Abstract
The mechanical force-induced activation of the adhesive protein von Willebrand Factor (VWF), which experiences high hydrodynamic forces, is essential in initiating platelet adhesion. The importance of the mechanical force-induced functional change is manifested in the multimeric VWF’s crucial role in blood coagulation, when high fluid shear stress activates plasma VWF (pVWF) multimers to bind platelets. Here we showed that a pathological level of high shear stress exposure of pVWF multimers results in domain conformational changes, and the subsequent shifts in the unfolding force allow us to use force as a marker to track the dynamic states of multimeric VWF. We found that shear-activated pVWF multimers (spVWF) are more resistant to mechanical unfolding than non-sheared pVWF multimers, as indicated in the higher peak unfolding force. These results provide insight into the mechanism of shear-induced activation of pVWF multimers.
Von Willebrand factor (VWF) is a large multimeric protein constructed from two identical VWF monomers linked by C-terminal disulfide bonds into dimers, and the dimers then polymerize via N-terminal disulfide bonds into long VWF multimers [1–3]. The domain organization of a 250 kDa, 60 nm long VWF monomer [2, 4] is shown in Fig. 1(a). The largest VWF multimers contain up to 200 monomers [5] and are concentrated after synthesis in Weibel-Palade bodies and α-granules, the storage compartments of endothelial cells and platelets, respectively [2]. In response to stimulation by cytokines and other agents, these ultra-large VWF (ULVWF) multimers are rapidly secreted in long string-like structures by endothelial cells (ECs), where they are anchored [6]. EC-anchored ULVWF multimeric strings are hyper-adhesive in their capacity to bind platelet glycoprotein (GP) Ib-IX-V complexes [3, 7]. Circulating pVWF multimers are hemostatically inactive toward platelets, but can be activated by exposure to high shear stress [2, 8, 9]. It has been proposed that, under high shear stress, pVWF multimers undergo a change in conformation from a globular to an elongated form (quaternary structure change) [2, 9–11]. More recently, it has been demonstrated that shear-activated pVWF multimers become laterally apposed into fibrils via multimer-to-multimer disulfide bonds [12].
FIG. 1.
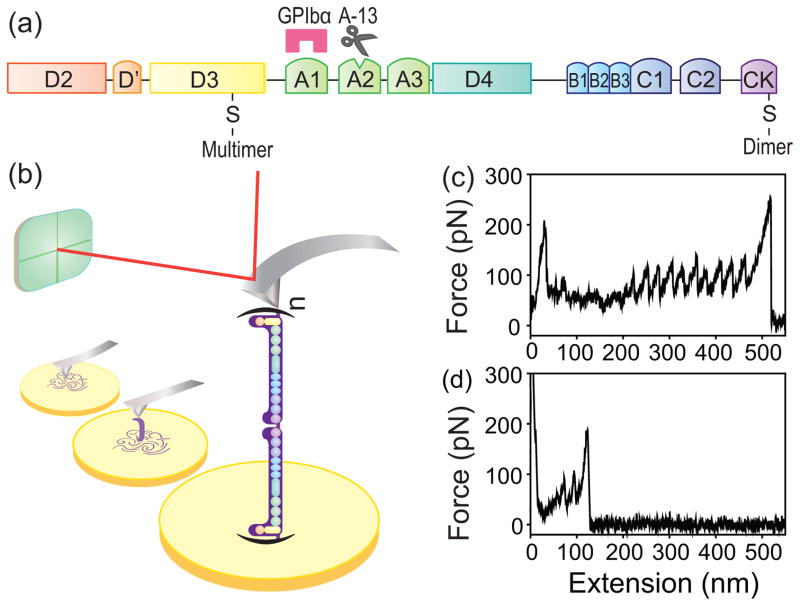
Multimeric VWF unfolding with AFM. (a) The domain organization of a VWF monomeric subunit, where A1 is the platelet GP Ib-IX-V receptor binding domain, A2 contains the cleavage site for ADAMTS-13, and D3 contains the binding site for the subendothelial matrix collagen. The locations of the disulfide bonds where the VWF connects to form dimers and multimers are presented. (b) Experimental setup of single-molecule pulling of a VWF multimer using AFM. A purified pVWF multimer, which is composed of n polymerized dimers of VWF, was pulled while the force was recorded. Typical force-extension curve of (c) a pVWF multimer, and (d) a pVWF dimer, pulled at a constant velocity of 1000 nm/s.
