Abstract
Alternatively activated macrophages express the pattern recognition receptor scavenger receptor A (SR-A). We demonstrated previously that co-culture of macrophages with tumour cells upregulates macrophage SR-A expression. We show here that macrophage SR-A deficiency inhibits tumour cell migration in a co-culture assay. We further demonstrate that co-culture of tumour-associated macrophages (TAMs) and tumour cells induces secretion of factors which are recognized by SR-A on TAMs. We tentatively identified several potential ligands for the SR-A receptor in tumour cell – macrophage co-cultures by mass spectrometry. Competing with the co-culture induced ligand in our invasion assay recapitulates SR-A deficiency and leads to similar inhibition of tumour cell invasion.
In line with our in vitro findings, tumour progression and metastasis is inhibited in SR-A-/- mice in two in vivo models of ovarian and pancreatic cancer. Finally, treatment of tumour-bearing mice with 4F, a small peptide SR-A ligand able to compete with physiological SR-A ligands in vitro, recapitulates the inhibition of tumour progression and metastasis observed in SR-A-/- mice. Our observations suggest that SR-A may be a potential drug target in the prevention of metastatic cancer progression.
Introduction
Solid tumours are comprised of neoplastic cells, non-malignant resident stromal cells and migratory haematopoietic cells. Complex interactions between the cell types in this microenvironment regulate tumour growth, progression, metastasis and angiogenesis (1). It is well established that stromal cells, including macrophages, within the microenvironment may contribute to tumour growth and spread (1).There is a body of pre-clinical and clinical evidence associating abundance of tumour-associated macrophages, TAM, with poor prognosis (2).
Macrophages exhibit marked phenotypic heterogeneity and have been broadly classified into M1 or M2 type (3, 4). M1 macrophages classically are activated by interferon gamma (IFN-γ), with or without microbial products, produce large amounts of proinflammatory cytokines, express high levels of MHC molecules, and are implicated in the killing of pathogens and tumour cells (3). M2 macrophages moderate the inflammatory response, eliminate cell debris, and promote angiogenesis and tissue remodelling (3, 5). Stimulation with IL-4, IL-13 and IL-10 drives macrophages towards the M2 phenotype. The macrophages present in neoplastic tissues (TAM) mainly display an M2-like phenotype with expression of classes of innate pattern recognition receptors (PRRs) such as mannose receptor (MR) and scavenger receptor class A (SR-A) (5, 6). However, the role of SR-A in TAMs is entirely unclear.
Scavenger receptors are broadly defined by their ability to bind modified low density lipoproteins (mLDL) and other polyanions, including proteins, lipids, carbohydrates and nucleic acids (7). SR-A in particular binds a wide range of polyanionic ligands from artificial, microbial and endogenous origin including polyribonucleotides, polysaccharides and glycated proteins, among which extracellular matrix proteoglycans, and oxidized/modified lipids and lipoproteins.
SR-A is restricted to the myeloid lineage and is expressed on most mature tissue macrophages and on bone marrow-derived dendritic cells (DC) and splenic DCs, but not on their immature precursor monocytes. In addition, SR-A is found on smooth muscle cells and on a small subpopulation of endothelial cells in the lung (8, 9).
SR-A, most well studied for its role in atherosclerosis, has been implicated in the metabolic changes that affect macrophages exposed to high fat loads and oxidative stress (10). Many of these SR-A functions may be relevant to other diseases with underlying metabolic and oxidative changes, including cancer (11, 12). SR-A-mediated adhesion of macrophages signals changes in oxidative output (13), and differential ligand binding to SR-A influences the inflammatory status of macrophages (14-16). Both phenomena may drive physiological changes in the microenvironment of SR-A expressing macrophages.
We have previously demonstrated that ovarian cancer cells switch co-cultured macrophages to a phenotype similar to that found in ovarian tumours (6). Tumour cells caused up-regulation of scavenger receptor SR-A, which is consistent with other publications identifying this receptor as a marker on alternatively activated macrophages. We confirmed that SR-A was expressed on TAMs in ovarian cancer patients (6). Recent data published by Bak et al. showed that targeting macrophages via the SR-A receptor in a murine orthotopic ovarian cancer model led to disease stabilization (17), assigning a pro-oncogenic role to SR-A. In addition, lack of SR-A was shown to improve stimulation of tumour immunity in vivo (18).
We therefore aimed to investigate the role of SR-A during tumour development and progression. Here we show that SR-A expression on macrophages is important for tumour progression and metastasis in vitro and in vivo. Upon co-culture of tumour cells with macrophages a SR-A ligand is secreted into the supernatant. Competition with the physiological SR-A ligand in vitro and possibly in vivo inhibits macrophage induced tumour cell invasion, and tumour burden was significantly reduced specifically in SR-A-/- mice. We conclude that the macrophage SR-A and its uncharacterised ligand(s) contribute significantly to the host-tumour relationship.
Materials and Methods
Cell lines and reagents
Unless otherwise indicated, all reagents were purchased from Sigma (Poole, UK). Acetylated LDL (AcLDL) was from Molecular Probes (Eugene, OR, USA), PMH-Liposorb was from Calbiochem. 4F and scr4F peptides were generous gifts from Dr Alan Fogelman (UCLA, Los Angeles, USA). Cell lines were grown in RPMI-1640 medium supplemented with 10% FCS. All experiments were performed under endotoxin-free conditions. The murine cancer cell lines, ID8 and Panc02 were cultured in DMEM medium supplemented with 10% FCS.
Co-culture
Tumour:macrophage co-cultures were performed as described previously (19). Briefly, luciferase-expressing tumour cells (2.5 × 105 per well) and bone-marrow-derived macrophages (5 × 105 per transwell insert) were grown without cell-cell contact in the upper compartment of a modified Boyden chamber, and invasiveness was measured by quantifying transmigrated tumour cells in the lower compartment using relative luciferase units (RLU). Cell viability was assessed using the Beckham Coulter ViCell XR Counter (Beckham, UK). To test for scavenging capacity in co-cultures, supernatants were repeatedly incubated for 1h on wt or SR-A-/- BMM monolayers and then tested for SR-A ligand activity by ELISA.
