Abstract
Surface plasmon resonance (SPR) is a powerful technique for monitoring the affinity and selectivity of biomolecular interactions. SPR allows for analysis of association and dissociation rate constants and modeling of biomolecular interaction kinetics, as well as for equilibrium binding analysis and ligand specificity studies. SPR has received much use and improved precision in classifying protein–protein interactions, as well as in studying small-molecule ligand binding to receptors; however, lipid–protein interactions have been underserved in this regard. With the field of lipids perhaps the next frontier in cellular research, SPR is a highly advantageous technique for cell biologists, as newly identified proteins that associate with cellular membranes can be screened rapidly and robustly for lipid specificity and membrane affinity. This technical perspective discusses the conditions needed to achieve success with lipid–protein interactions and highlights the unique lipid–protein interaction mechanisms that have been elucidated using SPR. It is intended to provide the reader a framework for quantitative and confident conclusions from SPR analysis of lipid–protein interactions.
LIPID–PROTEIN INTERACTIONS AND SURFACE PLASMON RESONANCE
The lipid bilayer has a highly polarized structure consisting of a central hydrocarbon core and two flanking interfacial regions that are highly dynamic and may contain >1000 different lipids. This dynamic variety of glycerophospholipids, sphingolipids, and sterols in membrane organelles provides spatial and temporal architecture to direct signaling processes through target proteins (Van Meer et al., 2008). Given that nearly half of all proteins are located in or on membranes, it is not surprising that there are a variety of conserved lipid-binding domains in eukaryotes, some of which rank in the top 15 modular domains in the human genome (Cho and Stahelin, 2005). Because high-affinity interactions between these peripheral proteins regulate lipid signaling and trafficking events from cellular membranes, accurately characterizing lipid specificity and membrane affinity of peripheral proteins can clearly establish how the membrane interface signals throughout the cell. A number of well-established biochemical and biophysical experiments have been used to determine the lipid specificity and affinity of proteins. Among these assays, surface plasmon resonance (SPR) has emerged as a robust technique for measuring both protein affinity and specificity for different lipids and has been used successfully in determining the Kd values of peripheral proteins harboring a diverse set of lipid-binding modules (Cho and Stahelin, 2005; Narayan and Lemmon, 2006). Most SPR instruments are based on the attenuated total reflectance configuration developed by Kretschmann and Raether (1968). This configuration relies on the phenomenon of total internal reflection that is observed when light traveling through an optically dense medium (e.g., glass) reaches an interface between this medium and a medium of lower optical density (e.g., air) and is reflected back (Figure 1A). A monochromatic, p-polarized light source is used in most SPR instruments, and the interface between two optically dense media is coated with a thin metal film, typically gold due to its inherent stability. The evanescent wave is a component of the incident light that is able to couple with free oscillating electrons (plasmons) in the metal film at a specific angle of incidence (resonance angle). This specific angle of incidence produces a shadow or SPR because of energy transfer between the evanescent wave and plasmons on the surface. The evanescent wave propagates into the medium opposite the gold interface and decays exponentially to ∼100 Å. The SPR signal is sensitive to the mass concentration on the gold surface, where the measure of mass concentration on the sensor chip surface is expressed in resonance units (RU). The mass change on the surface can be detected in a time-dependent manner by a two-dimensional array of photodiodes or charge-coupled detectors, which allows for real-time biomolecular interaction analysis.
