Abstract
The role of affinity in determining neutralizing potency of monoclonal antibodies directed against viruses is not well understood. We investigated the kinetic, structural, and functional advantage conferred by individual naturally-occurring somatic mutations in the antibody heavy chain variable region of Fab19, a well-described neutralizing human monoclonal antibody directed to respiratory syncytial virus (RSV). Comparison of the affinity-matured antibody Fab19 with recombinant Fab19 antibodies that were variants containing reverted amino acids from the inferred unmutated ancestor sequence revealed the molecular basis for affinity maturation of this antibody. Enhanced binding was achieved through mutations in the third heavy chain complementary determining region (HCDR3) that conferred a markedly faster on-rate and a desirable increase in antiviral neutralizing activity. In contrast, most somatic mutations in the HCDR1 and HCDR2 regions did not significantly enhance antigen binding or antiviral activity. We observed a direct relationship between the measured association rate (Kon) for F protein and antiviral activity. Modeling studies of the structure of the antigen-antibody complex suggested the HCDR3 loop interacts with the antigenic site A surface loop of the RSV F protein, previously shown to contain the epitope for this antibody by experimentation. These studies define a direct relationship of affinity and neutralizing activity for a viral glycoprotein-specific human monoclonal antibody.
Keywords: Human, antibodies, viral, comparative immunology/evolution
Introduction
Respiratory syncytial virus (RSV) is the leading cause of viral bronchiolitis and viral pneumonia in infants and children (1). The virus is classified in the genus Pneumovirus of the family Paramyxoviridae. Its genome is composed of single-stranded negative-sense RNA encoding 11 proteins, including the fusion (F) surface glycoprotein that is a target for protective neutralizing antibodies. By two years of age, nearly all children are seropositive for RSV, consistent with the widespread prevalence of the virus in the pediatric population (2). Lack of an effective antibody response to RSV made by infants likely contributes to the high morbidity caused by this virus during the first 6 months of life. Despite nearly universal infection at a young age, immunity following RSV infection is incomplete, and upper respiratory tract reinfection can occur throughout life. Serum and mucosal neutralizing antibodies are thought to be the principal immune effectors responsible for the prevention of severe lower respiratory tract infection (3). A central role for antibodies in the prevention of severe RSV disease has been supported by the clinical effectiveness of palivizumab, an FDA-approved humanized murine monoclonal antibody (mAb) licensed for prophylaxis of high-risk subjects. Passive immunization with palivizumab in selected high-risk infants reduces the rate of hospitalization due to severe RSV lower respiratory tract disease (4, 5, 6). Additional RSV-specific human mAbs have been studied in preclinical studies for prophylaxis or therapy, including the potent and broadly-reactive neutralizing antibody designated Fab19 that we previously described (7, 8).
A major feature of RSV infection in infants is the poor quality of the antibody response compared to that in adults (9), the likely result of a naïve response mediated by an immature immune system that is still developing. We have shown previously that the principal difference between virus-specific antibody sequences of infants and those of adults is a marked lack of somatic mutations in infant antibodies (10, 11). However, most of our previous studies investigated human antibodies to rotavirus VP6 protein, which is not a classic neutralizing antibody target. RSV F protein in contrast is a major target for neutralizing antibodies. The role of somatic hypermutation and affinity maturation in antibody-mediated virus neutralization is not clear. Some studies suggest that increased affinity enhances neutralizing potency (12, 13, 14, 15, 16), while others do not (17). We sought to explore the role of affinity in neutralizing potency for RSV F antibodies by studying affinity variants of an inhibitory human monoclonal antibody. Using the human RSV F-specific monoclonal antibody Fab19, we defined the structural and functional consequences of naturally-occurring somatic mutations.
To date there are limited studies that offer detailed kinetic analysis of the direct contribution of individual naturally-occurring somatic mutations to affinity of antibody binding in the context of antibody function (18, 19, 20, 21, 22). Many of the past binding studies focused on haptens or model protein antigens such as hen egg lysozyme (23, 24, 25, 26), and studies of pathogen-specific antibodies have examined the effect of random changes to mature antibody sequences rather than the enhancements gained through natural affinity maturation (12, 13, 14, 15). Although both approaches have produced important contributions, it is of interest to study how naturally-occurring somatic mutations in human antibody genes affect the response to an important biologically relevant protein, particularly an immunodominant protein from a major viral pathogen like RSV. We investigated here in detail the kinetic advantage and impact on antiviral activity of individual naturally-occurring somatic mutations in a human monoclonal antibody specific for the RSV F protein.
Materials and Methods
Construction of Fab19 expression vector
Previously isolated heavy and light chain gene fragments of Fab19 were sequenced-optimized for bacterial expression, and cDNA was synthesized (GENEART AG, Regensburg, Germany) (8). Synthesized cDNAs were designed to include 5′ XhoI – ApaI 3′ and 5′ SacI – EcoRV 3′ restriction sites at the ends of the heavy and light chain variable gene fragments, respectively. To construct a Fab fragment vector that expresses soluble Fab19, the sequenced-optimized Fab19 heavy and light chain variable gene cDNAs were ligated in step-wise fashion into the linearized pComb3XΔ19 vector that contains both bacterial sequence-optimized CH1 and Cκ regions in-frame with the VH and VL insert, as previously described (27). All cloned DNA constructs were verified by nucleotide sequence analysis.
Expression and purification of recombinant antibodies
The Fab19 expression vector was transformed into the nonsuppressor E. coli strain HB2151. A 100 mL overnight culture prepared from a single, transformed colony was inoculated into 1 liter of 2XYT broth (Research Products International, Prospect, IL) containing 100 μg/mL ampicillin and incubated in a bacterial shaker at 200 rpm at 30°C until the culture exhibited an OD600 of 0.5. Expression of soluble Fab fragments was induced by the addition of 5 mM IPTG (isopropyl-ß-D-thioglalctopyranoside (Research Products International) to the culture and continued incubation at 30°C for an additional 16 hrs. After incubation, cells were harvested from liquid culture by centrifugation at 3000 × g for 20 min at 4°C. For isolation of soluble Fab fragments from the bacterial periplasm, the cell pellet was resuspended vigorously in 25 mL chilled TS buffer (0.2 M Tris-HCl, 0.5 M sucrose, pH 7.4) and incubated on ice for 30 min. Insoluble cell debris was removed by centrifugation at 16,000 × g for 20 min at 4 °C. Fab fragments were purified from the periplasmic extract by immobilized metal ion affinity chromatography using pre-packed 5 mL HisTrap HP Ni-Sepharose columns (GE Healthcare Bio-Sciences AB, Uppsala, Sweden), according to the manufacturer’s recommended protocol. The eluted purified Fab product then was buffer exchanged with phosphate buffered saline and concentrated using an Amicon Ultra Centrifugal Filter Device with a 30 kD molecular weight cut off (Millipore, Billerica, MA). Purity was confirmed by visualizing on a denaturing, non-reducing SDS-PAGE gel a single, silver-stained band at the expected 50 kD migration. Protein concentration was determined by a Bradford-dye based assay (Bio-Rad Laboratories, Hercules, CA). Purified Fabs were stored at 4 °C until analysis.
