Abstract
Messenger ribonucleic acid (mRNA) turnover is a major control point in gene expression. In mammals, many mRNAs encoding inflammatory cytokines, oncoproteins, and G-protein-coupled receptors are destabilized by the presence of AU-rich elements (AREs) in their 3′-untranslated regions. Association of ARE-binding proteins (AUBPs) with these mRNAs promotes rapid mRNA degradation. ARE/poly(U)-binding/degradation factor 1 (AUF1), one of the best-characterized AUBPs, binds to many ARE-mRNAs and assembles other factors necessary to recruit the mRNA degradation machinery. These factors include translation initiation factor eIF4G, chaperones hsp27 and hsp70, heat-shock cognate protein hsc70, lactate dehydrogenase, poly(A)-binding protein, and other unidentified proteins. Numerous signaling pathways alter the composition of this AUF1 complex of proteins to effect changes in ARE-mRNA degradation rates. This review briefly describes the roles of mRNA decay in gene expression in general and ARE-mediated decay (AMD) in particular, with a focus on AUF1 and the different modes of regulation that govern AUF1 involvement in AMD.
INTRODUCTION
Regulation of gene expression is essential for all organisms as it permits proper development and adaptability to the environment.1 In eukaryotes, two vital processes that allow cells to respond to stimuli involve messenger ribonucleic acid (mRNA) stability and translation (reviewed in Ref 2). The balance between mRNA decay and mRNA synthesis, pre-mRNA splicing, mRNA transport, and translation contributes to the levels of mRNAs and thus protein levels.3 Many oncogenes and cytokine genes are regulated by mRNA stability to ensure correct levels and timing of expression, and aberrant control of their regulation has been implicated in diseases like cancer, chronic inflammatory diseases, and coronary disease.4
Multiple players of mRNA decay have been recognized and characterized, and some mechanisms involving them have been elucidated. Messenger RNA turnover has been linked to translation, as elements within the mRNA that confer stability (e.g., 7 mGpppG cap and poly(A) tail) are associated with translation initiation factors [e.g., eIF4F and poly(A)-binding protein (PABP)] (Ref 5, reviewed in Ref 6). Messenger RNA decay pathways in mammalian cells are either deadenylation dependent or deadenylation independent (Figure 1) (reviewed in Ref 7). We direct our attention to the deadenylation-dependent pathway, as it is the one most often observed in ARE-mediated decay (AMD). It involves exonucleolytic removal of the poly(A) tail, followed by degradation of the mRNA body, either by decapping and 5′ → 3′ degradation or by continued exonucleolytic degradation in the 3′ → 5′ direction (reviewed in Refs 8-10). Messenger RNA breakdown involves trans-acting factors that interact with cis-acting elements within the mRNA. These cis-acting determinants can be found in the 5′-untranslated region (UTR), the 3′-UTR, and/or the coding region of the transcript. Although a plethora of cis-acting elements have been identified, including the histone mRNA 3′-terminal stem loop, the iron-response element, nonsense mutations, and long-range stem loop of insulin-like growth factor II, we only describe those involving AU-rich elements (AREs)3 and briefly comment on a case involving a destabilizing determinant within the coding region of FOS. However, readers interested in these elements are directed to some first-rate reviews on the topic.3,7
FIGURE 1.
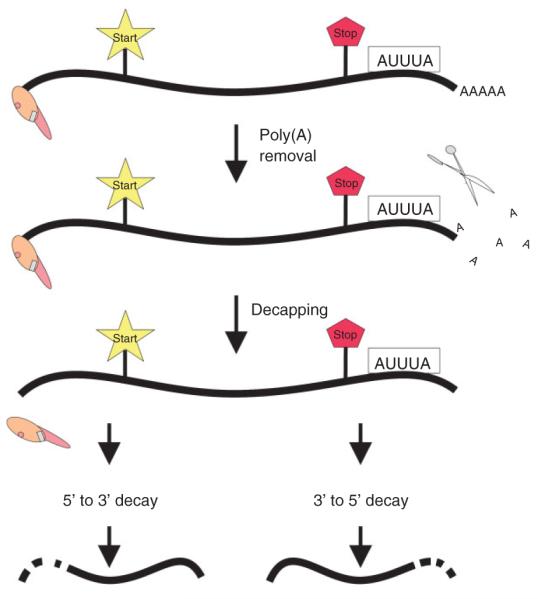
Schematic of deadenylation-dependent, AU-rich element (ARE)-mediated messenger ribonucleic acid (mRNA) degradation. The mRNA is depicted with a 5′-cap structure and 3′-poly(A) tail. The ARE is shown as the boxed AUUUA. The start and stop codons are highlighted. The sequential pathway involves exoribonucleolytic removal of the poly(A) tail, followed by decapping and subsequent 5′ → 3′ and/or 3′ → 5′ degradation.
Unstable mRNAs, including those encoding inflammatory mediators, cytokines, oncoproteins, and G-coupled receptors, contain AREs in their 3′-UTRs that promote their rapid degradation.11 AREs are typically 50–150 nucleotides and, as their name implies, are rich in adenosine and uridine bases (reviewed in Ref (2)). They often consist of one or more AUUUA motifs located within a uracil-rich region in the 3′-UTR.9,12 Two classes of AREs have been defined based on the number and distribution of the AUUUA pentamers within the mRNA; a third class has no AUUUA pentamers, but rather AU-rich sequence. Their decay is dependent upon active translation of the mRNA in many cellular systems.13,14 AMD is responsive to exogenous factors, including cytokines and growth factors. This is consistent with observations that AREs play an important role in the expression of genes during cell growth and differentiation.3
Twenty or so trans-acting factors, known as ARE-binding proteins (AUBPs), bind ARE-mRNAs to control AMD. AUBPs that affect mRNA degradation include ARE/poly(U)-binding/degradation factor 1 (AUF1)/hnRNP D, human antigen R (HuR), tristetrapolin, K-homology splicing regulatory protein, and butyrate-responsive factor-1. Their binding to ARE-mRNAs affects stability, translation, or sub-cellular localization of the mRNA.4
HuR is expressed in all proliferating cells and has a stabilizing effect (reviewed in Ref 15). Its overexpression causes an accumulation of target mRNAs16 and its nucleocytoplasmic export may be intrinsically coupled to mRNA stabilization.17 HuR shares several targets with AUF1, including mRNAs encoding p21 and cyclin D1,18 suggesting possible competitive binding or co-occupancy mechanisms that could dictate mRNA fate. For example, live-cell fluorescence resonance energy transfer (FRET) experiments revealed colocalization of the two in the nucleus and cytoplasm19 and co-occupancy of the two on select mRNAs.20 As well, HuR and AUF1 are developmentally expressed to similar levels among the same tissues from C57BL6 embryos and male mice, with increased levels in spleen, thymus, and fetal liver; moderate levels in lung; and low levels in adult tissue.21 Thus, the interplay of AUBPs may offer versatility for control of mRNA decay rates. The following section focuses on AUF1 and its destabilizing role.
ARE/POLY(U)-BINDING/DEGRADATION FACTOR
One of the first AUBPs identified was AUF1,22 also known as hnRNP D.23 It was isolated using an in vitro mRNA decay system that reproduced rapid MYC mRNA degradation observed in cells.24 AUF1 also binds selected AREs with high affinity and has a destabilizing effect on ARE-mRNAs.11,25 Moreover, the different binding affinities of AUF1 to a variety of AREs are proportional to the potency of the AREs to destabilize the mRNA.11,25,26 Numerous subsequent experiments indicated that the uridylate residues may be key for AUF1 to identify its target mRNAs. However, AUF1 is not solely a poly(U)- or AUBP, as in vitro experiments have demonstrated that it can also bind poly(A).27 This binding showed properties that suggested oligomeric potential and co-operative binding. In the following sections we focus on the description of the AUF1 family of proteins, how are they generated, their structure and function, and the regulation they undergo.
