Abstract
The microbial production of ethanol from lignocellulosic biomass is a multi-component process that involves biomass hydrolysis, carbohydrate transport and utilization, and finally, the production of ethanol. Strains of the genus Thermoanaerobacter have been studied for decades due to their innate abilities to produce comparatively high ethanol yields from hemicellulose constituent sugars. However, their inability to hydrolyze cellulose, limits their usefulness in lignocellulosic biofuel production. As such, co-culturing Thermoanaerobacter spp. with cellulolytic organisms is a plausible approach to improving lignocellulose conversion efficiencies and yields of biofuels. To evaluate native lignocellulosic ethanol production capacities relative to competing fermentative end-products, comparative genomic analysis of 11 sequenced Thermoanaerobacter strains, including a de novo genome, Thermoanaerobacter thermohydrosulfuricus WC1, was conducted. Analysis was specifically focused on the genomic potential for each strain to address all aspects of ethanol production mentioned through a consolidated bioprocessing approach. Whole genome functional annotation analysis identified three distinct clades within the genus. The genomes of Clade 1 strains encode the fewest extracellular carbohydrate active enzymes and also show the least diversity in terms of lignocellulose relevant carbohydrate utilization pathways. However, these same strains reportedly are capable of directing a higher proportion of their total carbon flux towards ethanol, rather than non-biofuel end-products, than other Thermoanaerobacter strains. Strains in Clade 2 show the greatest diversity in terms of lignocellulose hydrolysis and utilization, but proportionately produce more non-ethanol end-products than Clade 1 strains. Strains in Clade 3, in which T. thermohydrosulfuricus WC1 is included, show mid-range potential for lignocellulose hydrolysis and utilization, but also exhibit extensive divergence from both Clade 1 and Clade 2 strains in terms of cellular energetics. The potential implications regarding strain selection and suitability for industrial ethanol production through a consolidated bioprocessing co-culturing approach are examined throughout the manuscript.
Introduction
Consolidated bioprocessing (CBP), whereby microbial enzyme production, biomass hydrolysis and substrate conversion to ethanol all occurs in a single bioreactor, offers improved economic feasibility and process efficiencies compared with alternative approaches to lignocellulosic ethanol production [1]–[3]. However, to date, no single organism has been identified that is capable of performing all of these tasks at industrially significant levels [4]. The utilization of designer co-cultures, allowing for potential complementary or synergistic phenotypes between multiple organisms to improve process efficiencies and yields, is an alternative strategy to potentially overcome these limitations (reviewed by Brenner et al. [5] and Zuroff & Curtis [6]).
From a CBP standpoint, the use of thermophilic, anaerobes belonging to the Firmicutes offers many advantages. Growth at elevated temperatures reduces energy costs by avoiding repeated heating and cooling steps associated with cycling between microbial growth and both upstream pre-processing as well as downstream product recovery. Additionally, the native capacity for many strains to produce lignocellulose hydrolytic enzymes in situ may reduce or eliminate the need for enzymatic pre-treatment of biomass. Finally, the ability of multiple strains to ferment a broad range of lignocellulose constituent saccharides into ethanol allows for efficient conversion and utilization of the biomass.
To date, the organisms garnering the most attention for thermophilic CBP include strains in the orders Clostridiales or Thermoanaerobacteriales [7]. One of these organisms, Clostridium thermocellum, has been at the forefront of lignocellulosic ethanol research for decades due to its high cellulolytic capabilities [8]–[10]. However, the inability of C. thermocellum to ferment pentose sugars resulting from hemicellulose hydrolysis reduces potential biomass conversion efficiencies and represents a major limitation in its development as an industrial microorganism. Various strains of the genus Thermoanaerobacter, on the other hand, are known to hydrolyze hemicellulose and ferment hemicellulose constituent sugars [11]–[15], naturally produce [11], [16] and tolerate [13], [17] comparatively high ethanol concentrations, and are amenable to genetic manipulation for purposes of further improving biofuel yields [18], [19]. Furthermore, previous studies have reported that cellulose degradation rates and overall biofuel yields are improved by C. thermocellum-Thermoanaerobacter spp. co-cultures as compared to C. thermocellum mono-cultures [20]–[23]. As such, the use of a Thermoanaerobacter strain as an industrially relevant co-culture partner for C. thermocellum shows great potential as a CBP strategy.
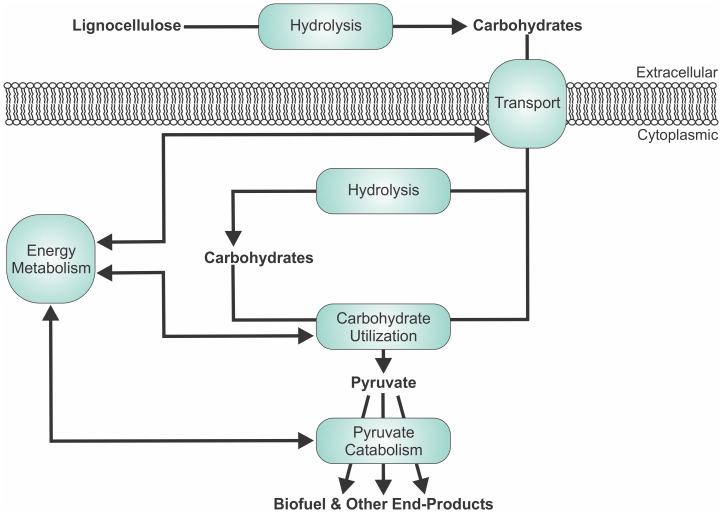
The genomes of multiple Thermoanaerobacter spp. are currently available publicly. The purpose of the current publication is to conduct genus wide comparative genomic analysis of all available genomes, including a de novo genome from Thermoanaerobacter thermohydrosulfuricus WC1 [15], [24], and evaluate the potential of each strain as a C. thermocellum co-culture partner. Factors evaluated include the capacity for lignocellulose hydrolysis, transport of the resulting hydrolysis products, carbohydrate utilization and the potential to produce ethanol relative to other fermentation end-products. Further, given that these processes are interconnected with cellular energy metabolism (Figure 1), the potential mechanisms for energy conservation will also be evaluated.
Figure 1. Schematic representation of key physiological processes pertinent to lignocellulosic ethanol production in a CBP system.
Physiological processes are identified as the text in blue boxes.
Methods
DNA Extraction, Genome Sequencing and Assembly
In lab glycerol stock cultures of T. thermohydrosulfuricus WC1 were revived, grown overnight and plated as previously described [15]. A single colony was picked and used to inoculate fresh ATCC 1191 liquid medium (10 mL) containing 2 g L−1 cellobiose. The resulting culture was allowed to grow overnight and gDNA was extracted using the Wizard® Genomic DNA Purification kit (Promega Corp, Madison, WI).
The genome of T. thermohydrosulfuricus WC1 was first sequenced at the McGill University and Genome Quebec Innovation Centre using shotgun 454 pyrosequencing [25] to get 121,690 reads with an average length of 626 bp. Genome assembly by Newbler v2.6 generated 123 contigs (size >100 bp) with the longest contig 182,175 bp with ∼15× depth coverage. To improve gap closure and perform scaffolding of the contigs, genome re-sequencing was conducted with Illumina HiSeq 2000 paired-end technologies generating 122,042,203 reads of 100 bp from each end with an average insert size of 277 bp. Illumina reads were pre-processed by adaptor clipping, quality trimming with the FASTX-Toolkit [26] and random subset data selection. Multiple assembly pipelines combining both 454 and Illumina data were evaluated including Optimized-Velvet [27], Ray [28] and Newbler v.2.6. Based on statistics metrics, Newbler v2.6 generated the longest contigs with the highest N50 and the resulting assembly was composed of 47 scaffolds plus an additional 15 unscaffolded contigs (size >100 bp with the longest contig 261,431 bp) with ∼56× depth coverage.
Genome Annotation and Proteogenomic Analysis
The assembled DNA scaffolds and contigs were submitted to the Joint Genome Institute’s (JGI) Integrated Microbial Genomes-Expert Review (IMG-ER) platform [29] for gene calling and annotation using their annotation pipeline (http://img.jgi.doe.gov/w/doc/img_er_ann.pdf). The annotated genome was subsequently submitted to the JGI’s GenePRIMP pipeline [30], which reported 358 anomalies. All anomalies were manually curated.
Proteogenomic analysis provided supporting evidence for manual curation decisions regarding reported GenePRIMP anomalies. A mechanism of genome assembly independent proteomics, similar to the approach by Krug et al. [31], was implemented for this analysis. In brief, raw genomic sequencing reads for T. thermohydrosulfuricus WC1, rather than an assembled genome, were transcribed into amino acid sequences in all six possible reading frames with all potential STOP codons being reported as new elements. These elements were subjected to in silico tryptic digestion to create a “naïve” peptide database. This database was used to search a 2D-HPLC-MS/MS experimental output, generated from mid-exponential phase T. thermohydrosulfuricus WC1 cells grown in liquid medium (as described above), using a high-performance GPU-based identification engine developed for this project [32]. Correlating observed peptide retention times against their computed hydrophobicities as described by Krokhin [33] further supported the assignment confidences. As the resultant in silico naïve peptide collection was wholly disconnected from their source proteins, the peptide database served as a validation for the genome assembly and annotation workflows. Observed peptides were compared against the annotation-derived protein database and those unique to the “naïve” database search were reported. Unique peptides (869), corresponding to unannotated protein coding sequences (CDS) within the genome, were used to support or refute GenePRIMP identified annotation anomalies. Modifications to the annotation based upon these peptide data are reported as “notes” within the GenBank file associated with the genome. The genome has been deposited at DDBJ/EMBL/GenBank under accession number AMYG00000000. The version described in this paper is the first version, AMYG01000000.
Comparative Genomic Analysis
Comparative genomic analysis was conducted on genomes and gene annotations available (as of July 2012) using the IMG-ER platform unless specified elsewhere. Genes of interest, with the exception of transporters and carbohydrate active enzymes (CAZymes), were identified within the IMG-ER annotated genomes using independent searches for the Clusters of Orthologous Groups (COG) [34], KEGG Orthology (KO) [35] and TIGRFAMs [36] unique identifiers. Transporters were identified using the assignment criteria given by the Transporter Classification Database (TCDB) [37] as part of the IMG-ER system and substrate specificities were inferred based upon KO annotations of the same genes. The annotation accuracy for all genes identified using the above search methods was manually assessed using a combination of genomic contextual analysis, sequence alignments and through literature and database searches. Gene annotations using additional databases made available through the IMG-ER annotation pipeline including Pfams [38], IMG Terms [29], the SEED [39] and Interpro [40] were used in the manual assessment of COG, KO and TIGRFAM designations when appropriate. Analysis of CAZymes was conducted by accessing the CAZy database [41] for all Thermoanaerobacter genomes available in the database or analysed de novo. For Thermoanaerobacter ethanolicus CCSD1 and Thermoanaerobacter sp. X561 the unfinished draft genomes were downloaded from the NCBI and were analyzed using the standard detection pipeline of the CAZy database. Substrate specificities for all CAZymes were not inferred unless specifically discussed (in Results & Discussion) and were instead limited to substrates reported for each enzyme class by the CAZy database. The subcellular localization of identified CAZymes was predicted by uploading fasta files of all identified gene amino acid sequences into the PSORTb 3.0 database [42] and using the final predictions.
Sequence and Phylogenetic Analysis
Sequence alignments were performed using BioEdit v.7.0.9.0 [43]. Phylogenetic analysis of individual sequences was performed using MEGA 4 [44]. Phylogeny was inferred using the neighbor-joining method with evolutionary distances calculated via the Poisson correction as described elsewhere [45]. Alignment gaps were deleted via pair wise sequence comparisons only and clusters grouped using the bootstrapping method (10,000 replicates).