Generation of variant Fab antibody fragments
Plasmid vectors, based on pComb3XΔ19, containing mutant derivatives of Fab19 were prepared by changing one of the naturally-occurring mutations in the affinity-matured Fab19 VH gene sequence back to the amino acid in the inferred germline sequence. The predicted germline amino acids residues were determined by comparing antibody nucleotide sequences by alignment software with the ImMunoGeneTics database (http://www.imgt.org/) (28). The predicted mutations were introduced using QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), per the manufacturer’s protocol. Briefly, complementary oligonucleotides that contained the desired nucleotide back-mutation sequence were synthesized according to the manufacturer’s recommendations. Each PCR reaction included 5 μL 10× Reaction Buffer, 25 ng Fab19/pComb3XΔ19 plasmid template, 125 ng of each complementary oligonucleotide primer, 1 μL of dNTP mix (100 mM), 1 μL PfuTurbo DNA polymerase (2.5 U/μL), and ddH2O to a final volume of 50 μL. PCR conditions consisted of a single melting cycle of 95 °C for 30 seconds followed by 16 cycles of 95 °C for 30 seconds, 60 °C for 1 minutes, and 68 °C for 5 minutes. Following PCR, 1 μl of DpnI restriction enzyme (New England Biosystems) was added directly to each PCR reaction and incubated at 37 °C for 1 hour. DpnI-treated DNA was transformed into E. coli strain DH5α competent cells and plasmids purified with the QIAprep Miniprep Kit (Qiagen). Plasmid constructs were screened by nucleotide sequence analysis to verify the presence of each intended back-mutation in the variant Fab19 VH segment. The mutant Fabs were named by the original amino acid at the position indicated, followed by the single amino acid code for the inferred germline residue to which it was changed.
Cloning of RSV fusion (F) protein ectodomain
The RSV F ectodomain construct (pcDNA3.1-FECTO-myc/His), was constructed by PCR amplification of the ectodomain, removing the transmembrane domain and cytoplasmic tail regions from full-length RSV Fopt(domains and construct previously described) (29). The PCR product was cloned directionally into pcDNA3.1/myc-His B (Invitrogen) using restriction sites 5′-BamHI-EcoRI - 3′. Ligated product was transformed into E. coli strain DH5α competent cells, and plasmids purified with the QIAprep Miniprep Kit (Qiagen). All plasmid constructs were sequenced to confirm in-frame cloning with the C-terminal c-myc epitope and polyhistidine (6xHis) tag of the expression vector.
RSV F protein expression and purification
The pcDNA3.1-FECTO-myc/His recombinant plasmid was transfected into suspension 293-F cells, as recommended by the manufacturer (Freestyle 293 Expression System, Invitrogen). Transfected cells were incubated for 4 days, then the cells were centrifuged for 10 minutes at 100 × g at 4 °C and supernatant filtered through 0.2 μm filters prior to purification and stored at 4 °C. Purification of 6xHis-tagged RSV FECTO protein was performed by immobilized metal ion affinity chromatography using pre-packed 5 mL HisTrap HP Ni-Sepharose columns (GE Healthcare Bio-Sciences AB, Uppsala, Sweden), according to the manufacturer’s recommended protocol. Purified protein was concentrated and dialyzed against phosphate buffered saline through Amicon Ultra centrifugal filters with a 30 kD molecular weight cutoff (Millipore, Billerica, MA). Purified preparations were identified by denaturing, non-reducing SDS-PAGE analysis as a single, silver-stained band at the expected 60 kD migration. Concentration was determined by the Bradford dye assay. Purified RSV F protein was stored at 4 °C until analyzed.
Measurement of kinetics of Fab fragment binding to RSV F protein
The association and dissociation rates of wild-type or mutant Fab19 antibody fragments with RSV F protein were measured on a Biacore 2000 biosensor (Biacore AB, Uppsala, Sweden). Purified recombinant RSV F protein was diluted to 30 μg/mL in 10 mM sodium acetate, pH 4.5, and covalently immobilized at 5 μL/min by amine coupling to the dextran matrix of a CM5 sensor chip (Biacore AB, Uppsala, Sweden) with a target RU density of 1,200. A surface density of 1,200 RU was used to yield an Rmax of ~100 RU during the binding experiments. Unreacted, active ester groups on the sensor chip were blocked with 1 M ethanolamine. For use as a reference, a blank surface, containing no protein, was prepared under identical immobilization conditions. Each purified Fab19 antibody variant was prepared at 5 different concentrations ranging from 5 to 500 nM in HBS/Tween-20 buffer (Biacore AB, Uppsala, Sweden), and injected over the immobilized RSV F protein and reference cell surfaces. Each Fab19 variant, at every concentration, was run in triplicate. Fab fragment binding was measured at a flow rate of 30 μL/min for 180 seconds and dissociation was monitored for an additional 360 seconds. Residual bound Fab fragment after each cycle was removed from the sensor chip by flowing 50 mM HCl at 100 μL/min for 30 seconds. Association rates (Kon) and dissociation rates (Koff) were calculated by aligning the binding curves globally to fit a 1:1 Langmuir binding model using BIAevaluation 4.1 software (Biacore AB, Uppsala, Sweden). Calculated rates were accepted only if the goodness of each fit based on the agreement between experimental data and the calculated fits had a χ2 value below 5. The KD values then were calculated for each Fab from the equation koff/kon = KD.
Measurement of Fab fragment neutralization activity
The neutralizing activity of the Fabs was measured by a plaque reduction assay using HEp-2 cell culture and the A2 strain of RSV. The activity was measured in μg/mL and calculated as the lowest concentration of Fab that reduced plaque number by 60%. Diluted RSV strain A2 suspended to yield 50 plaques per well was mixed with 1:4 dilutions of Fab, and incubated at 37 °C for 60 min. Cell monolayers in 24-well tissue culture plates at 80-90% confluency were inoculated in duplicate by replacing the medium in each well with 100 μL of virus-Fab mixture. After incubation at 37 °C for 1 hr, cell monolayers then were overlaid with 0.75% methylcellulose in Opti-MEM I (Invitrogen) supplemented with 2% FBS, 320 μg/mL l-glutamine, 2.7 μg/mL amphotericin B, and 45 μg/mL gentamicin. Cultures were incubated for 4 days at 37 °C in 5% CO2, after which the overlay was removed and the monolayers were fixed in 80% cold methanol. Plaques were stained and quantified by an immunoperoxidase staining procedure, as described (30). Plaques for each Fab dilution were counted, duplicate values averaged, and Fab dilution versus plaque number plotted. The 60% plaque reduction neutralizing activity (μg/mL) was determined by regression curve analysis.
Predicting the structure of Fab19 antibody fragments and the Fab19-RSV F complex
Blast (31) searches were performed using the Fab19 heavy and light chain variable regions as queries. The template heavy and light chains were selected from PDB 3OTE and 2UZI, respectively, based on sequence similarity with Fab19. The rigid body orientation of the heavy and light chain variable fragments in the homology model was selected to mimic the orientation of the chains in 2UZI owing to overall sequence similarity with Fab19. However, the 2UZI heavy chain variable sequence contained insertions and deletions relative to Fab19, therefore the heavy chain from PDB 3OTE was used instead.
