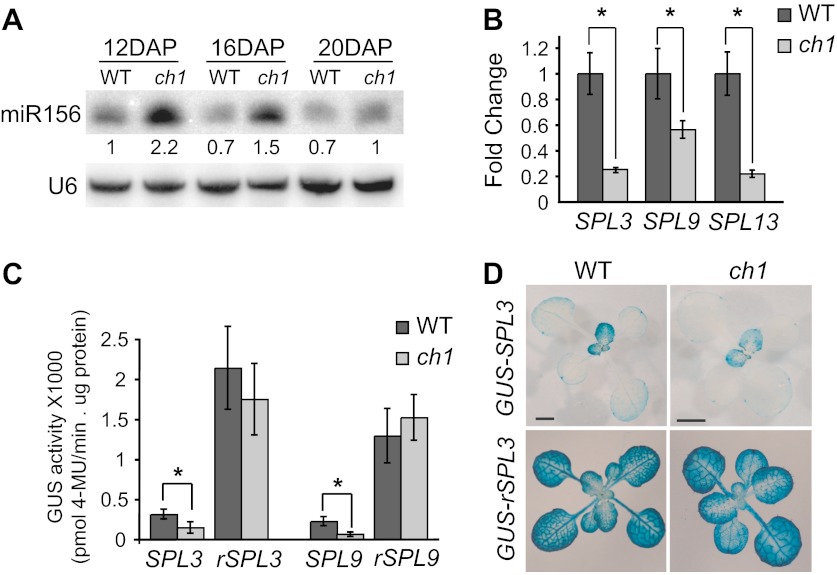
Figure 2. Expression of miR156 and SPL transcripts in ch1-4.
(A) Northern blot of mature miR156 in ch1-4 and Col reveals that miR156 is elevated in ch1-4 and declines at a slower rate in this mutant. U6 was used as a loading control. Hybridization intensities are compared to the value in WT 12 DAP. (B) qRT-PCR of the transcripts of SPL3, SPL9, and SPL13 in 16-day-old wild-type and ch1-4 demonstrates that these transcripts are present at a significantly lower level in ch1-4. (C) MUG assay of the GUS activity of miR156-sensitive and miR156-resistant SPL3 and SPL9 reporters in Col and ch1-4 demonstrates that the reduced expression of GUS-SPL3 and SPL9-GUS in ch1 is dependent on miR156. (D) The expression pattern of miR156-sensitive and miR156-resistant pSPL3::GUS-SPL3 in 14-day-old rosettes of Col and 18-day-old rosettes of ch1-4; older rosettes of ch1-4 were chosen to compensate for the slower growth rate of this mutant. pSPL3::GUS-SPL3 is expressed at a lower level in ch1-4. pSPL3::rSPL3-GUS is expressed at the same level and in the same pattern in Col and ch1-4. Scale bar = 2 mm. Asterisks in (B) and (C) indicate significant difference (Student's t-test), p<0.01, n = 3. Error bars indicate SEM.