The shear-induced conformational change exposes or alters the A1 domain in VWF monomeric subunits, enabling large VWF multimers to bind to platelet GP Ib-IX-V and initiate platelet adhesion and aggregation. The difference in the dynamic states of various forms of VWF multimers determines the on-off switching of VWF multimeric activation for platelet binding. In this study, we used single-molecule manipulation to monitor the force response of different forms of VWF multimers. The peak force was used as an indicator of the dynamic states of VWF monomeric subunit domains within VWF multimers.
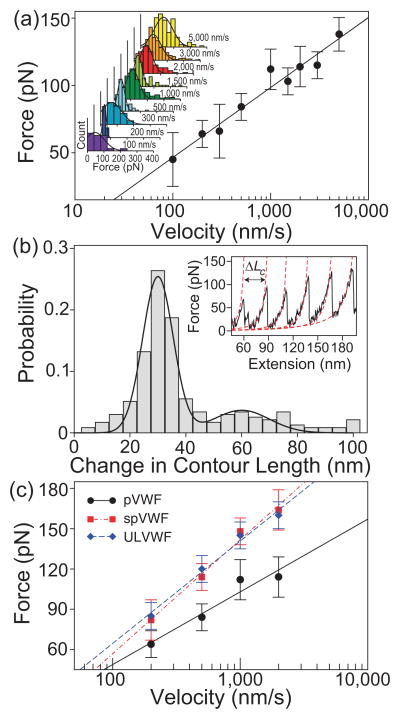
We pulled single VWF multimeric molecules using an atomic force microscope (AFM) [Fig. 1(b)] (see Supplemental Material [13]). The sawtooth patterns of force peaks in the force-extension curves of pVWF multimers [Fig. 1(c), (d)] are characteristic of multi-domain protein unfolding (tertiary structure change) [14–16]. The value of the peaks specifies the force required to unfold the domains, and is related linearly to the unfolding free energy barrier and logarithmically to the pulling speed [Fig. 2(a)] [16–18]. The change in contour length (ΔLc) histogram of pVWF [Fig. 2(b)] shows a major peak at 30(8) nm. The values in the parentheses are one standard deviation. This is a typical length for an unfolded domain of 90 amino acid residues assuming 0.34 nm per residue [5, 19]. In addition, there was a minor peak at 60(15) nm, corresponding to an unfolded domain of 180 amino acid residues, consistent with domain unfolding.
FIG. 2.
Unfolding of multimeric VWF. (a) Unfolding force as a function of pulling velocity for pVWF. The unfolding forces were binned by pulling velocity. The inset shows the binned histograms fitted with Gaussian curves. The peak of the the fitted Gaussian was plotted against the pulling velocity. The error bars indicate half bin width. The line is a linear fit to the logarithm of the velocity. (b) Histogram of the change in contour length of pVWF. The solid line indicates a double Gaussian fit to the distribution, which has a major peak at 30 nm and a minor peak at 60 nm. We analyzed all the data together, and therefore, the major contribution of the data comes from ΔLc 30 nm. Inset: The change in contour length, ΔLc, was the difference of the Lc determined by WLC model fit to the data (dashed red lines), , where F is the force, x is the distance, Lp and Lc are the persistence length and the contour length, respectively, T is the temperature, and kB is the Boltzmann constant [20–22]. The persistence length Lp was 0.4 nm, which is consistent with unfolded protein chains. (c) Velocity-dependent unfolding forces of different forms of multimeric VWF. The trend is similar to protein domain unfolding.
