Abstract
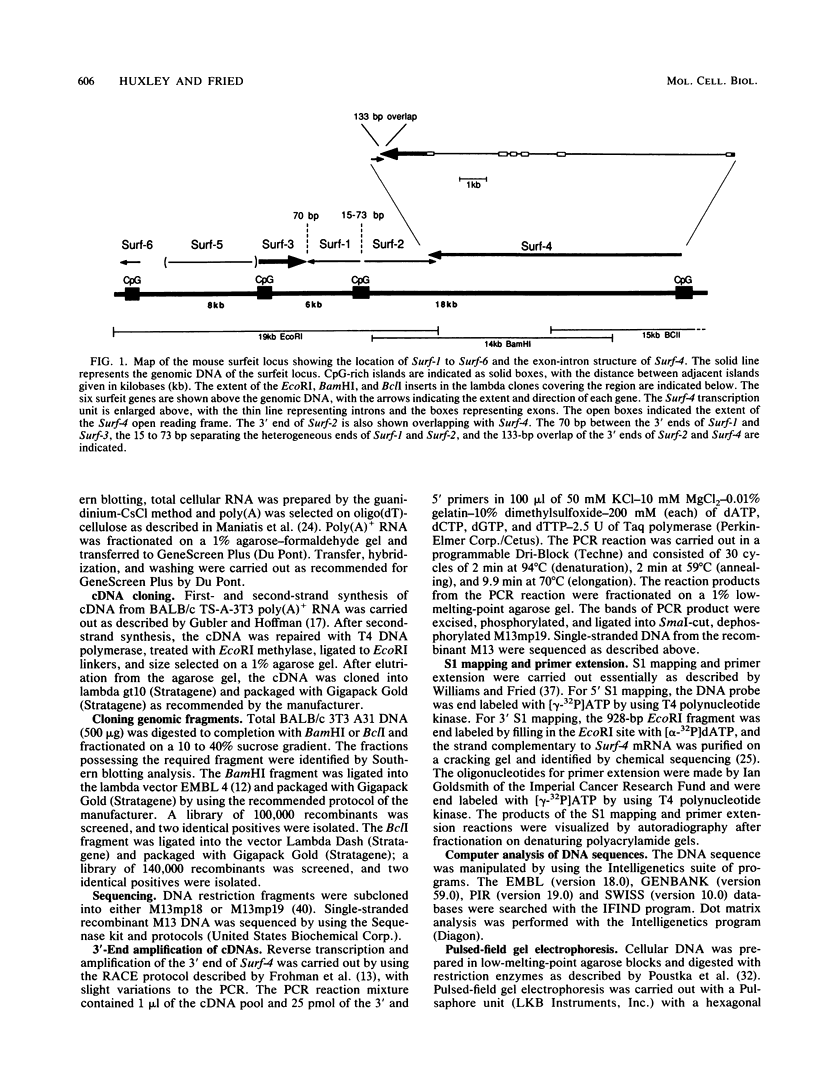
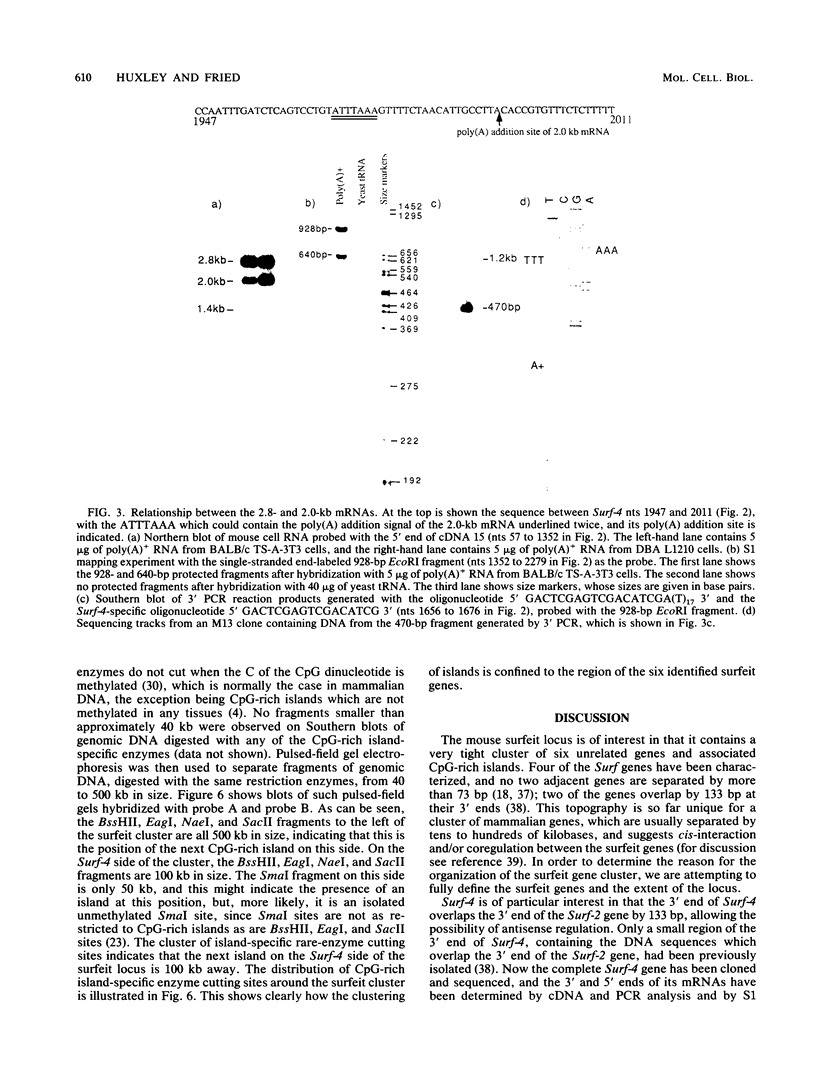
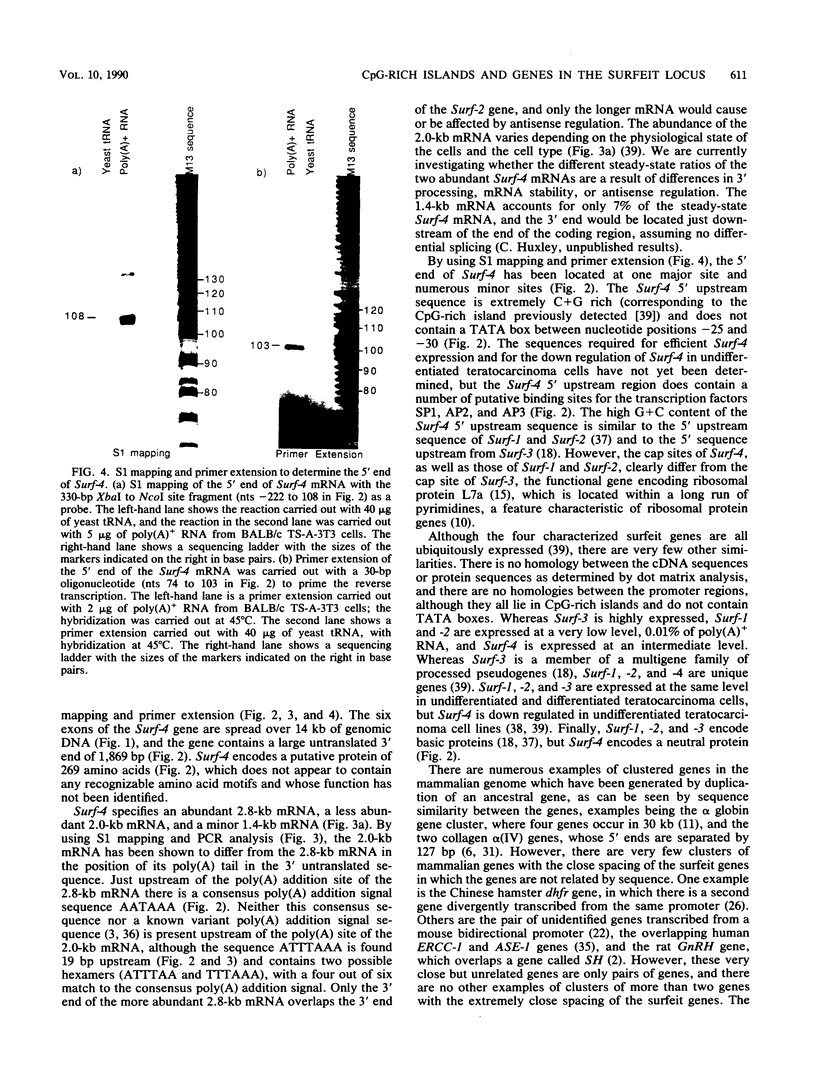
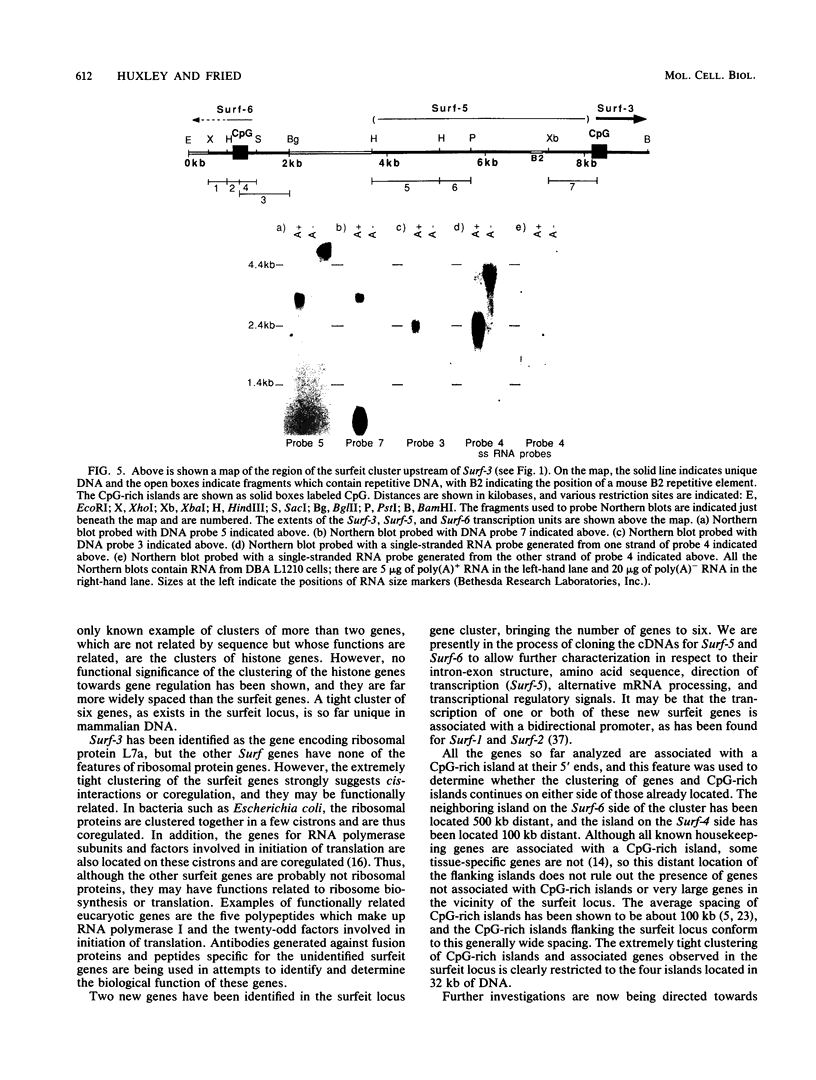
The clustered arrangement (no two adjacent genes are separated by more than 73 base pairs [bp] and two genes overlap by 133 bp at their 3' ends) of the four genes (Surf-1 to -4) identified so far in the mouse surfeit locus (T. Williams, J. Yon, C. Huxley, and M. Fried, Proc. Natl. Acad. Sci. USA 85:3527-3530, 1988) is the tightest gene clustering found in any mammalian genome to date and strongly suggests the possibility of cis-interaction and/or coregulation of gene expression. Thus, we are analyzing the surfeit genes in detail and are defining the extent of the cluster. Here we present the sequence of the entire Surf-4 gene and define the 3' and 5' extents of its mRNAs. The Surf-4 gene has heterogeneous transcriptional start sites, and its 5' end lies in a CpG-rich island. The gene specifies three mRNAs, with the two most abundant mRNAs differing in the locations of their 3' polyadenylation sites. Only the most abundant Surf-4 mRNA would overlap the 3' end of the Surf-2 gene by 133 bp. Two new genes (Surf-5 and Surf-6) have been identified in the surfeit gene cluster by Northern (RNA) blot analysis. The 5' end of Surf-6 lies within the CpG-rich island about 8 kilobases (kb) from the CpG-rich island containing the 5' end of Surf-3, and Surf-5 lies between Surf-3 and Surf-6. Thus, the cluster contains a unique arrangement of four CpG-rich islands within 32 kb associated with the 5' ends of the six surfeit genes. The neighboring CpG-rich islands have been located 500 and 100 kb distant on either side of the surfeit cluster, indicating that the end of the cluster of islands has been reached.
