Abstract
While it is now evident that the two Bacteroidales species Bacteroides fragilis and Tannerella forsythia both have general O-glycosylation systems and share a common glycosylation sequon, the ability of these organisms to glycosylate a protein native to the other organism has not yet been demonstrated. Here, we report on the glycosylation of heterologous proteins between these two organisms. Using genetic tools previously developed for Bacteroides species, two B. fragilis model glycoproteins were expressed in the fastidious anaerobe T. forsythia and the attachment of the known T. forsythia O-glycan to these proteins was demonstrated by liquid chromatography electrospray ionization tandem mass spectrometry. Likewise, two predominant T. forsythia glycoproteins were expressed in B. fragilis and glycosylation with the B. fragilis O-glycan was confirmed. Purification of these proteins from B. fragilis allowed the preliminary characterization of the previously uncharacterized B. fragilis protein O-glycan. Based on mass spectrometric data, we show that the B. fragilis protein O-glycan is an oligosaccharide composed of nine sugar units. Compositional and structural similarities with the T. forsythia O-glycan suggest commonalities in their biosynthesis. These data demonstrate the feasibility of exploiting these organisms for the design of novel glycoproteins.
Keywords: Bacteroides fragilis, glycoengineering, O-glycosylation, Tannerella forsythia
Introduction
Many different species of bacteria have been shown to glycosylate proteins (Nothaft and Szymanski 2010). During evolution they have developed complex systems allowing them to systematically transfer glycans onto proteins. General N- (targeting the amide nitrogen of Asn residues) and O- (targeting the hydroxyl oxygen of Ser, Thr or Tyr residues) glycosylation systems have been described in bacteria and knowledge about the molecular mechanisms underlying these processes is continuously expanding. A general N-glycosylation system was described for Campylobacter jejuni (Szymanski et al. 1999), where a heptasaccharide is transferred on to different proteins containing the conserved (D/E)X1NX2(S/T) glycosylation sequon (X1 and X2 can be any amino acid except for Pro) (Young et al. 2002; Kowarik et al. 2006). Recently, general O-glycosylation systems have been described in several bacterial species (e.g. Neisseria gonorrhoeae (Ku et al. 2009; Vik et al. 2009), Bacteroides fragilis (Fletcher et al. 2009), Tannerella forsythia (Posch et al. 2011, 2012) and Acinetobacter baumanii (Iwashkiw et al. 2012). In B. fragilis and T. forsythia, O-glycans are specifically attached to extracytoplasmic proteins containing the conserved D(S/T)(A/I/L/V/M/T) motif (Fletcher et al. 2009; Posch et al. 2011). To date, 20 glycoproteins have been experimentally confirmed in B. fragilis, but the actual number of proteins that are glycosylated in this organism is predicted to be much higher (Fletcher et al. 2011). Similarly, T. forsythia also synthesizes a wide repertoire of glycoproteins (Veith et al. 2009; Posch et al. 2011).
B. fragilis and T. forsythia both belong to the order Bacteroidales, within the Bacteroidetes phylum of bacteria. The Bacteroidales contain anaerobic species that associate with mammalian, vertebrate and invertebrate hosts, largely as commensals, symbionts or pathogens (Wexler 2007). Whereas T. forsythia inhabits the human oral cavity and is considered a periodontal pathogen (Holt and Ebersole 2005; Pihlstrom et al. 2005), B. fragilis colonizes the human intestine where it provides beneficial properties to the host (Mazmanian et al. 2008). Even though T. forsythia and B. fragilis colonize distinct ecosystems, they are phylogenetically close and thus might have evolved comparable protein O-glycosylation systems, which allow them to furnish proteins with glycan moieties using an identical glycosylation sequon (Fletcher et al. 2009; Posch et al. 2011).
In a previous study, we showed that T. forsythia synthesizes a complex oligosaccharide that is attached not only to the two surface (S-) layer proteins TfsA and TfsB of the organism but presumably also to numerous other proteins (Posch et al. 2011). The T. forsythia O-glycan is a complex, structurally unique oligosaccharide that is most likely involved in biofilm formation, as an isogenic mutant lacking a terminal portion of the glycan shows significantly altered biofilm formation compared with the wild-type strain (Honma et al. 2007).
The discovery of broad-spectrum bacterial glycosylation systems along with conserved glycosylation sequons prompted efforts to functionally exploit them for glycoengineering purposes. The use of bacteria for glycoengineering was successfully demonstrated when the C. jejuni N-glycosylation pathway was transferred into Escherichia coli (Wacker et al. 2002). Since then, several studies have shown that a well-directed transfer of glycans onto proteins in E. coli is feasible, which has also been used for the production of homogenous glycoproteins with eukaryotic N-glycans (Feldman et al. 2005; Schwarz et al. 2010; Lizak et al. 2011). Only recently, a bottom-up synthetic pathway in E. coli was engineered, allowing the production of eukaryotic trimannosyl chitobiose glycans and their transfer to specific residues in target proteins (Valderrama-Rincon et al. 2012).
The presence of a common glycosylation sequon in these Bacteroidales species raises the possibility of precisely targeting the addition of glycans onto proteins. In B. fragilis, it was previously demonstrated that introduction of a glycosylation sequon into a naturally unglycosylated B. fragilis protein brings about site-specific glycosylation at the engineered sites (Fletcher et al. 2011).
Here, we report on the transfer of the B. fragilis O-glycan onto heterologously expressed T. forsythia proteins and vice versa. Additionally, the first mass spectrometric characterization of the previously undescribed B. fragilis protein O-glycan is presented. We show that “cross-glycosylation” of proteins in Bacteroidales by utilizing the conserved D(S/T)(A/I/L/V/M/T) motif is feasible, allowing the design of novel glycoproteins.
Results
Expression of His-tagged (glycosylated) proteins in T. forsythia and B. fragilis
Expression of recombinant proteins in B. fragilis is well demonstrated (Smith et al. 1992; Bayley et al. 2000) using E. coli-Bacteroides shuttle plasmids, modification of which has allowed the production of C-terminally His-tagged fusion proteins (Fletcher et al. 2009).
Heterologous protein expression in T. forsythia has not yet been described, which may be attributed both to the inherently demanding nature of the organism's growth and to the lack of suitable genetic tools.
