The order Zingiberales comprises ∼2500 species of tropical to subtropical plants, including agriculturally (e.g. banana, ginger) and horticulturally (e.g. cannas, heliconias, bird-of-paradise) important plants. Throughout the evolution of this order, the stamens have been modified from the ancestral filamentous structures that produce pollen (seen in Banana flowers) to petal-like structures that no longer bear pollen sacs (seen in Canna flowers). This results in a reduction of pollen, but an effective increase in the overall size of the floral display and perhaps in the efficacy of specialized pollinators by converting stamens into ‘petals’. This study investigates the genetic mechanisms that are involved in making petal-like structures in place of pollen-producing stamens.
Keywords: Canna, evo-devo, floral development, MADS-box genes, petaloid stamens, petaloidy, Zingiberales
Abstract
Flowers of the order Zingiberales demonstrate a remarkable trend of reduction in the number of fertile stamens; from five or six fertile, filamentous stamens bearing two thecae each in Musaceae and Strelitziaceae to just a single petaloid stamen bearing a single theca in Cannaceae and Marantaceae. As one progresses from ancestral to derived floral forms, 5–6 fertile stamens are replaced by 4–5 petaloid staminodes. In Cannaceae and Costaceae, all members of the androecial whorls exhibit petaloidy, including the fertile stamen. In Costaceae, a single fertile stamen develops two thecae embedded on a broad petaloid appendage, while in Cannaceae the single fertile stamen is further reduced to a single theca with a prominent, expanded petaloid appendage. Whether petaloidy of the fertile stamen is a synapomorphy of the entire ginger clade (including Cannaceae, Costaceae, Zingiberaceae and Marantaceae), or the result of independent convergent evolution in Cannaceae, Costaceae, and some Zingiberaceae, is unclear. We combine a developmental series of the formation of the petaloid fertile stamen in Canna indica with data on the expression of B- and C-class floral organ identity genes to elucidate the organogenetic identity of the petaloid stamen and staminodes. Our data indicate that the single fertile theca in C. indica and its petaloid appendage are derived from one-half of the primordium of a single stamen, with no contribution from the remaining part of the stamen (i.e. the second theca primordium) which aborts early in development. The petaloid appendage expands later, and develops from the position of the filament/connective of the developing theca. Floral identity gene expression shows that petal identity genes (i.e. B-class genes) are expressed in all floral organs studied while C-class gene AG-1 is expressed in an increasing gradient from sepals to gynoecium, and AG-2 is expressed in all floral organs except the petals. The canonical model for molecular specification of floral organ identity is not sufficient to explain petaloidy in the androecial whorl in Canna sp. Further studies understanding the regulation of gene networks are required.
Introduction
The Zingiberales are a group of herbaceous tropical monocots comprising eight families and ∼2000 species. They diverged from their sister order Commelinales (Bremer et al. 2009) ∼80 million years ago. In Zingiberales, the flowers are organized into five distinct whorls of three organs each: calyx (consisting of three sepals), corolla (consisting of three petals), two androecial whorls for a total of six (three inner and three outer) stamens and the tripartite gynoecium (Kirchoff 1983).
The Zingiberales order has been traditionally divided into two groups based on overall floral morphology: the banana families, including families Musaceae, Lowiaceae, Strelitziaceae and Heliconiaceae, and the derived ginger families, a monophyletic lineage containing families Costaceae, Zingiberaceae, Marantaceae and Cannaceae (Fig. 1A). Most major evolutionary changes in floral morphology that define these two groups occur in the petal and stamen whorls. In particular, there is an impressive reduction in the number of fertile stamens across the order, from 5–6 fertile stamens in the banana families to a single fertile stamen in Costaceae and Zingiberaceae, and a half fertile stamen in Cannaceae and Marantaceae (Kirchoff et al. 2009). In the flowers of the ginger families, three to five infertile members of the androecial whorls develop as sterile petaloid structures (Kirchoff 1991).
