Abstract
Lanthanide-doped upconversion nanoparticles (UCNPs) have shown promise in biomedical applications. However, as the UCNPs are normally capped with hydrophobic ligands, it remains challenging to prepare biocompatible UCNPs with specific molecular recognition capabilities. We herein report an exceptionally simple strategy to prepare uniform DNA-modified upconversion nanoparticles (DNA-UCNPs) as versatile bioprobes. The approach can directly convert as-prepared hydrophobic UCNPs into water-soluble DNA-UCNPs without any chemical modification of UCNPs or oligonucleotides. Furthermore, DNA molecules on the DNA-UCNPs retain their biorecognition ability, allowing programmable assembly of hybrid nanostructures. More importantly, we show that these DNA-UCNPs are capable of crossing cell membranes without the need of transfection agents, and their use as agents for bioimaging and DNA delivery are also demonstrated. Finally, DNA aptamer-conjugated UCNPs can be readily used for targeted imaging of cancer cells.
Lanthanide–based upconversion nanoparticles (UCNPs), typically NaMF4:Yb3+/Ln3+ (M = Y or Gd, Ln = Er or Tm)), are capable of converting near-infrared (NIR) excitation to tunable shorter-wavelength luminescence from the deep-UV to the NIR range.1,2 These UCNPs have attracted attention from researchers in a variety of fields such as biological imaging, therapeutics, and photovoltaics, due to their unique properties including tunable multicolor emission, exceptional photostability, deep tissue penetration and suppression of autofluorescence, and low in vitro and in vivo toxicity.1,2 Despite their promise, preparation of water-dispersible, biocompatible and functionalized UCNPs remains a formidable challenge, as they are normally capped with inactive hydrophobic ligands.1 To overcome this limitation, much effort has been devoted to functionalizing UCNPs with other molecules (e.g. DNA or proteins).1 For instance, one-step solvothermal synthesis, 1g,h ligand exchange method,1i,j and silica coating,2d have recently been developed to render the UCNPs water-dispersible and functionalizable. To make the process even simpler and more accessible to biologists and others outside the field of UCNPs, it is desirable to avoid the extra step of bioconjugations using heterobifunctional cross-linkers (e.g., DCC or EDC), whose efficiency can be low.1a,f
As a well-known biomolecule, DNA is arguably the most attractive platform for bionanotechnology due to the ease of its synthesis, its excellent stability, and its unique programmable base pairing interactions.3 Since the landmark work in 1996 demonstrating attachment of thiolated DNA to gold nanoparticles (AuNPs),4 the interest in DNA-based bionanotechnology has expanded significantly due to their wide variety of applications ranging from sensitive biosensing,5 self-assembly of nanoparticles (NPs),6 and gene and drug delivery.7 To enable these applications, a crucial step is to attach DNA on solid NPs. AuNPs have been the most studied subjects in this field because of the well-established method for DNA attachment based on thiol adsorption on a gold surface.4–7 However, thiolate-Au chemistry is not generally applicable for other NPs. Therefore, significant effort have been expended towards the facile modification of other NPs with DNA.8 Despite much progress, simple and efficient strategies are far from fully developed. Development of new strategies for DNA functionalization of NPs with unique properties is greatly needed for various applications.
Considering the huge potentials of both UCNPs and DNA-based bionanotechnology, development of a simple, general, and versatile method to produce DNA-attached UCNPs will advance the biomedical application of UCNPs significantly. Herein, we report such an approach to prepare DNA-functionalized UCNPs based on ligand exchange at the liquid–liquid interface (Scheme 1). Importantly, the facile one-step approach can not only directly convert hydrophobic UCNPs into water-dispersible and biocompatible ones, but it also leads to the formation of bioconjugates that retain the advantageous properties of both DNA and UCNPs, including programmable NP assembly. Interestingly, the DNA-UCNPs are capable of crossing cell membranes without the use of transfection agents, and thus serve as an excellent agent for bioimaging and DNA delivery, including targeted imaging of breast cancer cells.
