Abstract
The ciliated protozoan Tetrahymena thermophila has been an important model system for biological research for many years. During that time a variety of useful strains, including highly inbred stocks, a collection of diverse mutant strains, and wild cultivars from a variety of geographical locations have been identified. In addition, thanks to the efforts of many different laboratories, optimal conditions for growth, maintenance, and storage of Tetrahymena have been worked out. To facilitate the efficient use of Tetrahymena, especially by those new to the system, this chapter presents a brief description of many available Tetrahymena strains, and lists possible resources for obtaining viable cultures of T. thermophila and other Tetrahymena species. Descriptions of commonly used media, methods for cell culture and maintenance, and protocols for short and long term storage are also presented.
I. Introduction
The increasing use of Tetrahymena for both research and educational purposes has been facilitated by the ease with which it can be grown and maintained in a wide range of conditions, ranging from single cells in hanging drops to multi-liter cultures grown in large bio-reactors. Sexual reproduction is dependably controlled by transfer to non-nutritive media and simple selection schemes are available for the identification of sexual progeny. Under optimal conditions, Tetrahymena has a rapid growth rate, with a doubling time of less than two hours. However, slowly growing vegetative cultures can be maintained on the bench for several months with very limited loss of function or fertility, and strains can be stored for years in liquid nitrogen. A number of mutant and inbred strains of Tetrahymena thermophila, the species most commonly used for physiological, biochemical, and molecular research, are readily available. A number of other Tetrahymena species, many of which can be maintained under conditions similar to those used to culture T. thermophila, are also easily obtainable. This chapter provides basic information on the most frequently utilized T. thermophila strains, current sources for obtaining T. thermophila strains and other Tetrahymena species, and methods for growing, maintaining, mating, and storing Tetrahymena cultures.
II. Tetrahymena thermophila Strains
T. thermophila provides a rich resource of useful strains, including highly inbred stocks derived from wild isolates over half a century ago (Allen and Gibson, 1973; and Ch. 2 in this volume), a collection of diverse mutant strains, and wild cultivars from a variety of geographical locations. T. thermophila strains are generally named according to location of origin. Wild type isolates are given a two letter prefix based on location of origin (e.g. WH for isolates originally collected at Woods Hole), while strains developed in individual laboratories are give a two letter prefix representing the location of the lab of origin followed by a strain number (e.g. CU428 is Cornell University strain number 428). Basic standards for describing micronuclear and macronuclear genotypes and phenotypes can be found in Allen (2000) and Allen et al. (1998). A more extensive revised version of the current preferences for Tetrahymena nomenclature can be found on the Tetrahymena Stock Center website (http://tetrahymena.vet.cornell.edu/extras/revised_tetrahymena_nomenclature.doc). A brief description of frequently utilized T. thermophila strain types is provided below. More detailed information about individual T. thermophila strains can be found on the Tetrahymena Stock Center website (http://tetrahymena.vet.cornell.edu).
A. Inbred Wild-Type Strains
The T. thermophila strains most commonly used in research labs are highly inbred B lines derived from fertile natural isolates collected in Woods Hole, MA in the early 1950’s by Elliott and co-workers (Nanney and Simon, 2000; and Ch. 2 in this volume). Inbred strains derived from other mating type families are also available, but are not generally used for physiological, genetic, or molecular research. One exception is inbred strain C3. Naturally occurring genetically polymorphisms inherent in inbred strains B and C3 have proven to be useful genetic tools for genome mapping (Brickner et al., 1996; Lynch et al., 1995; Orias, 1998), and for investigation of amplification, replication, and maintenance of the rDNA (Larson et al., 1986; Lovlie et al., 1988; Luehrsen et al., 1987). The American Type Culture Collection (ATCC; http://www.atcc.org) houses representative strains from inbred mating type families A and B, derived from cells collected at Woods Hole, Massachusetts, family C, derived from a strain originally collected in Vermont, and family D, derived from a strain originally isolated in Michigan (Nanney and Simon, 2000). ATCC T. thermophila strains are identified by mating type allele family, inbreeding history, and mating type. In many cases, multiple mating types of the same inbreeding cross are represented. ATCC Tetrahymena cultures are stored frozen in liquid nitrogen freezers and are shipped frozen on dry ice. A smaller collection of strain B, C, and D inbred lines are also available from the Tetrahymena Stock Center at Cornell University. Tetrahymena Stock Center cultures are stored in liquid nitrogen freezers, and shipped as viable cultures in proteose peptone media.
B. Star Strains
Star strains are specialized strains of T. thermophila that lack a functional germinal nucleus. The vestigial genetic material remaining in the micronucleus cannot contribute to the formation of viable sexual progeny. Matings involving star strains follow an alternative developmental program known as “genomic exclusion” (Allen, 1967a; Allen, 1967b). During the mating of a star cell and a cell with a functional micronucleus, mating partners separate prematurely, following unidirectional exchange of a gametic pronucleus from the non-star cell to the star partner. The results of this initial round of mating (RdI) are two cells with identical 100% homozygous micronuclei derived from one meiotic product from the non-star parent. Both RdI exconjugant cells retain their original macronucleus, and thus their original phenotypes, including mating type. The RdI exconjugants can immediately enter into a second round of mating. Since both partners now contain identical functional micronuclei, the second round of mating proceeds normally, giving rise to progeny that are whole genome homozygotes. Star strains are a valuable genetic tool, useful for an array of genetic applications, including the construction of homozygous strains (Allen, 1967b), the creation of functional heterokaryons including gene knock-out (KO) strains (Dave et al., 2009; Hai et al., 2000), conjugation rescue (Satir et al., 1986), short circuit genomic exclusion (Bruns et al., 1976), and uniparental cytogamy (Cole and Bruns, 1992).
Star lines derived from inbred strain A, B, and C genetic backgrounds are available in several different mating types (A*III, A*V, C*III, B*VI, and B*VII) from the Tetrahymena Stock Center. Any star strain can be used to construct homozygous strains following two rounds of mating. However, star strains show different utility for use in other genetic procedures. C*III is the strain of choice for short circuit genomic exclusion and can be used for uniparental cytogamy. A*III is not ideal for short circuit genomic exclusion or uniparental cytogamy progeny but is excellent for making homozygous heterokaryons and a good choice for conjugation rescue (Satir et al., 1986). A*V and B*VI are the best star strains for use in experiments involving uniparental cytogamy, and B*VII can also be used effectively in that procedure (Cole and Bruns, 1992). It should be noted that A*III carries resistance to 6-methylpurine in its macronucleus.
C. Mutant Strains
In Tetrahymena, the physical and functional separation of germinal and somatic nuclei (nuclear dimorphism; see Prescott, 1994; Ch. 3 in this volume) permits the creation of cells that are genetically different in the germinal micronucleus and the somatic macronucleus. Nuclear dimorphism, in conjunction with allelic assortment in the somatic macronucleus has fostered the creation of genetically useful strains that carry a non-expressed mutant or modified allele in the germinal nucleus but express only the non-mutant allele in the somatic nucleus (functional heterokaryons; Bruns and Brussard, 1974b). Such lines can be either homozygous or heterozygous in the micronucleus. Potentially lethal genetic constructs, including homozygous knockouts of essential genes (Dave et al., 2009; Hai et al., 2000), lethal mutations, and chromosome modifications such as deletions and loss of one or more entire micronuclear chromosomes (nullisomics; Bruns and Brussard, 1981; Bruns et al., 1983) can only be maintained as heterokaryons. Functional heterokaryons carrying such potentially lethal constructs in the micronucleus can produce viable progeny when mated with cells carrying the equivalent wild type sequence (except in the case of dominant lethal genes), although in many cases such matings exhibit a somewhat lower frequency of progeny production.
All work with mutant strains must take into consideration the natural phenotypic assortment that occurs in the macronucleus during vegetative growth. The macronucleus is highly polyploid, containing ~ 45 copies of each macronuclear chromosome (Doerder, 1979; Doerder et al., 1992; Larson et al., 1991), with the exception of the rDNA palindromic chromosome, which is present at about 9000 copies (Kapler, 1993; Yao and Gorovsky, 1974). Since assortment of macronuclear chromosomes is random, over time cultures initially heterozygous in the macronucleus will produce daughter cells that are homozygous for a given allele. Such assortment can result in the loss of a mutant allele within a clone if the cells are maintained under conditions that provide any selective advantage for cells expressing the wild type allele. This phenomenon is used to advantage to produce heterokaryons, but can lead to problems if continued expression of the mutant allele is desired. A similar problem can occur if wild type revertants arise in mutant cells maintained in conditions that favor the growth of wild type cells. Mutant strains should be maintained under the most permissive conditions possible, and frozen in liquid nitrogen as soon as possible (Bruns et al., 2000; Cassidy-Hanley, Smith, and Bruns, 1995; section VI.B). When working with stock cultures maintained on the bench, mutant phenotypes should be verified before undertaking any experimental protocol that requires the expression of a mutant allele that is heterozygous in the macronucleus to eliminate the possibility of allelic loss due to phenotypic assortment. If loss of the mutant phenotype occurs, new working stocks should be established from frozen cultures.
The Tetrahymena Stock Center houses a variety of T. thermophila mutant strains, including those carrying naturally occurring and induced mutations, defined chromosomal modifications, and genetically engineered modifications. A brief description of basic types of mutant and modified strains available to researchers is listed below. A complete listing of available strains, updated as new strains become available, can be found at http://tetrahymena.vet.cornell.edu/strains.php.
1. Drug resistant functional heterokaryons
Among the most frequently utilized T. thermophila mutants are a series of functional heterokaryons homozygous for mutations conferring resistance to either cycloheximide, 6-methylpurine (Byrne, 1978; Byrne et al., 1978), or paromomycin (Bruns et al., 1985) in the germinal micronucleus but expressing the wild type (drug sensitive) allele in the somatic macronucleus. Paromomycin and cycloheximide heterokaryons are available in both B and C3 backgrounds, while the 6-methylpurine mutation is currently limited to the B strain. Resistance heterokaryons greatly simplify genetic analyses in Tetrahymena, allowing for direct selection of progeny cells in a mass mating (Bruns and Brussard, 1974b).
2. Exocytosis mutants
T. thermophila strains carrying a variety of mutations that affect regulated mucocyst secretion have been characterized (Gutierrez and Orias, 1992; Haddad and Turkewitz, 1997; Melia et al., 1998; Orias et al., 1983). These strains have been used in the study of secretory granule biogenesis and regulated exocytosis (Turkewitz, 2004). Mutants blocked in exocytosis have also proven useful for the efficient purification of cell organelles and macromolecules (Dentler, 1995; Johnson, 1986; Lombillo et al., 1993; Williams, 2000) since secretion of the sticky mucocyst contents can interfere with the purification of cellular components (Tiedtke, 1985).
3. Temperature sensitive mutations
Temperature sensitive (ts) mutations affecting pathways as diverse as phagocytosis (Suhr-Jessen and Orias, 1979), morphological development (Frankel et al., 1993; Williams and Honts, 1987) and cell division (Frankel et al., 1976; Frankel et al., 1980) have been described in T. thermophila. Many of the original ts mutants developed by Joseph Frankel (University of Iowa) and Eduardo Orias (University of California, Santa Barbara) are available through the Tetrahymena Stock Center.
4. Conjugation mutants
Conjugation, the sexual stage in the Tetrahymena life cycle, is a complex developmental program that is highly conserved in a number of ciliate species (Raikov, 1976). In Tetrahymena, conjugation involves an array of activities including mating type recognition, pair attachment, and cell fusion, as well as nuclear events including meiosis, mitosis, nuclear transfer, fertilization, and developmental nuclear modification (Ch. 3 and Ch. 7 in this volume). A series of mutant strains have been developed which affect early, middle and late stage events in conjugation, from chromatin condensation to macronuclear anlagen development (Cole and Soelter, 1997; Cole et al., 1997). Morphological pattern mutants arising as a result of problems in conjugation have also been identified (Cole and Frankel, 1991; Cole, 1991). These strains provide a unique and valuable resource for examining factors influencing prezygotic, postzygotic, and exconjugant developmental.
5. Chromosomal modifications
Although the Tetrahymena germinal micronucleus is not expressed (Gorovsky and Woodard, 1969; Mayo and Orias, 1981), and many Tetrahymena species apparently lack a micronucleus (Elliot and Hayes, 1955), the T. thermophila strains commonly used in the lab appear to require at least a vestigial micronucleus for cell viability. With one possible exception (Kaney and Speare, 1983; Karrer et al., 1984), complete loss of the micronucleus in T. thermophila is lethal. However, cells missing large portions of the micronuclear genome are viable when grown vegetatively. A collection of single and multiple nullisomic strains missing both copies of one or more micronuclear chromosomes have been created (Bruns and Brussard, 1981; Bruns et al., 1983), as well as strains unisomic (containing only a single micronuclear chromosome) for each of the micronuclear chromosomes, in both a B and a C3 genetic background. A series of overlapping deletions have also been created for each of the five micronuclear chromosomes. Since monosomic and hemizygous strains containing a single copy of part or all of any micronuclear chromosome are viable, deletion and nullisomic strains are very useful for genetic mapping (Altschuler and Bruns, 1984).
D. Meiotic Segregation Panels
Panels of B-C3 meiotic segregants and terminal assortants derived from heterozygous progeny of matings of inbred B and C3 strains, developed by the Orias lab for use in mapping genes to micronuclear and macronuclear chromosomes (Brickner et al., 1996; Lynch et al., 1995), are available from the Tetrahymena Stock Center. These strains are useful for localizing mutant genes to the micronucleus to help determine relationships among mutants with similar phenotypes, and for identifying the macronuclear location associated with a specific phenotype to facilitate identification and cloning of mutant genes (Hamilton and Orias, 2000).
E. Genetically Engineered Lines
With the development of facile techniques for gene manipulation, genetic engineering of new Tetrahymena strains has become routine. Gene disruptions, gene replacements, knockouts and knock-ins can easily be accomplished in the micronucleus by biolistic transformation (Bruns and Cassidy-Hanley, 2000a; Cassidy-Hanley et al., 1997), and in the macronucleus by biolistic transformation, electroporation (Gaertig et al., 1994; Gaertig and Kapler, 2000), and microinjection (Chalker et al., 2000; Tondravi and Yao, 1986). Genetically modified strains are available from the labs of origin, or increasingly, through the Tetrahymena Stock Center. Among the T. thermophila strains currently available are significant portions of the original strain collections developed in the laboratories of Joseph Frankel, University of Iowa (IA strains), Peter Bruns (CU strains), Eduardo Orias (SB strains), and Martin Gorovsky (various genetically modified transformant strains), as well as a variety of genetically engineered strains developed in other labs (for a current list see http://tetrahymena.vet.cornell.edu/strains.php).
