Abstract
E12/E47 proteins (encoded by E2A gene) are members of the class I basic helix-loop-helix (bHLH) transcription factors (also known as E proteins). E47 has been described as repressor of E-cadherin and inducer of epithelial-mesenchymal transition (EMT). We reported previously that EMT mediated by E47 in MDCK cells occurs with a concomitant overexpression of Id1 and Id3 proteins. Id proteins belong to class V of HLH factors that lack the basic domain; they dimerise with E proteins and prevent their DNA interaction, thus, acting as dominant negative of E proteins. Here, we show that E47 interacts with Id1 in E47 overexpressing MDCK cells that underwent a full EMT as well as in mesenchymal breast carcinoma and melanoma cell lines. By conducting chromatin immunoprecipitation assays we demonstrate that E47 binds directly to the endogenous E-cadherin promoter of mesenchymal MDCK-E47 cells in a complex devoid of Id1. Importantly, our data suggest that both E47 and Id1 are required to maintain the mesenchymal phenotype of MDCK-E47 cells. These data support the collaboration between E47 and Id1 in the maintenance of EMT by mechanisms independent of the dominant negative action of Id1 on E47 binding to E-cadherin promoter. Finally, the analysis of several N0 breast tumour series indicates that the expression of E47 and ID1 is significantly associated with the basal-like phenotype supporting the biological significance of the present findings.
Introduction
Epithelial-mesenchymal transition (EMT) is presently recognised as a key process for tumour invasion and metastasis [1]–[3]. The hallmarks defining the EMT process are the loss of E-cadherin mediated cell-cell adhesion and epithelial cell polarity concomitant to the acquisition of mesenchymal markers and increased motility and invasiveness [2]–[4]. Several transcriptional factors, presently called EMT-TFs, have been identified as EMT inducers, many of them also acting as direct E-cadherin repressors. Among them, factors from the Snail (Snail1/Snail2), Zeb (ZEB1/ZEB2) and bHLH (E47, Twist1, E2-2) families have been described [1], [2], [5].
The molecular mechanisms underlying the action of the various EMT-TFs are unequally known for Snail1 [6]–[11], Snail2 and ZEB1 factors [12], [13]. Much less is known on the mechanism of bHLH factors participating in EMT [14].
Class I bHLH E12/E47 are two splice variants of the E2A (also known as TCF3) gene product. E2A proteins regulate gene target expression as homodimers or heterodimers with other bHLH factors [15]. Id proteins (Id1-4), which lack the basic DNA binding domain, associate with E2A proteins and prevent them from binding to DNA [16]. A role for Id1 in mediating EMT-induced by TGFbeta or other factors has been proposed. The action of Id1 in EMT appears to depend on its dominant negative action on E2A factors in some systems [17], [18], but mediated by interaction with cytosolic/membrane proteins in other situations [19]. Interestingly, Id1 expression in breast cancer cells has been correlated with poor prognosis and lung metastasis [20], [21], [22]. Moreover, mice lacking Id1/Id3 genes are resistant to tumour neoangiogenesis and tumour cells failed to grow and/or metastasize [23], [24]. More recently, Id1 has been shown to be required for self-renewal of adult neural stem cells and glioblastoma tumours [25], [26] and for the mobilization of endothelial precursor cells to lung metastasis [27], further reinforcing the relevance of Id1 in tumorigenesis.
Our previous studies demonstrated that E47 is a strong EMT inducer and E-cadherin repressor [28], [29], but the specific mechanisms mediating E47 actions and whether E47 expression is required for EMT induction and/or maintenance of the mesenchymal phenotype are still unknown. Our previous analysis showed that Id factors, particularly Id1 and Id3, are strongly upregulated in MDCK cells stably expressing E47 or E2-2 factors [30]–[32]. The present study analyse the interplay between E47 and Id1 in E-cadherin repression and EMT. Our results reveal that E47 mediates E-cadherin transcriptional repression by direct interaction with its promoter in a complex devoid of Id1. Sustained expression of E47 and Id1 is required to maintain the mesenchymal phenotype of MDCK-E47 cells and to preserve cell viability. Remarkably, E47 as well as ID1 mRNAs are more frequently expressed in basal-like breast carcinomas compared to non-basal tumours supporting the participation of these proteins in defining this aggressive breast tumour subtype.
Results
Id1 protein is upregulated and interacts with E47 in MDCK-EGFP-E47 cells
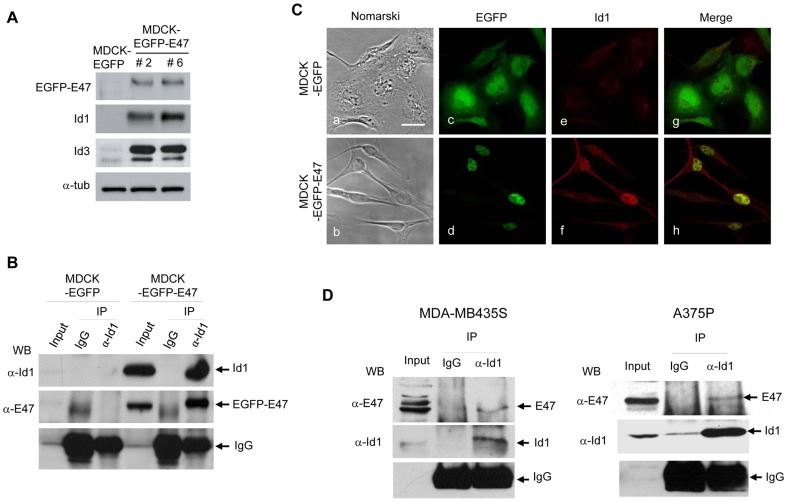
To further characterize the E47/Ids interplay during EMT, and because of the lack of reliable anti-E47 commercial antibodies for immunoprecipitation assays, we generated stable transfectant MDCK-EGFP-E47 cells. Extensive characterization of MDCK-EGFP-E47 cells was performed in three independent clones that showed almost complete repression of E-cadherin and the same mesenchymal phenotype and properties than previously described for MDCK-E47 cells including a full EMT conversion and overexpression of Id1 and Id3 factors (Figure 1A; Figure S1).
Figure 1. Id1 is upregulated and interacts in vivo with E47 in MDCK-EGFP-E47 cells.
(A) Cell extracts derived from MDCK-EGFP and MDCK-EGFP-E47 cells were analysed for expression of EGFP-E47, Id1 and Id3 proteins by Western blot; alpha-tubulin was used as loading control. Two independent clones (#2, #6) generated after stable expression of EGFP-E47 in MDCK cells are shown. (B) Cell extracts derived from MDCK-EGFP and MDCK-EGFP-E47 cells were immunoprecipitated with anti-Id1 antibodies and control IgGs and analysed by Western blotting with the indicated anti-Id1 and anti-E2A antibodies. (C) Nomarski (a,b) and confocal immunofluorescence images (c-h) of MDCK-EGFP and MDCK-EGFP-E47 cells. Localization of EGFP (c,g), EGFP-E47 (d,h) and Id1 (e,f) is shown. See the colocalization of Id1 with EGFP-E47 in the nuclei (merged image, h). Bars, 25 um. (D) Cell extracts derived from MDA-MB435S (left panels) and A375P (right panels) cells were immunoprecipitated with anti-Id1 antibodies and control IgGs and analysed by Western blotting with the indicated anti-Id1 and anti-E2A antibodies.
