Abstract
1. Rationale
The therapeutic promise of trans-resveratrol (tRes) is limited by poor bioavailability following rapid metabolism. We hypothesise that trans-arachidin-1 (tA1) and trans-arachidin-3 (tA3), peanut hairy root-derived isoprenylated analogs of tRes, will exhibit slower metabolism/enhanced bioavailability and retain biological activity via cannabinoid receptor (CBR) binding relative to their non-prenylated parent compounds trans-piceatannol (tPice) and tRes, respectively.
2. Results
The activities of eight human UDP-glucuronosyltransferases (UGTs) toward these compounds were evaluated. The greatest activity was observed for extrahepatic UGTs 1A10 and 1A7, followed by hepatic UGTs 1A1 and 1A9. Importantly, an additional isoprenyl and/or hydroxyl group in tA1 and tA3 slowed overall glucuronidation. CBR binding studies demonstrated that all analogs bound to CB1Rs with similar affinities (5–18 µM); however, only tA1 and tA3 bound appreciably to CB2Rs. Molecular modelling studies confirmed that the isoprenyl moiety of tA1 and tA3 improved binding affinity to CB2Rs. Finally, although tA3 acted as a competitive CB1R antagonist, tA1 antagonised CB1R agonists by both competitive and non-competitive mechanisms.
3. Conclusions
Prenylated stilbenoids may be preferable alternatives to tRes due to increased bioavailability via slowed metabolism. Similar structural analogs might be developed as novel CB therapeutics for obesity and/or drug dependency.
Keywords: UDP-glucuronosyltransferases, molecular modelling, natural products, G-protein activity, bioproduction, cannabinoid receptors
Introduction
Recently, much attention has focused on potential therapeutic uses for the naturally occurring plant stilbenoid, trans-resveratrol (trans-3,5,4′-trihydroxystilbene; tRes), based on many reports demonstrating significant anti-oxidant, anti-inflammatory, anti-carcinogenic and anti-aging properties (Baur & Sinclair 2006). Although initial pre-clinical studies are encouraging (Cucciolla et al. 2007), the oral bioavailability of tRes following rapid absorption is very poor due to swift and extensive phase II metabolism primarily to the 3-O-glucuronide and sulphate conjugates (Vitaglione et al. 2005; Wenzel & Somoza 2005). This observation presents a significant limitation to future clinical development of this potentially valuable drug.
One potential avenue to circumvent the effect of the unfavourable pharmacokinetics of tRes would be to employ naturally occurring or synthetic structural analogs and/or conjugates that exhibit more favourable metabolic profiles leading to enhanced bioavailability. Several plants, such as peanuts, naturally synthesise tRes and a number of its analogs. However, very few studies have investigated the biological activity of these phytochemicals because they are only produced in limited amounts from plant sources. To overcome this problem, we have developed an in vitro peanut hairy root culture system to easily biosynthesise relatively large quantities of stilbenoids such as tRes. Specifically, phytochemical production is induced upon treatment of cultures with abiotic elicitors, thereby providing a reproducible, controlled, sustainable and scalable bioproduction system for these polyphenolic compounds (Condori et al. 2010; Medina-Bolivar et al. 2010; Sivakumar et al. 2011). Along with tRes, its hydroxylated analog trans-piceatannol (trans 3,5,3′,4′-tetrahydroxystilbene; tPice) and several isoprenylated analogs such as arachidin-1 [trans-4-(3-methyl-1-butenyl)-3,5,3′,4′-tetrahydroxystilbene; tA1] and arachidin-3 [trans-4-(3-methyl-1-butenyl)-3,5,4′-trihydroxystilbene; tA3] (Figure 1) can be produced in this peanut tissue culture system. All three tRes analogs possess anti-inflammatory and anti-proliferative properties similar to, or even better than, tRes (Chang et al. 2006; Djoko et al. 2007).
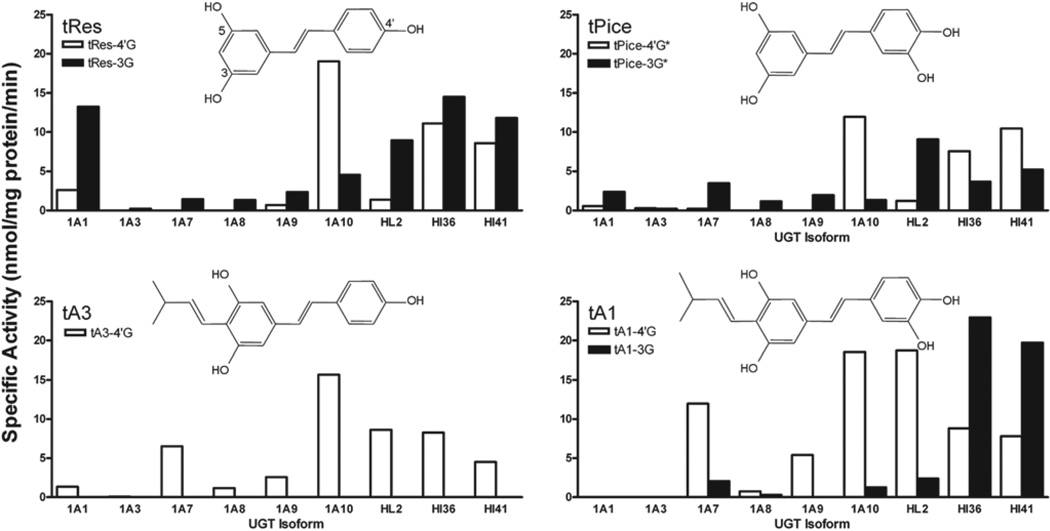
Figure 1.
Screening for trans-resveratrol (tRes) analog glucuronidation. Recombinant human UDP-glucuronosyltransferases (UGT) isoforms (5 µg) as well as HL2, HI36 and HI41 (50 µg) were evaluated for their ability to glucuronidate four compounds. Substrate and co-substrate (UDP-GlcUA) were 250 µM and 4 mM, respectively. No activity was seen for UGT1A4 or 1A6 toward any of the substrates analysed here. Assays were analysed by high-performance liquid chromatography.
The metabolism of tRes has been relatively well characterised. In this work, we focus only on the importance of glucuronidation. It has been shown that human hepatic and intestinal UDP-glucuronosyltransferases (UGTs) are actively involved in the glucuronidation of tRes and that both tissues contribute significantly to its biotransformation. The glucuronidation reactions are very regio- and stereo-specific, with the trans-3-O-glucuronide being the predominant product formed (Brill et al. 2006; Sabolovic et al. 2006; Vitaglione et al. 2005; Wenzel & Somoza 2005). Although a recent study has provided some limited information concerning the glucuronidation of tPice by human liver microsomes (HLMs) and a few recombinant UGTs (Miksits et al. 2010), no information is available regarding the metabolic profile of the naturally occurring tRes analogs, tA1 and tA3. Furthermore, information is lacking as to potential molecular target(s) mediating the beneficial effects of tRes or its natural analogs. Recently, it has been reported that tRes and several other polyphenols bind to a distinct, yet unidentified, plasma membrane bound receptor that occurs in high density throughout the brain (Han et al. 2006). Interestingly, cannabinoid receptors (CBRs) appear to share many characteristics with this newly discovered, uncharacterised tRes receptor.
This study was designed to characterise the glucuronidation activities of human recombinant UGTs toward tRes and its naturally synthesised analogs and, by in vitro and in silico methods, to determine whether these compounds and/or their conjugates might act as novel ligands at CBRs. Employing a multi-disciplinary approach, we demonstrate that isoprenylated stilbenoids produced and purified from a highly controlled peanut hairy root culture system exhibit slower metabolism and retain biological activity relative to their non-prenylated parent compounds. Perhaps more importantly, we also present compelling evidence that tRes, and its analogs, might produce a subset of their actions via interaction with CBRs as novel molecular targets.
Materials and methods
Materials
All chemicals used for this study were of at least reagent grade. Ethyl alcohol (95%, v/v) was purchased from AAPER (Shelbyville, KY, USA). tRes and tPice were of analytical grade (99%, w/w) and purchased from Sigma-Aldrich (St. Louis, MO, USA). Recombinant UGT2B4, 2B7, 2B15 and 2B17 were provided by BD Biosciences (San Diego, CA, USA). All other chemicals used in this study were obtained from Tocris Bioscience (Ellisville, MO, USA). [3H]CP-55,950 [(−)-cis-3-[2-Hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol] (168 Ci/mmol) and [35S]GTPγS (1250 Ci/mmol) were purchased from Perkin Elmer (Boston, MA, USA). Resveratrol 3-O-D-glucuronide (CAS 387372-17-0) and resveratrol 4′-O-D-glucuronide (CAS 387372-20-5) were purchased from Cayman Chemical (Ann Arbor, MI, USA). The additional reagents were purchased from Fisher Scientific Inc. (Pittsburgh, PA, USA).
Cell culture
Stably transfected CHO-hCB2 cells were generated in our laboratory (Shoemaker et al. 2005). Cells were incubated in a humidified atmosphere of 5% CO2, 95% air at 37°C in DMEM with 10% foetal calf serum, 100 units/mL penicillin, 100 mg/mL streptomycin and 2.5 mg/mL geneticin.
Bioproduction and purification of tA1 and tA3
tA1 and tA3 were purified from sodium acetate-elicited hairy root cultures of peanut as recently described (Abbott et al. 2010). Briefly, 9-day hairy root cultures of peanut line 3 (Condori et al. 2010) were elicited with 10 mM sodium acetate (Medina-Bolivar et al. 2007; Medina-Bolivar et al. 2010) for 48 h. During the elicitation period, packs containing DIAION HP-20 resin were included to trap the secreted polyphenols from the culture medium. The polyphenols were eluted from the resin with ethyl acetate and the organic phase was then dried in a rotavapour. The extract was fractionated by centrifugal partition chromatography as described in the study by Abbott et al. (Abbott et al. 2010). Fractions containing tA1 and tA3 were analysed by high-performance liquid chromatography (HPLC) and their identity was confirmed by UV spectra and mass spectrometry (MS) analyses as described earlier (Abbott et al. 2010; Condori et al. 2010). Purity of hairy root-derived tA1 and tA3 was >90% based on the HPLC area percent measurements performed at 320 nm.
Preparation of recombinant UGTs
Membrane fractions from baculovirus-infected insect cells expressing individual recombinant human UGTs were obtained from Dr. Moshe Finel and prepared as previously described (Kurkela et al. 2003; Kuuranne et al. 2003). Each enzyme tested in this study is known to be active toward substrates specific for that isoform. The expression level of individual recombinant UGTs was estimated by western blot analyses using monoclonal antibodies (Tetra-His antibodies; Qiagen, Hilden, Germany) against the His-tag they all carry (Kurkela et al. 2003). For activity comparison between individual UGTs, the enzyme level was normalised as previously described (Kuuranne et al. 2003).
Recombinant UGT isoform activity
UGT activity was assayed as described in detail previously (Little et al. 2004). Briefly, UGT recombinant membrane protein (5 µg) was incubated in 100 µM Tris–HCl (pH 7.4)/5 mM MgCl2/5 mM saccharolactone with 250 µM substrate for screenings and 100–2000 µM substrate for kinetic assays, in a total volume of 30 µL. Substrates were added in DMSO with a final concentration of 2%, and controls omitting substrates were run with each assay. All incubations were performed in duplicate, and no additional detergents or other activators were used in the incubations. All UGT2B enzymes were obtained from BD Biosciences and assayed according to manufacturer protocols. Reactions were started by the addition of the appropriate UDP-GlcUA co-substrate (4 mM) and incubated at 37°C. Reactions were stopped by addition of 40 µL of ethanol.