III. Other Tetrahymena Species
Although, as discussed above, T. thermophila is the primary species of choice for Tetrahymena research, significant work has been carried out using other Tetrahymena species. Tetrahymena ssp. are useful indicators for ecotoxicity tests (Gerhardt et al., 2010), and Tetrahymena pyriformis GL, an amicronucleate strain used for much of the early Tetrahymena research, is still frequently used for toxicological studies (Artemenko et al., 2011; Sauvant et al., 1995; Sauvant et al., 1997; Sauvant et al., 1999). Ribosomal RNA based phylogenies of various Tetrahymena species have been developed (Nanney et al., 1989; Preparata et al., 1989). In addition, phylogenetic relationships of many species have been analyzed by bar coding using the cytochrome c oxidase subunit 1 (COX1) (Kher et al., 2011), and the small subunit ribosomal RNA (SSrRNA) genes (Chantangsi et al., 2007; Chantangsi and Lynn, 2008). Phylogenetic relationships have also been examined using comparisons of telomerase RNA (Ye and Romero, 2002).
The American Type Culture Collection houses a collection of 38 different Tetrahymena species, including T. thermophila and 4 unidentified species. The Tetrahymena Stock Center houses a smaller collection of known Tetrahymena species, and a large collection of unknown Tetrahymena species derived from wild isolates collected by Paul Doerder, Cleveland State University. Fourteen different Tetrahymena species, as well as six T. thermophila strains, are available from the Culture Collection of Algae and Protozoa (CCAP) in the UK (http://www.ccap.ac.uk/index.htm). However, all of the Tetrahymena strains in the CCAP collection are maintained solely by serial sub-culture, and normally micronucleate strains are likely to have become germline-senescent and not useful for any research requiring integrity of the germline. Although fertility is not an issue, nonetheless, amicronucleate strains may also be adversely affected by continued serial transfer over long periods of time, and may exhibit decreased viability and eventual die-off of clones.
IV. Cell culture media
Originally cultured in bacterized hay or vegetable matter infusions, Tetrahymena was the first animal-like eukaryotic cell to be grown axenically (Lwoff, 1923). Tetrahymena has two separate nutrient uptake systems; phagocytosis, which in Tetrahymena involves intake of particulate matter via a highly specialized oral apparatus and subsequent nutrient digestion in food vacuoles, and a surface uptake system that transports nutrients in solution into the cell (Orias and Rasmussen, 1976; Rasmussen and Orias, 1975; Ch. 6 in this volume). Each of these uptake mechanisms is sufficient to support normal growth in appropriate media. In most standard media, phagocytosis is essential for cell growth (Rasmussen and Kludt, 1970; Rasmussen and Modeweg-Hansen, 1973). However, complete chemically defined media (CDM) (Hagemeister et al., 1999; Szablewski et al., 1991) can support normal growth without phagocytosis. Axenic proteose peptone based media are currently the most common choice for laboratory culture, but, as discussed below, Tetrahymena can be successfully grown in a wide variety of media, including bacterized peptone, bacterized infusions of lettuce or rye leaves (Cerophyll), skim milk based media, and chemically defined media.
A. Glassware
Tetrahymena is sensitive to even very low levels of some types of impurities in the media. To prevent problems, high purity distilled and/or de-ionized water should be used in making all media, and dedicated glassware should be set aside solely for use in making and storing media. It is critical to insure that no soap or acid residue remains on the surface of the glassware following washing. Although some automated dish washing protocols may yield adequate glassware, it is highly advisable wash and rinse bottles and flasks used for making and storing media by hand, ending with several careful rinses with high purity distilled/deionized water. To prevent build-up of water residue, glassware should be thoroughly drained in an inverted position before drying.
B. Proteose peptone based media
Proteose peptone (PP), an enzymatic digest of animal protein high in proteoses, is the traditional basis for most media used for growing Tetrahymena in the laboratory. In rich axenic media, PP is often supplemented by varying concentrations of yeast extract, glucose, and some form of iron. Bacto proteose peptone and Bacto yeast extract, originally manufactured by Difco, are now available directly from BD Sciences (BD Diagnostic Systems No.:211684 and BD Diagnostic Systems No.:212750 respectively) and through retailers like Fisher Scientific. It should be noted that not all grades of proteose peptone are suitable for making Tetrahymena media and care must be taken in selecting the appropriate peptone. PP media is occasionally supplemented with liver extract, primarily to maintain Tetrahymena species newly isolated from the wild, or other species that are difficult to maintain in standard PP media (Doerder, personal communication). Liver fraction L, which is no longer available, was originally used for Tetrahymena culture. A possible substitute is Sigma 03077 Liver Hydrolysate, which has proven useful in the culture of Trichomonas and other difficult to culture protozoa. The compositions of several common PP based media are shown in Table I. Regardless of the PP media chosen, a few basic considerations must be kept in mind.
Table I.
Rich Axenic Nutrient Media.
| Medium | Recipe |
|---|---|
| PPa | 2% proteose peptone |
| 10 μM FeCl3 or 90 μM sequestrene (Fe-EDTA) | |
| SP210b | 2% proteose peptone |
| 10 μM FeCl3 or 90 μM sequestrene (Fe-EDTA) | |
| 250 ug/ml streptomycin sulphate | |
| 250 ug/ml penicillin G | |
| Modified Neffc | 0.25% proteose peptone |
| 0.25% yeast extract | |
| 0.5% glucose | |
| 33.3 μM FeCl3 | |
| (To avoid precipitate formation, the FeCl3 is first dissolved in one quarter of the final H2O volume, and the glucose, yeast and PP are added and dissolved next. The remainder of the H2O is then added, bottled and autoclaved.) | |
| SSPd | 2% proteose peptone |
| 0.1% yeast extract | |
| 0.2% glucose | |
| 0.003% sequestrene (Fe-EDTA) (can be replaced with 33 μM FeCl3) | |
| PPYe | 1% proteose peptone |
| 0.15% yeast extract | |
| 0.01 mM FeCl3 | |
| PPYGf | 0.4% proteose peptone |
| 0.2% yeast extract | |
| 1.0% glucose | |
| PPYSe | 1% proteose peptone |
| 0.15% yeast extract | |
| 0.01 mM FeCl3 | |
| 0.2 M NaCl | |
| EPPg | 2% proteose peptone |
| 2 mM Na3 citrate·2H20 | |
| 1 mM FeCl3 | |
| 30 uM CuSO4·5H20 | |
| 1.7 uM Folinic acid, Ca salt | |
| Liver peptoneh | 1.5% proteose peptone |
| 0.1% yeast extract | |
| 0.25% bactotryptone | |
| 0.25% liver fraction L (liver hydrolysate, Sigma 03077) | |
| 0.5% glucose | |
| 0.1% KH2PO4 (7.35 mM) | |
| 0.1% Na2HPO4 (7.04 mM) | |
| Skim milk mediumi | 2% skimmed milk |
| 0.5% yeast extract | |
| 0.1% ferrous sulphate chelate solution | |
| 1% glucose | |
| MYE skim milk mediumj | 1%(w/v) skim milk |
| 1%(w/v) yeast extract |
Orias, E., Hamilton, E. P., and Orias, J. D. (2000). Tetrahymena as a laboratory organism: Useful strains, cell culture, and cell line maintenance. Methods Cell Biol, 62, 189–211.
Diaz, S., Amaro, F., Rico, D., Campos, V., Benitez, L., Martin-Gonzalez, A., et al. (2007). Tetrahymena metallothioneins fall into two discrete subfamilies. PLoS ONE, 2, e291.
Cassidy-Hanley, D., Bowen, J., Lee, J. H., Cole, E., VerPlank, L. A., Gaertig, J., et al. (1997). Germline and somatic transformation of mating tetrahymena thermophila by particle bombardment. Genetics, 146(1), 135–47.
Gorovsky, M. A., Yao, M. C., Keevert, J. B., and Pleger, G. L. (1975). Isolation of micro- and macronuclei of tetrahymena pyriformis. Methods Cell Biol, 9(0), 311–327.
Smith, D. L., and Doerder, F. P. (1992). Multiple effects of mutation on expression of alternative cell surface protein genes in tetrahymena thermophila. Genetics, 130(1), 97–104
Mori K., Yomo T., and Kashiwagi A. (2011). Single-cell isolation and cloning of tetrahymena thermophila cells with a fluorescence-activated cell sorter. J. Eukaryotic Microbiol. Journal of Eukaryotic Microbiology, 58(1), 37–42.
Orias, E., and Rasmussen, L. (1976). Dual capacity for nutrient uptake in tetrahymena. IV. growth without food vacuoles and its implications. Exp Cell Res, 102(1), 127–137.
(personal communication, P. Doerder)
Weide, T., Herrmann, L., Bockau, U., Niebur, N., Aldag, I., Laroy, W., et al. (2006). Secretion of functional human enzymes by tetrahymena thermophila. BMC Biotechnol, 6, 19.
De Coninck, J., Leclercq, B., Exbrayat, J. M., and Duyme, F. (2004). Factorial designs: An efficient approach to choosing the main factors influencing growth and hydrolase production by tetrahymena thermophila. J Ind Microbiol Biotechnol, 31(5), 204–208.
1. Composition
Growth in all PP media is limited by iron. The need for iron can be met by supplementation with iron salts (FeCl3 is most commonly used) at a final concentration as low as 10uM, or chelated iron salts like Fe-EDTA (Ethylenediaminetetraacetic acid iron(III) sodium salt hydrate, 12–14% Fe, Sigma # 03650), or sequestrene (Becker Underwood, Dayton, Ohio or Trilon B Fe 13% powder, BASF Corp., Mount Olive, NJ). Yeast extract contains some iron but it is a good policy to supplement media containing yeast extract with addition iron in one of the above forms. PP media containing liver extract generally does not require added iron. Addition of ferric or ferrous chloride can produce an iron precipitate if added prior to autoclaving. Although the precipitate does not affect growth, it can interfere with some downstream operations like electronic cell counting or collection of cells by high-speed centrifugation. Precipitation can be prevented by filter sterilizing a concentrated iron solution separately and adding it to the autoclaved media, immediately after it has cooled or just prior to use. A simpler method is to add the appropriate amount of iron chloride from a 1000x sterile concentrated stock solution (stored at 4°C) to about one fourth of the final volume of water, stir well before dissolving the other ingredients one at a time in the solution, bring up to final volume, and autoclave. Filter sterilization of any PP media is not recommended since some particulate matter appears to be necessary to induce the formation of food vacuoles (Rasmussen and Kludt, 1970; Rasmussen and Modeweg-Hansen, 1973). Alternatively, Fe-EDTA can be used as an iron source. Although somewhat more expensive, Fe-EDTA generally does not form a precipitate when added directly to the media before autoclaving. PP media should be autoclaved at 121°C and 15 psi for 30 minutes. Excessive autoclaving will decrease the ability of the media to support optimal growth.
Yeast extract (0.1% to 0.25%) and glucose (0.1% to 1%) are frequently included in rich axenic media used for growing Tetrahymena, with higher concentrations of each generally combined with lower concentrations of PP (Table I). In 2% PP both yeast extract and glucose can be omitted with little affect on growth rate. For routine work, cells may be grown in a simple 2% PP media supplemented with 10 μM FeCl3 or 90 μM Fe-EDTA. In media with lower PP concentrations like Neff (0.25% PP), the addition of yeast extract is recommended to maintain optimal growth rates. For short term expansion of cultures for non-genetic work, 1% PP supplemented with 10 μM FeCl3 will support normal doubling times, but long term maintenance in this media results in a greatly increased rate of infertility, and it is not recommended for work involving genetic analysis (Orias et al., 2000).
SPP (M. A. Gorovsky et al., 1975) and modified Neff’s medium (Cassidy-Hanley et al., 1997) represent the PP concentration extremes among commonly used PP media (Table I). Both efficiently support rapid growth in cell culture, even though there is an 8-fold difference in PP concentration, perhaps because the lower PP concentration in Neff is counterbalanced by a two and a half fold increase in the concentration of both yeast extract and glucose. There are, however, slight differences in the growth curve generated by cells growing in the two media. Cells double somewhat faster in SPP than in Neff (~2.5 hours vs. ~3 hours at 30 °C in shaking cultures), but cells plateau at slightly higher concentrations in Neff than in SPP (2–3 × 106 vs. 1–2 × 106 respectively). Once stationary phase is reached, Neff cultures maintain longer before crashing than equivalent SPP cultures, holding up to two weeks even in microtiter plates kept at room temperature. The longer holding time makes Neff an ideal media for maintaining cultures in stock tubes (Section V.B.1.), while SPP may be preferable for expanding cultures if a more rapid growth rate is required. For settings where cost is a major consideration (e.g. educational use), Neff media is a good all round media that is significantly less expensive than other PP based media. It should be noted that T. thermophila strains with different genetic backgrounds may grow differently in various PP media growth, and that many other Tetrahymena species grow more quickly and to higher densities in richer PP media.
2. Storage
PP media can be stored in various ways. Concentrated media can be made, aliquoted, and immediately stored frozen at −20°C until needed, at which time it is thawed, diluted to the appropriate concentration and autoclaved (Orias et al., 2000). If larger volumes are needed on a regular basis, sterile 1x PP media also can be stored in bottles for months at room temperature, although care should be taken to limit long-term exposure to bright light since some required vitamins exhibit light sensitivity over time.
C. Bacterized media
Bacterized media has historically been used as a method for inducing mating without the need to physically manipulate the Tetrahymena cells involved. Currently, growth in axenic media followed by replication into non-nutritive starvation media (Section IV.H) is the preferred protocol for inducing mating, but bacterized media can be useful when the mating of multiple clones without the need for further cell manipulation is desired, e.g. matings in 96 well microtiter plates. Bacterized media may also be useful for maintaining fastidious newly isolated wild Tetrahymena species that do not thrive in PP based media. The maximum Tetrahymena cell concentration achievable in bacterized media is relatively low relative to standard PP media, ~ 2 × 104 cells/ml (Ducoff, H. S. et al., 1964).
