SUMMARY
Huntington’s disease (HD) is caused by a CAG expansion in the huntingtin gene. Expansion of the polyglutamine tract in the huntingtin protein results in massive cell death in the striatum of HD patients. We report that human induced pluripotent stem cells (iPSCs) derived from HD patient fibroblasts can be corrected by the replacement of the expanded CAG repeat with a normal repeat using homologous recombination, and that the correction persists in iPSC differentiation into DARPP-32-positive neurons in vitro and in vivo. Further, correction of the HD-iPSCs normalized pathogenic HD signaling pathways (cadherin, TGF-β, BDNF, and caspase activation) and reversed disease phenotypes such as susceptibility to cell death and altered mitochondrial bioenergetics in neural stem cells. The ability to make patient-specific, genetically corrected iPSCs from HD patients will provide relevant disease models in identical genetic backgrounds and is a critical step for the eventual use of these cells in cell replacement therapy.
INTRODUCTION
Huntington’s disease (HD) is a progressive autosomal dominant neurodegenerative disorder caused by an expansion of polyglutamine (CAG) repeats in the huntingtin (HTT) protein. HD patients have progressive motor dysfunction, cognitive decline, and psychological problems. The age of onset for these symptoms is directly correlated with the number of repeats, where more than 36 repeats is considered a pathological threshold. With no effective disease modifying therapies yet developed for HD, regenerative medicine may offer new therapeutic approaches for treating the disease. While preliminary clinical trials using fetal neural grafts showed modest success (Bachoud-Lévi et al., 2006; Dunnett and Rosser, 2004), recent studies have looked to build upon progress in the human stem cell field to explore a potentially more expandable and adaptable source of cells for transplantation therapy (Dey et al., 2010; Schwarz and Schwarz, 2010; Snyder et al., 2010; Vazey et al., 2010).
The recent landmark discovery that somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) (Okita et al., 2007) sparked a confluence of studies to understand their growth and manipulation, as well as their potential use in regenerative medicine (Brennand et al., 2011; Park et al., 2008; Wu and Hochedlinger, 2011). In devastating neurological diseases such as HD, Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), and in stroke and spinal cord injury, it is the loss of neurons and the inability to efficiently mobilize inherent regenerative mechanisms to recover from the acute or progressive damage that underlies the pathology and prognosis. As such, stem-cell-based therapies hold promise for the future treatment of these diseases where currently no disease-modifying therapy exists. Complimentary to stem cell replacement therapies is the study of human stem cells as disease models to identify novel drugs by providing a platform for a better understanding of these diseases at the molecular and cellular level (Ku et al., 2010; Ruby and Zheng, 2009). In accord with this goal, we recently described an HD-iPSC-derived neural stem cell (NSC) model to understand disease pathogenesis and screen for drugs in HD research (Zhang et al., 2010).
The ability to genetically modify human stem cells is an invaluable approach for both modeling diseases and providing potential therapies. Gene targeting by homologous recombination is one feasible and surgically targeted modification that holds promise for providing normal cells that can be used to treat genetic diseases such as HD. Such methods have been well developed into highly optimized protocols for the genetic manipulation of mouse embryonic stem cells (ESCs). Yet due to a variety of technical challenges posed by human stem cell manipulation, gene targeting in human ESCs and iPSCs has not been as well established (Costa et al., 2007; Davis et al., 2009; Ruby and Zheng, 2009; Song et al., 2010; Urbach et al., 2004; Yusa et al., 2011; Zou et al., 2009; Zwaka and Thomson, 2003). Thus, gene targeting protocols in human pluripotent stem cells are still in their infancy, and require further development to establish their feasibility as tools for regenerative medicine.
As a critical step in advancing gene targeting strategies for the correction of disease mutations, we established methods to perform genetic correction in iPSCs derived from an HD patient. We obtained a line of HD-patient-derived iPSCs (HD-iPSC) established and characterized by G.Q. Daley’s group (Park et al., 2008) and us (Zhang et al., 2010). The results of our study are described below.
RESULTS
Targeted Correction of the Expanded HTT Gene in HD-iPSCs
To achieve successful gene targeting in HD-iPSCs, we developed efficient and reproducible methods for transfection and stable selection of human ESCs and iPSCs. We optimized a nucleofection-based protocol adapted to single-cell dissociated feeder free cultures of iPSCs to achieve maximal transfection efficiency and cell viability (Song et al., 2010; Zou et al., 2009; Zwaka and Thomson, 2003). In human cells, lower targeting efficiencies have led several groups to employ longer homologous arms (5–100 kb) with marked increases in efficiency reported (Frendewey et al., 2010; Song et al., 2010). Therefore we engineered longer homologous arms from a bacterial artificial chromosome (BAC) clone containing the entire HTT gene with normal CAG length (21 CAG). The targeting vector (Figure 1A) comprises a 4.5 kb short arm and a 10 kb long arm flanking the CAG repeat region plus a 1.5 kb upstream sequence of Htt. A removable cassette that contains a PGK-neo and the CMV-EGFP is incorporated at the 5′ end of the upstream sequence. Thus, the span of DNA that is targeted and replaced includes exon 1, the CAG repeat region, and the 1.5 kb of upstream sequence. The effective changes to the final targeted allele amount to the CAG repeat length, and insertion of the removable neomycin and EGFP cassette upstream of the reported promoter region.
