Abstract
This mini review covers the drug discovery aspect of both proteasome activators and inhibitors. The proteasome is involved in many essential cellular functions, such as regulation of cell cycle, cell differentiation, signal transduction pathways, antigen processing for appropriate immune responses, stress signaling, inflammatory responses, and apoptosis. Due to the importance of the proteasome in cellular functions, inhibition or activation of the proteasome could become a useful therapeutic strategy for a variety of diseases. Many proteasome inhibitors have been identified and can be classified into two groups according to their source: chemically synthesized small molecules and compounds derived from natural products. A successful case of developing a proteasome inhibitor as a clinically useful drug is that the peptide boronate, PS341 (Bortezomib), was approved for the treatment of multiple myeloma. In contrast to proteasome inhibitors, small molecules that can activate or enhance proteasome activity are rare and are not well studied. The fact that over-expression of the cellular proteasome activator PA28 exhibited beneficial effects on the Huntington’s disease neuronal model cells raised the prospect that small molecule proteasome activators could become useful therapeutics. The beneficial effect of oleuropein, a small molecule proteasome activator, on senescence of human fibroblasts also suggested that proteasome activators might have the potential to be developed into anti-aging agents.
Keywords: Proteasome, activator, inhibitor, natural products, betulinic acids, peptides, anti-cancer, bortezomib
Introduction
There are two major cellular proteolysis systems: lysosomes and proteasomes. While lysosomes are critical in breaking down proteins taken into cells through endocytosis, proteasomes are the main machinery for intracellular protein degradation in eukaryotes [1, 2]. In addition to degradation of damaged or misfolded proteins, evidences suggest that proteasomes are involved in many essential cellular functions, such as cell cycle control, cell differentiation, signal transduction, antigen processing in immune responses, stress signaling, inflammatory responses, and apoptosis. Proteins undergo polyubiquitination before proteasomal degradation [3–5]. Overall, the ubiquitin-proteasome pathway is essential for maintaining optimal cellular functions.
Structure of proteasomes
A model of the 20S proteasome is illustrated in Fig. (1). The proteasome (20S) has a cylindrical structure that contains four rings stacked on top of each other. Each ring is composed of seven subunits. The two outer rings contain α subunits that do not have enzymatic activity. The two inner rings are comprised of β subunits, where the proteolytic activities reside. The β subunits possess three major proteolytic activities: a chymotrypsin-like (β5), a trypsin-like (β2), and a caspase-like (β1) activity. These proteolytic activities enable the proteasome to cleave proteins into small peptides, usually 8–12 amino acids in length. The 20S proteasome is associated with 19S components to form the 26S proteasomes. The 19S components regulate the entry of polyubiquitin marked proteins into the 20S proteasome [6]. The 19S complex, also termed PA700, activates the proteasome for protein degradation. In addition to PA700, there are at least two other intracellular protein complexes, PA28 and PA200 that can also activate the 20S proteasome using certain types of peptide substrate as models [7, 8]. The ability to activate the proteasome and the evolutionary conservation of these two activators imply that they may play a key role in intracellular protein degradation and normal cellular functions. In addition, an isoform of the proteasome, immunoproteasome, has been described in cells under influence of cytokines, such as interferon γ [9]. A key role of the immunoproteasome is to generate peptide fragments that bind to MHC class I molecules in antigen presentation.
Figure 1.
Model of the 20S proteasome. Active sites are located at the N-termini of β1, β2 and β5. The substrate preferences of these sites are: β1 – caspase-like substrates; β2 – trypsin-like substrates; β5 – chymotrypsin-like substrates.
Due to the importance of the proteasome in the regulation of cellular function, targeting the proteasome has been used as strategies in developing new treatments against cancer, inflammatory disease, and ischemic stroke. Besides the potential clinical usefulness, specific proteasome inhibitors provide interesting and important tools for cell biologists to dissect cellular processes that involve the proteasome.
Proteasome Inhibitors
Many proteasome inhibitors were synthesized or discovered during the last two decades. Major progresses have been made in identifying potent and specific inhibitors as well as understanding the mechanisms of action. Reviews and perspectives providing insightful information on proteasome inhibitors had been previously published [10–13]. The current known proteasome inhibitors can be classified into two groups according to their sources: chemically synthesized small molecules and compounds derived from nature products.
a) Peptide derivatives
Majority of small molecule proteasome inhibitors are peptide derivatives that bind to the catalytic site of β1, β2, and β5 subunits of the proteasome. Based on chemical structures, synthetic peptidyl inhibitors include peptide aldehydes, peptide boronates, and peptide sulfones. The chemical structure of some peptide derivatives are shown in Fig. (2). The first generation of synthetic tri-peptide aldehydes, such as MG132, was widely used as biological probes to study the structure and function of the proteasome [14]. MG132 inhibits the β5 catalytic subunit of proteasome through interaction with threonine in the catalytic site to form a reversible hemiacetal intermediate. In addition to tri-peptide aldehydes, di-peptide aldehydes were also shown to inhibit the proteasome. CEP1612 is a dipeptide aldehyde proteasome inhibitor that was highly selective for the chymotrypsin-like proteolytic activity of the proteasome [15].
Figure 2.