Results and Discussion
Genome Properties of T. thermohydrosulfuricus WC1
The draft genome sequence of T. thermohydrosulfuricus WC1 is comprised of 2, 573, 514 bp and shows a G+C content of 34.35%, which is consistent with other sequenced Thermoanaerobacter strains (Table S1). There are 2,655 genes annotated with 2,552 predicted to be CDS. The chaperonin-60 universal target (cpn60 UT) nucleotide sequence, which has been shown to be a more reliable phylogenetic indicator than 16S rRNA encoding DNA for the Thermoanaerobacter genus [24], agrees 100% with the previously published sequence [15]. Based on the standards for genomic sequencing projects [46], the T. thermohydrosulfuricus WC1 permanent draft-genome belongs to the “Annotation-Directed Improvement” classification.
Whole Genome Comparative Analysis
The genomes of 11 Thermoanaerobacter strains (10 publicly available as of July 2012+ T. thermohydrosulfuricus WC1) were included in our genus wide comparison. The Thermoanaerobacter tengcongensis MB4 genome [47], corresponding to GenBank accession number: AE008691 was not included based upon the reclassification of the strain into the genus Caldanaerobacter [48]. The available genome sequences for the genus, in draft, permanent draft and finished states, range in size from 2.20 Mb –2.78 Mb and are annotated to contain anywhere from 2286–2800 CDS (Table S1).
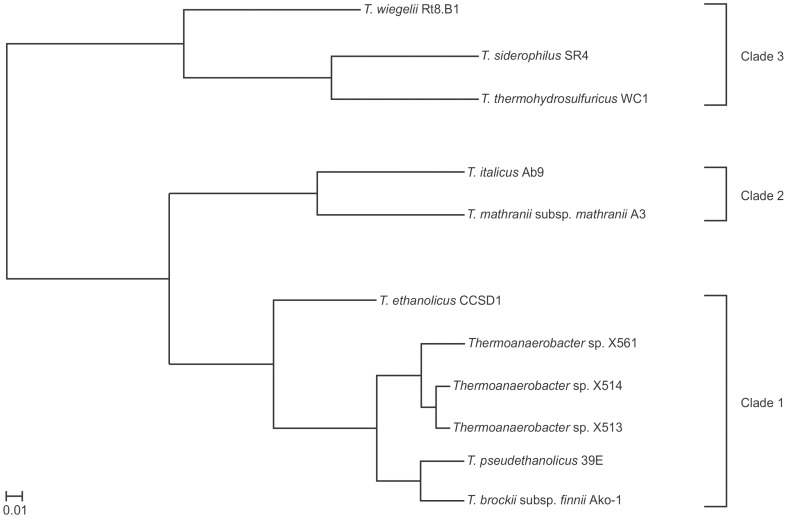
To determine the extent that similar functional profiles exist between strains, hierarchical clustering, based upon COG, KO and TIGRFAM qualifiers, was conducted independently for each qualifier. As shown in Figure 2, three distinct clades exist within the genus. Clade 1 contains the most genomes and could also potentially be divided into sub-clades. However, the study by Verbeke et al. [24] determined that all strains in Clade 1 share an average nucleotide identity (ANIm) score [49] greater than 0.97 and thus, may represent strains of the same species. Previous work by Hemme et al. [23] with Thermoanaerobacter pseudethanolicus 39E (formerly called Clostridium thermohydrosulfuricum 39E [16] and Thermoanaerobacter ethanolicus 39E [50]) and Thermoanaerobacter sp. X514, have shown that genomes with ANIm scores greater than 0.97 may have significant differences that impact relative strain suitability for lignocellulosic ethanol production. Thus, inter-clade functional divergence may have an even more drastic impact on co-culturing potential than intra-clade divergence. Specifics regarding strain suitability are discussed below.
Figure 2. Phylogram of annotated COG functional profiles for sequenced Thermoanaerobacter strains.
Cluster analysis, based on Cluster 3.0 analysis software [120], was conducted within the IMG-ER platform using the COG profiles for each genome. Branch lengths correspond to calculated distances between functional profiles. Similar clade architectures are observed when using KO or TIGRFAM descriptors (not shown).
CAZyme Analysis
The availability of fermentable sugars from lignocellulosic biomass is dependent on CAZymes capable of degrading the insoluble extracellular carbohydrate polymers (Figure 1). In designer co-cultures, the multiple hemicellulase enzymes in the C. thermocellum cellulosome may help to liberate hemicellulose constituent sugars from the polymeric backbone. The liberation of such saccharides, most notably pentoses, but also a mixture of hexoses, may subsequently be fermented to ethanol by a Thermoanaerobacter strain. However, expression analysis studies of C. thermocellum grown on various substrates by Raman et al. [10] have shown that xylanase expression levels were at their lowest when grown on a hemicellulose containing substrate (switchgrass) in comparison to growth on cellobiose or cellulose. Thus, under CBP conditions, the utilization of a Thermoanaerobacter strain which has xylan hydrolysis capabilities may help facilitate biomass hydrolysis similar to what has been reported in co-cultures of C. thermocellum and Caldicellulosiruptor bescii grown on switchgrass or cellulose+xylan [51].
To examine the potential that a Thermoanaerobacter strain may contribute to biomass hydrolysis, all CAZyme genes were identified within the genus. As shown in Table S2, between 45–61 independent CAZyme gene sequences, belonging to 27–40 distinct CAZyme classes, were identified within the sequenced strains.
A small subset (GH23 and various glycosyltransferases) of these genes encode proteins typically associated with cellular maintenance functions such as peptidoglycan processing and glycogen synthesis. Furthermore, based on the PSORTB 3.0 predictions, only a small fraction of each CAZyome is predicted to be localized extracellularly (Table 1) such that only a few genes per strain would be capable of contributing to the extracellular hydrolysis of complex polymers.
Table 1. Predicted extracellular CAZymes involved with lignocellulosic biomass hydrolysis within sequenced Thermoanaerobacter spp.
| Clade 1 | Clade 2 | Clade 3 | |||||||||
| CAZyme Designation&ModularStructure | T. brockiisubsp.finniiAko-1 | T. ethanolicusCCSD1 | T. pseudethanolicus39E | Thermoanaerobacter sp. X513 | Thermoanaerobacter sp. X514 | Thermoanaerobacter sp. X561 | T. italicusAb9 | T. mathraniisubsp.MathraniiA3 | T. siderophilusSR4 | T. thermohydrosulfuricusWC1 | T. wiegeliiRt8.B1 |
| GH10 | Thit_0188 | Tmath_0247 | |||||||||
| GH43 | Tmath_1695 | ||||||||||
| GH52 | Thit_0190 | Tmath_0249 | TthWC1_1012 | ||||||||
| CE4 | ThesiDRAFT1_1967 | TthWC1_1808 | Thewi_00017610 | ||||||||
| PL9 | Thit_1727 | ||||||||||
| GH66-CBM35-CBM35-GH15 | ThesiDRAFT1_0902 | TthWC1_0529 | |||||||||
| CBM22-CBM22-GH10-CBM9-CBM9-SLH-SLH-SLH | Thit_0192 | Tmath_0251 | TthWC1_1010 | ||||||||
Extracellular, lignocellulose hydrolyzing CAZymes
As is shown in Table 1, no predicted extracellular lignocellulose hydrolyzing CAZymes were identified in Clade 1 strains. Moreover, only 3 strains, T. italicus Ab9, T. mathranii subsp. mathranii A3 and T. thermohydrosulfuricus WC1, possess extracellular endo-xylanases capable of hydrolyzing the xylan backbone of many hemicelluloses. All three strains contain a secretable multi-component CDS with the modular structure CBM22-CBM22-GH10-CBM9-CBM9-SLH-SLH-SLH. To date, proteins belonging to the GH10 family are only known to act on xylan, while CBM22 and CBM9 modules are considered to be primarily xylan binding. All orthologs show high amino acid sequence similarity (>74%) and similar modular structure to the functionally characterized xynA gene in Thermoanaerobacterium saccharolyticum NTOU1 [52].
T. italicus Ab9 (Thit_0188) and T. mathranii subsp. mathranii A3 (Tmath_0247) also contain an additional extracellular GH10 family gene showing high amino acid sequence similarity (>77%) with the cloned and characterized xylanase/β-xylosidase from Caldicellulosiruptor saccharolyticus DSM 8903 [53].
The hydrolysis products resulting from GH10 mediated endo-xylanase activity would be xylo-oligomers. The generation of xylose monomers from xylo-oligomers requires extracellular and/or intracellular (see below) β-xylosidase activity. The T. italicus Ab9, T. mathranii subsp. mathranii A3 and T. thermohydrosulfuricus WC1 genomes also encode for an extracellular GH52 family enzyme (Table 1, Table S2), which, to date, are solely reported to have β-xylosidase activity. Additionally, unique to T. mathranii subsp. mathranii A3 is a putatively cell bound GH43 enzyme, which may have further pentose releasing hydrolysis capabilities.
The identification that only three of the strains evaluated contain GH10 family enzymes is consistent with the reports that only T. italicus Ab9 [12], T. mathranii subsp. mathranii A3 [13] and T. thermohydrosulfuricus WC1 (our lab, unpublished results) are capable of growing on xylan polymers. Other sequenced strains lacking extracellular xylanolytic enzymes, specifically T. siderophilus SR4 [54], Thermoanaerobacter sp. X514 [23] and T. pseudethanolicus 39E [20], have been specifically reported to not grow on xylan. The ability to use, or not use, xylan by the remaining Thermoanaerobacter strains has not yet been reported.
T. italicus Ab9 has the sole genome containing a pectate lyase [12], while genomes of T. thermohydrosulfuricus WC1 and T. wiegelii Rt8.B1 are the only two that encode a putative extracellular acetyl-xylan esterase for removal of acetyl-groups from xylan polymers. A final unique extracellular enzyme found in T. thermohydrosulfuricus WC1 and T. siderophilus SR4 (TthWC1_0529; ThesiDRAFT1_0902) has a modular structure of GH66-CBM35-CBM35-GH15. No database (GenBank – non redundant protein) entry yet is homologous over the entire length of these CDS, as <60% query coverage is observed when comparing all database entries with either the TthWC1_0529 or ThesiDRAFT1_0902 queries. Reported activities associated with GH66 and GH15 family proteins are reported to act on α-glucan linkages, while CBM35 modules reportedly bind β-linkages (xylans, mannans, galactans), but the functional role of these enzymes is unknown.
Intracellular, characterized lignocellulose hydrolyzing CAZymes
Only a few glycoside hydrolases with lignocellulose hydrolysis potential have been functionally characterized in any Thermoanaerobacter strain and these are all predicted to be localized intracellularly. One of these, a GH3 family protein xarB from T. ethanolicus JW200, was initially cloned and characterized and was reported to cleave β-1,4-xylobiose linkages as well as remove arabinosyl groups from xylo-oligomers [55]. Orthologous sequences, showing conserved genomic organization to T. ethanolicus JW200, are found in T. italicus Ab9 (Thit_0198), T. mathranii subsp. mathranii A3 (Tmath_0257) and T. thermohydrosulfuricus WC1 (TthWC1_1005) (also see Figure 6 in [23]). Alternatively, a near identical, though N-terminally truncated xarB ortholog, xglS, was characterized in T. brockii subsp. brockii HTD4 [56] and also shown to have β-xylosidase activity. Similarly to T. brockii subsp. brockii HTD4, the xglS orthologs in T. brockii subsp. finnii Ako-1 (Thebr_2099) and T. pseudethanolicus 39E (Teth39_2055) sequences are co-localized with a cglT (GH1) ortholog (Thebr_2098; Teth39_2054) immediately downstream. The xglS ortholog in T. wiegelii Rt8.B1 (Thewi_00009950) is separated from its cglT ortholog (Thewi_00009970) by a hypothetical protein, while the GH3 enzymes of Thermoanaerobacter spp. X513, X514 and X561, as well as T. siderophilus SR4 do not show conserved genomic organization to the experimentally characterized xarB or xglS sequences.