To confirm the interaction between EGFP-E47 and Id proteins, co-immunoprecipitation analyses were performed. EGFP-E47 protein strongly interacts with Id1 (Figure 1B) in MDCK-EGFP-E47 cells. Confocal analysis in MDCK-EGFP-E47 cells showed that Id1 protein was localized mainly in the nuclei; Id1 co-localized with EGFP-E47 in the majority of cells (Figure1C, panels f, h). Co-immunoprecitation analysis with Id3 showed a much weaker interaction with EGFP-E47 compared to Id1 (data not shown). Therefore, we focus our following studies on Id1. To confirm E47-Id1 interaction, co-immunoprecipitation assays were also performed in mesenchymal breast carcinoma MDA-MB435S and melanoma A375P cells, previously shown to be E-cadherin deficient and to overexpress E47 and Id1 factors ([29], [31] and data not shown). Results obtained indicated the interaction between E47 and Id1 both cell lines (Figure 1D) thus confirming our observations in MDCK-E47 cells.
Id1 is unable to overcome E-cadherin repression in MDCK-E47 cells
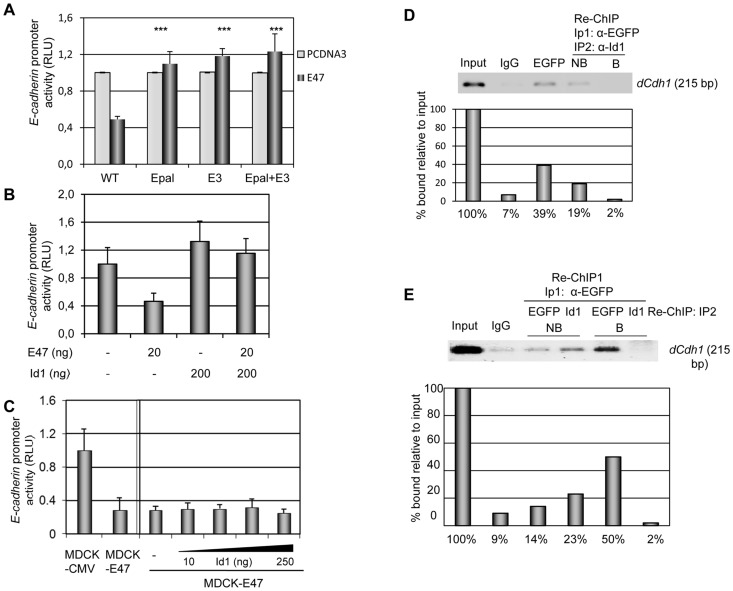
Since Id proteins have been extensively described as dominant negative of DNA binding of E47 and other class I bHLH factors [15], [16], we then analysed their effect on E47 repression of the E-cadherin promoter. We first confirmed that E47 repression depends on the proximal E-boxes, E-pal and E3, of the mouse E-cadherin promoter (Figure 2A), in agreement with previous observations on the dependence of the E-pal element for in vitro binding of E47 [28]–[33]. The E47-mediated repression of E-cadherin promoter in MDCK cells was fully overcome by transient co-transfection with Id1 (Figure 2B). We then analysed the effect of Id1 proteins in MDCK cells stably expressing E47 with a complete EMT and exhibiting very low E-cadherin promoter activity [29]. Transient Id1 overexpression was unable to de-repress E-cadherin promoter activity in MDCK-E47 cells (Figure 2C). Similar results were obtained in MDCK-EGFP-E47 cells (unpublished data). Moreover, Id1 overexpression did not de-repress the basal activity of E-cadherin promoter in MD-MDA435S and A375P cells (Figure S2). Altogether, these results demonstrate that E-cadherin repression by E47 is not sensitive to inhibition by Id1 protein once a full EMT is achieved.
Figure 2. E47 represses E-cadherin by direct binding to the endogenous promoter in complexes devoid of Id1.
(A) Dependence of E47 repression of E-cadherin promoter on proximal E-boxes. HEK293T cells transiently co-transfected with 200 ng of the proximal wild type mouse E-cadherin-Luciferase promoter (−178/+92 bp) or point mutants in the proximal E-boxes (E-pal +/− E-box3) and 10 ng of CMV-beta-gal plasmid in the presence of 50 ng of pcDNA3-E47 (dark grey bars) or control pcDNA3 (light grey bars) vectors, as indicated. Differences between the E47 mediated repression of the mutants versus the wild type promoter were statistically analysed by t-Student test (***p<0.001). (B) and (C) E-cadherin promoter activity in MDCK (B) and MDCK-CMV and MDCK-E47 (C) cells transiently co-transfected with 200 ng of the proximal mouse E-cadherin-Luciferase promoter (−178/+92 bp) and 20 ng of TK-Renilla (B) or 10 ng of CMV-beta-gal (C) plasmids, in combination with the indicated amounts of pcDNA3-E47 and/or pcDNA3-Id1 vectors. RLU is normalized to that detected with empty pcDNA3 vector (A, B) or in control MDCK-CMV cells (C, left); results represent the mean of at least two independent experiments +/− s.d. (D) and (E) Chromatin immunoprecipitation (ChIP) assays of EGFP-E47 at the endogenous E-cadherin promoter in MDCK-EGFP-E47 cells. (D) ChIP assays were first performed with an antibody specific for EGFP or control IgG and the bound fraction analysed for endogenous E-cadherin promoter sequences (dCdh1) (left three lanes), followed by re-ChIP with anti-Id1 antibodies (IP2) and analyses of the bound (B) and unbound (NB) fractions for E-cadherin promoter sequences (dCdh1) (right two lanes). The input fraction (1/100) is shown at the left. Quantification of the amplified endogenous E-cadherin promoter compared to the input fraction (1/100) is shown in the lower panel. (E) ChIP assays were first performed with anti-EGFP (IP1) followed by re-ChIP (IP2) with anti-EGFP or anti-Id1 antibodies, as indicated, followed by analyses of the bound (B) and unbound (NB) fractions for E-cadherin promoter sequences (dCdh1). Input (1/100) and control IgG ChIP are shown on the left two lanes. Quantification of the amplified endogenous E-cadherin promoter compared to the input fraction (1/100) is shown in the lower panel.
E47 interacts in vivo with the E-cadherin promoter in complexes devoid of Id1
The interaction of E47 with the endogenous E-cadherin promoter has not yet been studied at the chromatin level. To assess this point, we performed chromatin immunoprecipitation (ChIP) assays in MDCK-EGFP-E47 cells with anti-EGFP antibodies. EGFP-E47 factor binds specifically to the endogenous E-cadherin promoter in MDCK-EGFP-E47 cells compared to unspecific IgG control (Figure 2D). To further characterize the E47 complexes bound to the E-cadherin promoter, re-ChIP assays were carried out. A sequential immunoprecipitation with anti-EGFP followed by anti-Id1 antibodies showed the absence of Id1 binding to the E-cadherin promoter in the chromatin fraction bound by EGFP-E47 (Figure 2D, re-ChIP, B fraction), while the unbound fraction from the Id1 precipitates still retain a significant part of the endogenous E-cadherin promoter (Figure 2D, re-ChIP NB fraction). The specificity of the re-ChIP assay was further confirmed by parallel re-ChIP assays with anti-EGFP and anti-Id1 antibodies (Figure 2E) that showed the ability of anti-EGFP to effectively precipitate the remaining E-cadherin promoter from the Id1-ChIP fraction and the complete absence of Id1 binding to the EGFP-E47 bound fraction (Figure 2E, two most right lanes). These results indicate that E47 interacts in vivo directly with the endogenous E-cadherin promoter in MDCK-EGFP-E47 cells in complexes devoid of Id1.