ULVWF multimers contain a larger number of monomers than pVWF, and are more active in adhering to platelets and inducing platelet aggregation [23]. We observed differences in peak unfolding forces at high pulling speeds between ULVWF and pVWF multimers [Fig. 2(c)], indicating that pVWF multimers and ULVWF are in different conformational states. Similarly, at high levels of shear stress (60–120 dyn/cm2), the capacity of pVWF multimers to adhere to, and aggregate to platelets increases as previously shown [23] and re-affirmed in this study (Supplemental Fig. 1). That is, spVWF become functionally similar to ULVWF multimers. The peak unfolding force of pVWF multimers increased after exposure to high shear stress, but the force-extension curves are qualitatively similar to unsheared pVWF multimers and unsheared ULVWF multimers. However, the reaction of pVWF to spVWF may not be 100%, and the shift in the peak force is a qualitative observation. The difference in the peak unfolding forces between pVWF multimers and either spVWF or ULVWF multimers was more pronounced at high pulling velocities [Fig. 2(c)]. This finding is compatible with the shear-induced conformational change in pVWF (to spVWF) that increases the exposure of platelet-binding A1 domains in the VWF monomeric subunits of spVWF multimers. It has been shown that exposure to a 100 dyn/cm2, high fluid shear stress induces pVWF multimers to associate laterally and form VWF fibrils that have an increased capacity to bind to platelet GPIbα [8]. This fibrillar state of laterally-associated VWF multimers may be the conformation of spVWF multimers that is functionally similar to ULVWF (Supplemental Fig. 2).
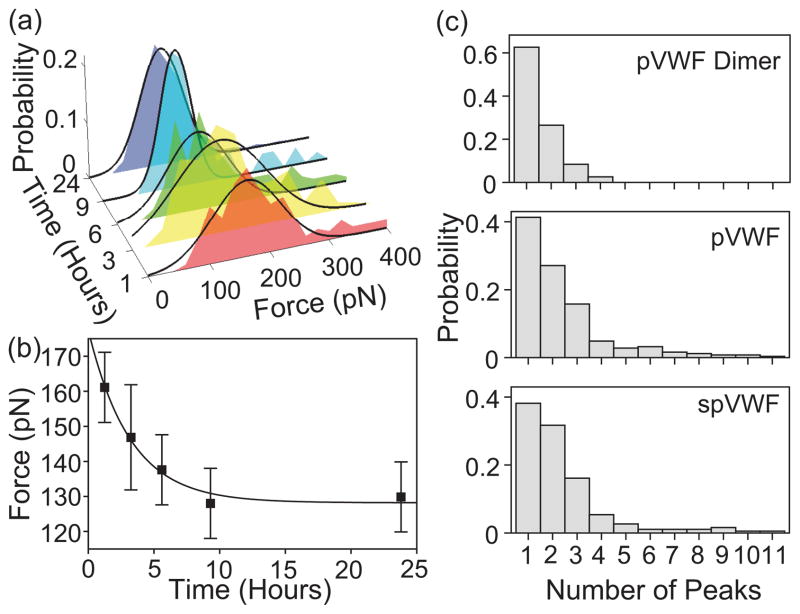
To measure the kinetics in spVWF force experiments, peak unfolding force measurements of pVWF were conducted at different delay times after shear exposure. The spVWF unfolding force decreased over time, and reached an equilibrium force after 10 hours [Figs. 3(a) and (b)]. Thus, the shear-induced change in pVWF to the spVWF multimeric conformation changes with time, with a prolonged relaxation time of several hours. Fitting the data to the exponential equation F(t) = Fp + (Fs − Fp) exp(−t/τ), where Fs is the peak force immediately after shear exposure, Fp is the equilibrium peak force, and τ is the time constant, yields Fs = 180 pN, Fp = 130 pN, and τ = 3 hours. The difference in the peak unfolding force between spVWF or ULVWF multimers and pVWF multimers is more pronounced at high pulling velocity.
FIG. 3.
Dynamics of VWF multimers. The data were taken at 1000 nm/s pulling velocity. (a) Peak force distributions of spVWF as a function of time since exposure to a pathological level of 100 dyn/cm2 fluid shear. (b) spVWF peak force decreases with time since shear exposure. The solid line is a fit to an exponential curve, F (t) = Fp + (Fs − Fp) exp(−t/τ) (see text). The error bars are half of the bin width in the histogram. (c) Number of unfolding peaks in a given force curve from different forms of VWF multimers. pVWF dimer has up to four force peaks, indicating that there are two unfolding events per monomer.