Synthetic peptide derivatives
Other reactive functional groups such as boronate and vinyl sulfones were used instead of the aldehyde in hope of obtaining compounds with enhanced potency and selectivity (Fig. (2)). The ZLVS and NLVS are examples of vinyl sulfones that can preferentially and irreversibly inhibit the chymotrypsin-like (β5) proteasome activity. Recently, peptide vinyl sulfones with phenolic substituents attached to the sulfone instead of a methyl group, were reported to be selective for the caspase-like (β1) proteasome activity [16]. Peptide boronates were shown to have significantly improved inhibitory activity over the aldehyde or vinyl sulfones on proteasome inhibition. MG262, a boronate analog of MG132, exhibited a 100-fold increase in anti-proteasome activity [17].
Due to involvement of the proteasome in many cellular functions, inhibition of the proteasome is predicted to cause severe side effects in potential clinical usage. Nevertheless, the peptide boronate, PS341 (Bortezomib), was successfully developed into an anti-cancer drug for the treatment of multiple myeloma [18]. Bortezomib is not as toxic as many people had predicted. It appeared that only the chymotrypsin-like activity of the proteasome was affected at therapeutic doses [19]. With the other two main proteolytic activities remaining, normal cells seem to suffer little consequence in the presence of the drug. Bortezomib is currently undergoing a number of clinical trials, including combinations with other anti-cancer drugs, for the treatment of hematologic malignancies other than multiple myeloma. Many mechanism of action studies indicated that bortezomib altered the level of cellular proteins associated with cell growth. Bortezomib stabilized IκB [20–25], activated c-Jun-N-terminal kinase [26–28], stabilized the CDK inhibitors p21 and p27 [20, 28, 29], stabilized the tumor suppressor p53, and stabilized pro-apoptotic proteins [30–32]. Stabilization of these proteins in cancer cells can result in apoptosis or inhibition of cancer cell proliferation.
b) Natural Products
Many natural product-derived proteasome inhibitors also contain peptidyl structures (Fig. (3)). Tyropeptin A is a tripeptide aldehyde natural product isolated from Kitasatospora sp. MK993-dF2. Like MG132, tyropeptin A preferentially inhibits the chymotrypsin-like proteasome activity through binding to the β5 subunit of the proteasome [33, 34]. Peptide epoxyketones, isolated from various microbials, are small peptides with a ketone epoxide functional group. For example, epoxomycin was derived from Streptomyces hygroscopicus [35]. TMC-86 and TMC-89 were isolated from Streptomyces sp. [36, 37] (Fig. (3)). Peptide epoxyketones inhibit the proteasome by covalently modifying the catalytic sites of theβ subunits. Carfilzomib (PR-171), an epoxyketone peptide structurally related to epoxomicin, is in Phase 2 clinical trials for patients with relapsed solid tumors including non-small cell lung cancer, small cell lung cancer, ovarian cancer, and renal cancer [38]. It is also in a phase 2 single-agent trial for patients with multiple myeloma and in a phase 1 study for lymphoma patients. Some peptide epoxyketone derivatives, such as dihydroeponemycin analogs, were shown to preferentially target the immunoproteasome [39].
Figure 3.
Natural product derived proteasome inhibitors
PR39 is a natural occurring antibacterial peptide containing 39 amino acid residues isolated from pig intestine. R39 was shown to inhibit the proteasome. Unlike small tripeptide proteasome inhibitors that bind to the proteolytic active site located at β5 subunit, PR39 binds to the non-proteolytic α7 subunit of the 20S proteasome [40]. It was also reported that PR11 (first 11 residues of PR39 sequence: RRRPRPPYLPR) and its analogs exhibited similar activity to that of PR39. Inhibition of the proteasome by PR11 and PR39 resulted in accumulation of IκB, a factor that regulates the NF-κB-dependent gene expression pathways [41].
Other natural product proteasome inhibitors include lactacystin and belactosins isolated from Streptomyces sp. [42–50]. Salinosporamide A was isolated from the marine actinomyces, Salinospora tropica [51, 52]. These peptide proteasome inhibitors contain a reactive β-lactone or β-lactone precursors that can form an ester complex with amino acids containing a hydroxyl group, such as threonine. For example, lactacystin was proposed to be converted non-enzymatically to omuralide that reacts with the threonine in the catalytic sites of the proteasome. Salinosporamide A (NPI-0052) is undergoing clinical Phase 1b trials in drug combination therapy for patients with non-small cell lung cancer, pancreatic cancer, or melanoma. TMC-95A and its diastereoisomers are cyclic polypeptides derived from Apiospora montagnei Sacc [53]. This class of compounds is very potent proteasome inhibitors with IC50 at low nanomolar range. Unlike the previously described proteasome inhibitors that contain a reactive functional group and form covalent adducts at catalytic sites, TMC-95A and related compounds do not carry any specific catalytic site reactive group. Instead, TMC-95A binds to the catalytic site through hydrophobic and multiple H-bond interaction. Recently, another cyclic peptide, argyrin A, derived from the myxobacterium Archangium gephyra was reported to be a potent proteasome inhibitor. Structurally, argyrin A is not related to TMC-95A (Fig. (3)). In comparison with bortezomib, argyrin A displayed similar proteasomal inhibition profiles on the caspase-, trypsin-, and chymotrypsin-like proteolytic activities of the proteasome [54].