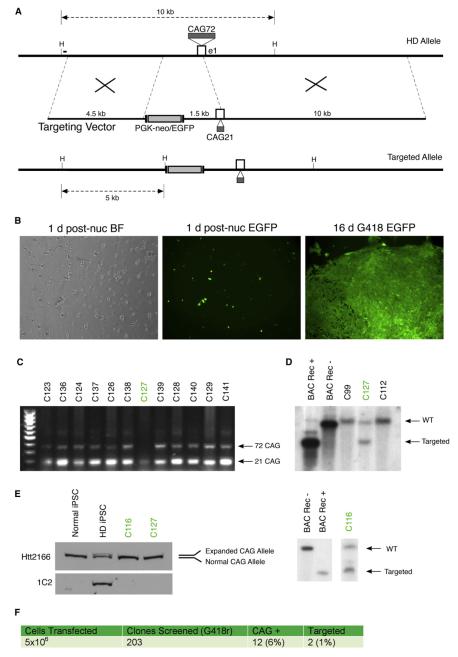
Figure 1. Targeted Correction of the Expanded Allele in HD-iPSCs.
(A) Exon 1 (e1) and surrounding regions of the expanded HD locus of patient-derived HD-iPSCs containing 72 polyglutamine repeats; the designed targeting vector from BAC RP11-866L6 with short and long arms of 4.5 and 10 kb respectively; and the resulting correctly targeted allele with 21 polyglutamine repeats and FLP flanked PGK-neo and EGFP cassettes inserted upstream of the reported promoter region. HindIII sites (H) and resulting targeted fragment sizes using the 300 bp external Southern probe (black bar) are shown.
(B) Brightfield (BF) and GFP fluorescent images of HD-iPSCs nucleofected with pEGFP plasmid at 1 day postnucleofection and after 3 weeks of G418 selection.
(C) Primary PCR-based screen of G418 resistant clones for loss of the expanded (72 CAG) allele. Targeted clones indicated in green were later verified by Southern blot as correctly targeted alleles.
(D) Southern blotting analysis of HindIII digested HD-iPSC clone genomic DNA and control samples. A P32 radiolabeled 300 bp external probe hybridizes with a 10 kb nontargeted (WT) or 5 kb targeted HindIII. Labeled are positive and negative BAC DNA controls for cassette insertion or noninsertion (BAC Rec+ and BAC Rec−, respectively); nontargeted HD-iPSC clones C99 and C112; and correctly targeted HD-iPSC clones C127 and C116. For further Southern blot analysis and sequencing of clones, see Figure S1.
(E) Expanded polyglutamine correction revealed by western blot analysis of lysates from HD-iPSCs and corrected clones C116 and C127 using antibodies for huntingtin (Htt2166) and expanded polyglutamine (1C2). A line of normal iPSCs (Park et al., 2008) derived from non-HD patient fibrobasts is included as a wild-type allele control.
(F) Summary of the gene targeting experiment and resulting targeting frequencies observed.
HD-iPSCs (Park et al., 2008) were nucleofected with both the targeting construct and pEGFP controls. These iPSCs were reprogrammed from HD patient fibroblast cells in which the expanded allele CAG repeat length is 72. EGFP-expressing cells are visible 1 day after nucleofection (Figure 1B). After a period of 2–3 weeks with selection on conditioned medium containing 50 μg/ml G418, discrete colonies were manually passaged into individual wells for expansion and screening.
In order to reduce the large screening population, we first designed a primary screen amplifying the CAG repeat region of the Htt gene. This screen was intended not as proof of gene targeting, but as a step by which we could reduce the screening pool for Southern analysis. Clones were determined to be targeted candidates if the PCR showed amplification of the normal allele in the absence of the expanded allele product (Figure 1C). While this screen does not exclude false positives such as nonspecific contraction, deletions, or PCR-based artifacts, it reduced our screening population to 6% of the original pool (Figure 1F). Candidate alleles were then screened using classic Southern blot analysis. A 300 bp probe external to the 5′ short arm hybridizes to a 10 kb HindIII fragment generated by a nontargeted allele, while a targeted recombination introduces a HindIII site 1.5 kb upstream of exon 1, resulting in a 5 kb HindIII fragment (Figures 1A and 1D). Clones that were shown by Southern blot analysis to be correctly targeted were verified again with independent genomic DNA preparations. In addition, a second Southern blot using an internal neomycin probe was performed to confirm the absence of other off-target integrants (Figure S1A available online). Among 203 clones screened, 12 clones were found to be CAG positive, and 2 (c-HD-iPSC-clone C127 and c-HD-iPSC-clone C116) of the 12 clones were further verified by Southern blot analysis to be correctly targeted (Figure 1D), roughly 1% of the colonies screened (Figure 1F). This rate of recombination is consistent with previous reports of homologous recombination in human ESCs (Costa et al., 2007; Sakurai et al., 2010; Zwaka and Thomson, 2003). Sequencing of exon 1 region was performed to determine CAG repeat lengths. The original HD-iPSC line sequenced has two alleles of 73 and 19 CAG repeats, while corrected lines C116 and C127 have sequences with 21 CAG repeats for C116 and 20 CAG repeats for C127 (Figure S1B).