FIGURE 1:
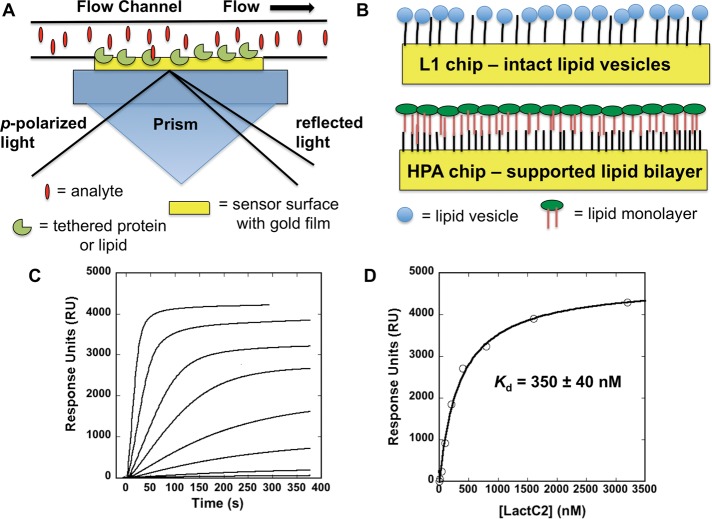
SPR system and lipid-coating setups, along with equilibrium binding analysis of the lactadherin C2 domain to a 1-palmitoyl-2-oleoylphosphatidylcholine (POPC):1-palmitoyl-2-oleoylphosphatidylserine (POPS; 70:30) vesicle surface. (A) A typical SPR setup is shown for one flow cell in a SPR instrument. The gold surface of a sensor surface can be modified chemically to allow attachment of a ligand, which could be a protein, a lipid, or another biomolecule of interest. The analyte is then injected and flowed over the ligand surface to detect binding of the analyte. Detection of binding to ligand on the gold sensor surface is possible as below the gold chip surface p-polarized light traveling through an optically dense prism reaches the interface between the prism and a medium of lower optical density and is reflected back and detected by an array of photodiodes or charge-coupled detectors. The specific angle of incidence of light produces a shadow or SPR because of energy transfer between the evanescent wave and plasmons on the surface of the gold chip. The evanescent wave propagates into the flow cell opposite the gold interface to ∼100 Å and is sensitive to the mass concentration on the surface. The mass change on the surface can be detected in a time-dependent manner as a change in angle that is reflected to the detectors and displayed on the instrument as change in signal, typically RU. (B) The two most common surfaces used for coating lipid vesicles are Biacore L1 and HPA chips. The L1 chip (top) allows for coating of intact lipid vesicles through insertion of alkane groups present on the carboxymethyldextran-coated gold surface. The HPA chip contains a hydrophobic alkanethiol surface, which promotes the docking of a lipid monolayer on the surface to form a supported bilayer. Either surface may be useful for SPR experiments, as outlined in the text. (C) A POPC:POPS surface coated on a L1 chip was used to test the binding of the lactadherin C2 domain with a Biacore X. A POPC surface was used as a control to correct for any refractive index changes upon injection of the protein at increasing concentrations. The C2 domain was injected at increasing concentrations (5, 10, 40, 100, 200, 400, 800, 1600, and 3200 nM) at a flow rate of 30 μl/min to determine the saturation response in signal (RU) at each respective protein concentration. (D) Saturation values determined from the RU at respective C2 domain concentrations (shown in C) were plotted and fit with a nonlinear least squares analysis using the equation Req = Rmax/(1 + Kd/C) to determine Kd as shown. Experiments were repeated in triplicate to determine the average Kd and SD for the lactadherin C2 domain (320 ± 90 nM) to POPC:POPS (70:30) vesicles.
RECOMMENDED SETUP FOR LIPID SURFACES
SPR is advantageous not only because interactions can be monitored in real time, but also because neither the ligand nor the analyte require labeling, instruments have high sensitivity, and high throughput of samples can be performed. To this end, SPR has been rigorously applied to protein–protein and small molecule–protein interactions. For these types of characterizations there are extensive reviews and technical perspectives (Myszka, 1997; Morton and Myszka, 1998; Rich et al., 2001). However, the field of lipid–protein interactions has been less extensively scrutinized with regard to experimental details and methods. Although SPR has had a number of success stories, it can be difficult to obtain reliable and reproducible binding data using SPR due to nonspecific binding to the sensor chip, mass transport effects, and protein or lipid surface stability. The first step in these experiments is to prepare a reliable membrane surface that can recapitulate membranes that the protein of interest would encounter in cells. For instance, one needs to use concentrations of the potential lipid ligand of interest that are close to physiological conditions but also at a level that the sensitivity of instrument can detect binding. For phosphoinositides (PIs) it is recommended that concentrations in the 1–3 mol% range be used in a phosphatidylcholine (PC) vesicle. In this way phosphatidylcholine can be used as a control to directly compare binding of the protein to PC or PC:PI (97:3) vesicles.