The force-extension curves showed that the unfolding force peaks correspond to the changes in the VWF multimeric conformation at the level of one or more domains within the VWF monomeric subunits. This conclusion is supported by i) the force-extension curves display a characteristic sawtooth pattern of repeated force peaks, resembling the known sequential unfolding of other multi-domain proteins [14]; ii) the increase in contour length after each peak, ΔLc, is 30 nm, which is comparable to the contour length of an unfolded domain or an intermediate state; and iii) at 1000 nm/s pulling velocity, the value of the peak unfolding force was distributed at 100–130 pN, and varied linearly with the natural logarithm of pulling velocity, as is typical of domain unfolding.
In addition, by stretching VWF dimers under similar conditions, we have observed up to four unfolding peaks per force-extension curve [Fig. 3(c)], suggesting that there are two unfolding events per monomer. The two force peaks can be from the unfolding of two different domains or from two partial domains. This conclusion is supported by when pulling eight serially linked titin I27 domain, (I27)8, up to eight unfolding peaks have been observed [14]. For comparison, pVWF curves have up to ten force peaks, suggesting that there are up to five monomers at a given pull. The number of peaks of ULVWF does not differ from pVWF or spVWF significantly, even though the ULVWF sample has more repeating units than pVWF [13]. Hence, it suggests a stronger interaction in ULVWF among monomers that may prevent the ULVWF polymer chain to be isolated to undergo domain unfolding. The dimer force peak and change in contour length, ΔLc, distributions are consistent with that of the multimer, further supporting the conclusion that the features in the multimeric VWF force-extension curves correspond to individual domain unfolding.
In the monomeric subunits of VWF multimers, the force peaks may be the combined result of unfolding of different domains. It is likely, however, that there is a major contributor to the force signal. A probable candidate is the VWF A2 domain, because it does not have disulfide bonds, and has been observed to unfold in the pico-newton force range [24, 25]. ΔLc of 60(15) nm determined from our experiment is similar to 57(5) nm observed by Zhang et al. [5]. The most frequent ΔLc observed, 30(8) nm, is consistent with the values reported in Zhang et al. [5], showing 40% of the unfolding peaks with ΔLc of 23(5) nm, which is attributed to partial unfolding of A2 [5, 24, 26]. The A1 and A3 domains contain disulfide bonds, which are unlikely to unfold during stretching experiments, because at a 100 nm/s pulling velocity, disulfide bonds typically rupture at 2 nN [27], a force much higher than the typical unfolding force (100–200 pN) observed in our study. Previous studies of the forced-unfolding of A1A2A3 triple domains also reveal that the VWF A2 domain can be partially or completely unfolded, possibly after inter-domain uncoupling [26, 28]. These findings suggest that the unfolding of a portion of the A2 domain in VWF monomeric subunits may be the main contributor to our unfolding force signals. We have ruled out that the change of unfolding force is simply due to more exposed A2 domains without intramolecular interactions, since such a configuration will only yield more unfolding peaks in a given pull [Fig. 3(c)], but not a significantly altered unfolding force [10, 17].
Our results suggest that high shear stress (100 dyn/cm2) converts spVWF multimers to a conformation that was metastable, probably due to the lateral association of spVWF multimers, with a long relaxation time. Over several hours, the metastable state of spVWF crossed the energy barrier and relaxes to a more stable state. Using the time constant τ = 3 hours determined from the relaxation curve shown in Fig. 3(b), we estimate the activation free energy barrier from spVWF to pVWF, using the Arrhenius equation, k = A exp(−ΔG/kBT), where k= 1/τ is the rate constant, ΔG is the free energy of barrier from spVWF to pVWF, and A is the pre-exponential factor. Assuming that A is between 105 s−1 and 1010 s−1, ΔG is 12–19 kcal/mol [29–31]. The barrier height from an active state to an inactive state is comparable to protein unfolding, further supporting the notion of domain conformational changes for the observed force peak change.