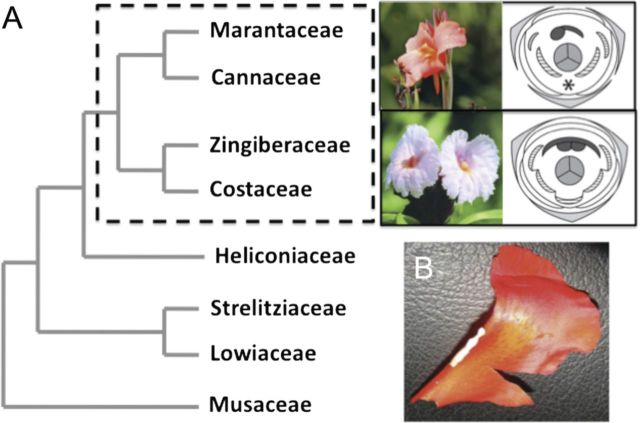
Figure 1.
Phylogenetic context for studying comparative organogenesis in Zingiberales. (A) Zingiberales phylogeny according to molecular and morphological characteristics (Kress 1990; Kress et al. 2001). The dashed square highlights the ginger clade, comprising a monophyletic group of four families (Costaceae, Zingiberaceae, Cannaceae and Marantaceae). Photographs: C. indica (top); Costus spicatus (bottom). On the right, floral diagrams representative of flowers of the Cannaceae (top) and Costaceae (bottom) families. Light grey, sepals; white, petals; hashed, petaloid staminodes; dark grey, fertile stamen; *, aborted stamen; centre grey, gynoecium. (B) Canna indica half fertile stamen with petaloid appendage.
In most Zingiberales flowers, the fertile stamens produce two mature pollen sacs or thecae. In the banana families, these fertile stamens have a narrow connective and thus are filamentous in form. Any petaloid members of the androecial whorls of the banana families are infertile, completely lacking thecae (Kirchoff et al. 2009). However, in the ginger clade a petaloid appendage can develop from the filament or connective of the fertile members of the androecial whorl (Fig. 1B) (Kirchoff 1991; Glinos and Cocucci 2011). This results in the potential for all members of the androecial whorls, whether fertile or sterile, to develop petaloidy.
In Costus scaber, the anther consists of two locules, positioned adjacent to each other on the ventral surface of a petaloid structure in the inner androecial whorl (Kirchoff 1988). Development of the petaloid component of the fertile stamen, which includes both filament and connective, is simultaneous with the development of the anther (Kirchoff 1988). The stamen primordium is divided into two parts—the ventral portion produces the anthers and the dorsal portion produces the petaloid filament and connective (Kirchoff 1988). Conversely, in the Zingiberaceae (sister to Costaceae; Fig. 1A), Leinfellner characterized the petaloid component of the fertile stamen as occurring late in development, thus classifying the petaloid portion as an accessory structure (Leinfellner 1956) and implying lack of homology between the petaloid structures in the fertile stamens of Costaceae and Zingiberaceae.
The concentric androecial whorls of Canna indica consist of 3–4 petaloid staminodes (sterile) and one-half of a single fertile petaloid stamen (Glinos and Cocucci 2011). The fertile stamen, labellum and inner staminode constitute the inner androecial whorl, while the outer androecial whorl is made up of the two (or sometimes one) remaining staminodes (Eichler 1875; Rao and Donde 1955; Pai 1963; Kirchoff 1983). According to Kirchoff (1988, 1991), the fertile stamen is always found in the inner androecial whorl, which develops before the outer androecial whorl. However, the developmental origin of the petaloid appendage of the fertile stamen in Cannaceae remains unclear.
Our understanding of the molecular basis of floral development has increased greatly since the first descriptions of the genes responsible for specifying the identity of floral organs in Antirrhinum and Arabidopsis (Bowman et al. 1991; Jack et al. 1992). According to the canonical ABC model of floral development (Weigel and Meyerowitz 1994), differential gene expression results in the specification of the identity of the various floral organs. In Arabidopsis, A-class genes [APETALA2 (AP2) and APETALA1 (AP1)] are involved in the specification of sepals (first whorl organ), and together with B-class genes [GLOBOSA (GLO) or PISTALLATA (PI), and DEFICIENS (DEF) or APETALA3 (AP3)] they specify petal identity (second whorl). B-class genes are also involved in the specification of stamen identity when expressed together with the C-class gene [AGAMOUS (AG)]. Furthermore, AG alone is responsible for the specification of carpel identity (Coen and Meyerowitz 1991). Although most components of the ABC model of floral development hold true for most model species studied so far, it is unclear to what extent this model can explain the morphological diversity and evolution of floral development across angiosperms. In the case of monocots, the most well-studied systems are among the grasses where the highly derived flower morphology of the Poaceae renders statements of homology a difficult task.