SR-A ELISA
The SR-A-specific ELISA was carried out as described (20). Supernatants were coated onto ELISA plates overnight at a protein concentration of 10 μg/ml and SR-A binding was detected using wild-type (wt, 129 ICR) or SR-A-/- BMMɸ lysate, followed by anti-SR-A monoclonal antibody 2F8 and appropriate HRP-coupled secondary antibody. AcLDL was used as a positive control. For lipid depletion before ELISA, supernatants were processed with PMH-Liposorb resin (Calbiochem) following the manufacturer’s instructions.
Animals
The following knock-out mouse strains were used: SR-A-/-, MARCO-/- and SR-A/MARCO double knock-out (db-/-), TLR2-/- and TLR4-/- – all on C57BL/6 background, along with their wild-type control Charles River strain. Bones from CD36-/- mice (21) were obtained from David Kluth, Edinburgh, UK with kind permission of Roy Silverstein. TLR3-/- mice were obtained from Richard Flavell/The Jackson Laboratory (22). TLR7-/- and TLR9-/- mice were kindly provided by Dr. Shizuo Akira (23, 24). For BMMɸ lysates used in ELISA and Far Western blot, SR-A-/- on 129 ICR background and the corresponding wt control strain were used. All animals were bred and housed under specific pathogen-free conditions and all procedures involving animals were conducted according to the requirements of the United Kingdom Home Office Animals (Scientific Procedures) Acts, 1986.
Radiation Chimaeras
Mice were irradiated by giving a single absorbed dose of 10 Gy to the whole body using a linear accelerator with 15 MV nominal photon energy (3.6 Gy/min). After the irradiation, the mice were rested for 4 to 6 hours and thereafter reconstituted by intravenous injection with 5 × 106 bone marrow cells from SR-A-/-, MARCO-/-, db-/- or wt mice. All mice were maintained in IVCs on enrofloxacin (Baytril) for 4 weeks after irradiation to minimize the risk for infection. After transplantation, the animals were maintained in IVCs on sterilized food and acidified sterile water.
Macrophages
Murine BMMɸ were obtained and cultured as described before (19). Mɸ were routinely cultured in RPMI (Gibco) supplemented with 50 IU/ml penicillin G, 50 μg/ml streptomycin and 2 mM L-glutamine (PSG), 10% foetal calf serum (FCS) in the presence of 10 ng recombinant murine MCSF/ml. For preparation of lysates, macrophages were washed five times in ice-cold PBS and cells were lysed using 1 ml NP-40 protein lysis buffer per 1 × 107 cells. Aliquots were stored at – 80°C until required.
Dose and route of 4F administration
In treatment experiments 4F and scr4F were injected intraperitoneally at 10 mg/kg once daily every 3 days.
Tumour Imaging and Quantitation
Tumour progression was assessed in situ by bioluminescent imaging as described previously (25). Lentiviral infection of tumour cells with luciferase reporter construct was performed as described before (25). Ascitic cells were counted using a haemocytometer; mice presented with approximately 10 ml of ascites at the end of the experiment. Cytospins were prepared from 500 μl of ascitic fluid and cells were differentiated with Wright’s staining.
Statistical analysis
Experiments were performed at least in triplicate unless otherwise indicated and representative data are shown. Results were tested for statistical significance using Student’s t-test or Mann-Whitney test with GraphPad Prism Version 5.0c software.
Results
Macrophage SR-A expression promotes tumour cell invasion
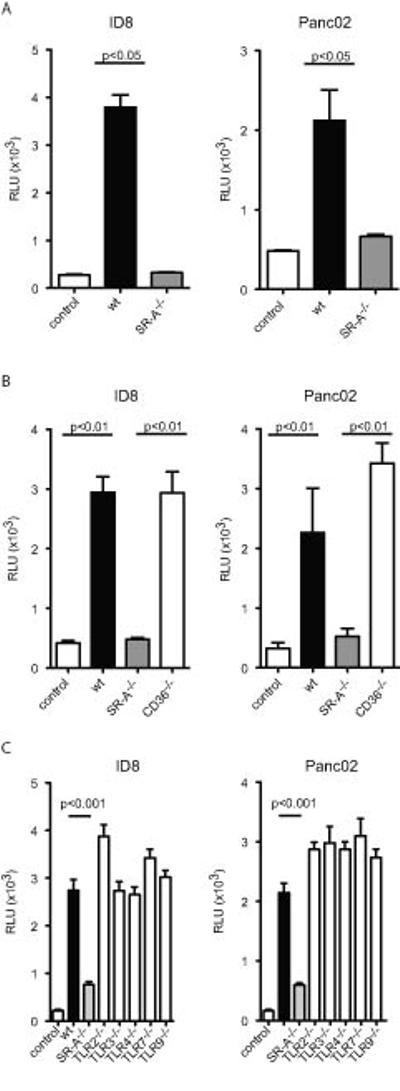
We have previously shown that co-culture of bone marrow-derived macrophages (BMMɸ) with human and murine ovarian cancer cells leads to increased tumour cell invasion (6). We used this in vitro co-culture system to investigate the role of scavenger receptor family members. First, we co-cultured macrophages from SR-A-/- mice with different tumour cell lines (ID8, Panc02). In a modified Boyden chamber we analysed tumour cell invasion through an artificial basal membrane. Compared to wild-type (wt) macrophages, SR-A-/- macrophages showed a reduced ability to induce tumour cell invasion upon co-culture with both tumour cell types (Figure 1A). We then tested the involvement of a class B scavenger receptor, CD36. There was no difference between wt and CD36-/- macrophages (Figure 1B), suggesting that induction of tumour cell invasion is a mechanism specific to SR-A.
Figure 1. SR-A is necessary and sufficient to promote invasion in a macrophage: tumour cell in vitro invasion assay.
A: Wild type or SR-A-/- BMMɸ were co-cultured with ID8-Luc or Panc02-Luc cells in a modified Boyden chamber without direct cell-cell contact for 72h. Invasion of ID8-Luc/Panc02-Luc cells was assessed by Luciferase activity in the lower part of the chamber. SR-A-/- macrophages had a reduced ability to promote ID8-Luc and Panc02-Luc invasion (p<0.05, t-test). B: Culture of ID8 or Panc02 cells alone or co-cultured with wt, SR-A-/- or CD36-/- macrophages. CD36-/- macrophages do not significantly reduce ID8-Luc invasion. C: Culture of ID8 or Panc02 cells alone or co-cultured with wt, SR-A-/- , TLR2-/-, TLR3-/-, TLR4-/-, TLR7-/- or TLR9-/- macrophages. Data are represented as mean ± SEM of n=6. Representative data are shown from at least 3 independent experiments.
