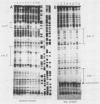
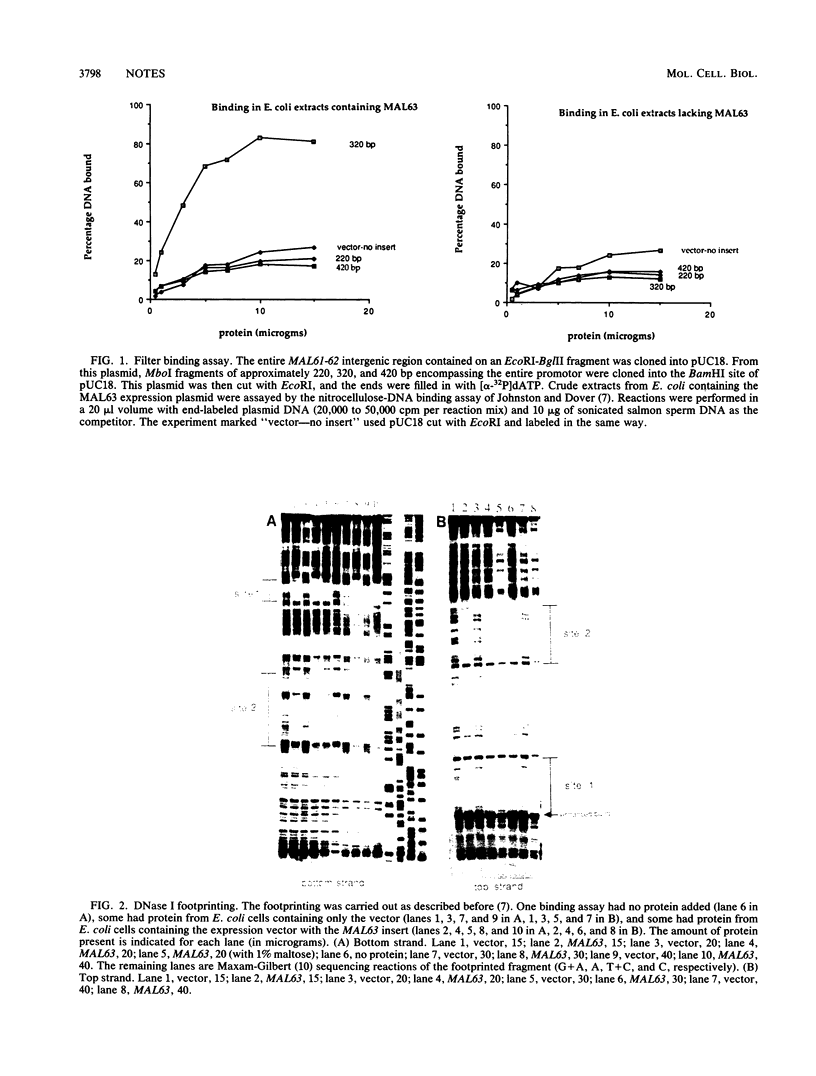
Abstract
Maltose fermentation in Saccharomyces species requires the presence of at least one of five unlinked MAL loci: MAL1, MAL2, MAL3, MAL4, and MAL6. Each of these loci consists of a complex of genes involved in maltose metabolism; the complex includes maltase, a maltose permease, and an activator of these genes. At the MAL6 locus, the activator is encoded by the MAL63 gene. While the MAL6 locus has been the subject of numerous studies, the binding sites of the MAL63 activator have not been determined. In this study, we used Escherichia coli extracts containing the MAL63 protein to define the binding sites of the MAL63 protein in the divergently transcribed MAL61-62 promotor. When a DNA fragment containing these sites was placed upstream of a CYC1-lacZ gene, maltose induced beta-galactosidase. These sites therefore constitute an upstream activating sequence for the MAL genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cerdan M. E., Zitomer R. S. Oxygen-dependent upstream activation sites of Saccharomyces cerevisiae cytochrome c genes are related forms of the same sequence. Mol Cell Biol. 1988 Jun;8(6):2275–2279. doi: 10.1128/mcb.8.6.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. S., Dubin R. A., Perkins E., Michels C. A., Needleman R. B. Identification and characterization of the maltose permease in genetically defined Saccharomyces strain. J Bacteriol. 1989 Nov;171(11):6148–6154. doi: 10.1128/jb.171.11.6148-6154.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin R. A., Needleman R. B., Gossett D., Michels C. A. Identification of the structural gene encoding maltase within the MAL6 locus of Saccharomyces carlsbergensis. J Bacteriol. 1985 Nov;164(2):605–610. doi: 10.1128/jb.164.2.605-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. H., Marmur J. Upstream regulatory regions controlling the expression of the yeast maltase gene. Mol Cell Biol. 1987 Jul;7(7):2477–2483. doi: 10.1128/mcb.7.7.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Michels C. A. The MAL63 gene of Saccharomyces encodes a cysteine-zinc finger protein. Curr Genet. 1988 Oct;14(4):319–323. doi: 10.1007/BF00419988. [DOI] [PubMed] [Google Scholar]
- Kopetzki E., Buckel P., Schumacher G. Cloning and characterization of baker's yeast alpha-glucosidase: over-expression in a yeast strain devoid of vacuolar proteinases. Yeast. 1989 Jan-Feb;5(1):11–24. doi: 10.1002/yea.320050104. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Kaback D. B., Dubin R. A., Perkins E. L., Rosenberg N. G., Sutherland K. A., Forrest D. B., Michels C. A. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc Natl Acad Sci U S A. 1984 May;81(9):2811–2815. doi: 10.1073/pnas.81.9.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Michels C. Repeated family of genes controlling maltose fermentation in Saccharomyces carlsbergensis. Mol Cell Biol. 1983 May;3(5):796–802. doi: 10.1128/mcb.3.5.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Prezant T., Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987 Apr 10;49(1):19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- Siliciano P. G., Tatchell K. Identification of the DNA sequences controlling the expression of the MAT alpha locus of yeast. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2320–2324. doi: 10.1073/pnas.83.8.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollitti P., Marmur J. Primary structure of the regulatory gene from the MAL6 locus of Saccharomyces carlsbergensis. Mol Gen Genet. 1988 Jul;213(1):56–62. doi: 10.1007/BF00333398. [DOI] [PubMed] [Google Scholar]
- ten Berge A. M., Zoutewelle G., van de Poll K. W. Regulation of maltose fermentation in Saccharomyces carlsbergensis. I. The function of the gene MAL6, as recognized by mal6-mutants. Mol Gen Genet. 1973 Jul 2;123(3):233–246. doi: 10.1007/BF00271242. [DOI] [PubMed] [Google Scholar]