Scheme 1.
Direct Synthesis of DNA-Functionalized UCNPs from As-prepared Hydrophobic Ones through a Facile One-Step Ligand Exchange Strategy.
High quality NIR-to-visible UCNPs with a composition of NaYF4:18%Yb/2%Er were synthesized using oleic acid as the capping agent.2d As shown in Figure 1a, these nanocrystals display hexagonal structure with uniform sizes of ~70 nm. High-resolution TEM (HRTEM) images show the UCNPs are single crystals (Figure 1b and Figure S1). We explored the use of DNA molecules to replace the original oleic acid ligands on UCNPs based on a ligand exchange process, to directly prepare DNA-functionalized UCNPs (Scheme 1). In a typical procedure, a chloroform solution containing hydrophobic UCNPs was slowly added to a water solution of DNA, and the solution was vigorously stirred for one day. Afterward, the UCNPs were observed to have transferred into the water layer from the chloroform layer due to DNA attachment through the interaction between the negatively charged phosphates of the DNA with surface lanthanide ions.9 The resulting DNA-UCNPs were separated after the phase transfer and re-dispersed in water.
Figure 1.
TEM images of the (a,b) as-prepared and (c,d) DNA–modified UCNPs (DNA layer was marked for DNA-UCNPs in HRTEM).
Representative TEM images (Figure 1c and Figure S1) of the resulting DNA-UCNPs using a 30-mer poly T oligonucleotide show that they are well-dispersed in water without change in shape, suggesting that these DNA-capped nanocrystals have become hydrophilic. A uniform 2-nm-thick DNA layer around the crystallized nanocrystals could be visualized by HRTEM after being stained with OsO4 (Figure 1d and Figure S1). This DNA attachment was also confirmed by the observation of typical absorbance at 260 nm after DNA functionalization (Figure S2). Attachment of DNA molecules onto UCNPs was further confirmed by the appearance of the characteristic vibrations of the thymine base (1714 cm−1) and the sugar-phosphate (1250–1000 cm−1) in the FTIR spectrum (Figure S3). Zeta potential measurements (Figure S4) showed that the UCNPs are highly negatively charged after DNA attachment (−28.5 mV). Using fluorescently labeled DNA, the surface density of DNA was estimated to be 80 DNA molecules per UCNPs.
As a result of the DNA coating, the hydrophobic UCNPs could be directly transformed into water-soluble ones. As shown in Figure 2a, the DNA-UCNPs possess excellent water solubility due to the existence of a dense layer of DNA molecules on the outer surface. Upon continuous excitation at 980 nm, the luminescence of the DNA-UCNPs in water appears predominantly green in color. As shown in Figure 2b, the corresponding upconversion spectrum of DNA-UCNPs in water is similar to that of the as-prepared UCNPs in cyclohexane, which showed that the characteristic upconversion optical property of the nanoparticles was retained. It shows DNA-UCNPs can also be well-dispersed in physiological buffers such as PBS and serum (Figure S5). Significantly, since this strategy only relies on the surface properties of the nanocrystal, it is generally applicable to many other lanthanide-based UCNPs with a variety of compositions, sizes, and shapes. As an example, ~40 nm NaYF4:Yb/Tm nanoparticles were also functionalized with DNA molecules in this fashion (Figure S6). Furthermore, DNA of other sequences (e.g. A30, C30, and G20) could also be used by the same method to obtain DNA-UCNPs (Figure S7).
Figure 2.
(a) Photographs of the transparent solution of DNA–UCNPs in water without laser illumination (left) and under 980 nm laser illumination (right). (b) Room-temperature upconversion luminescence spectra of as-prepared UCNPs in cyclohexane and DNA–UCNPs in water under excitation at 980 nm.
