Abstract
Herein, we describe a polymeric micellar nanoparticle capable of rendering nucleic acids resistant to nuclease digestion. This approach relies on utilizing DNA as the polar head group of a DNA-polymer amphiphile in order to assemble well-defined, discrete nanoparticles. Dense packing of DNA in the micelle corona allows for hybridization of complementary oligonucleotides while prohibiting enzymatic degradation. We demonstrate the preparation, purification and characterization of the nanoparticles, then describe their resistance to treatment with endo- and exonucleases including snake-venom phosphodiesterase (SVP) a common, general DNA digestion enzyme.
Keywords: DNA, nanotechnology, polymer, nuclease, resistance
Nucleic acids are unique, informational molecules with exceptional potential in the preparation of complex nanostructured materials1 with utility as potent and specific therapeutic agents in vivo and as powerful investigative tools in vitro.2–4 Despite this promise, unmodified nucleic acids are inherently susceptible to enzymatic degradation in biological milieu, limiting their practical utility in detection and as therapeutics in real-world applications. To mitigate these issues, considerable effort has been applied to the generation of DNA analogues capable of resisting attack.5,6 Unfortunately, in biological settings, these synthetic analogues exhibit unpredictable off-target effects7 and preclude advantageous interactions with key cellular machinery.8,9 Therefore, two nanotechnology-based approaches for packaging nucleic acids for resistance have been pursued whereby the base identity is preserved along with backbone chemistry; 1) utilizing gold nanoparticles as templates for arranging oligonucleotides as a spherical brush,10 2) packing nucleic acids as DNA-origami.11,12 Importantly, both approaches use well-defined nanostructures to arrange nucleic acids in a given pattern. Herein, we describe a third route that avoids two key limitations imposed by these current approaches: 1) the need for preparing nucleic acid-intensive DNA origami structures, that by necessity consist of large portions of double stranded DNA, unavailable for subsequent hybridization, and 2) the need for a metal nanoparticle template that precludes the incorporation of a chemically diverse core. Our strategy is based on the hypothesis that single-stranded oligonucleotides, consisting of natural nucleotide structures, may be made resistant to nuclease attack by densely packing them as organic polymeric micellar nanoparticles. The design is predicated on the idea that steric hindrance through dense packaging limits the accessibility of DNA to selective and non-selective nucleases. We sought to demonstrate this approach to resistant nucleic acids by utilizing a polymerization strategy that is known to be highly functional group tolerant, would allow efficient end-terminus functionalization, and provides polymers of low polydispersity.13 Nucleic acids were packed as DNA-polymer amphiphiles (DPAs)14 into micellar nanoparticles consisting of a high-density ssDNA corona with a hydrophobic organic polymer core.15 We demonstrate that this morphology allows free access to additional complementary DNA strands while preventing and/or inhibiting the activity of various types of nucleases.
RESULTS AND DISCUSSION
Preparation of DPA nanoparticles
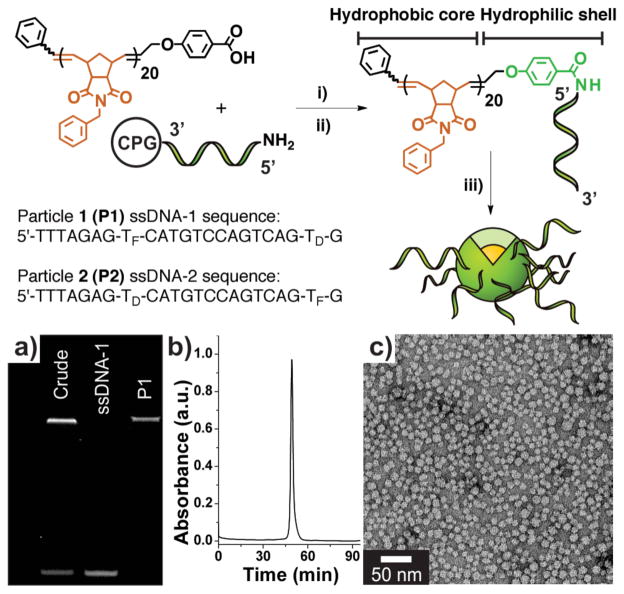
DPAs were prepared via conjugation of a hydrophobic polymer (prepared via ring-opening metathesis polymerization), terminally modified with a carboxylic acid moiety, to a 5′-amino-modified oligonucleotide on solid support (Figure 1). The resulting DNA-polymer conjugate was separated from unmodified polymer by thoroughly rinsing the support. Subsequent cleavage and dialysis gave a mixture of spherical micellar nanoparticles and free, non-conjugated single-stranded DNA (ssDNA, Figure 1a, lane 1). The particles were separated from ssDNA via size-exclusion chromatography (SEC-FPLC) to give purified material (Figure 1a, b). This procedure was utilized in the preparation of two micellar nanoparticles, P1 and P2, both exhibiting low polydispersity with diameters on the order of 20 nm, as determined by TEM (Figure 1c), and dynamic light scattering (DLS, see Supporting Information Figure S8). Static light scattering (SLS) was utilized to confirm aggregation numbers (Nagg) on the order of ~200 DNA strands per particle (see Supporting Information Figure S8). Therefore, DNA is at exceptionally high densities16 of approximately 0.2 DNA strands/nm2 on the surface of the micelles.
Figure 1.
Preparation of DNA-polymer amphiphiles (DPAs) and assembly of micelles. Synthesis: i) A hydrophobic polymer, terminally modified with a carboxylic acid moiety was mixed with a coupling agent and reacted with a 5′-amino modified oligonucleotide on solid support (controlled pore glass, CPG). ii) Deprotection and cleavage of the resulting DNA-polymer conjugate from solid support. iii) Dialysis of cleaved DPA into deionized water to form a mixture of micelles and free, non-conjugated nucleic acid. TF and TD correspond to fluorescein- and DABCYL-modified thymidine phosphoramidites. a) PAGE analysis. Lane 1: Crude material post-micelle (P1) formation showing conjugate (top band) and free ssD-NA (lower band). Lane 2: HPLC purified sample of ssDNA-1. Lane 3: Purified P1, isolated via size-exclusion chromatrography (SEC). b) SEC trace of purified P1 (λabs = 260 nm). c) Transmission electron micrograph of P1. See Supporting Information Figure S7 for P2 data.