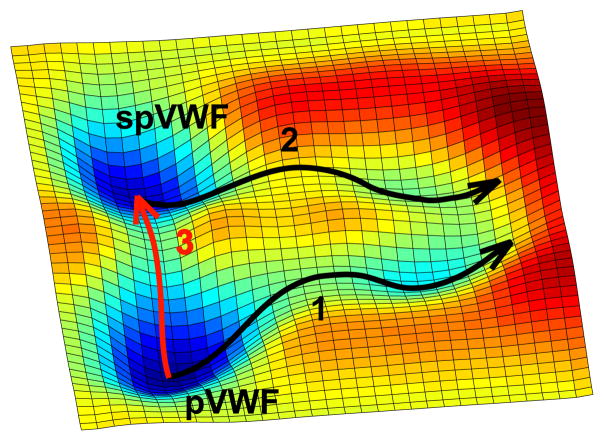
Figure 4 highlights the free energy landscape [32] of different forms of VWF multimers. Our results suggest that pVWF multimers have different conformational states before and after shear exposure that unfold through different pathways (pathways 1 and 2). Proteins with multiple conformational states of different activities have been observed by force measurements [33]. Here, pVWF multimers are in a native, inactive state, but can be converted to a metastable, active state (spVWF) by exposure to high shear stress. This state may be considered ‘misfolded’ since it is a non-native state [34]. The spVWF multimer’s peak unfolding force, which is related to the barrier height [17, 35], is likely to be higher than that of pVWF multimers, because high shear stress induces the lateral association of several pVWF multimers into a fibrillar form spVWF multimers. Thus, shear effects on VWF monomeric A2 domains causes an associated increase in the exposure of platelet-binding VWF A1 domains.
FIG. 4.
Free energy landscape of multimeric VWF. pVWF is the native, equilibrium state. Fast stretching of pVWF molecules results in domain unfolding (pathway 1), while the spVWF domain unfolds through a different pathway (pathway 2). High shear stress promotes the pVWF to the metastable spVWF state (pathway 3), which relaxes to a lower energy state, over several hours. The barriers of pathways 1 and 2 result in the force peaks observed for pVWF and spVWF, respectively. The barrier of pathway 3 contributes to the change in peak force in the time-dependent studies of spVWF shown in Fig. 3(b).
In summary, our results demonstrate that pVWF multimers have a different conformational state that unfold via a different pathway after exposure to high shear stress. pVWF is in a native, inactive state that can be converted to a metastable, active state, spVWF, by high shear stress. The peak unfolding force of spVWF multimers is higher than that of unsheared pVWF multimers, potentially because high shear stress induces the lateral association of pVWF multimers into a fibrillar form as indicated by the formation of large VWF aggregates after shear exposure. Thus, an increased intramolecular interaction shifts the domains to a different state that has a higher unfolding barrier. Shear-activated conformational changes in the A2 domains in VWF monomeric subunits of spVWF multimers may provoke an increased exposure of neighboring (platelet-binding) A1 domains. The effect decreases over several hours. It will be interesting to investigate if structural studies can resolve the two states and what external factors, whether physical, chemical, or biological, may affect the stability of and switching rate between these states.
Supplementary Material
Acknowledgments
We thank NIH HL71895 and HL85769 (J.F.D.); NSF 0907676, NIH GM008362, and Welch Foundation C-1632 (C.H.K.); the ANH (DOE/NASA), and NIGTP NIH Grant No. T32EB009379 (E.W.F.); the Mary R. Gibson Foundation and Everett Hinkson Fund (J.M.) for support.