Natural products derived from plant sources, such as celastrol isolated from the traditional herbal medicine “Thunder-god vine” and withaferin A isolated from Indian winter cherry, were shown to inhibit the proteasome at low micromolar concentrations [55, 56]. Celastrol is a triterpene and withaferin A is structurally related to steroids (Fig. (4)). Withaferin A was reported to induce apoptosis and inhibit the growth of human breast cancer cells [57]. Gliotoxin is a fungal metabolite structurally related to the epipolythiodioxo-piperazines with a broad range of biological activities and found to be particularly toxic to rodents. It was reported that gliotoxin inhibited the 20S protesaome [58, 59]. Green tea polyphenolic catechins such as (−)-epigallocatechin-3-gallate {(−)-EGCG} and its analogs have been widely studied for their possible benefits in cancer prevention. EGCG was reported to potently inhibit the chymotrypsin-like activity of the proteasome in vitro and in cultured tumor cells. It was suggested that free phenolic hydroxyl groups on the benzoate ring were essential for forming H-bond with the β5 subunit of the proteasome [60].
Figure 4.
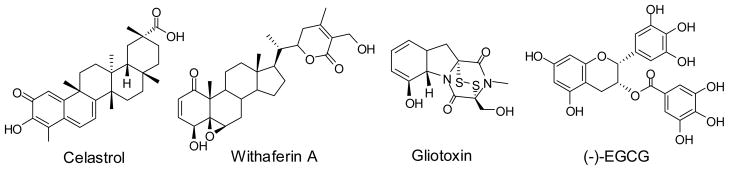
Non-peptide proteasome inhibitors
c) Other proteasome inhibiting agents
Disulfiram, a drug for the treatment of alcohol dependence, was shown to inhibit the proteasome (Fig. (5)). Because of proteasome inhibition, it inhibited the TNF-α-induced NF-κB translocation and caused the accumulation of the tumor suppressor protein p27kip1. Disulfiram was shown to have selective cytotoxicity against the chronic lymphocytic leukemia cells but not to normal peripheral blood mononuclear cells [61].
Figure 5.
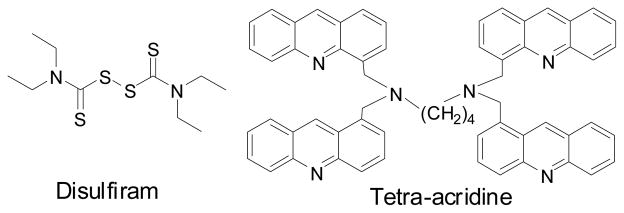
Other proteasome inhibitors
Acridine derivatives were known as a class of anti-cancer agents primarily targeting DNA and topoisomerase II. The structure of tetra-acridine is shown in Fig. (5). Recently, a study showed that tetra-acridine derivatives possessed proteasome inhibitory activity in addition to their effects on DNA [62]. These compounds displayed equal potency of cytotoxicity with/without down-regulation of topoisomerase II activity. On the other hand, other acridine derivatives that lack proteasome inhibition activity, such as massacring, showed weaker cytotoxicity toward cells with down-regulation of topoisomerase II. Proteasome inhibition by these compounds appeared to be partly responsible for their anti-tumor activity.
Proteasome activators
In contrast to the development of proteasome inhibitors, drug-like small molecules that can activate or enhance proteasome activity are rare and are not well studied [63]. On the other hand, there are three known types of cellular proteasome activators, PA28, PA200 and PA700 [6–8, 64, 65]. Over-expression of PA28 was shown to enhance the survival of Huntington’s disease (HD) neuronal model cells [66]. Dysfunction of the ubiquitin-proteasome system was indicated in patients with Huntington’s disease. The beneficial effects of PA28 over-expression on the HD cell model suggested that small molecule proteasome activators might be developed as useful therapeutic agents.
Several types of small molecules, including denaturing reagents (SDS), lipids and peptide-based activators were shown to activate the proteasome at relatively high concentrations. For example, SDS at 0.05%, polylysine at 1 mg/ml, and some other peptide-based activators were shown to activate 20S proteasome at 100 μM [67]. Fatty acids such as oleic, linoleic, and linolenic acids were also shown to activate the proteasome [68, 69]. Fatty acids modulated proteasome activity in a similar pattern to that of SDS by partially denaturing the proteasome. On the other hand, synthetic peptidyl alcohols, esters, p-nitroanilides, and nitriles were reported to activate the proteasome in a concentration range of 50–150 mM through binding to the same site as PA28 [70]. Cellular lipid components such as ceramides, lysophosphatidylinositol, and cardiolipin were found to activate the proteasome [71–73]. In addition, the arginine-rich histone H3, a protein found in chromatin, was reported to selectively enhance protein degradation by the proteasome [74].
In contrast to the above discussed proteasome activators with either relatively low potency or high molecular mass, oleuropein and betulinic acid (BA) are two small molecules that can activate the proteasome at low micromolar concentrations (Fig. (6)). Oleuropein is a natural product isolated from Olea europaea [75]. Oleuropein was shown to enhance all three proteasome activities and delay replicative senescence of human embryonic fibroblasts. Unlike PA28, SDS, or oleuropein that activates all three proteasome proteolytic activities, BA preferentially activated the chymotrypsin-like activity and with no or minimal effect on trypsin-like and caspase-like activities [76]. In addition to activation of the proteasome, BA and BA derivatives have been extensively studied for their potential as anti-HIV agents [77–82]. BA, a lupene-type pentacyclic triterpene derived from many plant species such as birch tree [83], was also extensively studied as a potential anti-tumor agent due to its selective cytotoxicity towards some cancer cell lines [84–86].
Figure 6.