Full text
PDF



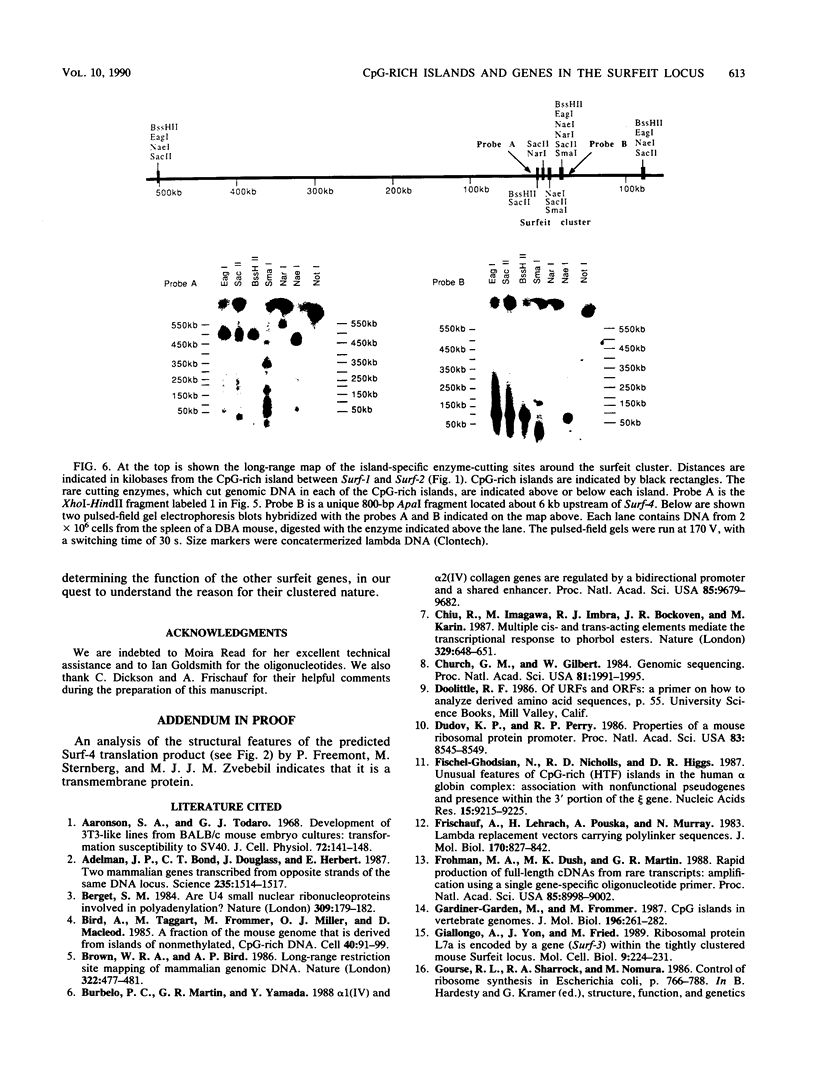

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Todaro G. J. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J Cell Physiol. 1968 Oct;72(2):141–148. doi: 10.1002/jcp.1040720208. [DOI] [PubMed] [Google Scholar]
- Adelman J. P., Bond C. T., Douglass J., Herbert E. Two mammalian genes transcribed from opposite strands of the same DNA locus. Science. 1987 Mar 20;235(4795):1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Brown W. R., Bird A. P. Long-range restriction site mapping of mammalian genomic DNA. 1986 Jul 31-Aug 6Nature. 322(6078):477–481. doi: 10.1038/322477a0. [DOI] [PubMed] [Google Scholar]
- Burbelo P. D., Martin G. R., Yamada Y. Alpha 1(IV) and alpha 2(IV) collagen genes are regulated by a bidirectional promoter and a shared enhancer. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9679–9682. doi: 10.1073/pnas.85.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R., Imagawa M., Imbra R. J., Bockoven J. R., Karin M. Multiple cis- and trans-acting elements mediate the transcriptional response to phorbol esters. Nature. 1987 Oct 15;329(6140):648–651. doi: 10.1038/329648a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudov K. P., Perry R. P. Properties of a mouse ribosomal protein promoter. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8545–8549. doi: 10.1073/pnas.83.22.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischel-Ghodsian N., Nicholls R. D., Higgs D. R. Unusual features of CpG-rich (HTF) islands in the human alpha globin complex: association with non-functional pseudogenes and presence within the 3' portion of the zeta gene. Nucleic Acids Res. 1987 Nov 25;15(22):9215–9225. doi: 10.1093/nar/15.22.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Giallongo A., Yon J., Fried M. Ribosomal protein L7a is encoded by a gene (Surf-3) within the tightly clustered mouse surfeit locus. Mol Cell Biol. 1989 Jan;9(1):224–231. doi: 10.1128/mcb.9.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Huxley C., Williams T., Fried M. One of the tightly clustered genes of the mouse surfeit locus is a highly expressed member of a multigene family whose other members are predominantly processed pseudogenes. Mol Cell Biol. 1988 Sep;8(9):3898–3905. doi: 10.1128/mcb.8.9.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavia P., Macleod D., Bird A. Coincident start sites for divergent transcripts at a randomly selected CpG-rich island of mouse. EMBO J. 1987 Sep;6(9):2773–2779. doi: 10.1002/j.1460-2075.1987.tb02572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay S., Bird A. P. Use of restriction enzymes to detect potential gene sequences in mammalian DNA. 1987 May 28-Jun 3Nature. 