The macrophage-specific growth factor, CSF-1, plays a major role in host-tumour interactions and is associated with a poor prognosis (26-29). It is secreted by many tumours, serves to recruit monocytes/macrophages to mouse and human cancers, and stimulates growth and differentiation of macrophages which lack cytocidal activity, thus promoting tumour establishment and expansion (30). CSF-1 is a potent and selective inducer of SR-A expression in macrophages, enhancing their adhesive and endocytic functions (31). We were able to detect CSF-1 in both single and co-cultures, but the levels did not differ, and by itself CSF-1 is not a ligand for SR-A (Supplementary Figure 1A).
Next we confirmed that tumour invasiveness was due to expression of SR-A on macrophages and not on tumour cells (Supplementary Figure 1B), and was not associated with other pattern recognition receptors, such as TLR2 and TLR4, which have been shown to interact with SR-A. Loss of TLR2 or TLR4 did not influence tumour cell invasion (Figure 1C); neither did loss of TLR3,-7,-9 receptors, indicating that SR-A is sufficient on its own to promote tumour invasiveness, and does not rely on innate immune signalling via TLR-dependent pathways.
Receptor competition with polyanionic ligands in the co-culture assay
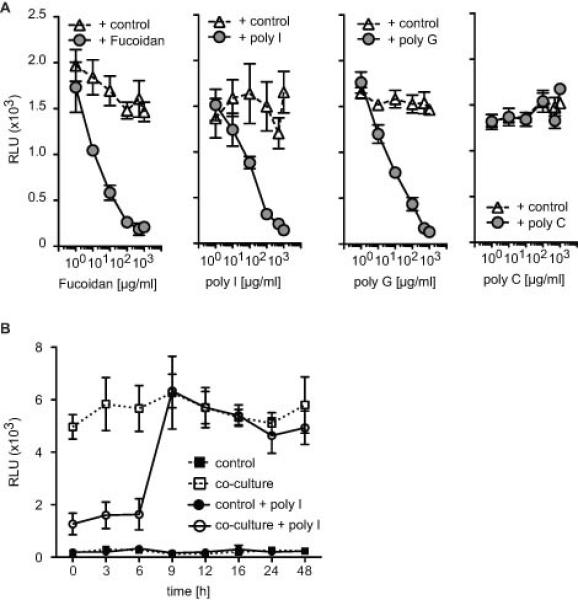
SR-A is an endocytic receptor, and signals upon ligand binding. If ligand-receptor interaction was necessary for SR-A to exert its tumour-promoting effect, then it should be possible to block tumour invasion by out-competing the tumour-promoting ligand with other known SR-A ligands. Therefore we co-cultured wild type murine macrophages with ID8 tumour cells and used polyanionic ligands to compete with the co-culture induced SR-A ligand (Figure 2). We screened a range of non-microbial polyanionic ligands and their closely related non-ligand controls. First, we established concentration curves for the respective ligands (Figure 2A). Whilst the addition of fucoidan, poly I and poly G inhibited ID8 tumour cell invasion in a dose dependent manner, the non-binding control poly C had no effect. Addition of fucoidan, poly I or poly G to ID8 or Panc02 cells alone did not induce invasion (Supplementary Figure 1C), and previously published data showed that stimulation of tumour cells with pro-inflammatory LPS has no invasion-promoting effect (19).
Figure 2. Competition with the co-culture induced ligand in the in vitro invasion assay by large polymeric SR-A ligands.
A. Macrophages and ID8 tumour cells were co-cultured in the presence of fucoidan, poly I, poly G or poly C. Addition of known SR-A ligands can inhibit tumour cell invasion (p<0.01, t-test). Poly C served as a negative control. Data are represented as mean ± SEM of n=6. Representative data are shown from at least 3 independent experiments. B. Addition of 100μg/ml of poly I up to 6hr after the start of the macrophage: ID8 co-culture inhibits tumour cell invasion. When added after 9h there is no impact on the invasion assay.
To investigate whether SR-A activity is required at early or late stages of tumour cell invasion, we added poly I to the ID8:macrophage co-culture at different time points after the start of the co-culture (Figure 2B). Addition of poly I up to 6h after the start of the experiment inhibited macrophage induced tumour cell invasiveness. Addition of poly I at later time points had no effect, indicating that SR-A is required early during invasion (Figure 2B). Similar results were obtained when we co-cultured macrophages with Panc02 tumour cells (data not shown).
Receptor recognition of a co-culture induced ligand
The observation that the tumour-promoting effect of SR-A can be blocked by a range of SR-A ligands suggests the presence of a co-culture-induced SR-A ligand at some stage during invasion. To assess the possibility of such an SR-A ligand, we co-cultured macrophages with ID8 and Panc02 tumour cells or embryonic mouse fibroblasts (EMF, benign control) and collected the supernatant after 24h. Single-culture supernatant from the respective cells, wt and SR-A-/- macrophages, ID8, Panc02 and EMF served as negative controls. We first screened all control and co-culture supernatants for differential SR-A binding activity by ELISA (Figure 3A) (32). This assay is non-specific for the molecular nature of the ligand. Our results showed that SR-A recognized ligand(s) in co-culture supernatants of wt or SR-A-/- macrophages with either ID8 or Panc02 but not in RPMI medium or supernatants from any single culture (Figure 3A). SR-A binding activity was also absent from macrophages co-cultured with EMFs, proving that ligand induction is a specific feature of co-culture with tumours. Ligand activity disappeared from wt-tumour cell supernatants after repeated passaging on wt but not on SR-A-/- BMM monolayers (Figure 3B), further corroborating the presence of an SR-A ligand which can be scavenged from the co-culture milieu in an SR-A-specific manner.
Figure 3. Ligand binding assay reveals SR-A ligand(s) in macrophage:tumour cell co-culture supernatants.