Proteasome activators
Triterpene derivatives as either proteasome inhibitors or activators
Although BA is a proteasome activator, it can be structurally modified to become an inhibitor. In contrast to BA, many BA derivatives inhibited the proteasome. For example, BA derivatives, such as DSB (3′,3′-dimethylsuccinyl betulinic acid) and compound 4, inhibited the proteasome (Table 1). In a cell-based assay, DSB effectively inhibited the chymotrypsin-like proteasome activity by 50% (EC50) at 5.5 μM [76]. DSB is a potent anti-HIV-1 agent targeting viral maturation [77]. However, the proteasome inhibition activity of this compound does not appear to be responsible for its anti-HIV-1 activity.
Table 1.
Regulation of the chymotrypsin-like proteasome activity by triterpene derivatives.

| ||||
|---|---|---|---|---|
| R | BA derivatives | EC50 (μM) | GLA derivatives | EC50 (μM) |
| H | BA | Activator | GLA | 22.3 |

|
DSB | 5.5 | 1 | 0.87 |
|
|
4 | 2.9 | 2 | 1.05 |

|
3 | 0.22 | ||
Some of the data presented in the review were from studies supported by the NIH grants: AI65310 to C H Chen. We thank Greg Ho and Xian Qiao for proof reading the manuscript.
Triterpenes other than BA can be used as scaffolds to synthesize potent proteasome inhibitors. For example, Glycyrrhetinic acid (GLA) is a β-amyrin type pentacyclic triterpene that shares structural similarities to BA and is better for the synthesis of proteasome inhibitors (Table 1). The major differences between GLA and BA are on the size of E-ring, and the extra α, β conjugated ketone on its C-ring. GLA and the 3-diglucuronide derivative glycyrrhizin can be extracted from Glycyrrhiza glabra (Leguminosae). Biological activities including anti-inflammatory, anti-ulcer, anti-tumor and anti-viral activity have been reported for GLA related compounds [87]. In contrast to BA, GLA did not activate proteasome, it inhibited the chymotrypsin-like activity of the proteasome (Table 1).
Various C-3 modified derivatives of GLA have been synthesized and tested for proteasome inhibition [88]. With the same C-3 side chain modifications, GLA derivatives were in general more potent than BA derivatives as seen in 1 versus DSB, and in 2 versus 4 (Table 1). Among the GLA derivatives, GLA monoisophthalate (3) exhibited a 2-log improvement in potency over GLA. Glycyrrhizic acid, the glycoside of GLA, is a major component from licorice species and has been used as food sweetener and clinical treatment of HBV with low toxicity [89]. Thus, GLA derivatives might have the potential to be developed as clinically useful proteasome inhibitors.
Summary
Due to the importance of proteasome in multiple cellular processes, activation or inhibition of the proteasome is expected to affect many vital cellular functions. Potential applications and biomedical implications of proteasome regulators could be interesting and significant in many areas including cell cycle control, apoptosis, stress and heat shock responses, regulation of gene expression, modulation of antigen presentation for appropriate immune responses, inhibition of virus replication by proteasome inhibitors [90–92], inflammatory diseases [93, 94], neurodegenerative diseases, and aging. Many factors such as cell type, proliferation status, and specificity could affect the ultimate effects of proteasome regulators. Although proteasome regulators may affect diverse biological responses, potent and specific activities can be developed for biological and clinical applications as seen with bortezomib.
References
- 1.Goldberg AL, Dice JF. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43:835–69. doi: 10.1146/annurev.bi.43.070174.004155. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AL, St John AC. Intracellular protein degradation in mammalian and bacterial cells: Part 2. Annu Rev Biochem. 1976;45:747–803. doi: 10.1146/annurev.bi.45.070176.003531. [DOI] [PubMed] [Google Scholar]
- 3.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 7.Hill CP, Masters EI, Whitby FG. The 11S regulators of 20S proteasome activity. Curr Top Microbiol Immunol. 2002;268:73–89. doi: 10.1007/978-3-642-59414-4_4. [DOI] [PubMed] [Google Scholar]
- 8.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–25. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16:76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001:739–58. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 11.Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21:245–73. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KB, Fonseca FN, Crews CM. Development and characterization of proteasome inhibitors. Methods Enzymol. 2005;399:585–609. doi: 10.1016/S0076-6879(05)99039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borissenko L, Groll M. 20S proteasome and its inhibitors: crystallographic knowledge for drug development. Chem Rev. 2007;107:687–717. doi: 10.1021/cr0502504. [DOI] [PubMed] [Google Scholar]