The present analysis indicates that none of the GH3 CDS are predicted to be secreted extracellularly is in agreement with the findings for T. thermohydrosulfuricus JW 102 [55]. The predicted cytosolic location of this enzyme suggests that xylo-oligosaccharides, and not just xylose, are capable of being transported into the cell. The presence of both extracellular GH52 and cytosolic GH3 β-xylosidase orthologs in the T. italicus Ab9, T. mathranii subsp. mathranii A3 and T. thermohydrosulfuricus WC1 genomes is an interesting redundancy regarding xylan/xylo-dextrin hydrolysis patterns within these organisms, though the potential impact it may have on xylan hydrolysis and utilization in raw substrates is unknown.
The study by Breves et al. [56] also characterized a GH1 family protein, designated cglT, which is also predicted to be localized intracellularly. In T. brockii subsp. brockii HTD4, cglT is capable of hydrolyzing β-1,4-glucosidic linkages (up to cellopentaose tested) as well as the β-1,3- and β-1,2-glucosidic linkages of laminaribose and sophorose respectively and is co-localized with xglS (see above). Only three sequenced strains, T. brockii subsp. finnii Ako-1, T. pseudethanolicus 39E and T. wiegelii Rt8.B1 share similar genomic organization. GH1 domain containing sequences, highly similar to the cglT gene in T. brockii subsp. brockii HTD4, can be identified in all other Thermoanaerobacter strains, but are not co-localized with xglS and thus, may simply represent cglT homologs, rather than orthologs.
The release of fermentable sugars from lignocellulosic biomass is often considered the rate limiting step in simultaneous saccharification and fermentation processes [57]–[59]. Enzymatic pre-treatment of biomass is a common strategy for improving lignocellulose saccharification [60], though the accumulation of soluble sugars generated through hydrolysis are well known to have an inhibitory effect on continued enzymatic activity [61], [62]. Simultaneous incubation of exogenous enzymes and sugar fermenting bacteria is one strategy to relieve this inhibition and continue driving hydrolysis. However, Podkaminer et al. [63] have recently shown that in some cases, exogenous enzyme activity is reduced when incubated under conditions amenable for bacterial growth. Thus, using the hydrolytic machinery native to ethanol producing strains is favorable in terms of maintaining enzymatic activity and limiting costs associated with exogenous enzyme addition. The present analysis of the Thermoanaerobacter spp. genomes has identified that only T. italicus Ab9, T. mathranii subsp. mathranii A3 and T. thermohydrosulfuricus WC1 possess the potential enzymatic machinery needed to hydrolyze complex xylan polymers and potentially facilitate lignocellulose hydrolysis under CBP conditions.
Carbohydrate Transport
The extracellular hydrolysis of insoluble carbohydrate polymers requires that the resulting soluble saccharides be transported into the cell prior to fermentation (Figure 1). Based upon genome annotations, Thermoanaerobacter spp. import carbohydrates via ABC-type transporters, phosphotransferase system (PTS) transporters and via cationic symporters (Table 2). Of the three annotated systems, ABC-type transporters, are the most abundant as Thermoanaerobacter spp. contain anywhere from 123–150 total genes belonging to the ATP-binding Cassette (ABC) Superfamily (TC:3.A.1) based upon TCDB designations. Only a subset of these will be involved with carbohydrate import, and to date, only a few of this subset have been characterized within any strain of the genus.
Table 2. Selected1 transporters associated with carbohydrate import identified within sequenced Thermoanaerobacter spp.
| Clade 1 | Clade 2 | Clade 3 | |||||||||
| T. brockii subsp. finnii Ako-1 | T. ethanolicus CCSD1 | T. pseudethanolicus 39E | Thermoanaerobacter sp. X513 | Thermoanaerobacter sp. X514 | Thermoanaerobacter sp. X561 | T. italicus Ab9 | T. mathranii subsp. mathranii A3 | T. siderophilus SR4 | T. thermohydrosulfuricus WC1 | T. wiegelii Rt8.B1 | |
| Cationic Symporters | |||||||||||
| 2-Keto-3-Deoxygluconate | + | + | + | − | − | − | − | − | − | − | − |
| Glycoside/Pentoside/Hexuronide Cation Symporter | + | + | + | + | + | + | + | + | + | + | + |
| ATP Binding Family | |||||||||||
| Lactose/L-arabinose | − | − | − | − | − | − | + | + | + | + | + |
| Maltose/Maltodextrin | + | + | + | + | + | + | + | + | + | + | + |
| Methyl-Galactoside | − | + | − | − | − | − | − | − | − | − | − |
| Multiple Sugar (unspecified) | + | + | + | + | + | + | + | + | + | + | + |
| Oligogalacturonide | − | − | − | − | − | − | + | − | + | − | − |
| Putative Multiple Sugar (unspecified) | − | − | − | + | + | + | − | − | + | + | + |
| Putative Sugar (unspecified) | − | − | − | − | − | − | + | − | − | − | − |
| Ribose | + | + | + | + | + | + | + | + | − | + | + |
| Simple Sugar (unspecified) | + | + | + | + | + | + | + | + | + | + | + |
| Xylose | − | + | − | + | + | + | + | + | + | + | + |
| PTS Family | |||||||||||
| Cellobiose | + | + | + | + | + | + | + | + | + | + | + |
| Fructose | + | + | + | + | + | + | + | − | + | + | + |
| Galactitol | + | + | + | + | + | + | + | + | + | + | + |
| Galactosamine | − | − | − | − | − | − | − | − | + | + | + |
| Glucitol/Sorbitol | − | − | − | − | − | − | − | − | + | + | + |
| Mannitol | + | + | + | + | + | + | + | + | + | + | + |
| Mannose | + | + | + | + | + | + | + | + | + | + | + |
| N-Acetylglucosamine | + | + | + | + | + | + | + | + | + | + | + |
| Sucrose | − | − | − | + | + | + | − | − | + | + | − |
Symbols denote the presence (+) or absence (−) of a particular annotated transporter within each genome. Substrate specificity is inferred based upon KO annotated specificity of the substrate binding protein (ABC transporters) or the membrane linked EIIC components (PTS transporters).
Transport systems presented are limited to complexes showing co-localization of all genes needed to form a functional complex. Complexes lacking annotation of a single component are not included. Redundancy in transport systems exists, but is not identified.
ABC-type transporters
Despite the presence of annotated poly- and oligosaccharide transporters within Thermoanaerobacter genomes (Table 2), few studies have investigated the ability of Thermoanaerobacter strains to utilize these saccharides. Working with Thermoanaerobacter strains not analyzed here, the study by Wiegel et al. [64] noted strain specific differences in xylo-oligomer utilization patterns and rates, yet to date, no correlation between genome content and poly- or oligosaccharide utilization has been established for any strain within the genus. The prediction that most Thermoanaerobacter CAZymes are intracellular also suggests that these strains have the innate capacity to transport complex soluble carbohydrates as there is little evolutionary advantage to maintain intracellular CAZymes if the saccharides upon which they act cannot be transported into the cell. However, experimental characterization of Thermoanaerobacter transport systems is needed. This is particularly true given that the mean degree of polymerization for cellulose hydrolysis products generated by C. thermocellum is four [65], and that C. thermocellum xylan hydrolysis yields principally xylo-oligomers [66]–[69]. The ability, or lack thereof, to transport, poly- and oligosaccharides in a Thermoanaerobacter strain may significantly impact carbohydrate utilization in a co-culture with C. thermocellum.
Xylose transport has been one of the most thoroughly investigated sugar import mechanisms within the genus and an ABC-type xylose binding protein was first identified in T. pseudethanolicus 39E by Erbeznik et al. [70] and a xylFGH operon, coding for the entire ABC-type transporter, was later determined in the same strain [71]. However, the amino acid sequences reported do not agree with those identified in the T. pseudethanolicus 39E genome. Nevertheless, the reported partial xylF sequence and complete xylGH sequences reported do show 99.5%, 100% and 100% sequence identity, respectively, and similar genomic architecture, with the annotated xylose ABC-transport system in T. italicus Ab9.
In T. italicus Ab9, as well as T. mathranii subsp. mathranii A3, T. thermohydrosulfuricus WC1 and T. wiegelii Rt8.B1, the annotated xylose transport genes are not co-localized with the xylose isomerase and xylulokinase (xylAB) operon. They are however, co-localized in Thermoanaerobacter spp. X513, X514 and X561, T. ethanolicus CCSD1 and T. siderophilus SR4 as is reported elsewhere [23]. Thermoanaerobacter spp. X513, X514, and X561, as well as T. siderophilus SR4, all contain a secondary gene cluster with an annotated xylose binding protein located further downstream. Recently, Lin et al. [72] report that in Thermoanaerobacter sp. X514, the xylose transport genes found co-localized with the xylAB operon are induced when grown on xylose giving support to the annotation of these genes as xylose transporters. The response of the secondary gene cluster was not discussed and thus, its role in xylose transport is not yet confirmed. The presence of xylose specific ABC-transport systems in some, but not all, Thermoanaerobacter strains (Table 2) has been proposed to potentially account for increased xylose uptake and utilization [23]. Assuming this translates throughout the genus, only two strains, T. brockii subsp. finnii Ako-1 and T. pseudethanolicus 39E, may be limited in xylose uptake efficiency based on their annotated genomes, as they may rely on non-specific mechanisms for xylose transport.
The transport of cellobiose and/or glucose has also been proposed to occur via an ABC-type transport system designated cglF-cglG(xarG) with the substrate specificity inferred based upon its proximal location to a glycoside hydrolase (cglT – discussed above) showing high activity towards cellobiose in Thermoanaerobacter brockii subsp. brockii HTD4 [56]. However, Mai et al. [55] note that in T. ethanolicus JW200 the orthologous gene is co-localized with a bifunctional xylosidase-arabinosidase (xarB/xglS) and not with a cglT ortholog. Thus, it may be involved with transporting xylan hydrolysis products. Only six Thermoanaerobacter genomes, T. brockii subsp. finnii Ako-1, T. pseudethanolicus 39E, T. italicus Ab9, T. mathranii subsp. mathranii A3, T. thermohydrosulfuricus WC1 and T. wiegelii Rt8.B1, contain the cglF-cglG/xarG gene cluster and in all cases, show similar genomic organization to T. ethanolicus JW200.
PTS-mediated transport
Genes annotated to transport nine different substrates via PTS-mediated mechanisms are identified within the genus (Table 2), though none of these gene sequences have been functionally characterized. Alternative to the ABC-dependent glucose uptake proposed (above), Lin et al. [72] have identified genes in Thermoanaerobacter sp. X514 which may form a glucose specific PTS complex. Expression analysis of the gene cluster identified (Teth514_0412-Teth514_0414) was specifically linked to mid-exponential phase growth of cells grown on glucose only. However, the KO annotation (KO:K02803/KO:K02804) suggests that this gene cluster is specific for N-Acetyl-glucosamine (GlcNAc) rather than glucose. The KO annotation is further supported by the fact that the phosphotransferase specificity of the orthologous EIICB gene in Caldanaerobacter subterraneus subsp tengcongensis MB4 (87.7% shared amino acid identity with Thermoanaerobacter sp. X514) has a Vmax 4-fold higher for GlcNAc than it does for glucose [73]. Additionally, the studies by Ng and Zeikus [74] on T. pseudethanolicus 39E and Cook et al. [75] on T. wiegelii Rt8.B1 have identified that glucose import is via non-PTS-mediated transporters. Thus, the genes responsible for glucose uptake in any Thermoanaerobacter strain are not yet confirmed.