HPLC analyses
HPLC methods were elaborated for the separation and quantification of stilbenoid glucuronides from enzymatic assays. Analyses were performed using an HP 1050 HPLC system equipped with a UV-Vis diode array detector. Instrument operation and data acquisition were controlled through the Agilent ChemStation software package. Samples were separated using a Supelcosil LC-18 (25cm × 4.6 mm, 5 µm) column maintained at 37°C. The solvent system consisted of 0.1% (v/v) acetic acid in water (A) and methanol (B) at a flow rate of 1 mL/min. The separation of the stilbenoid and its glucuronide(s) was achieved using the following elution gradient: 100% A (5 min), linear gradient from 100% A to 100% B (5–25 min). The column was then re-equilibrated at initial conditions for 10 min between runs. The elution of each metabolite was monitored at 313 nm (tRes and tPice) or 335 nm (tA1 and tA3). Primary standards for the glucuronidated metabolites were not available; therefore, product concentrations were semi-quantified using the responses for external standards of each stilbenoid substrate. It has been shown previously that the addition of the glucuronic acid moiety does not alter the extinction coefficients from that of the unreacted substrate (Doerge et al. 2000).
A second HPLC method was used to confirm the identity of reaction products using UV spectra. Aliquots of the glucuronidation reaction products containing glucuronides and their aglycons were analysed by reverse-phase HPLC as described previously (Condori et al. 2009). HPLC profiles were monitored at 320 nm and the UV spectra of the glucuronides and their aglycons were compared with that of authentic aglycon standards.
HPLC-MS analyses of glucuronides
The HPLC-PDA/MS analytical system employed in this study consisted of two Prostar™ 210 HPLC pumps (Varian Inc., Water Creek, CA, USA), which were serially connected to a Prostar™ 335 PDA detector and a Varian® 1200 L Quadrupole LC/MS/MS system (Varian Inc.). An auto sampler (Prostar™ model 420) from the same source was connected for sample delivery. The HPLC/MS system was operated and the data were collected using Prostar/Dynamax™ software (version 2.4) (Varian, Walnut Creek, CA), whereas the HPLC/PDA was managed by Polyview 2000 (version 6.9) (Varian, Walnut Creek, CA). This study used a Sunfire™ C18 column (4.6 mm i.d. × 250 mm) purchased from Waters Inc. (Taunton, Ireland). The mobile phase was composed of 0.05% (v/v) formic acid in water (A) and 99:1 (v/v) mixture of acetonitrile and water containing 0.05% (v/v) of formic acid (B) at a flow rate of 0.5 mL/min. Both the solvents were filtered through a 0.45-µm porous micro filter before use. The gradient run from A (90%) was changed to 82% in 8 min and maintained for 2 min (up to 10 min). Then, it was changed to reach 75% at 15 min followed by 65% at 18 min. This percentage was maintained till 40 min and was lowered to 40% at 99 min. At 105 min, it was raised to reach 90% and maintained the same until 110 min. The injection volume was 10 µL per sample. The HPLC-MS analysis was performed using electrospray ionisation (ESI) technique in negative ion mode. A scan mode at a range of m/z 120–700 was used. The temperatures of manifold, atomic pressure ionisation (API) housing and API drying gas were adjusted to 42°C and 65°C, respectively. Air was used as the API nebulising gas at a pressure of 42 psig. The heated capillary was set to 200°C, the electrospray voltage to 2.8 kV and the sheath and auxiliary gas to 75 and 20 arbitrary units, respectively.
Tandem MS analysis
The Varian 1200 L Quadrupole LC/MS/MS system was operated in the ESI mode to obtain both the mass spectra (MS1) and the product ion spectra (MS2) of the stilbenoid and glucuronidated products. Polypropylene glycol was used to calibrate the instrument for both the negative and positive ionisation. The tuning and MS/MS spectra were obtained for the target analytes by direct infusion of ~25 µmol/mL glucuronidated and aglycon standard solutions in mobile phase A, at a flow rate of 20 µL/min with a Harvard syringe pump (Bonaduz AG, Bonaduz, Switzerland). The ionised molecules were fragmented by collision-activated dissociation. Argon was used as the collision gas at a pressure of 2.5 mTorr. The MS/MS detections were carried out in a multiple reaction monitoring (MRM) mode. The ESI-MS/MS detections were carried out using a heated capillary set at 225°C, with the electrospray voltage at 2.8 kV and the sheath and auxiliary gases adjusted to 65 and 20 arbitrary units, respectively. The API source was maintained at 65°C. Initially, during the method development, the mass spectrometer was used in scan mode, followed by collision-induced dissociation method.
Membrane preparation
Brain tissue was collected from decapitated male and female B6SJL mice obtained from an in-house breeding colony. Whole brains were pooled before beginning homogenisation. Pellets of frozen/thawed cells or freshly harvested brain tissue were resuspended in a homogenisation buffer containing 50 mM Hepes pH 7.4, 3 mM MgCl2 and 1 mM EGTA. Using a 40-mL Dounce glass homogeniser (Wheaton, PA, USA), samples were subjected to 10 complete strokes and centrifuged at 40,000g for 10 min at 4°C. After repeating the homogenisation procedure twice, the samples were resuspended in Hepes buffer (50 mM, pH 7.4) and subjected to 10 strokes utilising a 7-mL glass homogeniser. Membranes were stored in aliquots of ~1 mg/mL at ~−80°C.
Competition receptor binding
Increasing concentrations of WIN-55,212-2 or different tRes analogs/conjugates were incubated with 0.2 nM of [3H]CP-55,940 in a final volume of 1 mL of binding buffer as described previously (Shoemaker et al. 2005), with slight modification. Each binding assay contained 100 (mouse brain) or 25 (CHO-hCB2) µg of membrane protein. tRes analogs/conjugates and membranes were preincubated in the dark for 15 min at 37°C in the absence of [3H]CP-55,940. After cooling for 30 min at room temperature, [3H]CP-55,940 was added and reactions were incubated for 90 min at room temperature, in the dark with mild agitation.
Non-specific binding was defined as binding observed in the presence of 1 µM of non-radioactive WIN-55,212-2. Reactions were terminated by rapid vacuum filtration through Whatman GF/B glass fibre filters followed by two washes with ice-cold binding buffer. Analysis of the binding data was performed using the non-linear regression (Curve Fit) function of GraphPad Prism® v4.0b (GraphPad Software Inc., San Diego, CA, USA) to determine the concentration of the drug that displaced 50% of [3H]CP-55,940 (IC50). A measure of affinity (Ki) was derived from the IC50 values utilising the Cheng–Prushoff equation (Cheng & Prusoff 1973).
[35S]GTPγS binding
[35S]GTPγS binding assays were performed with minor modifications as described previously (Shoemaker et al. 2005) in a buffer containing 20 mM Hepes (pH 7.4), 100 mM NaCl, 10 mM MgCl2 and 0.1% bovine serum albumin. Each binding reaction contained 25 µg of mouse brain membrane protein, cannabinoid ligands, 0.1 nM of [35S]GTPγS and 10 µM of GDP. Non-specific binding was defined by 10 µM of non-radioactive GTPγS. Following incubation in the dark at 30°C for 30 min, reactions were terminated by filtration and bound radioactivity determined by liquid scintillation counting.
Data analysis
Curve-fitting and statistical analyses were conducted utilising GraphPad Prism® v4.0b. Data obtained from three or more experimental groups were analysed by a one-way analysis of variance, followed by a Dunnett’s post-hoc comparison of individual groups. A non-paired Student’s t-test was employed to statistically compare data obtained from two experimental groups.
Kinetic constants for the glucuronidation of tRes, tPice, tA1 and tA3 by recombinant UGT1A1, -1A7 and 1A10 were obtained by fitting experimental data to the following kinetic models using the non-linear regression (Curve Fit) function of GraphPad Prism 4.
Michaelis–Menten (M–M) equation for one-enzyme model:
Hill equation, which describes sigmoidal autoactivation kinetics, where S50 is the substrate concentratoin at 50% Vmax (analogous to Km in M–M kinetics), and n is the Hill coefficient, which can be considered to be a measure of autoactivation and reflects the extent of co-operativity among multiple binding sites (Weiss 1997).
Uncompetitive substrate inhibition (USI) model, where Ki is the inhibition constant describing the reduction in rate:
Biphasic model, where CLint2 represents the slope of the linear portion:
Goodness of fit for each model to glucuronidation kinetic data was assessed from the standard error, 95% confidence intervals and R2 values. Kinetic curves were also analysed as Eadie–Hofstee plots to support kinetic models. Kinetic constants were reported as the mean ± standard error of the parameter estimated.
Molecular modelling
For molecular modelling, we made use of an extensively developed and tested protocol (RY Patel & RJ Doerksen, unpublished). We constructed multiple three-dimensional protein models for CB2R, based on a bovine rhodopsin X-ray structure (PDB:1F88), using Modeller software (Sali & Blundell 1993).
To select the best protein model, we first docked 12 compounds shown in Table 4 into 132 CB2R protein models, using the Glide docking program and Glide scoring function (2010). After the best model was found, three additional 4′-glucuronidated derivatives were docked into that model. For comparison, we also docked N′-[(3Z)-1-(4-chlorobenzyl)-2-oxo-1,2-dihydro-3H-indol-3-ylidene]benzohydrazide, a reported (Diaz et al. 2008) CB2R agonist. Some evidence has been reported of the particular importance of K109 and S285 for binding of CB2R ligands (Reggio 2010). In this work, we selected S285 as a central point for docking. During the docking, we used a hydrogen bonding restraint between S285 and the ligand to be docked. We calculated the root-mean-squared deviation (RMSD) of the common atoms of the ligands (the 12 carbon atoms of the two phenyl rings, the two carbons connecting these rings and the hydroxyl O at position 4′), relative to the binding pose of whichever docked ligand had the best G-score for a particular CB2R structure and used the RMSD to monitor how consistently the ligands bound to the various models. Low RMSD of the common atoms of the docked ligands means that all the ligands are docked in more or less the same orientation in a given protein model. (Note that in vivo the molecules do not necessarily all bind in a similar binding mode, but given the lack of reliable data on the binding mode, we feel it is a fair assumption that this RMSD should be minimised.) Using the RMSD and visual inspection of the docking poses, we selected one particularly good model and present the results for it.
Table 4.
Glide GScore docking scores and experimental Ki values.
| Ligand | Glide GScore | Kia |
|---|---|---|
| N-alkyl isatin benzohydrazide | −8.18 | b |
| 3-glucuronide-trans-arachidin-3c | −7.51 | — |
| trans-arachidin-2 (tA2)c | −7.48 | — |
| trans-arachidin-3 (tA3)c | −7.11 | 11.3 ± 0.6 |
| 3-glucuronide-trans-arachidin-2c | −7.00 | — |
| trans-arachidin-1 (tA1)c | −6.95 | 12.6 ± 1.7 |
| 3-glucuronide-trans-arachidin-1c | −6.88 | — |
| trans-resveratrol (tRes)c | −6.66 | 65.5 ± 12.4 |
| 4′-glucuronide-trans-arachidin-3 | −6.54 | — |
| trans-piceatannol (tPice)c | −6.27 | 114 ± 7.8 |
| 4′-glucuronide-trans-piceatannolc | −6.03 | — |
| 3-glucuronide-trans-resveratrolc | −5.95 | — |
| 3-glucuronide-trans-piceatannolc | −5.86 | >100 |
| 4′-glucuronide-trans-arachidin-2 | −5.80 | — |
| 4′-glucuronide-trans-resveratrolc | −5.71 | >100 |
| 4′-glucuronide-trans-arachidin-1 | −5.06 | — |
— No data.
In µM.
IC50 = 131.1 (±1.7) nM for N′-[(3Z)-1-(4-chlorobenzyl)-2-oxo-1,2-dihydro-3H-indol-3-ylidene] benzohydrazide (54 in Ref. [Diaz et al. 2008]).
Compounds docked for the selection of the protein model.
Results
Glucuronidation activity of human recombinant UGTs toward tRes, tPice, tA1 and tA3
As an initial screening for glucuronidation activity toward tRes, tA1, tA3 and tPice, 12 human recombinant UGTs from the 1A family (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9 and 1A10) expressed as His-tag proteins in baculovirus-infected Sf9 insect cells, as well as UGT2B4, 2B7, 2B15 and 2B17 from BD Biosciences, were evaluated for their ability to glucuronidate these four compounds (250 µM) under standard incubation conditions described in Section Methods. No activity was seen with UGT1A4, 1A6, 2B4, 2B7, 2B15 or 2B17 toward any substrate assayed.