Considering that T. forsythia and B. fragilis are closely related organisms, we set out to determine whether the existing B. fragilis genetic tools could be used in T. forsythia. As the recipient strain, we chose to use an S-layer-deficient T. forsythia mutant strain, to allow better conjugation efficiency compared with the wild-type strain. Two B. fragilis recombinant His-tagged protein encoding genes were chosen for transfer (BF2494, GI: 60681974; BF3567, GI 60683022) as both are glycosylated in their parent strain at the D-(S/T)-(A/I/L/V/M/T) motif (Fletcher et al. 2009). Our approach thus served a double purpose: First, to determine whether expression vectors and procedures developed for B. fragilis would function in T. forsythia and, secondly, to ascertain whether T. forsythia would recognize the provided glycosylation sequons and produce heterologously glycosylated proteins. Likewise, we wanted to investigate whether B. fragilis would attach glycans to the T. forsythia S-layer proteins TfsA and TfsB, thus allowing “cross-glycosylation” of proteins within these two species based on the shared glycosylation target motif.
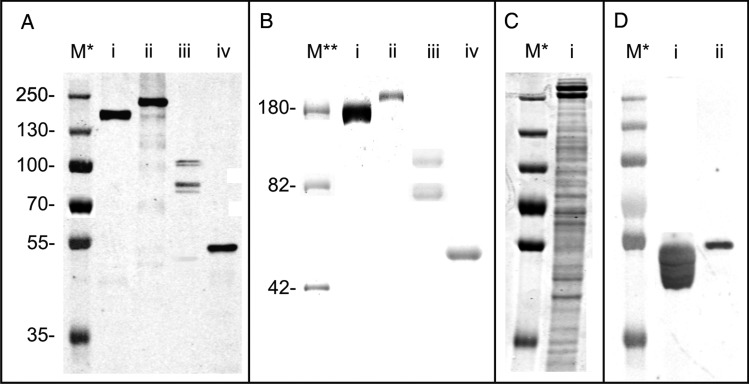
Western blot analysis of purified His-tagged proteins (Figure 1A, lanes i and ii) showed that TfsA and TfsB can be readily expressed in B. fragilis. Noticeably, the masses of both proteins on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) gels were higher than their predicted molecular masses. For TfsA-His (calculated molecular weight (Mw), 133.3 kDa with the signal peptide removed) an upshift to ∼170 kDa was observed, whereas TfsB-His (calculated Mw, 150.8 kDa with the signal peptide removed) was shifted to ∼200 kDa. These data indicated that both proteins are likely glycosylated in B. fragilis, which was further confirmed by glycostaining of the gel (Figure 1B, lanes i and ii). Additionally, comparing the migration behavior of both TfsA and TfsB recombinantly produced in B. fragilis with their native counterparts (Figure 1C) demonstrates that the glycosylation is different in these two species, as natively produced TfsA and TfsB glycoproteins show higher apparent molecular masses (∼230 and ∼270 kDa, respectively).
Fig. 1.
(A) Western blot analysis of (i) TfsA-His, (ii) TfsB-His, (iii) BF3567-His and (iv) BF2494-His purified from B. fragilis (i and ii) and T. forsythia (iii, iv), respectively. The observed Mw of all four proteins is larger than the one calculated, suggesting a posttranslational modification of these proteins. (B) Pro-Q Emerald-stained gel showing that both (i) TfsA-His and (ii) TfsB-His purified from B. fragilis as well as BF3567-His and BF2494-His purified from T. forsythia are glycosylated. (C) Coomassie Blue-stained SDS–PAGE gel of a T. forsythia crude extract (i), showing that native (glycosylated) TfsA (lower of the two broad bands migrating around 250 kDa) and TfsB migrate at higher apparent Mw than their heterologously expressed versions shown in A (lanes i and ii), indicating a difference in posttranslational modification. (D) Western blot probing BF2494-His expressed in (i) B. fragilis wild-type (Fletcher et al. 2009) as well as in (ii) T. forsythia with an anti-BF2494 antibody. BF2494-His from T. forsythia shows altered migration behavior compared with wild-type BF2494-His (which additionally shows a banding pattern indicating a consecutive transfer of monosaccharides), indirectly indicating altered glycosylation. M*, PageRuler Plus prestained protein ladder (Thermo Fisher Scientific, Vienna, Austria); M**, CandyCane glycoprotein molecular weight ladder (Invitrogen).
As shown in Figure 1A (lanes iii and iv), T. forsythia is also capable of expressing C-terminally His-tagged (glyco)proteins from the B. fragilis derived vector using the native B. fragilis promoter, demonstrating the feasibility of using existing Bacteroides genetic tools in this organism. His-tagged BF2494 and BF3567 were purified from T. forsythia and, interestingly, both proteins also exhibited a migration behavior different from that expected from their calculated molecular masses (BF2494-His calculated Mw, 45.9 kDa without the signal peptide, observed Mw ∼55 kDa; BF3567-His calculated Mw, 66.8 kDa without the signal peptide, observed Mw >75 kDa). Again, this suggested that both proteins are posttranslationally modified in T. forsythia. In a western blot, probing with an anti-BF2494-His antibody, an altered migration pattern of BF2494-His expressed in B. fragilis and T. forsythia can be deduced (Figure 1D, lanes i and ii), indirectly indicating altered glycosylation. Noteworthy, BF3567-His expressed in T. forsythia did not show one uniform band upon probing with anti-His antibody (Figure 1A, lane iii) but several bands >75 kDa gave a positive signal, potentially indicative of a consecutive transfer of glycans onto this protein or glycoprotein degradation.