Protein Family with Diverse Functions
AUF1 was identified as 37- and 40-kDa polypeptides that copurified with fractions of a 130,000g, postribosomal supernatant that accelerated degradation of MYC mRNA in a cell-free mRNA degradation system.24 Immunoblot analyses revealed a third, immunologically related 45-kDa polypeptide in nuclear extracts. This was the first clue that pointed at a family of proteins that could be expressed in different subcellular compartments, perhaps accounting for different activities. For example, yeast two-hybrid experiments identified AUF1 as a member of the α-globin mRNA stability complex.28 In NIH-3T3 cells, AUF1 functions as an mRNA-stabilizing factor upon overexpression.29,30 Specifically, the p40AUF1 and p45AUF1 isoforms have been proposed to have stabilizing roles. p45AUF1 binding to estrogen receptor mRNA is induced by estradiol, stabilizing the transcript.31 However, specific knockdown of these two isoforms by small interfering RNA (siRNA) also results in elevated levels of granulocyte-macrophage colony-stimulating factor (GM-CSF). These findings suggest a dual role as stabilizing and destabilizing proteins for the two isoforms.32 AUF1 immunoprecipitation experiments demonstrated a complex with lactate dehydrogenase (LDH).33 Affinity screening and nuclear magnetic resonance (NMR) suggested a role in telomere maintenance.34,35 Also, a role in transcriptional activation has been suggested for AUF1.36,37 It is found in a complex with the LR1 DNA-binding complex that activates the Epstein-Barr virus promoter,38 and more recently, we reported a role for AUF1 in MYC mRNA translation.39 Although in this review we focus on AMD, we note that there is at least one case in which the p37AUF1 isoform is in a protein complex involved in FOS mRNA degradation, directed by a major protein-coding region (mCRD) determinant of instability. The complex also includes PABP, PABP-interacting protein 1, and the RNA-binding proteins UNR and NSAP1.40 Today we know that this family consists of four proteins named according to their apparent molecular size p37AUF1, p40AUF1, p42AUF1, and p45AUF1; this section describes them and their involvement in AMD in detail.
The AUF1/HNRPD genes map to human chromosomes 4 and X, with sublocalization to chromosomal regions 4q21.1-q21.2 and Xq12; the X chromosome locus is a pseudogene.38,41,42 Amino acid sequence comparisons between murine and human showed 97% conservation, underscoring essential functions for the AUF1 family of proteins.36,38,43 Gene organization studies revealed four AUF1 isoforms exhibiting diverse RNA-binding activities and specificities. These isoforms are differentially expressed in some cell types41,44; this may affect the control of mRNA degradation rates. For example, p37AUF1 and p40AUF1 in mononuclear cells (MNCs) are expressed exclusively in newborns, whereas p42AUF1 is expressed in MNCs from both newborns and adults; p45AUF1 on the other hand is present only in adult MNCs.44 This might explain in part why neonatal myelopoiesis is undeveloped due to increased degradation of cytokine transcripts. Specifically, there is a higher turnover of GM-CSF mRNA within neonatal versus adult MNCs.45,46 This could be due to the exclusive expression of p37AUF1 in MNCs from cord, as this isoform exhibits the highest degradation potential.29 AUF1 is also differentially expressed in the submaxillary gland (SMG) of male and female mice and regulated by testosterone and dihydrotestosterone in a tissue-specific manner in mice. Specifically, the p45AUF1 isoform is predominant in the nucleus of both female and male SMG and appears to be the only isoform to be influenced by the hormones in this compartment.47 These observations, together with isoform-specific knockdown by siRNAs, suggest that ARE-mRNA stability might be differentially influenced by individual levels of the AUF1 isoforms, rather than the absolute levels of all isoforms.32
Structure and Function
All four AUF1 isoforms contain the same two, nonidentical RNA recognition motifs (RRMs), N-terminus, and glutamine-rich domain.24,25 Although there is no crystal structure of the AUF1 proteins, the NMR structure of RRM1 has been resolved.48 These data together with sequence comparisons to other hnRNP proteins suggest an RRM folding pattern involving four, antiparallel β-sheets with two α-helices packed within the faces of the β-sheets.25,49 However, the AUF1–RNA interaction appears to be more complex than that of other classic hnRNP proteins. Homology modeling of AUF1’s RRM2 to that of RRMs of other hnRNP proteins and electromobility shift assays demonstrated that AUF1 binding to the RNA is achieved mainly through stacking interactions (conserved amino acids) with no base-specific recognition (variable amino acids).50
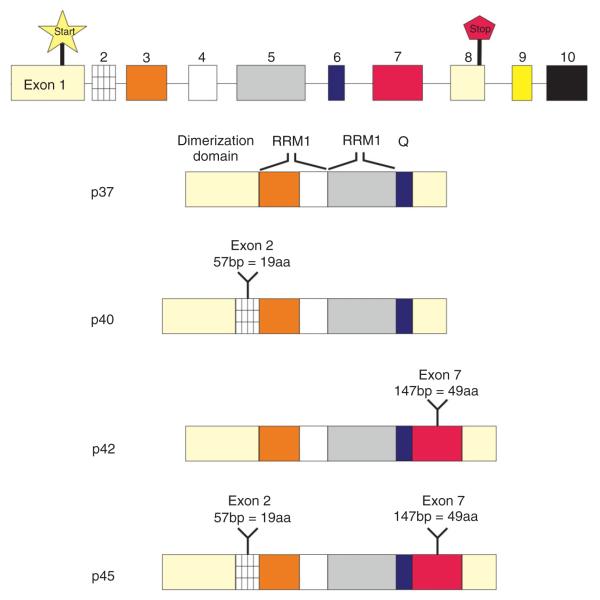
The isoforms differ in the presence or absence of two regions—an N-terminal, 19-amino acid insert or a C-terminal, 49-amino acid insert. In increasing order of length, the smallest isoform is p37AUF1, which lacks both amino acid inserts, followed by p40AUF1, which has the 19-amino acid insert; p42AUF1 has the 49-amino acid insert, and the largest isoform, p45AUF1, has both the 19- and 49-amino acid inserts (Figure 2). As shown by RNA-binding experiments with various AUF1 mutants, the two RRMs are required for binding to RNA; amino acids neighboring the RRMs also contribute to maximal RNA binding.25 The spliced isoforms likely provide functional versatility within the AUF1 family of proteins because they all exhibit different ARE-binding affinities44 (Table 1).
FIGURE 2.
ARE/poly(U)-binding/degradation factor 1 proteins are generated by alternative pre-messenger ribonucleic acid (mRNA) splicing. The AUF1/HNRPD gene has 10 exons (not shown to scale). The start codon in exon 1 and the stop codon in exon 8 are highlighted. The domain structures are shown in increasing order of apparent molecular weights. The dimerization domain, RRM1, RRM2, and Q-rich domain common to all isoforms are highlighted. Alternatively spliced exons 2 and 7 are represented by the stripped box and the red box, respectively. (Reprinted with permission from Ref. 44 Copyright 1998 Elsevier).
TABLE 1.
Binding Activities of Wild-type His6-AUF1 Isoforms for the FOS ARE
| AUF1 Isoform | Binding Affinity (nm) |
|---|---|
| His6-p37 | 9.5 ± 0.5 |
| His6-p40 | 330 ± 190 |
| His6-p42 | 40 ± 10 |
| His6-p45 | 90 ± 30 |
The isoforms are dictated by different exon–exon combinations. The AUF1/HNRPD gene has 10 exons; the first exon encodes the 5′-UTR and the N-terminus common to all isoforms; exon 2 contains the 19-amino acid insert found in p40AUF1 and p45AUF1. Exons 3–5 encode RRM1, RRM2, and the glutamine-rich domain; exon 7 encodes the 49-amino acid insert found in p42AUF1 and p45AUF1; exon 8 encodes the C-terminus and part of the 3′-UTR. Contrary to most other genes, the AUF1/HNRPD gene contains two extra exons (exons 9 and 10) after the stop codon. Exon 9 is a 107-nucleotide, alternatively spliced region of the 3′-UTR; exon 10 contains the remainder of the 3′-UTR and poly(A) signal. These additional exons (and introns) perhaps provide additional means of regulation.44,51 Indeed, AUF1 binds these intron sequences, suggesting self-regulatory ribonucleoprotein (RNP) interactions (Ref 51, reviewed in Ref 52). As well, analysis of spliced variants revealed an exon–exon junction 50 nucleotides after the translational stop codon (for two of the variants), suggesting that some of the AUF1 variants might be subject to nonsense-mediated decay (NMD). Experiments using knockdowns of the NMD factors Upf1 and Upf2 resulted in increased levels of these two variants, suggesting links between AMD and NMD.53 This result was further confirmed by accumulation of AUF1 splice variants in a splicing-sensitive microarray after blocking NMD.54
AUF1 is predominantly nuclear, but all isoforms shuttle between the nucleus and cytoplasm (reviewed in Ref 2). The C-terminal domains of p37AUF1 and p40AUF1 contain a nuclear import signal (NIS), while insertion of exon 7-encoded amino acids in p42AUF1 and p45AUF1 disrupt the NIS to promote cytoplasmic localization. Exon 7 contains a nuclear export signal instead55 and allows shuttling of the larger isoforms regardless of their interaction with the nuclear matrix.56 Deletion of the C-terminal domain of p37AUF1 suggested a mechanism by which p37 enters the nucleus, binds target mRNAs, and brings them to the cytoplasm where it effects its role.57 Later studies with p40AUF1 identified the C-terminal, 19-amino acids responsible for nucleocytoplasmic shuttling to be SGYGKVSRRGGHQNSYKPY and showed its interaction with the nuclear import receptor transportin (Trn-1).58 Later experiments identified 14-3-3σ, a member of a family of acidic proteins, as an AUF1-interacting protein. It interacts strongly with p37AUF1 and to a lesser extent with p40AUF1, but not with p42AUF1 and p45AUF1. The interaction between 14-3-3σ and p37AUF1 results in AUF1 retention in the cytoplasm.59 This demonstrates a very organized and tight control of the AUF1 protein localization and shuttling ensuring diverse expression of the different isoforms that guarantees versatile activities among them.