In C. indica, it is unclear whether the petaloid appendage of the half fertile stamen is produced by the secondary expansion of residual meristematic tissue from the filament of a single fertile theca, or whether it is a result of a homeotic transformation of one of the thecae into a petaloid structure. Here, we use developmental studies to characterize the origin of the petaloid tissue in the Canna stamen and investigate whether the combinatorial expression of MADS-box genes can explain petaloidy in C. indica androecial whorls.
Methods
Developmental series
Living material of Canna sp. was collected from the UC Berkeley Botanical Gardens, the Specht Lab diversity collection at the Oxford Tract Greenhouses, from residential neighbourhoods in the Berkeley hills (with consent from home-owners) and from the UC Berkeley Student Organic Garden (SOGA) (Table 1). In total, 30 inflorescences were collected from C. indica (18), Canna edulis (4), Canna tuerckheimii (4) and Canna sp. (4). Although several Canna species were observed in order to characterize any potential differences across Cannaceae, the developmental series portrayed and the molecular characterization focus specifically on the development of C. indica.
Table 1.
Accession of Canna sp. used in morphological and molecular studies of floral developmental evolution.
Inflorescences were dissected from living material, removing the outer bracts to expose most floral buds and floral organ primordia at the inflorescence apex. The apices were vacuum infiltrated for 10–20 min in formalin–acetic acid–alcohol (FAA) (3.7 % formaldehyde), and stored in cold FAA for up to 2 weeks. Tissue fixation was carried out using a standard microwave procedure (Schichnes et al. 1999) as follows: three rounds of 15–min microwave cycles at 37 °C, followed by an ethanol dehydration series (50, 70, 95 and 100 % ethanol) for 5 min at 67 °C for each ethanol concentration. Tissue was stained in 1 % w/v fast green FCF in 100 % ethanol for 2–3 days at 4 °C. Subsequently, tissue was destained with 100 % ethanol for 2–5 days at 4 °C, as necessary for final dissection, observation and photography (Sattler 1968).
Inflorescences were further dissected under an Olympus dissecting scope, and photographs were taken using a ×3.8 Ultrapak epi-illumination objective (Posluszny et al. 1980; Charlton et al. 1989) on a Leitz Orthoplan microscope equipped with a Nikon Digital Sight 5M digital camera, as described by Bartlett et al. (2008). NIS Elements software was used to process the images taken at different focal points (Bartlett et al. 2008) to expand the depth of focus.
Gene expression
Canna indica flowers were dissected from the same plants as used above. Fresh flowers were quickly dissected, separating sepals, petals, staminodes, petaloid part of the fertile stamen, anther of the fertile stamen and gynoecium into separate vials. RNA was extracted from each of the floral parts individually. RNA extraction was carried out from fresh tissue with Plant RNA Reagent (Invitrogen), according to the manufacturer's guidelines. cDNA was synthesized after DNase treatment of each sample (Fermentas) using a BIO-RAD iScript Reverse Transcription Supermix kit with poliT primers. Reverse transcription (RT) primers were designed for AGAMOUS-1 and AGAMOUS-2 (AG-1 and AG-2), DEFICIENS (DEF), and GLOBOSA-1 and GLOBOSA-2 (GLO-1 and GLO-2). GLO sequences were downloaded from NCBI (GU594924.1 and GU594945.1) and used for RT primer design. DEF and AG genes were first amplified using degenerate primers. Polymerase chain reaction (PCR) products were cloned into Top10 cells and sequenced using an ABI Big Dye Terminator kit on a 3700 sequencer. Recovered sequences were used to develop copy-specific RT primers.