A. An ELISA based ligand binding assay was used to screen for a possible SR-A ligand in co-culture supernatants. Supernatant from ID8, PANC02, wt or SR-A-/- macrophages or embryonic mouse fibroblasts (EMF) alone showed no ligand binding. Co-culture supernatant from wt or SR-A-/- macrophages with ID8 or Panc02 cells showed a significant increase in SR-A binding activity (p<0.01, t-test). There was no SR-A ligand binding detectable in the supernatant from macrophages with EMF. AcLDL was used as a positive control. Data are represented as mean ± SD of n=12. Representative data are shown from at least 3 independent experiments. B. SR-A binding activity is lost after repeated passaging of co-culture supernatants on wt but not SR-A-/- BMM monolayers. Co-culture supernatants were incubated twice or five times for 1h with wt or SR-A-/- BMMs, then SR-A ligand activity in the passaged supernatant was assessed by ELISA as in A. Data are represented as in A. C. Co-culture supernatants contain a lipid ligand. Supernatants as in B. were treated with PMH-Liposorb resin to remove all lipids or lipid-bound molecules, and then tested for SR-A binding activity by ELISA as in A.
Studies, not shown, indicated that the ligand was heat labile (56°C, 30 min), acid labile (pH6), with an apparent MW of 20-60 kDa as determined by cut-off filters, and RNAse and DNAse stable. In addition, lipid depletion of co-culture supernatants significantly decreased ligand activity (Figure 3C).
Having demonstrated the presence of SR-A ligand(s) in the supernatant of TAM/tumour co-cultures, we undertook provisional steps to isolate and identify potential protein ligand(s) by SR-A-specific Far Western blot, followed by mass spectrometry analysis of SR-A interacting proteins from corresponding gel bands (20). Potential candidates are listed in Supplementary Table I. Clustering of GO terms associated with ligands in this list suggests enrichment in extracellular matrix proteins, proteins involved in wound healing and proteolysis, and proteins of inflammatory pathways (Supplementary Table II).
The large number of candidate ligands retrieved reflects the known promiscuous nature of scavenger receptors. In view of a possible therapeutic intervention, we therefore reasoned that it would be more parsimonious to target the receptor rather than one out of many potential ligands.
SR-A deficiency delays tumour progression in the ID8 model and reduces lung metastasis in the Panc02 model
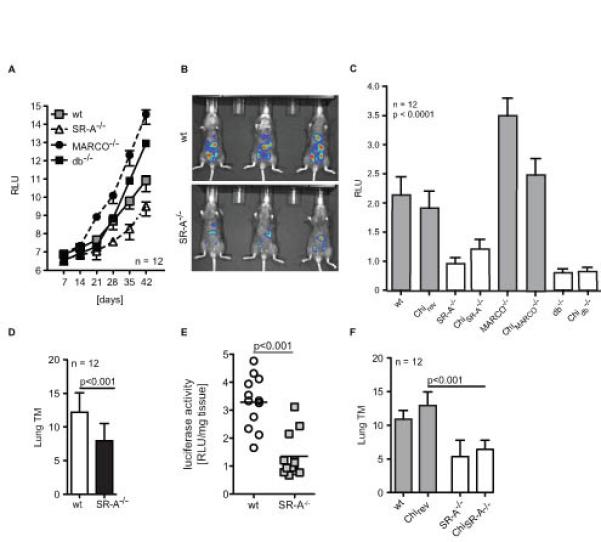
Based on our in vitro results we next investigated the role of SR-A during tumour progression in vivo. We previously reported a syngeneic model of ovarian cancer using a luciferase labelled cell line ID8 (33). In this model, ID8 cells are injected i.p. and form local tumours in the peritoneal cavity. SR-A deficiency did not prevent the development of peritoneal tumours; however tumour progression (growth of single tumours) was significantly slowed down in SR-A-/- compared to wt animals (Figure 4A, B; t-test p<0.05, n=12). Interestingly, mice deficient in another class A scavenger receptor, MARCO, showed a significant increase in disease burden compared to wt mice (Figure 4A), which was suppressed in db-/- mice, suggesting opposing effects of MARCO and SR-A. To exclude the contribution of non-haematopoietic SR-A receptor involvement in our observations we generated SR-A-/-, MARCO-/- and db-/- (SR-A-/-, MARCO-/-, db-/- bone marrow in wt mice) and reverse (wt bone marrow in SR-A-/-, MARCO-/- or db-/- mice) chimeras to study the role of haematopoietic/myeloid SR-A contribution. SR-A-/- and db-/-, but not the respective reverse chimeras, showed a similar growth delay in the ID8 model as described above (Figure 4C), proving that SR-A is specifically required on TAMs but not on other stromal cells.
Figure 4. Macrophage-specific loss of SR-A inhibits in vivo tumour growth.
A. 106 ID8-Luc cells were injected i.p. into syngeneic mice. Quantification of bioluminescence from primary tumours (n=12 each) was obtained weekly. ID8 tumours grew significantly slower in SR-A-/- mice compared to wt, MARCO-/- or db-/- mice (p<0.01). Data represented as mean ± SEM of n=12. Representative data are shown from 2 independent experiments. B. Representative bioluminescence picture: red, the highest photon flux; blue, the lowest photon flux. C. Ex vivo luminescence analysis of the ascitic cell population (106 cells/ml) of radiation chimeras injected with ID8-Luc cells as in A. to exclude the impact of non-haematopoietic SR-A expression. D. 106 Panc02 cells were injected s.c. into the flank of syngeneic C57BL/6 mice. Tumour multiplicity (TM) at end point was assessed by H&E immunohistochemistry. E. Quantification of luciferase activity per mg of lung tissue in lung tumours from mice in D. p<0.001 by Student’s t test. F. Tumour multiplicity in radiation chimeras using the Panc02 model.
The ID8 model only addresses the role of SR-A in primary tumour growth, not in metastasis to distant sites. As our in vitro co-culture assay showed reduced invasiveness in the absence of SR-A for ID8 as well as for the Panc02 pancreatic cancer cell line, we used the latter to study the role of SR-A during tumour progression/metastasis in vivo. In the Panc02 model, subcutaneous injection of tumour cells results in both primary tumours at the injection site and lung metastasis. We implanted 105 Panc02 cells subcutaneously into wt and SR-A-/- mice and primary tumour growth and lung metastasis (tumour multiplicity) were assessed after 28 days at which point there was no difference in the size of primary Panc02 tumours (data not shown). However, we observed a significant decrease in lung metastasis in SR-A-/- mice (Figure 4D, E), suggesting that in this model, SR-A does not affect local tumour growth, but specifically promotes migration of tumour cells to distant sites. To differentiate between haematopoietic/myeloid SR-A contribution we generated bone marrow chimeras. The respective SR-A-/- chimera but not the reverse chimera protected the mice from developing lung metastasis (Figure 4F).