Post-translational Modifications Regulate the AUF1 Family of Proteins
Numerous signal transduction events have been related to regulation, highlighting the importance of post-translational modifications. The coactions of the multiple cis- and trans-acting factors involved in AMD allow for multiple regulatory events to occur. Depending on the signal, AMD can be either promoted or inhibited. The various AUF1 proteins are modified post-translationally by methylation, phosphorylation, glycosylation, and ubiquitination. We will limit our discussion to phosphorylation and ubiquitination.
When initially performed, immunoprecipitation experiments using extracts of cell metabolically labeled with 32P showed phosphorylation of p40AUF1 to a high extent, suggesting that its activity might be controlled by protein kinases. PhosphoBase, a database of phosphorylation sites, predicted two serines, Ser83 and Ser87, within exon 2-encoded amino acids as possible kinase sites. Protein kinase A and glycogen synthase kinase 3-beta can phosphorylate p40AUF1 in vitro on Ser87 and Ser83, respectively36 (Figure 3). The in vivo action of these kinases on AUF1 has not been confirmed yet. Importantly, Ser83 phosphorylation may require prior Ser87 phosphorylation, pointing at a hierarchical mode of regulation. PhosphoBase also predicted seven additional phosphorylation sites, but as of now, these sites have not been fully investigated, except for a CKI site at Thr91; thus, we will restrict our discussion to Ser83 and Ser87. The phosphorylation state of AUF1 may affect mRNA decay kinetics. The possible effects this could have are numerous, including changes in ARE-binding affinity, protein oligomerization potential, mRNP conformation, and interaction with other proteins.60 The fact that these sites mapped to exon 2-encoded amino acids suggests that p45AUF1 might also be phosphorylated at these sites, allowing for more versatile regulation among this family of proteins. To our knowledge, this hypothesis has not been further investigated.
FIGURE 3.
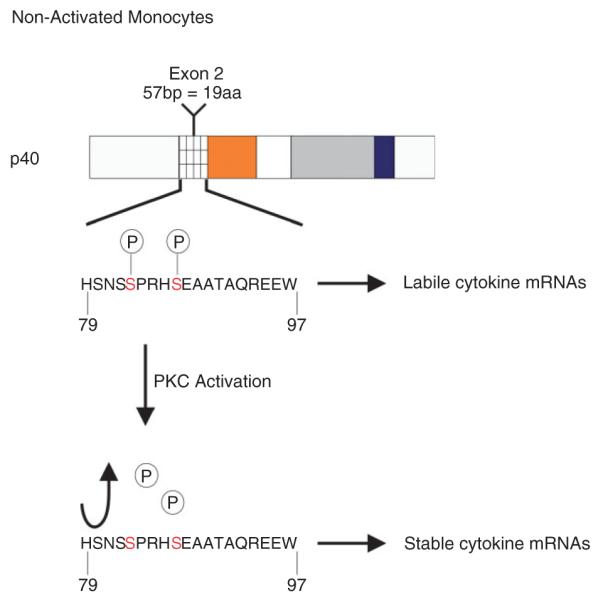
p40AUF1 phosphorylation changes upon PKC: protein kinase c activation. In nonactivated monocytes, Ser 83 and Ser 87 are phosphorylated, concomitant with labile cytokine messenger ribonucleic acids (mRNAs). PKC activation promotes loss of phosphates from Ser 83 and Ser 87, concomitant with cytokine mRNA stabilization.
The major system employed to address the effects of AUF1 phosphorylation on the fate of mRNA has been monocytes. While monocytes circulate in the bloodstream, cytokine mRNAs are very unstable, but upon adhesion of these cells at locations of tissue damage, they penetrate tissues by extravasation. Throughout the motility phase, cytokine mRNAs remain unstable, but upon their exposure to endotoxin, cell motility halts and the transcripts undergo stabilization. The stabilized transcripts are believed to localize to sites of contact between the monocyte and the extracellular matrix. Although the mechanism by which cytokine mRNA stabilization occurs in adhered monocytes remains elusive, one hypothesis to explain it is that AUBPs, mRNA decay factors, and cytokine transcripts relocate to distinct intracellular regions.61 AUF1 complexes have been found in circulating monocytes, but are altered upon adhesion.62 In the THP-1 pro-monocytic cell system, Ser83 and Ser87 of polyribosome-associated p40AUF1 are dephosphorylated after exposure of cells to the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA). TPA activates PKC signaling and mimics adhesion of monocytes to extracellular matrix60 (Figure 3). Dephosphorylation of p40AUF1 occurs concomitantly with stabilization of the IL1β and TNFα mRNAs. But how does the phosphorylation state of AUF1 affect its activity?
The phosphorylation state of p40AUF1 affects an ARE-RNA chaperone-like activity it possesses. In vitro FRET experiments revealed that nonphosphorylated p40AUF1 promotes formation of a condensed, less flexible structure within the TNFα ARE.63 While dually phosphorylated p40AUF1 has a slightly lower (twofold) binding affinity for the TNFα ARE, it maintains the RNA in a less condensed form.63 These observations suggest that AUF1 may control AMD by phosphorylation state-dependent alterations in ARE-RNA presentation, which might then affect assembly of other proteins with the ARE or mRNA.
Recently, the oncogenic tyrosine kinase known as nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) was identified as a new AUF1-interacting partner. AUF1 binding to NPM-ALK occurs through the ALK domain and results in hyperphosphorylation of AUF1. In vivo experiments demonstrated that NPM-ALK binds all the isoforms to some extent, but mass spectrometric analysis consistently revealed the p45AUF1 isoform bound to NPM-ALK in murine and human cell lines. Both proteins colocalized within a distinct type of cytoplasmic granule. The hyperphosphorylated state of AUF1 resulted in stabilization of ARE-mRNAs.64
Another important means of regulating AUF1 is protein degradation. This process facilitates critical homeostatic functions in organisms.65 Despite different protein degradation pathways, we focus on the ubiquitin-protesome degradation pathway (UPP). Experiments examining connections between mRNA degradation and heat shock, a process tightly associated with UPP, have established links between AMD, UPP, and AUF1 degradation.66 Heat shock stabilizes ARE-mRNAs and leads to perinuclear sequestration of AUF1 by hsp70, a chaperone protein that has also been described as an AUBP.67 AUF1 immunoprecipitation with lysates of heat-shocked cells resulted in pull-down of heat-shock cognate protein hsc70, a protein known to promote ubiquitin-dependent decay. This was the first clue that AUF1 might be ubiquitinated and degraded by the UPP. We know now from in vitro data that p37AUF1 and p40AUF1 are prime targets for ubiquitination and degradation by the proteasome. p40AUF1 is three to four times less ubiquitinated than p37AUF1, consistent with the higher mRNA degradation potential of p37AUF1. The p42AUF1 and p45AUF1 isoforms on the other hand are the least ubiquitinated. They both contain the exon 7-encoded amino acids that appear to block AUF1 ubiquitination in cis, perhaps by altering protein structure.68 These observations point to possibly concurrent degradation of AUF1 and ARE-mRNAs, though the exact mechanism remains elusive.
AUF1-INTERACTING PROTEINS
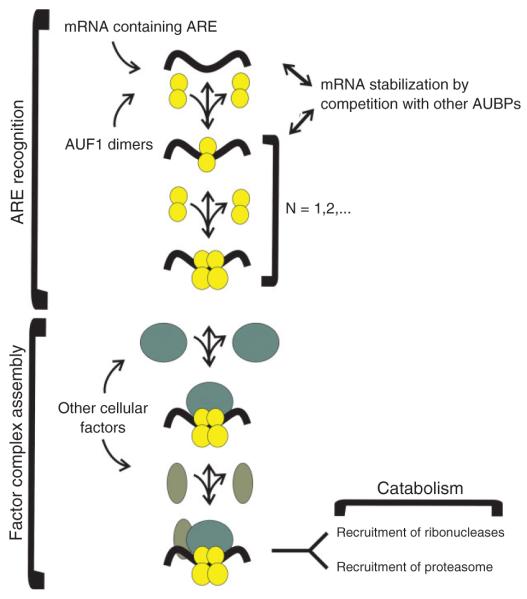
Rapid turnover of ARE-containing mRNAs is the result of protein–ARE interactions as well as numerous protein–protein interactions involving AUF1. A model of possible AUF1 function has been proposed69 (Figure 4). AUF1 first forms a dimer (either a homodimer or heterodimer) in the absence of an ARE and this requires the alanine-rich N-terminus; sequential dimer binding forms AUF1 oligomers on the ARE.25,26 We and others have shown that p37AUF1–p37AUF1 dimers also form in live cells in both the nuclear and cytoplasmic compartments, as demonstrated by live-cell FRET experiments.19,69 We believe that ARE-dependent AUF1 oligomerization may provide a large platform for other cellular factors to assemble.