Western blots of iPSCs derived from controls without disease (normal iPSCs), HD-iPSCs, c-HD-iPSC-clone C127, and c-HD-iPSC-clone C116 confirm the concordant loss of expanded-length Htt protein (Figure 1E). This is shown by the separation of mutant Htt from the wild-type Htt in the western blot (lane 2), as detected by Htt antibody 2166 (which recognizes both expanded and nonexpanded protein) and by the use of disease-specific antibody 1C2 (which recognizes the expanded polyglutamine stretch). As expected, both corrected (C116 and C127) and normal iPSCs express only the wild-type Htt protein.
Corrected HD-iPSCs Retain Pluripotent Characteristics
Next, we investigated whether the corrected HD-iPSCs maintained the pluripotent characteristics of the original HD-iPSC clone. The original HD-iPSC clone had been well characterized with a variety of assays for pluripotency (Park et al., 2008). In further characterizing the corrected HD-iPSCs, we chose to apply several widely accepted assays in order to evaluate pluripotency and genetic stability. Immunocytochemistry (ICC) staining for a panel of stem cell markers showed uniform expression of these classical pluripotency markers, including Nanog, Sox2, Oct4, SSEA4, and TRA-1-60 (Figure 2A). Further, immunostaining of attached embryoid bodies (EBs) derived from these clones showed cells differentiated into endoderm, ectoderm, and mesoderm layers (Figure 2B). Karyotyping and G-banding analysis of both clones were performed to confirm the absence of chromosomal abnormalities (Figure 2C).
Figure 2. Characterization of Corrected HD-iPSC Clones.
(A) Corrected HD-iPSC line C127 immunostained for stem cell markers (Oct4, Sox2, Nanog, SSEA4, and TRA-1-60), costained with DAPI.
(B) Immunostaining of C127-derived embryoid bodies shows differentiation into ectoderm (Nestin), mesoderm (smooth muscle actin, SMA), and endoderm (Sox17) fates.
(C) Karyotyping and G-banding analysis confirms the absence of genetic abnormalities in one line (C127) of corrected HD-iPSC clones.
(D) Genes altered as a function of genetic correction in the CAG locus of HTT in pathways and domain interactions.
(E) Gene array analysis indicates that the corrected clones remain similar to the HD-iPSC clones, suggesting that the genetic correction did not alter the gene expression profile of the cells. See Figure S2 and Table S1 for microarray analysis.
(F) RT-qPCR verification of altered pathways in uncorrected HD-iPSCs compared with corrected HD-iPSCs and normal iPSCs. RNA samples were harvested from HD-iPSCs, normal iPSCs, and two lines of corrected HD-iPSCs. RT-qPCR was performed to show upregulation of cadherin family genes and down-regulation of TGF-β pathway genes. One-way ANOVA is used for statistical analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
To establish that the genetically corrected HD-iPSCs were similar to our starting cells, we performed gene expression profiling (Figure 2D, accession number GSE37547). Strikingly, we found very few changes in mRNA expression levels in our genetically corrected HD-iPSCs when compared to the starting HD-iPSCs, suggesting that they maintained their original pluripotency and that the transfection process did not cause gross alterations in gene expression. The two groups do not separately cluster as demonstrated by hierarchical clustering and principal component analysis of the corrected HD-iPSCs and the starting HD-iPSCs (Figures S2A and S2B).
Htt CAG Contraction Corrects Cadherin and TGF-β Signaling Pathways
However, when we compared the corrected HD-iPSC group (n = 8) versus the starting HD-iPSC group (n = 8) for significant differential gene expression using a t test with a false discovery rate of 5% (Benjamini-Hochberg), we detected 259 differentially expressed genes (Table S1). These results suggest that any changes we observe are due to the CAG contraction because these cells have an identical genetic background. Pathway analysis of the genes that were changed suggested that cadherin family, TGF-β, and caspase-related signaling molecules were altered in the nonexpanded iPSCs (Figure 2E). More specifically, we detected that the Interpro protein domains “IPR013164:Cadherin, N-terminal,” “IPR002126:Cadherin,” and “IPR002190:MAGE protein” were overrepresented in genes that have a lower expression level in the HD-iPSCs, and that the KEGG pathway “hsa04350:TGF-beta signaling pathway” and Reactome pathway “REACT_604:Hemostasis” were statistically enriched in genes with a higher expression level in HD-iPSCs. These pathways are altered in HD models and postmortem tissue (Battaglia et al., 2011; Kandasamy et al., 2010; Reis et al., 2011). Further, reverse transcriptional quantitative PCR (RT-qPCR) analysis was used to verify the gene expression change on these targets using independent samples from HD-iPSCs, corrected HD-iPSCs (C116 and C127), and normal iPSCs (Figure 2F). When comparing corrected HD-iPSCs and normal iPSCs to HD-iPSCs, the RT-qPCR showed upregulation of cadherin family genes, including protocadherin 11 (PCDH11), protocadherin beta 13 (PCDHB13), and protocadherin gamma subfamily A10 and A2 (PCDHGA10 and PCDHGA2), and downregulation of TGF-β pathway genes, including cyclin-dependent kinase inhibitor 2B (CDKN2B), inhibitor of DNA binding 2 (ID2), inhibitor of DNA binding 4 (ID4), paired-like homeodomain transcription factor 2 (PITX2), thrombospondin 1 (THBS1), and left-right determination factor 2 (LEFTY2).