Cationic symporters
Only two distinct carbohydrate-relevant types of cationic symporters are annotated within the genus (Table 2). The study by Hemme et al. [23] has proposed that Na+ gradient-linked transport may be a mechanism for xylose uptake in T. pseudethanolicus 39E, which lacks an annotated xylose specific ABC-type transport system (above). While the substrate binding specificity of the annotated cationic symporters is not yet known, experimental characterization of these enzymes may shed valuable insights into Thermoanaerobacter spp. carbon transport, particularly in strains lacking annotated carbon-specific transport systems.
Carbohydrate Utilization
While cellulose is comparatively chemically homogenous (β-1,4-glucose linkages), hemicellulose fractions are chemically and structurally diverse. Thus, the products of lignocellulose hydrolysis will generate a mixed pool of saccharides available for utilization. While sugar composition ratios and linkages vary from hemicellulose to hemicellulose, the principal simple saccharides of hemicellulose are xylose, arabinose, mannose, glucose, galactose and glucuronic acid [76]–[78]. As the conversion of hemicellulose to ethanol plays an important role in making lignocellulosic biofuels economically feasible [79], [80], identifying a strain capable of fermenting multiple sugars, particularly multiple sugars simultaneously, into ethanol is essential.
Utilization of hexose sugars
The potential for all sequenced strains within the genus to utilize the seven primary constituent sugars of lignocellulose was evaluated (Table 3, Table S3). All strains contain a complete Embden-Meyerhoff-Parnas (EMP) pathway. Redundancy in genome annotations for the EMP pathway were only observed for genes encoding glucokinase, 6-phopshofructokinase, fructose-1,6-bisphosphate aldolase and phosphoglyceromutase (Table S3).
Table 3. Identification of the genomic potential for sequenced Thermoanaerobacter strains to utilize the major carbohydrate hydrolysis products of lignocellulose degradation.
| Clade 1 | Clade 2 | Clade 3 | |||||||||||
| T. brockii subsp. finnii Ako-1 | T. ethanolicus CCSD1 | T. pseudethanolicus 39E | Thermoanaerobacter sp. X513 | Thermoanaerobacter sp. X514 | Thermoanaerobacter sp. X561 | T. italicus Ab9 | T. mathranii subsp. mathranii A3 | T. siderophilus SR4 | T. thermohydrosulfuricus WC1 | T. wiegelii Rt8.B1 | |||
| Cellulose Hydrolysis Products1 | Glucose | Genes Present | + | + | + | + | + | + | + | + | + | + | + |
| Reported Phenotype | + | NR | + | + | + | + | + | + | + | + | + | ||
| Cellobiose | Genes Present | + | + | + | + | + | + | + | + | + | + | + | |
| Reported Phenotype | + | NR | + | NR | NR | NR | + | + | + | + | + | ||
| Hemicellulose Hydrolysis Products | Arabinose | Genes Present | − | − | − | − | − | − | + | + | − | − | − |
| Reported Phenotype | − | NR | – | NR | NR | NR | + | + | − | − | − | ||
| Galactose | Genes Present | + | + | + | + | + | + | + | + | + | + | + | |
| Reported Phenotype | + | NR | + | NR | NR | NR | + | − | NR | + | + | ||
| Mannose | Genes Present | + | + | + | + | + | + | + | + | + | + | + | |
| Reported Phenotype | + | NR | + | NR | NR | NR | + | + | NR | + | + | ||
| Xylose | Genes Present | + | + | + | + | + | + | + | + | + | + | + | |
| Reported Phenotype | + | NR | + | + | + | + | + | + | + | + | + | ||
| Glucuronic Acid | Genes Present | − | − | − | − | − | − | + | + | − | + | − | |
| Reported Phenotype | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR | ||
| Reference | [83] | −2 | [14] | [85] | [85] | [85] | [12] | [13] | [54] | [15] | [84] | ||
Symbols denote the presence (+) or absence (–) of all enzymes needed for hydrolysis to pyruvate or whether the substrate has been reported to be used (+) or not used (−) in the literature. NR denotes substrate utilization has not yet been reported for a particular strain.
Cellulose hydrolysis products have been limited to glucose and cellobiose and not higher order cellodextrins.
Physiological data has not yet been reported for this strain.
Genes for a complete Entner-Doudoroff pathway could not be identified in agreement with previous findings [23]. Additionally, a 6-phosphogluconolactonase encoding gene, potentially allowing for hexose utilization through the oxidative pentose phosphate pathway, was not identified in any of the genomes. This pathway has been shown to be non-functional in Thermoanaerobacter sp. X514 [81]. However, genes encoding enzymes common to both the Entner-Duodoroff and pentose phosphate pathways are identified within the genomes as they may serve as entry points into the EMP pathway for the catabolism of specific sugars.
All of the sequenced genomes contain a mannose-6-P isomerase responsible for converting mannose-6-P to fructose-6-P and needed for mannose utilization. Additionally, a conserved 5 gene cluster needed for conversion of galactose to glucose-6-P via the Leloir pathway was identified in all of the genomes. This cluster is orthologous to the novel gal operon characterized in Ca. subterraneus subsp. tengcongensis MB4 [82].
The entry points into the EMP pathway for the products of cellobiose hydrolysis are somewhat difficult to predict and would largely be dependent on the mode of transport into the cell (see above), which is not yet known. Transport via an ABC-type system would import cellobiose, while transport via a PTS transporter would yield cellobiose-6-P. In the former situation, cellobiose could be hydrolyzed to two glucose molecules or, if hydrolyzed using a cellobiose phosphorylase (Table S3), could yield 1 glucose +1 glucose-1-P using an inorganic phosphate for phosphorylation. If cellobiose import is via a PTS transporter, hydrolysis would yield 1 glucose +1 glucose-6-P and could occur via any of the GH1, GH3, GH4 or GH5 enzymes identified in all Thermoanaerobacter strains (Table S2, Table S3). To date, only the cglT gene characterized in T. brockii subsp. brockii HTD4, which has homologs in all other Thermoanaerobacter genomes (see above), has been shown to have β-glucosidase (including cellobiose) activity [56]. Transformation from glucose-1-P to glucose-6-P could occur via a phosphoglucomutase (COG0637; KO:K01838) found in all genomes.
Genes for glucuronic acid metabolism are not universally conserved throughout the genus. Only the Clade 2 strains, plus T. thermohydrosulfuricus WC1, have the necessary genes for conversion of glucuronic acid to glyceraldehyde-3-P+pyruvate in conserved 5-gene clusters. Multiple genes are annotated to encode for a 2-keto-3-deoxyphosphogluconate (KDPG) aldolase (COG0800; KO:K01625; TIGR01182) in all Thermoanaerobacter strains, though. This enzyme, which is common to the Entner-Doudoroff pathway, also serves as the entry point in glucuronic acid utilization into the EMP pathway. However, with the exception of the strains mentioned above, the necessary genes for glucuronic acid utilization are not identified and thus the role of these annotated KDPG aldolases is difficult to infer.
Utilization of pentose sugars
The present analysis confirms that in all strains, xylose is likely isomerized and phosphorylated via xylose-isomerase (xylA) and xylulose-kinase (xylB) reactions prior to entering the pentose phosphate pathway, in agreement with previous findings [23]. Arabinose utilization genes seem to be limited to Clade 2 strains. Both T. italicus Ab9 and T. mathranii subsp. mathranii A3 have a conserved 3-gene cluster (Table S3) annotated as L-arabinose-isomerase, L-ribulokinase and L-ribulose-5-P-4-epimerase needed to convert L-arabinose to D-xylulose-5-P prior to entering the pentose phosphate pathway.
Of the 77 in silico predictions for carbohydrate utilization (11 genomes x 7 carbohydrates), 45 agree with phenotypes reported in the literature [12]–[15], [54], [83]–[85] (Table 3). Thirty-one of the strain-substrate combinations have not yet been investigated experimentally to either confirm or refute these predictions and one prediction (galactose utilization by T. mathranii subsp. mathranii A3) disagrees with reported phenotypes [13]. T. mathranii subsp. mathranii A3 has orthologs to the functionally characterized gal operon (mentioned above) in Ca. subterraneus subsp. tengcongensis MB4. The arrangement of the genes is identical to Ca. subterraneus subsp. tengcongensis MB4 and the annotated genes share >89% amino acid sequence similarity to each respective ortholog. Thus, the reason galactose utilization was not observed by Larsen and coworkers [13] may be due to regulatory differences, a few select mutations affecting enzyme functionality or even an inability to transport galactose, but the exact reason is not clear at this time.
Only Clade 2 strains have the potential to utilize all of the major lignocellulose hydrolysis products (Table 3). However, the significance of this in CBP terms may vary dependent on the nature of the lignocellulosic feedstock. As the composition of hemicellulose varies between feedstocks, the inability to utilize substrates in low abundance may represent acceptable losses for any single CBP system. Alternatively, using a strain with diverse substrate utilization capabilities affords flexibility in designing a CBP system, independent of the nature of the biomass feedstock, not present in strains lacking the ability to utilize specific hemicellulose-relevant saccharides.
Pyruvate Catabolism and End-product Synthesis
Pyruvate catabolism
Fermentation of the above mentioned carbohydrates leads to the formation of pyruvate (Figure 1), a key branch point in Thermoanaerobacter carbohydrate metabolism. All Thermoanaerobacter strains are reported to have branched catabolic pathways from pyruvate, which yield both ethanol and non-ethanol end-products in varying end-product ratios (Table 4). As such, understanding pyruvate catabolism, and identifying mechanisms to maximize carbon flow towards ethanol, is an important component in making Thermoanaerobacter strains industrially relevant.