FIGURE 4.
Three-step model of ARE/poly(U)-binding/degradation factor 1 (AUF1)-dependent, AU-rich element (ARE)-mediated decay. The first step, dynamic AUF1 dimer binding and oligomerization, may be sufficient for this phase of assembly. Other association of ARE-binding proteins, including the Hu family of messenger ribonucleic acid (mRNA)-stabilizing proteins, may compete for AUF1 binding to the ARE at this stage, thus preventing AUF1 oligomerization and subsequent factor recruitment. The second step, trans-acting complex assembly, involves association of AUF1 with eIF4G, PABP, hsp/hsc70, hsp27, and additional unidentified proteins. The third step, mRNA catabolism, involves two linked catabolic steps—ubiquitin-dependent degradation of AUF1 by proteasomes and mRNA destruction by mRNA decay enzymes. (Reprinted with kind permission from Ref. 101 Copyright 2002 Springer Science and Business Media).
AUF1 by itself does not exhibit any ribonucleolytic activity. Therefore, in this model, the second step proposes that additional protein-protein interactions must be nucleated by AUF1 to form a trans-acting, mRNA-destabilizing complex in cells. Coimmunoprecipitation experiments with AUF1 have revealed its association with other proteins, including translation initiation factor eIF4G, PABP, hsp70, 70-kDa heat-shock cognate protein hsc70,66 LDH,33 and hsp27.69 This latter interaction was confirmed in live cells by FRET and suggests a role for hsp27-AUF1 complexes as sensors to pair cytokine induction with monocyte adhesion and motility, as noted in section ‘post-translational modifications regulate the AUF1 family of proteins’.
It is thought that the binding of the AUF1 dimer to the ARE nucleates assembly with these other proteins, forming an AUBP and signal transduction regulated complex (ASTRC)69 that recruits the mRNA degradation machinery.26 Thus, the last step in the model proposes two components: ubiquitination of AUF1 and its degradation by proteosomes66 and recruitment of exosomes for exonucleolytic mRNA degradation.70 Each of these steps is likely subject to regulation to manipulate stabilization and destabilization of ARE-mRNAs.
Although we have focused our attention on ASTRC, it is imperative to again note that the p37AUF1 isoform can form complexes with other proteins involved in mRNA decay. For example, yeast two-hybrid experiments identified the ubiquitin-conjugating enzyme E2I and the RNA-binding proteins NSEP-1, NSAP-1, and IMP-2 as putative AUF1-binding partners.71 NSAP1 is also a member of the AUF1 complex involved in mCRD-mediated decay, as noted in section ‘post-translational modifications regulate the AUF1 family of proteins’. Thus, the complex of proteins that AUF1 assembles may be dependent upon the specific mRNA to which it is bound.
Physiological Significance of These Protein–Protein Interactions
Protein–protein interactions are crucial for the vast majority of processes in a cell. This section focuses on the different protein–protein interactions within ASTRC and summarizes findings that point to possible links between AMD and other processes such as translation and UPP, as well as describing events that alter the stoichiometry of the various subunits within ASTRC and how these alterations affect mRNA fate.
Heat Shock
The subunit composition of ASTRC is dynamic. Heat shock of cells alters association of translation initiation factor eIF4G and hsp70 with ASTRC and stabilizes a β-gal reporter transcript containing the ARE from mRNA-encoding GM-CSF.66 eIF4G forms a ternary complex with PABP and cap-binding protein eIF4E; this may circularize mRNAs, protecting them from deadenylases and decapping enzymes. Hsp70 is a cytoplasmic heat-shock protein implicated in regulation of AMD. These findings and the more recent discovery of hsp70 binding to AREs67 posed the question as to how these interactions could link AMD with translation. Recent in vitro studies showed that AUF1 directly interacts with PABP, both independently of eIF4G and in a complex with the C-terminal region of eIF4G. Binding of AUF1 to an ARE or to hsp70 impairs the AUF1–PABP interaction, however. The ARE does not affect the AUF1–eIF4G interaction. These results led to a model in which AUF1 interacts simultaneously with eIF4G and the ARE, while PABP binds both eIF4G and the poly(A) tail. But with active translation of the ARE-mRNA, the ribosome might relocate AUF1, which could bind PABP, perhaps causing exposure of the poly(A) tail to deadenylases to initiate decay.72 Hsp70 appears to oppose the AUF1–PABP interaction in accordance with data showing that heat shock induces hsp70 and promotes ARE-mRNA stabilization by perinuclear–nuclear sequestration of AUF1 (Figure 5). It is important to note that these findings are in contradiction with other studies that were unable to show a direct interaction between AUF1 and PABP in the absence of RNA. This and the fact that AUF1 oligomers bind to poly(A) present another possible scenario in which AUF1 binding to the poly(A) tail might prevent PABP from binding to it making it susceptible to deadenylases.27
FIGURE 5.
Dynamics of the ARE/poly(U)-binding/degradation factor 1 (AUF1) complex of proteins and AU-rich element (ARE)-mediated mRNA decay. AUF1 interacts with eIF4G and the ARE. During ongoing translation, AUF1 is displaced from the ARE in a complex with poly(A)-binding protein (PABP), perhaps exposing the poly(A) tail to ribonucleases. This may require destruction of AUF1 by proteasomes. During heat shock, association of hsp70 with AUF1 may disrupt or block the AUF1-PABP interaction, leaving PABP free to remain bound to the poly(A) tail, thus masking it from ribonucleases.
Hsc70, another heat-shock protein within ASTRC, has been linked to UPP because its binding to proteins promotes ubiquitin-dependent degradation. Interestingly, under heat-shock conditions, which appear to decrease proteosome activity, hsc70 stoichiometry in ASTRC does not significantly change, while that of hsp70 increases highly.66 This can be explained as these two chaperons might have opposite roles within the complex—hsp70 as a negative regulator of AMD and hsc70 as a positive regulator of AMD through UPP-dependent degradation of AUF1.
Monocyte Activation
As previously mentioned in section ‘post-translational modifications regulate the AUF1 family of proteins’, monocytes are major contributors of the host defense system through the release of cytokines. Their adhesion at sites of tissue injury leads to elevated levels of transcripts and secretion of cytokines and other inflammatory mediators. The THP-1 cell line, upon treatment with TPA, recapitulates activated monocytes, and therefore serves as a useful model system. Upon activation of THP-1 cells with TPA, eIF4G and hsp27 both increase association with ASTRC. This occurs concomitantly with ARE-mRNA stabilization and dephosphorylation of p40AUF1, perhaps as a result of cytoskeletal protein remodeling.69 Hsp27 is a cytoplasmic protein that belongs to the ATP-independent family of small heat-shock proteins. It is involved in many cellular processes including actin polymerization and UPP. Hsp27 is also an AUBP and required for AMD.69 As hsp27 is required for cell motility, we have speculated that AUF1-hsp27 complexes may link monocyte motility and cytokine mRNA decay in monocytes.69
Addressing the Implication of Protein-Protein Rearrangements and mRNA Stabilization
As noted above, the stoichiometry of eIF4G within ASTRC increases either upon heat shock or monocyte activation, two conditions that induce stabilization of ARE-mRNAs. These conditions also differentially affect two different chaperone proteins, hsp70 and hsp27. Hsp70 association with ASTRC increases during heat shock, but not during monocyte activation. Association of hsp27 with ASTRC, on the other hand, does increase upon monocyte activation. These observations suggest regulatory mechanisms in which ASTRC undergoes subunit rearrangements that control AMD depending upon the stimulus and perhaps even cell type. Future work will undoubtedly address the significance of these intriguing observations.
AUF1 BINDS MANY mRNA TARGETS—BIOLOGICAL EFFECTS
AUF1 proteins bind a number of ARE-containing transcripts (Table 2). Contrary to the stabilizing protein HuR, AUF1 binds transcripts containing any of the three ARE classes, including those encoding early response gene products73 such as FOS and MYC, as well as bcl2, β-adrenergic receptors, cyclin D1, and many others.
TABLE 2.