Towards an SR-A specific therapeutic intervention
Since genetic ablation of SR-A resulted in reduced tumour invasiveness, we sought to determine whether SR-A could be therapeutically targeted in vivo to obtain similar results. Although an SR-A monoclonal antibody is available (2F8), and antibody therapy of cancer has proven successful in several models (34), small molecule inhibitors remain advantageous over monoclonal antibodies in ease of delivery and cost. We therefore investigated whether the small SR-A receptor inhibitor 4F could block tumour cell invasion in our model. 4F is a 16-amino acid amphipathic peptide which mimics the broad anti-atherogenic and anti-inflammatory properties of apo A-I (35, 36), and can compete with a range of SR-A ligands (20). In our in vitro invasion assay, 4F, but not the negative control scr4F, showed dose-dependent inhibition of co-culture induced ID8 or Panc02 invasion (Figure 5A). 100 μg/ml of 4F were sufficient to nearly completely block co-culture induced tumour cell invasion (Figure 5B).
Figure 5. The small molecule SR-A inhibitor 4F prevents tumour invasiveness in vitro and in vivo.
A. Dose response curve. Macrophages and ID8 tumour cells were co-cultured in the presence of 4F, an SR-A ligand. Scrambled 4F (scr4F), which contains the same amino acids as 4F but in random order to prevent amphipathic helix formation, served as a negative control. B. Invasion assay of macrophages with either ID8 or Panc02 cells in the presence of 100 μg/ml scr4F or 4F. Data in A. and B. are represented as mean ± SEM of n=6. Representative data are shown from at least 3 independent experiments. C. 4F prevents tumour growth in vivo. Animals were treated with 4F s.c. and tumour burden was assessed using IVIS imaging system. Loss of SR-A does not add to the effect of 4F on tumour cell invasion.
The SR-A inhibitor 4F prevents tumour progression in vivo
Since loss of SR-A seemed to prevent tumour growth and metastasis in vivo, and blocking SR-A with physiological ligands reduced tumour invasiveness in vitro, we next assessed whether SR-A could be successfully targeted with 4F in vivo. We used the ID8 model to quantify tumour growth in the presence of the SR-A inhibitor 4F or its scrambled control peptide scr4F. The inhibitor was administered by intra-peritoneal injection every three days throughout tumour development, and significantly delayed tumour growth (Figure 5C). This is consistent with previous studies testing the therapeutic effect of 4F in tumour models, although the mechanisms put forward in these models do not directly involve macrophages or SR-A (37-39). We reasoned that if 4F reduced tumour burden by a mechanism independent of SR-A, then additional loss of SR-A in the presence of 4F should decrease tumour development further. However, there was no significant difference in the reduction of tumour development between wt mice treated with 4F, SR-A-/- mice treated with 4F or SR-A-/- mice treated with scr4F, suggesting that 4F and SR-A act in the same pathway.
Discussion
In the present study, we demonstrate that SR-A is required on macrophages to support tumour invasiveness during co-culture in vitro and metastasis in vivo, and that competition with SR-A ligand(s) can reduce macrophage induced tumour cell invasion. We detected SR-A ligand binding activity in the co-culture medium but not in single culture controls, and identified a range of candidate protein ligands. Although TAMs have been associated with tumour promotion in many previous studies, the present study provides the first direct evidence that a specific scavenger receptor, the SR-A, is implicated in trophic tumour–host interactions. SR-A is necessary and sufficient to promote tumour invasiveness, and does not seem to require additional signalling via TLR pathways. Our experiments indicate that communication between TAMs and tumour cells requires SR-A at an early time point, and results in chimeric mice establish that SR-A is only needed on haematopoietic cells, consistent with its expression on macrophages. Importantly, loss of SR-A affected tumour metastasis, which was delayed and not prevented, rather than tumour growth at the initial inoculation site.
Two cancer-derived cell lines of ovarian and pancreatic origin, but not diploid embryonic fibroblasts, gave consistent results in migration assays of macrophage co-cultures. The SR-A was the only pattern recognition receptor tested which showed a phenotype in our in vitro assay; in particular excluding another SR, CD36, which is expressed in primary bone marrow-derived macrophages. MARCO is not expressed in bone marrow cultures as used here, but did not mimic the SR-A role in vivo. On the contrary, loss of MARCO seemed to enhance rather than reduce tumour multiplicity in vivo. Although these two Class A receptors share many common structural and functional features, they differ subtly on several points. SR-A and MARCO have distinct ligand binding domains and share overlapping but distinct ligand repertoires. SR-A is expressed broadly on most tissue resident macrophages while MARCO is restricted to subsets of macrophages. However, MARCO can be readily induced on most macrophage populations after inflammatory challenge (40, 41). Further studies are needed to establish the basis of their contrasting behaviour in host-tumour interactions.
The in vitro model did not require contact between tumour cells and macrophages, indicating that secretion products from either cell contributed to their interactions, but only when cultured together. The cytokine TNFα is one such co-culture induced molecule which has been implicated in tumour invasiveness before (19, 25). Although TNFα was induced in an SR-A-dependent fashion (Supplementary Figure 1D), and might partially explain the loss of invasiveness in SR-A-/- co-cultures, we hypothesised that there were additional factors present that acted as ligands for the SR-A; consistent with this was our ability to detect ligand activity in a specific ELISA used previously to characterize SR-A ligands. The tumour-promoting effect of SR-A could therefore be attributed to ligand-induced receptor signalling, switching TAMs towards a tumorigenic phenotype, or to receptor-mediated clearance of a tumour-inhibiting ligand. Ligands detected by ELISA were present at comparable levels in co-cultures of tumour cells with both wt and SR-A-/- macrophages, but could be readily scavenged from co-culture supernatants by incubation on BMM monolayers, tentatively favouring the latter hypothesis.