- 14.Kim KB, Crews CM. Chemical Genetics: Exploring the Role of the Proteasome in Cell Biology Using Natural Products and Other Small Molecule Proteasome Inhibitors. J Med Chem. 2008;51:2600–5. doi: 10.1021/jm070421s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iqbal M, Chatterjee S, Kauer JC, Das M, Messina P, Freed B, Biazoo W, Siman R. Potent inhibitors of proteasome. J Med Chem. 1995;38:2276–7. doi: 10.1021/jm00013a002. [DOI] [PubMed] [Google Scholar]
- 16.van Swieten PF, Samuel E, Hernandez RO, van den Nieuwendijk AMCH, Leeuwenburgh MA, van der Marel GA, Kessler BM, Overkleeft HS, Kisselev AF. A cell-permeable inhibitor and activity-based probe for the caspase-like activity of the proteasome. Bioorg Med Chem Lett. 2007;17:3402–5. doi: 10.1016/j.bmcl.2007.03.092. [DOI] [PubMed] [Google Scholar]
- 17.Adams J, Behnke M, Chen S, Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg Med Chem Lett. 1998;8:333–8. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 18.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508–13. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 19.Kisselev AF, Callard A, Goldberg AL. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem. 2006;281:8582–90. doi: 10.1074/jbc.M509043200. [DOI] [PubMed] [Google Scholar]
- 20.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61:3071–6. [PubMed] [Google Scholar]
- 21.Russo SM, Tepper JE, Baldwin ASJ, Liu R, Adams J, Elliott P, Cusack JCJ. Int J Radiat. doi: 10.1016/s0360-3016(01)01446-8. [DOI] [PubMed] [Google Scholar]; Enhancement of radiosensitivity by proteasome inhibition: implications for a role of NF-kappaB. Oncol Biol Phys. 2001;50:183–93. doi: 10.1016/s0360-3016(01)01446-8. [DOI] [PubMed] [Google Scholar]
- 22.Sunwoo JB, Chen Z, Dong G, Yeh N, Crowl BC, Sausville E, Adams J, Elliott P, Van Waes C. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin Cancer Res. 2001;7:1419–28. [PubMed] [Google Scholar]
- 23.Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, Munshi N, Dang L, Castro A, Palombella V, Adams J, Anderson KC. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277:16639–47. doi: 10.1074/jbc.M200360200. [DOI] [PubMed] [Google Scholar]
- 24.Tan C, Waldmann TA. Proteasome inhibitor PS-341, a potential therapeutic agent for adult T-cell leukemia. Cancer Res. 2002;62:1083–6. [PubMed] [Google Scholar]
- 25.Ma MH, Yang HH, Parker K, Manyak S, Friedman JM, Altamirano C, Wu ZQ, Borad MJ, Frantzen M, Roussos E, Neeser J, Mikail A, Adams J, Sjak-Shie N, Vescio RA, Berenson JR. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clin Cancer Res. 2003;9:1136–44. [PubMed] [Google Scholar]
- 26.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101:1530–4. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- 27.Chauhan D, Li G, Podar K, Hideshima T, Mitsiades C, Schlossman R, Munshi N, Richardson P, Cotter FE, Anderson KC. Targeting mitochondria to overcome conventional and bortezomib/proteasome inhibitor PS-341 resistance in multiple myeloma (MM) cells. Blood. 2004;104:2458–66. doi: 10.1182/blood-2004-02-0547. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Ikezoe T, Saito T, Kobayashi M, Koeffler HP, Taguchi H. Proteasome inhibitor PS-341 induces growth arrest apoptosis of non-small cell lung cancer cells via the JNK/c-Jun/AP-1 signaling. Cancer Sci. 2004;95:176–80. doi: 10.1111/j.1349-7006.2004.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah SA, Potter MW, McDade TP, Ricciardi R, Perugini RA, Elliott PJ, Adams J, Callery MP. 26S proteasome inhibition induces apoptosis and limits growth of human pancreatic cancer. J Cell Biochem. 2001;82:110–22. doi: 10.1002/jcb.1150. [DOI] [PubMed] [Google Scholar]
- 30.Williams SA, McConkey DJ. The proteasome inhibitor bortezomib stabilizes a novel active form of p53 in human LNCaP-Pro5 prostate cancer cells. Cancer Res. 2003;63:7338–44. [PubMed] [Google Scholar]
- 31.Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000;275:21648–52. doi: 10.1074/jbc.M001083200. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97:3850–5. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Momose I, Sekizawa R, Hashizume H, Kinoshita N, Homma Y, Hamada M, Iinuma H, Takeuchi T. Tyropeptins A and B, new proteasome inhibitors produced by Kitasatospora sp. MK993-dF2. I. Taxonomy, isolation, physico-chemical properties and biological activities. J Antibiotics. 2001;54:997–1003. doi: 10.7164/antibiotics.54.997. [DOI] [PubMed] [Google Scholar]
- 34.Momose I, Umezawa Y, Hirosawa S, Iinuma H, Ikeda D. Structure-based design of derivatives of tyropeptin A as the potent and selective inhibitors of mammalian 20S proteasome. Bioorg Med Chem Lett. 2005;15:1867–71. doi: 10.1016/j.bmcl.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 35.Hanada M, Sugawara K, Kaneta K, Toda S, Nishiyama Y, Tomita K, Yamamoto H, Konishi M, Oki T. Epoxomicin, a new antitumor agent of microbial origin. Epoxomicin, a new antitumor agent of microbial origin. J Antibiot (Tokyo) 1992;45:1746–52. doi: 10.7164/antibiotics.45.1746. [DOI] [PubMed] [Google Scholar]