B. fragilis O-glycan analysis
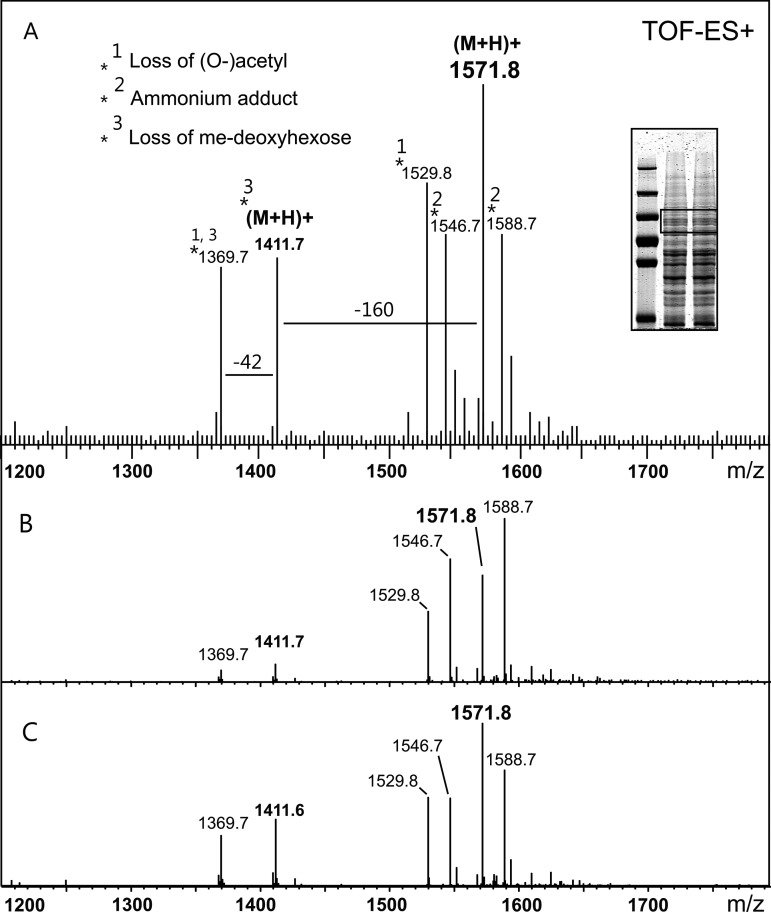
Protein bands originating from a crude extract of a B. fragilis culture were excised from SDS–PAGE gels (Figure 2A, inset) and served as the initial source of B. fragilis glycoproteins for the characterization of its yet undescribed O-glycan. In-gel β-elimination of glycans from these protein bands followed by porous graphitized carbon electrospray ionization mass spectrometry (PGC-ESI-MS) allowed the detection of a 1571.56 (molecular ion (mi), [M + H]+)–Da glycan structure (Supplementary data, Figure S1) showing in-source fragments corresponding to one methylated deoxyhexose and one potentially acetylated glycan constituent (Figure 2A). Collision-induced dissociation (CID) fragmentation analysis of the 1571.56-Da glycan revealed it to be a hetero-oligomer consisting of nine sugar residues (Figure 3). Mass increments for three hexoses (one reduced), two hexuronic acids, two N-acetyl hexosamines (one uncertain), one methylated deoxyhexose, as well as one deoxyhexose were identified (Supplementary data, Table S1). The terminating 203-Da residue, which is suspected to be one of the N-acetyl hexosamines, was found to be highly prone to deacetylation (loss of 42 Da during the strong basic reduction process). Such a propensity for deacetylation would, in our experience, indicate O-acetylation more strongly than N-acetylation. Thus, although the unit mass is appropriate for an N-acetyl hexosamine, the nature of this glycan constituent could not be fully resolved in this study.
Fig. 2.
(A) ESI-TOF-MS spectrum of the B. fragilis glycan as detected after in-gel reductive β-elimination of Coomassie Blue-stained SDS–PAGE bands (see inset, boxed area) originating from a B. fragilis crude protein extract. A dominant glycan structure of 1571.8 Da (M + H)+ was observed, along with an in-source fragment corresponding to one methylated deoxyhexose (−160 Da). Also, one acetyl group was found to be partially lost during β-elimination (−42 Da). (B) Summarized LC-ESI-MS spectrum of the O-glycan as isolated from the T. forsythia S-layer protein TfsA expressed in B. fragilis (see Figure 1A, i). (C) Summarized LC-ESI-MS spectrum of the O-glycan as isolated from the T. forsythia S-layer protein TfsB expressed in B. fragilis (see Figure 1A, ii). Both O-glycan structures observed in (B) and (C) correspond to the B. fragilis O-glycan as isolated in (A), confirming the attachment of the B. fragilis glycan to heterologously expressed T. forsythia proteins. Note: Glycan screening was done with the Micromass Global Ultima instrument (Waters), showing a slight mass deviation compared with high-resolution measurements (Supplementary data, Figure S1).
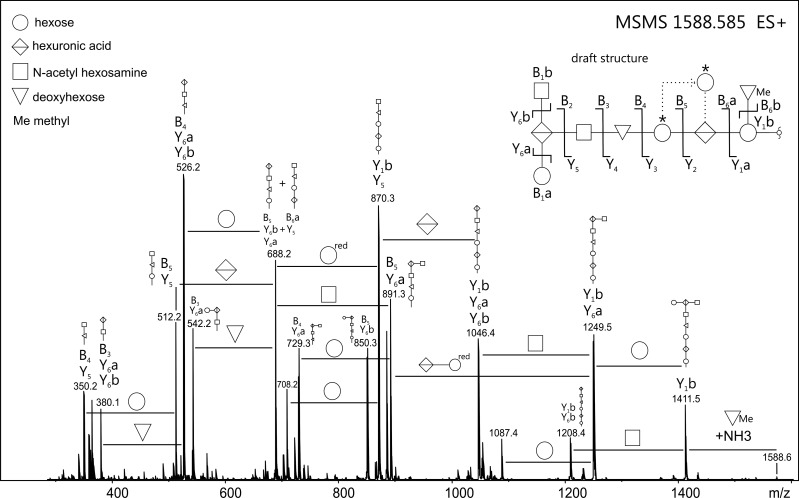
Fig. 3.
ESI-TOF-MS/MS spectrum of the borohydride-reduced B. fragilis O-glycan, as observed from protein bands excised from SDS–PAGE gels. The fragmentation pattern revealed the presence of a nine-sugar oligosaccharide comprising three hexoses, two hexuronic acids, two N-acetyl hexosamines as well as two deoxyhexoses (modified or free). Based on the observed fragment ions, the B. fragilis O-glycan structure was drafted (see inset top right). Fragments were labeled using the nomenclature as described (Domon and Costello 1988). The terminal N-acetyl hexosamine residue is found with considerable uncertainty, both by its accurate mass (see Supplementary data, Table S1) and the fact that it is prone to loss of a 42 Da fragment (indicating O-acetylation rather than N-acetylation) during β-elimination. In addition, the existence of a glycan isoform with the middle of the three hexose residues (marked with asterisk) branching from the hexuronic acid residue cannot be completely ruled out. Potential rearrangement artifacts upon CID were excluded upon measurement of sodium adducts of the B. fragilis O-glycan (Wuhrer et al. 2011; data not shown). Note: Line positions between residues of the draft B. fragilis O-glycan structure do not represent actual linkage types.