Primer sequences are as follows: GLO1 Forward, CCC TTC CAC GTT ATC GAC GAT T; GLO2 Forward, CGT CCA CCT CGT TGT CTG AG; GLO Reverse, TTG TGC ATC TTC CAA ATC TCC; DEF Forward, CCT CCA CTG AAA CAA AGA AGA TT; DEF Reverse, CAG TTC ATG CAG CAA GTT CC; AG1 Forward, AGC CTA TGA ATT GTC GGT CTT G; AG1 Reverse, AGC TGA GAG ACT CAC CCA TCA; AG2 Forward, CGT ACG AAT TGT CCG TGC TT; AG2 Reverse, TCT GCT CTC GAG TTG CTT CA. Reverse transcription-polymerase chain reactions were carried out using a Phire DNA Polymerase kit (Finnzymes) and the following: 2 μL of 5× Phire buffer; 0.2 μL of 10 mM dNTPs; 0.5 μL of each primer; 0.1 μl of Phire polymerase; 1 μL of [1:10]cDNA; and ddH2O, for a total volume of 10 μL. Thermocycling conditions followed the manufacturer's recommendations, and the following annealing temperatures for 30 cycles: GLO1, 66 °C; GLO2, 68 °C; DEF, 63 °C; AG1 and AG2, 70 °C.
Reverse transcription-polymerase chain reactions were visualized on 1 % agarose gels, and stained with GelRedTM (Phoenix Research Products) according to the manufacturer's protocol.
Results
Canna indica fertile stamen development
Canna indica early floral development has been described previously (Kirchoff 1983). Here, we present only our new developmental data focused on the fertile stamen in order to understand the origin of the petaloid appendage. Therefore, early stages of floral development are only briefly discussed.
The earliest discernible stage in C. indica floral development (Stage 1; Fig. 2A) is represented by the development of two meristematic bulges, previously described as the sepal primordia (Kirchoff 1983). As the floral bud continues to develop, the apex flattens out, forming a disc-shaped structure, the ‘floral cup’ (Stages 2, 3; Fig. 2B). The periphery of the floral cup continues to grow and differentiate, eventually becoming delineated into the distinct petal and stamen primordia (Stages 4, 5; Fig. 2C, D). At about Stage 6 (Fig. 2E–G), the young fertile stamen protrudes out of the floral cup, distinguishing itself from the young petals. These observations are consistent with Canna floral development that has been well documented and described until Stage 6 (Rao and Donde 1955; Pai 1963; Kirchoff 1983).
Figure 2.
Canna indica floral development series. (A) Floral initiation showing the protrusion of the sepal primordial. The arrowhead points to a floral primordium amplified in (B); (B, C) development of the ‘floral cup’; (D) sepal primordia already separated from the remaining floral primordium, and evident petal primordia; (E) early stages of fertile stamen development, with two theca primordia; (F, G) fertile stamen development; (H) later stages of fertile stamen development. A single theca has developed with its petaloid appendage, while the other theca arrests development (arrowhead); (I) an almost mature stamen with its petaloid appendage (arrowhead); and the aborted theca to its right. se, sepal; pe, petal; pe/sta, petal/stamen common primordium; std, staminode; the, fertile theca; app, fertile stamen appendage; x, aborted theca primordium.
Stages 7 and 8 (Fig. 2H, I) depict the continued growth of the fertile stamen and the determination of organ identity. By Stage 7, the rapid development of the fertile stamen and its accompanying petaloid appendage becomes evident, and becomes a distinct feature in the floral bud (Fig. 2H). One theca continues to develop, while the other becomes comparatively reduced in size and discontinues growth or expansion (Fig. 2H). The petaloid appendage is connected to the developing theca along the filament and apparently below where the connective would normally develop (Fig. 2I). Owing to the abortion of the second theca, no connective region is apparent.
At Stage 8 (Fig. 2I), the final stage of this developmental series, the nearly mature fertile stamen is represented by a single developed theca that is connected to a rapidly expanding petaloid appendage emerging from the filament. A line of cleavage separates the aborted theca from the growing fertile theca with its petaloid appendage.
Gene expression during floral development
Reverse transcription-polymerase chain reaction for C. indica was used in order to assess the expression pattern of B- and C-class MADS-box genes in various floral organs (Fig. 3). Sepals (sep), petals (pet), staminodes (std) and gynoecium (gyn) were studied in their entirety. For a better account of gene expression patterns on Canna organs, the fertile stamen was divided into petaloid appendage (pap) and theca (the), which were studied independently. Canna indica has at least one copy of DEFICIENS (DEF), two copies of GLOBOSA (herein referred to as GLO-1 and GLO-2) and two copies of AGAMOUS (AG-1 and AG-2) (A. M. R. Almeida et al., unpubl. data).