In addition to making hydrophobic UCNPs hydrophilic for biomedical applications, DNA-functionalization can endow the nanomaterials with target recognition abilities. To test the functionality of the DNA on the DNA-UCNPs, UNCPs covered with T30 oligonucleotides (T30-UCNP) were assembled with 5 nm AuNPs that were themselves modified with a complementary DNA strand (A27-AuNPs) via a 5′–thiol attachment (Figure 3a). As shown in Figure 3b and Figure S8, The T30-UCNPs were surrounded by a number of A27-AuNPs, forming a satellite structure. In contrast, when 5 nm AuNPs were functionalized with noncomplementary DNA (T27-AuNPs) and incubated with T30-UCNPs, no assembly of the nanoparticles was observed (Figure 3c and Figure S8). These results further confirm that the DNA was functionalized on UCNPs in large numbers. Furthermore, the DNA strands attached on UCNPs retain their ability to specifically hybridize to complementary DNA targets. Therefore, it should be possible to utilize the complementary nature of DNA hybridization to further functionalize DNA-covered UCNPs, which is of significant importance toward any future biomedical application. For the present strategy, although the attachment of specific DNA on UCNPs can be achieved without any chemical modification of UCNPs or oligonucleotides, it requires the pre-modification of T30 DNA for hybridization. It shows that the reversibility of DNA duplex still hold on the UCNPs (Figure S9).
Figure 3.
(a) Schematic illustration of DNA-directed assembly of UCNPs and AuNPs. TEM images of T30-UCNPs assembled with AuNPs bearing (b) cDNA and (c) noncomplementary DNA.
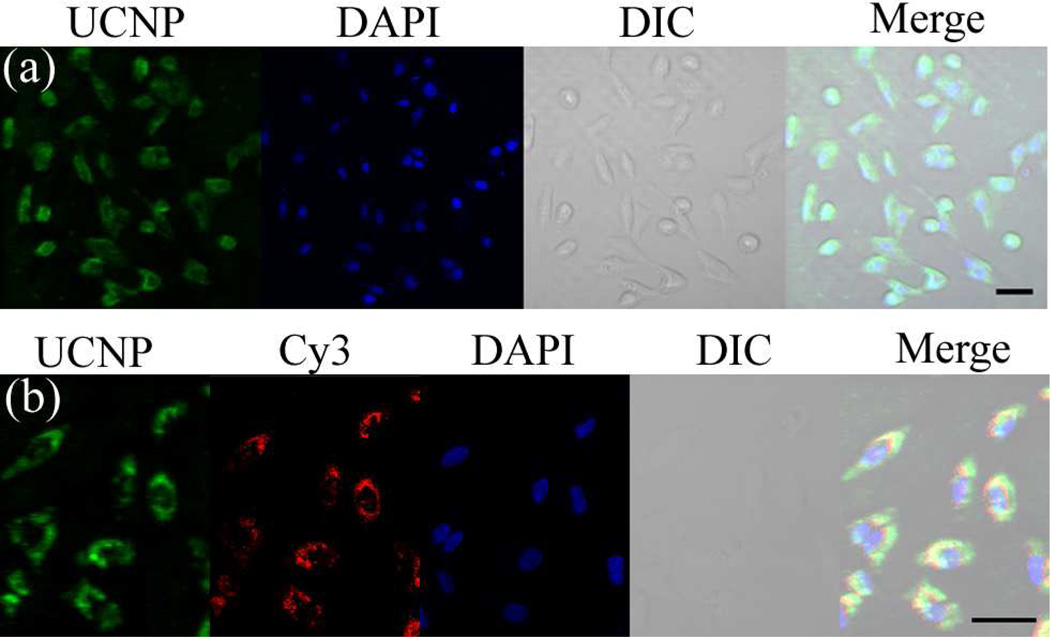
Having demonstrated an exceptionally simple strategy for preparing DNA-functionalized UCNPs, we then investigated their ability to enter live cells for bioimaging applications. The resulting T30-UCNPs was incubated with HeLa cells for 12 h in the absence of any cationic transfection agents. The resulting HeLa cells exhibited bright green up-converted fluorescence, primarily within the cytoplasm, indicating excellent internalization of nano-particles (Figure 4a and Figure S10). A series of Z-stack images confirmed that the up-converted luminescence signal was in the same focal plane as the nuclear stain (Figure S11), which further demonstrated that the DNA-UCNPs were indeed localized within the cell. Interestingly, the DNA-UCNPs reside mainly in the cytoplasm, probably because they maybe too large to cross the nuclear pores. This result is consistent with cellular uptake features observed in a previously reported polyvalent DNA-AuNP conjugates. 