Representative AUF1-Binding Targets and the Effects of AUF1 Binding on Half-life and Function
| Target | Determinant | Effect on Half-life | Functional Significance | Reference |
|---|---|---|---|---|
| mRNAs | ||||
| TNFα | ARE | Destabilizing | Proinflammatory cytokine regulation | 7,9,26,32,60,62,74–78 |
| IL-1, IL-2, IL-3, IL-6 | ARE | Destabilizing | ||
| GM-CSF | ARE | Destabilizing | ||
| IL-10 | ARE | Destabilizing | Anti-inflammatory cytokine regulation | 79,80 |
| MYC | ARE | Destabilizing | AUF1 destabilizes c-myctranscripts in a cell-free system |
7,10 |
| ARE | None | Translational regulation | 7,39 | |
| mcry1 | ARE | Destabilizing | Clock gene regulation | 81 |
| β1-AR | ARE | Destabilizing | Elevated AUF1 downregulates β1-ARt in failing human hearts |
7,82 |
| β2-AR | ARE | Destabilizing | Elevated AUF1 downregulates β2-ARt in hamsters DTTM1-MF2 cell |
7,82 |
| AT1R | ARE | Destabilizing | Binding of AUF1 to human AT1R | 83 |
| CR | Unclear | |||
| Kv4.3 | ARE | Destabilizing | AUF1 upregulation by angiotensin II destabilizes cardiac channel mRNA |
84 |
| DNMT1 | ARE | Destabilizing | Cell cycle-dependent regulation of DNA methylation |
85 |
| PEPCK | ARE | Destabilizing | PEPCK catalyzes a rate-limiting step in gluconeogenesis and its expression is regulated by mRNA stability in response to metabolic cues |
86 |
| CU-rich | Destabilizing | |||
| bcl-2 | ARE | Destabilizing | Apoptosis elevates AUF1 and results in a decrease in mRNA stability |
87 |
| Cyclin D1 | ARE | Destabilizing | Changes in extracellular environment alter AUF1-regulated turnover with implications for the cell cycle |
18,88 |
| Cyclin B2 | CPE | Unknown | Elr1 and AUF1 differentially bind cyclin B2 mRNA | 89 |
| PTH | ARE | Stabilizing | AUF1 binds to and parathyroid hormone transcript resulting in stabilization of the transcript which is regulated by ion and vitamins in serum |
90 |
| DNAs | ||||
| Telomeres | ssDNA | N/A | Involvement in telomere DNA maintenance. Possibility that AUF1 binding prevents telomerase elongation |
34,35 |
| Promoters | c-mycP1 | N/A | AUF1 is a component of the LR1 DNA-binding transcriptional activator complex that binds c-myc and EBV promoters |
38 |
| EBV Fp | N/A | |||
| EBV C | N/A | AUF1 binds ss and dsDNA containing CBF2 element |
89 | |
| CD-21 | N/A | AUF1 promotes transcription of the cell surface receptor CD-21 expressed on membranes of mature B-lymphocytes |
91 |
AUF1, ARE/poly(U)-binding/degradation factor 1; mRNA,messenger ribonucleic acid; ARE, AU-rich element; IL, interleukin; GM-CSF, granulocyte-macrophage colony-stimulating factor; DNMT1, DNA methyltransferase 1; ssDNA, single-stranded DNA; dsDNA, double-stranded DNA; EBV, Epstein-Barr virus.
This section focuses on the diverse groups of transcripts degraded by AUF1 and the biological consequences of their degradation.
AUF1 Binds Many Cytokine Transcripts
Many cytokine mRNAs contain AREs within their 3′-UTRs and are subject to regulation by AUF1 (Table 2) and other AUBPs. Today we know that AUF1 binds transcripts encoding immune regulators such as interleukin (IL)-1, IL-2, IL-3, IL-6, and GROα.7,62 Monocytes and macrophages are responsible for production of both pro- and anti-inflammatory cytokines, including IL-1β and TNFα. These drive idiopathic human diseases and responses to infections.92 The interplay of these cytokines can induce other cytokines as well as repress their production (e.g., IL-10). It is, therefore, important to maintain their balance to ensure proper immune responses; otherwise, serious complications such as septic shock can occur. This is nicely demonstrated by auf1-null mice, which are highly sensitive to endotoxin-induced septic shock. These animals present endotoxemia, diarrhea, tachypnea, lethargy, piloerection, and increased mortality due to an extended inflammatory response mediated by failure to properly degrade TNFα and IL1β mRNAs.74 More recently, experiments with these mice have underscored the role AUF1 plays in the complex inflammatory response in skin, as these animals also develop chronic pruritic inflammatory skin dermatitis.75
Human melanoma cells block tumor rejection and show increased levels of the anti-inflammatory cytokine IL-10. Localization experiments in normal melanocytes versus melanoma cells revealed AUF1 to be cytoplasmic in normal melanocytes, but restricted to the nucleus in melanoma.79 This restriction of all AUF1 proteins to the nucleus might explain the increased IL-10 levels as AMD takes place in the cytoplasm. Endotoxin and interferon (INF)-γ also modulate IL-10 levels in monocytes; endotoxin induces IL-10 levels while INF-γ blocks induction. Knockdown of AUF1 with siRNA blocks endotoxin-induced IL-10, suggesting a role for AUF1 in promoting induction of IL-10.80 Thus, AUF1 controls both the pro- and anti-inflammatory arms of innate immune responses.
Eosinophils, like monocytes, are a type of white blood cell, but are in charge of fighting parasites and controlling allergic reactions and asthma. In asthma, allergic inflammation requires extended survival of activated eosinophils, which is controlled by cytokines. GM-CSF ensures eosinophil survival. Stability of its mRNA is regulated by Pin1, one member of the peptidyl-prolyl family of isomerases. Pin1 interacts with all AUF1 isoforms, which contain Pin1 isomerization sites.76 More recent studies revealed that Pin1 association with AUF1 modulates GM-CSF mRNA stability in T-lymphocytes as well.77 In conclusion, AUF1 plays major roles in immune responses.
AUF1 and Cell Death
Apoptosis refers to the natural and controlled process of programmed cell death. This involves cells’ self-destruction and removal by phagocytic cells. Bcl-2 is an important protein that prevents cell death and other various processes, including cell growth and differentiation. Its mRNA contains an ARE that is bound by all AUF1 isoforms and targets it for rapid degradation. UVC-induced apoptosis resulted in higher levels of p45AUF1 in the cytoplasm, but these higher levels were decreased by exposure of cells to caspase inhibitor. AUF1 also binds to a cell death inhibiting RNA (CDIR) known for its antiapoptotic role; intriguingly hsp27, a known ASTRC subunit, was also found in this complex.93 Cells expressing CDIR showed increased levels of two AUF1 targets known for their antiapoptotic roles, p21 and bcl2.87 These observations demonstrate a role for AUF1 as a regulator of cell death and underscore the importance of hsp27 as an essential cofactor in AMD.
AUF1 and Other Systems
AUF1 and Cardiac Tissue
In eukaryotes, G-protein-coupled receptors play an important role in the activation of signal transduction pathways. As such, these receptors are subject to tight regulation, in some cases, by AUF1. For example, exposure of hamster DDT1-MF2 smooth muscle cells to receptor agonists reduces β2-adrenergic receptor mRNA levels.94 Congestive heart failure (CHF) is associated with elevated activity of the adrenergic nervous system. In failing hearts there is a decrease in β1-adrenergic receptor mRNA levels.82 The 3′-UTRs of β-adrenergic receptor mRNAs contain AREs bound by AUF1. Interestingly, AUF1 levels are elevated in cardiac tissue from patients with CHF, suggesting that elevated AUF1 might contribute to reduced β-adrenergic receptor mRNA levels.
A number of pathologies and stress conditions induce AUF1 expression, with concomitant effects upon gene expression and cardiac function. For example, AUF1 binds another G-protein-coupled receptor mRNA, AT1R, both within the 3′-UTR (distal ARE) and the coding region. Exposure of cells to angiotensin II elevates AUF1 levels and reduces AT1R mRNA levels by 50%.83 Activation of the renin-angiotensin system is also a consequence of myocardial infarction.95 Angiotensin II-induced AUF1 expression promotes its binding to, and destabilization of, the ARE-mRNA-encoding Kv4.3, a key contributor of the transient outward current (Ito) in human myocardium.84 Stress conditions also alter post-transcriptional gene regulation in cardiomyocytes. For example, JNK activation in neonatal and adult cardiomyocytes reduces by 70% the levels of protein phosphatase 2 catalytic subunit, B56α. Its mRNA contains an ARE which p37AUF1 binds to destabilize the mRNA.96 A large number of transcripts in cardiac tissue contains AREs and are thus likely targets of AUF1. We are currently identifying these in order to understand the many pathologies resulting from AUF1 overexpression.