Figure 3.
Expressions of B- and C-class MADS-box genes in the floral organs of C. indica as detected by RT-PCR. Each C. indica floral organ was dissected and RNA was extracted independently. The fertile stamen was divided into petaloid appendage and theca. sep, sepal; pet, petal; std, staminode; stm, stamen; pap, petaloid appendage of stamen; the, theca; gyn, gynoecium. Actin was used as an endogenous control for the cDNA synthesis. B-class genes: DEF, DEFICIENS; GLO-1, GLOBOSA-1; GLO-2, GLOBOSA-2. C-class genes: AG-1, AGAMOUS-1; AG-2, AGAMOUS-2.
B-class MADS-box genes (DEF, GLO-1 and GLO-2) are expressed in all the floral parts studied (Fig. 3). It is interesting to note that expression of these genes is reduced in sepals, especially for DEF and GLO-1. B-class gene expression shows an expanded pattern when compared with the Arabidopsis ABC model, where expression of the B-class genes is restricted to petals and stamens. C-class MADS-box genes (AG-1 and AG-2) also show an expanded pattern of expression when compared with the expected expression pattern based on the canonical ABC model (Fig. 3): AG-1 seems to be expressed in a gradient, increasing from sepals (low) to gynoecium (high), while AG-2 is evenly expressed in all floral parts studied with the exception of the petals, where no expression was observed.
Discussion
The initial stages of organogenesis in this developmental series confirm past studies on Canna floral development (Rao and Donde 1955; Pai 1963; Kirchoff 1983). Here we focus on the development of the fertile stamen with particular attention given to its petaloid appendage.
Petaloidy is a striking trend in the evolution of Zingiberales floral morphology, especially in the ginger clade where the number of fertile stamens is drastically reduced and the remaining infertile androecial members are petaloid. The extreme case is observed in Cannaceae flowers, in which all androecial elements are petaloid and the half fertile stamen has a marked petaloid appendage (Fig. 1B). In this case, only one theca is apparent at anthesis, and the question remains whether (i) the petaloid appendage of the fertile stamen develops from the filament and connective of the same primordium that gives rise to the single theca, or (ii) the appendage is the result of the growth and expansion of a separate theca primordium that undergoes homeotic transformation into a sterile, petaloid structure. In the first case, only half of the original stamen primordium would develop fully, forming both the anther and the petaloid appendage (see Fig. 4B, x). In the second case, the entire stamen primordium would grow and mature, with one-half giving rise to a petaloid structure and the other half forming an anther.
Figure 4.

Summary results for gene expression and the corresponding floral organ morphology in Arabidopsis and Canna. (A) Classical ABC model of floral development based on Arabidosis thaliana. Only B-class (DEFICIENS and GLOBOSA) and C-class (AGAMOUS) MADS-box genes are depicted, as the role of A-class MADS-box genes in floral development in monocots awaits further investigation. In the classical ABC model, petal identity is a result of A- and B-class MADS-box gene expression, while stamen identity results from concomitant expression of B- and C-class MADS-box genes. (B) Canna indica B- and C-class MADS-box gene expression pattern. Canna indica contains two GLOBOSA genes (GLO-1 and GLO-2) and two AGAMOUS genes (AG-1 and AG-2). B- and C-class MADS-box genes are expressed in most of the floral parts studied here, and when compared with the classical ABC model, show an expansion in their expression domains. x, position of the aborted theca primordium relative to the half fertile stamen.
The morphological series presented here (Fig. 2G–I) provides evidence for the first hypothesis: that the petaloid appendage of the Canna fertile stamen develops from the same primordium that produces the theca, emerging from the position of the filament. This finding has implications for understanding fertile stamen development in other genera within the ginger lineage. For instance, because it appears that the entire structure (theca and petaloid appendage) is produced from a single half (stamen) primordium, other fertile stamen configurations, such as those observed in Costaceae and Zingiberaceae, could very probably result from concerted laminar development of the filament and connective associated with both fertile thecae.