7a Furthermore, the uptake of the DNA-UCNPs was investigated by using the UCNPs functionalized with a dye-labeled DNA, with the results showing that the intracellular green up-converted fluorescence of UCNPs was co-localized with the red fluorescence of the dye (Figure S12). Significantly, these results show that, despite being capped with only negatively charged oligonucleotides, DNA-UCNPs readily enter cells without the need for transfection agents. More importantly, the upconverted signals of the DNA-UCNPs in HeLa cells are extremely stable during long-term continuous excitation under laser-scanning con-focal microscopy, while the fluorescent signals of a control group using CdTe/ZnS QDs as labels were greatly reduced after irradiation (Figure S13). Combined with various advantages of NIR excitation, the excellent photostability of DNA-UCNPs allows for long-term continuous imaging and tracking of living cells that is superior to many other DNA-nanomaterials.
Figure 4.
(a) Confocal microscopy images of HeLa cells treated with (a) T30-UCNPs and (b) T30-UCNPs functionalized with Cy3 dye-labeled double stranded DNA. For UCNP images, λex = 980 nm, and emission was collected in the range λ = 510–560 nm. For Cy3 images, λex = 560 nm, and emission was collected in the range λ = 575–625 nm. Scale bars are 50 µm.
After establishing their facile cellular uptake, we further explored the use of the DNA-UCNPs as nanocarriers for DNA delivery. A Cy3-labeled double stranded DNA sequence (dsDNA) with a poly A (A24) tail was attached to T30-UCNPs by hybridization. The resulting nanoparticles are well-dispersed in the buffer solution (Figure S14c). Successful functionalization was confirmed by the down-conversion fluorescence of Cy3 and the potential FRET between UCNPs and the dye (Figure S14b, d). After 12 h of incubation, both the upconverting and down-converting fluorescence from the UCNPs and Cy3, respectively, were visualized in the cell cytoplasm from each laser excitation channel (Figure 4b). The upconverted fluorescence of UCNPs co-localized well with Cy3 red fluorescence, indicating that DNA was readily delivered into cells using the UCNPs. As a control, no DNA-derived fluorescence was observed inside the cells when the cells were incubated with Cy3-labeled dsDNA alone (Figure S14e). Owing to the easy DNA attachment and unique optical properties of UCNPs, the present nanocarrier is expected to have widespread applications of DNA delivery with real-time imaging and tracking in vitro and in vivo.