Table 4. Reported end-product yields and related growth conditions for sequenced Thermoanaerobacter strains1 grown on glucose, xylose or cellobiose.
| End Products (mol/mol hexose equivalent) | Culture Conditions | |||||||||
| Clade | Strain | H2 | CO2 | Acetate | Ethanol | Lactate | Substrate | Concentration | Type | Reference |
| 1 | T. brockii subsp. finnii Ako-1 | 0.25 | 1.66 | 0.26 | 1.66 | 0.38 | Glucose | 5 g L−1 | B | [121] |
| T. pseudethanolicus 39E | NI | NQ | 0.38 | 1.11 | 0.30 | Glucose | NR | B | [23] | |
| NI | NQ | 0.29 | 1.52 | 0.10 | Xylose | 2 g L−1 | B | [23] | ||
| 0.45 | 1.72 | 0.10 | 1.41 | 0.15 | Glucose | 10 g L−1 | B | [90] | ||
| NI | 1.63 | 0.08 | 1.55 | 0.23 | Glucose | 4 g L−1 | B | [122] | ||
| NI | NI | 0.24 | 1.25 | 0.32 | Xylose | 5 g L−1 | B | [123] | ||
| NI | NI | 0.17 | 1.10 | 0.35 | Glucose | 5 g L−1 | B | [123] | ||
| Thermoanaerobacter sp. X513 | NI | NI | NI | NQ | NI | Xylose | 1.5 g L−1 | B | [85] | |
| Thermoanaerobacter sp. X514 | NI | NQ | 0.51 | 1.02 | 0.29 | Glucose | NR | B | [23] | |
| NI | NQ | 0.25 | 1.55 | 0.12 | Xylose | 2 g L−1 | B | [23] | ||
| NI | NQ | 0.33 | 1.25 | 0.17 | Glucose | 2.1 g L−1 | B | [81] 2 | ||
| Thermoanaerobacter sp. X561 | NI | NI | NI | NQ | NI | Xylose | 1.5 g L−1 | B | [85] | |
| 2 | T. italicus Ab9 | NQ | NC | NC | NC | NC | Glucose | 5 g L−1 | B | [12] 3 |
| T. mathranii subsp. mathranii A3 | 1.08 | 2.17 | 0.48 | 1.32 | 0.07 | Xylose | 2 g L−1 | B | [13] | |
| 3 | T. siderophilus SR4 | NQ | NQ | NI | NQ | NQ | Glucose | 5 g L−1 | B | [54] |
| T. thermohydrosulfuricus WC1 | 0.56 | 1.19 | 0.71 | 0.61 | 0.33 | Cellobiose | 2 g L−1 | B | [15] | |
| T. wiegelii Rt8.B1 | NI | NI | 0.50 | 0.88 | 0.17 | Glucose | 8.6 g L−1 | B | [109] 2 | |
| NI | 1.60 | 0.50 | 1.10 | 0.04 | Glucose | 9 g L−1 | P | [124] 4 | ||
| NI | 1.84 | 0.53 | 1.32 | 0.06 | Xylose | 7.5 g L−1 | P | [124] 4 | ||
| NI | 1.68 | 0.61 | 1.06 | ND | Cellobiose | 17 g L−1 | P | [124] 4 | ||
Abbreviations: NI = end-product not investigated; ND = end-product investigated, but not detected; NQ = end-product detected, but not quantified; NC = end-product quantified, but ratio not calculable due to missing substrate consumption data; B = batch culture; P = pH-controlled batch culture.
T. ethanolicus CCSD1 omitted as no physiological data is available.
End product and substrate utilization data used for calculations approximated from graphical data.
Succinate production reported as a minor product.
Propionate detected on xylose grown cells (molar ratio = 0.12) and cellobiose grown cells (molar ratio = 0.06).
Pyruvate decarboxylation in all Thermoanaerobacter strains, forming acetyl-CoA+CO2+ reducing equivalents, appears to proceed through the use of pyruvate:ferredoxin oxidoreductase (POR). This is supported by the identification of genes (Table S4) homologous to the single subunit characterized POR in the phylogenetically related Moorella thermoacetica [86], [87] and that multiple investigations of Clade 1 strains have reported a significant role for Fd, as well as ferredoxin:NAD(P)H reductase activity [88]–[90]. Four-gene clusters, annotated as the alpha, beta, gamma and delta subunits of a multi-subunit POR complex are also identified within all strains (Table S4). It is difficult to predict on an in silico basis which gene or gene clusters encode the primary POR responsible for pyruvate catabolism and which encode gene products that may act on alternative keto-acids such as indolepyruvate, 2-ketoisovalerate or 2-ketoglutarate.
Lactate synthesis
The production of lactate has been reported for all Thermoanaerobacter strains with physiological data available (Table 4) and occurs via the reduction of pyruvate using a lactate dehydrogenase (ldh) enzyme. Strains in all 3 clades have a single gene annotated as a ldh (KO:K00016; TIGR01771), though by COG annotation (COG0039), these same genes are designated as malate/lactate dehydrogenases. Distinguishing between ldh and malate dehydrogenase (mdh) genes in silico can be difficult, though the CDS identified in all genomes (Table S4), with the exception of Thermoanaerobacter sp. X561 (truncated due to contig break), share >86% amino acid sequence similarity with the characterized ldh from Thermoanaerobacterium saccharolyticum [91]. As no other obvious ldh is identified, and lactate production is reported throughout the genus, the genes identified in Table S4 are proposed to catalyze Thermoanaerobacter lactate formation.
Acetate synthesis
Strains of Thermoanaerobacter are also reported to produce acetate (Table 4). POR mediated pyruvate catabolism will yield acetyl-CoA, which can be converted to acetate +1 ATP via phosphotransacetylase (pta) and acetate kinase (ack). In all strains, the PTA and ACK enzymes are co-localized within the genome (Table S4). Three strains, Thermoanaerobacter spp. X513, X514 and X561 have additional ack genes annotated, though these are not co-localized with pta genes. Working with a non-sequenced Thermoanaerobacter strain, Thermoanaerobacter thermohydrosulfuricus DSM570, Mayer et al. [92] observed a severe reduction in acetate production and enzyme activity in pta − and ack − mutants and additionally proposed that the pta and ack genes were co-localized and formed an operon. The residual acetate production observed may be in part due to the fact that in all sequenced strains, additional gene sequences annotated as phosphate butyryltransferases and butyrate kinase genes are also identified (Table S4), and the substrate specificity of these genes is not yet known.
Ethanol synthesis
Ethanol production occurs via the reduction of acetyl-CoA to acetaldehyde via an acetaldehyde dehydrogenase followed by a second reduction to ethanol via an alcohol dehydrogenase. Three functionally characterized alcohol dehydrogenase (ADH) genes, adhA, adhB and adhE, have been reported to be principally involved with ethanol formation and a model describing the physiological roles of each gene has been proposed [92]. The adhA gene from T. ethanolicus JW200 is a reported Zn-binding NADPH-dependent primary alcohol dehydrogenase [94], [95]. In comparison, the adhA gene from T. pseudethanolicus 39E, which is capable of utilizing both NADH and NADPH, showed a higher catalytic efficiency for NADH oxidation over NADPH oxidation [96]. Pei et al. [93] demonstrated, in vitro, that the adhB and adhE gene products from T. ethanolicus JW200 displayed bifunctional acetaldehyde/alcohol dehydrogenase activity despite the fact that only the adhE gene contained two independent domains related to aldehyde dehydrogenase and alcohol dehydrogenase families, respectively. However, when assayed using measured intracellular concentrations of NAD(P)+ and NAD(P)H, the adhE gene product displayed only aldehyde dehydrogenase activity (NADH dependent), while the adhB gene strongly favored acetaldehyde reduction over acetyl-CoA reduction.
Only five Thermoanaerobacter genomes contain genes annotated as standalone aldehyde dehydrogenases (Table S4), but no evidence yet exists to suggest that these genes function as acetaldehyde dehydrogenases. Furthermore, genomic context provides no further insights into substrate specificity. As such, it is likely that the reduction of acetyl-CoA to acetaldehyde via adhE is a conserved physiological process throughout the genus. The adhB encoding gene in T. pseudethanolicus 39E is considered to be NADPH-dependent [97] and, upon ethanol accumulation, has shown a higher specific activity towards ethanol oxidation as opposed to ethanol formation [93]. Thus, its role in vivo is not yet confirmed.
The three ADH genes, adhA, adhB and adhE, in conjunction with a recently described redox-sensing transcriptional regulator in T. ethanolicus JW200 [98], have largely formed the basis for our understanding of Thermoanaerobacter ethanologenesis in a few select strains. However, given that the sequenced Thermoanaerobacter genomes have annotated anywhere from 5–9 putative alcohol dehydrogenases, most of which have unknown specificity, this model of ethanol metabolism may not fully encompass all ethanol producing reactions within the cell.
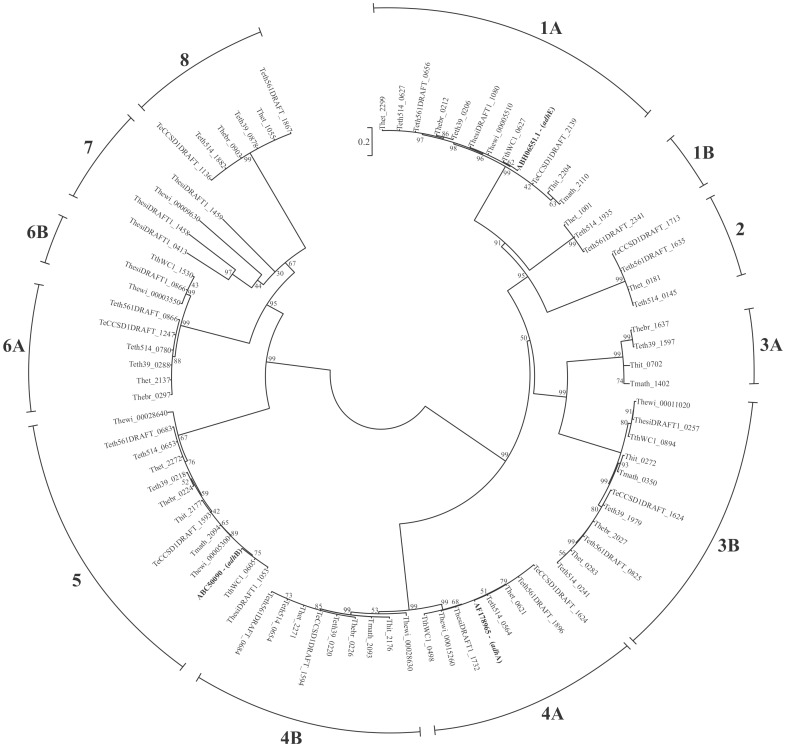
The three characterized Thermoanaerobacter alcohol dehydrogenase genes belong to COG1454-Class IV alcohol dehydrogenase (adhA, adhE) and COG1063-Threonine dehydrogenase and related Zn-dependent dehydrogenases (adhB). Additional sequences belonging to each COG designation were identified, as well as sequences belonging to COG1979-Uncharacterized oxidoreductase, Fe-dependent alcohol dehydrogenase family (Table S4). To identify whether the genomes encode potential additional ethanol producing alcohol dehydrogenases, we conducted phylogenetic analysis of all gene sequences identified in COG1063, COG1454 and COG1979 (Figure 3) as a means of inferring specificity.
Figure 3. Phylogenetic analysis of all annotated alcohol dehydrogenase genes within sequenced Thermoanaerobacter strains.
All included sequences belong to COG1063, COG1454 or COG1979. Tmath_0755 was excluded from analysis as the annotated sequence appears to be a CDS fragment. Sequences in bold correspond to the GenBank accession numbers for functionally characterized sequences from T. ethanolicus JW200 [93], [95]. Tree construction was as described in Materials and Methods. Bootstrapping support values are indicated by their respective nodes.
Of the 80 gene sequences analyzed, 8 distinct clades, and an additional 4 sub-clades were identified. Gene sequences homologous to adhA were identified in Clusters 4A and 4B. Additionally, 5 genomes contained paralogous pairs within these clusters that showed >90.3% amino acid sequence identity within each respective pair. Microarray data from Hemme et al. [23] have shown that in Thermoanaerobacter sp. X514, both genes in the paralagous pair (Teth514_0564– Cluster 4A; Teth514_0654– Cluster 4B) are expressed. Additionally, in one case (Teth514_0564), expression is dependent on growth conditions. All genomes contained sequences orthologous to the characterized adhB gene (Cluster 5) as well as the adhE gene (Cluster 1A). Three strains, Thermoanaerobacter spp. X513, X514 and X561 contained an additional adh gene that grouped near the adhE orthologs (Cluster 1B), though the annotation of these sequences suggests only alcohol dehydrogenase, and not aldehyde dehydrogenase, activity. Surprisingly, Clusters 3A and 3B, which belong to COG1979, group more closely to Clusters 1 and 2 (COG1454) than does Cluster 4 (also COG1454).