Sustained E47 expression is essential for the maintenance of EMT and tumour aggressiveness
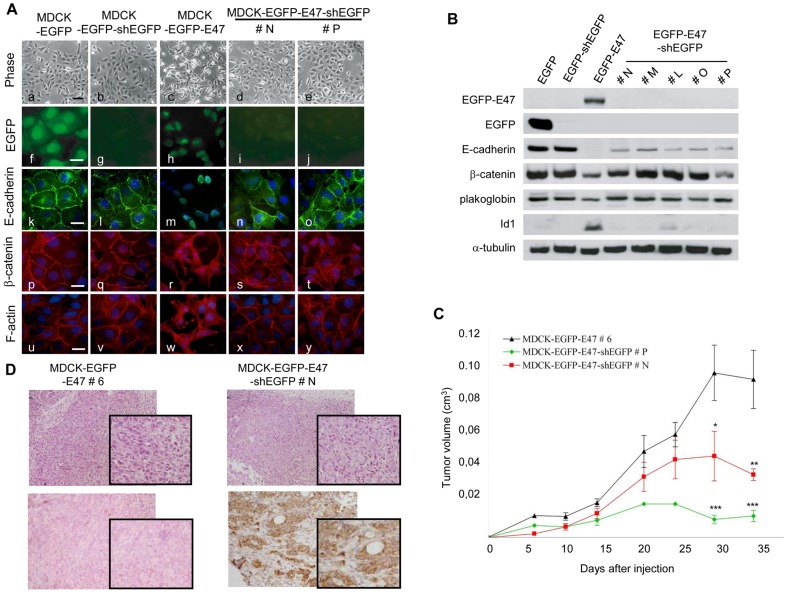
One important and still unresolved question is whether continued expression of E47 is necessary for maintenance or just required for induction of EMT in MDCK cells, as reported for E2-2 factors [32]. Since repeated attempts to knockdown E47 by shE47 in MDCK cells were unsuccessful, we decided to silence E47 expression by shRNA against EGFP in MDCK-EGFP-E47. As a control, MDCK-EGFP cells were used to discard side-off effects of the shEGFP. We first verified that EGFP-shRNA is effectively knocking down EGFP and EGFP-E47 in all analysed clones from MDCK-EGFP-shEGFP and MDCK-EGFP-E47-shEGFP, respectively (Figure 3A, panels g,i,j, and 3B). Control MDCK-EGFP cells after shEGFP remained with an epithelial phenotype, expressing high levels of E-cadherin and other epithelial markers and lacking Id1 expression (Figure 3A, B). In contrast, EGFP-E47 silencing by shEGFP induced a remarkable reversion to an epithelial phenotype in MDCK-EGFP-E47 cells (Figure 3A, panels d,e), associated with re-expression of E-cadherin and its re-localization at cell-cell contacts (Figure 3A and B). Upregulation and membrane re-localization of the epithelial markers beta-catenin and plakoglobin was also observed (Figure 3A and B). Moreover, EGFP-E47 silencing promoted the reorganization of the F-actin cytoskeleton from stress fibers and lamellipodia-like structures to cortical actin filaments (Figure 3A panels x,y, vs panel w). Significantly, EGFP-E47 silencing was associated to almost complete suppression of Id1 overexpression in MDCK-EGFP-E47-shEGFP clones (Figure 3B), further supporting the link between expression of E47, EMT and Id1 induction in the MDCK cell system.
Figure 3. E47-silencing in MDCK-EGFP-E47 cells induces a MET process, downregulation of Id1 and reduction of tumour growth.
(A) Phase contrast images (a-e) and immunofluorescence images (f–y) of the indicated cell lines showing localization of EGFP (f,g), EGFP-E47 (h–j), Ecadherin (k–o), beta-catenin (p–t) and F-actin organization (u–y). Nuclei were stained with DAPI (k–y). Black bar, 60 um (a–e); white bars, 25 um (f–y). (B) Western blot analyses of epithelial markers and Id1 in the indicated cell lines; alpha-tubulin was used as loading control. Two (#N, #P) and five independent clones (#N, #M, #L, #O, #P) generated after stable expression of shEGFP into MDCK-EGFP-E47 (clone #6) cells are shown in A and B, respectively. (C) The tumorigenic potential of MDCK-EGFP-E47#6 and MDCK-EGFP-E47-shEGFP#N and #P cells was analysed by subcutaneous injection into nude mice. ANOVA analysis: *p<0.05; **p<0.01; ***p<0.001. (D) Histology (upper panels) and immunohistochemical analysis for E-cadherin (lower panels) of tumours induced by MDCK-EGFP-E47#6 (left) and MDCK-EGFP-E47-shEGFP#N cells (right). Amplification x20. Insets show amplified fields at x40.
The relevance of E47 interference in the tumorigenic properties of MDCK-EGFP-E47 cells was then analysed. EGFP-E47-interfered clones were injected into nude mice, in parallel with control MDCK-EGFP-E47 cells that exhibit similar tumorigenic properties than previously reported for MDCK-E47 cells [28], [29], [31]. All cell lines gave rise to tumours at all injection sites. However, the shEGFP-E47 derived tumours grew at low rate with an overall 50% to 85% reduction of tumour volume compared to the tumours induced by control cells (Figure 3C). Tumours induced by MDCK-EGFP-E47 cells were highly undifferentiated spindle cell tumours, without evidence of epithelial differentiation (Figure 3D, left). In contrast, tumours generated by MDCK-EGFP-E47-shEGFP cells showed areas of glandular differentiation surrounded by undifferentiated spindle cells (Figure 3D, right) or were very small tumours (Figure 3C). Moreover, the MDCK-EGFP-E47-shEGFP xenografts express E-cadherin at differentiated areas in contrast to the expected absence of E-cadherin expression in control MDCK-EGFP-E47 tumours (Figure 3D, lower panels).
These results indicate that sustained E47 expression is required for the maintenance of the full phenotypic characteristics and markers of EMT, including Id1 expression and for aggressive tumorigenic properties of MDCK-E47 expressing cells.