Loop modeling simulations and structural clustering were performed within the Rosetta software package (32) to generate 25 distinct low-energy models for Fab19. Loop modeling (33) was used to generate 10,000 initial models for Fab 19. Diverse conformations for the CDR loops of heavy and light chains were sampled by fragment insertion and using a low-resolution energy function; subsequently each complete model, including the CDR and non-CDR regions, was subjected to rounds of full atom refinement. The 1,000 best (lowest energy) models then were clustered according to structural similarity. A cluster radius of 1.5 Å was employed, yielding a total of 25 clusters. The lowest energy model within each cluster was selected as a homology model for Fab19.
The 25 homology models for Fab19 were docked to the crystal structure of the post-fusion conformation of the RSV F trimer (PDB 3RRR). The initial Fab19-RSV trimer rigid-body orientation was similar to the orientation of the humanized and affinity-matured RSV F mAb motavizumab when bound to its epitope in the RSV-trimer. The motavizumab rigid-body orientation was derived from the alignment of the RSV peptide-epitope crystallized in complex with motavizumab (PDB 3IXT) to the RSV F trimer structure (34). This initial configuration of the Fab19 was considered reasonable given that the epitopes of Fab 19 and motavizumab are located in the same RSV segment. The docking simulations included a first stage of local full-atom docking (35, 36) that sampled diverse rigid-body orientations in which Fab19 models were docked locally relative to their starting configuration, a second stage of backrub refinement (37) at the antibody-antigen interface, and a final stage of local full-atom docking. A group of 25,000 docking models was generated for each of the 25 homology models, for a total of 625,000 docking models. Initially, the models were filtered based on binding energy (computed as Ecomplex – ERSVF – EFab19 where ERSVF and EFab19 were removed from the complex without any further relaxation), but this procedure was unsatisfactory because many of the lowest energy complexes did not have an interface involving the CDR loops of the antibody. Therefore, to restrict to models that were approximately consistent with the mutagenesis data, the models were instead filtered on the distance between RSV residue I266 (a Fab 19 escape mutation) and Fab 19 residue P210. A Cα-Cα distance cutoff of 10 Å left 140 models for further analysis. These models were evaluated based on buried surface area and on the fraction of contacts involving the CDR loops. Two models were selected with the largest buried surface areas and with the CDR loops interacting with RSV F. These two models were subjected to a final step of all-atom refinement in which stages of side-chain rotamer sampling and minimization were performed together with backbone minimization and rigid-body optimization. In total, 300 refined models were computed for each of the two selected models.
Three-dimensional images of each antibody were produced using PyMol, a molecular visualization system (The PyMOL Molecular Graphics System, Version 1.3, Schrodinger, LLC) (38).
Results
Analysis of Fabs with heavy chain variable region residues from the reverted unmutated ancestor of Fab19
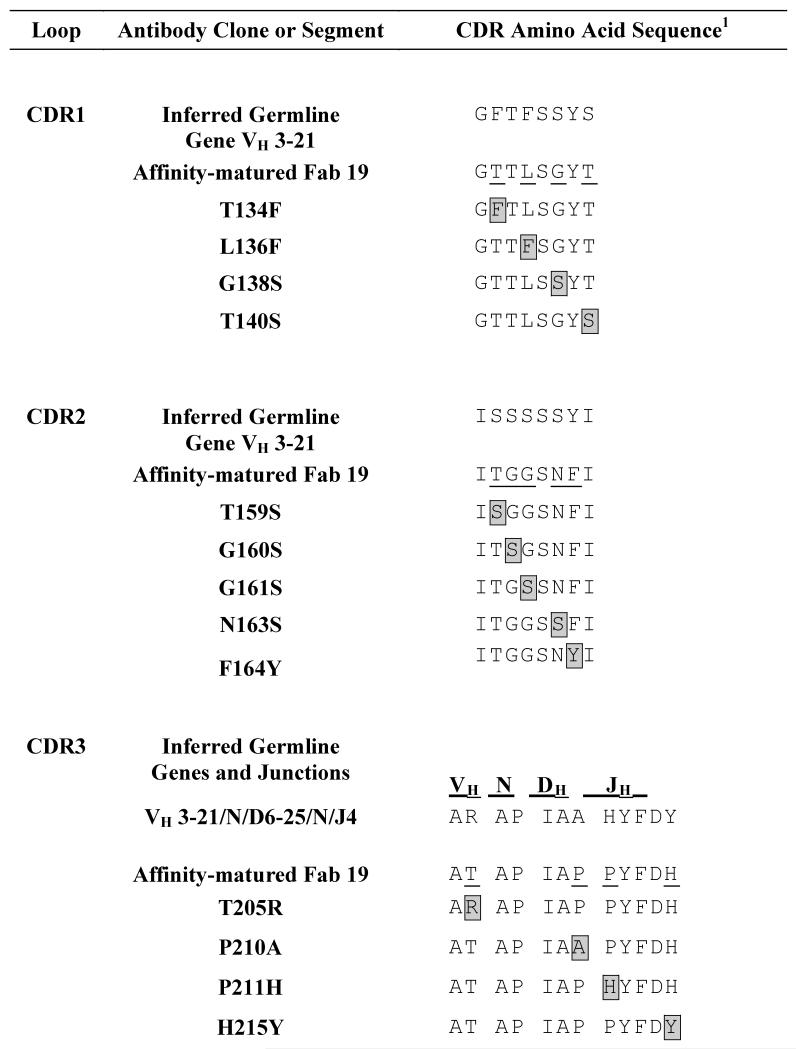
We hypothesized that a wide spectrum of affinity effects could be detected after reverting the mutated Fab19 heavy chain CDR residues to the amino acid in the inferred unmutated ancestor gene. Somatically mutated residues in the antibody variable region first were identified by comparing the Fab19 amino acid sequence to the inferred germline sequence of Fab19, specifically comparing the mature antibody to gene segments VH3-21*01, DH6-25*01 in reading frame 2, and JH4*02 (Supplemental Table 1). The first 20 nucleotides at the 5′ end of framework 1 of the Fab19 gene were not considered since they were encoded by PCR primers used for amplification. Thirteen amino acid changes within the three heavy chain CDR (HCDR) regions were identified by comparison with the inferred germline sequence (Table 1). The somatic mutations were distributed evenly through all three HCDR loops, with four in HCDR1, five in HCDR2, and four in HCDR3. Six of the thirteen somatic mutations occurred at positions in the inferred germline gene that originally encoded serine, a slightly polar, neutral amino acid. Four of these six original inferred germline gene serine residues were located in HCDR2.
Table 1. Mutated sequences of 13 different variant Fab 19 molecules.