The majority of our experiments have been performed on a Biacore X (GE Healthcare), but other Biacore systems and a new variety of SPR instruments (Bio-Rad, ForteBio, Horiba Scientific, Reichert Analytical Instruments, etc.) are available and can be applied. It is recommended, however, that in most cases only two flow cells be applied at a time to an experiment. In most SPR instruments the mobile phase flows sequentially over the flow cells, and migration of some lipid species, such as phosphoinositides (Narayan and Lemmon, 2006) and anionic sphingolipids, has been detected from the control flow cell to the active or “test“ flow cell surface. This can be detrimental when comparing lipid specificity or affinity, as both flow cell surfaces may contain an altered concentration or even type of phosphoinositide. Preparing flow cell 1 as the control and flow cell 2 as the active surface will prevent migration and sample loss of some lipids from flow cell 1 to flow cell 2. In our experience, this is necessary to obtain reproducible data over the course of 1 or 2 d of experimentation with a lipid surface. SPR should receive dedication once a system is working and reproducible. The lifetime of a lipid surface on an L1 chip can last from 12 to 48 h, so I always reiterate dedication, organization, and experimental planning during these times for lab members to collect robust reproducible data over a period of 1–2 d. Subsequently, lipid vesicles can be stripped from the surface and the process repeated, as discussed later.
LIPID COATING
A variety of methods have been used to capture lipids on the sensor surface of SPR instrumentation. The most popular and standardized methods use a supported bilayer (HPA chip) or intact lipid vesicles (L1 chip; Figure 1B). The HPA chip uses hydrophobic interactions between alkanethiol groups on the gold sensor surface, which will capture the hydrophobic tails of lipid molecules injected into the instrument. This forms a lipid monolayer on the alkanethiol referred to as a supported bilayer. The L1 chip captures intact lipid vesicles injected into the instrument using proprietary hydrophobic groups on the gold carboxymethyldextran sensor surface. In our experience both systems work well for coating and lipid-binding experiments, with the L1 chip providing more reproducibility and a longer lifetime of the sensor surface. On the other hand, the HPA chip is better for proteins that may or are known to cause vesicle fusion, as these interactions can change the appearance of the vesicles on the L1 chip surface. To coat the L1 surface, lipid vesicles are prepared at a concentration of 0.5 mg/ml in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, containing 0.16 M KCl, and are vortexed vigorously and passed through a 100-nm polycarbonate filter using an Avanti MiniExtruder (Avanti Polar Lipids, Alabaster, AL) according to the manufacturer's instructions. Before coating with lipids, the sensor surfaces are washed with 25 μl of 40 μM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), followed by 25 μl of β-octylglucoside at a flow rate of 30 μl/min. Residual detergent can then be washed away by increasing the flow rate to 100 μl/min for 10 min or injecting 10 μl of 30% ethanol.
The lipid vesicles can then be coated by injecting 80 μl of lipid vesicles at a flow rate of 5 μl/min, always coating the active flow cell first, followed by coating of the control flow cell. This also allows one to adjust the lipid coating of the control flow cell to match that of the active flow cell surface in RU. The coating ability of the lipid vesicles can vary based upon anionic lipid content and usually ranges from 5000 to 9000 RU, with pure zwitterionic vesicles giving the highest saturation value. The lipid layers are then stabilized with three injections of 20 μl of 0.1 M NaOH at 30 μl/min, which is also commonly used as a regeneration solution for removing protein from the lipid layer. The significance of lipid coating can be verified by injecting 0.1 mg/ml bovine serum albumin (BSA), as <100 RU of BSA should bind to a well-coated surface, whereas >1000 RU of BSA will bind to an uncoated or poorly coated lipid surface. We have demonstrated that BSA left on the sensor surface will not influence lipid-binding parameters and under some conditions can reduce nonspecific binding to the L1 chip should the protein of interest nonspecifically associate with the carboxylmethyldextran layer. Unlike SPR experiments with protein–protein or protein–nucleic acids, it is important not to include small amounts of detergents in the SPR buffers, as this can destabilize the lipid surfaces. One drawback to the absence of detergents in SPR buffers is that the instrument needs to be cleaned more frequently (every 2–3 d), as protein will be lost to the inner tube walls of the SPR during experimentation. After 1–2 d of experimentation the lipid surface can be removed with injection of 25 μl of 40 μM CHAPS, followed by 25 μl of β-octylglucoside at a flow rate of 30 μl/min, and the process of recoating can then be started. The L1 chip surface can generally support ∼40–60 lipid coatings before a reduction in the coating efficiency is observed.