It should be noted that when we used gene array analysis to compare iPSCs from normal individuals to either HD-iPSCs or corrected iPSC lines, we found that the number of differentially expressed genes differed by an order of magnitude more over the number resulting from the comparison of the isogenic HD-iPSCs and the corrected iPSC pair (data not shown). Thus, while correction of the Htt expansion may confer an allelic similarity to a normal cell line in terms of the Htt locus, it is clear that cell lines derived from normal but genetically diverse individuals are an inappropriate point of reference.
Reversal of HD Phenotype in Genetically Corrected HD-NSCs
We sought to establish whether genetic correction results in the reversal of previously established HD disease phenotypes. In other words, are we correcting both the genetic mutation and the phenotype causing the disease? This of course is particularly important if these cells are going to be eventually utilized in patient-based stem cell therapies.
Given that targeted correction of the HTT gene results in an isogenic background between the HD-iPSC and corrected iPSC lines, it is not surprising to find that correction of the expanded allele results in very few changes in expression at the undifferentiated iPSC state. Consistent with this, we observed that corrected iPSCs showed no significant changes in proliferation when compared to HD-iPSCs (Figures S3A and S3B). Therefore, we investigated the phenotype of NSCs derived from HD-iPSCs, corrected HD-iPSCs, and iPSCs derived from healthy individuals. One of the hallmarks of HD is massive cell death found in the striatum and cortex. Therefore, using the TUNEL assay, we tested whether, under growth factor withdrawal, the NSCs derived from HD patients were more vulnerable to cell death than the corrected NSCs or normal NSCs. We found that NSCs derived from uncorrected HD-iPSCs (HD-NSCs) had an ~2-fold increase in TUNEL-positive cells when compared to normal NSCs or NSCs from two corrected HD-iPSC lines (C116-NSCs and C127-NSCs) (Figures 3A and 3B).
Figure 3. Corrected HD-iPSC NSCs Have a Phenotype Similar to That of Normal iPSC NSCs.
(A and B) TUNEL staining and quantification of NSCs derived from uncorrected HD-iPSCs (HD-NSCs), two lines of corrected HD-iPSCs (C116-NSCs and C127-NSCs), and healthy iPSCs (normal NSCs).
(C) NSCs derived from uncorrected HD-iPSCs showed an elevated caspase-3/7 activity upon growth factor withdrawl. +GF, normal culture condition with growth factors; −GF, growth factor deprived condition. We did not detect phenotypes in iPSCs; see Figure S3.
(D) NSCs from corrected and uncorrected HD-iPSCs show that BDNF levels in corrected NSCs are consistent with those of a normal phenotype.
(E) NSCs derived from corrected cells show higher maximum respiration capacity than NSCs from uncorrected HD-iPSCs. Cells were analyzed on a Seahorse XF24 Extracellular Flux Analyzer for oxygen consumption rate (OCR). The average OCR after addition of FCCP (an uncoupler) reflects mitochondria maximum respiration capacity. Corrected NSCs and normal NSCs showed higher maximum respiration capacity than uncorrected HD-NSCs. Also see Figure S3D.
(F) RT-qPCR of TGF-β1 showed a decreased level of TGF-β1 mRNA in HD-NSCs compared with that in corrected and normal NSCs.
(G and H) RT-qPCR and western blotting of N-cadherin showed a decreased level of N-cadherin mRNA and protein in HD-NSCs compared with that in corrected and normal NSCs.
One-way ANOVA is used for statistical analysis (*p < 0.05, **p < 0.01, ***p < 0.001).
We previously found that HD-iPSC-derived NSCs had increased caspase activity compared with NSCs derived from either iPSCs without a CAG expansion or H9 ESCs (Park et al., 2008; Zhang et al., 2010). Elevated caspase activity upon stress has been observed in HD models and patients (Hermel et al., 2004; Kiechle et al., 2002; Squitieri et al., 2011). Therefore, we hypothesized that genetic correction of the CAG repeat in the HD-iPSC should normalize this pathway and HD-associated phenotypes. We tested the NSCs derived from uncorrected and corrected HD-iPSCs (clone 116 and clone 127) and iPSCs with normal repeat length for caspase-3/7 activity after growth factor deprivation. Indeed, we found that only HD-NSCs showed a significant increase in caspase-3/7 activity upon growth factor deprivation. In contrast, the two groups of corrected NSCs (C116-NSCs and C127-NSCs), as well as NSCs bearing a normal repeat length, did not show an increase in caspase-3/7 activity upon growth factor withdrawal (Figure 3C). We also tested for caspase-3/7 activity in uncorrected and corrected HD-iPSCs (clone 116 and clone 127) after serum/growth factor withdrawal. Similarly to the proliferation experiments, we found no difference in measured change of caspase-3/7 activity upon growth factor/serum withdrawal in the undifferentiated iPSC state (Figure S3C). Interestingly the difference between iPSC and NSC results suggests that polyglutamine expansion makes cells more vulnerable to cell death only after differentiation. This may be relate to Htt’s function in neural development where loss of Htt results in disturbed cell migration, reduced proliferation, and increased cell death (Tong et al., 2011).