Id1 expression is required for EMT induced by E47 and cell survival
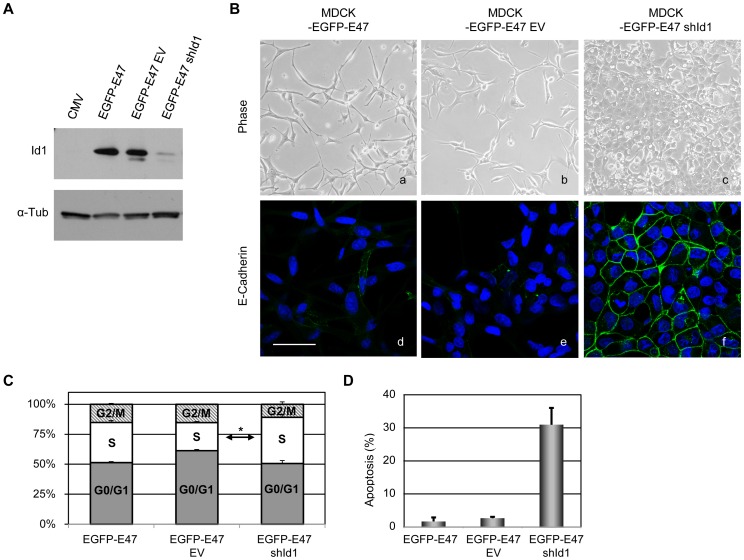
Once established that E47 is required to maintain the EMT phenotype, we decided to study whether EMT also depends on induction of Id1 in MDCK-E47 cells. To answer this point, we knocked down Id1 in MDCK-EGFP-E47 cells by lentiviral infection. Three independent shId1 lentiviral constructs were tested and results obtained with shId1 9032 are shown (Figure 4). It should be noted that the dog genome is only partially annotated and shId1 sequences were selected based on one Id1 sequence with strong human/dog homology. However, only shId1 9032 of the three analysed shId1 lentivirus blocked efficiently endogenous Id1 in dog MCDK cells (data not shown). To discard side-off effects, the action of the selected shId1 construct on MDCK-EGFP-E47 cells was compared with control lentivirus and with non-infected cells. Efficient knockdown of Id1 was detected 48 h after infection (about 90%) with no significant effect detected with the control lentivirus (Figure 4A). After 48 h of infection, Id1 knockdown induced a striking reversion to an epithelial-like phenotype in MDCK-EGFP-E47-shId1 cells (Figure 4A, panel c) compared to non-infected and control infected cells that remain fully mesenchymal (Figure 4B, panels a,b). The phenotypic Mesenchymal to Epithelial (MET)-like reversion was accompanied by strong organization of E-cadherin at cell-cell contacts in MDCK-EGFP-E47-shId1 cells (Figure 4B, panel f) compared to parental and control cells (Figure 4B, panels d,e). Knockdown of Id1 for longer time periods induced a massive cell death; indeed, repeated attempts to obtain stable infections with shId1 lentivirus were unsuccessful. Analysis of the cell cycle indicated no major alterations in MDCK-EGFP-E47-shId1 cells compared to parental cells (Figure 4C). However, a dramatic increase in the sub-G0 population was detected in MDCK-EGFP-E47-shId1 cells indicative of a high apoptotic rate compared to either parental and control cells (Figure 4D).
Figure 4. Id1 knockdown in MDCK-EGFP-E47 cells induces a partial MET and impairs cell survival.
(A) Western blot analysis of Id1 expression in MDCK-EGFP-E47 cells after 48 h infection with shId1 or control lentivirus (EV), compared to non-infected (EGFP-E47) and control MDCK-CMV (CMV) cells; alpha-tubulin was used as loading control. (B) Phase contrast (upper panels) and confocal immunofluorescence images of E-cadherin (lower panels) of EGFP-E47-shId1 (right) and EGFP-E47-EV control (middle) cells 48 h after lentiviral infection; non infected MDCK-EGFP-E47 cells (left) are shown as additional control. Nuclei were stained with DAPI; bars, 50 um. (C) Cell cycle analysis of the indicated cell lines by propidium iodide staining and FACS analysis. (D) Apoptosis rate of the indicated cells determined by the sub-G0 population. Results in C and D represent the mean +/− s.d. on triplicate samples.
Together, these results indicate that expression of Id1 is required for maintenance of the mesenchymal phenotype after EMT and cell viability of MDCK-EGFP-E47 cells.
E47 and ID factors expression in breast cancer
At present, very little is known about the expression of E47 and IDs and their correlation with specific subtypes of human breast carcinomas, although ID1 expression has been correlated with poor prognosis, metaplastic tumours and lung metastasis in breast carcinomas [20]–[22], [34], [35]. To investigate the biological significance of E47 and ID factors in human breast cancer we analyzed their expression together with markers defining the basal and non-basal subtypes in unsupervised clustering analysis in a series of 97 sporadic N0 breast carcinomas profiled by Vańt Veer et al. [36].
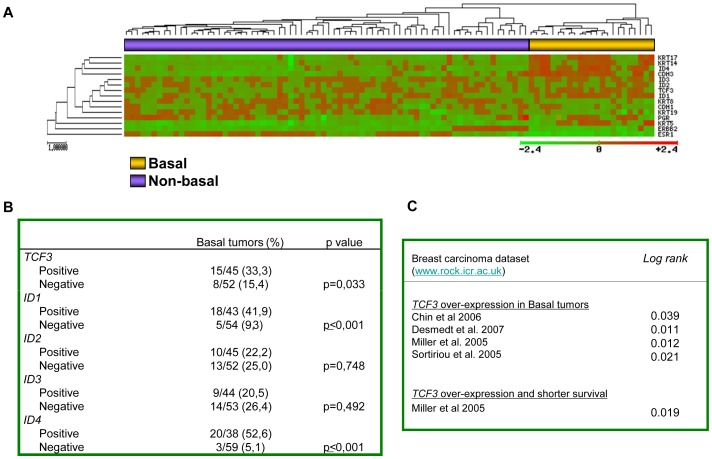
As shown in Figure 5A, tumours were sub-classified into two main clusters, indicating that TCF3 (E47/E12) and ID genes are differentially expressed among breast carcinomas. Attending to the markers defining tumour subgroups, the right cluster (yellow bar) contained the hormone-receptor-negative tumours; according to the low expression of ESR, PGR, ERBB2 and luminal keratins (KRT8 and KRT19) and high expression of basal keratins (KRT5, KRT14, KRT17) this subgroup corresponds to the basal-like tumours. The left-handed cluster (purple bar) includes the non-basal tumours (luminal and ErbB2+). In this dataset, positive TCF3, ID1 and ID4 expression were more frequent (Chi-square test, p = 0.033, p<0.001, and p<0.001, respectively) in the basal-like tumours (Figure 5B). Moreover, TCF3, ID1 and ID4 expression showed a strong association with the expression of basal markers, such as CK14, CK17, and P-cadherin (Table S1). These results agree with and complement previous studies on ID1 protein expression in metaplastic and triple negative breast cancer tumours [21], [34]. Furthermore, the analysis of TCF3 expression in several breast carcinomas dataset [37]–[40] showed that TCF3 upregulation associated to the basal-like phenotype (Figure 5C, upper) and exemplified in dedifferentiated breast carcinoma MDA-MB435 cells (Figure 1D). In addition, TCF3 upregulation was associated to shorter overall survival in the Miller's series [39] (Figure 5C, bottom). Taken together, these results suggest TCF3 as a new marker of basal-like breast tumors and support the interplay between E47 and ID factors in this tumour subtype.
Figure 5. E47 and ID factors are preferentially expressed in basal-like breast carcinomas.