In order to investigate the molecular mechanisms associated with androecial petaloidy in C. indica, the expressions of class B and classC MADS-box genes were analysed in various floral organs. We did not investigate A-class gene expression, as the role of the A function genes outside of Arabidopsis is unclear; alternatively, B and C function has been shown to predict the stamen and petal development model for several groups of monocots (Kim et al. 2006; Tang et al. 2007). The canonical expression pattern for B- and C-class MADS-box genes (Fig. 4A) does not appear to hold for Canna. We expected to find B-class genes in the petal and stamen whorls, and C-class genes in stamen and gynoecium whorls, with perhaps some changes in expression defining the differences between petaloid vs. fertile stamens within the androecial whorls. Instead, B-class (GLO and DEF) genes have expression domains that are expanded in both directions to include the first whorl and the gynoecium. C-class (AG) genes also show a broad expression pattern and are found in petals (AG-1) and sepals (AG-2) as well as the androecial and gynoecium whorls (Fig. 4B). There was no differentiation between fertile or sterile elements within the androecial whorl, nor was there a combination that seemed to define petaloidy regardless of its whorl of origin.
As petaloidy in C. indica is, however, not restricted to the corolla (petal) and androecial (stamen) whorls, the extension of B-class gene expression into the gynoecium might explain the laminar morphology of the carpels in Canna (Glinos and Cocucci 2011). Most of the Canna flower shows simultaneous B- and C-class MADS-box gene expression, which in the classical ABC model would result in the specification of stamen identity. Clearly, this combination is not functioning as stamen identity in the Canna flower, with its single half fertile stamen. This expression pattern implies that Canna petaloidy, whether in the petals, stamens or even the carpels, is probably not a simple result of re-deployment of the classical petal specification mechanisms (A- and B-class MADS-box gene expression), and potentially involves an as yet uncharacterized molecular basis.
Considering the origins of stamens from a petal-like organ (Goethe 1790), it is possible that the filamentous stamens that are ancestral to Zingiberales and that characterize Musa flowers are the result of a restriction of laminar growth associated with the development of fertility. The lack of pollen sac production in the majority of androecial members of the ginger families might cause a de-repression of laminar growth, resulting in the production of petaloid organs in the androecial whorls. When petaloidy is found in organs that do contain fertile thecae, it is unclear as to the mechanisms that enable laminar growth in the presence of pollen sac production. Current studies are focusing on the role of polarity genes that establish the abaxial/adaxial boundary and regulate laminar vs. radial morphology of lateral organs.
Conclusions
It is possible to conclude, based on the data from this study, that the developmental mechanisms resulting in petaloid floral organs are different even in closely related taxa such as Cannaceae and Costaceae. It appears that the development of a petaloid appendage on the filament of a single theca in C. indica might be the result of ectopic development resulting in the appearance of a half fertile, petaloid stamen. In contrast, in Costaceae the petaloid stamen might be the result of laminar growth of the filament and connective, returning to an ancestral leaf-like laminar development as seen in the petaloid stamens of early diverging angiosperms (e.g. Nymphaea). Investigations on candidate gene expression during development of the stamens in Costaceae and Cannaceae will be necessary to determine if the genetic mechanisms underlying the development of the petaloid stamens are indeed different in these two families, indicating that homoplasy can be at work even in closely related species.
Accession Numbers
Novel sequences have been submitted to GenBank (http://www.psc.edu/general/software/packages/genbank/genbank.php) and will be released upon publication. Accession numbers: Canna GLO1, GU594924.1; Canna GLO2, GU594945.1; Canna AGAMOUS1, JQ180191; Canna AGAMOUS2, KC763343; Canna DEF, KC763344.
Sources of Funding
Funding was provided by a UC Berkeley College of Natural Resources Sponsored Projects for Undergraduate Research (SPUR) student-initiated research award to A.B., a Fulbright/CAPES award to A.M.R.A. and NSF CAREER IOS 0845641 to C.D.S.
Contributions by the Authors
All authors contributed to the writing and editing of the manuscript. Dissections and microscopy were performed by A.B. following training and mentoring by A.M.R.A. Images were interpreted and edited by A.M.R.A. and C.D.S.
Conflict of Interest Statement
None declared.
Acknowledgements
We thank Roxana Yockteng for laboratory assistance and discussion of ideas associated with the manuscript.