The two particles (P1 and P2) were engineered to incorporate ssDNA sequences (ssDNA-1 and ssDNA-2) as substrates for a selective endonuclease and also a pair of indiscriminate exonucleases. ssDNA-1 and ssDNA-2 differ only in the location of dye- and quencher-labels. ssDNA-1 consists of a DABCYL-modifier located towards the 3′-terminus, and a fluorescein-modifier 13 bases away towards the 5′-terminus. By contrast, ssDNA-2 has the reverse arrangement with a fluorescein-modifier towards the 3′-terminus. This pair of sequences was designed to detect nuclease activity via a FRET assay in which the enzyme-triggered release of a DABCYL- (ssDNA-1), or fluorescein- (ssDNA-2) modified fragment from the oligonucleotide sequence results in an increase in fluorescence signal (vide infra). Additionally, to serve as independent controls, ssDNA-1 and ssDNA-2 were purified as non-polymer conjugated oligonucleotides.
Endonuclease activity against DPA nanoparticles
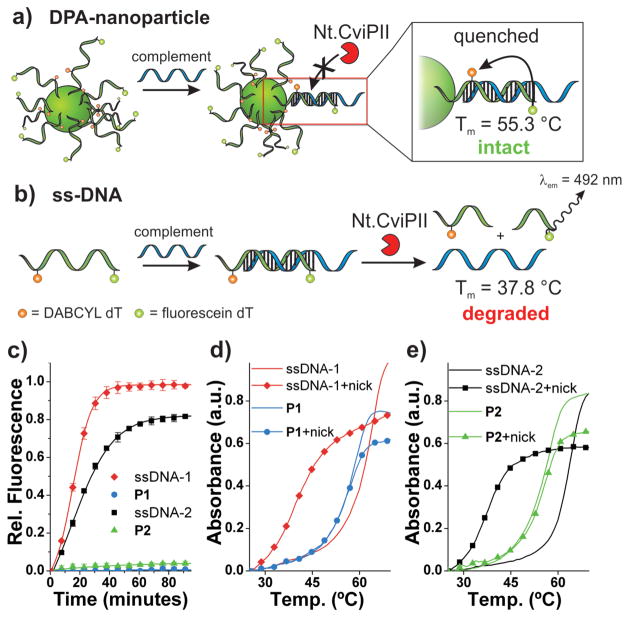
To examine how DPA-nanoparticles respond to sequence-selective endonucleases, we incorporated a substrate for nicking endonuclease Nt.CviPII (5′…CCA…3′, see Supporting Information Figure S4) between fluorescein- and DABCYL-labeled thymidine moieties of the oligonucleotide. Nt.CviPII is a nicking endonuclease that recognizes double-stranded DNA (dsDNA) and introduces a single-strand break on the 5′ side of the recognition site (5′…*CCX…3′, X = A, G, or T).17 The system was designed such that nucleolytic cleavage occurs on the sequence of the DPA-nanoparticle or ssDNA analogue while leaving the complementary sequence of the duplex fully intact. We reasoned that this design would facilitate a catalytic degradation of both the nanoparticle and ssDNA in response to small quantities of complementary DNA in the presence of the enzyme. Specifically, we programmed the nick site between bases 10 and 11 of the 20 bp duplex such that the melting temperature (Tm) of the nicked product would drop to approximately half that of the full 20 bp duplex (from ~ 60 to 30 °C). Through subsequent, thermodynamically favorable strand invasion, intact ssDNA or nanoparticle DNA would then be allowed to hybridize to its complement in order to recycle the target.
To monitor Nt.CviPII activity, we employed two complementary analytical techniques; a fluorescence assay and an assessment of DNA melting temperature with and without enzyme treatment (Figure 2). The first method involves a fluorescence dequenching experiment wherein the particles or ssDNA sequences are allowed to hybridize to complementary DNA and subsequently introduced to the endonuclease. Fluorescein fluorescence was monitored over time in order to assess the activity of the enzyme. In this case, an increase in fluorescein fluorescence corresponds to a nick in the oligonucleotide sequence and a dissociation of the quencher- and fluorophore-labeled fragments. Indeed, after hybridization to complementary DNA, the labeled ssDNA sequence is readily destroyed in the presence of the nicking endonuclease (Figure 2b, c). On the contrary, the DPA-nanoparticles show virtually no activity via fluorescence under identical conditions (Figure 2a, c). Notably, this observation is independent of the dye and quencher arrangement in the nanoparticle substrates. This is a critical observation, because for P1 there is the possibility that the fluorescein-labeled fragment may be quenched by neighboring, fully intact strands within the particle shell, whereas this is not possible for P2, as the fluorescein-labeled fragment should be free in solution following nicking (Tm ~ 37 °C). Alternatively, we reasoned that perhaps the lack of fluorescence increase for both P1 and P2 could be due to the possibility that a nicked sequence on the particle may not dissociate into solution due to the density of DNA in close proximity to the cleaved product. To rule out these possibilities, we analyzed the Tm of both single-stranded and nanoparticle-based systems following nuclease treatment (Figure 2d, e). This analysis confirms that the activity of the endonuclease on the ssDNA-complement duplex is accompanied by a significant decrease in the Tm of the duplex (Δ = −26.1 °C for ssDNA-1 and ssDNA-2), consistent with complete nicking of the oligonucleotide. By contrast, the Tm of the nanoparticle-complement duplex remains consistent (Δ = −0.5 °C for P1, −1.6 °C for P2) after nuclease treatment, thus indicating the presence of an intact 20-base oligonucleotide shell on the DPA nanoparticle.
Figure 2.