(A) Unsupervised hierarchical clustering of the vant’Veer dataset (ref. 36) for the expression of the indicated basal and luminal markers, together with estrogen (ESR1) and progesterone receptors (PGR) and ErbB2, identifies two clusters of basal (yellow) and non-basal (purple) tumours. Increased TCF3 and ID1/ID4 transcripts cluster with the basal-like subgroup. (B) Association between the expression of TCF3 and ID1/ID4 with the basal-like phenotype in the vańt Veer's dataset. To validate the association among specific genes and/or tumour subgroups, each variable was categorized using the percentile 75 value as the cut-off. (C) Overexpression of TCF3 associated to basal-like breast tumours (upper) and to decreased overall survival (bottom) in the indicated public datasets.
Discussion
Class I bHLH E2A factors (E12/E47) have been previously shown to promote E-cadherin repression and to induce EMT and invasive, tumorigenic and angiogenic or fibrotic properties when expressed in epithelial cells [17], [28], [29], [31], [41]. However, the mechanisms underlying E-cadherin repression and the specific role of E12/E47 in EMT have not yet been elucidated. We report here that E47 transcriptional repressor binds directly to the E-cadherin promoter in MDCK cells stably expressing E47 factor. We also show that continued expression of E47 and Id1 is necessary for maintenance of EMT in the MDCK-EGFP-E47 cell system.
Consistent with the dominant negative action of Ids on bHLH factors [15], [16] we show that Id1 protein efficiently overcome E47-induced repression of E-cadherin promoter in transient co-transfection assays in epithelial MDCK cells, similar to previously reported for Id3 [17]. However, overexpressed Id1 protein is unable to overcome E47-dependent repression of E-cadherin expression in mesenchymal MDCK-E47 cells; a similar situation was observed for Id2 and Id3 (unpublished data). Indeed, the ChIPs assays show that E47 binds to the endogenous E-cadherin promoter of MDCK-EGFP-E47 cells in complexes devoid of Id1. Nonetheless, this situation occurs despite the physical interaction between E47 and Id1 detected in mesenchymal MDCK-EGFP-E47 cells. The E47/Id1 interaction and inability of Id1 to reactivate the E-cadherin promoter was further confirmed in E47-expressing basal-like breast carcinoma and melanoma cells. One possible explanation to reconcile these apparently contradictory findings is that post-translational modifications might regulate E47 transcriptional activity, as previously described for site-specific phosphorylation and/or covalent disulfide bond dimerization [42]-[45]; such modifications in a subset of E47 molecules might attenuate their interaction with Id1 and allow E47 binding to the E-cadherin promoter in absence of Id1. It is tempting to speculate that this subset of E47 molecules can act as active homodimers promoting E-cadherin repression; in fact, our previous in vitro band-shift and in vivo yeast-one-hybrid analysis indicated that E47 interacts with the mouse E-cadherin promoter as a homodimer [29], [33]. Another possibility is the interaction of E47 with tissue-specific class II bHLH factors [15]. One potential candidate is Twist1, another EMT inducer [46]; we tested this hypothesis, but we could not find any functional E47/Twist1 interaction in E-cadherin promoter assays (unpublished data) in line with recent data suggesting an indirect role of Twist1 in E-cadherin repression [3], [47]. Alternatively, the presence of activating E proteins sequestered by Id1 induction in the presence of E47 could be another option. Although further studies are clearly required to clarify the mechanism, in the present scenario, we favour the hypothesis that a subset of E47 molecules with potential post-translational modifications could be responsible of E-cadherin repression likely in homodimeric form.
The present data also provide evidence for a strong link between E47 and Id1 in EMT. Sustained expression of E47 is required to maintain the mesenchymal phenotype after EMT, in contrast to the behaviour of E2-2 factors that act as EMT-inducers but are dispensable for maintenance of EMT [32]. Significantly, the MET induced by E47-silencing in MDCK-EGFP-E47 cells is associated to strong downregulation of Id1. Moreover, Id1 knockdown also induces a MET-like process in MDCK-EGFP-E47 cells, indicating that expression of Id1 is indeed required for maintenance of EMT induced by E47 and, importantly, for cell survival. These results agree with the pleiotropic effect of Id1 on cell proliferation and survival, partly independent of E2A proteins [16], [48], and support the collaboration between E47 and Id1 in EMT beyond E-cadherin repression. Interestingly, Id1 is induced by additional EMT-TFs, like Snail1, Snail2 and E2-2A/B [30]-[32], further supporting its participation in maintenance of EMT. Whether the specific action of Id1 in EMT maintenance is dependent of its interaction with other nuclear, cytosolic or membrane proteins remains to be established.
Many studies highlight distinct roles for Id proteins in tumorigenesis, invasiveness and metastasis in epithelial, endothelial and hematopoietic cells [16]. Furthermore, ID1 overexpression has been associated with malignancy, poor clinical outcome or lung metastasis in different tumour types [20]–[22], [25], [34], [35], [49]. Overexpression of E47 in some tumours and cell lines has also been reported [29], [50], [51]. Interestingly, our analysis of E47 (TCF3) and IDs expression in the N0 breast carcinoma series profiled by Van’t Veer et al. [36] indicates that the expression of E47 and ID1 is significantly associated with the basal-like phenotype in sporadic tumours, in agreement with previous observations on ID1 protein expression [21]. Overexpression of ID4 was also associated with basal-like tumours, in agreement with a recent report on ID4 protein overexpression in triple negative breast tumours (TNBC) [52], and also detected in the vant’ Veer series (Figure 5B). Rrecent studies have highlighted the connection between ID4 induction and downregulation of BRCA1 in TNBCs [53] as well as with p53 mutation in estrogen receptor negative (ER-) breast tumours [54] that might contribute to chemoresistance of p53 mutant breast tumours [55]. Interestingly, E47 has been also reported to contribute to doxorubicin chemoresistance [56]. Upregulation of Id4 transcripts were also previously observed in MDCK cells overexpressing E47, although to a lower extent than Id1 [31], supporting also the correlation between ID4 and TCF3 upregulation in basal breast tumours. Whether ID1 and ID4 play non-redundant or complementary actions on EMT and/or the basal phenotype and chemoresistance of breast tumours remains to be established in future studies. Importantly, our present study detected TCF3 upregulation in basal-like tumours in several additional public datasets and associated to poor survival (Figure 5C), reinforcing previous observations indicating that basal-like carcinomas are the most likely candidate breast tumours to suffer EMT processes [57]-[59]. A few pathways and molecules have been so far implicated in the regulation of epithelial plasticity in basal-like tumours, including the Wnt signaling, Forkhead 1 factor FOXC2, and lysyl oxidase-like 2 (LOXL2) [60]–[63]. The evidences here presented suggest that additional genes such as TCF3 and ID1 could also mediate EMT in basal-like carcinomas. Furthermore, our previous observation that upregulation of TCF8 (ZEB1) occurs in metastatic and more undifferentiated breast tumours of the Vańt Veer's series [64] suggest a potential link between E47/ID1 or ID4 and TCF8 expression in undifferentiated basal-like tumours. Although further studies are required, our present observations on the E47 and ID1 expression profile in the N0 breast carcinoma series provide biological basis to the interplay between E47 and ID1 here described.