1. Bacterized PP
Although no longer commonly used, Tetrahymena can be maintained in bacterized peptone. Bacterized peptone can be prepared by inoculating PP medium (essentially any 1% to 2% PP media will suffice), prepared without added antibiotics, with Klebsiella pneumoniae (formerly K. aerogenes), and shaking the culture at ~200 rpm overnight at either 30°C or 37°C (Orias et al., 2000). This 100% bacterized PP medium (100% BP media) can be maintained at 4°C for up to a week. The 100% BP medium is diluted 1:50 to 1:100 with sterile water just prior to use. If the bacterized medium is to be used for mating (Orias and Flacks, 1973; Simon and Whang, 1967), it is important to keep the final concentration of PP at a level that will allow starvation-induced mating once the bacteria have been consumed. To do this, the Tetrahymena culture used for inoculation must also be diluted at least 50-fold in sterile water. Following growth in bacterized media, any remaining bacteria can be eliminated without affecting the Tetrahymena cells by the addition of penicillin and streptomycin (250 μg/ml each) to the media.
2. Bacterized cereal grass (Cerophyll) or lettuce infusions
Bacterized infusions of lettuce (derived from early Paramecium culture media; D. L. Nanney, 1953; van Wagtendonk and Hackett, 1949) or rye leaves (Cerophyll; D. L. Nanney, 1953; Simon and Nanney, 1979) were commonly used in early work with Tetrahymena. Although rarely used for general culture or maintenance today, cereal grass based media are occasionally useful for specific experimental purposes. The original Cerophyll is no longer available, but Ward’s Natural Science -Hay Medium Solution, provided as a sterile 2x solution, is similar. If dry rye or cereal grass products are used, 0.6 gm should be added to 500 ml of boiling distilled H2O, boiled for 2 minutes, and filtered through Whatman filter paper #1 while still warm, and autoclaved. The sterile infusion media is innoculated with Klebsiella pneumoniae and incubated overnight at 37°C.
D. Chemically defined media
Consistent growth of Tetrahymena cells in synthetic, chemically defined media was first shown by Kidder and Dewey (1951), and provided a controlled means of examining cell nutritional requirements. A modified chemically defined media (CDM, Table II), initially described by Szablewski et al. (1991), supports rapid growth similar to that observed in PP media (2 hour doubling time at 37°C), and cell concentrations of up to 106 cells/ml. Amino acids in group A are required for growth, but a minimal chemically defined media (minimal CDM) can be prepared by omitting amino acids contained in solutions B–E in the CDMA recipe shown in Table II. Minimal CDM is useful for working with auxotrophic mutants (Sanford and Orias, 1981), but cell doubling time in minimal CDM is slightly slower than in complete CDM (~ 2.5 hr at 30°C). CDM will support indefinite propagation by serial transfer providing the initial inoculum concentration is at least 500 cells/ml. However, lower concentrations of cells transferred to CDM will not survive, perhaps as a result of a need for a critical initial cell density to condition the medium with required autocrine factors (Christensen et al., 1995; Christensen et al., 2001; Rasmussen et al., 1996). If an inoculum of less than 2500 cell/ml CDM medium is necessary, the medium should be supplemented with 7.5 μM hemin to insure cell viability. Hemin can be prepared as a stock solution by dissolving in 0.01 N NaOH and autoclaving (Christensen and Rasmussen, 1992). Alternatively, Hagemeister et al. (1999) suggested that Tetrahymena cell death in CDM innoculated with a low concentration of cells is not apoptotic, but is rather the result of accidental cell lysis at the medium-air interface. They provide an alternate CDM recipe (CDMC, Table II) that permits the growth of low cell inoculum or single cells in the presence of an air-medium interface without the addition of any supplements. However, the doubling time in CDMC is 3.5 hr at 36°C, considerably slower than in CDM. Initial cell concentration is not an issue in PP and bacterized media, which provide adequate amounts of any growth factors necessary for autocrine regulation of cell survival and support strong cell proliferation growth even in single cell subcultures.
Table II.
Chemically Defined Synthetic Media: Composition, Concentration of Stock Solutions, and Preparation.
| CDMA a | CMDC b | CDMA a | CMDC b | ||
|---|---|---|---|---|---|
| Amino Acid Solution A | mg/ml | Salts and Chelator Solution | mg/ml | ||
| L-Arg-HCl | 12 | 2.4 | K2HPO4 • 3H2O | 25 | 5 |
| L-His-HCl • H2O | 8 | 1.6 | KH2PO4 | 25 | 5 |
| L-Ile | 8 | 8 | MgSO4 • 7H2O | 50 | 10 |
| L-Leu | 8 | 8 | CaCl2 • 2H2O | 1 | 0.2 |
| L-Lys-HCl | 8 | 16 | Tri-Potassium Citrate | 65 | 13 |
| L-Met | 6 | 6 | Vitamins (Solution A) | ||
| L-Phe | 6 | 6 | Na Riboflavin Phosphate • 2H2O | 0.05 | 0.05 |
| L-Ser | 6 | 12 | Vitamins (Solution B) | ||
| L-Thr | 8 | 16 | DL-6, 8-Thioctic Acid | 0.01 | 0.01 |
| L-Trp | 6 | 12 | Vitamins (Solution C) | ||
| L-Val | 4 | 4 | Thiamin-HCl | 0.05 | 0.05 |
| Amino Acid Solution B | Prydoxal-HCl | 0.01 | 0.01 | ||
| L-Gln | 4 | 0.8 | Nicotinic acid | 0.09 | 0.09 |
| Amino Acid Solution C | D-Pantothenic Acid, hemi Ca-salt | 0.08 | 0.08 | ||
| L-Asn • H2O | 8 | 16 | Vitamins (Solution D) | ||
| L-Pro | 8 | 16 | Folinic acid, Ca salt | 0.01 | 0.01 |
| Amino Acid Solution D | Trace metals solution | ||||
| L-Ala | 6 | 12 | FeCl2 • 6H2O | 1 | 0.2 |
| L-Asp | 8 | 1.6 | MnSO4 • 4H2O | 0.16 | 0.032 |
| L-Glu | 16 | 0.8 | Co (NO3)2 • 6H2O | 0.05 | 0.01 |
| Gly | 16 | 32 | ZnSO4 • 7H2O | 0.45 | 0.09 |
| Amino Acid Solution E | CuSO4 • 5H2O | 0.03 | 0.006 | ||
| L-Tyr (Do not prepare ahead) | 8 | 8 | (NH4)6 Mo7O24 • 4H2O | 0.01 | 0.002 |
| Nucleoside solutions | Glucose solution | ||||
| Adenosine | 0.2 | 0.2 | Glucose | 250 | 250 |
| Cytidine | 0.2 | 0.2 | |||
| Guanosine | 0.2 | 0.2 | |||
| Uridine | 0.2 | 0.2 | |||
CDMA is modified chemically defined media that supports rapid growth similar to that observed in PP media (Szablewski et al., 1991).
CDMC is a modification of CDMA that permits growth of low concentration inoculum (including single cells) without additional supplements (Hagemeister, Grave, Kristiansen, Assaad, and Hellung-Larsen, 1999).
1. Media preparation:
- Amino acid solutions A – D are prepared as 40-fold concentrated stock solutions. Preparation notes: Solutions A and C: adjust pH to 7 and sterilize by filtration; Solution B: store frozen; Solution D: dissolve aspartic and glutamic acids in water with stirring, keep pH from dropping below 7 with 1N KOH, add alanine and glycine, adjust pH to 7, filter sterilize and store at 4°C.
- Nucleoside solutions are prepared as 10-fold concentrated stock solutions.
- Salts and Chelator solutions are prepared as 100-fold concentrated stock solutions.
- Vitamins are prepared as 100-fold concentrated stock solutions and stored frozen. To make Vitamin Solution B, dl-6, 8-Thioctic Acid is dissolved in 1 ml absolute ethanol, then diluted in 100 ml H2O and filter sterilized.
- Trace metals solution are prepared as a 100-fold concentrated solution, and adjusted to approximately pH 2 with 1 N HCl.
- Glucose solution is prepared as a 50-fold concentrated solution.
To make the complete media, dissolve Tyr at 60°C, adjust to give a concentration of 0.2 mg/ml in the final medium, cool and then proceed to add the remaining solutions. The pH may be adjusted as required. The medium may be sterilized by autoclaving or by filtration; if autoclaving is used, glucose should be added aseptically after cooling.
- Minimal defined media. Amino acid solution A contains all required amino acids. A minimal defined medium can be created by omitting amino acid solutions B – E.
- Low cell inoculum in CDMA. When inocula of less than 2500 cells/ml of final medium will be used, CDMA should be supplemented with hemin at a final concentration of 7.5 μM. To prepare a stock solution, dissolve hemin in 0.01 N NaOH and autoclave ((Christensen and Rasmussen, 1992).
- Phagocytosis deficient cells. For growing phagocytosis-deficient cells, the final concentrations of FeCl, CuSO4 and folinic acid should be increased to 1mM, 25 μM and 1 mg/ml respectively (Orias and Rasmussen, 1979).
Christensen, S. T., and Rasmussen, L. (1992). Evidence for growth factors which control cell multiplication in tetrahymena thermophila. Acta Protozoologica, 31(4), 215–219.
Hagemeister, J. J., Grave, M., Kristiansen, T. B., Assaad, F., and Hellung-Larsen, P. (1999). Interface-mediated death of unconditioned tetrahymena cells: Effect of the medium composition. J Eukaryot Microbiol, 46(1), 6–11.
Orias, E., and Rasmussen, L. (1979). Dual capacity for nutrient uptake in tetrahymena. V. utilization of amino acids and proteins. J Cell Sci, 36, 343–353.
Szablewski, L., Andreasen, P.H., Tiedtke, A., Florin-Christensen, J., Florin-Christensen, M., and Rasmussen, L. (1991). Tetrahymena thermophila: Growth in synthetic nutrient medium in the presence and absence of glucose. Journal of Eukaryotic Microbiology, 38(1), 62–65.
E. Skimmed milk media
PP media, while widely used, are an expensive option for large scale cultivation of Tetrahymena. Increasing interest in the industrial use of Tetrahymena (Ethuin et al., 1995; Jayaram et al., 2010; Kiy and Tiedtke, 1992; Weide et al., 2007) spurred the examination of less expensive media for large scale culture, in particular the use of skim milk based media (Table I). In a bioreactor under conditions of high cell density fermentation and cell retention, skim milk based medium has supported the culture of Tetrahymena at cell densities of more than 2.2 × 107 cells/ml, equivalent to 48 g dry weight (Weide et al., 2006). A more dilute skim milk based medium (MYE medium, 1%(w/v) skim milk, 1%(w/v) yeast extract) has also been successfully used in batch fermenters to support growth up to ~3 × 106 cells/ml (De Coninck et al., 2004).
F. Media for phagocytosis deficient cells
Phagocytosis is essential for cell growth in most media (Rasmussen and Kludt, 1970; Rasmussen and Modeweg-Hansen, 1973), and temperature sensitive mutants defective in phagocytosis cannot survive past two to three doublings in any standard PP medium (Orias and Pollock, 1975). EPP, a modified PP based axenic medium (Table I), allows indefinite cell growth in the absence of phagocytosis (Orias and Rasmussen, 1976). A specialized chemically defined media (Table II) for phagocytosis deficient cells has also been developed, based on modifications of standard CDM (Orias et al., 2000). Phagocytosis deficient cells cannot grow in bacterized media.
G. Media for long term stock culture
In situations where vegetative cells are only used infrequently and/or freezing is not an option, it is sometimes useful to maintain unfrozen Tetrahymena cultures long term (e.g. in classroom settings requiring only vegetatively propagated cells). Long term maintenance of growing cultures also presents an alternative for Tetrahymena species that are difficult to maintain in PP media or that are not amenable to freezing in liquid nitrogen. It should be noted that although these methods can successfully maintain viability for long periods, the effects on genetic stability have not been carefully analyzed, and it is likely that genetic deterioration and eventual sterility will occur in micronucleate Tetrahymena strains maintained under these conditions (Simon and Nanney, 1979).
1. Bean medium
The simplest media for prolonged storage in stock tube cultures uses a whole soy or garbanzo bean as a nutrient source (Sweet and Allis, 2010; Williams et al., 1980). To prepare the media, place a single bean in 10 ml distilled water and autoclave or boil ~5 minutes in a capped culture tube. Any tubes that show signs of significant evaporation or that become cloudy within 24 hours should be discarded. Streptomycin and penicillin (250 μg/ml each) and 0.25 μg/ml Amphotericin B (Fungizone; Fisher BioReagents, #BP2645-20) can be added after the media cools, but for general use neither are necessary. After inoculation with sterile cells, add 1 – 2 ml of sterile paraffin oil to the tubes to preventevaporation, cap lightly, and store at 15°C to 20°C. Cells should be transferred every 6 to 8 months. Direct transfer from bean medium tube to bean medium tube is possible, but it is much preferable to provide an intervening passage in PP media between bean tube transfers.
2. Rat gut medium
A second method with much more limited utility uses rat intestine as the primary nutrient source (Williams et al., 1980). While crude, this method is useful for the maintenance of Tetrahymena species that are difficult to freeze or maintain in other media. To make the growth media, place a one cm section of cleaned rat intestine in 8 ml of distilled water in a culture tube, layer with 8 mm of heavy paraffin oil, and autoclave. When the media is cool, Tetrahymena can be added by inoculating through the oil layer with a sterile small bore pipette. Cultures in rat gut medium can be kept at room temperature for at least a year. Cells should be briefly grown in a rich PP media like SPP between yearly rat gut tube transfers. Three sequential transfers in the PP media over several days will reinvigorate the culture and prepare it for re-inoculation into the rat gut media, where it can be maintained for another year.
H. Starvation media
A number of protocols, especially those involving mating reactivity, require starvation in non-nutritive media. Starved cells undergo rapid and extensive physiological, biochemical, and molecular changes, including starvation-induced proteolysis (Grinde and Jonassen, 1987), and changes in ribosome biosynthesis (Hallberg and Bruns, 1976) and gene expression (Miao et al., 2009; Song and Gorovsky, 2007; Xiong et al., 2011). Cells can be maintained in starvation media for several days, although viability is decreased if the cells are held at high density or in low surface to volume culture vessels that do not provide adequate aeration (e.g. culture tubes). Low ionic strength salt based media (see Table III for recipes) like Dryl’s medium (Dryl, 1959), 10 mM Tris, pH7.5 (Bruns and Brussard, 1974a), or NKC media (Sugai and Hiwatashi, 1974) are routinely used to induce sexual reactivity. Starvation without induction of mating reactivity can be accomplished by starving cells in 50 to 70 mM Tris, pH7.5. In this medium, cells starve but fail to undergo initiation, the first step in the activation of the sexual cycle, and do not become mating reactive (Bruns and Brussard, 1974a).