Endonuclease resistance of DPA nanoparticles. a) Scheme depicting DPA-nanoparticle (P2) resistance to nicking endonuclease (Nt.CviPII) and consequently intact, quenched duplex DNA at the particle surface. b) Scheme depicting dsDNA degradation by Nt.CviPII and consequently a decrease in duplex melting temperature and increase in fluorescein fluorescence. c) Nt.CviPII activity over time, monitored via fluorescein fluorescence dequenching (λex = 485 nm, λem = 535 nm). d) Thermal denaturation analysis with and without Nt.CviPII treatment for P1 and ssDNA-1. λabs = 260 nm. Sample subjected to enzyme for 100 minutes at 37 °C. ssDNA-1 + Complement: Tm = 63.9 °C; ssDNA-1 + Nt.CviPII + Complement: Tm = 37.8 °C; P1 + Complement: Tm = 58.8 °C; P1 + Nt.CviPII + Complement: Tm = 58.3 °C). e) Thermal denaturation analysis with and without Nt.CviPII treatment for P2 and ssDNA-2. λabs = 260 nm. Sample subjected to enzyme for 100 minutes at 37 °C. ssDNA-2 + Complement: Tm = 63.9 °C; ssDNA-2 + Nt.CviPII + Complement: Tm = 37.8 °C; P2 + Complement: Tm = 56.9 °C; P2 + Nt.CviPII + Complement: Tm = 55.3 °C). See Supporting Information Figure S12 for derivative plots of melting temperatures. Complement: 5′-TATTATATCTTTAGACACTGA CTGGACATGACTCT-3′
We note that unlike previously reported DNA-functionalized gold nanoparticle systems18 we do not see an enhanced melting temperature on the initial DPA-nanoparticles, which would have been indicative of cooperative hybridization of complementary DNA. Rather, we observe a slight depression in the melting temperature (Figure 2d, e); an observation consistent with steric hindrance at the interface between DNA and the hydrophobic polymer. This type of effect has been noted by others with respect to unusual DNA hybridization characteristics at interfaces.19
Exonuclease activity against DPA nanoparticles
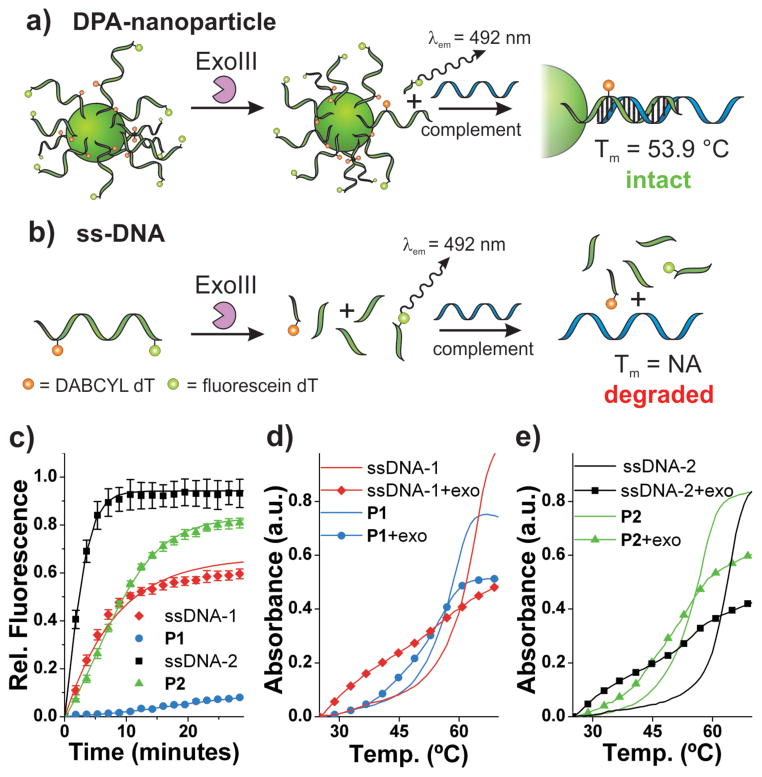
Given that DPA-nanoparticles exhibit a high level of resistance against a sequence-specific nicking endonuclease, we were interested in determining how they would respond as substrates to a non-specific 3′ exonuclease (Figure 3). Exonuclease III (Exo III, from E. coli) is reported to catalyze the stepwise removal of mononucleotides from the 3′-hydroxyl termini of duplex DNA with preferred substrates being blunt or recessed 3′ termini.20 However, in our hands, the enzyme exhibits indiscriminate activity on both ssDNA and dsDNA substrates (see Supporting Information Figure S9). Therefore, we analyzed the activity of Exo III against ssDNA and corresponding DPA-nanoparticles in the absence of any additional complementary DNA. Exo III activity against ssDNA and DPA-nanoparticles was monitored via fluorescein fluorescence dequenching over time. Upon initial observation, P1 appears to be highly resistant to Exo III digestion, while P2 appears to be degraded at a substantial rate. Indeed, a detailed kinetic analysis of P2 with respect to Exo III reveals that it is a substrate, albeit a significantly poorer one than ssDNA-2 with a 3-fold difference in the magnitude of the second order rate constant and a greater than 4-fold difference in initial rates (Figure 3c, Table 1). However, these kinetic data, derived from fluorescence measurements (see Supporting Information Figure S10), reveal only that P2 is indeed a substrate with respect to removal of the first few bases at the 3′-terminus; that is, at the location where fluorescein is liberated and hence detectable. Therefore, we reasoned that the apparent discrepancy between Exo III activity on P1 and P2 (as monitored by fluorescence) is most likely due to the fact that for P2, the fluorescein-labeled nucleotide is located only one base from the 3′ hydroxyl terminus. Therefore, liberation of the fluorescent product into solution (i.e. detection of fluorescence) only requires the removal of two bases. In the case of P1, the liberation of a DABCYL-labeled nucleotide does not have the same effect. Here, we conclude the fluorescein-labeled nucleotide is not liberated into solution but remains in an environment surrounded by DABCYL quencher molecules still present on intact, neighboring DNA strands, as well as neighboring guanosine bases, which are also known to quench fluorescein fluorescence.21 Therefore, the discrepancy between P1 and P2 response to Exo III is consistent with the nuclease digesting, or “shaving” away a limited fraction of 3′-terminal bases.
Figure 3.
Exonuclease resistance of DPA nanoparticles. a) Scheme depicting DPA-nanoparticle resistance to Exonuclease III and consequently, intact DNA at the particle surface available for hybridization with complementary ssDNA. b) Scheme depicting ssDNA being degraded by ExoIII and consequently, no intact DNA available for hybridization with complementary ssDNA. c) Exonuclease III activity over time monitored by fluorescein fluorescence dequenching (λex = 485 nm, λem = 535 nm). d) Thermal denaturation analysis with and without Exo III treatment for P1 and ssDNA-1. λabs = 260 nm. Samples subjected to enzyme for 60 minutes at 37 °C. P1 + Complement: Tm = 58.8 °C; P1 + Exo III + Complement: Tm = 55.8 °C). e) Thermal denaturation analysis with and without Exo III treatment for P2 and ssDNA-2. λabs = 260 nm. Samples subjected to enzyme for 60 minutes at 37 °C. P2 + Complement: Tm = 56.9 °C; P2 + Exo III + Complement: Tm = 53.9 °C). See Supporting Information Figure S13 for derivative plots of melting temperatures. Complement: 5′-TATTATATCTTTAGACAC TGACTGGACATGACTCT-3′
Table 1.