Materials and Methods
Plasmid constructs
Enhanced green fluorescent protein EGFP-E47 construct was generated by PCR using the full-length mouse E47 cDNA as template [29] with the following primers (restriction sites indicated in bold): 5′-CTCGAGGGATGATGAACCAGTCTCA-3′ (forward) and 5′-GAATTCGGCTCACAGGTGCCCGGCTGGGT -3′ (reverse). The PCR product was cloned in the pEGFP-C1 vector (Clontech, Mountain View; CA). Mouse Id1 construct (pcDNA3-Id1) was provided by Dr. M. Saitoh (Tokio University, Japan).
Cell culture and gain of function studies
MDCK-II (Madin Darby canine kidney type II) and human HEK293T (human embryonic kidney 293 transformed with T-antigen), MDA-MB435S whose origin has been firmly established as dedifferentiated breast carcinoma [65], [66] and melanoma A375P cells were obtained from the American Type Cell Culture (ATCC) (LGC Standards-SLU; Barcelona, Spain) and cultured as previously described [29]. Stable transfectants were obtained from parental MDCK-II (Madin Darby canine kidney type II) cells (obtained from the ATCC) (LGC Standards-SLU; Barcelona, Spain) after transfection with pEGFP-C1-E47, or the corresponding control vector, using Lipofectamine (Invitrogen; Carlsbad, CA). Cells were grown as previously described [67] in the presence of G418 (400 µg/ml; Gibco; Madrid, Spain) for 3–4 weeks and individual clones isolated with cloning rings. At least ten independent clones were isolated and results from two representative clones from each setting are shown. The origin and characterization of MDCK-CMV and MDCK-E47 cells has been previously described [29], [67].
RNA interference
The generation of shRNA containing specific oligonucleotide sequences against EGFP cloned into the pSuperior-Puro vector (Oligoengine; Seattle, WA) was previously described [30]. MDCK-EGFP and MDCK-EGFP-E47 (clone #6) cells were transfected with shEGFP vector using Lipofectamine 2000 (Invitrogen) and sorted using a FACSVantage SE (Becton Dickinson; Madrid, Spain) to obtain populations with more than 90% EGFP-negative cells. At least ten clones were isolated from each pool by limited dilution and results from two to five independent clones from each setting are shown.
Lentiviral infection
Id1 knockdown was performed by lentiviral infection with specific shRNA vectors (TRCN0000019032; Sigma-Aldrich; St Louis, MO). Briefly, HEK293T cells were transfected with a mix containing VSV-G envelope and ΔR 8.91 packaging genes and either control vector (pLKO: EV) or Id1 specific shRNA using Lipofectamine 2000 and incubated for 48 h. MDCK-EGFP-E47 cells were infected with the lentiviral supernatant with polybrene (8 ng/ml) during 48 h.
E-cadherin promoter assays
The activity of the mouse E-cadherin (Cdh1) promoter on HEK293T, MDCK and MDCK-derived clones, MDA-MB435S and A375P cells was determined as described [67]. Mutants in the E-pal and/or E3-box were previously described [32]. Statistical analyses were performed by the two paired t-Student’s test.
Northern Blotting
15 to 30 ug of total RNA from the different cell lines were analysed in Northern blot using a specific P32-cDNA Cdh1 probe as described [30].
Cell extracts, Western blot analysis and immunoprecipitations
Whole cell extracts, lysates, immunoprecipitation conditions and Western blot analyses were performed as described [11], [12], [67]. The primary and secondary antibodies used are described in Table S2.
Chromatin immunoprecipitation (ChIP) and Sequential ChIP assays
ChIP assays were performed in MDCK-EGFP-E47 (clone #6) cells, using formaldehyde before sonication, as described [11], [32]. For detection of interaction between the tagged factors and the endogenous Cdh1 promoter, an anti-EGFP antibody (Molecular Probes; Grand Island, NY) or unspecific rabbit IgG (Jackson ImmunoResearch Laboratories; Suffolk, UK) were used. A 215 bp fragment of the dog Cdh1 promoter (–85/+130) was amplified using the primers and amplification conditions previously described [32]. For re-ChIP assays, the immune complexes were harvested with elution buffer (TE 1X, 2% SDS, 15 mM DTT and protease inhibitors), diluted 1∶10 with dilution buffer (10 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, 0.01% SDS, 0.5% Nonidet P-40 and protease inhibitors), and the steps of the ChIP assay repeated with a second round of immunoprecipitation using anti-EGFP (Molecular Probes) or anti-Id1 (Santa Cruz; California, CA) antibodies.
Cell cycle and apoptosis assays
Cell cycle analysis was performed by propidium iodide staining. Semi-confluent cells were trypsinized and washed twice with PBS, fixed with cold 70% ethanol, washed, resuspended in PBS with 10 ug/ml of propidium iodide, and analysed in a FACScanI (Becton Dickinson) using a ModFIT LT v3.2 software. Assays were performed on triplicate samples.
Immunofluorescence
Immunofluorescence analysis and F-actin stain was performed as described [67]. The primary and secondary antibodies used are described in Table S2. Preparations were visualized using a Zeiss Axiophot or a Leica confocal TCSSP2 microscope.
Induction of xenografted tumours and histological studies
Subconfluent cells were trypsinised, washed and resuspended at 1×107 cells/ml in PBS. 1×106 cells were injected into the flanks of 8-week old male BALB/c nude mice (Charles River; Wilmington, MA). Tumours were measured every 3–4 days using a calliper by determination of the two orthogonal external diameters; statistical comparisons were made by ANOVA analysis. Mice were sacrificed when the tumours reach 0.1 cm3 size; tumours were surgically excised and processed for histology as described [67]. Mice were housed and maintained under specific pathogen-free conditions and used in accordance with institutional guidelines and approved by the Use Committee for Animal Care. The protocol was approved by the Committee on the Ethics of the University Autonomous of Madrid (UAM. Permit Number: CEI-25-587) and Spanish National Research Council (CSIC), according with present Spanish law (R.D. 1201/2005, 10th October, BOE 21st October 2005). All animals were monitored for ill effects and sacrificed at 32 days post-injection; all efforts were made to minimize suffering according to UAM/CSIC guidelines. A minimum of 4 tumours from each cell line were generated and analysed by histology and immunohistochemistry.
Human breast cancer expression data set and statistical analysis
Microarray and clinical data from 97 sporadic breast tumours, reported by van’t Veer et al. [36] were used. The sporadic samples included primary invasive carcinoma <5 cm (T1 or T2), with no axillary lymph node metastases (N0). To classify the breast samples according to basal and non-basal (luminal and ErbB22+) phenotypes, we extracted the log ratio values of ESR1 (estrogen receptor), PGR (progesterone receptor), ERBB2, and luminal (CDH1, KRT8, and KRT19) and basal markers (CDH3, KRT5, KRT14, and KRT17) present in the Vańt Veer dataset; in addition, we select the TCF3 (E47) and ID (ID1–4) factors expression. Hierarchical cluster analysis was performed assuming euclidean distances between genes. For significance associations Yates correction or Fisher’s exact test were used. To evaluate TCF3 association to breast carcinoma subtypes, several independent dataset of carcinoma series were also analysed [37]–[40]. The clinical data were obtained from the ICR database (www.rock.icr.ac.uk). TCF3 expression values were categorized using upper quartile (25% against rest).