Table III.
Starvation media.
| Medium | Recipe |
|---|---|
| Dryl’sa | 0.59 g of Na citrate-2H2O (2 mM) |
| 0.14 g of NaH2PO4 • H2O (1 mM) | |
| 0.14 g of Na2HPO4 (1 mM) | |
| 0.13 g of CaCl2 (1.5 mM) | |
| (To avoid precipitation of the CA phosphate, the CaCl2 solution is autoclaved separately from the mixture of sodium salts, and the two solutions are mixed aseptically after cooling.) | |
| Tris bufferb | 10 mM Tris HCl, pH 7.5 |
| NKC solutionc | 0.2% NaCl |
| 0.008% KCl | |
| 0.12% CaCl2 |
Dryl, S. (1959). Antigenic transformation in paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J. Protozool., 6(25)
Bruns, P. J., and Brussard, T. B. (1974). Pair formation in tetrahymena pyriformis, an inducible developmental system. J Exp Zool, 188(3), 337–44.
Sugai, T., and Hiwatashi, K. (1974). Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in tetrahymena pyriformis. J Protozool, 21(4), 542–58.
If large numbers of individual matings must be performed, e.g. in microtiter plates, initial growth in Neff medium simplifies the starvation process. Since Neff contains only 0.25% PP, a simple 1:10 dilution decreases nutrient availability to the point where cells become mating reactive. Replicating clones grown in Neff from microtiter plates into microtiter plates containing enough starvation medium to ensure at least a 1:10 dilution of the original Neff will produce mating reactive cells in the replicate plates. Growth in bacterized PP medium (Section IV.C.1) can also be used to carryout matings since once bacteria in the medium have been consumed, Tetrahymena will undergo starvation and become mating reactive.
V. Cell culture
A. Basic information
Tetrahymena can be easily cultured using a wide variety of media, containers, and conditions, as long as basic requirements for nutrition, aeration, temperature, and cell concentration are met. Culture vessels must be kept meticulously clean, and the use of dedicated flasks for cell growth is strongly recommended. The same stringent criteria described above (Section IV.A) for preparing glassware used in making media should be applied to all culture vessels. High surface to volume ratios should be maintained in standing cultures. If volume requirements preclude the use of shallow conditions, culture vessels should be shaken or rotated to provide necessary aeration. When very large volumes are required, as in a fermenter or multi-liter bottle, forced aeration and agitation must be supplied to insure sufficient oxygenation and gas exchange for optimal growth. If aeration and/or agitation are violent enough to cause foaming, an anti-foam agent (e.g. 0.001% (v/v) Sigma AntiFoam 204, A6426) should be added to the culture to minimize cell damage. Since aeration stimulates growth and maximizes final cell concentration, but excessive agitation can cause cell damage, overall conditions for each large scale culture unit must be optimized individually (De Coninck et al., 2004).
It is important to note that, regardless of the culture method employed, care must be taken to maintain cell viability when harvesting or manipulating cells. Tetrahymena cells are extremely sensitive to the changes in aeration and cell concentration resulting from centrifugation, much more so than bacteria or yeast. The force necessary to efficiently pellet Tetrahymena cells may vary depending on culture volume, tube and centrifuge type. For routine work, wild type cells can be pelleted in 50 ml conical tubes at ~ 600g to 1000g for 1 minute with no adverse effects. However, the supernate must be immediately removed from the pellet since Tetrahymena are strong swimmers, and significant numbers of cells may be lost from the pellet if the supernate is not swiftly removed. High centrifugal velocities, especially if accompanied by sudden, dramatic temperature changes, can cause massive cell lysis, as can too vigorous resuspension of cell pellets. Vortexing should never be used to resuspend intact Tetrahymena. Rapid, massive cell death, accompanied by the release of high amounts of proteases and nucleases, can also occur if cells are allowed to remain in unfavorable conditions (e.g. in a pellet following centrifugation) for more than a few minutes. Cell pellets must be either immediately resuspended in fresh medium and returned to an appropriate culture vessel (if maintenance of viable cells is desired), or processed without delay.
When Tetrahymena cells must be cultured for a large number of generations requiring repeated serial transfer, care should be taken that cells are not transferred in a manner that constantly maintains continuous exponential growth resulting from the repeated transfer of a very small initial inoculum into a highly dilute culture. The creation of clones exhibiting reduced growth rate or the establishment of variant cells incapable of net telomere elongation can occur as a result of coordinate telomere lengthening in vegetatively growing log phase cells. To avoid this possibility, cultures should periodically be allowed to remain at stationary phase between sustained bursts of log phase growth (Larson et al., 1987). One simple solution is to refrain from transferring cell cultures on weekends to provide an opportunity for cells to regularly undergo a brief transition to stationary phase. Maintaining cultures at room temperature will prevent the long-telomere phenotype, but also extends the time necessary for long term growth procedures like maturation of immature clones or terminal phenotypic assortment of mixed macronuclei.
B. Methods of cell culture
1. Stock cultures
For routine daily use, Tetrahymena cultures are generally maintained out of direct light, between 18°C and 20°C – 24°C (room temperature) in slow growing stock tube cultures. Stock tubes then provide a constant source of cells from which to establish larger working cultures. Neff medium is especially useful for stock tubes since cell cultures generally maintain longer in Neff than in richer PP media. Serial transfer of a small cell inoculum into fresh stock tubes every 2 to 4 weeks is an efficient, economical way to maintain cultures for routine short term use (up to 6 months). However, stock tubes are not suitable for longer term maintenance of cell lines in which stable genetic characteristics are required. Serial transfer of viable cells in liquid culture should only be used for long term maintenance (> 6 months) if the integrity of the germ line is not essential for downstream use. Genetically important strains should be stored frozen in liquid nitrogen (Section VI.B), and new stock tubes established from frozen cultures about every 6 months to insure genetic stability within working clones. If freezing in liquid nitrogen is not an option, fresh clones of many strains can be obtained from the Tetrahymena Stock Center at a relatively inexpensive cost.
Methods of maintaining stock tubes vary among labs (e.g. Orias et al., 2000 provides an alternate approach), but the following method, used by the Tetrahymena Stock Center, works well for general maintenance. Ten ml of Neff medium, with no antibiotics, are pipetted into 18 mm by 150 mm culture tubes, loosely capped, and autoclaved. Tubes are stored out of direct light and used as needed. A sterile 9 inch Pasteur pipette (Fisher Scientific #136786B) is used to inoculate fresh stock tubes with a few drops (~ 100 – 300 μl) of cells taken from the top region of the original stock tube. It is preferable not to disturb the bottom of a stock tube when removing samples since the upper two thirds of a well grown stock tube culture contains healthy, very slowly dividing cells, but over time the bottom third becomes densely littered with dead and dying cells. Long Pasteur pipettes are ideal for transfer between stock tubes since the extra length prevents any inadvertent contact of unsterile surfaces with the inside of the stock tube, which can be a problem if 5 inch Pasteur pipettes or micropipettors and tips are used. It is good practice to keep at least 2 sequential transfer tubes of every clone to insure against contamination or accidental loss.
When a larger volume of cells (working culture) is needed, a 1:10 dilution of cells taken from the top third of a healthy stock tube culture and innoculated into fresh PP medium (1 ml inoculum to 9 ml PP medium in a culture plate or 100 ml flask) will result in a well grown mid-log phase culture following overnight (16 – 20 hr) incubation at 30°C. If larger working cultures are desired, to insure healthy, rapid growth in the larger volume, it is good practice to first establish a fresh 10 ml culture from a stock tube, and use that log phase culture to seed the larger volume culture.
2. Growth in liquid culture
Tetrahymena will thrive under conditions as diverse as standing cultures, shaken cultures, rotated bottles or tubes, industrial fermenters, microtiter plates, or hanging drops. In the lab, working cultures of Tetrahymena are generally established from slowly growing cultures maintained in stock tubes at room temperature (Section V.B.1). When grown in stationary containers, maximal growth rates are obtained if the depth of the working culture is limited. As a general rule, standing cultures in flasks should be limited to about 1/10 the flask volume, e.g. 100 ml in a one liter flask. Somewhat larger total volumes are possible in Fernbach-style culture flasks designed for culturing organisms requiring a large surface area to volume ratio (e.g. PYREX® 2800mL Fernbach-Style Culture Flask #4420). In smaller containers like Petri plates, depth should be limited to ~ 5 mm. If volume requirements make these limitations impractical, then cultures should be shaken or rotated. Cells in PP media can be shaken up to 200 rpm, although generally shaking speeds of ~ 100 – 150 rpm provide sufficient aeration to support optimal growth without foaming or bubble formation. Baffled Fernbach flasks (e.g. PYREX® 2800mL Fernbach-Style Culture Flask with Baffles #4423) can be used to provide maximal oxygen transfer to shaken cultures. The temperature range for sustained growth in liquid media varies considerably among Tetrahymena species. T. thermophila exhibits sustained growth at 40.7°C, higher than many other Tetrahymena species and shows very slow but consistent growth at 15°C (NYBERG, 1981). Optimal doubling time for T. thermophila occurs at ~ 35°C, with a generation time of about 2 hours (Frankel and Nelsen, 2001; Orias et al., 2000), but for routine laboratory use, where factors in addition to doubling time are often important, cultures are most frequently grown between 27°C and 32°C.
Successful large scale fermentation strategies using low cost nutrient media have been developed using T. thermophila (Hellenbroich et al., 1999; Kiy and Tiedtke, 1992; Noseda et al., 2007). However, conditions for growing Tetrahymena in fermenters or bioreactors can vary considerably depending on instrument type and experimental design, and optimal settings need to be worked out specifically for each large scale set-up (De Coninck et al., 2004).
3. Growth in micro volumes
A number of procedures frequently used in genetic analysis and the establishment of clonal lines require isolation of single cells into hanging drops and/or the growth of clonal cultures in 96 well microtiter plates. Clonal lines can be initiated by physically separating single cells into individual hanging drops, letting the cells replicate to high concentration within the drop, and replicating into microtiter plates. Individual mating pairs and exconjugants arising from individual mating pairs can also be cloned using the hanging drop system (Bruns and Cassidy-Hanley, 2000b). When isolating mating pairs or ex-conjugants, it is important to note that mating type in T. thermophila is not inherited across the sexual stage of the life cycle, and that progeny from a single mating pair frequently give rise to clones expressing different mating types. The segregation of different mating types among the descendants of a given pair appears to be correlated with the segregation of new macronuclei to the four karyonides produced by the first post-zygotic division of the two exconjugant cells (Orias, 1981). The mating type of a T. thermophila clone is not uniquely expressed until sexual maturity, ~ 50 – 80 fissions after conjugation. If the creation of a pure clone expressing a single mating type is needed, individual cells must be isolated after exconjugant clones become sexually mature, and tested for expression of a single mating type using a panel of known mating type testers (available from the Tetrahymena Stock Center). Immature cells cannot mate, adolescent cells can mate, but are not exclusive, i.e., they fail to form pairs with more than one mating type tester strain. Sexually mature clones express a unique mating type, forming pairs with all but one mating type tester strain. All of the necessary sub cloning, growth, and mating can be carried out in hanging drops and microtiter plates, as long as care is taken to prevent contamination. Mature clones expressing a single mating type are stable, and generally will not give rise to cells of different mating types. Clonal lines established in microtiter plates can easily be expanded into stock tubes that can be used to provide working cultures of any volume, and cells for freezing in liquid nitrogen.
Drop plate culture
Drop plates can be created in 100 mm × 15 mm standard Petri plates in several ways. Regardless of the method used, the drop array should match the wells of half of a 96 well microtiter plate. i.e. a 6 × 8 grid array spaced to correspond to microtiter plate wells (Fig. 1). The use of a “drop maker” device is the simplest method, especially if the creation of drop plates is a frequent, routine procedure in the lab, but requires the initial construction of drop maker (Bruns and Cassidy-Hanley, 2000b). This device (Fig. 2) consists of 48 aluminum prongs, each 6-mm in diameter, separated by 9-mm center to center (matching the arrangement of wells in a half of a 96 well microtiter plate), and can be easily constructed by a local machine or metal working shop. To make the drops, the drop maker is sterilized by dipping in distilled water to remove any residual material clinging to the prongs, blotting the prongs on several layers of paper towels to completely remove the water, and dipping the prongs into acetone (or 95% alcohol) placed in a large (15 × 150 mm) covered glass petri dish, preferably placed on a separate metal cart outside the hood and away from any possible contact with the Bunsen burner. The acetone should be deeper then the depth of the medium that will be used to form the drops to insure sterility. The glass lid should be replaced on the Petri plate containing the acetone as soon as the drop maker is removed to protect the contents from accidental contact with flames or hot liquid. The acetone covered prongs are briefly flamed in a Bunsen burner to remove all residual acetone (even a slight residue is potentially lethal to Tetrahymena cells), and allowed to cool. When completely cool, the drop maker is dipped into 25–30 ml of sterile PP media containing penicillin and streptomycin (250 μg/ml each) and 0.25 μg/ml Amphotericin B in a 15 × 100 mm Petri plate, rapidly lifted straight up out of the medium and immediately touched down on the inside of a fresh sterile Petri plate, where each prong deposits a 40 – 50 μl drop of culture medium. As long as standard precautions are taken and work is done in a sterile hood, multiple drop plates can be made from the same medium without resterilizing the drop maker. When finished, the drop maker should be rinsed in distilled water to prevent the build up of medium on the surface, and dried on a paper towel. Drops made using this protocol are quite flat, optically clear, and very stable, adhering more firmly to the plate surface than drops made using other methods. If a drop maker is not available, or if only occasional drop plates are needed, drops can be placed on a Petri plate in the appropriate array by hand using a Pasteur pipette or a single or multichannel pipettor. Drop plates should be stored in plastic boxes (TriState Plastics, #079C or #195C) on a raised platform above a small amount of water to decrease the rate of evaporation. Clean, empty microtiter plates work well for supporting the drop plates above the water level.