Exonuclease III kinetics on ssDNA-2 and P2 substrates
| Substrate | Initial Rate/× 10−9 M/s | Vmax/× 10−9 M/s | Km/× 10−6 M | kcat/s−1 | kcat/Km/× 105 s−1 •M−1 |
|---|---|---|---|---|---|
| ssDNA-2 | 18.6 | 50 | 10.6 | 1.42 | 1.3 |
| P2 | 4.4 | 12.5 | 7.0 | 0.36 | 0.5 |
To confirm observations and conclusions drawn from fluorescence kinetic studies (Table 1), and to determine the extent of digestion, a hybridization study via DNA duplex melting analyses was required. Briefly, DPA-nanoparticles or ssDNA analogues were allowed to react with Exo III for one hour before deactivating the enzyme with EDTA and heat. Following enzyme deactivation, an equimolar quantity of complementary DNA was allowed to hybridize to the nanoparticle or ssDNA. Thermal denaturation analysis reveals the absence of a melting transition in the case of both ssDNA strands indicating complete degradation following enzyme treatment (Figure 3d, e, see Supporting Information Figure S11, S13). By contrast, in the case of enzyme-treated DPA-nanoparticles, P1 and P2, we observe a sharp melting transition of the particle-duplex indicative of an intact DPA-nanoparticle. However, we do observe a slight decrease in the particle-duplex Tm (Δ = −3 °C in each case, Figure 3d, e, see Supporting Information Figure S13), consistent with fluorescence evidence suggesting that the enzyme digests several bases of the nanoparticle nucleic acid shell at the outer edge and is subsequently sterically hindered thus preventing complete digestion. Indeed, the data following this partial digestion are consistent with a duplex on the order of approximately 18 base pairs compared to 20 base pairs for the full-length sequence without enzymatic treatment. Indeed, encouraged by our results demonstrating DPA-nanoparticle resistance to Exo III, we aimed to determine whether a nuclease routinely used for complete digestion of synthetic oligonucleotides would yield similar results. In addition, we sought to answer whether DPA nanoparticles serve to protect DNA against general exonuclease digestion and not just specifically Exo III digestion. Therefore, we subjected the single-stranded particles and corresponding ssDNA analogues to Snake Venom Phosphodiesterase (Phosphodiesterase I from Crotalus adamanteus), an enzyme known for its aggressive 3′-exonuclease activity and routinely utilized for complete digestion of synthetic oligonucleotides.22,23
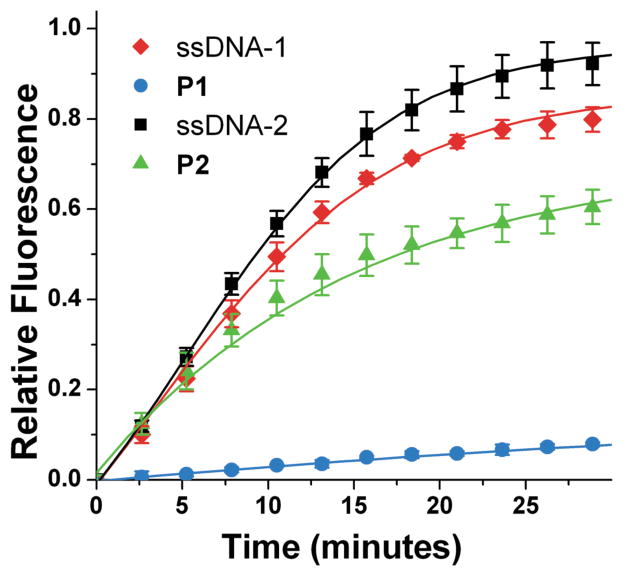
Indeed, based on fluorescence dequenching experiments identical to those for Exo III activity analysis, the DPA-nanoparticles exhibit exceptional resistance consistent with observations made utilizing Exo III (Figure 4). Although the relative initial rates differ between SVP and ExoIII depending on substrate, it is clear that the trends in activity are consistent between the two nucleases, that is, P2 is resistant compared to ssDNA-2 and P1 is resistant compared to ssDNA-1.
Figure 4.
SVP activity over time monitored by fluorescein fluorescence dequenching (λex = 485 nm, λem = 535 nm).
CONCLUSION
In summary, we have described a novel approach for rendering DNA resistant to two key classes of nuclease that are otherwise capable of rapidly degrading substrates in a sequence-selective or non-selective fashion. We propose steric hindrance due to dense packing is the simplest explanation for the observation that the endonuclease has undetectable activity on P1 and P2, whereas 3′-nucleases show some activity, but only at the outer few bases.
Inspiration for this investigation is drawn from the increasing interest in novel approaches for packaging and delivering nucleic acids for in vivo applications.3,4,24–26 This interest has led to an array of materials designed to facilitate potent and selective communication with important cellular machinery.27–34 Our approach is predicated on the idea that a key requirement for any enabling technology of this type is a well-defined nucleic-acid based material that maintains the integrity of the base-sequence in nuclease-rich environments. The utilization of DPA-nanoparticles for targeted delivery of intact hybridization competent nucleic acids in vitro and in vivo is currently underway in our laboratories. This approach, together with other well-defined DNA-based nanomaterials,10–12 constitutes a concerted effort to move away from amorphous, poorly defined, multicomponent, and cytotoxic polyplexed transfection agents.35,36 Finally, we note that this approach is likely general in terms of particle core chemistry, as other polymerization strategies are amenable to the incorporation of DNA, and potentially the preparation of resistance micellar particle systems.15,19,37
METHODS
Monomer, Termination Agent, and Polymer Synthesis
Synthesis of (N-Benzyl)-5-norborene-exo-2,3-dicarboximide (1)
See Supporting Information Figure S1 for chemical structure. Compound 1 was prepared according to a modification of a previously reported procedure.38 To a stirred solution of N-benzylamine (2.85 g, 26.6 mmol) in dry toluene (125 mL) was added 5-norbornene-exo-2,3-dicarboxylic anhydride (4.10 g, 25.0 mmol) and triethylamine (3.83 mL, 27.5 mmol). The reaction was heated to reflux overnight under an atmosphere of N2. The reaction was cooled to room temperature and washed with 10% HCl (3 × 50 mL) and brine (2 × 50 mL). The aqueous layers were combined and extracted with ethyl acetate (60 mL). The combined organic layers were dried with MgSO4, filtered and concentrated to dryness yielding a pale yellow solid that was then recrystallized from ethyl acetate/hexanes to give 1 (4.98 g, 79%) as white crystals. 1H NMR (CDCl3): δ (ppm) 1.07 (d, 1H, CH2, J=9.6 Hz,), 1.42 (d, 1H, CH2, J=9.6 Hz), 2.69 (s, 2H, 2 x CH), 3.26 (s, 2H, 2 x CH), 4.61 (s, 2H, CH2), 6.28 (s, 2H, CH=CH), 7.25–7.40 (m, 5H, Ar). 13C NMR (CDCl3): δ (ppm) 42.18, 42.28, 45.13, 47.62, 127.74, 128.48, 135.76, 137.76, 177.48. LRMS (CI), 253.99 [M+H]+. HRMS, theo: 254.1176 [M+H]+, found: 254.1175 [M+H]+.