Literature Cited
- Bartlett ME, Kirchoff BK, Specht CD. Epi-illumination microscopy coupled to in situ hybridization in non-model species. Development Genes and Evolution. 2008;218:273–279. doi: 10.1007/s00427-008-0211-6. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development. 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- Bremer B, Bremer K, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. The Angiosperm Phylogeny Group. [Google Scholar]
- Charlton WA, MacDonald AD, Posluszny U, Wilkins CP. Additions to the technique of epi-illumination light microscopy for the study of floral and vegetative apices. Canadian Journal of Botany. 1989;67:1739–1743. [Google Scholar]
- Coen E, Meyerowitz E. The war of the whorls—genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Eichler AW. Blüthendiagramme construirt und erläutert. Vol. 1. Leipzig: W. Engelmann; 1875. [Google Scholar]
- Glinos E, Cocucci AA. Pollination biology of Canna indica (Cannaceae) with particular reference to the functional morphology of the style. Plant Systematics and Evolution. 2011;291:49–58. [Google Scholar]
- Goethe JW. Versuch die Metamorphose der Pflanzen zu erklären. Gotha, Germany: Ettinger; 1790. [Google Scholar]
- Jack T, Brockman L, Meyerowitz E. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- Kim S, Soltis PS, Wall K, Soltis DE. Phylogeny and domain evolution in the APETALA2-like gene family. Molecular Biology and Evolution. 2006;23:107–120. doi: 10.1093/molbev/msj014. [DOI] [PubMed] [Google Scholar]
- Kirchoff B. Floral organogenesis in five genera of the Marantaceae and in Canna (Cannaceae) American Journal of Botany. 1983;70:508. [Google Scholar]
- Kirchoff B. Inflorescence and flower development in Costus scaber (Costaceae) Canadian Journal of Botany. 1988;66:339–345. [Google Scholar]
- Kirchoff B. Homeosis in the flowers of the Zingiberales. American Journal of Botany. 1991;78:833. [Google Scholar]
- Kirchoff B, Lagomarsino LP, Newman WH, Bartlett ME, Specht CD. Early floral development of Heliconia latispatha (Heliconiaceae), a key taxon for understanding the evolution of flower development in the Zingiberales. American Journal of Botany. 2009;96:580–593. doi: 10.3732/ajb.0800305. [DOI] [PubMed] [Google Scholar]
- Kress J. The phylogeny and classification of the Zingiberales. Annals of the Missouri Botanical Garden. 1990;77:698–721. [Google Scholar]
- Kress J, Prince L, Hahn WJ, Zimmer EA. Unraveling the evolutionary radiation of the families of the Zingiberales using morphological and molecular evidence. Systematic Biology. 2001;50:926–944. doi: 10.1080/106351501753462885. [DOI] [PubMed] [Google Scholar]
- Leinfellner W. Die blattartig flachen Staubblätter und ihre gestaltliche Beziehung zum Bautypus des Angiospermenstaubblattes. Oesterreichische Botanische Zeitschrift. 1956;103:267–290. [Google Scholar]
- Pai RM. The floral anatomy of Canna indica L. Bulletin of the Botanical Society, College of Science, Nagpur. 1963;4:45–53. [Google Scholar]
- Posluszny UM, Scott MG, Sattler R. Revisions in the technique of epi-illumination light microscopy for the study of floral and vegetative apices. Canadian Journal of Botany. 1980;54:2491–2495. [Google Scholar]
- Rao VS, Donde N. The floral anatomy of Canna flaccida. Journal of the University of Bombay. 1955;24:1–10. [Google Scholar]
- Sattler R. A technique for the study of floral development. Canadian Journal of Botany. 1968;46:720–722. [Google Scholar]
- Schichnes D, Nemson JA, Slohberg L, Ruzin S. Microwave protocols for paraffin microtechinique and in situ localization in plants. Microscopy and Microanalysis. 1999;4:491–496. doi: 10.1017/s1431927698980461. [DOI] [PubMed] [Google Scholar]
- Tang M, Li G, Chen M. The phylogeny and expression pattern of APETALA2-like genes in rice. Journal of Genetics and Genomics. 2007;34:930–938. doi: 10.1016/S1673-8527(07)60104-0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes—a review. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]