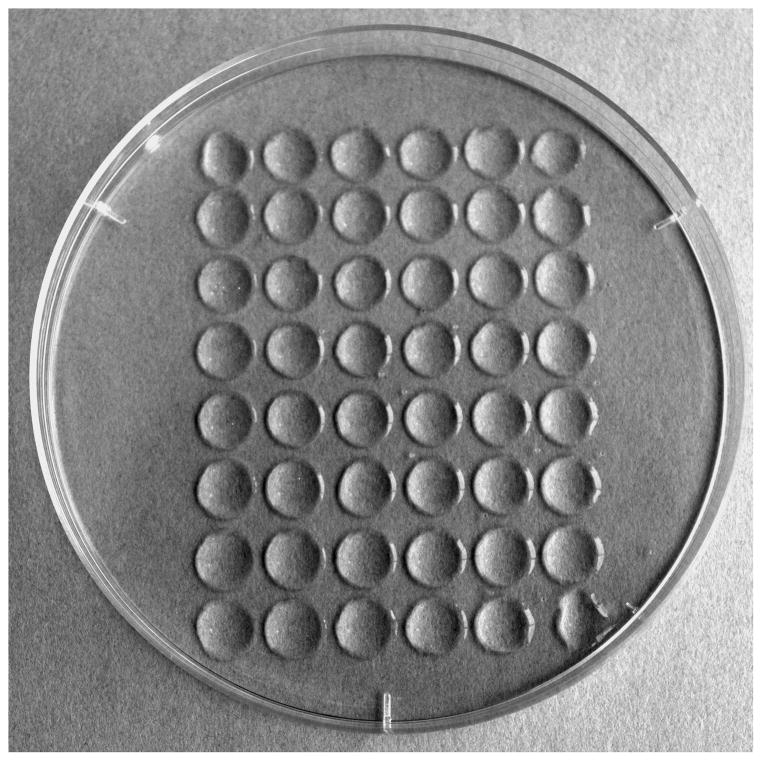
Figure 1. Drop plate array.
The drop array shown is created in a standard 100 mm × 15 mm Petri plate using a 6 × 8 grid drop maker (Fig. 2). The 6 × 8 drop array matches half of a 96 well microtiter plate. Each drop is about 50μl.
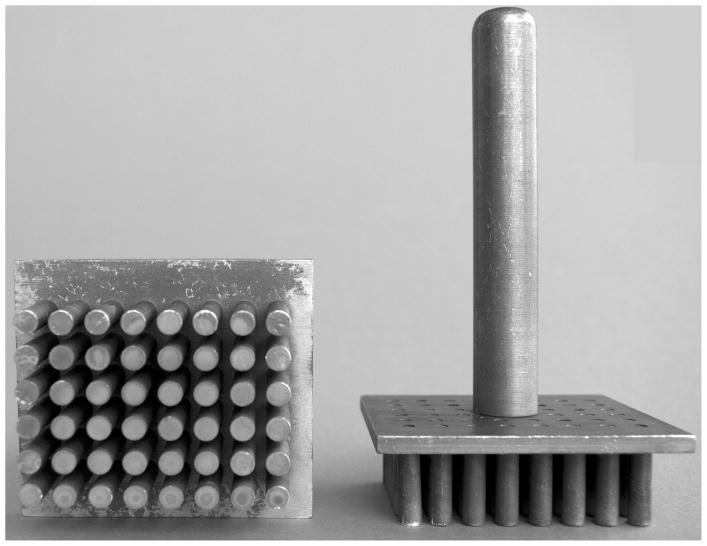
Figure 2. Drop maker.
The drop maker shown consists of 48 aluminum prongs, each 6-mm in diameter, separated by 9-mm center to center, matching the arrangement of wells in one half of a 96 well microtiter plate. The device can be easily constructed by a local machine or metal working shop.
A drawn-out glass micropipette is used to transfer single cells into individual drops while viewing the transfer under a dissecting microscope. Micropipettes of appropriate size can be made by hand by drawing the end of a Pasteur pipette or thin glass tubing through the flame of a Bunsen burner, or by using a micropipette puller. Suction to control cell deposition can be controlled manually using a pipette bulb, or by mouth using tubing to connect a mouthpiece to the pipette (Orias and Bruns, 1976). A much more easily controlled pipette system utilizes braking pipettes (Bruns and Cassidy-Hanley, 2000b), in which thin pieces of capillary tubing (Kimble Chase Capillary tube, # 34500-99, 1.5 –1.8 mm × 100mm) are drawn out at both ends to an inner diameter of ~ 0.1 – 0.15 mm (Fig. 3a). Both ends are clipped with fine forceps under a dissecting microscope to make sure they are open, and one end is carefully inserted about half way into an aspirator tube assembly (Sigma #A5177)(Fig. 3b). The attached mouthpiece is used to control flow, which is fairly easy since pulling out both ends of the capillary tube gives a pipette with neutral action with the back constriction acting as a brake. Once the delivery end is filled with liquid by capillary action, the rest of the tube does not fill, providing much more exact control than that provided by pipettes with a single drawn end. In all cases, the pipette tip is sterilized by repeatedly drawing in and expelling boiling distilled water maintained on a hot plate adjacent to the microscope, and allowed to cool before use. A few microliters of the cell culture from which cells are to be isolated are placed in a marked corner drop, and under the microscope 50 or so cells are picked up in the pipette tip. Individual cells are deposited into separate drops by gently moving the drop plate under the microscope to position each drop sequentially in the field of view, carefully monitoring deposition to insure that only one cell is released into each drop. When isolating pairs, the initial cell inoculum placed in the source drop should result in a dilute cell concentration to make it easier to pick out mating pairs and avoid single cells. With a little practice, it is relatively easy to isolate cells into 10 or more drop plates in an hour. Once cells have been transferred, the drop plates should be placed in a humid chamber (covered plastic box with distilled water on the bottom and a raised platform to hold the plates above the water) to prevent drying out, and placed at 30°C in a stationary incubator for 2 to 3 days, or until the cells have grown enough to allow replication to microtiter plates.
Figure 3.

a. Braking pipette To make the braking pipette shown, thin pieces of capillary tubing (Kimble Chase Capillary tube, # 34500-99, 1.5–1.8 mm × 100mm) are drawn out at both ends to an inner diameter of ~ 0.1 – 0.15 mm. Both ends are clipped with fine forceps under a dissecting microscope to make sure they are open.
b. Braking pipette in aspirator tube assembly. To assemble the aspirator, one end of a braking pipette is carefully inserted about half way into the aspirator tube assembly (Sigma #A5177) as shown.
Microtiter plate culture
Tetrahymena is well adapted to rapid growth in very small volumes, e.g. 100 μl cultures in 96 well microtiter plates. Clear U-bottom microtiter plates with 100 μl PP medium per well are used for most routine procedures, although for fixation and cytology, flat bottom plates give better optics. Two types of replicators are useful for manipulating Tetrahymena grown in microtiter plate cultures: a 48-prong replicator, used to transfer cultures from drop plates to microtiter plates, and a 96–prong replicator for transfer from microtiter plate to microtiter plate. Replicators can be made in a variety of ways, using either a wooden block with straight metal rods ~ 1/16 in diameter arrayed to match either a half (48 prong) or full (96 prong) microtiter well array (Orias et al., 2000) or an aluminum assembly similar to the drop maker described above, except that the diameter of each prong is 4 mm, to easily fit into microtiter plate wells (Bruns and Cassidy-Hanley, 2000b). When using the larger pronged aluminum replicator, care must be taken to insure that drops do not run together when touched by the prongs. This is best done by first dipping the sterile replicator into fresh, sterile growth medium, so that each prong tip is covered by a drop of medium, and then touching the ends of the prongs to the drops on the drop plate. This insures that each prong transfers a reasonable cell sample and decreases the risk of drop to drop contact during the transfer process. A 96-prong replicator (Fig. 4) is available commercially (Nunc replicator #250520, available from Fisher Scientific). For occasional use with small numbers of samples, a pipettor (multichannel or single channel) can be used for both transfer from drop plates and replication between microtiter plates, but for large numbers of samples the process is labor and cost intensive. Replicators are sterilized using the same technique described for drop makers, paying particular attention to rinsing the replicators in distilled water between uses to prevent the build-up of peptone residue on the prongs. When large numbers of transfers must be done, it is useful to have several replicators of each size that can be used in an alternating fashion. One replicator is rinsed, sterilized, flamed, and then allowed to cool while a second replicator is used, rinsed, sterilized and flamed. This process insures that repeated flaming will not cause the replicator to become too hot.
Figure 4. 96-prong replicator.
The 96 prongs of the replicator shown match the well distribution on a 96 well microtiter plate. The replicator can be used to transfer small volumes (~5 μl) of cells to new microtiter plates for screening purposes, to set up small volume plate matings, and for serial plate transfer.
It is possible to re-use microtiter plates, as long as they are not allowed to dry out before washing. Dried-on cells and medium can be difficult to remove, and may complicate subsequent analyses. Plates should be decontaminated by submerging in 25% Clorox (1.3% sodium hypochlorite) for 5 minutes, followed by washing in a dishwashing machine or by hand. After rinsing to insure removal of all traces of bleach, plates can also be sonicated for 10 minutes to insure removal of all organic material from the wells. In all cases, after washing, the plates must be carefully rinsed by hand in high purity distilled water. All wells must be completely filled and emptied by vigorous shaking with the plate inverted over a sink at least 3 times. Covers should be washed and rinsed in a similar manner. Plates are dried inverted standing on edge at an angle to optimize draining. Following air drying, the open plates and lids can be sterilized by exposure to 20 minutes of UV irradiation at a distance of 15 cm from a bank of six 40-watt sunlamps (Bruns and Cassidy-Hanley, 2000b), or by a 1 hour exposure to a germicidal UV lamp 6490 (Orias et al., 2000). In both cases UV lamps are enclosed in a light tight container to protect eyes and skin form UV exposure. Since UV lamp output can decrease with use, UV lamps should occasionally be monitored using a UV dosimeter. Following irradiation, lids are placed on the plates, and closed plates are stored until needed.
Cells grown in microtiter plates at 30°C can generally be replicated after 1 to 3 days. For longer term maintenance (up to a week) plates should be kept at room temperature out of direct light. If maximum growth rate is desired, e.g. when attempting to quickly mature progeny clones, plates can be replicated daily, but long term continuous daily replication can sometimes cause clonal loss (Section V.A). For optimal clonal health, it is advisable to allow the clones to periodically reach stationary stage, most simply by replicating daily Monday through Friday but letting the clones rest each weekend.
4. Growth on solid medium
Although Tetrahymena is generally grown in liquid culture, in certain circumstances (e.g.. the isolation of mutant clones following mutagenesis or cytotoxicity assays) it is useful to grow cells on solid medium. Several methods have been developed for obtaining clones on solid medium. Gardonio et al., (1973) developed the following agar plate method using 1.5% bottom agar and a thin layer of 0.3% top agar, both made up in either 2% PP and 0.1% liver extract, or a defined medium supplemented with 0.04% PP, and containing 250 μg each of penicillin and streptomycin sulfate. Plates are dried for 2 days at 37°C or a week at room temperature before adding cells. To inoculate the plate, about 0.5 ml of a very dilute Tetrahymena culture (~100–150 cells) is placed on the dried agar surface. After the liquid is partially absorbed by the agar, sterile G-25 fine Sephadex is sprinkled onto the plate. Individual clones, established around individual Sephadex beads, can then be isolated into liquid culture. Dobra and Ehret (1980) developed an alternate technique for culturing Tetrahymena as continuous monolayers on the surface of nutrient agar. Cells grown at low densities on solid agar are motile and have normal structural characteristics, including production of food vacuoles and an oral apparatus. At higher densities (2 – 5 × 105/cm2), cells are relatively non-motile and form a tightly packed continuous monolayer. A modification of this method has also been used to cultivate Tetrahymena on the surface of sterile cellulose nitrate filters placed on top of PP agar.
C. Culture Contamination: prevention and treatment
Basic sterile techniques should be employed whenever possible when working with all Tetrahymena cultures. All glass and plasticware should be sterile, and media should be monitored for possible signs of contamination like clouding prior to each use. Whenever feasible cell work should be carried out in a sterile hood. However, some techniques like isolation of cells into hanging drops are more easily done under a microscope on an open lab bench. In these cases contamination can be minimized by judicious use of antibiotics and fungizone. Additionally, anything coming into direct contact with the cells or media (pipettes, drop makers, etc) must be carefully sterilized, and exposure time of open plates minimized. With these basic precautions, contamination of drops or microtiter wells is rarely a problem.
If contamination occurs in working cultures or stock tubes, the simplest response is to discard the contaminated cells and re-establish the culture from a back-up stock tube or a frozen stock. Contaminated cultures should immediately be autoclaved and discarded to prevent the spread of potential contaminants within the lab area. Since most contaminants grow more quickly than Tetrahymena, contaminated cultures are generally easy to spot fairly early on by the macroscopic appearance of the culture. Bacterial contaminants make flask cultures look cloudier and denser than pure Tetrahymena cultures, and generally release an off odor when the flask is opened. Fungal and mold contaminants generally produce obvious mats of growth. Contamination in individual wells of a microtiter plate presents a special problem since contamination can quickly spread within a plate. At the first sign of contamination, the contents of the affected well should be removed by aspiration with a sterile pipette tip, and the well cleaned with a cotton swab soaked in 95% ethanol. Any remaining alcohol must be aspirated from the well, and the plate left uncovered in a sterile hood until the alcohol is completely evaporated. If the plate is covered before the alcohol evaporates completely, alcohol vapors will kill the entire contents of the plate. If multiple wells are affected, the plate should be decontaminated and discarded. If a contaminated culture is irreplaceable, possible methods for generating healthy, clean cultures are discussed below.
1. Antibiotics and Fungizones
Tetrahymena cultures grown in the laboratory using basic sterile techniques generally do not require the addition of antibiotics or fungizones to maintain sterility. However, when cells must be knowingly exposed to potentially contaminating conditions, penicillin G and streptomycin sulfate should be routinely added to the PP media at a final concentration of 250 μg/ml each. A 1000x stock solution containing both antibiotics can be prepared by filter sterilization and stored in 1 ml aliquots in cryovials at −20°C. One microliter of the drug cocktail is added aseptically per ml culture media immediately prior to use. The fungizone Amphotericin B can also be added to minimize fungal contamination, which is often a major problem in Tetrahymena cultures exposed to unsterile conditions. Tetrahymena is quite resistant to Amphotericin B and concentrations from 0.025 to 25 μg/ml have been used without problems. For general preventative use, 0.25 μg/ml is usually sufficient.