We employed a previously validated protocol, SDS-PAGE and Far Western blotting, to identify ligand–containing bands on gels, which we subsequently analysed by mass spectrometry (Supplementary Tables I and II). Candidate protein ligands identified in our study are significantly enriched in extracellular matrix components, and SR-A like many other SRs has been shown to bind a variety of these proteins (7). Remarkably, an expression profiling study of TAMs co-migrating with metastasizing tumour cells in vivo showed a specific enrichment in tissue- and organ-development genes, suggesting that TAMs help to shape the environment for the spreading tumour cells (42). This is in agreement with the enrichment of extracellular matrix components found in our SR-A candidate list. Indeed, another SR, stabilin-2, exhibits pro-tumour activity through its ability of scavenging hyaluronic acid (HA), an abundant ECM component. Loss of stabilin-2 increased circulating levels of HA, and led to reduced tumour invasiveness in the lungs (43). It is conceivable that SR-A might similarly scavenge ECM breakdown products from the tumour microenvironment, thereby clearing the road for tumour cells to emigrate and metastasize to distant sites. Even though we did not observe significantly enhanced ligand activity in SR-A-/- co-cultures, which would have reflected a clearance role for SR-A, the ELISA conditions may not be quantitative enough to detect subtle differences in ligand availability.
Another aspect of SR-A function in relation to tumour biology is its role in the clearance of apoptotic cells, which downregulates macrophage activation by production of TGF-β and prostaglandin E. The natural ligand for SR-A in apoptotic cells has not been defined, but may be related to phosphatidylserine and oxidised lipoprotein, as is the case with other scavenger receptors. Such an additional pathway may reinforce and amplify the benefit to the tumour, while sparing inflammatory injury. Given the molecular nature of known SR-A ligands, and based on our observation that lipid depletion from co-culture supernatants reduces its ligand content, it is also possible that the co-culture induced ligand is of lipid origin. Bioactive lipids are known to contribute to tumour progression, and SR-A promotes uptake of a range of oxidized lipoproteins and lipids, including lysophosphatidic acid, the precursor for tumour-promoting lysophosphatidic acid (44). It is noteworthy that we did not detect any lipoprotein-related candidate SR-A ligand, including apolipoprotein E, which has been implicated in cancer. Further studies are needed to identify the SR-A natural ligand(s) in tumour-macrophage interactions.
While identification of tumour-environment derived SR-A ligands is highly desirable for our understanding of the mechanism underlying TAM-dependent promotion of metastasis, our results using an SR-A inhibitor may provide a more direct therapeutic handle.
The ability of 4F peptide, an apolipoprotein A1 mimic, to inhibit the interaction of tumour cells and macrophages in vitro and to slow tumour burden in vivo, provides support for SR-A involvement. Although 4F peptide is not a specific inhibitor for SR-A, we have previously demonstrated its efficacy in inhibiting SR-A-mediated adhesion and endocytosis (20). 4F is easier to deliver than SR-A antibodies or large polymeric ligands and has successfully passed safety trials for clinical use in humans (45), arguments in favour of its potential therapeutic use in blocking SR-A-enhanced tumour invasiveness. Two recent studies already present promising anti-tumour effects of 4F in an ovarian cancer model in vitro and in vivo (ID8 cells). Subcutaneous or orally administered 4F was able to decrease ID-8 cell tumour burden in mice, a mechanism attributed to its scavenging capacity of pro-oncogenic lysophosphatidic acid from the serum of tumour-bearing mice (39). In the same model, 4F inhibited viability and proliferation of tumours in vitro and in vivo through targeted upregulation of Mn-superoxide dismutase in tumour cells, improving the antioxidant profile of ID-8 cells (37). It could be hypothesized that a reduction of oxidative stress in the tumour milieu would also decrease bioavailability of oxidized scavenger receptor ligands. Unfortunately, we were unable to detect LPA in our co-culture assay, and therefore cannot conclude on its function in our model. Importantly, in our model 4F treatment and loss of SR-A did not show cumulative effects on tumour invasiveness in vivo, suggesting that 4F and SR-A act in the same pathway.
Association studies present only weak evidence for a causative role of SR-A in the development of human cancer. Germline mutations in the human MSR1 gene have been associated with hereditary prostate cancer (HPC), and were found to be enriched in individuals with non-hereditary prostate cancer (46). Importantly, among the non-synonymous changes, several affect highly conserved amino acids in both the cytoplasmic signalling domain, as well as in the extracellular ligand binding domain, potentially altering SR-A function. However, a large-scale meta-analysis did not find any significant risk associated with the most prevalent of these mutations, a truncation of the ligand-binding domain (47). Likewise, when specific mutations were assessed for association with metastatic progression of prostate tumours, no significant association was found (48). Results from the Framingham cohort also found no significant genome-wide association of SNPs with either breast cancer or prostate cancer (49). More recently, germline mutations in MSR1 were associated with incidence of Barrett’s oesophagus, a relatively common condition (1-10% of general population) that may evolve into oesophageal adenocarcinoma (50). In summary, further studies are required, based on defined molecules involved in SR-A function and regulation in tumour-associated macrophages. One possibility is that soluble SR-A, shed by macrophages (our unpublished observations), may provide a useful diagnostic and prognostic biomarker.
In conclusion, we have described previously unexplored pathways by which tumours can subvert macrophages in an unholy alliance. Further studies in mouse and man may provide another drug target to intervene therapeutically.
Supplementary Material
Acknowledgments
We thank Dr Benjamin Thomas at the Central Proteomics Facility (www.proteomics.ox.ac.uk), University of Oxford, for advice on mass spectrometry and sample analysis; Roy Silverstein for permission and David Kluth, Spike Clay for providing CD36-/- bones; Dr. Richard Flavell, Yale School of Medicine, Immunobiology, for providing TLR3-/- mice; and Dr. Shizuo Akira, WPI-Immunology Frontier Research Center, Osaka University, for providing TLR7-/- and TLR9-/- mice.
Grant support This work was supported by a project grant from the Ovarian Cancer Action (TH, EM, MB), the Wellcome Trust (Grant 076256/Z/04/Z to C.N.), the E.P. Abraham Trust, Oxford, UK (S.M.) and the Medical Research Council, UK (A.P., S.G.).
Footnotes
Abbreviations: SR-A, scavenger receptor A; MARCO, macrophage receptor with collagenous structure; BMMɸ, bone marrow-derived macrophage; TAM, tumour-associated macrophage.