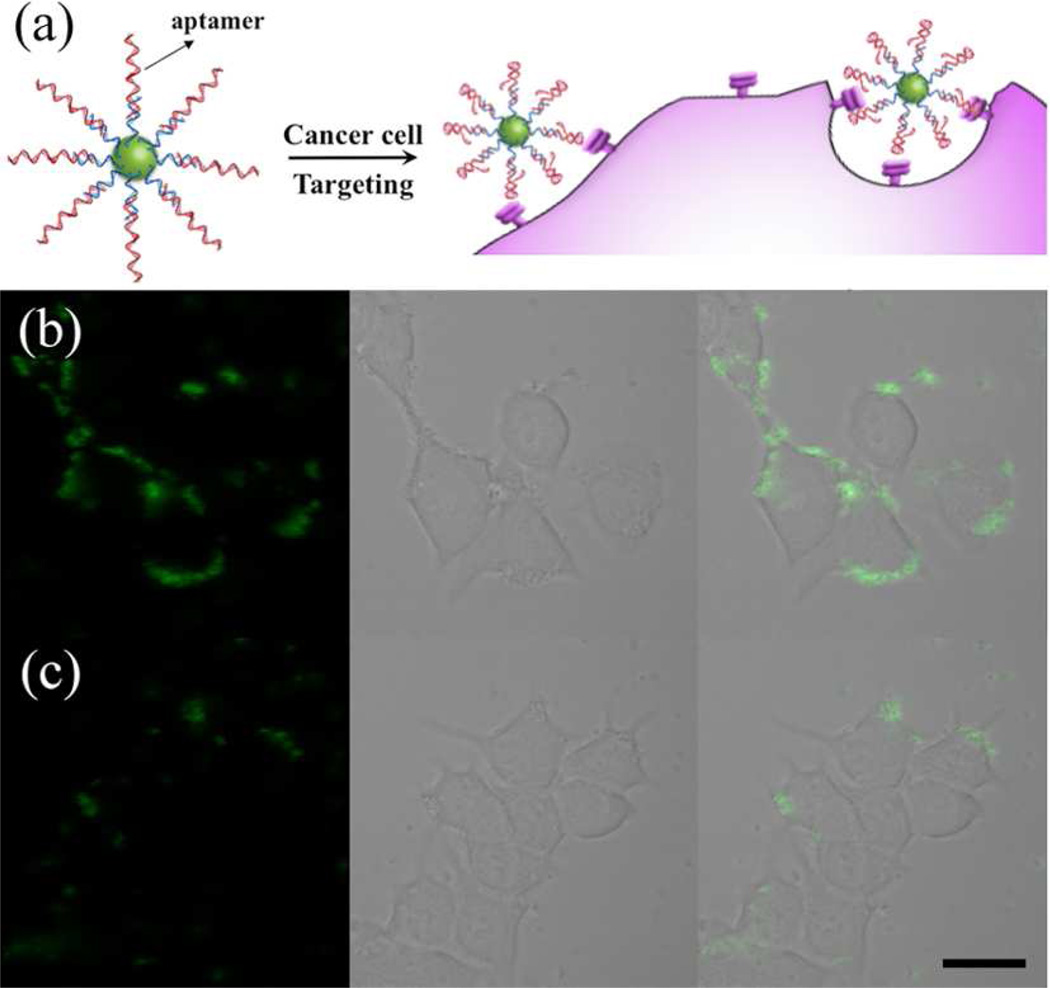
Finally, we explored the use of the DNA-functionalized UCNPs for targeted imaging of cancer cells based on aptamer-UCNP bioconjugates (Figure 5a). Aptamers are short single stranded DNA or RNA sequences that are selected to bind to a wide array of biological targets with high affinity and specificity.5,7b To provide cancer-targeting capabilities to UCNPs, a 26-mer DNA aptamer AS1411, which is capable of targeting nucleolin, was conjugated to UCNPs for targeted imaging of the nucleolin-overexpressed breast cancer cell line MCF-7.10 The sequence of the aptamer used (5–GGT GGT GGT GGT TGT GGT GGT GGT GG A24–thiol–3) contains 24 extra A bases at the 3′–terminus to facilitate the attachment of the aptamer on T30-UCNPs based on hybridization. This further ensures that the binding domains of the aptamer extend away from the UCNP surface, to allow enough flexibility for the aptamer folding. The successful conjugation of AS1411 on UCNPs was confirmed by the test of its hybridization with corresponding cDNA functionalized AuNPs (Figure S15). The Apta-UCNPs were incubated with MCF-7 cells for 1 h and excess nanoparticles were removed by washing. Subsequent analysis revealed that the cells showed strong up-converting fluorescence (Figure 5b and Figure S16). Overlays of confocal fluorescence and bright-field images demonstrated that the fluorescence was mainly on the surface of the cells, confirming the aptamer-mediated accumulation of the Apta-UCNPs on the nucleolin-overexpressed surface of MCF-7 cells. In comparison, the T30-UCNPs modified with control DNA with a randomized sequence (Rdm-UCNPs) showed much less binding to the MCF-7 cells (Figure 5c and Figure S16), confirming the role of the specific aptamer sequence in cell targeting. As the incubation time was increased to 4 h, the aptamer–receptor–mediated endocytosis of the bioconjugates increased (Figure S17). Both the Apta-UCNPs and Rdm-UCNPs exhibited negligible binding with a nucleolin negative cell line (NIH-3T3 cells) (Figure S18), which further validated that the targeting was indeed mediated by the aptamer. The availability of various aptamers specific for different molecular markers and metabolites and the ease of conjugation to UCNPs makes the current system very promising for applications in targeted imaging, diagnosis, and therapy.