Glucuronides of the two prenylated stilbenoids, tA1 and tA3, were formed (Figure 1). A single glucuronide product of tA3 was formed, and the site of the glucuronidation was suggested by MS to be at the–OH group on the 4′ carbon forming tA3–4′G. Two glucuronide products were seen with tA1: the major product was found to be in the 4′ position, tA1–4′G, and a second product seen in reactions with UGT1A7 and HLMs (minor product) and with human intestinal microsomes (HIMs) reactions (major product) was suggested to be tA3–3G.
The major isoforms involved in the glucuronidation of both compounds were extrahepatic UGT1A7 and UGT1A10, with higher activity shown toward tA1 for these isoforms. The contribution of hepatic isoforms was much less pronounced with UGT1A9 glucuronidating both substrates and UGT1A1 producing only a small amount of tA3–4′G. The HLMs used in these studies showed high activity and preferential glucuronidation of tA1 over tA3.
Glucuronidation reactions with tRes and tPice each resulted in the production of two distinct products (Figure 1). On the basis of the literature (Brill et al. 2006; Sabolovic et al. 2006), the major product of tRes glucuronidation was assigned as tRes-4′G and the minor as tRes-3G. The position of the two tPice glucuronide moieties could not be assigned by MS due to the similar nature of all the possible positions; however, we tentatively assigned the major and minor products as tPice-4′G* and tPice-3G*, respectively, based on their similarity to the tRes products seen.
The major isoforms involved in the glucuronidation of tRes and tPice were UGT1A1, which is predominantly hepatic, and UGT1A7 and UGT1A10, which are exclusively extrahepatic. For all active enzymes, other than UGT1A10, which favoured the production of the–4′G form (Figure 1), the–3G glucuronide products were the most highly produced.
Glucuronidation activity of hepatic and intestinal microsomes toward tRes, tPice, tA1 and tA3
As an initial screening for glucuronidation activity toward tRes, tA1, tA3 and tPice, HLMs (from one donor; HL2) and HIMs (from two donors; HI36 and HI41) were evaluated for their ability to glucuronidate these four compounds (250 µM) under standard incubation conditions described in the Methods section.
The activities for tRes and tPice for both HLM and HIM are high and vary in the proportions of 3-O- and 4′-O-Glucs produced. For each, the products produced fit well with the products expected based on the screenings of liver- and intestine-specific isoforms (i.e. UGT1A1 and 1A10). As with the individual isoforms, glucuronidation of tA3 yields a single product.
Our HIM show the highest activity toward tA1. However, the major product observed is not the major product of any of the UGT isoforms under investigation in this work, suggesting that another yet unidentified isoform is involved. Further experiments will be needed to reveal the source of this activity.
Steady-state kinetics for tRes, tPice, tA1 and tA3 glucuronidation by selected recombinant UGTs
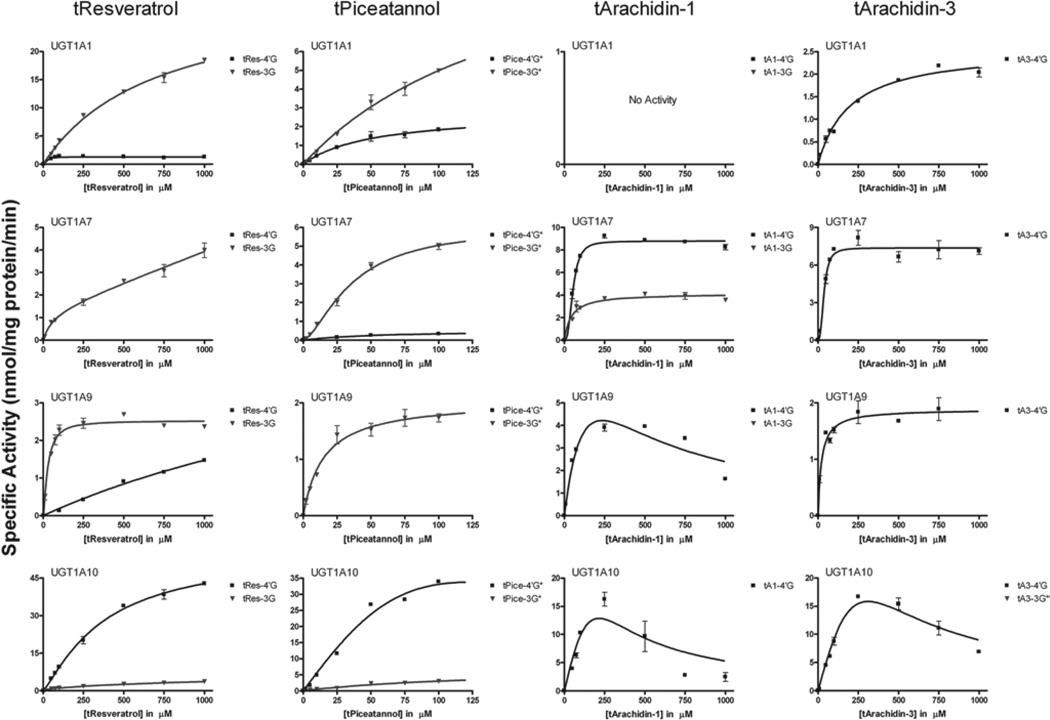
On the basis of our specific activity screen, UGT1A1, -1A7, -1A9 and -1A10 were selected for further catalytic studies to determine the respective steady-state parameters for tRes, tA1, tA3 and tPice glucuronidation (Figure 2). Eadie–Hofstee plots were analysed (data not shown) and kinetic parameters of glucuronidation and mathematical models used to fit each data set are presented in Table 1.
Figure 2.
Kinetic constants for glucuronidation. Recombinant human UGT1A1 and -1A10 were assayed with increasing concentrations of substrate. Black indicates 4′-O-Gluc and blue indicates 3-O-Gluc.
Table 1.
Steady-state parameters for glucuronidation of tRes, tPice, tA1 and tA3 by UGT1A1, -1A7, -1A9 and -1A10. (See Equations in Data Analysis section).
| 1A1 | 1A7 | 1A9 | 1A10 | |
|---|---|---|---|---|
| tResveratrol | ||||
| tRes-4'G | ||||
| Km (µM) or S50 (µM) | 25.3 ± 4.8 | 792 ± 240 | 300 ± 54 | |
| Vmax (nmol/mg/min) | 1.3 ± 0.05 | 2.5 ± 0.5 | 49 ± 4.2 | |
| N | 2.2 | 1.4 | 1.3 | |
| Kinetic model | Hill | Hill | Hill | |
| R2 | 0.9396 | 0.9945 | 0.9883 | |
| tRes-3G | ||||
| Km (µM) or S50 (µM) | 708 ± 87 | 52.3 ± 42 | 28.9 ± 3.3 | 489 ± 72 |
| Vmax (nmol/mg/min) | 31 ± 2.0 | 1.4 ± 0.5 | 2.5 ± 0.07 | 12 ± 0.8 |
| N | 1.5 | |||
| CLint2 | 2.7 ± 0.4 | |||
| Kinetic model | M-M | Biphasic | Hill | M-M |
| R2 | 0.9941 | 0.9734 | 0.9692 | 0.9852 |
| tPiceatannol | ||||
| tPice-4'G* | ||||
| Km (µM) or S50 (µM) | 51 ± 16 | 43.5 ± 8.1 | 15.1 ± 2.7 | 14.1 ± 5.7 |
| Vmax (nmol/mg/min) | 2.8 ± 0.4 | 0.46 ± 0.03 | 2.1 ± 0.1 | 26 ± 0.8 |
| N | 4.1 | |||
| Kinetic model | M-M | M-M | M-M | Hill |
| R2 | 0.9584 | 0.9705 | 0.9635 | 0.9804 |
| tPice-3G* | ||||
| Km (µM) or S50 (µM) | 171 ± 67 | 32.8 ± 2.4 | 44.5 ± 10.9 | |
| Vmax (nmol/mg/min) | 13.5 ± 3.8 | 5.7 ± 0.2 | 3.9 ± 0.3 | |
| N | 1.7 | |||
| Kinetic model | M-M | Hill | M-M | |
| R2 | 0.9796 | 0.9915 | 0.9381 | |
| tArachidin-1 | ||||
| tA1–4'G | ||||
| Km (µM) or S50 (µM) | 52.9 ± 3.2 | 188 | 23082 | |
| Vmax (nmol/mg/min) | 8.8 ± 0.2 | 11 | 2766 | |
| N | 2.8 | |||
| Ks | 296 | 2.0 | ||
| Kinetic model | Hill | USI | USI | |
| R2 | 0.9799 | 0.9075 | 0.7467 | |
| tA1–3G | ||||
| Km (µM) or S50 (µM) | 54.4 ± 4.5 | |||
| Vmax (nmol/mg/min) | 3.9 ± 0.1 | |||
| N | 1.8 | |||
| Kinetic model | Hill | |||
| R2 | 0.9642 | |||
| tArachidin-3 | ||||
| tA3–4'G | ||||
| Km (µM) or S50 (µM) | 196 ± 24 | 36.7 ± 5.9 | 21.3 ± 5.2 | 81173 |
| Vmax (nmol/mg/min) | 2.6 ± 0.1 | 3.7 ± 0.1 | 1.9 ± 0.08 | 8266 |
| N | 2.6 | |||
| Ks | 1.2 | |||
| Kinetic model | M-M | Hill | M-M | USI |
| R2 | 0.9829 | 0.9473 | 0.9324 | 0.9490 |
Parameters determined using the software GraphPad Prism 4.
tRes, trans-Resveratrol; tPice, trans-piceatannol; tA1, trans-arachidin-1; tA3, trans-arachidin-3; UGT, UDP-glucuronosyltransferases; USI, uncompetitive substrate inhibition.
Comparison of the steady-state kinetic parameters for each of the compounds with UGT1A1 demonstrates dramatic differences in both capacity as well as affinity for these substrates. There is no visible glucuronidation of tA1 by this isoform, and only the 4′-O-Gluc of tA3 is formed (Vmax = 2.6 nmol/mg protein/min). This is in striking contrast with the glucuronidation of both tRes and tPice where two products are formed with the predominant one being the 3-O-Gluc (Vmax = 31 µM and 13.5 nmol/mg protein/min, respectively). Interestingly, the Vmax values of the 3-O-products for these two compounds are very similar to that for tA1 (1.3 nmol/mg protein/min and 2.6 nmol/mg protein/min, respectively). The presence of the prenyl group in tA1 totally prevents the glucuronidation of this compound. In tA3, the addition of the prenyl group appears to block only the formation of the major product of tRes glucuronidation. The rank order of the Km values for these compounds with UGT1A1 is tRes < tPice < tA3, indicating decreased affinity for the latter compounds. Both of these parameters indicate that the addition of the prenyl group would be predicted to slow the metabolism of these compounds in vivo as compared with tRes.
The contribution of the intestinal isoform, UGT1A10, to the metabolism of these compounds is also striking. UGT1A10 metabolises all four of these compounds with the highest activity observed thus far for recombinant isoforms; however, the major product of this reaction is the 4′-O-Gluc. For tRes and tPice, the Vmax values for the formation of the major products are 49 and 26 nmol/mg protein/min, respectively. Reactions of tA1 and tA3 with this isoform exhibited strong USI. A maximum activity of ~15 nmol/mg protein/min was observed for each reaction at ~250 µM. At higher concentrations, marked substrate inhibition was observed.
Extrahepatic UGT1A7 is also involved in the glucuronidation of our model compounds. The capacity and affinity of this isoform for all of the substrates is similar (Vmax = 1.4–8.8 nmol/mg protein/min; Km 36.7–54.4 µM), although for tRes and tPice, the 3-O-Gluc is the major/only product, and for tA1 and tA3, the 4′-O-Gluc is preferred.
A small contribution from hepatic UGT1A9 is also observed with a pattern similar to that seen for UGT1A1. The biggest difference in these two isoforms is the production of the tA1–4′Gluc by UGT1A9, which is not produced by UGT1A1. In fact, the activity of UGT1A9 toward this substrate is the highest of the four substrates assayed. Also, significant substrate inhibition is observed in this reaction with a maximum activity of ~4 nmol/mg protein/min and inhibition seen at a concentration >500 µM.