References
- 1.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 2.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 5.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 6.Hagemann T, Wilson J, Burke F, Kulbe H, Li NF, Pluddemann A, Charles K, Gordon S, Balkwill FR. Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J Immunol. 2006;176:5023–5032. doi: 10.4049/jimmunol.176.8.5023. [DOI] [PubMed] [Google Scholar]
- 7.Pluddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Horvai A, Palinski W, Wu H, Moulton KS, Kalla K, Glass CK. Scavenger receptor A gene regulatory elements target gene expression to macrophages and to foam cells of atherosclerotic lesions. Proc Natl Acad Sci U S A. 1995;92:5391–5395. doi: 10.1073/pnas.92.12.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes DA, Fraser IP, Gordon S. Murine macrophage scavenger receptor: in vivo expression and function as receptor for macrophage adhesion in lymphoid and non-lymphoid organs. Eur J Immunol. 1995;25:466–473. doi: 10.1002/eji.1830250224. [DOI] [PubMed] [Google Scholar]
- 10.Horiuchi S, Sakamoto Y, Sakai M. Scavenger receptors for oxidized and glycated proteins. Amino Acids. 2003;25:283–292. doi: 10.1007/s00726-003-0029-5. [DOI] [PubMed] [Google Scholar]
- 11.Chiurchiu V, Maccarrone M. Chronic inflammatory disorders and their redox control: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011;15:2605–2641. doi: 10.1089/ars.2010.3547. [DOI] [PubMed] [Google Scholar]
- 12.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Maxeiner H, Husemann J, Thomas CA, Loike JD, El Khoury J, Silverstein SC. Complementary roles for scavenger receptor A and CD36 of human monocyte-derived macrophages in adhesion to surfaces coated with oxidized low-density lipoproteins and in secretion of H2O2. J Exp Med. 1998;188:2257–2265. doi: 10.1084/jem.188.12.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu HY, Chiu SL, Wen MH, Chen KY, Hua KF. Ligands of macrophage scavenger receptor induce cytokine expression via differential modulation of protein kinase signaling pathways. J Biol Chem. 2001;276:28719–28730. doi: 10.1074/jbc.M011117200. [DOI] [PubMed] [Google Scholar]
- 15.Jozefowski S, Kobzik L. Scavenger receptor A mediates H2O2 production and suppression of IL-12 release in murine macrophages. J Leukoc Biol. 2004;76:1066–1074. doi: 10.1189/jlb.0504270. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura T, Suzuki H, Wada Y, Kodama T, Doi T. Fucoidan induces nitric oxide production via p38 mitogen-activated protein kinase and NF-kappaB-dependent signaling pathways through macrophage scavenger receptors. Biochem Biophys Res Commun. 2006;343:286–294. doi: 10.1016/j.bbrc.2006.02.146. [DOI] [PubMed] [Google Scholar]
- 17.Bak SP, Walters JJ, Takeya M, Conejo-Garcia JR, Berwin BL. Scavenger receptor-A-targeted leukocyte depletion inhibits peritoneal ovarian tumor progression. Cancer Res. 2007;67:4783–4789. doi: 10.1158/0008-5472.CAN-06-4410. [DOI] [PubMed] [Google Scholar]
- 18.Wang XY, Facciponte J, Chen X, Subjeck JR, Repasky EA. Scavenger receptor-A negatively regulates antitumor immunity. Cancer Res. 2007;67:4996–5002. doi: 10.1158/0008-5472.CAN-06-3138. [DOI] [PubMed] [Google Scholar]
- 19.Hagemann T, Robinson SC, Schulz M, Trumper L, Balkwill FR, Binder C. Enhanced invasiveness of breast cancer cell lines upon co-cultivation with macrophages is due to TNF-alpha dependent up-regulation of matrix metalloproteases. Carcinogenesis. 2004;25:1543–1549. doi: 10.1093/carcin/bgh146. [DOI] [PubMed] [Google Scholar]
- 20.Neyen C, Pluddemann A, Roversi P, Thomas B, Cai L, van der Westhuyzen DR, Sim RB, Gordon S. Macrophage scavenger receptor A mediates adhesion to apolipoproteins A-I and E. Biochemistry. 2009;48:11858–11871. doi: 10.1021/bi9013769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, Silverstein RL. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 22.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 23.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 24.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 25.Kulbe H, Thompson R, Wilson JL, Robinson S, Hagemann T, Fatah R, Gould D, Ayhan A, Balkwill F. The inflammatory cytokine tumor necrosis factor-alpha generates an autocrine tumor-promoting network in epithelial ovarian cancer cells. Cancer Res. 2007;67:585–592. doi: 10.1158/0008-5472.CAN-06-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groblewska M, Mroczko B, Wereszczynska-Siemiatkowska U, Mysliwiec P, Kedra B, Szmitkowski M. Serum levels of granulocyte colony-stimulating factor (G-CSF) and macrophage colony-stimulating factor (M-CSF) in pancreatic cancer patients. Clin Chem Lab Med. 2007;45:30–34. doi: 10.1515/CCLM.2007.025. [DOI] [PubMed] [Google Scholar]
- 27.Ide H, Hatake K, Terado Y, Tsukino H, Okegawa T, Nutahara K, Higashihara E, Horie S. Serum level of macrophage colony-stimulating factor is increased in prostate cancer patients with bone metastasis. Hum Cell. 2008;21:1–6. doi: 10.1111/j.1749-0774.2007.00042.x. [DOI] [PubMed] [Google Scholar]
- 28.Kluger HM, Dolled-Filhart M, Rodov S, Kacinski BM, Camp RL, Rimm DL. Macrophage colony-stimulating factor-1 receptor expression is associated with poor outcome in breast cancer by large cohort tissue microarray analysis. Clin Cancer Res. 2004;10:173–177. doi: 10.1158/1078-0432.ccr-0699-3. [DOI] [PubMed] [Google Scholar]