Figure 5.
(a) Schematic illustration of the targeted imaging of cancer cells with Aptamer-UCNP bioconjugates. Confocal microscopy images of MCF-7 cells treated with (b) Apta-UCNPs and (c) Rdm-UCNPs. λex = 980 nm, emission was collected in the range λ = 510–560 nm. Scale bars are 20 µm.
In conclusion, we have developed a very simple method for direct fabrication of DNA-modified UCNPs from as-prepared hydrophobic UCNPs through a one-step strategy that is generally applicable to all lanthanide-based UCNPs with a variety of compositions, sizes, and shapes. The approach does not require preparation of functionalizable UCNPs or chemical modification of oligonucleotides. The simplicity, cost efficiency, and versatility of this approach afforded hybridizable DNA-UCNPs that can be readily taken up by cells and visualized under NIR light excitation with high photostability. We have also shown that the DNA-UCNPs can be used for DNA delivery and targeted imaging of cancer cells. This novel approach and the resulting outstanding combination of properties will advance both the fields of UCNPs and DNA-based bionanotechnology, and will find wide application in areas including bioinspired nanoassembly, biomedicine, and biological assays.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the U.S. National Institute of Health (Grant ES016865).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Detailed synthesis and characterization data as well as supplementary results. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.(a) Wang F, Liu X. Chem. Soc. Rev. 2009;38:976–989. doi: 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]; (b) Haase M, Schäfer H. Angew. Chem. Int. Ed. 2011;50:5808–5829. doi: 10.1002/anie.201005159. [DOI] [PubMed] [Google Scholar]; (c) Auzel F. Chem. Rev. 2003;104:139–174. doi: 10.1021/cr020357g. [DOI] [PubMed] [Google Scholar]; (d) Feng W, Sun L, Zhang Y, Yan C. Coord. Chem. Rev. 2010;254:1038–1053. [Google Scholar]; (e) Mader HS, Kele P, Saleh SM, Wolfbeis OS. Curr. Opin. Chem. Biol. 2010;14:582–596. doi: 10.1016/j.cbpa.2010.08.014. [DOI] [PubMed] [Google Scholar]; (f) Li L, Zhang R, Yin L, Zheng K, Qin W, Selvin PR, Lu Y. Angew. Chem. Int. Ed. 2012;51:6121–6125. doi: 10.1002/anie.201109156. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Ju Q, Tu D, Liu Y, Li R, Zhu H, Chen J, Chen Z, Huang M, Chen X. J. Am. Chem. Soc. 2012;134:1323–1330. doi: 10.1021/ja2102604. [DOI] [PubMed] [Google Scholar]; (h) Zhou J, Yao L, Li C, Li F. J. Mater. Chem. 2010;20:8078–8085. [Google Scholar]; (i) Liu Y, Tu D, Zhu H, Li R, Luo W, Chen X. Adv. Mater. 2010;22:3266–3271. doi: 10.1002/adma.201000128. [DOI] [PubMed] [Google Scholar]; (j) Liu Y, Zhou S, Tu D, Chen Z, Huang M, Zhu H, Ma E, Chen X. J. Am. Chem. Soc. 2012;134:15083–15090. doi: 10.1021/ja306066a. [DOI] [PubMed] [Google Scholar]; (k) Wu S, Duan N, Wang Z, Wang H. Analyst. 2011;136:2306–2314. doi: 10.1039/c0an00735h. [DOI] [PubMed] [Google Scholar]; (l) Yang Y, Shao Q, Deng R, Wang C, Teng X, Cheng K, Cheng Z, Huang L, Liu Z, Liu X, Xing B. Angew. Chem. Int. Ed. 2012;51:3125–3129. doi: 10.1002/anie.201107919. [DOI] [PubMed] [Google Scholar]