The observed susceptibility to growth factor withdrawal in HD-NSCs may correlate with the well-documented alteration in BDNF levels in HD patients and models of HD (Zuccato et al., 2001; Zuccato et al., 2008). Strikingly, we found that the genetic correction of the CAG expansion in HD-iPSCs (C116 and C127) corrected the altered BDNF levels found in NSCs derived from these cells (Figure 3D).
Another phenotypic difference between HD and normal cells is reduced bioenergetics of mitochondria in HD (Mochel and Haller, 2011). We investigated a key bioenergetic property, oxygen consumption rate (OCR), in uncorrected and corrected NSCs. The XF24 extracellular flux analyzer (Seahorse Biosciences) measured OCR in real time and we were able to compare uncorrected, corrected, and normal NSCs for respiratory rate. We found that maximum respiration (average OCR after addition of FCCP, an uncoupler) was higher in corrected NSCs or normal NSCs as compared with uncorrected HD-NSCs (Figure 3E), suggesting that genetic correction of CAG repeat resulted in enhanced maximum respiration and mitochondrial function in NSCs. The OCR changes in response to sequentially added oligomycin, FCCP, and rotenone/antimycin A are indicative of the appropriate response in this assay (Figure S3D).
Our gene array analysis pointed out altered expression of genes belonging to cadherin and TGF-β signaling pathways in uncorrected HD-iPSCs compared with corrected or normal iPSC cells (Figure 2F). Previous work has also demonstrated altered cadherin and TGF-β signaling in cellular and mouse models of HD (Battaglia et al., 2011; Kandasamy et al., 2010; Reis et al., 2011). Therefore, we investigated whether these signaling pathways were altered in NSCs. We found that the level of TGF-β1 mRNA was lower in the HD-NSCs compared with that in normal NSCs or corrected NSCs (C116-NSCs or C127-NSCs) (Figure 3F). We also found that mRNA and protein levels of N-cadherin were lower in the HD-NSCs compared with those of normal NSCs or corrected NSCs (Figures 3G and 3H).
Striatal Differentiation of Corrected HD-iPSCs In Vitro and In Vivo
Using the differentiation protocol we previously described (Zhang et al., 2010) with slight modifications, we were able to direct the corrected HD-iPSCs along the differentiation pathway into NSCs, as shown by immunocytochemical staining of the NSC-specific markers Nestin (Figure 4A) and Sox1 (Figure S4A). Then these NSCs were further induced into striatal neurons, as shown by the expression of DARPP-32, calbindin, and gamma-aminobutyric acid (GABA) (Figures 4B and 4C).
Figure 4. Directed Differentiation of Corrected HD-iPSC Clones.
(A) NSCs were derived from corrected HD-iPSCs with the EB method and were stained for the NSC marker Nestin.
(B) NSCs were further induced to striatal neurons with expression of DARPP-32 and βIII-tubulin.
(C) Calbindin and GABA is shown by ICC staining.
(D) Two weeks after striatal injection of corrected NSCs (C116-NSCs), immunofluorescence of R6/2 mouse brain sections detects human-specific STEM121 and DAPI.
(E–G) STEM121 and MAP2 R6/2 mouse brain sections counterstained with DAPI.
(H and I) STEM121 and GABA R6/2 mouse brain sections counterstained with DAPI.
(J and K) STEM121 and DARPP-32 R6/2 mouse brain sections counterstained with DAPI.
(L) STEM121 and GFAP R6/2 mouse brain sections counterstained with DAPI.
For further analysis see Figure S4.
Next the ability of corrected iPSC-derived NSCs to populate the striatum was assessed given recent stem cell replacement studies with human ESCs supporting functional recovery in a quinolinic acid model of HD (Ma et al., 2012). NSCs derived from corrected iPSCs were transplanted into the striatum of HD mouse model R6/2 brains at 6 weeks of age. Two weeks after injection, brain sections were stained for markers specific for human cytoplasmic protein (STEM121) or human nuclear antigen (HuNu). Confocal imaging reveals an abundant number of STEM121-positive cells in the striatum of these mice (Figure 4D), suggesting that the corrected NSC survived transplantation. Further, we found human cells colabeled with neuronal marker MAP2 (Figures 4E–4G), medium spiny neuron marker DARPP-32 (Figures 4J and 4K), and GABAergic neuron marker GABA (Figures 4H and 4I). We also found glial-specific marker GFAP (Figure 4L) colabeled with STEM121, although at a lower percentage. Individual panels of the staining are shown in Figure S4B. These results suggest that corrected HD-iPSC-derived NSCs are able to populate the striatum of HD mouse model brains and undergo differentiation in vivo to DARPP-32-expressing neurons and glial cells.