MS confirmation of glucuronide formation and conjugation position prediction
The glucuronidated products of tRes, tPice and their corresponding 4-isoprenylated analogs tA3 and tA1, after incubation with UGTs, were identified by reverse-phase HPLC–photodiode array and MS analyses. Our HPLC method allowed for the separation of the non-polar stilbenoids from their more polar glucuronidated products. UV spectral analysis was performed as an initial confirmation of the glucuronidates since similar UV spectra are expected for the glucuronides and their parental stilbenoid aglycons. After these initial studies, LC-MS analyses were conducted to confirm the presence of monoglucuronides. A second MS analysis was performed to identify the most abundant fragmentation ions and potentially predict the position of the glucuronic acid in the conjugated product. tRes was used as a control because its glucuronidation pattern has been extensively studied in vitro. Similar to previous observations (Brill et al. 2006; Sabolovic et al. 2006), two glucuronidated products were identified using HPLC and MS after incubation of tRes with UGT1A7 and UGTA10.
tPice glucuronide analysis
The HPLC chromatogram of the glucuronidated products of tPice after incubation with UGT1A7 and UGT1A10 is shown in Supplementary Figure S1. After incubation with UGT1A7, one major product (Rt 26.03 min), along with the non-glucuronidated tPice (Rt 28.77 min), were observed, whereas the reaction of tPice with UGT1A10 resulted in the emergence of two products (Rt 23.6 and 26.03 min) along with tPice (28.77 min). The new peak (Rt 23.6 min) formed by UGT1A10 appeared to be unique and was higher in response indicating an increased quantity. The UV spectra of the peaks observed were similar (Figure S1) suggesting that they all share the tPice structure as a backbone. MS fragmentation was carried out using the developed method. The peak eluting at 28.77 min gave a base peak m/z ion (243.1), indicating [M-H]− of tPice (S1). When the standard solution of tPice prepared in 0.1% formic acid was directly injected into the MS, four prominent fragment ions were observed as presented in Table 2. From the fragments, a possible fragmentation pattern was charted out. The fragmentation of the peaks eluting at 23.60 and 26.03 min were found to give similar m/z ions at 419.2 indicating them to be monoglucuronides of piceatannol. A direct injection of a tPice sample, incubated with UGT1A10 for 3 h, was slowly infused into the MS using the MRM mode. The fragments formed during MS3 fragmentation studies were charted into a scheme as given as supplementary Figure S2. The parent ion m/z = 419 gave fragments at m/z = 175 and 113, which were typical of the glucuronic acid moiety.
Table 2.
Most frequent abundant ions of selected stilbenoid and their glucuronides upon ionisation.
| Compound | MW | UGT | Parent ions | (m/z−) most abundant ions | |||||
|---|---|---|---|---|---|---|---|---|---|
| tPice | 244 | 242.5 | 224.5 | 200.5 | 158.6 | ||||
| tPice-Gluc | 420 | 1A10 | 418.8 | 242.6 | 208.3 | 174.2 | 112.8 | ||
| 289.0 | 242.9 | 174.2 | 112.6 | ||||||
| 242.8 | 200.6 | 199.4 | 158.7 | ||||||
| tA1 | 312 | 310.7 | 266.6 | 254.6 | 241.8 | 240.6 | 158.6 | ||
| tA1-Gluc | 488 | 1A10 | 486.9 | 311.0 | 255.0 | 240.6 | 174.7 | 112.6 | |
| 311.8 | 293.7 | 231.7 | 190.4 | 175.6 | |||||
| 1A7 | 487.2 | 310.5 | 254.4 | 174.5 | 114.7 | 112.9 | |||
| 293.7 | 190.4 | 175.5 | 132.6 | ||||||
| tA3 | 296 | 294.7 | 250.7 | 239.8 | 238.6 | 194.7 | 175.0 | 112.8 | |
| tA3-Gluc | 472 | 1A10 | 470.8 | 299.6 | 294.7 | 112.7 | 100.7 | ||
| 340.7 | 178.6 | 118.7 | |||||||
| 1A7 | 470.8 | 294.6 | 238.9 | 174.5 | 112.8 | ||||
| 340.7 | 178.6 | 160.5 | 118.7 | 112.7 | |||||
tPice, trans-piceatannol; tA1, trans-arachidin-1; tA3, trans-arachidin-3; MW, molecular weight; UGT, UDP-glucuronosyltransferases.
tA1 glucuronide analysis
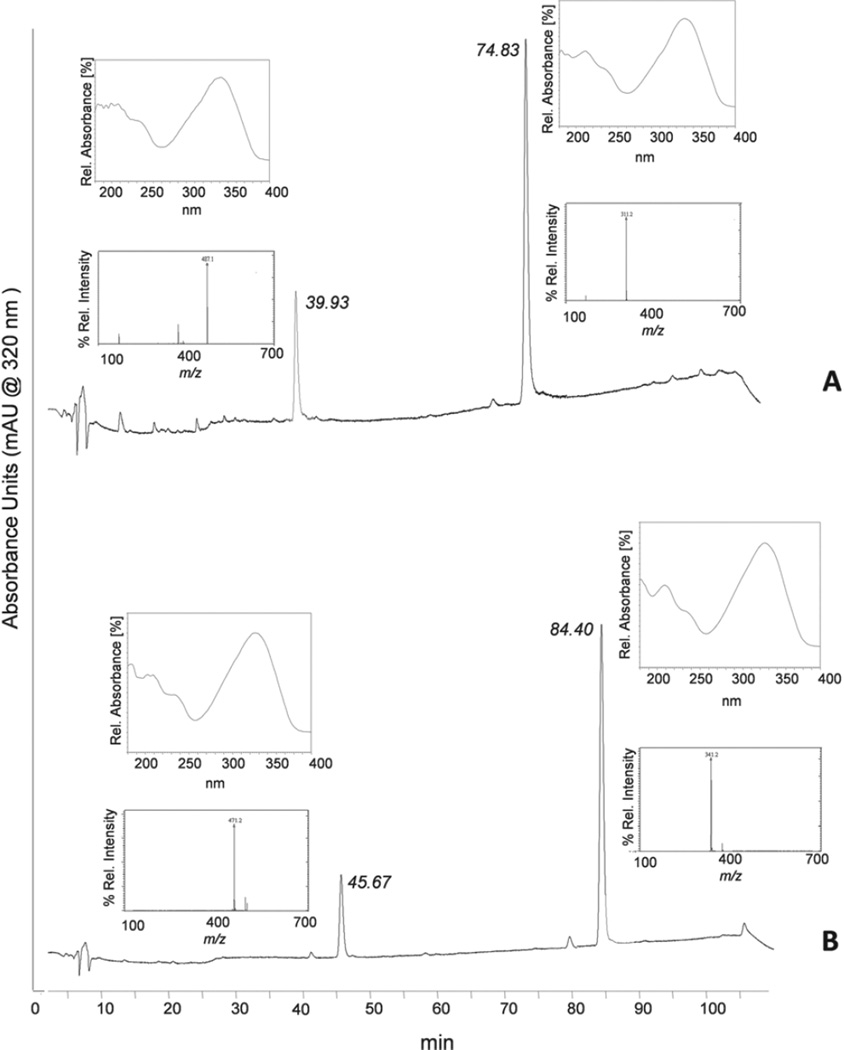
The tA1 sample was incubated separately with UGT1A7 and UGT1A10 for 3 h and aliquots were analysed using the developed HPLC method. The chromatogram shown in Figure 3A indicates the appearance of tA1 at Rt = 74.83 min and a second peak at Rt = 39.93 min. The UV spectra of these peaks were similar indicating a similar tA1 backbone. The MS fragmentations of the corresponding peaks were performed as shown in Figure 3A. The MS spectrum of the peak at Rt = 74.8 min gave an ion at m/z = 311.2 corresponding to tA1, whereas the peak at Rt = 39.9 yielded an ion at m/z = 487.1, which was indicative of a monoglucuronide of tA1. Each of the samples of tA1 incubated with UGT1A7 or UGT1A10 gave similar concentrations of monoglucuronide peaks with almost the same retention time. Hence, glucuronidation samples were incubated for 3 h and separately infused into the MS under the MRM mode to study their fragmentation pattern. The fragments formed by the parent ions at m/z = 487 were similar to those presented in Table 2. On the basis of the fragments obtained, a tentative scheme was charted as shown in Figure 4A, which suggests that tA1 is glucuronidated at the 4′ position.
Figure 3.
Analysis of trans-arachidin-1 (tA1) and trans-arachidin-3 (tA3) glucuronidated products produced by UGT1A7. Aliquots of the glucuronidation reaction products were analysed by reverse-phase HPLC-PDA and LC-MS. HPLC profiles were monitored at 320 nm. The MS analysis was carried out using electrospray ionisation (ESI) under negative ion mode. HPLC profiles show peaks of (A) tA1 (Rt: 74.83 min) and tA1 glucuronide (Rt: 39.93 min) and (B) tA3 (Rt: 84.40 min) and tA3 glucuronide (Rt: 45.67). Inserts next to these peaks show their UV spectrum (above) and mass spectrum (below).
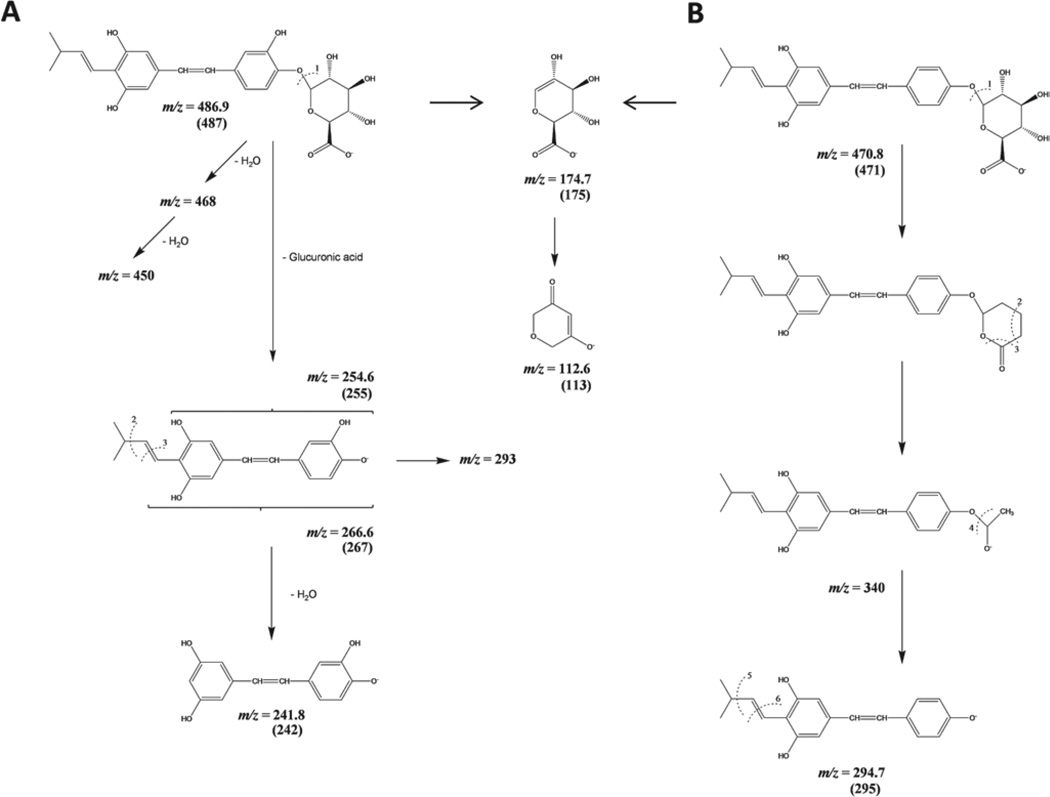
Figure 4.
Proposed MS fragmentation pattern of (A) trans-arachidin-l 4′-glucuronide and (B) trans-arachidin-3 4′-glucuronide.