Synthesis of (Z)-4,4′-(but-2-ene-1,4-diylbis(oxy)) dibenzoic acid (2)
See Supporting Information Figure S1 for chemical structure. To a stirred solution of ethyl 4-hydroxybenzoate (5.5 g, 33.1 mmol) in 100 mL dry DMF was added potassium carbonate (7.28 g, 52.7 mmol). To this stirred suspension was added cis 1,4-dichlorobutene (2.0 g, 16 mmol). The solution turned brown within minutes and the reaction was allowed to stir under an atmosphere of N2 at 90 °C overnight. The mixture was then cooled to room temperature, filtered, and concentrated to dryness. The resulting solid was dissolved in ethyl acetate and washed three times with H2O. The organic layer was dried over magnesium sulfate and concentrated to dryness to yield solid white crystalline needles. This solid was recrystallized from ether to yield the pure diester (2.18 g, 35%). 1H NMR (CDCl3): δ (ppm) 1.38 (t, 6H, 2 x CH3), 4.35 (q, 4H, 2 x CH2), 4.74 (d, 4H, 2 x CH2), 5.96 (t, 2H, CH=CH), 6.92 (d, 4H, 4 x ArH), 8.0 (d, 4H, 4 x ArH). The diester (2.18 g, 5.66 mmol) was dissolved in 95% ethanol and potassium hydroxide was added (12.0 g, 215 mmol). The reaction was heated to reflux for 5 hours, cooled to room temperature, diluted with an equal volume of H2O and acidified with HCl to form a white precipitate. The precipitate was filtered off to yield 2 as an orange-tan solid (1.78 g, 100%). 1H NMR (DMSO-d6, residual 1H = 2.50 ppm): δ (ppm) 3.38 (s broad, 2H, 2 x COOH), 4.80 (d, 4H, 2 x CH2), 5.89 (t, 2H, CH=CH), 7.03 (d, 4H, 4 x ArH), 7.87 (d, 4H, 4 x ArH). 13C NMR (DMSO-d6, residual 13C = 39.51 ppm): δ (ppm) 64.11, 114.50, 123.18, 128.33, 131.34, 161.72, 166.98. LRMS, 327.03 [M−H]−, HRMS, theo: 327.0874 [M−H]−, obs: 327.0877 [M−H]−.
Synthesis of Polymer (120-2)
See Supporting Information Figure S1 for chemical structure. Monomer 1 (870 mg, 3.4 mmol) was dissolved in 5 mL CDCl3 and cooled to −78 °C. Ruthenium catalyst (IMesH2)(C5H5N)2(Cl)2Ru=CHPh (124 mg, 0.17 mmol) was added as a powder, followed by 1 mL additional CDCl3 to solubilize the catalyst. The reaction was then allowed to warm to room temperature and stir under N2 for 35 minutes (NMR confirms the absence of the original olefin resonance from monomer 1 at 6.28 ppm, and the presence of broad cis and trans polymer backbone olefin resonances at 5.45 and 5.71 ppm). At this point, 200 μL of the reaction mixture was removed and quenched with an excess of ethyl vinyl ether to provide a homopolymer for molecular weight determination (SEC-MALS: Mn = 5221 g/mol, PDI = 1.075, Figure S2). Termination agent 2 (111 mg, 0.34 mmol) was dissolved in 2.0 mL DMF-d7, added to the reaction mixture, and the mixture was allowed to stir at room temperature for 20 minutes. The ruthenium alkylidene proton resonance was monitored in order to track the completion of the polymer termination event (Figure S3). Once termination was determined to be complete, excess ethyl vinyl ether was added to quench the ruthenium catalyst. The crude polymer was precipitated from cold methanol and further purified by column chromatography in order to eliminate any traces of unreacted termination agent. The crude precipitated polymer was dry loaded onto a silica column, the column was washed with 200 mL CH2Cl2, and the polymer was eluted with 3% methanol in CH2Cl2 to yield a glassy yellow-brown solid as the pure polymer (905 mg, 97%, rf = 0.56).
DNA Synthesis
Oligonucleotides ssDNA-1 and ssDNA-2 were synthesized in house using automated phosphoramidite chemistry and saccharin 1-methylimidazole as an activator. Standard 2-cyanoethyl protected phosphoramidites include dA (N-Bz), dG (N-dmf), (N-acetyl) dC, and T. Oligonucleotides were synthesized on a 1.0 μmol scale using columns packed with 1000 Å CPG beads. A 5′-amino modifer was incorporated into each synthetic oliognucleotide through use of 5′-amino modifer C12 phosphoramidite (Glen Research). In the case of ssDNA-1 and ssDNA-2, the 5′-amino terminus was acetylated on solid support using the automated synthesizer. The MMT group was removed by treatment with 3% trichloroacetic acid in CH2Cl2 for two minutes (until the yellow color due to the MMT+ cation was no longer visible in the eluting deblock solution) followed by a standard capping cycle to acetylate the free amine with acetic anhydride. Fluorescein and DABCYL labels were incorporated into the oligonucleotides via use of Fluorescein dT and DABCYL dT phosphoramidites (Glen Research). Oligos were cleaved from solid support and deprotected by treatment with AMA (concentrated NH4OH/40% methylamine, 1:1 v/v) at 55 ° C for 20 minutes, purified by HPLC, and characterized by MALDI-TOF MS. Complement DNA was purchased from Integrated DNA Technologies (purified by HPLC, confirmed by ESI-MS). Detailed sequences and enzyme recognition/cleavage sites are shown in Figure S4.
MALDI-TOF MS of Oligonucleotides
See Supporting Information Figure S5 for corresponding spectra. A MALDI target plate was spotted with 1 μL of matrix solution A for each sample to be analyzed and allowed 20 minutes to dry completely (matrix solution A was prepared by dissolving 50 mg of 3-HPA in 500 μL of HPLC acetonitrile/Nanopure H2O (1:1 v/v). 454 μL of this solution was mixed with 45 μL of 100 mg/ml diammonium hydrogen citrate in Nanopure H2O). Oligonucleotide samples were prepared for MALDI-TOF MS analysis using Zip-Tip C18 pipette tips. Oligos were loaded onto the C18 tips from concentrated stock solutions (ca. 50–100 μM) and eluted with matrix solution B (matrix solution B was prepared as follows: dissolve 50 mg of THAP in 500 μL of HPLC grade acetonitrile, assist dissolution by sonication and centrifuge the resulting solution to pellet any solid remaining, mix 250 μL of the supernatant with 250 μL of 23 mg/mL diammonium hydrogen citrate in Nanopure H2O). 1 μL of the oligonucleotide in matrix solution B was mixed with 1 μL of Oligonucleotide Calibration Standard (Bruker) dissolved in Nanopure H2O. 1 μL of this solution was then spotted onto the MALDI plate on top of crystallized matrix A. The samples were allowed to dry for 15–30 minutes before analyzing via MALDI-TOF MS.