- 2.(a) Wang F, Han Y, Lim CS, Lu YH, Wang J, Xu J, Chen HY, Zhang C, Hong M, Liu X. Nature. 2010;463:1061–1065. doi: 10.1038/nature08777. [DOI] [PubMed] [Google Scholar]; (b) Deng R, Xie X, Vendrell M, Chang Y-T, Liu X. J. Am. Chem. Soc. 2011;133:20168–20171. doi: 10.1021/ja2100774. [DOI] [PubMed] [Google Scholar]; (c) Mai H, Zhang Y, Si R, Yan Z, Sun L, You L, Yan C. J. Am. Chem. Soc. 2006;128:6426–6436. doi: 10.1021/ja060212h. [DOI] [PubMed] [Google Scholar]; (d) Li Z, Zhang Y, Jiang S. Adv. Mater. 2008;20:4765–4769. [Google Scholar]; (e) Wang L, Yan R, Huo Z, Wang L, Zeng J, Bao J, Wang X, Peng Q, Li Y. Angew. Chem. Int. Ed. 2005;44:6054–6057. doi: 10.1002/anie.200501907. [DOI] [PubMed] [Google Scholar]; (f) Zhou J, Liu Z, Li F. Chem. Soc. Rev. 2012;41:1323–1349. doi: 10.1039/c1cs15187h. [DOI] [PubMed] [Google Scholar]; (g) Abel KA, Boyer JC, van Veggel FCJM. J. Am. Chem. Soc. 2009;131:14644–14645. doi: 10.1021/ja906971y. [DOI] [PubMed] [Google Scholar]; (h) Cheng L, Yang K, Li Y, Chen J, Wang C, Shao M, Lee S, Liu Z. Angew. Chem. Int. Ed. 2011;50:7385–7390. doi: 10.1002/anie.201101447. [DOI] [PubMed] [Google Scholar]; (i) Nam SH, Bae YM, Il Park Y, Kim JH, Kim HM, Choi JS, Lee KT, Hyeon T, Suh YD. Angew. Chem. Int. Ed. 2011;50:6093–6097. doi: 10.1002/anie.201007979. [DOI] [PubMed] [Google Scholar]; (j) Nyk M, Kumar R, Ohulchanskyy TY, Bergey EJ, Prasad PN. Nano Lett. 2008;8:3834–3838. doi: 10.1021/nl802223f. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Wu S, Han G, Milliron DJ, Aloni S, Altoe V, Talapin DV, Cohen BE, Schuck PJ. Proc Natl. Acad. Sci. USA. 2009;106:10917–10921. doi: 10.1073/pnas.0904792106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Bogdan N, Vetrone F, Ozin GA, Capobianco JA. Nano Lett. 2011;11:835–840. doi: 10.1021/nl1041929. [DOI] [PubMed] [Google Scholar]; (m) Ye X, Collins J, Kang Y, Chen J, Chen DTN, Yodh AG, Murray CB. Proc. Natl. Acad. Sci. USA. 2010;107:22430–22435. doi: 10.1073/pnas.1008958107. [DOI] [PMC free article] [PubMed] [Google Scholar]; (n) Zhang F, Braun GB, Shi Y, Zhang Y, Sun X, Reich N, Zhao D, Stucky G. J. Am. Chem. Soc. 2010;132:2850–2851. doi: 10.1021/ja909108x. [DOI] [PubMed] [Google Scholar]; (o) Tu D, Liu L, Ju Q, Liu Y, Zhu H, Li R, Chen X. Angew. Chem. Int. Ed. 2011;50:6306–6310. doi: 10.1002/anie.201100303. [DOI] [PubMed] [Google Scholar]