DISCUSSION
The recent rapid progress in the advancement of reprogramming technology facilitating the more efficient and accessible generation of patient-specific iPSCs has opened the door to the development of strategies for iPSC-based disease modeling and regenerative therapy (Ye et al., 2009; Yu et al., 2007; Zou et al., 2009). In this study, we have demonstrated the use of gene targeting techniques to achieve a targeted correction of the expanded Htt allele in iPSCs derived from an HD patient. Targeted correction by our strategy results in the reduction of the expanded allele of the HTT gene to normal 21 polyglutamine repeats, while maintaining the pluripotent characteristics and unique gene profile of the original patient-specific iPSC line. We further demonstrate the ability for these corrected iPSCs to undergo directed differentiation into DARPP-32-positive striatal neurons in vitro and in vivo. Most significantly, we show that correction results in reversal of disease phenotype in NSCs derived from the corrected HD-iPSCs.
Despite recent success in using zinc-finger nuclease (ZFN)-mediated gene targeting (Hockemeyer et al., 2009; Zou et al., 2009), we chose not to pursue this strategy due to the time and resources required for experimentally optimizing the binding specificity of zinc-finger motifs, and, particularly relevant to CAG mutations, we wanted to avoid “off-target” effects. Our positive results further indicated that targeting frequencies as observed here and previously reported are within the reasonable range of achievability, given efficient protocols and a well designed targeting strategy. In the context of regenerative therapeutics, removal of cassette DNA containing selection markers would be a clear requirement, and as such, we designed our vectors with this capability in mind, and they can be utilized with multiple patient iPSCs.
With the inherent limitations of previous clinical xenograft transplantation approaches, which are prone to immunorejection and limited tissue supply, focus has shifted to stem-cell-based sources for transplantation (Dunnett and Rosser, 2004; Kelly et al., 2009). The ability to combine genetic manipulation technology with disease-specific iPSCs creates the potential for generation of autologous cells for cell-replacement therapy. While gene targeting approaches have been demonstrated previously in iPSCs, our current study provides an important proof of principle experiment demonstrating the genetic correction of the HD-causing mutation in patient-derived iPSCs. It is of particular significance that we find that genetic correction reverses disease phenotypes in these cells. Further, we provide evidence for the capability of these cells to populate and differentiate in the diseased striatum in vivo, which complements recent studies showing functional rescue in a quinolinic acid model of HD (Ma et al., 2012). As such these findings represent a significant contribution to ongoing efforts and future studies aiming to harness the potential of iPSCs for regenerative therapy.
Much focus has been invested on the use of disease-specific iPSCs to develop in vitro cell models for the study of disease pathway mechanisms and, in some cases, toward a platform for drug screening and discovery. In our study, an important goal was the creation of genetically corrected HD-iPSCs, which provides us with a human HD model in which the normal and disease cell lines carry identical genetic backgrounds with respect to each other. Gene-corrected iPSCs may serve as a more closely matched control for the study of HD-mediated gene changes because they are derived from the same patient and thus have a matched genetic background. With this in mind, we performed a preliminary analysis of gene expression differences between HD and corrected iPSCs, which revealed changes to cadherin family genes and TGF-β signaling pathways. Adding further utility is the ability to direct the differentiation of these HD and corrected iPSCs into NSCs and DARPP-32-positive striatal neurons.
In summary, we found that we could correct phenotypes associated with HD using homologous recombination at frequencies of homologous recombination comparable to those in previous reports (Costa et al., 2007; Song et al., 2010; Zwaka and Thomson, 2003). The corrected cells retain pluripotent characteristics, as monitored by ICC and microarray analysis, and could be differentiated into DARPP-32-expressing striatal neurons. One could easily have hypothesized that, due to epigenetic alterations in the diseased cells caused by genetic mutation and aging, the disease process could not be reversed. However, we found that the replacement of the disease gene mutation reversed the phenotype of the diseased cells. The observed elevation of capase-3/7 activity in HD NSCs upon growth factor deprivation recapitulates a well-characterized disease phenotype observed in various rodent and human HD models, and correction of the genetic mutation reversed this phenotype. Further, we found that the altered mitochondrial deficits, the lower levels of BDNF, and the altered cadherin and TGF-β signaling associated with HD were corrected. The NSCs derived from these corrected cells could populate the striatum in an HD mouse model. Our results are a significant advance in working with HD-iPSCs for stem cell replacement therapy and constructing human HD cellular models with the same human genetic background.
EXPERIMENTAL PROCEDURES
Construction of Targeting Vector
A 240 kb BAC (RP11-866L6) containing the 170 kb human HTT locus was modified stepwise using a Ret/ET-based recombineering kit (Genebridges). First, PGK-neo and CMV-EGFP expression cassettes flanked by FRT recognition sites was inserted 1.5 kb upstream of exon 1. The 20 kb fragment, including exon 1, inserted expression cassettes, a 4.5 kb upstream short arm, and a 10 kb downstream long arm, were inserted into a modified pPNT vector (neo cassette removed) adjacent to the HSV-TK.