The tA1 sample was also incubated with two HIMs (HI36 and HI41) for 3 h and aliquots were analysed by HPLC. The chromatogram indicated the appearance of tA1 as well as two product peaks with the minor peak being the one identified with UGT1A7 and 1A10, and a new major peak eluting later. The UV spectra of these peaks were similar indicating they are both products of the reaction and contain a tA1 backbone.
tA3 glucuronide analysis
The samples of tA3, incubated separately for 3 h with UGT1A7 and UGT1A10, were analysed by employing HPLC and MS. Both enzyme treatments formed a new peak (Rt 45.67 min) of almost similar ratio (Figure 3B). The peak formed in both the samples at Rt = 84.53 min gave an ion at m/z = 341.2, which corresponded to the formic acid adduct of tA3. The MS spectral analysis of the new peak indicated the formation of a tA3 monoglucuronide as indicated by the ion at m/z = 471.2.
The sample of tA3, incubated with UGT1A7 for 3 h, was slowly infused into the MS and fragments were observed in the MRM mode. The prominent fragments formed are listed in Table 1 and are based on the schematic fragmentation pattern presented. Figure 4B illustrates the possible fragmentation pathway of tA3 4′-glucuronide with typical fragments of the glucuronic acid moiety.
Affinity of trans-arachidins, tRes and tPice for CB1Rs and CB2Rs
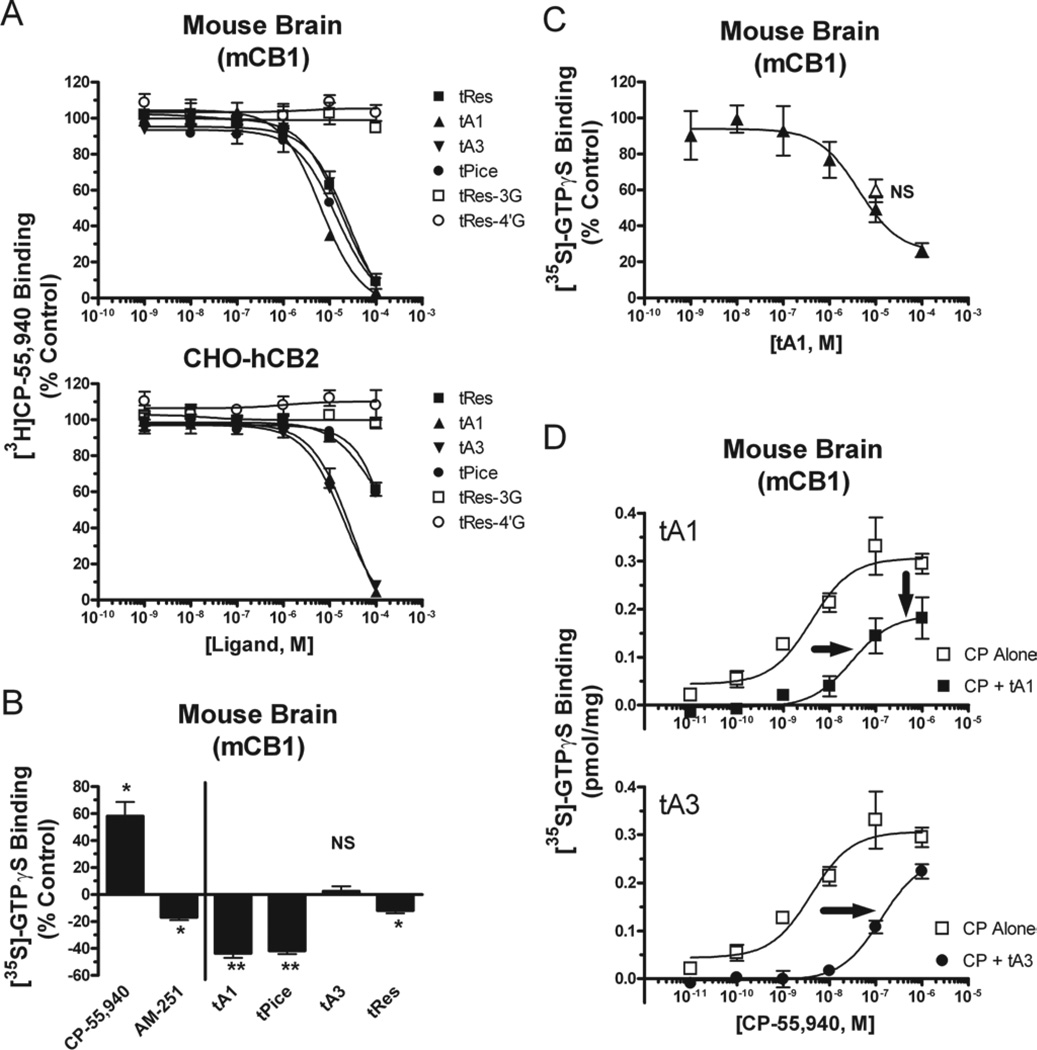
Cannabinoids acting at both types of CBRs produce antioxidant (Hampson et al. 1998) and anti-inflammatory (Klein 2005) effects similar to those reported for tRes. We, therefore, hypothesised that tRes and structurally related analogs might produce a subset of the reported effects through actions at CBRs. As such, we first conducted competition receptor binding studies to determine the affinity (Ki) of tRes and all analogs for mouse brain CB1Rs (mCB1Rs) and human CB2Rs (hCB2Rs) stably expressed in CHO cells. The well-characterised CBR agonist WIN-55,212-2 (Mackie & Hille 1992) binds non-selectively to both mCB1Rs and hCB2Rs with high affinity in the 3–6 nM range (Table 3). tRes and all analogs also bind to mCB1Rs, but with lower affinity relative to WIN-55,212, exhibiting Ki values in the 5–18 µM range (Figure 5A, upper panel) and a rank order of tA1 > tPice ≥ tRes = tA3 (Table 3). Interestingly, while the isoprenylated tRes derivatives tA1 and tA3 also bind to hCB2Rs with similar affinity to that observed for mCB1Rs (Figure 5A, lower panel), the affinity of tRes and tPice for hCB2Rs is 5- to 10-fold lower than that observed for mCB1Rs. In addition, all compounds except for tA3 exhibit approximately 2- to 10-fold selectively for binding to mCB1Rs relative to hCB2Rs (Table 3). Neither the 3- nor the 4′-O-D-glucuronidated metabolite of tRes binds to either mCB1Rs or hCB2Rs when tested at up to 100 µM concentrations (Figure 5A, Table 3). These studies clearly establish that while the isoprenylated tRes analogs tA1 and tA3 bind to both mCB1Rs and hCB2Rs with low µM affinity, the parent compounds tRes and tPice bind appreciably only to mCB1Rs.
Table 3.
Affinity of trans-arachidins for mCB1 and hCB2 receptors.
| Ki (µM) | ||||
|---|---|---|---|---|
| Drug | mCB1 | hCB2 | hCB2/mCB1 | Selectivity |
| WIN-55,212-2 | 0.0034 ± 1.6 (5) | 0.0058 ± 1.2 (4) | 1.7 | Non-selective |
| tA1 | 5.6 ± 0.4 (3) | 12.6 ± 1.7 (3) | 2.3 | Non-selective |
| tA3 | 17.5 ± 3.4 (3) | 11.3 ± 0.6 (3) | 0.6 | Non-selective |
| tRes | 14.3 ± 1.6 (5) | 65.5 ± 12.4 (3) | 4.6 | CB1 |
| tPice | 11.7 ± 1.0 (3) | 114 ± 7.8 (3) | 9.7 | CB1 |
| tRes-3-OD-Glu | >100 (3) | >100 (3) | NA | NA |
| tRes-4′-OD-Glu | >100 (3) | >100 (3) | NA | NA |
mCB1, mouse brain CB1; hCB2, human CB2; tPice, trans-piceatannol; tA1, trans-arachidin-1; tA3, trans-arachidin-3.
Figure 5.
(A) Affinity (Ki) of trans-resveratrol (tRes) and analogs for mouse brain CB1Rs (mCB1) and human CB2Rs (hCB2) receptors. Membranes prepared from mouse brains (containing mCBR1s, upper panel) or CHO-hCB2 cells (expressing hCB2Rs, lower panel) were incubated with 0.2 nM 3H-CP-55,940 and increasing concentrations (0.1–100 µM) of tRes, trans-arachidin-1 (tA1), trans-arachidin-3 (tA3), trans-piceatannol (tPice), trans-resveratrol-3-O-D-glucuronide (tRes-3G) or trans-resveratrol-4′-O-D-glucuronide (tRes-4′G) for 90 min at room temperature and filtered. (B) The effect of tRes and analogs on basal G-protein activity in mouse brain membranes. Mouse brain membranes were incubated with 0.1 nM 35S-GTPγS and CB1R agonist CP-55,940 (1 µM), CB1R inverse agonist AM-251 (1 µM), tA1 (10 µM), tPice (10 µM), tA3 (10 µM) or tRes (10 µM) for 30 min at 30°C and filtered. *,**Significantly different (p < 0.05, 0.01) from 0%, NS = Not significantly different from 0%. (C) The tRes analog tA1 reduces basal G-protein activity in mouse brain membranes in a concentration-dependent manner that is not reversed by a neutral CB1R antagonist. Mouse brain membranes were incubated with 0.1 nM 35S-GTPγS and increasing concentrations of tA1 alone (1 nM–100 µM, closed triangles) or co-incubated with the neutral CB1R antagonist O-2050 (1 µM, open triangle) for 30 min at 30°C and filtered. NS = Not significantly different from tA1 (10 µM) alone. (D) tRes analogs tA1 and tA3 act as antagonists at CB1Rs. Mouse brain membranes were incubated with 0.1 nM 35S-GTPγS and increasing concentrations of the CB1R agonist CP-55,940 (0.01–1,000 nM) alone (open squares, both panels) or in the presence of tA1 (10 µM, closed triangles, upper panel) or tA3 (30 µM, closed circles, lower panel) for 30 min at 30°C and filtered. Co-incubation with tA1 results in a right- and down-ward shift, while tA3 produces only a rightward shift, in the concentration-effect curve for CP-55,940.
Intrinsic activity of trans-arachidins, tRes and tPice at CB1Rs
Since tRes and all analogs bind to mCB1Rs, while only the isoprenylated analogs bind appreciably to hCB2Rs, subsequent studies were conducted to determine the intrinsic activity of tRes and its analogs at the mCB1R subtype (Figure 5B–D). [35S]GTPγS binding assays demonstrated that the well-characterised non-selective CBR agonist CP-55,940 (Deadwyler et al. 1993) (1 µM) stimulates, while the established CB1R inverse agonist AM-251 (Lan et al. 1999) inhibits, G-protein activity in membranes prepared from mouse brains expressing mCB1Rs (Figure 5B). Both stimulation and inhibition are blocked by co-incubation with the selective CB1R neutral antagonist O-2050 (Pertwee 2005) (data not shown). Interestingly, tA1 (10 µM) and tPice (10 µM) markedly reduce, while tA3 (10 µM) and tRes (10 µM) have little effect on, basal G-protein activity. Although the potential inverse activity produced by tA1 is concentration-dependent (Figure 5C, closed triangles), it does not appear to be mediated by mCB1Rs because co-incubation with a selective CB1R neutral antagonist O-2050 (1 µM) (that produced no effect on basal G-protein activity when administered alone, data not shown) did not significantly attenuate the ability of tA1 (10 µM) to reduce basal G-protein activity (Figure 5, open triangle). In any case, co-administration with tA1 (10 µM) or tA3 (30 µM) results in mCB1R antagonism, producing a 7- (4.3 vs 28 nM) or 30-fold increase (4.3 vs 127 nM; shift-to-the-right) in the ED50 required to activate G-proteins by the CB1R agonist CP-55,940, respectively (Figure 5D). In addition to increasing the ED50, tA1 but not tA3 also significantly reduces the maximal response produced by CP-55,940 from 0.34 ± 0.03 to 0.22 ± 0.04 pmol/mg protein. This indicates that tA1 may produce mCB1R antagonism by both competitive and non-competitive mechanisms.