The fragment spectra gave sufficient information to putatively assign the sequence and branching pattern of the B. fragilis glycan (Figure 3, Supplementary data, Figure S2). Interestingly, β-elimination and subsequent glycan analysis of TfsA-His and TfsB-His glycoprotein bands purified from B. fragilis gave identical mass spectra, indicating the successful O-glycan transfer onto those proteins (Figure 2B and C).
“Cross-glycosylation” of B. fragilis and T. forsythia proteins
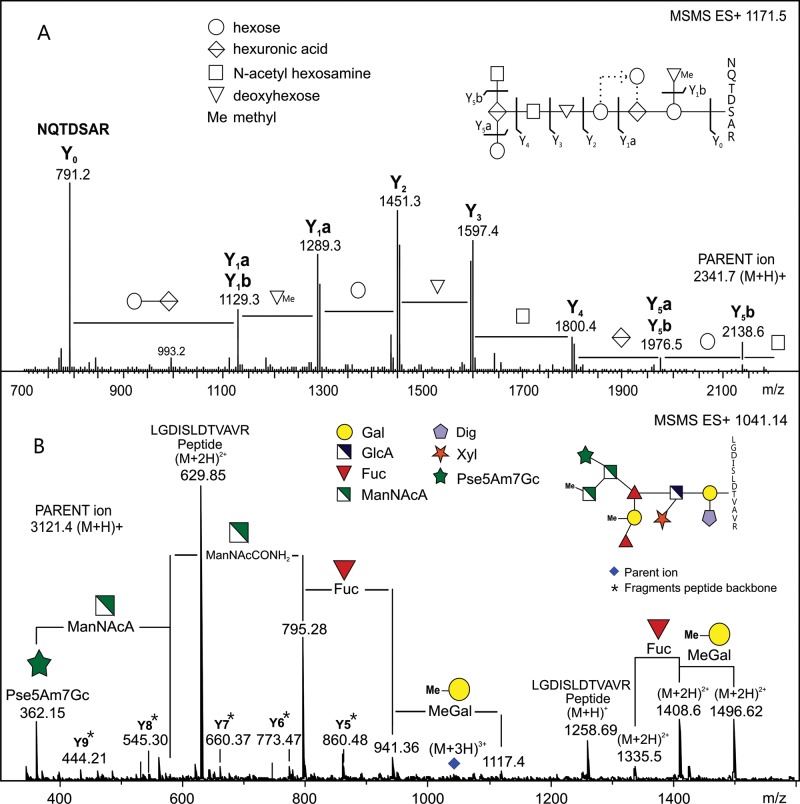
To analyze for heterologous glycosylation events, we excised Coomassie Blue-stained bands of our selected target proteins from SDS–PAGE gels and performed in-gel tryptic digests followed by reverse phase ESI-MS/MS (glyco)peptide mapping. Modified peptides of TfsA and TfsB synthesized in B. fragilis were identified by the observed masses not matching those of the predicted tryptic peptide masses. We subjected one of the putative glycopeptides derived from the TfsA protein to CID MS/MS and observed a series of singly charged fragment ions within an m/z range between 2341.7 and 791 (Figure 4A). Detailed analysis of the fragmentation pattern revealed the presence of a peptide component corresponding to the sequence NQTDSAR, which is the predicted product of tryptic cleavage at R938 and R945 within the TfsA protein, spanning the potential glycosylation site S943 within a DSA glycosylation sequon. The modification of this peptide was determined to perfectly match all constituents of the B. fragilis O-glycan moiety described above.
Fig. 4.
(A) Tandem ESI-MS of a glycopeptide derived from the T. forsythia S-layer protein TfsA expressed in B. fragilis confirming the heterologous attachment of the B. fragilis O-glycan to the protein (attachment to S943 within the glycosylation sequon DSA is shown). The fragmentation pattern of the O-glycan and the deduced glycan structure correlate with data presented in Figure 3. Attachment to the protein backbone occurs via hexose-hexuronic acid with one methylated deoxyhexose branching from the hexose at the reducing end. (B) Tandem ESI-MS of a glycopeptide derived from the B. fragilis protein BF3567 expressed in T. forsythia showing the heterologous attachment of the T. forsythia O-glycan to the protein (binding to T207 within the glycosylation sequon DTV is shown). Fragment ions marked with an asterisk correspond to peptide backbone fragments. Note: Line positions between residues of the B. fragilis glycan (A, inset top right) do not represent actual linkage types.
Analysis of tryptic peptides of BF2494 and BF3567 from T. forsythia ΔtfsAB yielded a very similar result. In addition to the predicted tryptic peptides, we also found potentially modified doubly and triply charged cleavage products. Again, we selected one of those cleavage products derived from BF3567 for CID MS/MS and observed a series of fragment ions beginning at m/z 1496.6 (M + 2H)2+ and ending at m/z 1258.7 (M + H)+ (Figure 4B). This perfectly fits peptide LGDISLDTVAVR, produced upon tryptic cleavage at K199 and R211 of BF3567, substituted with the T. forsythia O-glycan at T207 within the DTV glycosylation motif. By analyzing the fragment spectrum of the O-glycan, we found all known constituents including their modifications with the exception of the O-methyl group of the N-acetyl mannosaminuronamide residue. For yet unknown reasons, this lack of methylation is commonly observed on O-glycans of glycoproteins isolated from T. forsythia ΔtfsAB (Gerald Posch, unpublished data), and thus it may be specific to this mutant strain.
In Supplementary data, Table S2, all glycopeptides that were identified as being heterologously glycosylated are listed. In total, three glycopeptides of TfsA and TfsB were identified as being modified with the B. fragilis glycan. One representative “cross-glycosylated” peptide of BF2494 and two representative glycopeptides of BF3567 modified with the T. forsythia glycan were identified, demonstrating that the O-glycans of T. forsythia/B. fragilis can be added to non-native proteins. In general, the glycosylation of heterologous proteins appears to occur with high efficiency, as all of the peptides containing a glycosylation motif were either observed to be glycosylated or were not detected in their unglycosylated form.