Culture and Nucleofection of HD-iPSCs
Cell culture reagents were from Invitrogen unless otherwise mentioned. HD-iPSC (HD-iPS4) and normal iPSCs (hFib2-iPS5) (Park et al., 2008) were cultured on either Matrigel (BD) or irradiation inactivated mouse embryonic fibroblasts (MEFs). Further details are in the Supplemental Experimental Procedures.
Karyotyping Analysis and Sequencing
Karyotyping of the targeted iPSC was carried out by Applied Stemcell Inc. (Menlo Park, CA). Sequencing was performed by Laragen. Multiple primer sets were used to amplify the exon 1 region of the HTT gene. For each clone, the two CAG length allele variants were gel isolated and sequenced separately.
Immunocytochemical Staining
Cells grown on coverslips were fixed with 4% paraformaldehyde (Sigma) for 20 min at room temperature (RT), then permeabilized with 0.1% Triton X-100 (Sigma) for 10 min and blocked with 5% donkey serum (Millipore) for 45 min. See Supplemental Experimental Procedures.
Neuronal Differentiation of HD-iPSCs
NSCs were derived from iPSCs with the EB method as previously published and with some modifications. Normal NSCs (HIP Neural Stem Cells) derived from healthy iPSCs using a similar EB method were from GlobalStem. See Supplemental Experimental Procedures.
Western Blotting Analysis
HD-iPSCs, normal iPSCs, and corrected HD-iPSCs were detached by 1 mg/ml collagenase (Invitrogen) incubation at 37°C for 1 hr. See Supplemental Experimental Procedures.
Gene Arrays and RT-qPCR
RNA was extracted from pellets of cell cultures using an automated homogenization and extraction apparatus (QiaCube, QIAGEN, Valencia, CA) according to the manufacturer’s protocols for the RNeasy mini kit (QIAGEN). See Supplemental Experimental Procedures.
Caspase-3/7 Activity Assay
The caspase activity assay was performed with Apo3 HTS kit (Cell Technology).
NSC Injection and Analysis
105 corrected NSCs (C116) were injected bilaterally into the striatum of 6-week-old R6/2 transgenic HD model mice (via the following stereotaxic coordinates: bregma +0.86 AP, ±1.8 ML, and at −3.5, −2.5 DV depths) in a 1 μl volume using a 22G Hamilton syringe. Mice were transcardially perfused with 4% paraformaldehyde at 2 weeks postinjection, brains were processed in a sucrose gradient (10%–30%), and coronal sections (30 μm thick) through the striatum were serially collected. Injected cells were visualized by confocal imaging after immunostaining with human-specific nuclear antigen (1:100, clone 235-1, Millipore) or STEM121 (1:100, Stem Cells Inc.). Colabeling with other neuronal and glial markers was done with antibodies against DARPP-32 (1:20, Cell Signaling), MAP2 (1:100, Millipore), GABA (1:100, Abcam or Millipore), and GFAP (1:100, Sigma).
Statistical Analysis
For all comparisons we used one-way ANOVA analysis. For microarray and enrichment, statistical analysis is noted in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Daley for providing us with the normal iPSCs (hFib2-iPS5) and HD-iPSCs (HD-iPS4). Thanks to Laragen for sequencing of iPSC clones. This work was funded by the Buck Institute for Research on Aging and NIH T32 training grant AG000266 (to M.C.A.).
Footnotes
ACCESSION NUMBERS The accession number for the microarray data gathered in the course of this experiment is GSE37547.
SUPPLEMENTAL INFORMATION Supplemental Information for this article includes four figures, Supplemental Experimental Procedures, and one table and can be found with this article online at doi:10.1016/j.stem.2012.04.026.