- 36.Koguchi Y, Kohno J, Suzuki S, Nishio M, Takahashi K, Ohnuki T, Komatsubara S. TMC-86A, B and TMC-96, new proteasome inhibitors from Streptomyces sp. TC 1084 and Saccharothrix sp. TC 1094. II. Physico-chemical properties and structure determination. J Antibiot (Tokyo) 2000;53:63–5. doi: 10.7164/antibiotics.53.63. [DOI] [PubMed] [Google Scholar]
- 37.Koguchi Y, Nishio M, Suzuki S, Takahashi K, Ohnuki T, Komatsubara S. TMC-89A and B, new proteasome inhibitors from streptomyces sp. TC 1087. J Antibiot (Tokyo) 2000;53:967–72. doi: 10.7164/antibiotics.53.967. [DOI] [PubMed] [Google Scholar]
- 38.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FWB, Chanan-Khan AA, Orlowski RZ. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–90. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho YK, Bargagna-Mohan P, Wehenkel M, Mohan R, Kim KB. LMP2-specific inhibitors: chemical genetic tools for proteasome biology. Chem Biol. 2007;14:419–30. doi: 10.1016/j.chembiol.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao Y, Lecker S, Post MJ, Hietaranta AJ, Li J, Volk R, Li M, Sato K, Saluja AK, Steer ML, Goldberg AL, Simons M. Inhibition of ubiquitin-proteasome pathway-mediated I kappa B alpha degradation by a naturally occurring antibacterial peptide. J Clin Invest. 2000;106:439–48. doi: 10.1172/JCI9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anbanandam A, Albarado DC, Tirziu DC, Simons M, Veeraraghavan S. Molecular basis for proline- and arginine-rich peptide inhibition of proteasome. J Mol Biol. 2008;384:219–27. doi: 10.1016/j.jmb.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omura S, Matsuzaki K, Fujimoto T, Kosuge K, Furuya T, Fujita S, Nakagawa A. Structure of lactacystin, a new microbial metabolite which induces differentiation of neuroblastoma cells. J Antibiot (Tokyo) 1991;44:117–8. doi: 10.7164/antibiotics.44.117. [DOI] [PubMed] [Google Scholar]
- 43.Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. Lactacystin, a novel microbial metabolite, induces neuritogenesis of neuroblastoma cells. J Antibiot (Tokyo) 1991;44:113–6. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi S, Uchida K, Nakagawa A, Miyake Y, Kainosho M, Matsuzaki K, Omura S. Biosynthesis of lactacystin. J Antibiot (Tokyo) 1995;48:1015–20. doi: 10.7164/antibiotics.48.1015. [DOI] [PubMed] [Google Scholar]
- 45.Fenteany G, Standaert RF, Reichard GA, Corey EJ, Schreiber SL. A beta-lactone related to lactacystin induces neurite outgrowth in a neuroblastoma cell line inhibits cell cycle progression in an osteosarcoma cell line. Proc Natl Acad Sci U S A. 1994;91:3358–62. doi: 10.1073/pnas.91.8.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katagiri M, Hayashi M, Matsuzaki K, Tanaka H, Omura S. The neuritogenesis inducer lactacystin arrests cell cycle at both G0/G1 and G2 phases in neuro 2a cells. J Antibiot (Tokyo) 1995;48:344–6. doi: 10.7164/antibiotics.48.344. [DOI] [PubMed] [Google Scholar]
- 47.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–31. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 48.Imajoh-Ohmi S, Kawaguchi T, Sugiyama S, Tanaka K, Omura S, Kikuchi H. Lactacystin, a specific inhibitor of the proteasome, induces apoptosis in human monoblast U937 cells. Biochem Biophys Res Commun. 1995;217:1070–7. doi: 10.1006/bbrc.1995.2878. [DOI] [PubMed] [Google Scholar]
- 49.Asai A, Haseqawa A, Ochiai K, Yamashita Y, Mizukami T. Belactosin A, a novel antitumor antibiotic acting on cyclin/CDK mediated cell cycle regulation, produced by Streptomyces sp. J Antibiot (Tokyo) 2000;53:81–3. doi: 10.7164/antibiotics.53.81. [DOI] [PubMed] [Google Scholar]
- 50.Asai A, Tsujita T, Sharma SV, Yamashita Y, Akinage S, Funakoshi M, Kobayashi H, Mizukami T. A new structural class of proteasome inhibitors identified by microbial screening using yeast-based assay. Biochem Pharmacol. 2004;67:227–34. doi: 10.1016/j.bcp.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 51.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source a marine bacterium of the new genus salinospora. Angew Chem Int Ed Engl. 2003;42:355–7. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 52.Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55(Pt 5):1759–66. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 53.Koguchi Y, Kohno J, Nishio M, Takahashi K, Okuda T, Ohnuki T, Komatsubara S. TMC-95A, B, C, and D, novel proteasome inhibitors produced by Apiospora montagnei Sacc. TC 1093. Taxonomy, production, isolation, and biological activities. J Antibiot (Tokyo) 2000;53:105–9. doi: 10.7164/antibiotics.53.105. [DOI] [PubMed] [Google Scholar]
- 54.Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sorensen I, Steinmeta H, Kubicka S, Carlomagno T, Menche D, Gutgemann I, Buer J, Gossler A, Manns MP, Kalesse M, Frank R, Malek NP. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Celastrol, a triterpene extracted from the Chinese “Thunder of God Vine,” is a potent proteasome inhibitor and suppresses human prostate cancer growth in nude mice. Cancer Res. 2006;66:4758–65. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 56.Yang H, Shi G, Dou QP. The tumor proteasome is a primary target for the natural anticancer compound Withaferin A isolated from “Indian winter cherry”. Mol Pharmacol. 2007;71:426–37. doi: 10.1124/mol.106.030015. [DOI] [PubMed] [Google Scholar]