Discussion
In recent years, bacterial protein O-glycosylation has rapidly expanded from “curiosity” to “ubiquity” (Messner 2009) with the discoveries of broad-spectrum glycosylation systems. Recent scientific efforts are directed at exploiting these systems to create novel biosynthetic pathways resulting in tailor-made glycans that can be specifically attached to target proteins (Steiner et al. 2007; Lizak et al. 2011; Valderrama-Rincon et al. 2012). Considering the manifold properties glycans provide to proteins, including stabilizing functions (Krapp et al. 2003), improved thermal stability (Mimura et al. 2000) and protection from proteases (Langsford et al. 1987), this approach seems to be promising for the future generation of glycoproteins with improved characteristics.
The present study focused on the feasibility of heterologously glycosylating proteins in the two Bacteroidales species B. fragilis and T. forsythia. Both organisms are known to possess general O-glycosylation systems sharing a conserved amino acid motif that is targeted for glycosylation. We first partially characterized the native B. fragilis O-glycan in order to determine whether the same glycan was added to T. forsythia proteins expressed in B. fragilis. Mass spectrometric analysis of native B. fragilis glycoproteins subjected to β-elimination revealed the glycan to be an oligosaccharide consisting of nine sugar residues. Notably, the proposed structure based on MS data resembles that of the T. forsythia O-glycan in several aspects. In both glycans, attachment to the protein occurs via a hexose residue which is succeeded by a hexuronic acid. Additionally, a nonpolar constituent branches from the first sugar of the B. fragilis O-glycan. Also, both glycans contain a deoxyhexose residue in their linear structure. As in the T. forsythia glycan, the deoxyhexose of the B. fragilis glycan is most likely a fucose (Fuc), as glycoproteins of B. fragilis are readily detected with Aleuria aurantia lectin and depend on GDP-Fuc biosynthesis genes (Fletcher et al. 2009).
Knowing the size and rough composition of the B. fragilis glycan, we sought to determine whether transfer of the respective glycans on to non-native proteins occurs in B. fragilis and T. forsythia. We chose to analyze the well-characterized S-layer proteins TfsA and TfsB from T. forsythia (Lee et al. 2006; Sekot et al. 2012) for heterologous glycosylation in B. fragilis. In addition, two model glycoproteins of B. fragilis, BF2494 and BF3567, both of unknown function (Fletcher et al. 2009), were selected for heterologous expression in T. forsythia. Successful transfer of the O-glycans was expected to result in novel glycoproteins with the respective O-glycans linked via S/T within the Bacteroides glycosylation sequon.
Our analyses of the heterologously expressed proteins revealed electrophoretic migrations different from those predicted according to their molecular masses. This observation strongly supported our concept of “cross-glycosylation” of proteins within the two species. This was confirmed by analyzing glycopeptides of the respective proteins, ultimately showing that the O-glycans had been properly transferred. Furthermore, glycan transfer appeared to be highly efficient, as none of the peptides spanning a putative glycosylation sites was observed as unmodified.
Protein glycosylation in T. forsythia and B. fragilis is ubiquitous and seems to be an inherent feature of these organisms. Their well-established systems allow decoration of supposedly any extracytoplasmic protein bearing a glycosylation sequon (Fletcher et al. 2011). The reasons for modification of such an extensive number of proteins are still unclear. In T. forsythia, the protein O-glycan may be involved in the bacterium-host cross-talk, mediate cell adhesion (Sakakibara et al. 2007) and influence the biofilm formation capability of the organism (Honma et al. 2007). Also, it was shown that the glycosylated S-layer of T. forsythia displayed on wild-type cells delays recognition of the immune system in a macrophage cell culture model compared with the S-layer deficient T. forsythia ΔtfsAB strain (Sekot et al. 2011). Recently, the suppression of T-helper 17 responses in dendritic cells as well as increased periodontal bone loss in mice could be specifically attributed to the terminal 5-acetimidol-7-N-glycolylpseudaminic acid (Pse5Am7Gc)-N-acetylmannosamiuronamide (ManNAcCONH2)-N-acetylmannosamiuronic acid (ManNAcA) trisaccharide branch of the T. forsythia O-glycan (Settem et al. 2012).
As for B. fragilis, protein glycosylation has been shown to be essential for normal in vitro growth and for colonization of the mammalian intestine (Fletcher et al. 2009). Additionally, surface glycoproteins isolated from an outer membrane protein preparation are proposed to confer interaction with the extracellular matrix component laminin-1 (de O. Ferreira et al. 2006). However, the identities of the glycoproteins involved in this proposed interaction are not described.
Considering the vast amount of glycoproteins synthesized by B. fragilis—and most likely also by T. forsythia—it is likely that O-glycosylation of proteins has a more general function, for instance in protein stability. As secreted proteins of B. fragilis do not form intramolecular disulfide bonds (Dutton et al. 2008), protein glycosylation may serve a compensatory stabilizing role. Analyzing the influence of (heterologous) glycosylation on protein stability in T. forsythia and B. fragilis will contribute to a better understanding of protein glycosylation in general, as well as trigger efforts to specifically improve protein stability through glycoengineering.
Material and methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table I. E. coli strains were grown at 37°C overnight in Luria–Bertani (LB) broth supplemented with ampicillin (Amp, 100 µg mL−1), kanamycin (Km, 50 µg mL−1) or both.