Given the number and diversity of adh genes annotated, and that expression patterns of homologous genes in different strains are distinct [23] suggesting differential regulation, the current 3-gene model [93] proposed for ethanol formation in Thermoanaerobacter spp. may not translate across all strains. This is supported by the fact that the study by Hemme et al. [23] identified varied expression of 8 of the 9 adh annotated sequences in Thermoanaerobacter sp. X514 under different growth conditions. However, given that multiple Thermoanaerobacter spp. have been reported to grow on sugar alcohols [12], [13], [16], [58], [84], it is possible that some of these may have catabolic functions and are not involved with ethanol synthesis. Thus, understanding Thermoanaerobacter ethanologenesis requires more in depth functional and expression analysis studies across multiple strains.
Hydrogen synthesis
Hydrogen production, similar to lactate production, competes for reducing equivalents with ethanol synthesis to a greater or lesser extent in all strains of Thermoanaerobacter (Table 4), but in-depth cross-species analysis of Thermoanaerobacter hydrogenases has yet to be conducted.
[Fe-Fe] hydrogenases
Gene sequences homologous to the cytosolic NADH-dependent Fe-only enzyme characterized in Ca. subterraneus subsp. tengcongensis MB4 [99] are conserved in all strains of the Thermoanaerobacter genus (Table 5). Sequence analysis of the hydrogenase conserved domains suggests that these hydrogenases are most similar to the heterotrimeric A1 group exhibiting a TR(M3) modular structure described by Calusinska et al. [45]. These gene products are thought to be NAD-dependent due to the presence of NADH-binding domains in the accessory subunits. However, in Ca. subterraneus subsp. tengcongensis MB4, these same genes, as well as the orthologs in T. pseudethanolicus 39E have recently been proposed to function as potential bifurcating [Fe-Fe] hydrogenases [100], which couple the thermodynamically unfavourable oxidation of NADH to H2 production through utilization of the exergonic oxidation of Fdred.
Table 5. Annotated hydrogenases within sequenced Thermoanaerobacter spp.
| FeFe hydrogenases | Ni-Fe hydrogenase | ||||
| Bifurcating | Fd-linked | PAS-sensory | Ech | ||
| Clade 1 | T. brockii subsp. finnii Ako-1 | Thebr_1491–Thebr_1495 | Thebr_1498 | Thebr_0227 | |
| T. ethanolicus CCSD1 | TeCCSD1DRAFT_0391–TeCCSD1DRAFT_0395 | TeCCSD1DRAFT_0398 | TeCCSD1DRAFT_1595 | ||
| T. pseudethanolicus 39E | Teth39_1456–Teth39_1460 | Teth39_1463 | Teth39_0221 | ||
| Thermoanaerobacter sp. X513 | Thet_0793–Thet_0797 | Thet_0790 | Thet_2270 | ||
| Thermoanaerobacter sp. X514 | Teth514_2138–Teth514_2142 | Teth514_2145 | Teth514_0655 | ||
| Thermoanaerobacter sp. X561 | Teth561DRAFT_0433–Teth561DRAFT_0437 | Teth561DRAFT_0440 | Teth561DRAFT_0685 | ||
| Clade 2 | T. italicus Ab9 | Thit_0826–Thit_0830 | Thit_0823 | Thit_2175 | Thit_1612–Thit_1617 |
| T. mathranii subsp. mathranii A3 | Tmath_0865–Tmath_0869 | Tmath_0862 | Tmath_2092 | Tmath_1603–Tmath_1608 | |
| Tmath_1048 | |||||
| Clade 3 | T. siderophilus SR4 | ThesiDRAFT1_1833–ThesiDRAFT_1837 | ThesiDRAFT1_1830 | ThesiDRAFT1_1402–ThesiDRAFT1_1407 | |
| T. thermohydrosulfuricus WC1 | TthWC1_1780–TthWC1_1784 | TthWC1_1787 | TthWC1_1092–TthWC1_1097 | ||
| T. wiegelii Rt8.B1 | Thewi_00017870–Thewi_00017920 | Thewi_00017850 | Thewi_00028620 | Thewi_00009040–Thewi_00009090 | |
| Thewi_00028740 | Thewi_00005270 | ||||
Upstream of the genes in the A1 grouping in all Thermoanaerobacter strains is a histidine kinase protein as well as another putative hydrogenase gene which shows similar domain architecture to group D hydrogenases. This is consistent with the genomic organization described for bifurcating hydrogenases [45], which is also observed in Ca. subterraneus subsp. tengcongensis [99].
PAS-domain (pfam00989) containing sensory hydrogenases are also conserved throughout the genus with the exception of T. siderophilus SR4 and T. thermohydrosulfuricus WC1 (Table 5). Sensory hydrogenases have been reported to be linked with histidine kinase based signal transduction mechanisms and could play a role in regulating cellular redox levels [45], [101]. T. wiegelii Rt8.B1 and T. mathranii subsp. mathranii A3 both contain additional [Fe-Fe]-hydrogenases showing modular structures similar to the B1 or B3 monomeric hydrogenases described by Calusinska et al. [45].
[Ni-Fe] hydrogenases
[Ni-Fe] hydrogenase encoding genes can be identified for strains belonging to Clade 2 and Clade 3, but not to strains in Clade 1 (Table 5). Strains in Clades 2 and 3 both have the conserved 6-gene cluster coding for a membrane-bound cation transporting Fd-consuming energy conserving hydrogenase (Ech) directly followed by the 6-gene hypABFCDE gene cluster responsible for assembly of the [Ni-Fe] center. Fdred generated via POR in pyruvate catabolism is thought to provide the electrons needed for the evolution of H2 using the Ech complex.
Fd-dependence has been shown for the orthologous gene sequences in Ca. subterraneus subsp tengcongensis MB4 [99]. Furthermore, cell extracts of Ca. subterraneus subsp. tengcongensis MB4 were shown to favour H2-evolution over H2-consumption and Ech was additionally proposed to play a role in “proton respiration” [99]. However, to date, the physiological role of Ech has not been determined for any Clostridia and the exact nature of the exported cation (H+ of Na+) has not yet been determined. It is interesting to note that the absence of Ech encoding homologs (Table 5) in T. pseudethanolicus 39E (Clade 1) correlates with the lowest reported molar H2 yield (and highest molar ethanol yield) of strains with data available (Table 4). This is perhaps indicative of an important physiological role for Ech in Clade 2 and/or Clade 3 strains. Apart from Ech, no other [Ni-Fe] hydrogenases are identified within any Thermoanaerobacter spp. genome.
Energy Metabolism
Energy metabolism is interconnected with carbon and electron flux and governs many of the physiological processes involved with lignocellulosic ethanol production (Figure 1). The principal forms of metabolic energy in bacteria include ion motive force, ATP and/or in some cases, pyrophosphate (PPi). Understanding inter-strain differences in energy metabolism may help provide insight into the mechanisms governing observed physiological differences (Table 4).
Transmembrane ion gradient generating/consuming reactions
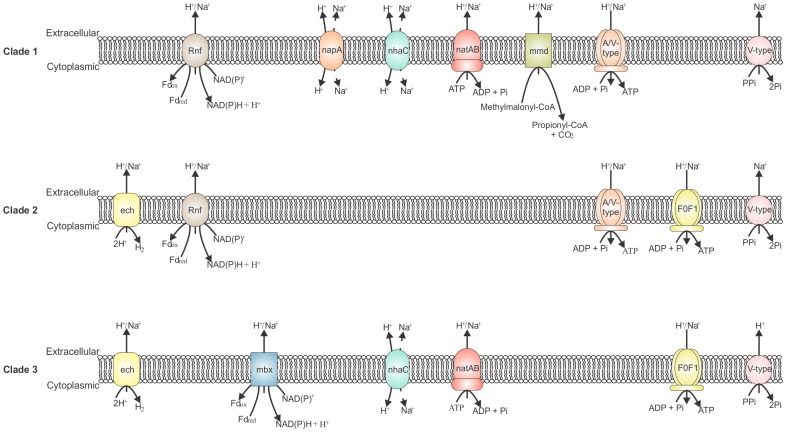
Transmembrane ion gradients can be used to drive endergonic reactions such as solute transport, including carbohydrate transport (Figure 1, Table 2), and ATP synthesis. The mechanisms of balancing ion motive force with the other energy currencies are poorly characterized in Thermoanaerobacter spp., but the present in silico analysis indicates that the potential energy conserving mechanisms within the genus show significant intra-clade conservation (Figure 4).
Figure 4. Transmembrane ion gradient generating and consuming reactions involved with Thermoanaerobacter cellular energetics.
With the exception of the natAB complex (see text, Table S5), all enzyme complexes show intra-clade conservation. Cation specificity is not inferred unless specifically discussed within the text. Dashed lines associated with napA and nhaC antiporters indicate a counter-directional flow of Na+ ions in relation to H+.
Strains of Clade 3 contain a 13-gene cluster (Figure 4) similar in genomic structure to the mbx genes of Pyrococcus furiosus proposed to encode for a complex with Fdred:NAD(P)+ oxidoreductase activity [102], with energy released via the oxidation of Fdred being conserved via translocation of a cation. In P. furiosus, the mbx gene cluster has been proposed to play a role in the reduction of elemental sulfur, where it transfers electrons from Fdred to NAD(P)H, which is subsequently oxidized via a NAD(P)H elemental sulfur oxidoreductase [102]. However, the role in sulfur metabolism for P. furiosus has been inferred based upon microarray data indicating increased gene expression of the mbx gene cluster in response to the addition of elemental sulfur to the growth medium. Assuming the proposed Fd:NAD(P)+ oxidoreductase activity of mbx is also observed in the Clade 3 strains, there is no evidence yet that suggests it is connected with sulfur reduction in these Thermoanaerobacter strains.
Clade 1 strains contain apparent remnants of the mbx gene cluster (Table S5) though not all 13-genes could be identified. Five genes, the mbxMJKLM cluster, are not found immediately following orthologs of the mbxABCDGGHH’ gene cluster as observed in Clade 3. Thus, Clade 1 strains do not appear to contain the genes necessary for a functional mbx complex. No orthologs were identified in the Clade 2 strains.
A functionally analogous Fd:NAD(P)+ oxidoreductase system, the ion-translocating Rnf complex, is present in all Clade 1 and Clade 2 strains (Figure 4, Table S5). The genomic organization is identical to what is reported for Acetobacterium woodii and multiple subunits of the T. pseudethanolicus 39E complex have been reported to be genetically similar to the partially characterized protein complex in A. woodii [103], [104]. In A. woodii, the complex is thought to translocate Na+ ions, yet definitive proof has not yet been determined.
Na+ energetics may play a prominent role in multiple Thermoanaerobacter strains. Of the five classes of recognized primary Na+ pumps [105], four of the classes are observed within the genus (Table S5). One class, comprised of Na+-translocating decarboxylation reactions, is limited to strains belonging to Clade 1. Hemme et al. [23] identify both a methylmalonyl-CoA decarboxylase and an oxaloacetate decarboxylase in the genomes of T. pseudethanolicus 39E and Thermoanaerobacter sp. X514. However, the present analysis is unable to find locus tags supportive of a membrane-associated oxaloacetate decarboxylase complex.
Annotation of the methylmalonyl-CoA decarboxylase encoding genes (Figure 4, Table S5) is supported by the presence of genes annotated to encode methylmalonyl-CoA mutase and methylmalonyl-CoA epimerase immediately adjacent to the methylmalonyl-CoA decarboxylase annotated gene sequences in all Clade 1 genomes. In all sequenced Thermoanaerobacter strains, genes annotated as oxaloacetate decarboxylase subunits (α, β, γ) can be identified, though they are never co-localized into a single gene cluster, as is observed with other Clostridia [106], and a functional membrane associated oxaloacetate decarboxylase complex may not exist in any of the Thermoanaerobacter strains.