HCDR residues reverted to the inferred germline sequence are boxed and highlighted in grey and aligned with the naturally-occurring mutated sequence (affinity matured Fab 19, somatic mutations underlined) or reverted unmutated ancestor (inferred germline) sequences of the VH, D and JH gene segments.
|
Binding kinetics of the wild-type Fab19 antibody
Prior to testing the panel of Fab19 inferred germline-reversion variants, the binding rate constants were determined for the fully matured Fab19 that contained all 13 naturally-occurring somatic mutations. The association and dissociation constants of Fab19 with RSV F protein were determined using surface plasmon resonance. The Kon (1/Ms), was 2.1 × 105 and the Koff (1/s), was 5.7 × 10−3. The Koff divided by the Kon determined the predicted steady state affinity of Fab19 with our recombinant RSV F protein, and was calculated to be 2.8 × 10−8 M, or 28 nM. This level of affinity is in the range of other highly active RSV antibodies, such as the clinically licensed RSV F mAb palivizumab (39).
Kinetic analysis of binding of Fab19 inferred germline-reversion variant antibodies
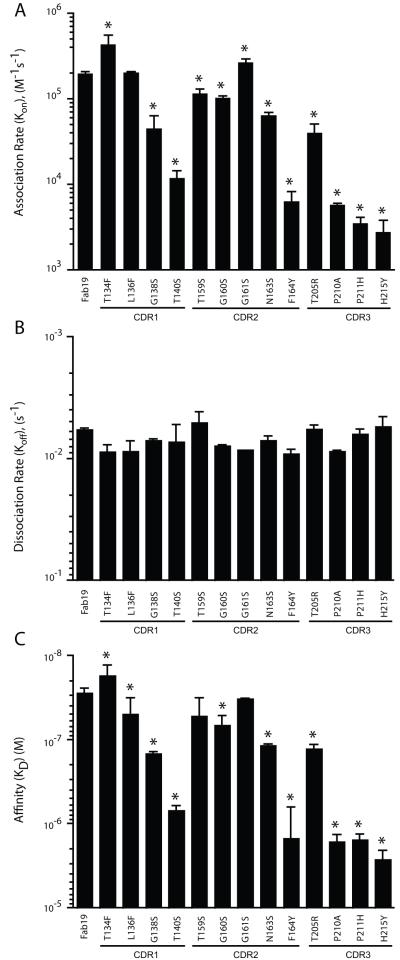
We determined the changes in kinetics of binding to the RSV F protein caused by HCDR somatic mutations in the fully matured Fab19 antibody by measuring the effect of back-mutating each predicted residue to its inferred germline amino acid. Using this mutation strategy, a decrease in the affinity of the inferred germline-reversion variant Fab suggests that the somatically mutated residue would be advantageous for a germline-encoded antibody to acquire. The association rate of each Fab19 inferred germline-reversion variant for binding to the RSV F protein is shown in Figure 1A. Compared to the association rate of the fully-matured Fab19 (2.1 × 105 1/Ms), the association rates with RSV F of the HCDR1 inferred germline-reversion variants ranged from 5.2 × 105 to 1.1 × 104 1/Ms, HCDR2 variants ranged from 2.6 × 105 to 4.3 × 103 1/Ms and HCDR3 variants ranged from 4.3 × 104 to 2.4 × 103 1/Ms. All but three variants (T134F, L136F, and G161S) showed reduced association rates with RSV F when compared to the fully-matured Fab19 antibody. Remarkably, the measured dissociation rates were similar among the Fab19 germline-reversion variants (Figure 1B). Compared to the fully matured Fab19 antibody steady state affinity of 2.8 × 10−8 M, all but four germline-reversion variants (T134F, L136F, T159S, and G161S) exhibited significantly lower calculated steady-state affinities for RSV F protein (Figure 1C).
Figure 1. Kinetics of Fab 19 or Fab 19 inferred germline-reversion variant binding to RSV F protein, measured by surface plasmon resonance.
Each variant is labeled to indicate the amino acid position with the somatically mutated residue from the matured antibody (to left) and the inferred germline residue to which it was reverted (to right). Association (A) or dissociation (B) rate of Fab 19 or Fab19 inferred germline-reversion variant binding to RSV F protein. Steady-state affinity [KD] of Fab19 or Fab19 inferred germline-reversion variants for binding to the RSV F protein (C). Each bar represents the mean rate constant or affinity (± standard deviation) calculated from experiments run in triplicate. * Indicates p < 0.05 compared to Fab19, as calculated by an unpaired t test.
Neutralization by the fully-matured Fab19 or the germline reversion-variant Fabs
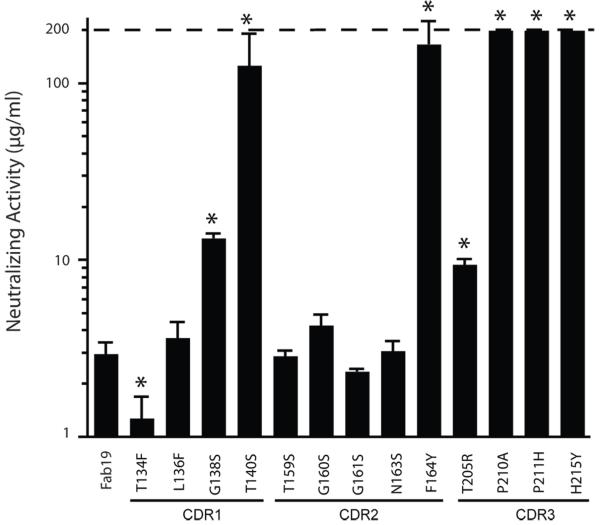
We determined the functional activity of each Fab19 inferred germline-reversion variant using a 60% viral plaque reduction assay. The inferred germline-reversion variant Fab neutralization activities varied greatly from 1.1 μg/mL to ≥ 200 μg/mL (Figure 2). Two of four inferred germline reversions in HCDR1 significantly reduced the antiviral activity of Fab19, one of five HCDR2 reversion variants exhibited reduced activity, and three of four HCDR3 variants showed reduced activity. Variant T134F, which had the highest association rate, also had the greatest virus neutralization activity.
Figure 2. Neutralizing activity of Fab19 or Fab19 inferred germline-reversion variants.
Activity was determined by plaque reduction (antibody concentration needed to reduce the number of RSV plaques by 60%). Greater values indicate less potent neutralizing activity. The range of antibody concentration tested was from 0.1 μg/mL to 200 μg/mL. Dotted line indicates upper limit of detection. Each bar represents the mean (± standard deviation) neutralization activity calculated from plaque reduction assays run in triplicate. * Indicates p < 0.05 compared to Fab19, as calculated by an unpaired t test.