Molecular docking of trans-arachidins, tRes, tPice and their glucuronidated metabolites to CB2R
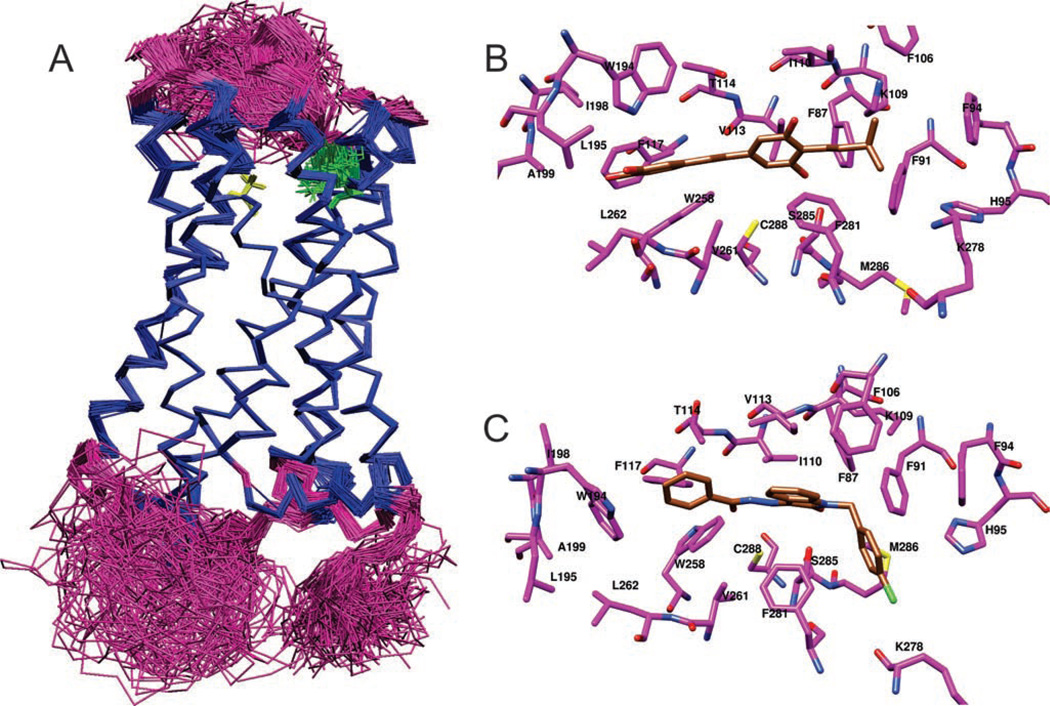
The isoprenylated analogs tA1 and tA3 bind with reasonable affinity to both mCB1R and hCB2Rs. In marked contrast, while tRes and tPice also bind to mCB1Rs, they lack appreciable affinity for hCB2Rs. To reveal a potential molecular basis for these marked differences in affinities for hCB2Rs, docking analyses of parent, isoprenylated analogs and several of their glucuronidated metabolites to CB2Rs were conducted. The three-dimensional protein models for CB2R were found to have a considerable degree of difference in side-chain and backbone conformation (Figure 6A).
Figure 6.
The conformational diversity observed in models generated using Modeller software, represented by a superposition of 132 CB2R structures. (A) The helical and non-helical regions of the backbone are rendered as blue and magenta trace, respectively. Variations in the side-chain conformation of K109 and S285 are shown as green and yellow sticks, respectively. Snapshot rendered using VMD (Visual Molecular Dynamics) (Humphrey et al. 1996). Putative binding pose of (B) tA1 and (C) an N-alkyl isatin benzohydrazide, bound to CB2R. The carbon atoms of the ligand and CB2R are rendered in gold and magenta, respectively, with nitrogen in blue, oxygen in red and sulphur in yellow. Snapshots rendered using the UCSF Chimera package from the Resource for Biocomputing, Visualisation and Informatics at the University of California, San Francisco (Pettersen et al. 2004).
We first performed docking of 12 resveratrol-like ligands on 132 models of CB2R. The selected best model, which had an RMSD for common atoms of 12 ligands of 2.41 Å (1.28 Å without two 4′-substituted derivatives) and passed visual inspection, was used to rationalise observed differences in binding of the parent compounds and metabolites. Three 4′-substituted compounds were docked on the selected model after the initial round of docking. The Glide G-scores and experimentally observed activities are given in Table 4. For comparison, an N-alkyl isatin benzohydrazide, a known nanomolar inhibitor of CB2R (Diaz et al. 2008), was also docked into the model and is included in Table 4. The binding poses of some of the ligands are shown in Figure 6A and in the Supporting Information (Table S3).
Roughly, the binding pocket of interest in CB2R contains three main regions. First, there is a central portion capable of hydrogen bonding, featuring K109 and S285 on opposite sides of the central portion of the binding site. Then, at each end is a largely hydrophobic pocket. In the docked poses, the ligands interact with all three regions of the pocket, forming a hydrogen bond with S285 (such a hydrogen bond was used as a restraint for docking the ligand) and hydrophobic interactions with two pockets, one formed by F87, F91, F281, K109 and M286, and another formed by F117, W258, W194 and I198. The ring with primed numbering orients into the latter hydrophobic pocket, while a 4-substituent orients into the former hydrophobic pocket. Either a 3-OH or 5-OH group forms a hydrogen bond with S285. The isoprenyl moiety of arachidins helped to improve the binding, with a hydrophobic substituent at position 4 that extends into the former hydrophobic pocket (better binding affinity and G-score for tA1 and tA3 than for tRes and tPice). An increase in hydrophilicity of the primed ring (additional OH), which interacts with the latter hydrophobic pocket, reduces the binding affinity (lower affinity and G-score for tA1 and tPice compared with for tA3 and tRes, respectively).
Docking of the glucuronidated metabolites predicts that in some cases they will have enhanced binding compared with their parent compound. For some of the ligands, the glucuronide group appeared to be sticking out of the binding pocket into the pocket’s entrance channel. This protrusion is more prominent for 4′-glucuronidated-tA3 and -tA1, while less prominent for 4′-glucuronidated-tRes and -tPice. The glucuronide group of three-substituted compounds docked in a similar fashion to that observed for 4′-glucuronidated compounds, but with flipped orientation of two rings, leading to the glucuronide group binding to the same site for 3- or 4′-glucuronidated compounds. It is normal to assume that since the glucuronide is hydrophilic, it is most likely to be exposed to solvent, saving the energetic penalty of burying the hydrophilic group in the protein. Although the in vitro competition receptor binding assay found no CBR affinity for two commercially available glucuronidated derivatives of tRes (resveratrol 3- or -4′-O-D-glucuronide), many other known glucuronidated derivatives of tRes have not yet been examined. Therefore, the three glucuronides with the highest Glide G-scores (e.g. 3-glucuronide-trans-arachidin-3, 3-glucuronide-trans-arachidin-2 and 3-glucuronide-trans-arachidin-1) are predicted to bind well to CB2Rs and should be synthesised and tested in vitro.
Discussion
The active constituents isolated from many medicinal plants exhibiting anti-carcinogenic, anti-inflammatory and anti-protozoal activities have been identified as prenylated polyphenols and prenylation appears to enhance their biological activity (Yazaki et al. 2009). The increased therapeutic activity of these natural products likely results from increased lipophilicity imparted by single or multiple prenyl groups present within their structure. It has been proposed that the greater lipophilicity of prenylated tRes analogs may allow for easier interaction between these polyphenols and cell membranes, enhancing access and association with potential membrane-bound molecular targets responsible for the beneficial actions of these compounds (Huang et al. 2010). This suggestion is supported by studies showing that in vivo administration of pterostilbene (a methylated, but non-prenylated, stilbene also exhibiting enhanced lipophilicity) results in markedly higher serum concentrations and slower metabolism relative to tRes (Kapetanovic et al. 2010). In agreement with this hypothesis, results from this study demonstrate that only the isoprenylated analogs tA1 and tA3 bind appreciably to hCB2Rs, and tA1 displays significantly higher affinity for CB1Rs than its non-isoprenylated analog tPice.
Slowed metabolism, leading to improved bioavailability, may be an additional major benefit provided by isoprenylation of tRes and its analogs. To confirm the slower metabolism of the isoprenylated stilbenoids examined, it was important to compare the kinetic properties of native tRes and tPice to those of their prenylated counterparts, tA3 and tA1. Our data importantly show that tA3 and tA1 do indeed demonstrate similar Km values, have significantly decreased Vmax values and are metabolised to fewer glucuronidated products than tRes and tPice. Of particular note, this metabolic profile occurs in response to in vitro metabolism by UGT isoforms occurring in the liver and intestine, the major tissues responsible for the metabolism of these compounds in vivo. Slower in vivo metabolism suggested by these data, coupled with potential enhanced measures of biological activity, indicate that isoprenylated tRes analogs might represent a novel class of compounds with significantly improved biological activity that might be exploited for future development as efficacious pharmaceutical drugs.
Extensive efforts have been directed toward evaluation of the in vitro effects of native tRes. However, in vivo pharmacokinetic studies, both in animal and humans, have demonstrated that tRes is readily absorbed and rapidly glucuronidated and sulphated in liver and intestinal epithelial tissues (Brill et al. 2006; De Santi et al. 2000; Vitaglione et al. 2005; Walle et al. 2004; Wenzel & Somoza 2005). tRes has also been shown to induce phase II enzymes, further contributing to its rapid clearance (Lancon et al. 2007). In spite of this well-documented poor bioavailability, many beneficial effects are nevertheless observed following in vivo tRes administration. Therefore, it is often suggested that tRes metabolites may possess biological activity (Lancon et al. 2007), although only one report has actually investigated the activity of such conjugates themselves in vivo (Calamini et al. 2010). The authors of this study conclude that “the interplay between resveratrol and its metabolites with different molecular targets may be responsible for the overall beneficial health effects previously attributed only to resveratrol itself.”(Calamini et al. 2010). On the basis of this information, we hypothesise that glucuronides and sulphates of tRes may also possess pharmacological activity, although the specific cellular mechanisms responsible are unknown.
In this study, we examined whether CBRs might represent novel molecular targets responsible for at least some of the pharmacological activity produced by tRes, its analogs and/or metabolites. Originally isolated from the marijuana plant (Cannabis sativa), both synthetic and naturally occurring cannabinoids such as Δ9-tetrahydrocannabinol produce their effects by acting at two G-protein coupled receptors; CB1Rs (Matsuda et al. 1990) and CB2Rs (Munro et al. 1993). CB1Rs are expressed in high abundance throughout the central nervous system, while CB2Rs are expressed predominantly in immune cells and non-neuronal tissues. Cannabinoids acting at both receptors produce anti-oxidant (Hampson et al. 1998) and anti-inflammatory (Klein 2005) effects, similar to those reported for tRes (Fraga 2007; Surh et al. 2005). Therefore, in this study, we initiated in vitro and in silico experiments to directly assess whether tRes, several analogs and glucuronides bind to and modulate the activity of CBRs.
We demonstrate that while the isoprenylated tRes analogs tA1 and tA3 bind to both mCB1Rs and hCB2Rs with low µM affinity, the parent compounds tRes and tPice bind appreciably only to mCB1Rs. Although the reported affinities (e.g. Ki values) for tRes and its analogs may seem relatively low, they are nevertheless likely to be physiologically relevant because the endogenously produced cannabinoid agonist 2-arachidonoylglycerol exhibits similar affinities of ~0.5–1.5 µM for CB1Rs and CB2Rs, respectively (Mechoulam et al. 1995). Furthermore, the affinities of tRes and its analogs determined here are also similar to the concentrations required to produce many of their previously demonstrated in vitro protective effects (Pervaiz & Holme 2009).