AUF1 and Cell Cycle
The cell cycle controls both DNA replication and cell division. Deregulation of the cell cycle is a hallmark of tumor formation and progression. AUF1 controls expression of numerous cell cycle proteins and oncogenes, including cyclin D1, p16, p21, and MYC (Table 2). For example, AUF1 binds mRNA-encoding cyclin B2 in Xenopus embryos. This binding is competitive with a Xenopus homolog of HuR, ElrA, and occurs at a specific stage of development within the midblastula.97
AUF1 itself is subject to regulation in a cell cycle-specific manner. DNA methyltransferase 1 (DNMT1) levels are altered in tumorigenesis.98,99 DNMT1 mRNA contains a noncanonical ARE bound by AUF1, which promotes mRNA degradation by recruiting the exosome to the mRNA. Degradation of AUF1 accelerates during S phase; this stabilizes the mRNA and elevates DNMT1 levels during S phase. This ensures sufficient DNA methylation of replicated DNA.85
AUF1 also binds the ARE-mRNA encoding a member of the c-Src family of tyrosine kinases, c-Yes. Hsp27, a subunit of the AUF1 complex of proteins, promotes degradation of the mRNA when overexpressed. The role of c-Yes in tumor invasiveness and metastasis underscores the significance of these observations.100
AUF1 and Circadian Rhythm
Many endogenous body processes like temperature homeostasis, cardiovascular function, metabolism, sleep, and others exhibit circadian rhythmicity. This rhythmicity is also observed in behavioral processes including feeding, excretion, learning, and sensory ability. Circadian rhythms or daily rhythms are defined as the body’s endogenous cycle that is affected by the 24-h environmental cycle. A pacemaker is responsible for controlling these rhythms, and in the case of the circadian rhythms, the major pacemaker is found in the suprachiasmatic nucleus.81 Many clock genes are involved in transcriptional/translational loops to regulate these rhythms. A recent study has identified AUF1 as a post-transcriptional regulator of the mcry1 gene, an important clock gene. This mRNA contains an ARE with no AUUUA pentamer, but U-rich sequence instead, within its 3′-UTR. Binding of AUF1 to the 3′-UTR of mcry1 promotes its degradation and appears to alter the oscillation rhythm. Oscillation of cytoplasmic AUF1 levels appears to control oscillation of mcry1 mRNA levels.81
Role of AUF1 in Pathology
Many experiments to elucidate a role for AUF1 in post-transcriptional regulation of gene expression have been performed using in vitro and ex vivo systems. At least two groups have performed experiments with transgenic mice. The first in vivo experiments using mice involved only the p37AUF1 isoform. Transgenic mice that overexpressed only this isoform were developed. The results confirmed AUF1 as a destabilizing factor for GM-CSF and TNFα mRNAs, which contain class II AREs. On the other hand, mRNAs containing other ARE classes like that of FOS, MYC, and JUN, accumulated.78 This suggested that AUF1 might play a role as both a stabilizing and destabilizing protein. Consistent with this idea, one of the p37AUF1 transgenic lines, which presented the highest levels of the isoform, developed sarcomas and these tumors expressed high levels of cyclin D1, whose mRNA is a known AUF1 target. Elevated cyclin D1 was somewhat surprising, as in some cell types; AUF1 is a destabilizer of its mRNA. Thus, AUF1 as an mRNA destabilizer or stabilizer may depend somewhat on the particular cell type in question. AUF1 null mice are another recent example of a transgenic mouse system that should prove useful to examine the roles of AUF1 in pathology. These animals are highly susceptible to endotoxic shock as noted in section ‘AUF1 binds many cytokine transcripts.’ When challenged with endotoxin, these mice presented a fivefold increase in mortality. This is due to their inability to properly degrade IL1β and TNFα mRNAs.74 Although many experiments still need to be conducted, these two transgenic systems clearly point to AUF1 as a key regulator of cancer and inflammatory diseases.
CONCLUSION
Messenger RNA turnover plays a major role in gene expression. The rates at which many mRNAs are degraded determine their steady-state levels. The degradation rates of individual mRNAs can also vary by an order of magnitude or more as a consequence of differentiation, a particular stage of the cell cycle, or response to infection by pathogens. Such variations comprise part of the normal pleiotropic responses to cell proliferation and differentiation factors and pathogens and usually involve only a subset of mRNAs. These include proto-oncogenes, cell cycle genes, inflammatory response genes, and developmentally regulated genes. Their rapid degradation ensures proper levels and timing of expression. A common feature of many labile mRNAs is the presence of one or more AREs, in the 3′-UTR, which is responsible for their rapid degradation. ARE-mediated mRNA decay requires association of one or more AUBPs, and in some cases microRNAs, with target mRNAs. AUBPs act to recruit the mRNA decay machinery to the transcript in an orchestrated fashion to initiate deadenylation followed by decapping and 5′ → 3′ and/or 3′ → 5′ degradation. These myriad decay activities are quite complex themselves, and many consist of numerous subunits. We identified the AUBP AUF1 in 1991 as an activity that could accelerate AMD in a cell-free mRNA decay system. Cloning revealed a family consisting of four, highly conserved proteins. The AUF1 complex of proteins includes translation initiation factor eIF4G, chaperones hsp/hsc70 and hsp27, and PABP, among others. AMD requires dissociation of eIF4G from AUF1, ubiquitination of AUF1, and degradation of AUF1 by proteasomes. Genetic experiments have firmly established its participation in AMD. As well, dissection of a number of diverse biological systems has revealed that proper regulation of AUF1 is essential for normal cellular, tissue, and organ homeostasis. For the future, it is important to continue focusing on the molecular mechanisms by which AUF1 exerts its actions, to identify the signaling pathways to which it responds, and to examine to how its deregulation leads to pathologies, particularly in the immune and cardiovascular systems.
ACKNOWLEDGEMENTS
We thank Kristina Sinsimer for assistance with Table 2. We also thank Jorge Benavides and Kelvin Caban for assistance with figure design and Estelle Ruidiaz for assistance with Figure 3. Work from the authors’ laboratory was supported by NIH grants CA052443 and AI057596 to G.B. F.M.G. was supported by Integrative Graduate Education and Research Traineeship DGE0333196 from the NSF to Prabhas Moghe (Department of Biomedical Engineering, Rutgers University). F.M.G. was also supported by Initiative for Minority Students R25 GM058389 from the NIH to Michael Leibowitz (UMDNJ).