Supporting Information
Stable expression of EGFP-E47 in MDCK cells triggers a complete EMT. (A) Phase contrast (a–c) and immunofluorescence (d–l) images of the indicated cell lines for EGFP (d), EGFP-E47 (e,f), E-cadherin (g–i) and vimentin (j–l). Nuclei were stained with DAPI (g–l). Bars, 60 mm (a–c); 25 mm (d–l). (B and C). Western blot (B) and Northern blot analysis (C) analyses of the indicated markers in E47 expressing cells and controls. alpha-tubulin and GAPDH were used as loading controls.
(TIF)
E-cadherin promoter activity in A375P (left) and MDA-435S (right) cells transiently cotransfected with 200 ng of the proximal mouse E-cadherin promoter and increasing amounts of Id1 and 10 ng of CMV-beta-gal. RLU is normalized to activity detected in the presence of pcDNA3. Results represent the mean +/- s.d. of three independent experiments performed on triplicate samples.
(TIF)
Association of TCF3 positive expression with basal and luminal markers in N0 breast tumours. The categorical expression of progesterone receptor (PGR), estrogen receptor (ESR1), ERBB2, and basal and luminal markers in the van’t Veer’s dataset (ref. 38) is shown.
(DOC)
Antibodies used in Western-blot, immunofluorescence immunoprecipitation and IHQ assays.
(DOC)
Acknowledgments
We thank all the people in A. Canós lab for their invaluable help and discussion. We also thank Thomas Look (Dana-Farber Cancer Institute, Boston, MA) for anti-E2A antibodies and Diego Megías (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain) for useful assistance with confocal microscopy.
Funding Statement
This work was supported by the Spanish Ministry of Education and Science (SAF2007-63051; SAF2010-21143; Consolider Ingenio 2010 CDS07/00017) to A.C.; (SAF2007-63075 and SAF2010-20175) to G.M.B.; Comunidad de Madrid (S2010/BMD-2303) to A.C and G.M.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Nieto MA (2011) The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 27: 347–376. [DOI] [PubMed] [Google Scholar]
- 2. Thiery JP, Acloque H, Huang RY, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139: 871–890. [DOI] [PubMed] [Google Scholar]
- 3. Yang J, Weinberg RA (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14: 818–829. [DOI] [PubMed] [Google Scholar]
- 4. Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428. [DOI] [PubMed] [Google Scholar]
- 6. Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, et al. (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol 2: 84–89. [DOI] [PubMed] [Google Scholar]
- 7. Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, et al. (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E cadherin expression. Nat Cell Biol 2: 76–83. [DOI] [PubMed] [Google Scholar]
- 8. Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, et al. (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28: 4772–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Langer EM, Feng Y, Zhaoyuan H, Rauscher FJ 3rd, Kroll KL, et al (2008) Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell 14: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin Y, Wu Y, Li J, Dong C, Ye X, et al. (2010) The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 29: 1803–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peinado H, Ballestar E, Esteller M, Cano A (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24: 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Molina-Ortiz P, Villarejo A, MacPherson M, Santos V, Montes A, et al. (2012) Characterization of the SNAG and SLUG domains of Snail2 in the repression of E-cadherin and EMT induction: modulation by serine 4 phosphorylation. PLoS One 7: e36132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, et al. (2010) ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 29: 3490–3500. [DOI] [PubMed] [Google Scholar]
- 14. Cano A, Portillo F (2010) An emerging role for class I bHLH E2-2 proteins in EMT regulation and tumor progression. Cell Adh Migr 4: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perk J, Iavarone A, Benezra R (2005) Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer 5: 603–614. [DOI] [PubMed] [Google Scholar]
- 17. Kondo M, Cubillo E, Tobiume K, Shirakihara T, Fukuda N, et al. (2004) A role for Id in the regulation of TGF-beta-induced epithelial-mesenchymal transdifferentiation. Cell Death Differ 11: 1092–1101. [DOI] [PubMed] [Google Scholar]
- 18. Kowanetz M, Valcourt U, Bergstrom R, Heldin CH, Moustakas A (2004) Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol 24: 4241–4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang X, Ling MT, Wang Q, Lau CK, Leung SC, et al. (2007) Identification of a novel inhibitor of differentiation-1 (ID-1) binding partner, caveolin-1, and its role in epithelial-mesenchymal transition and resistance to apoptosis in prostate cancer cells. J Biol Chem 282: 33284–33294. [DOI] [PubMed] [Google Scholar]
- 20. Fong S, Itahana Y, Sumida T, Singh J, Coppe JP, et al. (2003) Id-1 as a molecular target in therapy for breast cancer cell invasion and metastasis. Proc Natl Acad Sci U S A 100: 13543–13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta GP, Perk J, Acharyya S, de Candia P, Mittal V, et al. (2007) ID genes mediate tumor reinitiation during breast cancer lung metastasis. Proc Natl Acad Sci U S A 104: 19506–19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, et al. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436: 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Candia P, Solit DB, Giri D, Brogi E, Siegel PM, et al. (2003) Angiogenesis impairment in Id-deficient mice cooperates with an Hsp90 inhibitor to completely suppress HER2/neu-dependent breast tumors. Proc Natl Acad Sci U S A 100: 12337–12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, et al. (1999) Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677. [DOI] [PubMed] [Google Scholar]
- 25. Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, et al. (2010) TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell 18: 655–668. [DOI] [PubMed] [Google Scholar]
- 26. Nam HS, Benezra R (2009) High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell 5: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao D, Nolan DJ, Mellick AS, Bambino K, McDonnell K, et al. (2008) Endothelial progenitor cells control the angiogenic switch in mouse lung metastasis. Science 319: 195–198. [DOI] [PubMed] [Google Scholar]
- 28. Peinado H, Marin F, Cubillo E, Stark HJ, Fusenig N, et al. (2004) Snail and E47 repressors of E-cadherin induce distinct invasive and angiogenic properties in vivo. J Cell Sci 117: 2827–2839. [DOI] [PubMed] [Google Scholar]
- 29. Perez-Moreno MA, Locascio A, Rodrigo I, Dhondt G, Portillo F, et al. (2001) A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J Biol Chem 276: 27424–27431. [DOI] [PubMed] [Google Scholar]
- 30. Jorda M, Vinyals A, Marazuela A, Cubillo E, Olmeda D, et al. (2007) Id-1 is induced in MDCK epithelial cells by activated Erk/MAPK pathway in response to expression of the Snail and E47 transcription factors. Exp Cell Res 313: 2389–2403. [DOI] [PubMed] [Google Scholar]
- 31. Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, et al. (2006) Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res 66: 9543–9556. [DOI] [PubMed] [Google Scholar]
- 32. Sobrado VR, Moreno-Bueno G, Cubillo E, Holt LJ, Nieto MA, et al. (2009) The class I bHLH factors E2-2A and E2-2B regulate EMT. J Cell Sci 122: 1014–1024. [DOI] [PubMed] [Google Scholar]
- 33. Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, et al. (2003) The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci 116: 499–511. [DOI] [PubMed] [Google Scholar]