If preventative measures fail, and cultures become contaminated, if possible they should be autoclaved and discarded, and new cultures established from the freezer or a non-contaminated stock tube (Section V.B.1). Penicillin/streptomycin treatment is generally not effective in cleaning up already contaminated cultures. If a contaminated culture must be rescued, 100 μg/ml neomycin, kanamycin or tetracycline can be used to try to eliminate bacterial contamination. These drugs may also be useful in cleaning up newly collected isolates, since most Tetrahymena collected from the wild grow well even at these elevated levels (Clifford Brunk, personal communication; Ch. 9 in this volume)). However, the overall most effective, albeit expensive, drug treatment for retrieving contaminated cultures is Normocin (Invivogen, Cat. # ant-nr-1), which can be used at 2 μl/ml culture media directly as shipped. Normocin can also be used in conjunction with penicillin and streptomycin with no adverse effects on Tetrahymena, and has proved remarkably effective in eliminating both bacterial and fungal contamination. In extreme cases, the Normocin concentration can be increased 2 to 3 fold without affecting the Tetrahymena. To prevent the development of Normocin resistance, routine use of Normocin in standard cultures is not recommended.
2. Serial subculture
If all else fails, as may be the case with some fungal or antibiotic resistant bacterial contaminants, serial dilution (Lwoff, 1923) can be used to create contamination free clones. Individual cells are placed in the top 8 drops of a 6 × 8 drop array in several drop plates. Every 30 minutes a single cell is moved with a minimum of media into the drop directly below, for a total of 5 transfers. Since some bacteria can survive in Tetrahymena food vacuoles (Berk et al., 2008; Gourabathini et al., 2008; Meltz et al., 2007; Rehfuss et al., 2011), the timing between transfers must be long enough to insure that original food vacuole contents are fully excreted prior to the final transfer so as to avoid contaminating the final drop. Drops are incubated at 30°C overnight, and drops containing growing Tetrahymena and visually free of contamination are transferred to individual stock tubes for further incubation at 30°C. After 2 days, stock tubes with no sign of contamination can be further tested by spotting a small sample of the tube contents on a PP nutrient agar plate and incubating for 2 days. Since Tetrahymena can form small colony-like plaques on the agar surface of agar plates, the agar plate should be examined under a dissecting scope to differentiate bacterial colonies, which are generally rounded, dense, and opaque, from Tetrahymena plaques, which are more translucent, flat, and irregular in shape.
VI. Long term storage
A. Serial transfer
Tetrahymena can be maintained for years by serial transfer, providing a reasonable cell inoculum (a minimum of ~ 1000 cells) is used for each transfer. However, prolonged vegetative growth can lead to both micronuclear and macronuclear genetic changes over time. In micronucleate strains like T. thermophila, the transcriptionally inactive germinal micronucleus is not subject to direct selection and can accumulate chromosomal changes, including whole chromosome loss, deletions and lethal point mutations, that have no effect on vegetative viability but eventually result in the inability to produce true sexual progeny (clonal sterility; Allen et al., 1984; Nanney, 1974; Simon and Nanney, 1979). It is important to note that sterile cells generally maintain the ability to form physical pairs that may give rise to viable exconjugants. However exconjugants arising from matings between normal and sterile cells are not true progeny in that both exconjugant cells retain their parental macronuclei and parental phenotypes, and contain micronuclei of unknown genotype. Loss of fertility within a clone occurs gradually, and timing will vary with different strains and maintenance conditions.
Changes during prolonged vegetative growth can also effect macronuclear composition. Strains in which the macronucleus is not homozygous for alleles of interest will undergo random macronuclear phenotypic assortment during vegetative growth, eventually leading to the production of cells that are homozygous for a given allele (Bruns and Brussard, 1974b). In terms of strain utility, the importance of these changes depends on the type of strain and the intended use. Wild type strains not intended for genetic use can be maintained by serial transfer, as can strains homozygous in the macronucleus for a specific mutation of interest, as long as downstream use does not require fertility (e.g. for expression of a specific gene product in vegetative cells). However, the macronuclear composition of strains with macronuclei that are heterozygous for a particular gene of interest cannot be guaranteed to remain stable following prolonged serial vegetative transfer. To insure continued fertility and optimize genetic stability, important clones should be stored frozen in a liquid nitrogen freezer as soon as possible after construction. It should be noted that maintenance of a mixed genotype in the macronucleus cannot be absolutely guaranteed even if the clone is stored frozen in a liquid nitrogen freezer. During thawing, only a relatively small population of cells is recovered, and there is a chance that the subpopulation that survives thawing may not be representative of the pre-freezing population, especially if assortment gives rise to a subclone better able to withstand the freezing process, as may be the case for some wild type clones assorting from a deleterious mutation in a heterozygous macronucleus. It is therefore good practice to check the relevant phenotype of clones heterozygous in the macronucleus after thawing, if retention of a mixed genotype expressing a specific phenotype or gene product is essential.
B. Storage in liquid nitrogen
There are several protocols available for freezing Tetrahymena (Flacks, 1979; Orias et al., 2000; Simon, 1982). A number of factors shown to affect successful freezing of eukaryotic cells, including the physiological state of the cells (Rauen et al., 1994), cryoprotectant (Anchordoguy et al., 1987), rate of cooling (Farrant and Morris, 1973), storage temperature and rate and temperature of thawing (McGann and Farrant, 1976a; McGann and Farrant, 1976b) have been optimized for Tetrahymena (Cassidy-Hanley et al., 1995). The method described below is based on these optimized parameters (Bruns et al., 2000), and is used routinely by the Tetrahymena Stock Center. This technique has proven successful with multiple T. thermophila strains as well as a wide variety of other Tetrahymena species. All cell work is carried out in a sterile hood, and all equipment and materials used in the procedure are sterilized
1. Growth of cultures for freezing
Cells designated for freezing should be grown at 30°C to log phase (100mls @ ~ 5 × 105/ml) on a shaking incubator. It is important to start with a fresh, healthy, uncontaminated culture to insure consistently high rates of recovery. For optimal aeration and culture health, it is recommended that the flask culture conditions recommended above (Section V.B.2) be used.
2. Starvation
To insure efficient freezing, the cells must be starved prior to freezing. Unstarved cells give very poor recovery following freezing. Starve 100 mls of cells at ~ 1.5 – 2 × 105 per ml in sterile 10 mM Tris pH 7.5 for 2–3 days at 30°C in a flask with a high surface area to volume ratio for adequate aeration. Cell concentration and starvation temperature are important for optimal recovery of live cells.
3. Freezing
Concentrate 100 ml of starved cells by centrifugation (2 minutes at ~1100 g at room temperature) followed by aspiration of the supernate to 1 ml total volume (cells plus Tris). Immediately add 4 mls of high-quality 10% DMSO (Fisher #D1281) and gently resuspend cells into a total volume of 5 mls (final DMSO concentration 8%). Dispense 0.3 ml of the cell:DMSO mixture to individual cryovials (e.g. Nalgene® #5000-0020 or #5000-0012). The number of tubes frozen is a matter of individual preference, but in all cases one additional tube should be frozen for a test thaw. Incubate the cryovials at room temperature ~ 30 minutes, transfer the cryovials to a Nalgene® Cryo 1°C controlled rate freezing container (Nalgene #5100-0001) and place the container in a −80°C freezer overnight. Cryovials should not be left in the low temperature freezer for extended periods and care should be taken that the temperature in the freezer does not fluctuate or rise above −70°C since either occurrence can decrease recovery of viable cells. Cells maintained long term in a −80°C freezer often exhibit increasingly poor recovery over time, perhaps as a result of temperature changes when the freezer is opened and closed. Following initial freezing in a −80°C freezer, cells should be transferred to a liquid nitrogen freezer, using a nitrogen dewar or dry ice to prevent the cells from warming up during the transfer process. Do not use wet ice for transfer. For safety reasons, it is recommended that cryovials be frozen in the vapor phase of the liquid nitrogen tank, to reduce the risk of cryotube explosion during thawing. If tubes must be frozen in the liquid phase, it is recommended that tubes to be thawed be moved into the vapor phase of the freezer for at least 24 hours before thawing. One possibility is to leave an empty box in the top slot of one of the freezer racks, above the liquid level. Move the tubes to be thawed to that box at least 24 hours before thawing, being very careful to not let them warm up.
4. Thawing
Thaw tubes individually in a 42°C water bath, going directly from liquid nitrogen or dry ice into the waterbath. After ~15 seconds, using a glass transfer (Pasteur) pipette, add ~1 ml of NEFF prewarmed to 42°C. Gently move the tube in the water bath to speed thawing. When the pellet is dissolved, pipette the contents of the cryovial into a Petri plate containing 10 ml of NEFF plus penicillin and streptomycin (250 μg/ml each) and 0.25 μg/ml Amphotericin B, prewarmed to 30°C. Keep the pipette tip under the liquid to avoid bubbles, and swirl the plate contents gently. Culture the cells at 30°C. Live cells can often be observed within 30–60 minutes, and should be visible within 24 hours. Once the culture is established, transfer to a stock tube or use as needed.
Acknowledgments
DCH is supported by NIH NCRR SEPA grant 5 R25 RR025126-03 and by NIH grant # P40 RR019688-01A1. The Tetrahymena Stock Center is funded by NIH grant # P40 RR019688-01A1. Past support by NIH, NSF, and USDA is gratefully acknowledged for providing funding for the development of many of these techniques. DCH is a founding member of Tetragenetics, Inc., Ithaca, NY, a company developing and utilizing Tetrahymena as a platform for the manufacture of biotechnological products. Neither this connection, nor any financial, personal or professional interest of the author, has influenced this paper.
References
- Allen SL. Cytogenetics of genomic exclusion in tetrahymena. Genetics. 1967a;55(4):797–822. doi: 10.1093/genetics/55.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SL. Genomic exclusion: A rapid means for inducing homozygous diploid lines in tetrahymena pyriformis, syngen 1. Science. 1967b;155(762):575–57. doi: 10.1126/science.155.3762.575. [DOI] [PubMed] [Google Scholar]
- Allen SL. Genetic nomenclature rules for tetrahymena thermophila. Methods Cell Biol. 2000;62:561–563. [PubMed] [Google Scholar]
- Allen SL, Ervin PR, McLaren NC, Brand RE. The 5S ribosomal RNA gene clusters in tetrahymena thermophila: Strain differences, chromosomal localization, and loss during micronuclear ageing. Mol Gen Genet. 1984;197(2):244–253. doi: 10.1007/BF00330970. [DOI] [PubMed] [Google Scholar]
- Allen SL, Altschuler MI, Bruns PJ, Cohen J, Doerder FP, Gaertig J, et al. Proposed genetic nomenclature rules for tetrahymena thermophila, paramecium primaurelia and paramecium tetraurelia. the seventh international meeting on ciliate molecular biology genetics nomenclature. Genetics. 1998;149(1):459–462. doi: 10.1093/genetics/149.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SL, Gibson I. Genetics of tetrahymena. In: Elliott AM, editor. Biology of tetrahymena. Stroudsburg, Pa: Dowden, Hutchinson and Ross; 1973. pp. 307–373. [Google Scholar]
- Altschuler MI, Bruns PJ. Chromosome-designated mutation selection in tetrahymena thermophila. Genetics. 1984;106(3):387–401. doi: 10.1093/genetics/106.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchordoguy TJ, Rudolph AS, Carpenter JF, Crowe JH. Modes of interaction of cryoprotectants with membrane phospholipids during freezing. Cryobiology. 1987;24(4):324–331. doi: 10.1016/0011-2240(87)90036-8. [DOI] [PubMed] [Google Scholar]