How the lipid vesicles form on the L1 surface is still under debate, with most studies suggesting that vesicles are retained intact on the L1 chip surface. One study suggested that the vesicles fuse and form a lipid bilayer (Erb et al., 2000), whereas several others, using imaging and dye leakage assessment, found strong evidence that the lipid vesicles are intact on the sensor surface (Stahelin and Cho, 2001; Höning et al., 2005). The type of surface that forms may be specific to the types and origins of the lipids and lipid mixtures used, as well as to the pH and osmolarity of the running buffer. In any case, vesicles anchored to the L1 chip adopt a structure that is relevant for examining lipid–protein interactions. Recently membrane curvature has been shown to play an important role in recruitment of some peripheral proteins to membrane surface (Bigay and Antonny, 2012). In principle, this assay is feasible with SPR, as vesicles can be extruded through different-size polycarbonate filters (e.g., 50–1000 nm) to create vesicles of distinct diameters that can be assessed with dynamic light scattering and electron microscopy. In our experience vesicles of different diameters can be coated to similar densities on the L1 chip; however, how different vesicles appear on the surface (i.e., intact vesicles or fused bilayer) has not been well characterized. Thus data collected from SPR to assess curvature dependence should at minimum ensure that vesicles were retained intact on the chip surface by loading vesicles with fluorescent dye that can be detected in the running buffer should vesicles fuse. Finally, if at first your protein doesn't bind to the lipid ligand of interest, don't fret! It is coming to light that many peripheral proteins are coincidence detectors (Moravcevic et al., 2012; Scott et al., 2012) and actually bind to two or more distinct lipids. Thus some proteins require both lipids to be present in vesicles to have significant membrane affinity. This means that lipid screening by SPR or another assay may need to be performed to investigate the probable combination of lipids that could possibly bind.
LIPID-BINDING ANALYSIS
Kinetic analysis of SPR data can be quite cumbersome and requires careful consideration before publishing results. In brief, the rate of adsorption and desorption are dependent on intrinsic kinetics and mass transport of the system. Diffusion through the boundary layer is usually much slower than the intrinsic adsorption kinetics and is, therefore, the rate-determining factor. The best method of detecting a mass transport limitation is to vary the flow rate of the system and calculate rate constants under these varying conditions. If mass transport is not rate limiting, then rate constants will be consistent over a broad range of flow rates. This holds true because diffusion kinetics is dependent on the flow rate, whereas intrinsic kinetics is not. To eliminate potential mass transport effects, the rate of diffusion must be increased and the rate of binding reduced. Thus increasing the flow rate and decreasing the ligand density so as to reduce the number of available binding sites are two ways of minimizing the mass transport limitations of a system. The mass transfer effect and its kinetic interpretation have been studied at length but not for lipid–protein interactions. Some previous work addressed these issues to demonstrate that kinetics was independent of flow rate for some lipid-binding proteins (Stahelin and Cho, 2001; Stahelin et al., 2002). The mass transfer effect is not the only variable that can hinder accurate kinetic interpretation, as other factors to be considered include matrix effects, sample purity, and ligand homogeneity.