Genes homologous to the natAB genes described in Bacillus subtilis [107] are identified in multiple Thermoanaerobacter strains, but do not show intra-clade conservation (Table S5). A third primary Na+ pump includes the Rnf complex (discussed above). The fourth and final class of potential Na+ pumps identified in Thermoanaerobacter are the V-type inorganic pyrophosphatases (Table S5). All V-type pyrophosphatases identified in Clades 1 and 2 appear to be orthologous to each other. Genes in Clade 3 though, are significantly different than the V-type pyrophosphatases identified in Clades 1 and 2. Key residues, as determined by Luoto et al. [108], were identified for all annotated genes via sequence alignments. The sequences present in Clades 1 and 2 share key residues identical to reported K+-dependent, Na+-exporting V-type pyrophosphatases, while Clade 3 sequences share identical residues with K+-independent, H+-exporting versions (Table S6).
Given that the cation specificity for any of the above mentioned ion-translocating processes discussed have not yet been determined experimentally for any Thermoanaerobacter strain, it is impossible to predict their role. Cook [109] reports inhibited growth by T. wiegelii Rt8.B1 in the presence of the Na+ ionophore monensin, thus suggesting the importance on maintaining a transmembrane Na+ gradient for cell viability. However, genomic analysis identifies that a H+ motive force is also expected in T. wiegelii Rt8.B1 (eg. V-type pyrophosphatase). Both gradients may exist, and cellular demands are balanced by the use of H+/Na+ antiporters. Cation exchange antiporters, showing homology to the NhaC family, can be identified though in all strains except Clade 2 (Figure 4). Clade 1 also contains genes homologous to NapA type antiporters. Surprisingly, analysis of the annotated transport systems for Clade 2 strains does not reveal any cation exchangers. Thus, if Clade 2 strains generate both H+ and Na+ ion-motive forces, it is unclear how they balance these forces.
ATP and pyrophosphate (PPi) as energy currencies
Synthesis of ATP as an energy currency, which is closely linked with carbohydrate and pyruvate metabolism (Figure 1), can occur via glycolysis and through the production of acetate via acetate kinase in all Thermoanaerobacter strains. According to its annotation, both Clade 1 and Clade 3 genomes also encode an ATP-linked (in contrast to GTP-linked) PEP carboxykinase based on the enzyme commission number assigned to the annotated sequences (EC: 4.1.1.49) [110]. This bidirectional enzyme could either carboxylate PEP yielding 1 ATP+oxaloacetate or decarboxylate oxaloacetate at the expense of ATP. The flux models for T. pseudethanolicus 39E and Thermoanaerobacter sp. X514 [23] suggest that, in these two strains, oxaloacetate decarboxylation occurs, but it is unknown if this translates universally throughout the genus.
ATP synthesis via ATP synthase genes can occur through two distinct means within Thermoanaerobacter spp. Strains of Clade 1 contain a 9-gene cluster designated as an A/V-type ATP synthase (Figure 3, Table S5), while Clade 3 strains contain the F0F1-ATP synthase in a conserved 8-gene cluster. Strains of Clade 2 contain both the AV-type and the F0F1-ATP synthase gene clusters in identical genomic organization as Clade 1 or Clade 3, respectively.
PPi as an alternative energy carrier to ATP has been observed during exponential phase for the phylogenetically related strains M. thermoacetica [111] and Cal. saccharolyticus DSM 8903 [112]. Many of the key genomic elements to potentially generate and utilize PPi as a central energy carrier are similarly identified within all Thermoanaerobacter spp. Pyruvate kinase, which is identified in all Thermoanaerobacter strains, is typically considered to be responsible for the conversion of phosphoenolpyruvate to pyruvate. However, two distinct pyruvate phosphate dikinase (PPDK) genes (566–567 and 877 amino acids) are also universally conserved throughout the genus (Table S4). The longer of these genes shows >77.7% amino acid identity with the annotated PPDK gene (Csac_1955) from Cal. saccharolyticus DSM 8903, whose genome also encodes a pyruvate kinase [106]. In Cal. saccharolyticus DSM 8903, the PPDK has been proposed to function in a catabolic role during exponential growth whereby the conversion of PEP to pyruvate is coupled to the conversion of AMP+PPi to ATP+Pi [112]. Its presence in Thermoanaerobacter spp. suggests that PPi may also play a role in cellular energetics.
Additionally, sequence alignments (Figure S1) of the annotated 6-phosphofructokinase (PFK) genes (Table S7) identifies that Clade 1 and Clade 3 strains possess one copy of PFK that contains the conserved Asp104+ Lys124 residues (Escherichia coli numbering) associated with PPi-dependence [113] and also found in Cal. saccharolyticus DSM 8903 [112]. Energy may additionally be conserved as an ion-motive force through the use of a membrane linked cation-translocating V-type pyrophosphatase (see above). Strains reported to use PPi as an energy currency during exponential growth are expected to have relatively high intracellular PPi/ATP ratios and low cytosolic PPiase activity. While, the intracellular ATP concentrations of exponential T. wiegelii Rt8.B1 cells [109] are reportedly higher than those observed for Cal. saccharolyticus DSM 8903 [112], PPi levels and PPiase activity has not yet been investigated for any strain of the Thermoanaerobacter genus.
Conclusions
The analyses presented here have identified inter-strain differences at the genomic level within the Thermoanaerobacter genus that may account for differences in the industrial application of these bacteria in a CBP system. Based on genomic content, Clade 2 strains seem most well suited to biomass hydrolysis and utilization, though these strains have not yet been reported to have as high of ethanol yields as some Clade 1 strains do (Table 4). Conversely, despite the ethanologenic capabilities of Clade 1 strains, their genomes encode the fewest extracellular CAZymes of all strains within the genus (Table 1), which may potentially limit their hydrolytic capabilities. The genomes of Clade 3 strains show intermediate hydrolytic and substrate utilization capabilities, but also represent the most divergent lineage of the genus (Figure 2) and may have yet unexamined potential.
The use of a specific strain for development of a universal approach to lignocellulosic ethanol production may represent an idealistic concept. Rather, strain selection may be specific to a single CBP system and dependent on the nature of the feedstock. Efficient conversion of arabinose to ethanol is of little value for bioenergy feedstocks such as eucalyptus, which contains comparatively low amounts of arabinan (0.3%), in contrast to switchgrass (3.0%) [114]. Alternatively, extracellular xylan hydrolysis may not be an essential component in the microbial conversion of feedstocks such as softwoods, which have a low xylan and high glucomannan hemicellulose content [76], [115]. This is particularly true given that no extracellular glucomannanases were identified in any strain of Thermoanaerobacter (Table 1).
Development of biocatalysts with desired physiological characteristics using a strain with diverse, rather than specialized capabilities may be advantageous for constructing a robust and dynamic CBP system. For example, the construction of a single mutant (Δldh) in T. mathranii BG1, a platform organism of BioGasol (http://www.biogasol.com) [116], has shown to improve ethanol yields and still maintain substrate utilization capabilities similar to T. mathranii subsp. mathranii A3 (Clade 2). Alternatively, strategies that broaden the capabilities of a relatively specialized strain have also shown to be successful. The cloning and expression of a functional endoglucanase into Thermoanaerobacter sp. X514 [117], a comparatively good ethanol producer, has improved that strain’s hydrolytic capabilities.
The purpose of this paper was to evaluate genomic differences within members of the Thermoanaerobacter genus which may influence strain suitability in a CBP co-culture system. Also, correlating genome content with the reported physiologies is a first step to help shape molecular engineering strategies for strain improvement. It is important to consider though that the analysis presented here is of the genomic potential of sequenced Thermoanaerobacter strains and is not of the observed phenotypes. Future experiments such as expression profiling studies and enzymatic characterization, which can supplement the data presented here, will help to improve our understanding of the extent that the genomic potential is achieved within these strains. Experiments targeted towards improving the hydrolysis of raw lignocellulosic biomass, understanding carbon transport and simultaneous utilization of mono-, oligo-, and polysaccharides, evaluating the genomic and regulatory basis for the observed differences in end-product synthesis ratios and improving the correlation between energy metabolism and end-product yields will all help develop Thermoanaerobacter spp. into more efficient CBP microorganisms.
This study is focused on components associated with lignocellulosic biofuel production and does not investigate the genomics associated with other phenotypes reported within the genus such as peptide and amino acid oxidation [118], metal-reduction [54], [85], [119] or sulfur reduction [50]. The potential impact that these phenotypes may have on biofuel production is not yet known. For example, vitamin B12 biosynthesis, associated with co-factor metabolism, has recently been shown to play an important role in improving observed ethanol yields [22], [23] in select Thermoanaerobacter strains. Therefore, we cannot discount the possibility that additional components of cellular physiology may similarly influence the lignocellulosic ethanol production capabilities of these strains. However, this work does identify key genomic criteria pertinent to strain evaluation for the development of a C. thermocellum-Thermoanaerobacter sp. co-culture and represents the most comprehensive comparative genomic analysis of the genus to date. Furthermore, comparative genomic analysis such as this can be useful in identifying important physiological questions to address through experimentation not only for Thermoanaerobacter spp., but also in other organisms of interest for lignocellulosic ethanol production through CBP.
Supporting Information
Partial sequence alignment of selected PFK genes in different bacteria.
(PDF)
Selected genome metadata for sequenced Thermoanaerobacter spp.
(XLS)
All CAZyme designated gene sequences within sequenced Thermoanaerobacter strains as are available within the CAZy database or identified through de novo analysis.
(XLS)
Identified genes associated with the utilization of the major carbohydrates produced through lignocellulose hydrolysis in sequenced Thermoanaerobacter strains.
(XLS)
Genes associated with pyruvate metabolism in sequenced Thermoanaerobacter strains.
(XLS)
Genes associated with transmembrane ion gradient generating and consuming reactions involved with cellular energetics in sequenced Thermoanaerobacter genomes.
(XLS)
Key amino acid residues responsible for imparting predicted substrate specificity and K+ dependence in annotated Thermoanaerobacter V-type pyrophosphatases.
(XLS)
Identification of key residues characteristic of ATP or PPi dependent 6-phosphofructokinase genes in Thermoanaerobacter .
(XLS)
Acknowledgments
We would like to thank John A. Wilkins of the Manitoba Centre for Proteomics and Systems Biology. The use of his facilities and equipment made our proteogenomic analysis possible.
Funding Statement
This work was supported by funds provided by a Genome Canada grant titled “Microbial Genomics for Biofuels and Co-Products from Biorefining Processes”, the Natural Sciences and Engineering Research Council Strategic Grant (STPGP 365076) and by the University of Manitoba. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Lynd LR, van Zyl WH, McBride JE, Laser M (2005) Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotech 16: 577–583. [DOI] [PubMed] [Google Scholar]
- 2. Cardona CA, Sánchez ÓJ (2007) Fuel ethanol production: process design trends and integration opportunities. Bioresour Technol 98: 2415–2457. [DOI] [PubMed] [Google Scholar]
- 3. Olson DG, McBride JE, Shaw AJ, Lynd LR (2012) Recent progress in consolidated bioprocessing. Curr Opin Biotech 23: 396–406. [DOI] [PubMed] [Google Scholar]
- 4. Desvaux M (2006) Unravelling carbon metabolism in anaerobic cellulolytic bacteria. Biotechnol Prog 22: 1229–1238. [DOI] [PubMed] [Google Scholar]
- 5. Brenner K, You L, Arnold FH (2008) Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol 26: 483–489. [DOI] [PubMed] [Google Scholar]
- 6. Zuroff TR, Curtis WR (2012) Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol 93: 1423–1435. [DOI] [PubMed] [Google Scholar]
- 7.Lynd LR, Currie D, Ciazza N, Herring C, Orem N (2008) Consolidated bioprocessing of cellulosic biomass to ethanol using thermophilic bacteria. In: Wall JD, Harwood CS, Demain A, editors. Bioenergy. Washington: ASM Press. 55–74.