Table I.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| T. forsythia ATCC 43037 | Wild-type strain | ATCC |
| T. forsythia ΔtfsAB | Double knock-out mutant, devoid of the S-layer proteins | Sakakibara et al. (2007) |
| B. fragilis NCTC9343 | Wild-type strain | NCTC |
| E. coli DH5α | Cloning strain | Invitrogen |
| RK231 | broad-host-range mobilizing IncP plasmid, RK2 derivative; Km® | Guiney et al. (1984) |
| pJET 1.2 | Cloning vector, Amp® | Thermo Fisher Scientific |
| pCMF118 | E. coli-Bacteroides shuttle vector, pFD340 derivative; Amp®, Em® | Coyne et al. (manuscript in preparation) |
| pMJC95 | C-His10 tagged tfsA from T. forsythia cloned into pCMF118; Amp®, Em® | This study |
| pMJC94 | C-His10 tagged tfsB from T. forsythia cloned into pCMF118; Amp®, Em® | This study |
| pGP21 | C-His10 tagged BF2494 from B. fragilis cloned into pCMF118; Amp®, Em® | This study |
| pGP22 | C-His10 tagged BF3567 from B. fragilis cloned into pCMF118; Amp®, Em® | This study |
B. fragilis and T. forsythia strains were grown anaerobically in brain heart infusion (BHI) broth or on agar plates (1% w/v) supplemented with yeast extract (5 g L−1), cysteine (1 g L−1), hemin (50 µg mL−1) and menadione (10 µg mL−1). The T. forsythia medium was additionally supplemented with N-acetylmuramic acid (10 µg mL−1). Gentamicin (Gm, 200 µg mL−1) and erythromicin (Em, 5 µg mL−1) were added to the media when appropriate.
Cloning and transformation of constructs
DNA of T. forsythia and B. fragilis, respectively, was prepared as follows. 1 mL of stationary bacterial culture was harvested (6000 × g, 2 min) and the supernatant was discarded. The cells were resuspended in 50 µL of sterile distilled water and boiled for 5 min. Cell debris was removed by centrifugation (20,000 × g, 2 min) and the supernatant containing chromosomal DNA was used as template for all polymerase chain reactions (PCRs). All oligonucleotides used are listed in Table II. TfsA was PCR-amplified, digested with BglII and ligated into the unique BamHI site of vector pCMF118 (Coyne et al., manuscript in preparation), creating pMJC95. Similarly, PCR-amplified tfsB was digested with BamHI and ligated into BamHI digested pCMF118, creating pMJC94.
Table II.
Oligonucleotide primers used in this study
| Purpose | Sequencea |
|---|---|
| Amplification of T. forsythia genes for expression with His-tags in B. fragilis | |
| tfsA-for | AAATAGATCTGCGGTTTATAAGAGGAAGAAAATAAA |
| tfsA-rev | CTTAAGATCTTTTTACACAGCTTTCACTGCATTC |
| tfsB-for | CCTCGGATCCATCTCTTGCCTGCCACTCC |
| tfsB-rev | CGACGGATCCCTTCACCATCGCTTTTACAGC |
| Amplification of B. fragilis genes for expression with His-tags in T. forsythia | |
| BF2494-for | ATCAGGATCCACAATCATGAAAAGAGTATTATTTTC |
| BF2494-rev | ATCAGGATCCCATCATTTTCTCGATTTCTTCGAATTC |
| BF3567-for | ATCAGGATCCTACTAACTAAACGTGATTAATTTATG |
| BF3567-rev | ATCAGGATCCACGGGTTACTTCCAAATACTTCACC |
aSequences are given 5′–3′ with restriction sites underlined.
BF2494 and BF3567 PCR-amplified products were blunt-end cloned into pJET1.2 (Thermo Fisher Scientific, Vienna, Austria) and the inserts from positive clones were cut with BamHI and cloned into the unique BamHI site of pCMF118 creating pGP21 and pGP22, respectively. Transformants with the correct insert orientation were selected by PCR. All constructs (pMJC94, pMJC95, pGP21 and pGP22) were also confirmed by sequencing.
Plasmids were transferred from E. coli to T. forsythia ΔtfsAB and B. fragilis by conjugative transfer. First, 200 µL of E. coli RK231 overnight culture was mixed with 200 µL of E. coli DH5α cells containing the respective plasmid constructs. Cells were centrifuged (6000 × g, 2 min) and plated on LB agar without antibiotics. Following overnight incubation at 37°C, the growth was struck to LB agar plates supplemented with Amp and Km to select for clones containing both RK231 and the respective expression plasmids. Subsequently, positive clones were used in a second conjugation experiment to transform T. forsythia ΔtfsAB or B. fragilis by combining 3 mL of an overnight culture of the respective clones with 50 mL of T. forsythia ΔtfsAB or B. fragilis culture (OD600 ∼ 0.3–0.6) and collecting the cells by centrifugation (6000 × g, 10 min). The cell pellet was resuspended in a small volume of medium and plated to BHI agar plates without antibiotics. Following aerobic incubation (37°C, overnight), the growth was struck to Gm/Em-containing BHI agar plates and incubated anaerobically at 37°C for 2 days (B. fragilis) or up to 14 days (T. forsythia). Em-resistant transconjugants were confirmed by PCR. The resulting recombinant proteins were modified by the addition of glutamine-serine-10x histidine (GSH10) at the C-terminus, except TfsA, which was modified with arginine-serine-10x histidine (RSH10).
Purification of His-tagged proteins from T. forsythia and B. fragilis
Immobilized metal affinity chromatography (IMAC) was performed to purify His-tagged proteins. Briefly, cultures (2 L) of T. forsythia ΔtfsAB and B. fragilis harboring plasmids encoding His-tagged proteins were grown to stationary phase, harvested (6000 × g, 4°C, 15 min) and washed with buffer A (20 mM NaH2PO4, 20 mM imidazole; adjusted to pH 7.5 with 4 M NaOH). The bacterial pellets were resuspended at a ratio of 1:5 in buffer A (w/v; if necessary, 4 M urea was added to allow for denaturing purification). Following sonication (3 cycles of 30 pulses, each, with 1 min of cooling between the cycles), the cellular debris was removed by ultracentrifugation (50,000 × g, 4°C, 30 min) and the supernatant was loaded on to a HiTrap HP column (GE Healthcare, Vienna, Austria V = 1 mL) connected to a BioLogic DuoFlow FPLC system (BioRad, Vienna, Austria). Bound proteins were eluted in a linear gradient of 0–0.5 M imidazole in buffer A within 10 column volumes. Fractions containing the desired proteins were pooled, concentrated via Amicon Ultra-15 centrifugal filter units (30 kDa cutoff; Millipore, Vienna, Austria) and subjected to SDS–PAGE and western blot analysis.