REFERENCES
- 1.Nestler EJ, Hyman SE. Regulation of gene expression. In: Keneth DC, Davis L, Coyle Joseph T, Nemeroff Charles, editors. Neurophsycopharmacology: The Fifth Generation of Progress. American College of Neuropharmacology; Nashville, TN: 2002. pp. 217–228. [Google Scholar]
- 2.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. doi:33/22/7138 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. doi:10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 5.Tarun SZ, Jr, Sachs AB. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. doi: S0959-437X(00)00063-0 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. doi:S037811190100350X [pii] [DOI] [PubMed] [Google Scholar]
- 8.Fritz DT, Bergman N, Kilpatrick WJ, Wilusz CJ, Wilusz J. Messenger RNA decay in mammalian cells: the exonuclease perspective. Cell Biochem Biophys. 2004;41:265–278. doi: 10.1385/CBB:41:2:265. doi:CBB:41:2:265 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. doi:S0968000400891021 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Brewer G. Evidence for a 3′ → 5′ decay pathway for c-myc mRNA in mammalian cells. J Biol Chem. 1999;274:16174–16179. doi: 10.1074/jbc.274.23.16174. [DOI] [PubMed] [Google Scholar]
- 11.DeMaria CT, Brewer G. AUF1 binding affinity to A + U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 12.Zubiaga AM, Belasco JG, Greenberg ME. The non-amer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savant-Bhonsale S, Cleveland DW. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 14.Aharon T, Schneider RJ. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma WJ, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. doi:gka577 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. doi:10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanakahi LA, Dempsey LA, Li MJ, Maizels N. Nucleolin is one component of the B cell-specific transcription factor and switch region binding protein, LR1. Proc Natl Acad Sci USA. 1997;94:3605–3610. doi: 10.1073/pnas.94.8.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. doi:10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. doi:rna.501707 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, Cabo R, et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. emboj200967 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafon I, Carballes F, Brewer G, Poiret M, Morello D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. doi:10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- 22.Brewer G. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinol-Roma S, Choi YD, Matunis MJ, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein. AUF1 Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeMaria CT, Sun Y, Long L, Wagner BJ, Brewer G. Structural determinants in AUF1 required for high affinity binding to A + U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 26.Wilson GM, Sun Y, Lu H, Brewer G. Assembly of AUF1 oligomers on U-rich RNA targets by sequential dimer association. J Biol Chem. 1999;274:33374–33381. doi: 10.1074/jbc.274.47.33374. [DOI] [PubMed] [Google Scholar]
- 27.Sagliocco F, Laloo B, Cosson B, Laborde L, Castroviejo M, Rosenbaum J, Ripoche J, Grosset C. The ARE-associated factor AUF1 binds poly(A) in vitro in competition with PABP. Biochem J. 2006;400:337–347. doi: 10.1042/BJ20060328. doi:BJ20060328 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiledjian M, DeMaria CT, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loflin P, Chen CY, Shyu AB. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu N, Chen CY, Shyu AB. Versatile role for hnRNP D isoforms in the differential regulation of cytoplasmic mRNA turnover. Mol Cell Biol. 2001;21:6960–6971. doi: 10.1128/MCB.21.20.6960-6971.2001. doi:10.1128/MCB.21.20.6960-6971.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ing NH, Massuto DA, Jaeger LA. Estradiol up-regulates AUF1p45 binding to stabilizing regions within the 3′-untranslated region of estrogen receptor alpha mRNA. J Biol Chem. 2008;283:1764–1772. doi: 10.1074/jbc.M704745200. doi:M704745200 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. doi:10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pioli PA, Hamilton BJ, Connolly JE, Brewer G, Rigby WF. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem. 2002;277:35738–35745. doi: 10.1074/jbc.M204002200. doi:10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- 34.Eversole A, Maizels N. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol Cell Biol. 2000;20:5425–5432. doi: 10.1128/mcb.20.15.5425-5432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enokizono Y, Konishi Y, Nagata K, Ouhashi K, Uesugi S, Ishikawa F, Katahira M. Structure of hnRNP D complexed with single-stranded telomere DNA and unfolding of the quadruplex by heterogeneous nuclear ribonucleoprotein D. J Biol Chem. 2005;280:18862–18870. doi: 10.1074/jbc.M411822200. doi:M411822200 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Tolnay M, Baranyi L, Tsokos GC. Heterogeneous nuclear ribonucleoprotein D0 contains transactivator and DNA-binding domains. Biochem J. 2000;348:151–158. [PMC free article] [PubMed] [Google Scholar]
- 37.Hanakahi LA, Maizels N. Transcriptional activation by LR1 at the Emu enhancer and switch region sites. Nucleic Acids Res. 2000;28:2651–2657. doi: 10.1093/nar/28.14.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dempsey LA, Hanakahi LA, Maizels N. A specific isoform of hnRNP D interacts with DNA in the LR1 heterodimer: canonical RNA binding motifs in a sequence-specific duplex DNA binding protein. J Biol Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 39.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat Struct Mol Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. doi:nsmb1249 [pii] [DOI] [PubMed] [Google Scholar]
- 40.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. doi:S0092-8674(00)00102-1 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Wagner BJ, Long L, Rao PN, Pettenati MJ, Brewer G. Localization and physical mapping of genes encoding the A + U-rich element RNA-binding protein AUF1 to human chromosomes 4 and X. Genomics. 1996;34:219–222. doi: 10.1006/geno.1996.0269. doi:S0888-7543(96)90269-4 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Kamei D, Tsuchiya N, Yamazaki M, Meguro H, Yamada M. Two forms of expression and genomic structure of the human heterogeneous nuclear ribonucleoprotein D-like JKTBP gene (HNRPDL) Gene. 1999;228:13–22. doi: 10.1016/s0378-1119(99)00020-7. [DOI] [PubMed] [Google Scholar]
- 43.Ehrenman K, Long L, Wagner BJ, Brewer G. Characterization of cDNAs encoding the murine A + U-rich RNA-binding protein AUF1. Gene. 1994;149:315–319. doi: 10.1016/0378-1119(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 44.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. doi:S0888-7543(97)95142-9 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Buzby JS, Brewer G, Nugent DJ. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J Biol Chem. 1999;274:33973–33978. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- 46.Buzby JS, Lee SM, Van Winkle P, DeMaria CT, Brewer G, Cairo MS. Increased granulocyte-macrophage colony-stimulating factor mRNA instability in cord versus adult mononuclear cells is translation-dependent and associated with increased levels of A + U-rich element binding factor. Blood. 1996;88:2889–2897. [PubMed] [Google Scholar]
- 47.Sheflin LG, Spaulding SW. Testosterone and dihydrotestosterone regulate AUF1 isoforms in a tissue-specific fashion in the mouse. Am J Physiol Endocrinol Metab. 2000;278:E50–E57. doi: 10.1152/ajpendo.2000.278.1.E50. [DOI] [PubMed] [Google Scholar]
- 48.Nagata T, Kurihara Y, Matsuda G, Saeki J, Kohno T, Yanagida Y, Ishikawa F, Uesugi S, Katahira M. Structure and interactions with RNA of the N-terminal UUAG-specific RNA-binding domain of hnRNP D0. J Mol Biol. 1999;287:221–237. doi: 10.1006/jmbi.1999.2616. doi:S0022283699926165 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J Biol Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 50.Moraes KC, Lee WH, Kobarg J. Analysis of the structural determinants for RNA binding of the human protein AUF1/hnRNP D. Biol Chem. 2002;383:831–837. doi: 10.1515/BC.2002.087. doi:10.1515/BC.2002.087. [DOI] [PubMed] [Google Scholar]
- 51.Wilson GM, Sun Y, Sellers J, Lu H, Penkar N, Dillard G, Brewer G. Regulation of AUF1 expression via conserved alternatively spliced elements in the 3′ untranslated region. Mol Cell Biol. 1999;19:4056–4064. doi: 10.1128/mcb.19.6.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullmann R, Jr., Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. doi:MCB.00500-07 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banihashemi L, Wilson GM, Das N, Brewer G. Upf1/Upf2 regulation of 3′ untranslated region splice variants of AUF1 links nonsense-mediated and A + U-rich element-mediated mRNA decay. Mol Cell Biol. 2006;26:8743–8754. doi: 10.1128/MCB.02251-05. doi:MCB.02251-05 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. doi:21/6/708 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 56.Weighardt F, Cobianchi F, Cartegni L, Chiodi I, Villa A, Riva S, Biamonti G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]
- 57.Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki M, Iijima M, Nishimura A, Tomozoe Y, Kamei D, Yamada M. Two separate regions essential for nuclear import of the hnRNP D nucleocytoplasmic shuttling sequence. FEBS J. 2005;272:3975–3987. doi: 10.1111/j.1742-4658.2005.04820.x. doi:EJB4820 [pii] [DOI] [PubMed] [Google Scholar]