Our in vitro data further indicate that the tRes analogs tA1 and tA3 act as antagonists of CB1R function. The lack of any CB1R agonist activity (e.g. G-protein stimulation) observed for tRes is consistent with the established absence of psychotropic activity or abuse potential reported for this polyphenol (Cottart et al. 2010). It is important to note that while CBR agonists increase G-protein activation, because CBRs are constitutively active (Nie & Lewis 2001), inverse agonists have been shown to produce a reduction in basal G-protein activity by stabilising constitutively active CB1Rs (Bouaboula et al. 1997). As such, our initial data indicated that tA1 and tPice might act as inverse agonists at CB1Rs. However, subsequent experiments showed that the reduction in basal G-protein activity produced by tA1 could not be significantly attenuated by co-incubation with the neutral CB1R antagonist O-2050 (e.g. a selective CB1R antagonist producing no effect on basal G-protein activity when administered alone). These combined observations indicate that the action of tA1 (and by extrapolation, perhaps tPice) on basal G-protein activity is apparently not mediated via CB1Rs. However, it is also possible that the lack of tA1 attenuation by O-2050 might occur because these structurally distinct compounds bind to different domains of CB1Rs such that receptor occupation by O-2050 does not preclude the binding of tA1. In any case, our current observations that tA1 and tA3 markedly shift the dose-effect curve for G-protein activation produced by a CB1R agonist to the right, clearly demonstrate that these isoprenylated tRes analogs nevertheless do act as CB1R antagonists. These observations are significant because many recent studies demonstrate that CB1R antagonists (and inverse agonists) produce a variety of therapeutic effects, acting as very efficacious anti-inflammatory (Li et al. 2010), anti-obesity (Lee et al. 2009) and anti-carcinogenic (Bifulco et al. 2009) agents.
Although relatively poor bioavailability of tRes and its natural analogs, particularly in the central nervous system (CNS), might preclude observation of significant effects mediated by central CB1 receptors, peak serum concentrations in mice of approximately 1–2 µM of parent drug following a single, acute intra-peritoneal injection of moderate doses (~20–100 mg/kg) of tRes (Asensi et al. 2002) have been reported. Furthermore, very low doses of tRes protect against neuronal damage following cerebral ischaemia in rats (Wang et al. 2003). Similarly, the lipophilic resveratrol analogue pterostilbene was effective in reversing cognitive behavioural deficits in rats and working memory was correlated with pterostilbene levels in the hippocampus (Joseph et al. 2008). Altogether, these findings provide evidence that tRes and its lipophilic analogs, such as arachidin-1 and -3, are able to cross the blood–brain barrier in sufficient concentrations to provide a physiological effect.
However, even if tRes and its analogs fail to achieve therapeutic concentrations in the CNS, physiologically relevant levels of CB1 receptors are also present in the periphery (Kunos et al. 2009) and activation of these peripheral CB1 receptors is effective at suppressing inflammation that leads to chronic pain states (Gutierrez et al. 2007). Furthermore, several studies also indicate that the metabolic benefits of CB1 antagonists/inverse agonists in obese animals is due to action at peripheral, but not central, CB1 receptors (Pavon et al. 2008). Consistent with this hypothesis, although not attributed to action at CB1 receptors, others report that tRes reduces body weight in Zucker obese rats (Lekli et al. 2008). Therefore, polyphenol-derived, peripherally restricted CB1 agonists or antagonists might be developed as a novel class of non-toxic cannabinoids.
Our in vitro experiments with CBRs were complemented by in silico homology modelling and docking studies. If we consider the nature of the binding pocket with two end hydrophobic pockets, then it makes sense, as shown by the scores of the ligands docked into our selected model, that the arachidins should bind better than tPice or tRes, because the arachidin isoprenyl side chain is hydrophobic and will fit well into one of the end hydrophobic pockets of the binding site. The extra hydroxyl group in tA1 should lead to somewhat unfavourable binding compared with tA2 or tA3, as is also found in the model. Similarly, tRes should bind better than tPice because the former has one fewer–OH. This suggests that the CB2R model we have prepared is an appropriate one and will also be suitable for prediction of binding for novel ligands.
There is certainly scope to increase the affinity for CB2Rs of this class of compounds. For example, the positioning of the primed ring in a hydrophobic pocket allows us to predict that replacing the 4′-OH with a hydrophobic group such as methyl should enhance binding to CB2Rs. A comparison of the occupancy of the binding site of CB2R by tA2 with that of the N-alkyl isatin benzohydrazide, a highly potent CB2R binder, is given in Figure 6B and C, indicating that significant improvements to the binding of the resveratrol derivatives are possible.
Consistent with our hypothesis that metabolites of tRes may also possess pharmacological activity, our in silico molecular modelling analyses predict that some glucuronidated conjugates of tRes analogs (other than the 3- or 4′-O conjugates examined) may still be shown to bind CB2Rs with higher affinity than any of the compounds tested in this study. If these predictions are correct, this would be consistent with other reports demonstrating that the metabolite morphine-6-glucuronide also possesses higher receptor affinity and potency for μ-opioid receptors than its parent compound morphine (Wittwer & Kern 2006). Therefore, to adequately evaluate new tRes-based derivatives, our data suggest that future studies should importantly determine both CB1R and CB2R affinity of not only novel tRes analogs but also of their respective metabolic conjugates as well. Therefore, observations reported in this study suggest that minor modifications to the basic tRes structure result in marked alterations in the affinity of various analogs for CBRs. As such, tRes may likely represent a useful scaffold for future design of highly selective and efficacious CB1R and CB2R ligands.
Finally, based on the observations presented that prenylated stilbenoids may be preferable alternatives to tRes, it will be crucial to conduct future experiments to determine whether additional prenylated stilbenoids produced in the peanut plant share similar or additional biological activities. However, stilbenoids such as tRes and its analogs are a growing group of polyphenolic compounds found only in a limited number of plant families (Shen et al. 2009) and thus available in limited quantities. Therefore, future studies aimed at understanding the biological activities of these naturally occurring polyphenols may be hindered by their unavailability. Techniques presented in this study and our recent developments to scale-up peanut hairy root culture production in airlift balloon-type bioreactors (Sivakumar et al. 2011) will importantly allow us (and others) to continue, and even broaden, the study of the biological activities of these and other prenylated stilbenoids produced in peanut axenic cultures.
Conclusions
Findings presented in this study importantly indicate that prenylated stilbenoids may be preferable alternatives to tRes due to increased bioavailability via slowed metabolism. This is supported by observations that tA1 and tA3, the more lipophilic isoprenylated analogs of tRes and tPice, exhibit a markedly reduced rate of glucuronidation relative to the parent compounds. Furthermore, this metabolic profile occurs in response to in vitro metabolism by UGT isoforms occurring in the liver and intestine, the major tissues responsible for the metabolism of these compounds in vivo. A second major finding of this study is that both parent and isoprenylated compounds bind to CB1Rs, a novel cellular target. Furthermore, tA1 and tA3 not only bind to CB1Rs but they also antagonise actions produced by CB1R agonists. In contrast to CB1R binding, only analogs possessing an isoprenyl group, tA1 and tA3, demonstrate significant affinity for CB2Rs. Results presented for CB2R binding are consistent with predictions derived from molecular docking analyses. Finally, although elucidating the biological activities of these and other naturally occurring polyphenols has been previously hindered by their limited availability, the in vitro peanut hairy root culture system employed in this study offers a relatively straightforward biosynthetic method able to produce large amounts of these stilbenoids and their analogs for future studies.
Supplementary Material
Acknowledgments
The authors would like to thank Anna Dineva for her technical assistance in carrying out the screening experiments with the UGT2B isoforms. FMB was supported by the Arkansas Biosciences Institute, National Science Foundation-EPSCoR (grant # EPS–0701890; Center for Plant-Powered Production-P3), Arkansas ASSET Initiative and the Arkansas Science and Technology Authority. RJD was supported by USA National Institutes of Health (NIH) Grant Number 5P20RR021929 from the National Center for Research Resources (NCRR) and his research group’s investigations were conducted in a facility constructed with support from research facilities improvement program C06 RR-14503-01 from the NIH NCRR. AR-P was supported by USA National Institutes of Health (NIH) Grant Number GM075893 from the National Institute of General Medical Sciences (NIGMS).
Declaration of interest
The content in this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of NIH, NIGMS or NCRR.
References
- Abbott JA, Medina-Bolivar F, Martin EM, Engelberth AS, Villagarcia H, Clausen EC, Carrier DJ. Purification of resveratrol, arachidin-1, and arachidin-3 from hairy root cultures of peanut (Arachis hypogaea) and determination of their antioxidant activity and cytotoxicity. Biotechnol Prog. 2010;26:1344–1351. doi: 10.1002/btpr.454. [DOI] [PubMed] [Google Scholar]
- Asensi M, Medina I, Ortega A, Carretero J, Baño MC, Obrador E, Estrela JM. Inhibition of cancer growth by resveratrol is related to its low bioavailability. Free Radic Biol Med. 2002;33:387–398. doi: 10.1016/s0891-5849(02)00911-5. [DOI] [PubMed] [Google Scholar]
- Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- Bifulco M, Santoro A, Laezza C, Malfitano AM. Cannabinoid receptor CB1 antagonists state of the art and challenges. Vitam Horm. 2009;81:159–189. doi: 10.1016/S0083-6729(09)81007-8. [DOI] [PubMed] [Google Scholar]
- Bouaboula M, Perrachon S, Milligan L, Canat X, Rinaldi-Carmona M, Portier M, Barth F, Calandra B, Pecceu F, Lupker J, Maffrand JP, Le Fur G, Casellas P. A selective inverse agonist for central cannabinoid receptor inhibits mitogen-activated protein kinase activation stimulated by insulin or insulin-like growth factor 1. Evidence for a new model of receptor/ligand interactions. J Biol Chem. 1997;272:22330–22339. doi: 10.1074/jbc.272.35.22330. [DOI] [PubMed] [Google Scholar]
- Brill SS, Furimsky AM, Ho MN, Furniss MJ, Li Y, Green AG, Bradford WW, Green CE, Kapetanovic IM, Iyer LV. Glucuronidation of trans-resveratrol by human liver and intestinal microsomes and UGT isoforms. J Pharm Pharmacol. 2006;58:469–479. doi: 10.1211/jpp.58.4.0006. [DOI] [PubMed] [Google Scholar]
- Calamini B, Ratia K, Malkowski MG, Cuendet M, Pezzuto JM, Santarsiero BD, Mesecar AD. Pleiotropic mechanisms facilitated by resveratrol and its metabolites. Biochem J. 2010;429:273–282. doi: 10.1042/BJ20091857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, Lai YH, Djoko B, Wu PL, Liu CD, Liu YW, Chiou RY. Biosynthesis enhancement and antioxidant and anti-inflammatory activities of peanut (Arachis hypogaea L.) arachidin-1, arachidin-3, and isopentadienylresveratrol. J Agric Food Chem. 2006;54:10281–10287. doi: 10.1021/jf0620766. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Condori J, Medrano G, Sivakumar G, Nair V, Cramer C, Medina-Bolivar F. Functional characterization of a stilbene synthase gene using a transient expression system in planta. Plant Cell Rep. 2009;28:589–599. doi: 10.1007/s00299-008-0664-0. [DOI] [PubMed] [Google Scholar]