REFERENCES
- Bachoud-Lévi AC, Gaura V, Brugières P, Lefaucheur JP, Boissé MF, Maison P, Baudic S, Ribeiro MJ, Bourdet C, Remy P, et al. Effect of fetal neural transplants in patients with Huntington’s disease 6 years after surgery: a long-term follow-up study. Lancet Neurol. 2006;5:303–309. doi: 10.1016/S1474-4422(06)70381-7. [DOI] [PubMed] [Google Scholar]
- Battaglia G, Cannella M, Riozzi B, Orobello S, Maat-Schieman ML, Aronica E, Busceti CL, Ciarmiello A, Alberti S, Amico E, et al. Early defect of transforming growth factor β1 formation in Huntington’s disease. J. Cell. Mol. Med. 2011;15:555–571. doi: 10.1111/j.1582-4934.2010.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat. Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- Davis RP, Grandela C, Sourris K, Hatzistavrou T, Dottori M, Elefanty AG, Stanley EG, Costa M. Generation of human embryonic stem cell reporter knock-in lines by homologous recombination. Curr. Protoc. Stem Cell Biol. 2009 doi: 10.1002/9780470151808.sc05b01s11. Chapter 5, Unit 5B 1 1 1-34. [DOI] [PubMed] [Google Scholar]
- Dey ND, Bombard MC, Roland BP, Davidson S, Lu M, Rossignol J, Sandstrom MI, Skeel RL, Lescaudron L, Dunbar GL. Genetically engineered mesenchymal stem cells reduce behavioral deficits in the YAC 128 mouse model of Huntington’s disease. Behav. Brain Res. 2010;214:193–200. doi: 10.1016/j.bbr.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Dunnett SB, Rosser AE. Cell therapy in Huntington’s disease. NeuroRx. 2004;1:394–405. doi: 10.1602/neurorx.1.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D, Chernomorsky R, Esau L, Om J, Xue Y, Murphy AJ, Yancopoulos GD, Valenzuela DM. The loss-of-allele assay for ES cell screening and mouse genotyping. Methods Enzymol. 2010;476:295–307. doi: 10.1016/S0076-6879(10)76017-1. [DOI] [PubMed] [Google Scholar]
- Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE, Hackam AS, Logvinova AV, Peel AL, Chen SF, et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington’s disease. Cell Death Differ. 2004;11:424–438. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M, Couillard-Despres S, Raber KA, Stephan M, Lehner B, Winner B, Kohl Z, Rivera FJ, Nguyen HP, Riess O, et al. Stem cell quiescence in the hippocampal neurogenic niche is associated with elevated transforming growth factor-beta signaling in an animal model of Huntington disease. J. Neuropathol. Exp. Neurol. 2010;69:717–728. doi: 10.1097/NEN.0b013e3181e4f733. [DOI] [PubMed] [Google Scholar]
- Kelly CM, Dunnett SB, Rosser AE. Medium spiny neurons for transplantation in Huntington’s disease. Biochem. Soc. Trans. 2009;37:323–328. doi: 10.1042/BST0370323. [DOI] [PubMed] [Google Scholar]
- Kiechle T, Dedeoglu A, Kubilus J, Kowall NW, Beal MF, Friedlander RM, Hersch SM, Ferrante RJ. Cytochrome C and caspase-9 expression in Huntington’s disease. Neuromolecular Med. 2002;1:183–195. doi: 10.1385/NMM:1:3:183. [DOI] [PubMed] [Google Scholar]
- Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, Gottesfeld JM. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAA·TTC triplet repeat instability. Cell Stem Cell. 2010;7:631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, Sun Y, Zhang X, Zhang SC. Human Embryonic Stem Cell-Derived GABA Neurons Correct Locomotion Deficits in Quinolinic Acid-Lesioned Mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochel F, Haller RG. Energy deficit in Huntington disease: why it matters. J. Clin. Invest. 2011;121:493–499. doi: 10.1172/JCI45691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germlinecompetent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis SA, Thompson MN, Lee JM, Fossale E, Kim HH, Liao JK, Moskowitz MA, Shaw SY, Dong L, Haggarty SJ, et al. Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum. Mol. Genet. 2011;20:2344–2355. doi: 10.1093/hmg/ddr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- Sakurai K, Shimoji M, Tahimic CG, Aiba K, Kawase E, Hasegawa K, Amagai Y, Suemori H, Nakatsuji N. Efficient integration of transgenes into a defined locus in human embryonic stem cells. Nucleic Acids Res. 2010;38:e96. doi: 10.1093/nar/gkp1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz SC, Schwarz J. Translation of stem cell therapy for neurological diseases. Transl. Res. 2010;156:155–160. doi: 10.1016/j.trsl.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Snyder BR, Chiu AM, Prockop DJ, Chan AW. Human multi-potent stromal cells (MSCs) increase neurogenesis and decrease atrophy of the striatum in a transgenic mouse model for Huntington’s disease. PLoS ONE. 2010;5:e9347. doi: 10.1371/journal.pone.0009347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Squitieri F, Maglione V, Orobello S, Fornai F. Genotype-, aging-dependent abnormal caspase activity in Huntington disease blood cells. J. Neural. Transm. 2011;118:1599–1607. doi: 10.1007/s00702-011-0646-1. [DOI] [PubMed] [Google Scholar]
- Tong Y, Ha TJ, Liu L, Nishimoto A, Reiner A, Goldowitz D. Spatial and temporal requirements for huntingtin (Htt) in neuronal migration and survival during brain development. J. Neurosci. 2011;31:14794–14799. doi: 10.1523/JNEUROSCI.2774-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Vazey EM, Dottori M, Jamshidi P, Tomas D, Pera MF, Horne M, Connor B. Comparison of transplant efficiency between spontaneously derived and noggin-primed human embryonic stem cell neural precursors in the quinolinic acid rat model of Huntington’s disease. Cell Transplant. 2010;19:1055–1062. doi: 10.3727/096368910X494632. [DOI] [PubMed] [Google Scholar]
- Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat. Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc. Natl. Acad. Sci. USA. 2009;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rashid ST, Strick-Marchand H, Varela I, Liu PQ, Paschon DE, Miranda E, Ordóñez A, Hannan NR, Rouhani FJ, et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, An MC, Montoro D, Ellerby LM. Characterization of Human Huntington’s Disease Cell Model from Induced Pluripotent Stem Cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Conforti P, MacDonald ME, Tartari M, Cattaneo E. Systematic assessment of BDNF and its receptor levels in human cortices affected by Huntington’s disease. Brain Pathol. 2008;18:225–238. doi: 10.1111/j.1750-3639.2007.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.