- 59.He C, Schneider R. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 2006;25:3823–3831. doi: 10.1038/sj.emboj.7601264. doi:7601264 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson GM, Lu J, Sutphen K, Sun Y, Huynh Y, Brewer G. Regulation of A U-rich element-directed mRNA turnover involving+reversible phosphorylation of AUF1. J Biol Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. doi:10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 61.Pomorski P, Watson JM, Haskill S, Jacobson KA. How adhesion, migration, and cytoplasmic calcium transients influence interleukin-1beta mRNA stabilization in human monocytes. Cell Motil Cytoskeleton. 2004;57:143–157. doi: 10.1002/cm.10159. doi:10.1002/cm.10159. [DOI] [PubMed] [Google Scholar]
- 62.Sirenko OI, Lofquist AK, DeMaria CT, Morris JS, Brewer G, Haskill JS. Adhesion-dependent regulation of an A + U-rich element-binding activity associated with AUF1. Mol Cell Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. doi:10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- 64.Fawal M, Armstrong F, Ollier S, Dupont H, Touriol C, Monsarrat B, Delsol G, Payrastre B, Morello D. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 65.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. doi:ASN.2006010083 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 67.Wilson GM, Sutphen K, Bolikal S, Chuang KY, Brewer G. Thermodynamics and kinetics of Hsp70 association with A + U-rich mRNA-destabilizing sequences. J Biol Chem. 2001;276:44450–44456. doi: 10.1074/jbc.M108521200. doi:10.1074/jbc.M108521200. [DOI] [PubMed] [Google Scholar]
- 68.Laroia G, Schneider RJ. Alternate exon insertion controls selective ubiquitination and degradation of different AUF1 protein isoforms. Nucleic Acids Res. 2002;30:3052–3058. doi: 10.1093/nar/gkf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinsimer KS, Gratacos FM, Knapinska AM, Lu J, Krause CD, Wierzbowski AV, Maher LR, Scrudato S, Rivera YM, Gupta S, et al. Chaperone Hsp27, a novel subunit of AUF1 protein complexes, functions in AU-rich element-mediated mRNA decay. Mol Cell Biol. 2008;28:5223–5237. doi: 10.1128/MCB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. doi:S0092-8674(01)00578-5 [pii] [DOI] [PubMed] [Google Scholar]
- 71.Moraes KC, Quaresma AJ, Maehnss K, Kobarg J. Identification and characterization of proteins that selectively interact with isoforms of the mRNA binding protein AUF1 (hnRNP D) Biol Chem. 2003;384:25–37. doi: 10.1515/BC.2003.004. doi:10.1515/BC.2003.004. [DOI] [PubMed] [Google Scholar]
- 72.Lu JY, Bergman N, Sadri N, Schneider RJ. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA. 2006;12:883–893. doi: 10.1261/rna.2308106. doi: rna.2308106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bhattacharya S, Giordano T, Brewer G, Malter JS. Identification of AUF-1 ligands reveals vast diversity of early response gene mRNAs. Nucleic Acids Res. 1999;27:1464–1472. doi: 10.1093/nar/27.6.1464. doi: gkc281 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu JY, Sadri N, Schneider RJ. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev. 2006;20:3174–3184. doi: 10.1101/gad.1467606. doi: gad.1467606 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sadri N, Schneider RJ. Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J Invest Dermatol. 2009;129:657–670. doi: 10.1038/jid.2008.298. doi:jid2008298 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen ZJ, Esnault S, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates the stability of granulocyte-macrophage colony-stimulating factor mRNA in activated eosinophils. Nat Immunol. 2005;6:1280–1287. doi: 10.1038/ni1266. doi: ni1266 [pii] [DOI] [PubMed] [Google Scholar]
- 77.Esnault S, Shen ZJ, Whitesel E, Malter JS. The peptidyl-prolyl isomerase Pin1 regulates granulocyte-macrophage colony-stimulating factor mRNA stability in T lymphocytes. J Immunol. 2006;177:6999–7006. doi: 10.4049/jimmunol.177.10.6999. doi:177/10/6999 [pii] [DOI] [PubMed] [Google Scholar]
- 78.Gouble A, Grazide S, Meggetto F, Mercier P, Delsol G, Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 79.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S, Brewer G, et al. Increased interleukin-10 mRNAstability in melanoma cells is associated with decreased levels of A + U-rich element binding factor AUF1. J Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. doi:10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 80.Sarkar S, Sinsimer KS, Foster RL, Brewer G, Pestka S. AUF1 isoform-specific regulation of anti-inflammatory IL10 expression in monocytes. J Interferon Cytokine Res. 2008;28:679–691. doi: 10.1089/jir.2008.0028. doi: 10.1089/jir.2008.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woo KC, Ha DC, Lee KH, Kim DY, Kim TD, Kim KT. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol Cell Biol. 2010;30:197–205. doi: 10.1128/MCB.01154-09. doi:MCB.01154-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pende A, Tremmel KD, DeMaria CT, Blaxall BC, Minobe WA, Sherman JA, Bisognano JD, Bristow MR, Brewer G, Port J. Regulation of the mRNA-binding protein AUF1 by activation of the betaadrenergic receptor signal transduction pathway. J Biol Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- 83.Pende A, Giacche M, Castigliola L, Contini L, Passerone G, Patrone M, Port JD, Lotti G. Characterization of the binding of the RNA-binding protein AUF1 to the human AT(1) receptor mRNA. Biochem Biophys Res Commun. 1999;266:609–614. doi: 10.1006/bbrc.1999.1862. doi:10.1006/bbrc.1999.1862. [DOI] [PubMed] [Google Scholar]
- 84.Zhou C, Vignere CZ, Levitan ES. AUF1 is upregulated by angiotensin II to destabilize cardiac Kv4.3 channel mRNA. J Mol Cell Cardiol. 2008;45:832–838. doi: 10.1016/j.yjmcc.2008.08.004. doi:S0022-2828(08)00573-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torrisani J, Unterberger A, Tendulkar SR, Shikimi K, Szyf M. AUF1 cell cycle variations define genomic DNA methylation by regulation of DNMT1 mRNA stability. Mol Cell Biol. 2007;27:395–410. doi: 10.1128/MCB.01236-06. doi:MCB.01236-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hajarnis S, Schroeder JM, Curthoys NP. 3′-Untranslated region of phosphoenolpyruvate carboxykinase mRNA contains multiple instability elements that bind AUF1. J Biol Chem. 2005;280:28272–28280. doi: 10.1074/jbc.M501204200. doi:M501204200 [pii] [DOI] [PubMed] [Google Scholar]
- 87.Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Nicolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J Biol Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. doi:10.1074/jbc.M201377200M201377200 [pii] [DOI] [PubMed] [Google Scholar]
- 88.Lin S, Wang W, Wilson GM, Yang X, Brewer G, Holbrook NJ, Gorospe M. Down-regulation of cyclin D1 expression by prostaglandin A(2) is mediated by enhanced cyclin D1 mRNA turnover. Mol Cell Biol. 2000;20:7903–7913. doi: 10.1128/mcb.20.21.7903-7913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuentes-Panana EM, Peng R, Brewer G, Tan J, Ling PD. Regulation of the Epstein-Barr virus C promoter by AUF1 and the cyclic AMP/protein kinase A signaling pathway. J Virol. 2000;74:8166–8175. doi: 10.1128/jvi.74.17.8166-8175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J Biol Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- 91.Tolnay M, Vereshchagina LA, Tsokos GC. Heterogeneous nuclear ribonucleoprotein. D0 is a sequence-specific DNA-binding protein. Biochem J. 1999;338:417–425. [PMC free article] [PubMed] [Google Scholar]
- 92.Seymour RM, Henderson B. Pro-inflammatory-anti-inflammatory cytokine dynamics mediated by cytokine-receptor dynamics in monocytes. IMA J Math Appl Med Biol. 2001;18:159–192. [PubMed] [Google Scholar]
- 93.Shchors K, Yehiely F, Kular RK, Kotlo KU, Brewer G, Deiss LP. Cell death inhibiting RNA (CDIR) derived from a 3′-untranslated region binds AUF1 and heat shock protein 27. J Biol Chem. 2002;277:47061–47072. doi: 10.1074/jbc.M202272200. doi:10.1074/jbc.M202272200. [DOI] [PubMed] [Google Scholar]
- 94.Hadcock JR, Wang HY, Malbon CC. Agonist-induced destabilization of beta-adrenergic receptor mRNA. Attenuation of glucocorticoid-induced up-regulation of beta-adrenergic receptors. J Biol Chem. 1989;264:19928–19933. [PubMed] [Google Scholar]
- 95.Cleland JG. The renin-angiotensin system in heart failure. Herz. 1991;16:68–81. [PubMed] [Google Scholar]
- 96.Glaser ND, Lukyanenko YO, Wang Y, Wilson GM, Rogers TB. NK activation decreases PP2A regulatory subunit B56alpha expression and mRNA stability and increases AUF1 expression in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2006;291:H1183–H1192. doi: 10.1152/ajpheart.01162.2005. doi:01162.2005 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo X, Gourronc F, Audic Y, Lyons-Levy G, Mitchell T, Hartley RS. ElrA and AUF1 differentially bind cyclin B2 mRNA. Biochem Biophys Res Commun. 2008;377:653–657. doi: 10.1016/j.bbrc.2008.10.029. ISSN:1090-2104 (Electronic). doi:S0006-291X(08)02002-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. doi:10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 99.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 100.Sommer S, Cui Y, Brewer G, Fuqua SA. The c-Yes 3′-UTR contains adenine/uridine-rich elements that bind AUF1 and HuR involved in mRNA decay in breast cancer cells. J Steroid Biochem Mol Biol. 2005;97:219–229. doi: 10.1016/j.jsbmb.2005.09.002. doi:S0960-0760(05)00389-4 [pii] [DOI] [PubMed] [Google Scholar]
- 101.Wilson GM, Brewer G. Regulation of mRNA stability by AUF1. In: Sandberg K, Mulroney SE, editors. RNA Binding Proteins: New Concepts in Gene Regulation, Endocrine Updates. Vol. 16. Kluwer Academic Publishers; Norwell, MA: 2002. pp. 101–117. [Google Scholar]
FURTHER READING
- Wilson GM, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- Naveh-Many T, Bell O, Silver J, Kilav R. Cis and trans-acting factors in the regulation of parathyroid hormone (PTH) mRNA stability by calcium and phosphate. FEBS Lett. 2002;529:60–64. doi: 10.1016/s0014-5793(02)03259-3. doi:S0014579302032593 [pii] [DOI] [PubMed] [Google Scholar]
- Misquitta CM, Chen T, Grover AK. Control of protein expression through mRNA stability in calcium signalling. Cell Calcium. 2006;40:329–346. doi: 10.1016/j.ceca.2006.04.004. doi:S0143-4160(06)00070-4 [pii] [DOI] [PubMed] [Google Scholar]