The validity of using rate constants to calculate equilibrium constants has often been questioned due to neglect of factors in the binding model such as mass transport, steric hindrance, possibility of more-complex binding schemes, or ligand homogeneity. For these reasons it may be advantageous and more accurate to analyze the equilibrium binding constant (Kd) using saturation measurements. In fact, in some cases kinetic analysis has overestimated the Kd of some lipid-binding proteins (Narayan and Lemmon, 2006), whereas other studies have clearly demonstrated consistency between kinetic analysis and saturation measurements (Stahelin and Cho, 2001; Stahelin et al., 2002, 2007). The apparent Kd can be determined by first assuming that a protein molecule binds to one binding site present on a lipid vesicle (may be one lipid molecule or many). The protein is then injected at increasing concentrations to reach a steady-state signal for each respective concentration (Figure 1C), where it is assumed that steady state occurs and the rate of protein association and dissociation are equal. These steady-state RU values are then plotted versus protein concentration (Figure 1D) to calculate the apparent Kd using the equation Req = Rmax/(1 + Kd/C), where Req is the binding signal obtained at saturation for each respective free protein concentration (C) and Rmax is the binding signal at saturation. For newly discovered proteins a secondary method should be used to verify the Kd determined by SPR analysis. In addition, it his highly recommended that appropriate positive and negative controls be used for lipid ligand specificity and membrane affinity. For instance, for a new phosphatidylinositol-3-phosphate–binding protein a FYVE or PX domain could be used as a positive control, and a PH or C2 domain with different PI specificity could be used as a negative control. This will help to demonstrate the validity of the lipid specificity but also demonstrate that the PI is behaving appropriately.
SUMMARY
SPR has greatly affected the ability to characterize lipid specificity and the affinity of a variety of lipid-binding proteins. This technique provides a robust platform with which to come to quantitative and confident conclusions regarding lipid specificity and a Kd for different compositions of lipid vesicles. To this end, equilibrium binding analysis is recommended when determining Kd values. At a minimum, SPR should be able to decipher lipid specificity for a protein when only small protein sample sizes are available When large protein and mutant samples are available, SPR, in conjunction with other biochemical and cellular experiments, can be used to generate schematic models of lipid–protein and other biomolecular interactions.
Acknowledgments
The Stahelin lab is supported by grants from the National Institutes of Health (AI081077), the National Science Foundation (1122068), and the American Heart Association (GRNT12080254). I thank Jordan L. Scott for helpful discussions.
Abbreviations used:
- BSA
bovine serum albumin
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- POPC
1-palmitoyl-2-oleoylphosphatidylcholine
- POPS
1-palmitoyl-2-oleoylphosphatidylserine, RU, resonance unit
- SPR
surface plasmon resonance
Footnotes
REFERENCES
- Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Erb EM, Chen X, Allen S, Roberts CJ, Tendler SJ, Davies MC, Forsen S. Characterization of the surface generated by liposome binding to the modified dextran matrix of a surface plasmon resonance sensor chip. Anal Biochem. 2000;280:29–35. doi: 10.1006/abio.1999.4469. [DOI] [PubMed] [Google Scholar]
- Höning S, Ricotta D, Krauss M, Spate K, Spolaore B, Motley A, Robinson M, Robinson C, Haucke V, Owen DJ. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Kretschmann E, Raether H. Radiative decay of non-radiative surface plasmons excited by light. Z Naturforsch. 1968;230:2135–2136. [Google Scholar]
- Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton TA, Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Methods Enzymol. 1998;295:268–294. doi: 10.1016/s0076-6879(98)95044-3. [DOI] [PubMed] [Google Scholar]
- Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA. Determining selectivity of phosphoino sitide-binding domains. Methods. 2006;39:122–133. doi: 10.1016/j.ymeth.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich RL, Day YS, Morton TA, Myszka DG. High-resolution and high throughput protocols for measuring drug/human serum albumin interactions using BIACORE. Anal Biochem. 2001;296:197–207. doi: 10.1006/abio.2001.5314. [DOI] [PubMed] [Google Scholar]
- Scott JL, Musselman CA, Adu-Gyamfi E, Kutateladze TG, Stahelin RV. Emerging methodologies to investigate lipid-protein interactions. Integr Biol (Camb) 2012;4:247–258. doi: 10.1039/c2ib00143h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Cho W. Differential roles of ionic, aliphatic, and aromatic residues in membrane-protein interactions: a surface plasmon resonance study on phospholipases A2. Biochemistry. 2001;40:4672–4678. doi: 10.1021/bi0020325. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Murray D, Williams RL, Cho W. Structural and membrane binding analysis of the Phox homology domain of Bem1p: basis of phosphatidylinositol 4-phosphate specificity. J Biol Chem. 2007;282:25737–25747. doi: 10.1074/jbc.M702861200. [DOI] [PubMed] [Google Scholar]
- Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, Cho W. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]