- 29.Scholl SM, Bascou CH, Mosseri V, Olivares R, Magdelenat H, Dorval T, Palangie T, Validire P, Pouillart P, Stanley ER. Circulating levels of colony-stimulating factor 1 as a prognostic indicator in 82 patients with epithelial ovarian cancer. Br J Cancer. 1994;69:342–346. doi: 10.1038/bjc.1994.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 31.de Villiers WJ, Fraser IP, Hughes DA, Doyle AG, Gordon S. Macrophage-colony-stimulating factor selectively enhances macrophage scavenger receptor expression and function. J Exp Med. 1994;180:705–709. doi: 10.1084/jem.180.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluddemann A, Neyen C, Gordon S, Peiser L. A sensitive solid-phase assay for identification of class A macrophage scavenger receptor ligands using cell lysate. J Immunol Methods. 2008;329:167–175. doi: 10.1016/j.jim.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Hagemann T, Lawrence T, McNeish I, Charles KA, Kulbe H, Thompson RG, Robinson SC, Balkwill FR. Re-educating" tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 35.Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Fogelman AM. Apo A-1 mimetic peptides as atheroprotective agents in murine models. Curr Drug Targets. 2008;9:204–209. doi: 10.2174/138945008783755584. [DOI] [PubMed] [Google Scholar]
- 36.Van Lenten BJ, Navab M, Anantharamaiah GM, Buga GM, Reddy ST, Fogelman AM. Multiple indications for anti-inflammatory apolipoprotein mimetic peptides. Curr Opin Investig Drugs. 2008;9:1157–1162. [PMC free article] [PubMed] [Google Scholar]
- 37.Ganapathy E, Su F, Meriwether D, Devarajan A, Grijalva V, Gao F, Chattopadhyay A, Anantharamaiah GM, Navab M, Fogelman AM, Reddy ST, Farias-Eisner R. D-4F, an apoA-I mimetic peptide, inhibits proliferation and tumorigenicity of epithelial ovarian cancer cells by upregulating the antioxidant enzyme MnSOD. Int J Cancer. 2012;130:1071–1081. doi: 10.1002/ijc.26079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su F, Grijalva V, Navab K, Ganapathy E, Meriwether D, Imaizumi S, Navab M, Fogelman AM, Reddy ST, Farias-Eisner R. HDL mimetics inhibit tumor development in both induced and spontaneous mouse models of colon cancer. Mol Cancer Ther. 2012;11:1311–1319. doi: 10.1158/1535-7163.MCT-11-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su F, Kozak KR, Imaizumi S, Gao F, Amneus MW, Grijalva V, Ng C, Wagner A, Hough G, Farias-Eisner G, Anantharamaiah GM, Van Lenten BJ, Navab M, Fogelman AM, Reddy ST, Farias-Eisner R. Apolipoprotein A-I (apoA-I) and apoA-I mimetic peptides inhibit tumor development in a mouse model of ovarian cancer. Proc Natl Acad Sci U S A. 2010;107:19997–20002. doi: 10.1073/pnas.1009010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arredouani MS, Kobzik L. The structure and function of marco, a macrophage class a scavenger receptor. Cell Mol Biol (Noisy-le-grand) 2004;50 Online Pub:OL657-665. [PubMed] [Google Scholar]
- 41.Jozefowski S, Arredouani M, Sulahian T, Kobzik L. Disparate regulation and function of the class A scavenger receptors SR-AI/II and MARCO. J Immunol. 2005;175:8032–8041. doi: 10.4049/jimmunol.175.12.8032. [DOI] [PubMed] [Google Scholar]
- 42.Ojalvo LS, Whittaker CA, Condeelis JS, Pollard JW. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184:702–712. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirose Y, Saijou E, Sugano Y, Takeshita F, Nishimura S, Nonaka H, Chen YR, Sekine K, Kido T, Nakamura T, Kato S, Kanke T, Nakamura K, Nagai R, Ochiya T, Miyajima A. Inhibition of Stabilin-2 elevates circulating hyaluronic acid levels and prevents tumor metastasis. Proc Natl Acad Sci U S A. 2012;109:4263–4268. doi: 10.1073/pnas.1117560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang CL, Hsu HY, Lin HY, Chiang W, Lee H. Lysophosphatidic acid-induced oxidized low-density lipoprotein uptake is class A scavenger receptor-dependent in macrophages. Prostaglandins Other Lipid Mediat. 2008;87:20–25. doi: 10.1016/j.prostaglandins.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Bloedon LT, Dunbar R, Duffy D, Pinell-Salles P, Norris R, DeGroot BJ, Movva R, Navab M, Fogelman AM, Rader DJ. Safety, pharmacokinetics, and pharmacodynamics of oral apoA-I mimetic peptide D-4F in high-risk cardiovascular patients. J Lipid Res. 2008;49:1344–1352. doi: 10.1194/jlr.P800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Zheng SL, Komiya A, Mychaleckyj JC, Isaacs SD, Hu JJ, Sterling D, Lange EM, Hawkins GA, Turner A, Ewing CM, Faith DA, Johnson JR, Suzuki H, Bujnovszky P, Wiley KE, DeMarzo AM, Bova GS, Chang B, Hall MC, McCullough DL, Partin AW, Kassabian VS, Carpten JD, Bailey-Wilson JE, Trent JM, Ohar J, Bleecker ER, Walsh PC, Isaacs WB, Meyers DA. Germline mutations and sequence variants of the macrophage scavenger receptor 1 gene are associated with prostate cancer risk. Nat Genet. 2002;32:321–325. doi: 10.1038/ng994. [DOI] [PubMed] [Google Scholar]
- 47.Hope Q, Bullock S, Evans C, Meitz J, Hamel N, Edwards SM, Severi G, Dearnaley D, Jhavar S, Southgate C, Falconer A, Dowe A, Muir K, Houlston RS, Engert JC, Roquis D, Sinnett D, Simard J, Heimdal K, Moller P, Maehle L, Badzioch M, Eeles RA, Easton DF, English DR, Southey MC, Hopper JL, Foulkes WD, Giles GG. Macrophage scavenger receptor 1 999C>T (R293X) mutation and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:397–402. doi: 10.1158/1055-9965.EPI-04-0202. [DOI] [PubMed] [Google Scholar]
- 48.Noonan-Wheeler FC, Wu W, Roehl KA, Klim A, Haugen J, Suarez BK, Kibel AS. Association of hereditary prostate cancer gene polymorphic variants with sporadic aggressive prostate carcinoma. Prostate. 2006;66:49–56. doi: 10.1002/pros.20320. [DOI] [PubMed] [Google Scholar]
- 49.Murabito JM, Rosenberg CL, Finger D, Kreger BE, Levy D, Splansky GL, Antman K, Hwang SJ. A genome-wide association study of breast and prostate cancer in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S6. doi: 10.1186/1471-2350-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orloff M, Peterson C, He X, Ganapathi S, Heald B, Yang YR, Bebek G, Romigh T, Song JH, Wu W, David S, Cheng Y, Meltzer SJ, Eng C. Germline mutations in MSR1, ASCC1, and CTHRC1 in patients with Barrett esophagus and esophageal adenocarcinoma. JAMA. 2011;306:410–419. doi: 10.1001/jama.2011.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.