Relationship of neutralizing activity to binding constants
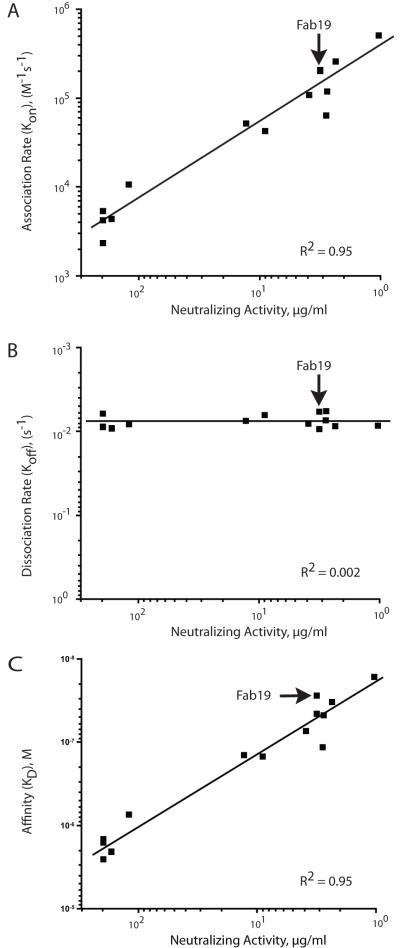
We next analyzed whether or not there was an effect of the kinetics of binding on neutralizing potency of the Fab19 inferred germline-reversion variants. We found that the association rate (Kon) exhibited a clear and direct relationship with neutralizing activity (Figure 3A); faster association rate was associated with decreased concentration needed to neutralize (i.e., higher potency). Dissociation rates (Koff) were similar among the Fab19 variants. Consequently, a correlation between changes in dissociation rate and neutralization could not be detected (Figure 3B). The positive correlation between Kon and neutralization also was reflected in the direct relationship between the steady-state affinity (KD) and neutralizing potency (Figure 3C). A summary of affinity and neutralizing activity for Fab 19 or related Fab mutants possessing reverted unmutated ancestor residues is shown in Table 2.
Figure 3. Relationship between association rate (A), dissociation rate (B), or steady-state affinity (B) and neutralizing activity of Fab19 inferred germline reversion mutations.
The trend lines are shown with the correlation coefficients (R2).
Table 2. Summary of affinity and neutralizing activity for Fab 19 or related Fab mutants possessing reverted unmutated ancestor residues.
| Location of reversion to unmutated ancestor |
Antibody | Steady-state affinity (KD) (M) |
Neutralizing specific
activity (μg/mL needed to neutralize) |
|---|---|---|---|
| None | wt Fab 19 | 2.9 × 10−8 | 3 |
| CDR1 | T134F | 1.8 × 10−8 | 1 |
| L136F | 5.0 × 10−8 | 4 | |
| G138S | 1.5 × 10−7 | 13 | |
| T140S | 7.2 x 10−7 | 120 | |
| CDR2 | T159S | 5.2 x 10−8 | 3 |
| G160S | 7.0 × 10−8 | 4 | |
| G161S | 3.3 × 10−8 | 2 | |
| N163S | 1.2 × 10−7 | 3 | |
| F164Y | 1.6 × 10−6 | 130 | |
| CDR3 | T205R | 1.3 × 10−7 | 9 |
| P210A | 1.7 × 10−6 | >200 | |
| P211H | 1.6 × 10−6 | >200 | |
| H215Y | 2.8 × 10−6 | >200 |
Predicted interactions between Fab 19 and RSV F protein
To predict the molecular interaction between RSV F and Fab 19, we created a homology model of Fab19 and docked the two molecules. We previously showed that a Fab19-selected escape mutant virus with a single F protein change (I266M) can no longer be neutralized by Fab19 mAb (7). The mutational data of the RSV escape mutant and of the Fab 19 inferred germline-reversion variants was used to select models on which residues shown to be important for binding affinity and virus neutralization were located in the interface of the modeled Fab 19–RSV F complex. The selected model shown in Figure 4A had the best binding energy among the final set of refined models; the binding energy was −11.4 Rosetta energy units, and the interfacial buried surface area on the RSV F-Fab 19 model was 1,072 Å2. For comparison, the complex of motavizumab with the RSV F trimer structure was refined using the same procedure; the binding energy of that complex was −13.2 Rosetta energy units and the interfacial buried surface area was 1,354 Å2. Certainly validation with atomic resolution structural data would be needed for certainty about the interactions at the interface, however we consider this predicted model a plausible binding mode by energetic and buried area considerations and because of its consistency with the biochemical data.
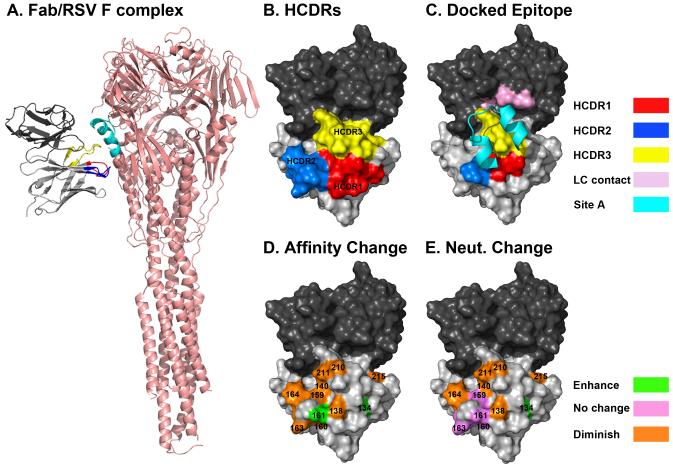
Figure 4. Docking model of Fab19 bound to the RSV F protein predicts interaction between the known RSV F protein antigenic site A and the HCDRs of Fab19.
(A) Surface projection of the predicted structural model of Fab19 showing the heavy chain (light gray), light chain (dark gray), HCDR1 (red), HCDR2 (blue), and HCDR3 (yellow) (B). The surface of Fab19, which the model predicts to be within 5Å of RSV F protein antigenic site A (cyan) is colored, as described above. Light chain residues within 5 Å of antigenic site A are shown in pink (C). Inferred germline reversion mutations are color coded according to their effect on the neutralizing activity of Fab19 (green – enhanced activity, violet – no effect, orange – reduced activity) (D). Inferred germline reversion mutations are color coded according to their effect on the Kon of Fab19, as described above (E).
The HCDRs of Fab19 are shown in Figure 4B, and the surface area of Fab19 with a predicted proximity to antigenic site A of RSV F of 5 Å is shown in Figure 4C. Surface areas of Fab19 greater than 5 Å from antigenic site A were considered to be too far from the epitope to interact directly with the antigen. The region of Fab19 close enough to interact directly with antigenic site A includes portions of HCDR1, HCDR2, and HCDR3 as well as residues from the light chain. HCDR1 and HCDR3 lie in the middle of the contact region.
The locations of the 11 inferred ancestor reversion variant residues predicted to lie on the surface of the Fab are represented on a modeled structure of the Fab19 protein (Figure 4D and 4E). Two different features of the experimental data were compared with the topography in the three-dimensional models. The magnitude of change of steady-state affinity for each germline-reversion residue variant reported in Figure 1 is projected onto the Fab structure in Figure 4D. Most back-mutations to the germline nucleotide resulted in decreased association rates. However, L136G had no effect, and T134F and G161S resulted in increased the association rates relative to Fab19. The effect of mutations on neutralizing activity for the panel of inferred germline-reversion variants reported in Figure 2 is projected onto the Fab structure in Figure 4E. Seven of the inferred ancestor reversion mutations resulted in significantly reduced virus neutralizing activity, while five mutations had no significant effect. The T134F mutation significantly enhanced the ability of Fab19 to neutralize virus. The models revealed the central combining surface of the antibody to be the region most sensitive to change for both affinity of binding to antigen and viral neutralization. Within this region, there was a bias towards mutations in the HCDR3 having greater effects than mutations in HCDR1 and HCDR2.
