.
Bivalent genes form gradually whereas DNA methylation changes occur late.
Refractory cells can be rescued by elevated reprogramming factor expression.
Acknowledgements
We thank members of the Hochedlinger and Ramaswamy labs for helpful suggestions and critical reading of the manuscript. We thank Laura Prickett and Kat Folz-Donahue at the MGH/HSCI flow cytometry core and Flowcore Monash for expert cell sorting, Paul Lacaze, Danielle Evans and Michelle Garred at Millenium Science for support with the Fluidigm experiments and Lucy Dagostino from Lifetech Australia for help in miRNA assays. Support to J.P. was from an ECOR postdoctoral fellowship, Monash Larkins Program and a NHMRC CDF. J.Z. was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH, USA. K.H. was supported by the NIH (DP2OD003266 and R01HD058013). S.R. was supported by an HHMI Physican-Scientist Early Career Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential Expression of Pluripotency Markers during Direct Reprogramming of Mouse Somatic Cells. Cell stem cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell stem cell. 2012 doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Ding Z, Hung YS, Fung PC. Fast network component analysis (FastNCA) for gene regulatory network reconstruction from microarray data. Bioinformatics. 2008;24:1349–1358. doi: 10.1093/bioinformatics/btn131. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu J, Yang J, Chen Y, Chen J, Ni S, Song H, Zeng L, Ding K, Pei D. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell research. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Reports. 2012 doi: 10.1016/j.celrep.2012.10.014. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo JS, Zambidis ET. Pivots of pluripotency: The roles of non-coding RNA in regulating embryonic and induced pluripotent stem cells. Biochimica et biophysica acta. 2012 doi: 10.1016/j.bbagen.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell stem cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Kaneko M, Iwasaki H, Togayachi A, Nishihara S, Abe K, Narimatsu H. Normal embryonic and germ cell development in mice lacking alpha 1,3-fucosyltransferase IX (Fut9) which show disappearance of stage-specific embryonic antigen 1. Molecular and cellular biology. 2004;24:4221–4228. doi: 10.1128/MCB.24.10.4221-4228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell stem cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell stem cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nature biotechnology. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Nachman I, Regev A, Meissner A. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nature biotechnology. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & development. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell stem cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanyam D, Blelloch R. From microRNAs to targets: pathway discovery in cell fate transitions. Current opinion in genetics & development. 2011;21:498–503. doi: 10.1016/j.gde.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nature cell biology. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Yang CS, Lopez CG, Rana TM. Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem cells (Dayton, Ohio) 2011;29:1528–1536. doi: 10.1002/stem.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.