SDS–PAGE, Coomassie Blue staining and western blot analysis
Purified His-tagged proteins or crude cell extracts were run on 10% SDS–PAGE gels according to standard protocols (Laemmli 1970). Proteins were directly visualized with Coomassie Blue staining or transferred to polyvinylidene difluoride (PVDF) membranes for western immunoblotting using an anti-His antibody (produced in mice; Invitrogen, Vienna, Austria) in combination with an anti-mouse secondary antibody labeled with IRDye 800CW (LI-COR, Bad Homburg, Germany). Membranes were visualized using the Odyssey Infrared Imaging system (LI-COR Biosciences) at 785 nm. Glycostaining of SDS–PAGE gels was performed using the ProQ-Emerald fluorescent stain (Invitrogen).
Glycopeptide preparation
For preparation of glycopeptides, gel slices containing the protein bands were excised from Coomassie Blue-stained gels, chopped into small pieces and destained (2 cycles of 50 and 100% acetonitrile, followed by reswelling of the gel pieces in 100 mM ammonium bicarbonate, with 10 min incubation time, each). S-carbamidomethylation, trypsin digestion and extraction of (glyco-)peptides were performed by routine methods (Stadlmann et al. 2008). Briefly, cysteine bonds were reduced by treatment with 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate for 1 h at 56°C. Cysteine residues were S-alkylated with 55 mM iodoacetamide in ammonium bicarbonate at room temperature for 1 h. Following subsequent washing, (glyco-)proteins were digested with trypsin (Roche, Vienna, Austria, 50 ng µL−1 in 25 mM ammonium carbonate) overnight at 37°C and the resulting peptides were extracted with alternating washes of water, bicarbonate buffer and acetonitrile (100%) and lyophilized prior to analysis.
In-gel reductive β-elimination
O-Glycan release for further LC-ESI-MS/MS analyses of the B. fragilis glycoprotein glycan was performed by in-gel reductive β-elimination of Coomassie Blue-stained protein bands. Briefly, bands of purified His-tagged proteins as well as bands of miscellaneous proteins originating from a crude B. fragilis protein extract were excised from gels, transferred to plastic reaction tubes, covered with 1 M NaBH4 in 500 mM NaOH and incubated at 50°C overnight. Excess salt was removed using a 10 mg HyperSep Hypercarb solid phase extraction (SPE) cartridge (Thermo Fisher Scientific) according to published protocols (Packer et al. 1998; Pabst and Altmann 2008).
LC ESI-MS/MS (IonTrap and Q-TOF)
Borohydride-reduced O-glycans were analyzed by PGC (Hypercarb, 0.32 × 150 mm, particle size 5 µm)-ESI-MS/MS as recently described (Pabst et al. 2007). Ammonium formate buffer (0.3% formic acid, adjusted to pH 3.0 with ammonia solution) was used as buffer A, and a 0–35% acetonitrile gradient was performed within 35 min using a Dionex Ultimate 3000 (cap flow, 8 µL min−1). Glycan mass screening was performed using a Global Ultima Q-TOF from Micromass (Waters, Eschborn, Germany). Data were evaluated using the MassLynx 4.0 software. High-resolution mass spectrometry experiments with direct infusion of purified glycans were performed using a Bruker Maxis 4G Q-TOF.
Glycoproteomics analyses were performed by reversed phase LC coupled to ESI-MS/MS either on a Bruker IonTrap AmaZon speed ETD or on the high-resolution Maxis 4G Q-TOF. The X! Tandem algorithm (Craig and Beavis 2004) as implemented by the Global Proteome Machine Organization website (http://www.thegpm.org) was used for peptide identification and estimating sequence coverage. Results were further evaluated using log(e) values to estimate correctness of peptide assignments (Fenyö and Beavis 2003).
Supplementary data
Supplementary data for this article are available online at http://glycob.oxfordjournals.org/.
Funding
Financial support came from the Austrian Science Fund FWF, projects P20605-B12 and P24317-B22 (to C.S.) and the PhD programme “BioToP—Biomolecular Technology of Proteins” (Austrian Science Fund, FWF project W1224), and National Institute of Health/National Institute of Allergy and Infectious Diseases grant R01AI067711 (to L.C.).
Conflict of interest
None declared.
Abbreviations
BHI, brain heart infusion; CID, collision induced dissociation; Dig, digitoxose; DTT, dithiothreitol; FPLC, fast protein liquid chromatography; Fuc, fucose; GSH10, glutamine-serine-10x histidine; IMAC, immobilized metal affinity chromatography; LC-ESI-MS/MS, liquid chromatography electrospray ionization tandem mass spectrometry; ManNAcA, N-acetylmannosamiuronic acid; ManNAcCONH2, N-acetylmannosamiuronamide; MeGal, 4-methyl-galactose; mi, molecular ion; Mw, molecular weight; PGC-ESI-MS/MS, porous graphitized carbon electrospray ionization mass spectrometry; Pse5Am7Gc, 5-acetimidol-7-N-glycolylpseudaminic acid; PCR, polymerase chain reaction; PVDF, polyvinylidene difluoride; RSH10, arginine-serine-10x histidine; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SPE, solid phase extraction.