- 57.Stan SD, Hahm ER, Warin R, Singh SV. Withaferin A causes FOXO3a- and Bim-dependent apoptosis and inhibits growth of human breast cancer cells in vivo. Cancer Res. 2008;68:7661–9. doi: 10.1158/0008-5472.CAN-08-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kroll M, Arenzana-Seisdedos F, Bachelerie F, Thomas D, Friguet B, Conconi M. The secondary fungal metabolite gliotoxin targets proteolytic activities of the proteasome. Chem Biol. 1999;6:89–98. doi: 10.1016/s1074-5521(00)80016-2. [DOI] [PubMed] [Google Scholar]
- 59.Hatabu T, Hagiwara M, Taguchi N, Kiyozawa M, Suzuki M, Kano S, Sato K. Plasmodium falciparum: the fungal metabolite gliotoxin inhibits proteasome proteolytic activity and exerts a plasmodicidal effect on P. falciparum. Exp Parasitol. 2006;112:179–83. doi: 10.1016/j.exppara.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 60.Landis-Piwowar KR, Wan SB, Wiegand RA, Kuhn DJ, Chan TH, Dou QP. Methylation suppresses the proteasome-inhibitory function of green tea polyphenols. J Cell Physicl. 2007;213:252–60. doi: 10.1002/jcp.21124. [DOI] [PubMed] [Google Scholar]
- 61.Lovborg H, Oberg F, Rickardson L, Gullbo J, Nygren P, Larsson R. Inhibition of proteasome activity, nuclear factor-KappaB translocation and cell survival by the antialcoholism drug disulfiram. Int J Cancer. 2006;118:1577–80. doi: 10.1002/ijc.21534. [DOI] [PubMed] [Google Scholar]
- 62.Vispe S, Vandenberghe I, Robin M, Annereau JP, Creancier L, Pique V, Galy JP, Ruczynski AK, Barret JM, Bailly C. Novel tetra-acridine derivatives as dual inhibitors of topoisomerase II and the human proteasome. Biochem Pharmaco. 2007;73:1863–72. doi: 10.1016/j.bcp.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 63.Dahlmann B, Becher B, Sobek A, Ehlers C, Kopp F, Kuehn L. In vitro activation of the 20S proteasome. Enzyme Protein. 1993;47:274–84. doi: 10.1159/000468685. [DOI] [PubMed] [Google Scholar]
- 64.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem J. 2000;345(Pt 1):1–15. [PMC free article] [PubMed] [Google Scholar]
- 65.DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–6. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 66.Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of huntington’s disease neuronal model cells. PLoS ONE. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tanaka K, Ii K, Ichihara A, Waxman L, Goldberg AL. A high molecular weight protease in the cytosol of rat liver. I. Purification, enzymological properties, and tissue distribution. J Biol Chem. 1986;261:15197–203. [PubMed] [Google Scholar]
- 68.Dahlmann B, Rutschmann M, Kuehn L, Reinauer H. Activation of the multicatalytic proteinase from rat skeletal muscle by fatty acids or sodium dodecyl sulphate. Biochem J. 1985;228:171–7. doi: 10.1042/bj2280171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe N, Yamada S. Activation of 20S proteasomes from spinach leaves by fatty acids. Plant Cell Physiol. 1996;37:147–51. doi: 10.1093/oxfordjournals.pcp.a028925. [DOI] [PubMed] [Google Scholar]
- 70.Wilk S, Chen WE. Synthetic peptide-based activators of the proteasome. Mol Biol Rep. 1997;24:119–24. doi: 10.1023/a:1006851428691. [DOI] [PubMed] [Google Scholar]
- 71.Ohkubo I, Gasa S, Namikawa C, Makita A, Sasaki M. Human erythrocyte multicatalytic proteinase: activation and binding to sulfated galacto- and lactosylceramides. Biochem Biophys Res Commun. 1991;174:1133–40. doi: 10.1016/0006-291x(91)91538-n. [DOI] [PubMed] [Google Scholar]
- 72.Matsumura K, Aketa K. Proteasome (multicatalytic proteinase) of sea urchin sperm and its possible participation in the acrosome reaction. Mol Reprod Dev. 1991;29:189–99. doi: 10.1002/mrd.1080290215. [DOI] [PubMed] [Google Scholar]
- 73.Ruiz de Mena I, Mahillo E, Arribas J, Castaňo JG. Kinetic mechanism of activation by cardiolipin (diphosphatidylglycerol) of the rat liver multicatalytic proteinase. Biochem J. 1993;296(Pt 1):93–7. doi: 10.1042/bj2960093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orlowski M. Selective activation of the 20 S proteasome (multicatalytic proteinase complex) by histone h3. Biochemistry. 2001;40:15318–26. doi: 10.1021/bi0116240. [DOI] [PubMed] [Google Scholar]
- 75.Katsiki M, Chondrogianni N, Chinou I, Rivett AJ, Gonos ES. The olive constituent oleuropein exhibits proteasome stimulatory properties in vitro and confers life span extension of human embryonic fibroblasts. Rejuvenation Res. 2007;10:157–72. doi: 10.1089/rej.2006.0513. [DOI] [PubMed] [Google Scholar]