- Artemenko AG, Muratov EN, Kuz’min VE, Muratov NN, Varlamova EV, Kuz’mina AV, et al. QSAR analysis of the toxicity of nitroaromatics in tetrahymena pyriformis: Structural factors and possible modes of action. SAR and QSAR in environmental research. 2011;22(5–6):575–601. doi: 10.1080/1062936X.2011.569950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk SG, Faulkner G, Garduno E, Joy MC, Ortiz-Jimenez MA, Garduno RA. Packaging of live legionella pneumophila into pellets expelled by tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Applied and Environmental Microbiology. 2008;74(7):2187–2199. doi: 10.1128/AEM.01214-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Lynch TJ, Zeilinger D, Orias E. Identification, mapping and linkage analysis of randomly amplified DNA polymorphisms in tetrahymena thermophila. Genetics. 1996;143(2):811–821. doi: 10.1093/genetics/143.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB. Pair formation in tetrahymena pyriformis, an inducible developmental system. J Exp Zool. 1974a;188(3):337–44. doi: 10.1002/jez.1401880309. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB. Positive selection for mating with functional heterokaryons in tetrahymena pyriformis. Genetics. 1974b;78(3):831–41. doi: 10.1093/genetics/78.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB. Nullisomic tetrahymena: Eliminating germinal chromosomes. Science. 1981;213:549–551. doi: 10.1126/science.213.4507.549. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB, Kavka AB. Isolation of homozygous mutants after induced self-fertilization in tetrahymena. Proc Natl Acad Sci U S A. 1976;73(9):3243–327. doi: 10.1073/pnas.73.9.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods in cell biology. 2000a;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D. Methods for genetic analysis. Methods in cell biology. 2000b;62:229–240. doi: 10.1016/s0091-679x(08)61533-2. [DOI] [PubMed] [Google Scholar]
- Bruns PJ, Katzen AL, Martin L, Blackburn EH. A drug-resistant mutation in the ribosomal DNA of tetrahymena. Proc Natl Acad Sci U S A. 1985;82(9):2844–286. doi: 10.1073/pnas.82.9.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Smith HR, Cassidy-Hanley D. Long-term storage. Methods in cell biology. 2000;62:213–218. [PubMed] [Google Scholar]
- Bruns PJ, Brussard TB, Merriam EV. Nullisomic tetrahymena. II. a set of nullisomics define the germinal chromosomes. Genetics. 1983;104(2):257–270. doi: 10.1093/genetics/104.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne BC. Induced resistance to 6-methylpurine and cycloheximide in tetrahymena. II. variation within vegetative cultures of micronucleate T. thermophila and amicronucleate T. pyriformis. C R Seances Soc Biol Fil. 1978;172(2):314–319. [PubMed] [Google Scholar]
- Byrne BC, Brussard TB, Bruns PJ. Induced resistance to 6-methylpurine and cycloheximide in tetrahymena. I. germ line mutants of T. thermophila. Genetics. 1978;89(4):695–702. doi: 10.1093/genetics/89.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, et al. Germline and somatic transformation of mating tetrahymena thermophila by particle bombardment. Genetics. 1997;146(1):135–47. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Smith HR, Bruns PJ. A simple, efficient technique for freezing tetrahymena thermophila. J Eukaryot Microbiol. 1995;42(5):510–55. doi: 10.1111/j.1550-7408.1995.tb05897.x. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Ward JG, Randolph C, Yao MC. Microinjection of tetrahymena thermophila. Methods Cell Biol. 2000;62:469–484. doi: 10.1016/s0091-679x(08)61551-4. [DOI] [PubMed] [Google Scholar]
- Chantangsi C, Lynn DH, Brandl MT, Cole JC, Hetrick N, Ikonomi P. Barcoding ciliates: A comprehensive study of 75 isolates of the genus tetrahymena. International Journal of Systematic and Evolutionary Microbiology. 2007;57:2412–2425. doi: 10.1099/ijs.0.64865-0. [DOI] [PubMed] [Google Scholar]
- Chantangsi C, Lynn DH. Phylogenetic relationships within the genus tetrahymena inferred from the cytochrome c oxidase subunit 1 and the small subunit ribosomal RNA genes. Molecular phylogenetics and evolution. 2008;49(3):979–987. doi: 10.1016/j.ympev.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Sorensen H, Beyer NH, Kristiansen K, Rasmussen L, Rasmussen MI. Cell death in tetrahymena thermophila: New observations on culture conditions. Cell Biol Int. 2001;25(6):509–519. doi: 10.1006/cbir.2000.0689. [DOI] [PubMed] [Google Scholar]
- Christensen ST, Wheatley DN, Rasmussen MI, Rasmussen L. Mechanisms controlling death, survival and proliferation in a model unicellular eukaryote tetrahymena thermophila. Cell Death Differ. 1995;2(4):301–308. [PubMed] [Google Scholar]
- Christensen ST, Rasmussen L. Evidence for growth factors which control cell multiplication in tetrahymena thermophila. Acta Protozoologica. 1992;31(4):215–219. [Google Scholar]
- Cole ES, Bruns PJ. Uniparental cytogamy: A novel method for bringing micronuclear mutations of tetrahymena into homozygous macronuclear expression with precocious sexual maturity. Genetics. 1992;132(4):1017–131. doi: 10.1093/genetics/132.4.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ES, Cassidy-Hanley D, Hemish J, Tuan J, Bruns PJ. A mutational analysis of conjugation in tetrahymena thermophila. 1. phenotypes affecting early development: Meiosis to nuclear selection. Dev Biol. 1997;189(2):215–32. doi: 10.1006/dbio.1997.8648. [DOI] [PubMed] [Google Scholar]
- Cole ES, Soelter TA. A mutational analysis of conjugation in tetrahymena thermophila. 2. phenotypes affecting middle and late development: Third prezygotic nuclear division, pronuclear exchange, pronuclear fusion, and postzygotic development. Dev Biol. 1997;189(2):233–45. doi: 10.1006/dbio.1997.8649. [DOI] [PubMed] [Google Scholar]
- Cole ES. Conjugal blocks in tetrahymena pattern mutants and their cytoplasmic rescue. I. broadened cortical domains (bcd) Dev Biol. 1991;148(2):403–19. doi: 10.1016/0012-1606(91)90260-a. [DOI] [PubMed] [Google Scholar]
- Cole ES, Frankel J. Conjugal blocks in tetrahymena pattern mutants and their cytoplasmic rescue. II. janus A. Dev Biol. 1991;148(2):420–428. doi: 10.1016/0012-1606(91)90261-z. [DOI] [PubMed] [Google Scholar]
- Dave D, Wloga D, Gaertig J. Manipulating ciliary protein-encoding genes in tetrahymena thermophila. Methods in cell biology. 2009;93:1–20. doi: 10.1016/S0091-679X(08)93001-6. [DOI] [PubMed] [Google Scholar]
- De Coninck J, Leclercq B, Exbrayat JM, Duyme F. Factorial designs: An efficient approach to choosing the main factors influencing growth and hydrolase production by tetrahymena thermophila. J Ind Microbiol Biotechnol. 2004;31(5):204–208. doi: 10.1007/s10295-004-0135-8. [DOI] [PubMed] [Google Scholar]
- Dentler WL. Isolation of cilia from tetrahymena thermophila. Methods Cell Biol. 1995;47:13–15. doi: 10.1016/s0091-679x(08)60784-0. [DOI] [PubMed] [Google Scholar]
- Dobra KW, Ehret CF. Growth kinetics of three species of tetrahymena on solid agar*. Journal of Eukaryotic Microbiology. 1980;27(2):226–230. [Google Scholar]
- Doerder FP. Regulation of macronuclear DNA content in tetrahymena thermophila. Journal of Protozoology. 1979;26:28–35. doi: 10.1007/BF00327377. [DOI] [PubMed] [Google Scholar]
- Doerder FP, Deak JC, Lief JH. Rate of phenotypic assortment in tetrahymena thermophila. Dev Genet. 1992;13(2):126–132. doi: 10.1002/dvg.1020130206. [DOI] [PubMed] [Google Scholar]
- Dryl S. Antigenic transformation in paramecium aurelia after homologous antiserum treatment during autogamy and conjugation. J Protozool. 1959;6 (Supplement):25. [Google Scholar]
- Ducoff HS, Williams SM, Syn TB, Butler BB. Quantitative studies of metabolism in phagotrophic ciliates. I. growth of tetrahymena pyriformis W on viable and on killed bacteria. J Protozool. 1964;11(3):309–313. [Google Scholar]
- Elliot AM, Hayes RE. Tetrahymena from mexico, panama, and colombia, with special reference to sexuality. J Eukaryotic Microbiology The Journal of Eukaryotic Microbiology. 1955;2(2):75–80. [Google Scholar]
- Ethuin P, De Coninck J, Dhulster P, Guillochon D. Comparison of complex organic media for the cultivation of the temperature-sensitive mutant tetrahymena thermophila SJ180. Enzyme and microbial technology. 1995;17(11):998–1002. [Google Scholar]
- Farrant J, Morris GJ. Thermal shock and dilution shock as the causes of freezing injury. Cryobiology. 1973;10(2):134–140. doi: 10.1016/0011-2240(73)90019-9. [DOI] [PubMed] [Google Scholar]
- Flacks M. Axenic storage of small volumes of tetrahymena cultures under liquid nitrogen: A miniaturized procedure. Cryobiology. 1979;16(3):287–291. doi: 10.1016/0011-2240(79)90040-3. [DOI] [PubMed] [Google Scholar]
- Frankel J, Jenkins LM, DeBault LE. Causal relations among cell cycle processes in tetrahymena pyriformis. an analysis employing temperature-sensitive mutants. J Cell Biol. 1976;71(1):242–260. doi: 10.1083/jcb.71.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J, Jenkins LM, Nelsen EM, Stoltzman CA. Hypoangular: A gene potentially involved in specifying positional information in a ciliate, tetrahymena thermophila. Dev Biol. 1993;160(2):333–354. doi: 10.1006/dbio.1993.1311. [DOI] [PubMed] [Google Scholar]
- Frankel J, Mohler J, Frankel AK. Temperature-sensitive periods of mutations affecting cell division in tetrahymena thermophila. J Cell Sci. 1980;43:59–74. doi: 10.1242/jcs.43.1.59. [DOI] [PubMed] [Google Scholar]
- Frankel J, Nelsen EM. The effects of supraoptimal temperatures on population growth and cortical patterning in tetrahymena pyriformis and tetrahymena thermophila: A comparison. J Eukaryot Microbiol. 2001;48(2):135–146. doi: 10.1111/j.1550-7408.2001.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in tetrahymena. Nucleic Acids Res. 1994;22(24):5391–538. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Kapler G. Transient and stable DNA transformation of tetrahymena thermophila by electroporation. Methods Cell Biol. 2000;62:485–500. doi: 10.1016/s0091-679x(08)61552-6. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in tetrahymena thermophila. Proc Natl Acad Sci U S A. 1994;91(10):4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardonio E, Crerar M, Pearlman RE. Growth and cloning of tetrahymena pyriformis on solid meduium. J Bacteriol. 1973;116(3):1170–1176. doi: 10.1128/jb.116.3.1170-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt A, Ud-Daula A, Schramm K. Tetrahymena spp. (protista, ciliophora) as test species in rapid multi-level ecotoxicity tests. Acta Protozool. 2010;49(4):271–280. [Google Scholar]
- Gorovsky MA, Yao MC, Keevert JB, Pleger GL. Isolation of micro- and macronuclei of tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gorovsky MA, Woodard J. Studies on nuclear structure and function in Tetrahymena pyriformis: I. RNA synthesis in macro- and micronuclei. The Journal of cell biology. 1969;42(3):673–682. doi: 10.1083/jcb.42.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourabathini P, Brandl MT, Redding KS, Gunderson JH, Berk SG. Interactions between food-borne pathogens and protozoa isolated from lettuce and spinach. Applied and Environmental Microbiology. 2008;74(8):2518–2525. doi: 10.1128/AEM.02709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinde B, Jonassen TO. Methionine is a regulator of starvation-induced proteolysis in tetrahymena. Exp Cell Res. 1987;173(2):496–503. doi: 10.1016/0014-4827(87)90289-8. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Orias E. Genetic characterization of tetrahymena thermophila mutants unable to secrete capsules. Dev Genet. 1992;13(2):160–166. doi: 10.1002/dvg.1020130210. [DOI] [PubMed] [Google Scholar]
- Haddad A, Turkewitz AP. Analysis of exocytosis mutants indicates close coupling between regulated secretion and transcription activation in tetrahymena. Proc Natl Acad Sci U S A. 1997;94(20):10675–10680. doi: 10.1073/pnas.94.20.10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeister JJ, Grave M, Kristiansen TB, Assaad F, Hellung-Larsen P. Interface-mediated death of unconditioned tetrahymena cells: Effect of the medium composition. J Eukaryot Microbiol. 1999;46(1):6–11. doi: 10.1111/j.1550-7408.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Hai B, Gaertig J, Gorovsky MA. Knockout heterokaryons enable facile mutagenic analysis of essential genes in tetrahymena. Methods Cell Biol. 2000;62:513–531. doi: 10.1016/s0091-679x(08)61554-x. [DOI] [PubMed] [Google Scholar]
- Hallberg RL, Bruns PJ. Ribosome biosynthesis in tetrahymena pyriformis. regulation in response to nutritional changes. J Cell Biol. 1976;71(2):383–94. doi: 10.1083/jcb.71.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Orias E. Genetically mapping new mutants and cloned genes. Methods Cell Biol. 2000;62:265–280. doi: 10.1016/s0091-679x(08)61536-8. [DOI] [PubMed] [Google Scholar]
- Hellenbroich D, Valley U, Ryll T, Wagner R, Tekkanat N, Kessler W, et al. Cultivation of tetrahymena thermophila in a 1.5-m3 airlift bioreactor. Appl Microbiol Biotechnol. 1999;51(4):447–455. doi: 10.1007/s002530051415. [DOI] [PubMed] [Google Scholar]
- Jayaram J, Papoyan A, Zhang X, Bisharyan Y, Colussi P, Cassidy-Hanley D, et al. An alternative platform for rapid production of effective subunit vaccines. BioPharm Int BioPharm International. 2010;23(10 SUPPL):6–13. [Google Scholar]
- Johnson KA. Preparation and properties of dynein from tetrahymena cilia. Methods Enzymol. 1986;134:306–317. doi: 10.1016/0076-6879(86)34098-9. [DOI] [PubMed] [Google Scholar]
- Kaney AR, Speare VJ. An amicronucleate mutant of tetrahymena thermophila. Exp Cell Res. 1983;143(2):461–47. doi: 10.1016/0014-4827(83)90074-5. [DOI] [PubMed] [Google Scholar]
- Kapler GM. Developmentally regulated processing and replication of the tetrahymena rDNA minichromosome. Curr Opin Genet Dev. 1993;3(5):730–735. doi: 10.1016/s0959-437x(05)80091-7. [DOI] [PubMed] [Google Scholar]
- Karrer K, Stein-Gavens S, Allitto BA. Micronucleus-specific DNA sequences in an amicronucleate mutant of tetrahymena. Dev Biol. 1984;105(1):121–129. doi: 10.1016/0012-1606(84)90267-7. [DOI] [PubMed] [Google Scholar]
- Kher CP, Doerder FP, Cooper J, Ikonomi P, Achilles-Day U, Kupper FC, et al. Barcoding tetrahymena: Discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox-1) barcode. Protist. 2011;162(1):2–13. doi: 10.1016/j.protis.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Kidder GW, Dewey VC. The biochemistry of ciliates in pure culture. In: Lwoff A, Hutner SH, editors. Biochemistry and physiology of protozoa. New York: Academic Press; 1951. pp. 323–400. [Google Scholar]