Supplementary Material
References
- Bayley DP, Rocha ER, Smith CJ. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. doi:10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- Craig R, Beavis RC. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. doi:10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- de O. Ferreira E, Araújo Lobo L, Barreiros Petrópolis D, dos S, Avelar KE, Ferreira MC, e Silva Filho FC, Domingues RMCP. A Bacteroides fragilis surface glycoprotein mediates the interaction between the bacterium and the extracellular matrix component laminin-1. Res Microbiol. 2006;157:960–966. doi: 10.1016/j.resmic.2006.09.005. doi:10.1016/j.resmic.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. doi:10.1007/BF01049915. [Google Scholar]
- Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. doi:10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. doi:10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyö D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75:768–774. doi: 10.1021/ac0258709. doi:10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Comstock LE. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J Bio Chem. 2011;286:3219–3226. doi: 10.1074/jbc.M110.194506. doi:10.1074/jbc.M110.194506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. doi:10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney DG, Hasegawa P, Davis CE. Plasmid transfer from Escherichia coli to Bacteroides fragilis: Differential expression of antibiotic resistance phenotypes. Proc Natl Acad Sci USA. 1984;81:7203–7206. doi: 10.1073/pnas.81.22.7203. doi:10.1073/pnas.81.22.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. doi:10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 2007;42:156–166. doi: 10.1016/j.micpath.2007.01.003. doi:10.1016/j.micpath.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8:e1002758. doi: 10.1371/journal.ppat.1002758. doi:10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. doi:10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J Mol Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. doi:10.1016/S0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- Ku SC, Schulz BL, Power PM, Jennings MP. The pilin O-glycosylation pathway of pathogenic Neisseria is a general system that glycosylates AniA, an outer membrane nitrite reductase. Biochem Biophys Res Commun. 2009;378:84–89. doi: 10.1016/j.bbrc.2008.11.025. doi:10.1016/j.bbrc.2008.11.025. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. doi:10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langsford ML, Gilkes NR, Singh B, Moser B, Miller RC, jr, Warren RAJ, Kilburn DG. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 1987;225:163–167. doi: 10.1016/0014-5793(87)81150-x. doi:10.1016/0014-5793(87)81150-X. [DOI] [PubMed] [Google Scholar]
- Lee SW, Sabet M, Um HS, Yang J, Kim HC, Zhu W. Identification and characterization of the genes encoding a unique surface (S-) layer of Tannerella forsythia. Gene. 2006;371:102–111. doi: 10.1016/j.gene.2005.11.027. doi:10.1016/j.gene.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Lizak C, Fan Y-Y, Weber TC, Aebi M. N-linked glycosylation of antibody fragments in Escherichia coli. Bioconj Chem. 2011;22:488–496. doi: 10.1021/bc100511k. doi:10.1021/bc100511k. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. doi:10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Messner P. Prokaryotic protein glycosylation is rapidly expanding from “curiosity” to “ubiquity”. ChemBioChem. 2009;10:2151–2154. doi: 10.1002/cbic.200900388. doi:10.1002/cbic.200900388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura Y, Church S, Ghirlando R, Ashton PR, Dong S, Goodall M, Lund J, Jefferis R. The influence of glycosylation on the thermal stability and effector function expression of human IgG1-Fc: Properties of a series of truncated glycoforms. Mol Immunol. 2000;37:697–706. doi: 10.1016/s0161-5890(00)00105-x. doi:10.1016/S0161-5890(00)00105-X. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: Sweeter than ever. Nat Rev Micro. 2010;8:765–778. doi: 10.1038/nrmicro2383. doi:10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Pabst M, Altmann F. Influence of electrosorption, solvent, temperature, and ion polarity on the performance of LC-ESI-MS using graphitic carbon for acidic oligosaccharides. Anal Chem. 2008;80:7534–7542. doi: 10.1021/ac801024r. doi:10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- Pabst M, Bondili JS, Stadlmann J, Mach L, Altmann F. Mass + retention time = structure: A strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal Chem. 2007;79:5051–5057. doi: 10.1021/ac070363i. doi:10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconj J. 1998;15:737–747. doi: 10.1023/a:1006983125913. doi:10.1023/A:1006983125913. [DOI] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–1820. doi: 10.1016/S0140-6736(05)67728-8. doi:10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. Characterization and scope of S-layer protein O-glycosylation in Tannerella forsythia. J Biol Chem. 2011;286:38714–38724. doi: 10.1074/jbc.M111.284893. doi:10.1074/jbc.M111.284893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch G, Sekot G, Friedrich V, Megson Z, Koerdt A, Messner P, Schäffer C. Glycobiology aspects of the of the periodontal pathogen Tannerella forsythia. Biomolecules. 2012;2:467–482. doi: 10.3390/biom2040467. doi:10.3390/biom2040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara J, Nagano K, Murakami Y, Higuchi N, Shimozato K, Yoshimura F. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–876. doi: 10.1099/mic.0.29275-0. doi:10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Huang W, Li C, Schulz BL, Lizak C, Palumbo A, Numao S, Neri D, Aebi M, Wang L-X. A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. Nat Chem Biol. 2010;6:264–266. doi: 10.1038/nchembio.314. doi:10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Messner P, Matejka M, Rausch-Fan X, Andrukhov O, Schäffer C. Potential of the Tannerella forsythia S-layer to delay the immune response. J Dent Res. 2011;90:109–114. doi: 10.1177/0022034510384622. doi:10.1177/0022034510384622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekot G, Posch G, Oh Y, Zayni S, Mayer H, Pum D, Messner P, Hinterdorfer P, Schäffer C. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia. Arch Microbiol. 2012;194:525–539. doi: 10.1007/s00203-012-0792-3. doi:10.1007/s00203-012-0792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, Sharma A. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2012 doi: 10.1038/mi.2012.85. doi:10.1038/mi.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. doi:10.1016/0147-619X(92)90014-2. [DOI] [PubMed] [Google Scholar]
- Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics. 2008;8:2858–2871. doi: 10.1002/pmic.200700968. doi:10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- Steiner K, Novotny R, Patel K, Vinogradov E, Messner P, Schäffer C. Functional characterization of the initiation enzyme of S-Layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus NRS 2004/3a. J Bacteriol. 2007;189:2590–2598. doi: 10.1128/JB.01592-06. doi:10.1128/JB.01592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. doi:10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- Valderrama-Rincon JD, Fisher AC, Merritt JH, Fan Y-Y, Reading CA, Chhiba K, Heiss C, Azadi P, Aebi M, DeLisa MP. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol. 2012;8:434–436. doi: 10.1038/nchembio.921. doi:10.1038/nchembio.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith PD, O'Brien-Simpson NM, Tan Y, Djatmiko DC, Dashper SG, Reynolds EC. Outer membrane proteome and antigens of Tannerella forsythia. J Proteome Res. 2009;8:4279–4292. doi: 10.1021/pr900372c. doi:10.1021/pr900372c. [DOI] [PubMed] [Google Scholar]
- Vik Å, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. doi:10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. doi:10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- Wexler HM. Bacteroides: The good, the bad, and the nitty-gritty. Clinical Microbiol Rev. 2007;20:593–621. doi: 10.1128/CMR.00008-07. doi:10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M, Deelder AM, van der Burgt YEM. Mass spectrometric glycan rearrangements. Mass Spectrom Rev. 2011;30:664–680. doi: 10.1002/mas.20337. doi:10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- Young NM, Brisson J-R, Kelly J, Watson DC, Tessier L, Lanthier PH, Jarrell HC, Cadotte N, St. Michael F, Aberg E, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. doi:10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.