Three of the top five somatically mutated residues that most affected neutralization in the fully matured Fab19 antibody (T140S, P210A, P211H) fall into this region and appeared to be capable of direct interactions with the viral protein (Figure 4C). Other mutations with less effect on neutralization also reside in the predicted interface region. However, F164Y and H215Y, both of which greatly affected neutralization and association rate, fall outside this region and do not appear to directly interact with antigenic site A. These mutations likely cause changes in the conformation of the antibody loops, which negatively affect binding of the Fab to F.
Discussion
In this study, we identified the molecular features that determine the basis for the high affinity and neutralizing potency of a human mAb to the human pathogen RSV. The central finding of this study is that there was a direct correlation between faster Kon and neutralizing potency for this neutralizing antibody to the major antigenic site on the F protein. By re-introducing the original inferred germline residues into the nucleotide sequence of this potent inhibitory RSV F human mAb, the functional enhancement of virus neutralization mediated by somatic hypermutation during the development of the humoral immune response was demonstrated at a fine level.
Although it is well established that somatic hypermutation leads to affinity maturation, relatively little primary data exists regarding the effect of somatic mutations in human antibody sequences on neutralization activity of viral pathogens. Increased affinity and on-rates of antibodies have been postulated to provide increased in vivo effectiveness and protection but rarely has this correlation been analyzed for antibodies specific for, and protective against, infectious agents (40). It is not clear whether or not higher antibody steady state affinity or improved Kon or Koff parameters always results in greater functional activity of antiviral antibodies (41). The relationship between antibody affinity for viral proteins and antibody antiviral function is complex and likely varies among viruses and antigenic sites. Previous studies of vesicular stomatitis virus antibodies demonstrated a lack of effect of affinity maturation on in vivo antibody-mediated protection and neutralization (42). Other studies have noted an inconsistent effect of affinity maturation on the activity of anti-HIV antibodies (43, 44, 45). Given that HIV infection often does not generate a high-titer, neutralizing antibody response to primary isolates (46), the HIV model has limitations when investigating the role of affinity maturation on antibody function. Our previous studies with rotavirus VP6-specific human antibodies that inhibit transcription revealed that prolonged dissociation rate (and not faster association rate) was the determining feature in antibody potency for that interaction (47). The RSV-specific human Fab used in the current study was chosen based on studies that previously demonstrated a high neutralizing potency of this Fab against a wide range of RSV isolates. The antibody binds to the immunodominant antigenic loop on the fusion protein, designated antigenic site A, which is the target for most RSV neutralizing antibodies including the licensed antibody palivizumab (7).
For many antibodies, the CDR that is most often involved in direct antigen contact is HCDR3, which usually is situated in the center of the antigen interacting site (48). We found that most HCDR3 mutations in Fab19 did have a major effect on binding. The central importance of mutations in the HCDR3 region on antigen binding affinity and neutralization supports our predicted model of RSV F protein in complex with the fully-matured Fab19 mAb. Mapping of escape mutant virus F protein sequence changes also showed the antigenic site A (approximately residues 260-278) and specifically the residues near I266M to comprise a contact region on the viral protein for Fab19 (8).
However, the somatic mutations in Fab19 are distributed through all three HCDRs. This finding suggests somatically mutated residues of Fab19 exhibit cooperativity in the enhancement of binding that is not strictly limited to a small loop or region. In contrast to previous data with antibody interactions with small hapten antigens (49), these data suggest the importance of a relatively large antibody contact region for interaction with viral proteins.
Previous studies by others with an RSV-specific mAb measured the binding rate changes caused by randomly changing CDR amino acid residues (39). That study showed that selected mutations in the HCDRs of the humanized murine RSV mAb palivizumab resulted in an increased affinity for the RSV F protein achieved by both faster Kon and slower Koff parameters for Fabs. Detailed studies of those variant antibodies made as bivalent IgG variants however showed that an increase in antibody antiviral activity (potency of virus neutralization) was achieved only by mutations in the CDRs that led to faster Kon. However, mutations that reduced Koff enhanced the virus neutralization activity of monovalent Fab proteins. Our study reveals a similar relationship with antibody kinetics for naturally-occurring mutations in a human antibody specific for antigenic site A. Over the full range of measured values, the affinity of Fab19 increased with amino acid changes that enhanced association rate with the RSV F protein and resulted in an increase in neutralization activity. However, the neutralizing activity reached a threshold once the association rate of the Fab reached ~5 × 104 or the affinity reached ~2 × 10−7. Beyond these values, the correlation of association rate or affinity with neutralizing activity was weaker. These threshold values likely vary among epitopes and in other settings may be a function of the Koff as well. The relatively similar Koff values for the Fab19 variants using naturally occurring somatic mutations we tested, in combination with the broad range of Kon values, make it impossible to directly assess the effect of Koff on neutralization activity in this setting.
The most striking finding of these studies is that mutated residues that affected association rate also regulated functional activity. The inferred germline-reversion variants that exhibited the greatest effect on reducing the measured neutralization activity also were clustered in the HCDR3 loop. Specifically, P201A, P211H, and H215Y inferred germline-reversion variants located in the HCDR3 loop all had lost neutralization activity (i.e. had measured neutralization activities > 200 μg/mL). The mutated residues that abolished neutralization were restricted to the HCDR3. However many of the germline-reversion variants exhibited neutralizing activity >10 μg/mL. Antibodies with such low potency likely would not contribute significantly to viral immunity (50, 51).
A caveat of interpretation of these studies is that it is not possible to determine retrospectively the sequence of accumulation of somatic mutations that occurred during the development of the final affinity-matured Fab19. Nevertheless, measuring the effect of changes in affinity and neutralization activity by altering one somatically-mutated residue back to the inferred germline amino acid one at a time provides insight into the magnitude of change that occurred during each somatic mutation event. The wide range of neutralization activities (from as weak as >200 μg/mL to as high as 1 μg/mL) revealed the type of changes that Fab19 experienced during the somatic hypermutation response following repeated RSV exposures. The back-mutated residues that had the greatest effect on affinity and neutralization were located in the central antigen-binding surface of the antibody. This region likely represents the paratope of the antibody.
Taken together, these results suggest that the HCDRs specified by germline configuration of the heavy chain germline gene sequences of Fab19 were structurally fit for binding to RSV F protein with a low affinity, but somatic mutations in the three HCDRs of the fully-matured antibody increased the affinity of the antibody through faster association rate. These changes in affinity led to a desirable increase in antiviral function. These structure-function studies have implications for understanding the natural immune response to RSV.