HPLC Purification of Oligonucleotides
See Supporting Information Figure S5 for corresponding chromatograms. Synthetic oligonucleotides ssDNA-1 and ssDNA-2 were purified via reverse-phase HPLC using a binary gradient as indicated on each chromatogram in Figure S5 (Solvent A: 10% methanol in 50 mM triethylammonium acetate (TEAA) pH 7.1, solvent B: methanol). For ssDNA-2, weak anion-exchange (WAX) HPLC was also necessary to purify the oligonucleotide. A quaternary gradient was used for WAX HPLC analyses and purification (Solvent A: nanopure H2O, solvent B: methanol, solvent C: 100 mM Tris(hydroxymethyl)aminomethane (Tris) pH 8.0, solvent D: 2 M NaCl). Oligonucleotides were desalted post WAX HPLC purification using Sep-Pak Plus C18 Environmental Cartridges.
DNA-Polymer Amphiphile Synthesis and Micellar Nanoparticle Formation
To a solution of polymer (120-2) (150 mg, 27.8 μmol) dissolved in 250 μL DMF was added N,N-diisopropylethylamine (48 μL, 280 μmol) and HATU (10.6 mg, 28 μmol). The solution was vortexed for 10 minutes at room temperature in order to activate the polymer carboxylic acid terminus. At this point, 5′-amino modified DNA on CPG solid support (ca. 1 μmol, MMT deprotected) was added. The mixture was allowed to vortex at room temperature overnight. The CPG beads were filtered away from the solution using an empty synthesis column and then washed with DMF (2 × 20 mL) and CHCl3 (2 × 20 mL). The DNA-polymer amphiphile (DPA) was cleaved from solid support via treatment with AMA at 65 ° C for 30 minutes. The CPG beads were filtered off using glass wool and subsequently washed consecutively with H2O (2.0 mL), DMSO (2.0 mL), Formamide (2.0 mL), H2O (3.0 mL), and DMSO (1.0 mL). To form micellar nanoparticles this solution of DPA was transferred to 3,500 MWCO snakeskin dialsysis tubing (Thermo Scientific) and 2.0 mL H2O, used to wash the filtrate container, was added. The resulting solution was dialyzed against 2.0 L of Nanopure H2O overnight. This dialyzed solution was then concentrated to 3.0 mL via Speed Vac evaporation. The resulting crude DPA-nanoparticle/ssDNA mixture was analyzed by denaturing PAGE and agarose gel electrophoresis to confirm the presence of DPA conjugates and free ssDNA. It is important to note that low molecular weight ssDNA impurities (≤8295 g/mol) remained present despite extensive dialysis attempts (20k MWCO slide-a-lyzer dialysis cassette). Therefore, the crude mixture was purified via SEC FPLC (mobile phase: 10 mM Tris, 0.5 mM EDTA pH 8.3, flow rate: 2 ml/min, λabs = 260nm). The DPA-nanoparticles (P1/P2) elute at ca. 50 minutes (Figure S6). Crude P1/P2 samples were purified using HiLoad 16/60 Superdex 200 prep grade SEC media and exhibit a retention time differing from that of pure P1/P2 (Figure 6a, inset) as subsequent purifications and reinjections of pure material were performed using HiPrep 26/60 Sephacryl S-200 High Resolution SEC media.
Gel Electrophoresis
Denaturing PAGE was accomplished using Bio-Rad Criterion 15% TBE-Urea precast gels (#345-0091) and loading 200 ng of DNA per lane for each sample to be analyzed. In the case of crude conjugate, 400 ng of DNA was loaded per lane. Samples were prepared to load by mixing 1:1 (v/v) with TBE-Urea Sample Buffer (#161-0768, Bio-Rad) and heating to 90 °C for 2 minutes followed by rapid cooling on ice. The gels were run in 1x Tris/Boric Acid/EDTA (TBE) buffer pH 8.4 at 200V for 70 minutes, stained with ethidium bromide (200 ng/L) for 30 minutes and visualized using a Bio-Rad Fluor-S MultiImager.
DNA Concentration Determination
Nucleic acid concentrations were determined by UV absorbance at 260 nm using a quartz cuvette (Fisher # 14-385-928A, pathlength = 10 mm). An extinction coefficient of 294,554.58 L/mol•cm was used for ssDNA-1, ssDNA-2, P1, and P2. This coefficient was calculated as the extinction coefficient of the entire sequence without the two thymine modified bases (226,654.58 L/mol•cm, OligoCalc) plus the extinction coefficients for each dye-lableled base at 260 nm (38,800 L/mol•cm for Fluorescein dT, and 29,100 L/mol•cm for DABCYL dT, Glen Research). Due to the fact that P1 and P2 contain additional aromatic groups capable of absorbing UV radiation, a slight correction factor was introduced. This correction factor was calculated as the ratio of absorbance of ssDNA-1 or ssDNA-2 at 492 nm versus 260 nm (A260/A492). This correction factor was multiplied by P1 or P2 absorbance at 492 nm in order to calculate what the absorbance at 260 nm would be if the system behaved as the standard ssDNA analogues. This corrected absorbance at 260 nm was then averaged with the actual DPA-nanoparticle absorbance at 260 nm and used to determined nucleic acid concentration. For example, P1 A260 = 0.168 (0.57 μM) and A260 corrected = 0.130 (0.44 μM). Therefore, A260 average = 0.149 (0.50 μM).
Transmission Electron Microscopy
Copper grids (formvar/carbon-coated, 400 mesh copper, Ted Pella # 01754) were prepared by glow discharging the surface at 20 mA for 1.5 minutes followed by treatment with 3.5 μL 250 mM MgCl2 in order to prepare the surface for DPA nanoparticle adhesion. The MgCl2 solution was wicked away with filter paper and 3.5 μL of DPA nanoparticle (ca 50 μM DNA in 10 mM Tris pH 8.5) solution was deposited on the grid surface. This solution was allowed to sit for 5 minutes before being washed away with 4 drops of glass distilled H2O and subsequent staining with 3 drops of 1% w/w uranyl acetate. The stain was allowed to sit for 30 seconds before wicking away with filter paper. All grid treatments and sample depositions were on the dark/shiny/glossy formvar-coated face of the grid (this side face up during glow discharge). Samples were then imaged via TEM.