- Kiy T, Tiedtke A. Continuous high-cell-density fermentation of the ciliated protozoon tetrahymena in a perfused bioreactor. Appl Microbiol Biotechnol. 1992;38(2):141–16. doi: 10.1007/BF00174458. [DOI] [PubMed] [Google Scholar]
- Larson DD, Blackburn EH, Yaeger PC, Orias E. Control of rDNA replication in tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell. 1986;47(2):229–240. doi: 10.1016/0092-8674(86)90445-9. [DOI] [PubMed] [Google Scholar]
- Larson DD, Spangler EA, Blackburn EH. Dynamics of telomere length variation in tetrahymena thermophila. Cell. 1987;50(3):477–83. doi: 10.1016/0092-8674(87)90501-0. [DOI] [PubMed] [Google Scholar]
- Larson DD, Umthun AR, Shaiu WL. Copy number control in the tetrahymena macronuclear genome. J Protozool. 1991;38(3):258–263. doi: 10.1111/j.1550-7408.1991.tb04439.x. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Coue M, McIntosh JR. In vitro motility assays using microtubules tethered to tetrahymena pellicles. Methods Cell Biol. 1993;39:149–165. doi: 10.1016/s0091-679x(08)60168-5. [DOI] [PubMed] [Google Scholar]
- Lovlie A, Haller BL, Orias E. Molecular evidence for somatic recombination in the ribosomal DNA of tetrahymena thermophila. Proc Natl Acad Sci U S A. 1988;85(14):5156–5160. doi: 10.1073/pnas.85.14.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen KR, Baum MP, Orias E. A restriction fragment length polymorphism in the 5′ non-transcribed spacer of the rDNA of tetrahymena thermophila inbred strains B and C3. Gene. 1987;55(2–3):169–178. doi: 10.1016/0378-1119(87)90277-0. [DOI] [PubMed] [Google Scholar]
- Lwoff A. Sur la nutrition des infusoires. C R Acad Sci. 1923;176:928–930. [Google Scholar]
- Lynch TJ, Brickner J, Nakano KJ, Orias E. Genetic map of randomly amplified DNA polymorphisms closely linked to the mating type locus of tetrahymena thermophila. Genetics. 1995;141(4):1315–125. doi: 10.1093/genetics/141.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo KA, Orias E. Further evidence for lack of gene expression in the tetrahymena micronucleus. Genetics. 1981;98(4):747–62. doi: 10.1093/genetics/98.4.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann LE, Farrant J. Survival of tissue culture cells frozen by a two-step procedure to −196 degrees C. I. holding temperature and time. Cryobiology. 1976a;13(3):261–268. doi: 10.1016/0011-2240(76)90106-1. [DOI] [PubMed] [Google Scholar]
- McGann LE, Farrant J. Survival of tissue culture cells frozen by a two-step procedure to −196 degrees C. II. warming rate and concentration of dimethyl sulphoxide. Cryobiology. 1976b;13(3):269–273. doi: 10.1016/0011-2240(76)90107-3. [DOI] [PubMed] [Google Scholar]
- Melia SM, Cole ES, Turkewitz AP. Mutational analysis of regulated exocytosis in tetrahymena. J Cell Sci. 1998;111 (Pt 1):131–140. doi: 10.1242/jcs.111.1.131. [DOI] [PubMed] [Google Scholar]
- Meltz Steinberg K, Levin BR. Grazing protozoa and the evolution of the escherichia coli O157: H7 shiga toxin-encoding prophage. Proceedings of the Royal Society Biological Sciences Series B. 2007;274(1621):1921–1929. doi: 10.1098/rspb.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, et al. Microarray analyses of gene expression during the tetrahymena thermophila life cycle. PloS one. 2009;4(2):e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney DL. Aging and long-term temporal regulation in ciliated protozoa. A critical review. Mech Ageing Dev. 1974;3(2):81–105. doi: 10.1016/0047-6374(74)90008-6. [DOI] [PubMed] [Google Scholar]
- Nanney DL, Meyer EB, Simon EM, Preparata RM. Comparison of ribosomal and isozymic phylogenies of tetrahymenine ciliates. J Protozool. 1989;36(1):1–8. doi: 10.1111/j.1550-7408.1989.tb02661.x. [DOI] [PubMed] [Google Scholar]
- Nanney DL, Simon EM. Laboratory and evolutionary history of tetrahymena thermophila. Methods Cell Biol. 2000;62:3–25. doi: 10.1016/s0091-679x(08)61527-7. [DOI] [PubMed] [Google Scholar]
- Nanney DL. Nucleo-cytoplasmic interaction during conjugation in tetrahymena. Biological Bulletin. 1953;105(1):133–148. [Google Scholar]
- Noseda DG, Gentili HG, Nani ML, Nusblat A, Tiedtke A, Florin Christensen J, et al. A bioreactor model system specifically designed for tetrahymena growth and cholesterol removal from milk. Applied Microbiology and Biotechnology. 2007;75(3):515–520. doi: 10.1007/s00253-007-0843-x. [DOI] [PubMed] [Google Scholar]
- NYBERG D. Three new “biological” species of tetrahymena (T. hegewischi n. sp., T. sonneborni n. sp., T. nipissingi n. sp.) and temperature tolerance of members of the “pyriformis” Complex1. Journal of Eukaryotic Microbiology. 1981;28(1):65–69. [Google Scholar]
- Orias E. Probable somatic DNA rearrangements in mating type determination in tetrahymena thermophila: A review and a model. Dev Genet Developmental Genetics. 1981;2(2):185–202. [Google Scholar]
- Orias E. Mapping the germ-line and somatic genomes of a ciliated protozoan, tetrahymena thermophila. Genome Res. 1998;8(2):91–9. doi: 10.1101/gr.8.2.91. [DOI] [PubMed] [Google Scholar]
- Orias E, Bruns PJ. Induction and isolation of mutants in tetrahymena. Methods Cell Biol. 1976;13:247–282. [PubMed] [Google Scholar]
- Orias E, Flacks M. Use of genomic exclusion to isolate heat-sensitive mutants in tetrahymena. Genetics. 1973;73(4):543–549. doi: 10.1093/genetics/73.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Flacks M, Satir BH. Isolation and ultrastructural characterization of secretory mutants of tetrahymena thermophila. J Cell Sci. 1983;64:49–67. doi: 10.1242/jcs.64.1.49. [DOI] [PubMed] [Google Scholar]
- Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: Useful strains, cell culture, and cell line maintenance. Methods Cell Biol. 2000;62:189–211. doi: 10.1016/s0091-679x(08)61530-7. [DOI] [PubMed] [Google Scholar]
- Orias E, Pollock NA. Heat-sensitive development of the phagocytotic organelle in a tetrahymena mutant. Exp Cell Res. 1975;90(2):345–356. doi: 10.1016/0014-4827(75)90324-9. [DOI] [PubMed] [Google Scholar]
- Orias E, Rasmussen L. Dual capacity for nutrient uptake in tetrahymena. IV. growth without food vacuoles and its implications. Exp Cell Res. 1976;102(1):127–137. doi: 10.1016/0014-4827(76)90307-4. [DOI] [PubMed] [Google Scholar]
- Preparata RM, Meyer EB, Preparata FP, Simon EM, Vossbrinck CR, Nanney DL. Ciliate evolution: The ribosomal phylogenies of the tetrahymenine ciliates. J Mol Evol. 1989;28(5):427–441. doi: 10.1007/BF02603078. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58(2):233–67. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raikov IB. Evolution of macronuclear organization. Annu Rev Genet. 1976;10:413–440. doi: 10.1146/annurev.ge.10.120176.002213. [DOI] [PubMed] [Google Scholar]
- Rasmussen L, Christensen ST, Schousboe P, Wheatley DN. Cell survival and multiplication. the overriding need for signals: From unicellular to multicellular systems. FEMS Microbiol Lett. 1996;137(2–3):123–18. doi: 10.1016/0378-1097(96)00053-5. [DOI] [PubMed] [Google Scholar]
- Rasmussen L, Kludt TA. Particulate material as a prerequisite for rapid cell multiplication in tetrahymena cultures. Exp Cell Res. 1970;59(3):457–463. doi: 10.1016/0014-4827(70)90654-3. [DOI] [PubMed] [Google Scholar]
- Rasmussen L, Modeweg-Hansen L. Cell multiplication in tetrahymena cultures after addition of particulate material. J Cell Sci. 1973;12(1):275–286. doi: 10.1242/jcs.12.1.275. [DOI] [PubMed] [Google Scholar]
- Rasmussen L, Orias E. Tetrahymena: Growth without phagocytosis. Science. 1975;190(4213):464–465. doi: 10.1126/science.1166313. [DOI] [PubMed] [Google Scholar]
- Rauen U, Noll T, Piper HM, Lauchart W, Becker HD, De Groot H. Endothelial cell toxicity of preservation solutions: Comparison of endothelial cells of different origin and dependence on growth state. Cryobiology. 1994;31(2):144–153. doi: 10.1006/cryo.1994.1018. [DOI] [PubMed] [Google Scholar]
- Rehfuss MY, Parker CT, Brandl MT. Salmonella transcriptional signature in tetrahymena phagosomes and role of acid tolerance in passage through the protist. The ISME journal. 2011;5(2):262–273. doi: 10.1038/ismej.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford YM, Orias E. Phenylketonuric tetrahymena: Phenylalanine hydroxylase mutants and other tyrosine auxotrophs. Proc Natl Acad Sci U S A. 1981;78(12):7614–7618. doi: 10.1073/pnas.78.12.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir BH, Reichman M, Orias E. Conjugation rescue of an exocytosis-competent membrane microdomain in tetrahymena thermophila mutants. Proc Natl Acad Sci U S A. 1986;83(21):8221–8225. doi: 10.1073/pnas.83.21.8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvant MP, Pepin D, Bohatier J, Groliere CA, Guillot J. Toxicity assessment of 16 inorganic environmental pollutants by six bioassays. Ecotoxicol Environ Saf. 1997;37(2):131–140. doi: 10.1006/eesa.1997.1519. [DOI] [PubMed] [Google Scholar]
- Sauvant MP, Pepin D, Groliere CA, Bohatier J. Effects of organic and inorganic substances on the cell proliferation of L-929 fibroblasts and tetrahymena pyriformis GL protozoa used for toxicological bioassays. Bull Environ Contam Toxicol. 1995;55(2):171–178. doi: 10.1007/BF00203006. [DOI] [PubMed] [Google Scholar]
- Sauvant MP, Pepin D, Piccinni E. Tetrahymena pyriformis: A tool for toxicological studies. A review. Chemosphere. 1999;38(7):1631–169. doi: 10.1016/s0045-6535(98)00381-6. [DOI] [PubMed] [Google Scholar]
- Simon EM. Breeding performance of tetrahymena thermophila following storage for 5 to 6 years in liquid nitrogen. Cryobiology. 1982;19(6):607–612. doi: 10.1016/0011-2240(82)90190-0. [DOI] [PubMed] [Google Scholar]
- Simon EM, Nanney DL. Germinal aging in tetrahymena thermophila. Mech Ageing Dev. 1979;11(4):253–268. doi: 10.1016/0047-6374(79)90005-8. [DOI] [PubMed] [Google Scholar]
- Simon EM, Whang SW. Tetrahymena: Effect of freezing and subsequent thawing on breeding performance. Science (New York, NY) 1967;155(763):694–696. doi: 10.1126/science.155.3763.694. [DOI] [PubMed] [Google Scholar]
- Song X, Gorovsky MA. Unphosphorylated H1 is enriched in a specific region of the promoter when CDC2 is down-regulated during starvation. Molecular and cellular biology. 2007;27(5):1925–1933. doi: 10.1128/MCB.01619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugai T, Hiwatashi K. Cytologic and autoradiographic studies of the micronucleus at meiotic prophase in tetrahymena pyriformis. J Protozool. 1974;21(4):542–58. doi: 10.1111/j.1550-7408.1974.tb03695.x. [DOI] [PubMed] [Google Scholar]
- Suhr-Jessen PB, Orias E. Mutants of tetrahymena thermophila with temperature sensitive food vacuole formation. II. physiological and morphological studies. Exp Cell Res. 1979;124(2):317–327. doi: 10.1016/0014-4827(79)90207-6. [DOI] [PubMed] [Google Scholar]
- Sweet MT, Allis CD. Long-term storage of unfrozen tetrahymena cultures in soybean medium. Cold Spring Harbor Protocols, 2006. 2010;(3) doi: 10.1101/pdb.prot4472. pdb.prot4472. [DOI] [PubMed] [Google Scholar]
- Szablewski L, Andreasen PH, Tiedtke A, Florin-Christensen J, Florin-Christensen M, Rasmussen L. Tetrahymena thermophila: Growth in synthetic nutrient medium in the presence and absence of glucose. Journal of Eukaryotic Microbiology. 1991;38(1):62–65. [Google Scholar]
- Tiedtke A. Isolation of pure pellicles containing intact basal bodies of tetrahymena pyriformis. J Cell Sci. 1985;77:155–165. doi: 10.1242/jcs.77.1.155. [DOI] [PubMed] [Google Scholar]
- Tondravi MM, Yao MC. Transformation of tetrahymena thermophila by microinjection of ribosomal RNA genes. Proc Natl Acad Sci U S A. 1986;83(12):4369–4373. doi: 10.1073/pnas.83.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkewitz AP. Out with a bang! tetrahymena as a model system to study secretory granule biogenesis. Traffic. 2004;5(2):63–68. doi: 10.1046/j.1600-0854.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- van Wagtendonk WJ, Hackett PL. The culture of paramecium aurelia in the absence of other living organisms. Proceedings of the National Academy of Sciences of the United States of America. 1949;35(3):155–159. doi: 10.1073/pnas.35.3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide T, Herrmann L, Bockau U, Niebur N, Aldag I, Laroy W, et al. Secretion of functional human enzymes by tetrahymena thermophila. BMC Biotechnol. 2006;6:19. doi: 10.1186/1472-6750-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide T, Bockau U, Rave A, Herrmann L, Hartmann MWW. A recombinase system facilitates cloning of expression cassettes in the ciliate tetrahymena thermophila. BMC Microbiology. 2007;7:12. doi: 10.1186/1471-2180-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NE. Preparation of cytoskeletal fractions from tetrahymena thermophila. Methods Cell Biol. 2000;62:441–447. doi: 10.1016/s0091-679x(08)61548-4. [DOI] [PubMed] [Google Scholar]
- Williams NE, Honts JE. The assembly and positioning of cytoskeletal elements in tetrahymena. Development. 1987;100(1):23–30. doi: 10.1242/dev.100.1.23. [DOI] [PubMed] [Google Scholar]
- Williams NE, Wolfe J, Bleyman LK. Long-term maintenance of tetrahymena spp. J Protozool. 1980;27(3):327. doi: 10.1111/j.1550-7408.1980.tb04270.x. [DOI] [PubMed] [Google Scholar]
- Xiong J, Lu X, Lu Y, Zeng H, Yuan D, Feng L, et al. Tetrahymena gene expression database (TGED): A resource of microarray data and co-expression analyses for tetrahymena. Science China Life sciences. 2011;54(1):65–67. doi: 10.1007/s11427-010-4114-1. [DOI] [PubMed] [Google Scholar]
- Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of tetrahymena pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Ye AJ, Romero DP. Phylogenetic relationships amongst tetrahymenine ciliates inferred by a comparison of telomerase RNAs. International Journal of Systematic and Evolutionary Microbiology. 2002;52(Pt 6):2297–2302. doi: 10.1099/00207713-52-6-2297. [DOI] [PubMed] [Google Scholar]