The studies presented here on the role of somatic mutations in improvement of affinity and neutralizing activity are of interest because they correlate well with observations of the global nature of the infant immune response. We have shown previously that the human infant B cell response to viruses can be distinguished from that of adults by a marked lack of somatic mutations (10). Infant and adult responses to RSV differ in several ways, most notably because infants are immunologically immature and infants experience primary infection while adults suffer reinfection. Our studies cannot distinguish whether the lack of somatic mutations in virus-specific antibody genes from infants is characteristic of the normal, primary (antigen-naïve) response to RSV or stems from immunologic immaturity characteristic of an immune system still under development. It is impossible to determine which of these factors is most important, because seroconversion is widespread by one year of age and therefore older antigen-naïve subjects are difficult to identify for study. Interpretation of such studies also would be clouded by the fact that some infants seroconvert following primary infection early in life but later revert to a seronegative state because of lack of durability of response. In this setting, identifying antigen-naïve subjects is nearly impossible. A recent animal study has shown that a lack of T follicular helper cells in very early life correlates with reduced germinal center formation following immunization (52). Inefficient germinal center formation in infants might also explain the poor quality of antibodies made following virus infection or immunization. Regardless of the cause of the lack of somatic mutations in infant antibody genes, the studies here showing reduced activity by some antibodies containing unmutated ancestor residues suggest a molecular basis for the poor RSV-specific humoral immunity previously observed in infants. Such naturally-occurring antibodies specified by unmutated antibody genes likely possess a decreased capacity to neutralize virus, when compared to those of adults that possess affinity-matured antibodies.
Supplementary Material
Acknowledgments
We thank Frances House, Emily Deckleman, and Sunny Mok for excellent technical assistance and John Williams for helpful comments.
References
- 1.Hacking D, J Hull. Respiratory syncytial virus--viral biology and the host response. J. Infect. 2002;45:18–24. doi: 10.1053/jinf.2002.1015. [DOI] [PubMed] [Google Scholar]
- 2.Handforth J, Friedland JS, Sharland M. Basic epidemiology and immunopathology of RSV in children. Paediatr Respir Rev. 2000;1:210–214. doi: 10.1053/prrv.2000.0050. [DOI] [PubMed] [Google Scholar]
- 3.Domachowske JB, Rosenberg HF. Respiratory syncytial virus infection: immune response, immunopathogenesis, and treatment. Clin. Microbiol. Rev. 1999;12:298–309. doi: 10.1128/cmr.12.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The IMpact-RSV Study Group Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 5.Groothuis JR, Nishida H. Prevention of respiratory syncytial virus infections in high-risk infants by monoclonal antibody (palivizumab) Pediatr. Int. 2002;44:235–241. doi: 10.1046/j.1442-200x.2002.01558.x. [DOI] [PubMed] [Google Scholar]
- 6.Top FH, Jr., Connor EM, Carlin DA, IMpact-RSV Study Group Prophylaxis against respiratory syncytial virus in premature infants. Lancet. 2000;355:1014. doi: 10.1016/s0140-6736(05)74754-1. [DOI] [PubMed] [Google Scholar]
- 7.Crowe JE, Firestone CY, Crim R, Beeler JA, Coelingh KL, Barbas CF, Burton DR, Chanock RM, Murphy BR. Monoclonal antibody-resistant mutants selected with a respiratory syncytial virus-neutralizing human antibody fab fragment (Fab 19) define a unique epitope on the fusion (F) glycoprotein. Virology. 1998;252:373–375. doi: 10.1006/viro.1998.9462. [DOI] [PubMed] [Google Scholar]
- 8.Crowe JE, Jr., Murphy BR, Chanock RM, Williamson RA, Barbas CF, 3rd, Burton DR. Recombinant human respiratory syncytial virus (RSV) monoclonal antibody Fab is effective therapeutically when introduced directly into the lungs of RSV-infected mice. Proc Natl Acad Sci U S A. 1994;91:1386–1390. doi: 10.1073/pnas.91.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandenburg AH, Groen J, van Steensel-Moll HA, Claas EC, Rothbarth PH, Neijens HJ, Osterhaus AD. Respiratory syncytial virus specific serum antibodies in infants under six months of age: limited serological response upon infection. J Med Virol. 1997;52:97–104. doi: 10.1002/(sici)1096-9071(199705)52:1<97::aid-jmv16>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 10.Weitkamp JH, Lafleur BJ, Greenberg HB, Crowe JE., Jr. Natural evolution of a human virus-specific antibody gene repertoire by somatic hypermutation requires both hotspot-directed and randomly-directed processes. Hum Immunol. 2005;66:666–676. doi: 10.1016/j.humimm.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Weitkamp JH, Kallewaard N, Kusuhara K, Bures E, Williams JV, LaFleur B, Greenberg HB, Crowe JE., Jr. Infant and adult human B cell responses to rotavirus share common immunodominant variable gene repertoires. J Immunol. 2003;171:4680–4688. doi: 10.4049/jimmunol.171.9.4680. [DOI] [PubMed] [Google Scholar]
- 12.Rani M, Bolles M, Donaldson EF, Van Blarcom T, Baric R, Iverson B, Georgiou G. Increased antibody affinity confers broad in vitro protection against escape mutants of severe acute respiratory syndrome coronavirus. J Virol. 2012;86:9113–9121. doi: 10.1128/JVI.00233-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Keck ZY, Saha A, Xia J, Conrad F, Lou J, Eckart M, Marks JD, Foung SK. Affinity maturation to improve human monoclonal antibody neutralization potency and breadth against hepatitis C virus. J Biol Chem. 2011;286:44218–44233. doi: 10.1074/jbc.M111.290783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustchina E, Li M, Louis JM, Anderson DE, Lloyd J, Frisch C, Bewley CA, Gustchina A, Wlodawer A, Clore GM. Structural basis of HIV-1 neutralization by affinity matured Fabs directed against the internal trimeric coiled-coil of gp41. PLoS Pathog. 2010;6:e1001182. doi: 10.1371/journal.ppat.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustchina E, Louis JM, Frisch C, Ylera F, Lechner A, Bewley CA, Clore GM. Affinity maturation by targeted diversification of the CDR-H2 loop of a monoclonal Fab derived from a synthetic naive human antibody library and directed against the internal trimeric coiled-coil of gp41 yields a set of Fabs with improved HIV-1 neutralization potency and breadth. Virology. 2009;393:112–119. doi: 10.1016/j.virol.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steckbeck JD, Orlov I, Chow A, Grieser H, Miller K, Bruno J, Robinson JE, Montelaro RC, Cole KS. Kinetic rates of antibody binding correlate with neutralization sensitivity of variant simian immunodeficiency virus strains. J Virol. 2005;79:12311–12320. doi: 10.1128/JVI.79.19.12311-12320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson CD, Palermo LM, Hafenstein SL, Parrish CR. Different mechanisms of antibody-mediated neutralization of parvoviruses revealed using the Fab fragments of monoclonal antibodies. Virology. 2007;361:283–293. doi: 10.1016/j.virol.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Genst E, Handelberg F, Van Meirhaeghe A, Vynck S, Loris R, Wyns L, Muyldermans S. Chemical basis for the affinity maturation of a camel single domain antibody. J Biol Chem. 2004;279:53593–53601. doi: 10.1074/jbc.M407843200. [DOI] [PubMed] [Google Scholar]