Fluorescence Measurements
Each experiment was measured in triplicate and plotted as a normalized average (ie time point zero was set to zero fluorescence) with standard deviation plotted as error bars. Sigmoidal fits were performed for each data set. Fluorescein fluorescence dequenching was monitored over time using a plate-reader and a 96 well plate (Corning, flat bottom non-binding surface #29110009). Time points were collected in 15-second intervals integrating three flashes per measurement. Identical gain and filter settings were used in every case. For measuring Nt.CviPII activity, the following conditions were used in each experiment: 5 μM ssDNA or DPA-nanoparticle, 300 nM ssDNA complement, 25 mM NaCl, 1X NE Buffer 4, 10 mM Tris pH 8.5, and 5 units of Nt.CviPII (100 units in 20 μL was diluted to 100 μL with 80 μL of Diluent A, 5 μL of this solution was used per reaction) all in 50 μL total volume. NE Buffer 4 and enzyme were mixed and added to each well. All other components were mixed and added to each enzyme/buffer-containing well simultaneously using a multi-channel pipettor. The plate reader was set to 37 °C for the duration of the 100 minute experiment. For measuring Exo III activity, the following conditions were used in each experiment: 5 μM ssDNA or DPA-nanoparticle, 25 mM NaCl, 50 mM Potassium Acetate, 1X NE Buffer 1, 10 mM Tris pH 8.5, and 10 units of Exo III (5,000 units in 50 μL was diluted to 500 μL with 450 μL of Diluent A, 1 μL of this solution was used per reaction) all in 50 μL total volume. NE Buffer 1 and enzyme were mixed and added to each well. All other components were mixed and added to each enzyme/buffer-containing well simultaneously using a multi-channel pipettor. The plate reader was set to 37 °C for the duration of the 60 minute experiment. For measuring SVP activity, the following conditions were used in each experiment: 5 μM ssDNA or DPA-nanoparticle, 25 mM NaCl, 50 mM Potassium Acetate, 12.5 mM MgCl2, 10 mM Tris pH 8.5, and 0.6 units of SVP (1.58 mg of lyophilized SVP powder (63 units/mg) was dissolved in 1.58 mL of buffer containing 100 mM Tris-HCl, pH 8.9, 110 mM NaCl, 15 mM MgCl2, and 50% glycerol, 10 μL of this solution was used per reaction) all in 50 μL total volume. MgCl2 and enzyme were mixed and added to each well. All other components were mixed and added to each enzyme/buffer-containing well simultaneously using a multi-channel pipettor. The plate reader was set to 37 °C for the duration of the 60 minute experiment.
DNA Melting Temperature Analysis
Melting temperature analyses were performed by heating each sample from 25 °C (5 minute equilibration time) to 70 °C using a temperature gradient of 1 °C/minute. Melting temperatures were calculated as first derivatives of the curve. Each strand was at a concentration of 0.83 μM. For melting analysis after Nt.CviPII treatment, the reaction mixture was heated to 70 °C for 20 minutes in order to denature the enzyme. The mixture was then cooled to room temperature and 228.1 μL of 10 mM Tris pH 8.5 was added, followed by 12.5 μL 2M NaCl and 9.4 μL of 24.8 μM complementary DNA in 10 mM Tris pH 8.5. Final concentrations of each strand are 0.83 μM and final NaCl concentration is 87.5 mM all in a total volume of 300 μL. At this point, the sample was heated at 90 °C for 5 minutes and then allowed to cool to room temperature over a period of 2 hours. The sample was refrigerated at 8 °C for 15 minutes and subsequently analyzed. For melting temperature analysis after Exo III treatment, after the reaction was complete (60 minutes), 10 μL of 0.5 M EDTA was added to inhibit the enzyme. The reaction was heated at 70 °C for 20 minutes and allowed to cool to room temperature. 217.5 μL of 10 mM Tris, followed by 12.5 μL of 2M NaCl and 10 μL of 24.8 μM complementary DNA was added. At this point the sample was treated identical to those in the case of Nt.CviPII.
Supplementary Material
Acknowledgments
We acknowledge support of this work from ARO (W911NF-11-1-0264), AFOSR (FA9550-11-1-0105), NIH (NIBIB - 1R01EB011633), NIH (DP2OD008724), and NIH (DA031591). N.C.G. acknowledges the Henry and Camille Dreyfus Foundation for a New Faculty Award and the Alfred P. Sloan Foundation. We acknowledge the UCSD Cryo-Electron Microscopy Facility (NIH, Agouron) and the UCSD Light Microscopy Facility (P30 grant NS047101). We thank N. Olson, J. Santini, A. Mrse, and Y. Su for their technical assistance.