- Condori J, Sivakumar G, Hubstenberger J, Dolan MC, Sobolev VS, Medina-Bolivar F. Induced biosynthesis of resveratrol and the prenylated stilbenoids arachidin-1 and arachidin-3 in hairy root cultures of peanut: Effects of culture medium and growth stage. Plant Physiol Biochem. 2010;48:310–318. doi: 10.1016/j.plaphy.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Cottart CH, Nivet-Antoine V, Laguillier-Morizot C, Beaudeux JL. Resveratrol bioavailability and toxicity in humans. Mol Nutr Food Res. 2010;54:7–16. doi: 10.1002/mnfr.200900437. [DOI] [PubMed] [Google Scholar]
- Cucciolla V, Borriello A, Oliva A, Galletti P, Zappia V, Della Ragione F. Resveratrol: From basic science to the clinic. Cell Cycle. 2007;6:2495–2510. doi: 10.4161/cc.6.20.4815. [DOI] [PubMed] [Google Scholar]
- De Santi C, Pietrabissa A, Spisni R, Mosca F, Pacifici GM. Sulphation of resveratrol, a natural compound present in wine, and its inhibition by natural flavonoids. Xenobiotica. 2000;30:857–866. doi: 10.1080/004982500433282. [DOI] [PubMed] [Google Scholar]
- Deadwyler SA, Hampson RE, Bennett BA, Edwards TA, Mu J, Pacheco MA, Ward SJ, Childers SR. Cannabinoids modulate potassium current in cultured hippocampal neurons. Recept Channels. 1993;1:121–134. [PubMed] [Google Scholar]
- Diaz P, Xu J, Astruc-Diaz F, Pan HM, Brown DL, Naguib M. Design and synthesis of a novel series of N-alkyl isatin acylhydrazone derivatives that act as selective cannabinoid receptor 2 agonists for the treatment of neuropathic pain. J Med Chem. 2008;51:4932–4947. doi: 10.1021/jm8002203. [DOI] [PubMed] [Google Scholar]
- Djoko B, Chiou RY, Shee JJ, Liu YW. Characterization of immunological activities of peanut stilbenoids, arachidin-1, piceatannol, and resveratrol on lipopolysaccharide-induced inflammation of RAW 264.7 macrophages. J Agric Food Chem. 2007;55:2376–2383. doi: 10.1021/jf062741a. [DOI] [PubMed] [Google Scholar]
- Doerge DR, Chang HC, Churchwell MI, Holder CL. Analysis of soy isoflavone conjugation in vitro and in human blood using liquid chromatography-mass spectrometry. Drug Metab Dispos. 2000;28:298–307. [PubMed] [Google Scholar]
- Fraga CG. Plant polyphenols: How to translate their in vitro antioxidant actions to in vivo conditions. IUBMB Life. 2007;59:308–315. doi: 10.1080/15216540701230529. [DOI] [PubMed] [Google Scholar]
- Gutierrez T, Farthing JN, Zvonok AM, Makriyannis A, Hohmann AG. Activation of peripheral cannabinoid CB1 and CB2 receptors suppresses the maintenance of inflammatory nociception: A comparative analysis. Br J Pharmacol. 2007;150:153–163. doi: 10.1038/sj.bjp.0706984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson AJ, Grimaldi M, Axelrod J, Wink D. Cannabidiol and (−)Delta9-tetrahydrocannabinol are neuroprotective antioxidants. Proc Natl Acad Sci USA. 1998;95:8268–8273. doi: 10.1073/pnas.95.14.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YS, Bastianetto S, Dumont Y, Quirion R. Specific plasma membrane binding sites for polyphenols, including resveratrol, in the rat brain. J Pharmacol Exp Ther. 2006;318:238–245. doi: 10.1124/jpet.106.102319. [DOI] [PubMed] [Google Scholar]
- Huang CP, Au LC, Chiou RY, Chung PC, Chen SY, Tang WC, Chang CL, Fang WH, Lin SB. Arachidin-1, a Peanut Stilbenoid, Induces Programmed Cell Death in Human Leukemia HL-60 Cells. J Agric Food Chem. 2010 doi: 10.1021/jf102993j. <Volume>:. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Joseph JA, Fisher DR, Cheng V, Rimando AM, Shukitt-Hale B. Cellular and behavioral effects of stilbene resveratrol analogues: Implications for reducing the deleterious effects of aging. J Agric Food Chem. 2008;56:10544–10551. doi: 10.1021/jf802279h. [DOI] [PubMed] [Google Scholar]
- Kapetanovic IM, Muzzio M, Huang Z, Thompson TN, McCormick DL. Pharmacokinetics, oral bioavailability, and metabolic profile of resveratrol and its dimethylether analog, pterostilbene, in rats. Cancer Chemother Pharmacol. 2010 doi: 10.1007/s00280-010-1525-4. <Volume>:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein TW. Cannabinoid-based drugs as anti-inflammatory therapeutics. Nat Rev Immunol. 2005;5:400–411. doi: 10.1038/nri1602. [DOI] [PubMed] [Google Scholar]
- Kunos G, Osei-Hyiaman D, Bátkai S, Sharkey KA, Makriyannis A. Should peripheral CB(1) cannabinoid receptors be selectively targeted for therapeutic gain? Trends Pharmacol Sci. 2009;30:1–7. doi: 10.1016/j.tips.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela M, García-Horsman JA, Luukkanen L, Mörsky S, Taskinen J, Baumann M, Kostiainen R, Hirvonen J, Finel M. Expression and characterization of recombinant human UDP-glucuronosyltransferases (UGTs). UGT1A9 is more resistant to detergent inhibition than other UGTs and was purified as an active dimeric enzyme. J Biol Chem. 2003;278:3536–3544. doi: 10.1074/jbc.M206136200. [DOI] [PubMed] [Google Scholar]
- Kuuranne T, Kurkela M, Thevis M, Schänzer W, Finel M, Kostiainen R. Glucuronidation of anabolic androgenic steroids by recombinant human UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31:1117–1124. doi: 10.1124/dmd.31.9.1117. [DOI] [PubMed] [Google Scholar]
- Lan R, Liu Q, Fan P, Lin S, Fernando SR, McCallion D, Pertwee R, Makriyannis A. Structure-activity relationships of pyrazole derivatives as cannabinoid receptor antagonists. J Med Chem. 1999;42:769–776. doi: 10.1021/jm980363y. [DOI] [PubMed] [Google Scholar]
- Lançon A, Hanet N, Jannin B, Delmas D, Heydel JM, Lizard G, Chagnon MC, Artur Y, Latruffe N. Resveratrol in human hepatoma HepG2 cells: Metabolism and inducibility of detoxifying enzymes. Drug Metab Dispos. 2007;35:699–703. doi: 10.1124/dmd.106.013664. [DOI] [PubMed] [Google Scholar]
- Lee HK, Choi EB, Pak CS. The current status and future perspectives of studies of cannabinoid receptor 1 antagonists as anti-obesity agents. Curr Top Med Chem. 2009;9:482–503. doi: 10.2174/156802609788897844. [DOI] [PubMed] [Google Scholar]
- Lekli I, Szabo G, Juhasz B, Das S, Das M, Varga E, Szendrei L, Gesztelyi R, Varadi J, Bak I, Das DK, Tosaki A. Protective mechanisms of resveratrol against ischemia-reperfusion-induced damage in hearts obtained from Zucker obese rats: The role of GLUT-4 and endothelin. Am J Physiol Heart Circ Physiol. 2008;294:H859–H866. doi: 10.1152/ajpheart.01048.2007. [DOI] [PubMed] [Google Scholar]
- Li YY, Li YN, Ni JB, Chen CJ, Lv S, Chai SY, Wu RH, Yüce B, Storr M. Involvement of cannabinoid-1 and cannabinoid-2 receptors in septic ileus. Neurogastroenterol Motil. 2010;22:350-e88. doi: 10.1111/j.1365-2982.2009.01419.x. [DOI] [PubMed] [Google Scholar]
- Little JM, Kurkela M, Sonka J, Jäntti S, Ketola R, Bratton S, Finel M, Radominska-Pandya A. Glucuronidation of oxidized fatty acids and prostaglandins B1 and E2 by human hepatic and recombinant UDP-glucuronosyltransferases. J Lipid Res. 2004;45:1694–1703. doi: 10.1194/jlr.M400103-JLR200. [DOI] [PubMed] [Google Scholar]
- Mackie K, Hille B. Cannabinoids inhibit N-type calcium channels in neuroblastoma-glioma cells. Proc Natl Acad Sci USA. 1992;89:3825–3829. doi: 10.1073/pnas.89.9.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- Mechoulam R, Ben-Shabat S, Hanus L, Ligumsky M, Kaminski NE, Schatz AR, Gopher A, Almog S, Martin BR, Compton DR. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol. 1995;50:83–90. doi: 10.1016/0006-2952(95)00109-d. [DOI] [PubMed] [Google Scholar]
- Medina-Bolivar F, Condori J, Rimando AM, Hubstenberger J, Shelton K, O’Keefe SF, Bennett S, Dolan MC. Production and secretion of resveratrol in hairy root cultures of peanut. Phytochemistry. 2007;68:1992–2003. doi: 10.1016/j.phytochem.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Medina-Bolivar F, Dolan M, Bennett S, Condori J, Hubstenberger J. Production of stilbenes in plant hairy root cultures. Patent 7,666,677. patent U.S. 2010
- Miksits M, Maier-Salamon A, Vo TP, Sulyok M, Schuhmacher R, Szekeres T, Jäger W. Glucuronidation of piceatannol by human liver microsomes: Major role of UGT1A1, UGT1A8 and UGT1A10. J Pharm Pharmacol. 2010;62:47–54. doi: 10.1211/jpp.62.01.0004. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Nie J, Lewis DL. Structural domains of the CB1 cannabinoid receptor that contribute to constitutive activity and G-protein sequestration. J Neurosci. 2001;21:8758–8764. doi: 10.1523/JNEUROSCI.21-22-08758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavón FJ, Serrano A, Pérez-Valero V, Jagerovic N, Hernández-Folgado L, Bermúdez-Silva FJ, Macías M, Goya P, de Fonseca FR. Central versus peripheral antagonism of cannabinoid CB1 receptor in obesity: Effects of LH-21, a peripherally acting neutral cannabinoid receptor antagonist, in Zucker rats. J Neuroendocrinol. 2008;20(Suppl 1):116–123. doi: 10.1111/j.1365-2826.2008.01693.x. [DOI] [PubMed] [Google Scholar]
- Pertwee RG. Inverse agonism and neutral antagonism at cannabinoid CB1 receptors. Life Sci. 2005;76:1307–1324. doi: 10.1016/j.lfs.2004.10.025. [DOI] [PubMed] [Google Scholar]
- Pervaiz S, Holme AL. Resveratrol: Its biologic targets and functional activity. Antioxid Redox Signal. 2009;11:2851–2897. doi: 10.1089/ars.2008.2412. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reggio PH. Endocannabinoid binding to the cannabinoid receptors: What is known and what remains unknown. Curr Med Chem. 2010;17:1468–1486. doi: 10.2174/092986710790980005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabolovic N, Humbert AC, Radominska-Pandya A, Magdalou J. Resveratrol is efficiently glucuronidated by UDP-glucuronosyltransferases in the human gastrointestinal tract and in Caco-2 cells. Biopharm Drug Dispos. 2006;27:181–189. doi: 10.1002/bdd.498. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shen T, Wang XN, Lou HX. Natural stilbenes: An overview. Nat Prod Rep. 2009;26:916–935. doi: 10.1039/b905960a. [DOI] [PubMed] [Google Scholar]
- Shoemaker JL, Joseph BK, Ruckle MB, Mayeux PR, Prather PL. The endocannabinoid noladin ether acts as a full agonist at human CB2 cannabinoid receptors. J Pharmacol Exp Ther. 2005;314:868–875. doi: 10.1124/jpet.105.085282. [DOI] [PubMed] [Google Scholar]
- Sivakumar G, Medina-Bolivar F, Lay JO, Jr, Dolan MC, Condori J, Grubbs SK, Wright SM, Baque MA, Lee EJ, Paek KY. Bioprocess and bioreactor: Next generation technology for production of potential plant-based antidiabetic and antioxidant molecules. Curr Med Chem. 2011;18:79–90. doi: 10.2174/092986711793979724. [DOI] [PubMed] [Google Scholar]
- Surh YJ, Kundu JK, Na HK, Lee JS. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J Nutr. 2005;135:2993S–3001S. doi: 10.1093/jn/135.12.2993S. [DOI] [PubMed] [Google Scholar]
- Vitaglione P, Sforza S, Galaverna G, Ghidini C, Caporaso N, Vescovi PP, Fogliano V, Marchelli R. Bioavailability of trans-resveratrol from red wine in humans. Mol Nutr Food Res. 2005;49:495–504. doi: 10.1002/mnfr.200500002. [DOI] [PubMed] [Google Scholar]
- Walle T, Hsieh F, DeLegge MH, Oatis JE, Jr, Walle UK. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- Wang YJ, He F, Li XL. The neuroprotection of resveratrol in the experimental cerebral ischemia. Zhonghua Yi Xue Za Zhi. 2003;83:534–536. [PubMed] [Google Scholar]
- Weiss JN. The Hill equation revisited: Uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- Wenzel E, Somoza V. Metabolism and bioavailability of transresveratrol. Mol Nutr Food Res. 2005;49:472–481. doi: 10.1002/mnfr.200500010. [DOI] [PubMed] [Google Scholar]
- Wittwer E, Kern SE. Role of morphine’s metabolites in analgesia: Concepts and controversies. AAPS J. 2006;8:E348–E352. doi: 10.1007/BF02854905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Sasaki K, Tsurumaru Y. Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry. 2009;70:1739–1745. doi: 10.1016/j.phytochem.2009.08.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.