References
- 1.Ruggeri ZM. Curr Opin Hematol. 2003;10:142. doi: 10.1097/00062752-200303000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Sadler JE. Annu Rev Biochem. 1998;67:395. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 3.Moake JL, Rudy CK, Troll JH, Weinstein MJ, Colannino NM, Azocar J, Seder RH, Hong SL, Deykin D. New Engl J Med. 1982;307:1432. doi: 10.1056/NEJM198212023072306. [DOI] [PubMed] [Google Scholar]
- 4.Fowler WE, Fretto LJ, Hamilton KK, Erickson HP, McKee PA. J Clin Invest. 1985;78:1491. doi: 10.1172/JCI112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Halvorsen K, Zhang C-Z, Wong WP, Springer TA. Science. 2009;324:1330. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Choi H, Nolasco L, Pownall HJ, Voorberg J, Moake JL, Dong J-F. J Thromb Haemost. 2008;6:1135. doi: 10.1111/j.1538-7836.2008.02991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong JF, Moake JL, Nolasco L, Bernardo A, Arceneaux W, CNSL, McIntire V, Fujikawa K, Lopez JA. Blood. 2002;100:4033. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 8.Choi H, Aboulfatova K, Pownall HJ, Cook R, Dong J-F. J Biol Chem. 2007;282:35604. doi: 10.1074/jbc.M704047200. [DOI] [PubMed] [Google Scholar]
- 9.Schneider SW, Nuschele S, Wixforth A, Gorzelanny C, Alexander-Katz A, Netz RR, Schneider MF. Proc Natl Acad Sci USA. 2007;104:7899. doi: 10.1073/pnas.0608422104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siedlecki CA, Lestini BJ, Kottke-Marchant K, Eppell SJ, Wilson DL, Marchant RE. Blood. 1996;88:2939. [PubMed] [Google Scholar]
- 11.Alexander-Katz A, Schneider MF, Schneider SW, Wixforth A, Netz RR. Phys Rev Lett. 2006;97:138101. doi: 10.1103/PhysRevLett.97.138101. [DOI] [PubMed] [Google Scholar]
- 12.Yeh H-C, Zhou Z, Choi H, Tekeoglu S, WM, Wang C, Turner N, Scheiflinger F, Moake JL, Dong J-F. J Thromb Haemost. 2010;8:2778. doi: 10.1111/j.1538-7836.2010.04094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.See Supplemental Material for sample preparation and experimental methods.
- 14.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 15.Marszalek PE, Lu H, Li H, Carrion-Vazquez M, Oberhauser AF, Schulten K, Fernandez JM. Nature. 1999;402:100. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 16.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Proc Natl Acad Sci USA. 1999;96:3694. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris NC, Song Y, Kiang C-H. Phys Rev Lett. 2007;99:068101. doi: 10.1103/PhysRevLett.99.068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris NC, Kiang C-H. Phys Rev E. 2009;79:041912. doi: 10.1103/PhysRevE.79.041912. [DOI] [PubMed] [Google Scholar]
- 19.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Müller DJ. Science. 2000;288:143. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 20.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265:1599. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 21.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Jr, Bustamante C. Science. 2003;299:1892. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakajin OB, Duke TAJ, Chou CF, Chan SS, Austin RH, Cox EC. Phys Rev Lett. 1998;80:2737. doi: 10.1103/PhysRevLett.86.1378. [DOI] [PubMed] [Google Scholar]
- 23.Moake JL, Turner NA, Stathopoulos NA, Nolasco LH, Hellums JD. J Clin Invest. 1986;78:1456. doi: 10.1172/JCI112736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldauf C, Schneppenheim R, Stacklies W, Obser T, Pienconka A, Schneppenheim S, Budde U, Zhou J, Grater F. J Thromb Haemost. 2009;7:2096. doi: 10.1111/j.1538-7836.2009.03640.x. [DOI] [PubMed] [Google Scholar]
- 25.Jakobi AJ, Mashaghi A, Tans SJ, Huizinga EG. Nat Commun. 2011;2:385. doi: 10.1038/ncomms1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Lin J, Cruz MA, Dong J-F, Zhu C. Blood. 2010;115:370. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iozzi MF, Helgaker T, Uggerud E. J Phys Chem A. 2011;115:2308. doi: 10.1021/jp109428g. [DOI] [PubMed] [Google Scholar]
- 28.Ying J, Ling Y, Westfield LA, Sadler JE, Shao J-Y. Biophys J. 2010;98:1685. doi: 10.1016/j.bpj.2009.12.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popa I, Fernandez JM, Garcia Manyes S. J Biol Chem. 2011;286:31072. doi: 10.1074/jbc.M111.264093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. Nat Struct Biol. 2009;16:281. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 31.Yang WY, Gruebele M. Nature. 2003;423:193. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 32.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Annu Rev Phys Chem. 1997;48:545. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 33.Gore J, Brant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. Nature. 2006;439:100. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 34.S̈ali A, Shakhnovich E, Karplus M. Nature. 1994;369:248. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 35.Wuite GJL, Smith SB, Young M, Keller D, Bustamante C. Nature. 2000;404:103. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.