Footnotes
Supporting Information Available. Experimental details and additional supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and Self-Assembly of Two-Dimensional DNA Crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar S, Hughes MD, Khan A, Bibby M, Hussain M, Nawaz Q, Double J, Sayyed P. The Delivery of Antisense Therapeutics. Adv Drug Deliv Rev. 2000;44:3–21. doi: 10.1016/s0169-409x(00)00080-6. [DOI] [PubMed] [Google Scholar]
- 3.Thomas M, Klibanov AM. Non-Viral Gene Therapy: Polycation-Mediated DNA Delivery. App Microbiol Biotechnol. 2003;62:27–34. doi: 10.1007/s00253-003-1321-8. [DOI] [PubMed] [Google Scholar]
- 4.Davis ME. Non-Viral Gene Delivery Systems. Curr Opinion Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 5.Leumann CJ. DNA Analogues: From Supramolecular Principles to Biological Properties. Bioorg Med Chem. 2002;10:841–854. doi: 10.1016/s0968-0896(01)00348-0. [DOI] [PubMed] [Google Scholar]
- 6.Eschenmoser A. Searching for Nucleic Acid Alternatives. Chimia. 2005;59:836–850. [Google Scholar]
- 7.Shi FX, Hoekstra DJ. Effective Intracellular Delivery of Oligonucleotides in Order to Make Sense of Antisense. Controlled Release. 2004;97:189. doi: 10.1016/j.jconrel.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Crooke ST, Lemonidis KM, Neilson L, Griffey R, Lesnik EA, Monia BP. Kinetic Characteristics of Escherichia-Coli Rnase H1 - Cleavage of Various Antisense Oligonucleotide-RNA Duplexes. Biochem J. 1995;312:599. doi: 10.1042/bj3120599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veal GJ, Agrawal S, Byrn RA. Sequence-Specific RNase H Cleavage of gag mRNA from HIV-1 Infected Cells by an Antisense Oligonucleotide in Vitro. Nucleic Acids Res. 1998;26:5670. doi: 10.1093/nar/26.24.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keum JW, Bermudez H. Enhanced Resistance of DNA Nanostructures to Enzymatic Digestion. Chem Commun. 2009:7036–7038. doi: 10.1039/b917661f. [DOI] [PubMed] [Google Scholar]
- 12.Hamblin GD, Carneiro KMM, Fakhoury JF, Bujold KE, Sleiman HF. Rolling Circle Amplification-Templated DNA Nanotubes Show Increased Stability and Cell Penetration Ability. J Am Chem Soc. 2012;134:2888–2891. doi: 10.1021/ja2107492. [DOI] [PubMed] [Google Scholar]
- 13.Leitgeb A, Wappel J, Slugovc C. The ROMP Toolbox Upgraded. Polymer. 2010;51:2927–2946. [Google Scholar]
- 14.Chien MP, Rush AM, Thompson MP, Gianneschi NC. Programmable Shape-Shifting Micelles. Angew Chem, Int Ed. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alemdaroglu FE, Herrmann A. DNA Meets Synthetic Polymers - Highly Versatile Hybrid Materials. Org Biomol Chem. 2007;5:1311–1320. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]
- 16.Hurst SJ, Lytton-Jean AKR, Mirkin CA. Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal Chem. 2006;78:8313–8318. doi: 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia YN, Morgan R, Schildkraut I, Vanetten JL. A Site-Specific Single-Strand Endonuclease Activity Induced by Nys-1 Virus-Infection of a Chlorella-Like Green-Alga. Nucleic Acids Res. 1988;16:9477–9487. doi: 10.1093/nar/16.20.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lytton-Jean AKR, Mirkin CA. A Thermodynamic Investigation into the Binding Properties of DNA Functionalized Gold Nanoparticle Probes Molecular Fluorophore Probes. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 19.Chen XJ, Sanchez-Gaytan BL, Hayik SEN, Fryd M, Wayland BB, Park SJ. Self-Assembled Hybrid Structures of DNA Block-Copolymers Nanoparticles with Enhanced DNA Binding Properties. Small. 2010;6:2256–2260. doi: 10.1002/smll.201001185. [DOI] [PubMed] [Google Scholar]
- 20.Putney SD, Benkovic SJ, Schimmel PR. A DNA Fragment with an Alpha-Phosphorothioate Nucleotide at One End Is Asymmetrically Blocked from Digestion by Exonuclease-III and Can Be Replicated in Vivo. Proc Natl Acad Sci USA. 1981;78:7350–7354. doi: 10.1073/pnas.78.12.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A. Effect of Primary and Secondary Structure of Oligodeoxyribonucleotides on the Fluorescent Properties of Conjugated Dyes. Nucleic Acids Res. 2002;30:2089–2095. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho NWY, Gilham PT. Analysis of Polydeoxyribonucleotides by Digestion with Phosphatase Phosphodiesterases. Biochim Biophys Acta. 1973;308:53–58. doi: 10.1016/0005-2787(73)90121-4. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Wang RS, Ding L, Sha RJ, Seeman NC, Canary JW. Templated Synthesis of Nylon Nucleic Acids Characterization by Nuclease Digestion. Chem Sci. 2012;3:1930–1937. doi: 10.1039/C2SC20129A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ledley FD. Nonviral Gene-Therapy - the Promise of Genes as Pharmaceutical Products. Hum Gene Ther. 1995;6:1129–1144. doi: 10.1089/hum.1995.6.9-1129. [DOI] [PubMed] [Google Scholar]
- 25.Jeong JH, Park TG, Kim SH. Self-Assembled and Nanostructured siRNA Delivery Systems. Pharm Res. 2011;28:2072–2085. doi: 10.1007/s11095-011-0412-y. [DOI] [PubMed] [Google Scholar]
- 26.Troiber C, Wagner E. Nucleic Acid Carriers Based on Precise Polymer Conjugates. Bioconjugate Chem. 2011;22:1737–1752. doi: 10.1021/bc200251r. [DOI] [PubMed] [Google Scholar]
- 27.Citti L, Rainaldi G. Synthetic Hammerhead Ribozymes as Therapeutic Tools to Control Disease Genes. Curr Gene Therapy. 2005;5:11–24. doi: 10.2174/1566523052997541. [DOI] [PubMed] [Google Scholar]
- 28.Ulijn RV. Enzyme-Responsive Materials: A New Class of Smart Biomaterials. J Mater Chem. 2006;16:2217–2225. [Google Scholar]
- 29.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Cellular Uptake of DNA block Copolymer Micelles with Different Shapes. Macromol Rapid Commun. 2008;29:326–329. [Google Scholar]
- 30.Lin CX, Rinker S, Wang X, Liu Y, Seeman NC, Yan H. In Vivo Cloning of Artificial DNA Nanostructures. Proc Natl Acad Sci USA. 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston APR, Lee L, Wang YJ, Caruso F. Controlled Degradation of DNA Capsules with Engineered Restriction-Enzyme Cut Sites. Small. 2009;5:1418–1421. doi: 10.1002/smll.200900075. [DOI] [PubMed] [Google Scholar]
- 32.Alvarez-Erviti L, Seow YQ, Yin HF, Betts C, Lakhal S, Wood MJA. Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 33.Cutler JI, Auyeung E, Mirkin CA. Spherical Nucleic Acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 34.Lee H, Lytton-Jean AKR, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, et al. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo siRNA Delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neu M, Fischer D, Kissel T. Recent Advances in Rational Gene Transfer Vector Design Based on Poly(ethylene imine) and its Derivatives. J Gene Medicine. 2005;7:992. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- 36.Lv HT, Zhang SB, Wang B, Cui SH, Yan J. Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J Controlled Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Zhang Y, Fullhart P, Mirkin CA. Reversible and Chemically Programmable Micelle Assembly with DNA Block-Copolymer Amphiphiles. Nano Lett. 2004;4:1055–1058. [Google Scholar]
- 38.Ku TH, Chien MP, Thompson MP, Sinkovits RS, Olson NH, Baker TS, Gianneschi NC. Controlling and Switching the Morphology of Micellar Nanoparticles with Enzymes. J Am Chem Soc. 2011;133:8392–8395. doi: 10.1021/ja2004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.