- 19.England P, Nageotte R, Renard M, Page AL, Bedouelle H. Functional characterization of the somatic hypermutation process leading to antibody D1.3, a high affinity antibody directed against lysozyme. J Immunol. 1999;162:2129–2136. [PubMed] [Google Scholar]
- 20.Lavoie TB, Drohan WN, Smith-Gill SJ. Experimental analysis by site-directed mutagenesis of somatic mutation effects on affinity and fine specificity in antibodies specific for lysozyme. J Immunol. 1992;148:503–513. [PubMed] [Google Scholar]
- 21.Lavoie TB, Mohan S, Lipschultz CA, Grivel JC, Li Y, Mainhart CR, Kam-Morgan LN, Drohan WN, Smith-Gill SJ. Structural differences among monoclonal antibodies with distinct fine specificities and kinetic properties. Mol Immunol. 1999;36:1189–1205. doi: 10.1016/s0161-5890(99)00130-3. [DOI] [PubMed] [Google Scholar]
- 22.Sagawa T, Oda M, Ishimura M, Furukawa K, Azuma T. Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten. Mol Immunol. 2003;39:801–808. doi: 10.1016/s0161-5890(02)00282-1. [DOI] [PubMed] [Google Scholar]
- 23.Altobelli G, Subramaniam S. Kinetics of association of anti-lysozyme monoclonal antibody D44.1 and hen-egg lysozyme. Biophys. J. 2000;79:2954–2965. doi: 10.1016/S0006-3495(00)76532-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldbaum FA, Cauerhff A, Velikovsky CA, Llera AS, Riottot MM, Poljak RJ. Lack of significant differences in association rates and affinities of antibodies from short-term and long-term responses to hen egg lysozyme. J. Immunol. 1999;162:6040–6045. [PubMed] [Google Scholar]
- 25.Furukawa K, Manabe A, Furukawa A, Kuba H, Okajima T, Azuma T. Initial repertoire of anti-(4-hydroxy-3-nitrophenylacetyl) antibodies as potential donors for effective affinity maturation. Mol. Immunol. 2006;43:1751–1760. doi: 10.1016/j.molimm.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Xavier KA, Willson RC. Association and dissociation kinetics of anti-hen egg lysozyme monoclonal antibodies HyHEL-5 and HyHEL-10. Biophys. J. 1998;74:2036–2045. doi: 10.1016/s0006-3495(98)77910-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitkamp JH, Kallewaard N, Kusuhara K, Feigelstock D, Feng N, Greenberg HB, Crowe JE., Jr. Generation of recombinant human monoclonal antibodies to rotavirus from single antigen-specific B cells selected with fluorescent virus-like particles. J Immunol Methods. 2003;275:223–237. doi: 10.1016/s0022-1759(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 28.Ruiz M, Giudicelli V, Ginestoux C, Stoehr P, Robinson J, Bodmer J, Marsh SG, Bontrop R, Lemaitre M, Lefranc G, Chaume D, Lefranc MP. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000;28:219–221. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brock SC, Heck JM, McGraw PA, Crowe JE., Jr. The transmembrane domain of the respiratory syncytial virus F protein is an orientation-independent apical plasma membrane sorting sequence. J Virol. 2005;79:12528–12535. doi: 10.1128/JVI.79.19.12528-12535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Bradley P, Baker D. Protein-protein docking with backbone flexibility. J Mol Biol. 2007;373:503–519. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 34.McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010;17:248–250. doi: 10.1038/nsmb.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleishman SJ, Leaver-Fay A, Corn JE, Strauch EM, Khare SD, Koga N, Ashworth J, Murphy P, Richter F, Lemmon G, Meiler J, Baker D. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 37.Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol. 2008;380:742–756. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Molecular Graphics System. Delano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 39.Wu H, Pfarr DS, Tang Y, An LL, Patel NK, Watkins JD, Huse WD, Kiener PA, Young JF. Ultra-potent antibodies against respiratory syncytial virus: effects of binding kinetics and binding valence on viral neutralization. J Mol Biol. 2005;350:126–144. doi: 10.1016/j.jmb.2005.04.049. [DOI] [PubMed] [Google Scholar]
- 40.Foote J, Milstein C. Kinetic maturation of an immune response. Nature. 1991;352:530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- 41.Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- 42.Kalinke U, Bucher EM, Ernst B, Oxenius A, Roost HP, Geley S, Kofler R, Zinkernagel RM, Hengartner H. The role of somatic mutation in the generation of the protective humoral immune response against vesicular stomatitis virus. Immunity. 1996;5:639–652. doi: 10.1016/s1074-7613(00)80277-0. [DOI] [PubMed] [Google Scholar]
- 43.Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- 44.Parren PW, Moore JP, Burton DR, Sattentau QJ. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13(Suppl A):S137–162. [PubMed] [Google Scholar]
- 45.Saphire EO, Montero M, Menendez A, van Houten NE, Irving MB, Pantophlet R, Zwick MB, Parren PW, Burton DR, Scott JK, Wilson IA. Structure of a high-affinity “mimotope” peptide bound to HIV-1-neutralizing antibody b12 explains its inability to elicit gp120 cross-reactive antibodies. J Mol Biol. 2007;369:696–709. doi: 10.1016/j.jmb.2007.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci U S A. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kallewaard NL, McKinney BA, Gu Y, Chen A, Prasad BV, Crowe JE., Jr. Functional maturation of the human antibody response to rotavirus. J Immunol. 2008;180:3980–3989. doi: 10.4049/jimmunol.180.6.3980. [DOI] [PubMed] [Google Scholar]
- 48.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 49.Mizutani R, Miura K, Nakayama T, Shimada I, Arata Y, Satow Y. Three-dimensional structures of the Fab fragment of murine N1G9 antibody from the primary immune response and of its complex with (4-hydroxy-3-nitrophenyl)acetate. J. Mol. Biol. 1995;254:208–222. doi: 10.1006/jmbi.1995.0612. [DOI] [PubMed] [Google Scholar]
- 50.Gimenez HB, Chisholm S, Dornan J, Cash P. Neutralizing and enhancing activities of human respiratory syncytial virus-specific antibodies. Clin Diagn Lab Immunol. 1996;3:280–286. doi: 10.1128/cdli.3.3.280-286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson LJ, Heilman CA. Protective and disease-enhancing immune responses to respiratory syncytial virus. J Infect Dis. 1995;171:1–7. doi: 10.1093/infdis/171.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Mastelic B, Kamath AT, Fontannaz P, Tougne C, Rochat AF, Belnoue E, Combescure C, Auderset F, Lambert PH, Tacchini-Cottier F, Siegrist CA. Environmental and T cell-intrinsic factors limit the expansion of neonatal follicular T helper cells but may be circumvented by specific adjuvants. J Immunol. 2012;189:5764–5772. doi: 10.4049/jimmunol.1201143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.