327(6120):336–338. doi: 10.1038/327336a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Wang C., Tjian R. Positive and negative regulation of transcription in vitro: enhancer-binding protein AP-2 is inhibited by SV40 T antigen. Cell. 1987 Sep 11;50(6):847–861. doi: 10.1016/0092-8674(87)90512-5. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1989;17 (Suppl):r389–r415. doi: 10.1093/nar/17.suppl.r389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka A., Pohl T. M., Barlow D. P., Frischauf A. M., Lehrach H. Construction and use of human chromosome jumping libraries from NotI-digested DNA. Nature. 1987 Jan 22;325(6102):353–355. doi: 10.1038/325353a0. [DOI] [PubMed] [Google Scholar]
- Pöschl E., Pollner R., Kühn K. The genes for the alpha 1(IV) and alpha 2(IV) chains of human basement membrane collagen type IV are arranged head-to-head and separated by a bidirectional promoter of unique structure. EMBO J. 1988 Sep;7(9):2687–2695. doi: 10.1002/j.1460-2075.1988.tb03122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruley H. E., Lania L., Chaudry F., Fried M. Use of a cellular polyadenylation signal by viral transcripts in polyoma virus transformed cells. Nucleic Acids Res. 1982 Aug 11;10(15):4515–4524. doi: 10.1093/nar/10.15.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell G. J., Walker P. M., Elton R. A., Subak-Sharpe J. H. Doublet frequency analysis of fractionated vertebrate nuclear DNA. J Mol Biol. 1976 Nov;108(1):1–23. doi: 10.1016/s0022-2836(76)80090-3. [DOI] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Williams T. J., Fried M. The MES-1 murine enhancer element is closely associated with the heterogeneous 5' ends of two divergent transcription units. Mol Cell Biol. 1986 Dec;6(12):4558–4569. doi: 10.1128/mcb.6.12.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3' ends. Nature. 1986 Jul 17;322(6076):275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- Williams T., Yon J., Huxley C., Fried M. The mouse surfeit locus contains a very tight cluster of four "housekeeping" genes that is conserved through evolution. Proc Natl Acad Sci U S A. 1988 May;85(10):3527–3530. doi: 10.1073/pnas.85.10.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Duin M., van Den Tol J., Hoeijmakers J. H., Bootsma D., Rupp I. P., Reynolds P., Prakash L., Prakash S. Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol Cell Biol. 1989 Apr;9(4):1794–1798. doi: 10.1128/mcb.9.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]