- 34. Perk J, Gil-Bazo I, Chin Y, de Candia P, Chen JJ, et al. (2006) Reassessment of id1 protein expression in human mammary, prostate, and bladder cancers using a monospecific rabbit monoclonal anti-id1 antibody. Cancer Res 66: 10870–10877. [DOI] [PubMed] [Google Scholar]
- 35. Yang HY, Liu HL, Ke J, Wu H, Zhu H, et al. (2010) Expression and prognostic value of Id protein family in human breast carcinoma. Oncol Rep 23: 321–328. [PubMed] [Google Scholar]
- 36. van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536. [DOI] [PubMed] [Google Scholar]
- 37. Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, et al. (2006) Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541. [DOI] [PubMed] [Google Scholar]
- 38. Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, et al. (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13: 3207–3214. [DOI] [PubMed] [Google Scholar]
- 39. Miller LD, Smeds J, George J, Vega VB, Vergara L, et al. (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A 102: 13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, et al. (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272. [DOI] [PubMed] [Google Scholar]
- 41. Slattery C, McMorrow T, Ryan MP (2006) Overexpression of E2A proteins induces epithelial-mesenchymal transition in human renal proximal tubular epithelial cells suggesting a potential role in renal fibrosis. FEBS Lett 580: 4021–4030. [DOI] [PubMed] [Google Scholar]
- 42. Benezra R (1994) An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell 79: 1057–1067. [DOI] [PubMed] [Google Scholar]
- 43. Johnson SE, Wang X, Hardy S, Taparowsky EJ, Konieczny SF (1996) Casein kinase II increases the transcriptional activities of MRF4 and MyoD independently of their direct phosphorylation. Mol Cell Biol 16: 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lluis F, Ballestar E, Suelves M, Esteller M, Munoz-Canoves P (2005) E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J 24: 974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sloan SR, Shen CP, McCarrick-Walmsley R, Kadesch T (1996) Phosphorylation of E47 as a potential determinant of B-cell-specific activity. Mol Cell Biol 16: 6900–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, et al. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117: 927–939. [DOI] [PubMed] [Google Scholar]
- 47. Casas E, Kim J, Bendesky A, Ohno-Machado L, Wolfe CJ, et al. (2011) Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res 71: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Asirvatham AJ, Carey JP, Chaudhary J (2007) ID1-, ID2-, and ID3-regulated gene expression in E2A positive or negative prostate cancer cells. Prostate 67: 1411–1420. [DOI] [PubMed] [Google Scholar]
- 49. Coppe JP, Itahana Y, Moore DH, Bennington JL, Desprez PY (2004) Id-1 and Id-2 proteins as molecular markers for human prostate cancer progression. Clin Cancer Res 10: 2044–2051. [DOI] [PubMed] [Google Scholar]
- 50. Kim J, Hong SJ, Park JY, Park JH, Yu YS, et al. (2010) Epithelial-mesenchymal transition gene signature to predict clinical outcome of hepatocellular carcinoma. Cancer Sci 101: 1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Patel D, Chaudhary J (2012) Increased expression of bHLH transcription factor E2A (TCF3) in prostate cancer promotes proliferation and confers resistance to doxorubicin induced apoptosis. Biochem Biophys Res Commun 422: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wen YH, Ho A, Patil S, Akram M, Catalano J, Eaton A, et al. (2012) Id4 protein is highly expressed in triple-negative breast carcinomas: possible implications for BRCA1 downregulation. Breast Can Res Treat 135: 93–102. [DOI] [PubMed] [Google Scholar]
- 53. Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, et al. (2007) BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene 26: 2126–2132. [DOI] [PubMed] [Google Scholar]
- 54. Fontemaggi G, Dell’Orso S, Trisciuoglio D, Shay T, Melucci E Francesco F, et al. (2009) The execution of the transcriptional axis mutant p53, E2F1 and ID4 promotes tumor neo-angiogenesis. Nat Str Mol Biol 16: 1086–1093. [DOI] [PubMed] [Google Scholar]
- 55. Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G (2010) ID4: a new player in the cancer arena. Oncotargets 1: 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gyorffy A, Vásárhelyi B, Szöke D, Dietel M, Tulassay T, et al. (2006) Comparative promoter analysis of doxorubicin resistance-associated genes suggests E47 as a key regulatory element. Anticancer Res 26: 2971–2976. [PubMed] [Google Scholar]
- 57. Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, et al. (2008) Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res 68: 989–997. [DOI] [PubMed] [Google Scholar]
- 58. Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, et al. (2009) Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res 69: 4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, et al. (2010) Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci U S A 107: 15449–15454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bergamaschi A, Kim YH, Kwei KA, La Choi Y, Bocanegra M, et al. (2008) CAMK1D amplification implicated in epithelial-mesenchymal transition in basal-like breast cancer. Mol Oncol 2: 327–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, et al. (2009) A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res 69: 5364–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, et al. (2007) Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A 104: 10069–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moreno-Bueno G, Salvador F, Martin A, Floristan A, Cuevas EP, et al. (2011) Lysyl oxidase-like 2 (LOXL2), a new regulator of cell polarity required for metastatic dissemination of basal-like breast carcinomas. EMBO Mol Med 3: 528–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Moreno-Bueno G, Portillo F, Cano A (2008) Transcriptional regulation of cell polarity in EMT and cancer. Oncogene 27: 6958–6969. [DOI] [PubMed] [Google Scholar]
- 65. Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, et al. (2006) Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25: 2273–2284. [DOI] [PubMed] [Google Scholar]
- 66. Chambers AF (2009) MDA-MB-435 and M14 cell lines: identical but not M14 melanoma? Cancer Res 69: 5292–5293. [DOI] [PubMed] [Google Scholar]
- 67. Moreno-Bueno G, Peinado H, Molina P, Olmeda D, Cubillo E, et al. (2009) The morphological and molecular features of the epithelial-to-mesenchymal transition. Nat Protoc 4: 1591–1613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stable expression of EGFP-E47 in MDCK cells triggers a complete EMT. (A) Phase contrast (a–c) and immunofluorescence (d–l) images of the indicated cell lines for EGFP (d), EGFP-E47 (e,f), E-cadherin (g–i) and vimentin (j–l). Nuclei were stained with DAPI (g–l). Bars, 60 mm (a–c); 25 mm (d–l). (B and C). Western blot (B) and Northern blot analysis (C) analyses of the indicated markers in E47 expressing cells and controls. alpha-tubulin and GAPDH were used as loading controls.
(TIF)
E-cadherin promoter activity in A375P (left) and MDA-435S (right) cells transiently cotransfected with 200 ng of the proximal mouse E-cadherin promoter and increasing amounts of Id1 and 10 ng of CMV-beta-gal. RLU is normalized to activity detected in the presence of pcDNA3. Results represent the mean +/- s.d. of three independent experiments performed on triplicate samples.
(TIF)
Association of TCF3 positive expression with basal and luminal markers in N0 breast tumours. The categorical expression of progesterone receptor (PGR), estrogen receptor (ESR1), ERBB2, and basal and luminal markers in the van’t Veer’s dataset (ref. 38) is shown.
(DOC)
Antibodies used in Western-blot, immunofluorescence immunoprecipitation and IHQ assays.
(DOC)