- 3.(a) Pinheiro AV, Han D, Shih WM, Yan H. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dhar S, Daniel WL, Giljohann DA, Mirkin CA, Lippard SJ. J. Am. Chem. Soc. 2009;131:14652–14653. doi: 10.1021/ja9071282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; (b) Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Jr, Schultz PG. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 5.(a) Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhao W, Chiuman W, Lam JCF, McManus SA, Chen W, Cui Y, Pelton R, Brook MA, Li Y. J. Am. Chem. Soc. 2008;130:3610–3618. doi: 10.1021/ja710241b. [DOI] [PubMed] [Google Scholar]; (c) Guo S, Wang E. Acc. Chem. Res. 2011;44:491–500. doi: 10.1021/ar200001m. [DOI] [PubMed] [Google Scholar]; (d) Wang H, Yang R, Yang L, Tan W. ACS Nano. 2009;3:2451–2460. doi: 10.1021/nn9006303. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Li D, Song SP, Fan CH. Acc. Chem. Res. 2010;43:631–641. doi: 10.1021/ar900245u. [DOI] [PubMed] [Google Scholar]
- 6.(a) Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]; (b) Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]; (c) Nykypanchuk D, Maye MM, van der Lelie D, Gang O. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]; (d) He Y, Ye T, Ribbe AE, Mao C. J. Am. Chem. Soc. 2011;133:1742–1744. doi: 10.1021/ja1060092. [DOI] [PubMed] [Google Scholar]; (e) Zhao W, Gao Y, Kandadai SA, Brook MA, Li Y. Angew. Chem. Int. Ed. 2006;45:2409–2413. doi: 10.1002/anie.200600061. [DOI] [PubMed] [Google Scholar]; (f) Pal S, Deng Z, Wang H, Zou S, Liu Y, Yan H. J. Am. Chem. Soc. 2011;133:17606–17609. doi: 10.1021/ja207898r. [DOI] [PubMed] [Google Scholar]
- 7.(a) Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AKR, Han MS, Mirkin CA. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]; (b) Lee JH, Yigit MV, Mazumdar D, Lu Y. Adv. Drug Delivery Rev. 2010;62:592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Wang Q, Liu Y, Ke Y, Yan H. Angew. Chem. Int. Ed. 2008;47:316–319. doi: 10.1002/anie.200703648. [DOI] [PubMed] [Google Scholar]; (b) Ma N, Sargent EH, Kelley SO. Nat. Nano-technol. 2009;4:121–125. doi: 10.1038/nnano.2008.373. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Liu Y. Nat. Nanotechnol. 2011;6:463–464. doi: 10.1038/nnano.2011.126. [DOI] [PubMed] [Google Scholar]; (d) Dave N, Liu J. ACS Nano. 2011;5:1304–1312. doi: 10.1021/nn1030093. [DOI] [PubMed] [Google Scholar]
- 9.Costa D, Burrows HD, da Graça Miguel M. Langmuir. 2005;21:10492–10496. doi: 10.1021/la051493u. [DOI] [PubMed] [Google Scholar]
- 10.(a) Soundararajan S, Chen W, Spicer EK, Courtenay-Luck N, Fernandes DJ. Cancer Res. 2008;68:2358–2365. doi: 10.1158/0008-5472.CAN-07-5723. [DOI] [PubMed] [Google Scholar]; (b) Cao Z, Tong R, Mishra A, Xu W, Wong GCL, Cheng J, Lu Y. Angew. Chem. Int. Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.