- 76.Huang L, Ho P, Chen CH. Activation and inhibition of the proteasome by betulinic acid and its derivatives. FEBS Letters. 2007;581:4955–9. doi: 10.1016/j.febslet.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kashiwada Y, Hashimoto F, Cosentino LM, Chen CH, Garrett PE, Lee KH. Betulinic acid and dihydrobetulinic acid derivatives as potent anti-HIV agents. J Med Chem. 1996;39:1016–7. doi: 10.1021/jm950922q. [DOI] [PubMed] [Google Scholar]
- 78.Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee KH, Aiken C, Chen CH. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol. 2004;78:922–9. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holz-Smith SL, Sun IC, Jin L, Matthews TJ, Lee KH, Chen CH. Role of human immunodeficiency virus (HIV) type 1 envelope in the anti-HIV activity of the betulinic acid derivative IC9564. Antimicrob Agents Chemother. 2001;45:60–6. doi: 10.1128/AAC.45.1.60-66.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mayaux JF, Bousseau A, Pauwels R, Huet T, Henin Y, Dereu N, Evers M, Soler F, Poujade C, De Clercq E, Le Pecq JB. Triterpene derivatives that block entry of human immunodeficiency virus type 1 into cells. Proc Natl Acad Sci U S A. 1994;91:3564–8. doi: 10.1073/pnas.91.9.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soler F, Poujade C, Evers M, Carry JC, Henin Y, Bousseau A, Huet T, Pauwels R, De Clercq E, Mayaux JF, Le Pecq JB, Dereu N. Betulinic acid derivatives: a new class of specific inhibitors of human immunodeficiency virus type 1 entry. J Med Chem. 1996;39:1069–83. doi: 10.1021/jm950669u. [DOI] [PubMed] [Google Scholar]
- 82.Huang L, Yuan X, Aiken C, Chen CH. Bifunctional anti-human immunodeficiency virus type 1 small molecules with two novel mechanisms of action. Antimicrob Agents Chemother. 2004;48:663–5. doi: 10.1128/AAC.48.2.663-665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cînt Pînzaru S, Leopold N, Kiefer W. Vibrational spectroscopy of betulinic acid HIV inhibitor and of its birch bark natural source. Talanta. 2002;57:625–31. [PubMed] [Google Scholar]
- 84.Kessler JH, Mullauer FB, de Roo GM, Medema JP. Broad in vitro efficacy of plant-derived betulinic acid against cell lines derived from the most prevalent human cancer types. Cancer Lett. 2007;25:132–45. doi: 10.1016/j.canlet.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 85.Pisha E, Chai H, Lee IS, Chagwedera TE, Farnsworth NR, Cordell GA, Beecher CW, Fong HH, Kinghorn AD, Brown DM, Wani MC, Wall ME, Hieken TJ, Das Gupta TK, Pezzuto JM. Discovery of betulinic acid as a selective inhibitor of human melanoma that functions by induction of apoptosis. Nat Med. 1995;1:1046–51. doi: 10.1038/nm1095-1046. [DOI] [PubMed] [Google Scholar]
- 86.Fulda S, Jeremias I, Pietsch T, Debatin KM. Betulinic acid: a new chemotherapeutic agent in the treatment of neuroectodermal tumors. Klin Padiatr. 1999;211:319–22. doi: 10.1055/s-2008-1043808. [DOI] [PubMed] [Google Scholar]
- 87.Farina C, Pinza M, Pifferi G. Synthesis and anti-ulcer activity of new derivatives of glycyrrhetic, oleanolic and ursolic acids. Farmaco. 1998;53:22–32. doi: 10.1016/s0014-827x(97)00013-x. [DOI] [PubMed] [Google Scholar]
- 88.Huang L, Yu D, Ho P, Quan K, Lee KH, Chen CH. Synthesis proteasome inhibition of glycyrrhetinic acid derivatives. Bioorg Med Chem. 2008;16:6696–701. doi: 10.1016/j.bmc.2008.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sato H, Goto W, Yamamura J, Kurokawa M, Kageyama S, Takahara T, Watanabe S, Shiraki K. Therapeutic basis of glycyrrhizin on chronic hepatitis B. Antiviral Res. 1996;30:171–7. doi: 10.1016/0166-3542(96)00942-4. [DOI] [PubMed] [Google Scholar]
- 90.Lindenthal C, Weich N, Chia YS, Heussler V, Klinkert MQ. The proteasome inhibitor MLN-273 blocks exoerythrocytic and erythrocytic development of Plasmodium parasites. Parasitology. 2005;131(Pt 1):37–44. doi: 10.1017/s003118200500747x. [DOI] [PubMed] [Google Scholar]
- 91.Schubert U, Ott DE, Chertova EN, Welker R, Tessmer U, Princiotta MF, Bennink JR, Krausslich HG, Yewdell JW. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc Natl Acad Sci U S A. 2000;97:13057–62. doi: 10.1073/pnas.97.24.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ott DE, Coren LV, Sowder RC, 2nd, Adams J, Schubert U. Retroviruses have differing requirements for proteasome function in the budding process. J Virol. 2003;77:3384–93. doi: 10.1128/JVI.77.6.3384-3393.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kalogeris TJ, Laroux FS, Cockrell A, Ichikawa H, Okayama N, Phifer TJ, Alexander JS, Grisham MB. Effect of selective proteasome inhibitors on TNF-induced activation of primary and transformed endothelial cells. Am J Physiol. 1999;276(4 Pt 1):C856–64. doi: 10.1152/ajpcell.1999.276.4.C856. [DOI] [PubMed] [Google Scholar]
- 94.Palombella VJ, Conner EM, Fuseler JW, Destree A, Davis JM, Laroux FS, Wolf RE, Huang J, Brand S, Elliott PJ, Lazarus D, McCormack T, Parent L, Stein R, Adams J, Grisham MB. Role of the proteasome and NF-kappaB in streptococcal cell wall-induced polyarthritis. Proc Natl Acad Sci U S A. 1998;95:15671–6. doi: 10.1073/pnas.95.26.15671. [DOI] [PMC free article] [PubMed] [Google Scholar]