- 8. Lynd LR, Grethlein HE, Wolkin RH (1989) Fermentation of cellulosic substrates in batch and continuous culture by Clostridium thermocellum . Appl Environ Microbiol 55: 3131–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levin DB, Islam R, Cicek N, Sparling R (2006) Hydrogen production by Clostridium thermocellum 27405 from cellulosic biomass substrates. Int J Hydrogen Energy 31: 1496–1503. [Google Scholar]
- 10. Raman B, Pan C, Hurst GB, Rodriguez M Jr, McKeown CK, et al. (2009) Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4: e5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wiegel J, Ljungdahl LG (1981) Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Arch Microbiol 128: 343–348. [Google Scholar]
- 12. Kozianowski G, Canganella F, Rainey FA, Hippe H, Antranikian G (1997) Purification and characterization of thermostable pectate-lyases from a newly isolated thermophilic bacterium, Thermoanaerobacter italicus sp. nov. Extremophiles 1: 171–182. [DOI] [PubMed] [Google Scholar]
- 13. Larsen L, Nielsen P, Ahring BK (1997) Thermoanaerobacter mathranii sp. nov., an ethanol-producing, extremely thermophilic anaerobic bacterium from a hot spring in Iceland. Arch Microbiol 168: 114–119. [DOI] [PubMed] [Google Scholar]
- 14. Onyenwoke RU, Kevbrin VV, Lysenko AM, Wiegel J (2007) Thermoanaerobacter pseudethanolicus sp. nov., a thermophilic heterotrophic anaerobe from Yellowstone National Park. Int J Syst Evol Microbiol 57: 2191–2193. [DOI] [PubMed] [Google Scholar]
- 15. Verbeke TJ, Dumonceaux TJ, Wushke S, Cicek N, Levin DB, et al. (2011) Isolates of Thermoanaerobacter thermohydrosulfuricus from decaying wood compost display genetic and phenotypic microdiversity. FEMS Microbiol Ecol 78: 473–487. [DOI] [PubMed] [Google Scholar]
- 16. Zeikus JG, Ben-Bassat A, Hegge PW (1980) Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol 143: 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Georgieva TI, Skiadas IV, Ahring BK (2007) Effect of temperature on ethanol tolerance of a thermophilic anaerobic ethanol producer Thermoanaerobacter A10: modeling and simulation. Biotechnol Bioeng 98: 1161–1170. [DOI] [PubMed] [Google Scholar]
- 18. Shaw AJ, Hogsett DA, Lynd LR (2010) Natural competence in Thermoanaerobacter and Thermoanaerobacterium species. Appl Environ Microbiol 76: 4713–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao S, Mikkelsen MJ (2010) Metabolic engineering to improve ethanol production in Thermoanaerobacter mathranii . Appl Microbiol Biotechnol 88: 199–208. [DOI] [PubMed] [Google Scholar]
- 20. Ng TK, Ben-Bassat A, Zeikus JG (1981) Ethanol production by thermophilic bacteria: fermentation of cellulosic substrates by cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum . Appl Environ Microbiol 41: 1337–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saddler JN, Chan MKH (1984) Conversion of pretreated lignocellulosic substrates to ethanol by Clostridium thermocellum in mono- and co-culture with Clostridium thermosaccharolyticum and Clostridium thermohydrosulphuricum . Can J Microbiol 30: 212–220. [Google Scholar]
- 22. He Q, Hemme CL, Jiang H, He Z, Zhou J (2011) Mechanisms of enhanced cellulosic bioethanol fermentation by co-cultivation of Clostridium and Thermoanaerobacter spp. Bioresour Technol 102: 9586–9592. [DOI] [PubMed] [Google Scholar]
- 23. Hemme CL, Fields MW, He Q, Deng Y, Lin L, et al. (2011) Correlation of genomic and physiological traits of Thermoanaerobacter species with biofuel yields. Appl Environ Microbiol 77: 7998–8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verbeke TJ, Sparling R, Hill JE, Links MG, Levin D, et al. (2011) Predicting relatedness of bacterial genomes using the chaperonin-60 universal target (cpn60 UT): application to Thermoanaerobacter species. Syst Appl Microbiol 34: 171–179. [DOI] [PubMed] [Google Scholar]
- 25. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pearson WR, Wood T, Zhang Z, Miller W (1997) Comparison of DNA sequences with protein sequences. Genomics 46: 24–36. [DOI] [PubMed] [Google Scholar]
- 27. Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boisvert S, Laviolette F, Corbeil J (2010) Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Markowitz VM, Mavromatis K, Ivanova NN, Chen IMA, Chu K, et al. (2009) IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25: 2271–2278. [DOI] [PubMed] [Google Scholar]
- 30. Pati A. Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, et al. (2010) GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 7: 455–457. [DOI] [PubMed] [Google Scholar]
- 31. Krug K, Nahnsen S, Macek B (2011) Mass spectrometry at the interface of proteomics and genomics. Mol Biosyst 7: 284–291. [DOI] [PubMed] [Google Scholar]
- 32. McQueen P, Spicer V, Rydzak T, Sparling R, Levin D, et al. (2012) Information-dependent LC-MS/MS acquisition with exclusion lists potentially generated on-the-fly: case study using a whole cell digest of Clostridium thermocellum . Proteomics 12: 1160–1169. [DOI] [PubMed] [Google Scholar]
- 33. Krokhin OV (2006) Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: application to 300- and 100-Å pore size C18 sorbents. Anal Chem 78: 7785–7795. [DOI] [PubMed] [Google Scholar]
- 34. Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haft DH, Selengut JD, White O (2003) The TIGRFAMs database of protein families. Nucleic Acids Res 31: 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saier MH Jr, Yen MR, Noto K, Tamang DG, Elkan C (2009) The Transporter Classification Database: recent advances. Nucleic Acids Res 37: D274–D278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37: D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, et al. (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37: D233–D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu NY, Wagner JR, Laird MR, Melli G, Rey S, et al. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26: 1608–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 44. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 45. Calusinska M, Happe T, Joris B, Wilmotte A (2010) The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156: 1575–1588. [DOI] [PubMed] [Google Scholar]
- 46. Chain PSG, Grafham DV, Fulton RS, FitzGerald MG, Hostetler J, et al. (2009) Genome project standards in a new era of sequencing. Science 326: 236–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bao Q, Tian Y, Li W, Xu Z, Xuan Z, et al. (2002) A complete sequence of the T. tengcongensis genome. Genome Res 12: 689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fardeau ML, Bonilla Salinas M, L’Haridon S, Jeanthon C, Verhé F, et al. (2004) Isolation from oil reservoirs of novel thermophilic anaerobes phylogenetically related to Thermoanaerobacter subterraneus: reassignment of T. subterraneus, Thermoanaerobacter yonsiensis, Thermoanaerobacter tengcongensis and Carboxydibrachium pacificum to Caldanaerobacter subterraneus gen. nov., sp. nov., comb. nov. as four novel subspecies. Int J Syst Evol Microbiol 54: 467–474. [DOI] [PubMed] [Google Scholar]
- 49. Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106: 19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee YE, Jain MK, Lee C, Zeikus JG (1993) Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp., nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and Clostridium thermohydrosulfuricum E100–69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus . Int J Syst Bacteriol 43: 41–51. [Google Scholar]
- 51. Kridelbaugh DM, Nelson J, Engle NL, Tschaplinski TJ, Graham DE (2013) Nitrogen and sulfur requirements for Clostridium thermocellum and Caldicellulosiruptor bescii on cellulosic substrates in minimal nutrient media. Bioresour Technol 130: 125–135. [DOI] [PubMed] [Google Scholar]
- 52. Hung KS, Liu SM, Fang TY, Tzou WS, Lin FP, et al. (2011) Characterization of a salt-tolerant xylanase from Thermoanaerobacterium saccharolyticum NTOU1. Biotechnol Lett 33: 1441–1447. [DOI] [PubMed] [Google Scholar]
- 53. Lüthi E, Love DR, McAnulty J, Wallace C, Caughey PA, et al. (1990) Cloning, sequence analysis, and expression of genes encoding xylan-degrading enzymes from the thermophile “Caldocellum saccharolyticum”. Appl Environ Microbiol 56: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Slobodkin AI, Tourova TP, Kuznetsov BB, Kostrikina NA, Chernyh NA, et al. (1999) Thermoanaerobacter siderophilus sp. nov., a novel dissimilatory Fe(III)-reducing, anaerobic, thermophilic bacterium. Int J Syst Bacteriol 49: 1471–1478. [DOI] [PubMed] [Google Scholar]
- 55. Mai V, Wiegel J, Lorenz WW (2000) Cloning, sequencing, and characterization of the bifunctional xylosidase-arabinosidase from the anaerobic thermophile Thermoanaerobacter ethanolicus . Gene 247: 137–143. [DOI] [PubMed] [Google Scholar]
- 56. Breves R, Bronnenmeier K, Wild N, Lottspeich F, Staudenbauer WL, et al. (1997) Genes encoding two different β-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl Environ Microbiol 63: 3902–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Piccolo C, Bezzo F (2009) A techno-economic comparison between two technologies for bioethanol production from lignocellulose. Biomass Bioenergy 33: 478–491. [Google Scholar]
- 58. Xu C, Qin Y, Li Y, Ji Y, Huang J, et al. (2010) Factors influencing cellulosome activity in consolidated bioprocessing of cellulosic ethanol. Bioresour Technol 101: 9560–9569. [DOI] [PubMed] [Google Scholar]
- 59. Das SP, Ravindran R, Ahmed S, Das D, Goyal D, et al. (2012) Bioethanol production involving recombinant C. thermocellum hydrolytic hemicellulase and fermentative microbes. Appl Biochem Biotechnol 167: 1475–1488. [DOI] [PubMed] [Google Scholar]
- 60. Sun Y, Cheng J (2002) Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83: 1–11. [DOI] [PubMed] [Google Scholar]
- 61. Ramos LP, Breuil C, Saddler JN (1993) The use of enzyme recycling and the influence of sugar accumulation on cellulose hydrolysis by Trichoderma cellulases. Enzyme Microb Technol 15: 19–25. [Google Scholar]
- 62. Xiao Z, Zhang X, Gregg DJ, Saddler JN (2004) Effects of sugar inhibition on cellulases and β-glucosidase during enzymatic hydrolysis of softwood substrates. Appl Biochem Biotechnol 113: 1115–1126. [DOI] [PubMed] [Google Scholar]
- 63. Podkaminer KK, Kenealy WR, Herring CD, Hogsett DA, Lynd LR (2012) Ethanol and anaerobic conditions reversibly inhibit commercial cellulose activity in thermophilic simultaneous saccharification and fermentation (tSSF). Biotechnol Biofuels 5: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wiegel J, Mothershed CP, Puls J (1985) Differences in xylan degradation by various noncellulolytic thermophilic anaerobes and Clostridium thermocellum . Appl Environ Microbiol 49: 656–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang YHP, Lynd LR (2005) Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci U S A 102: 7321–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Morag E, Bayer EA, Lamed R (1990) Relationship of cellulosomal and noncellulosomal xylanase of Clostridium thermocellum to cellulose-degrading enzymes. J Bacteriol 172: 6098–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hayashi H, Takehara M, Hattori T, Kimura T, Karita S, et al. (1999) Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum . Appl Microbiol Biotechnol 51: 348–357. [DOI] [PubMed] [Google Scholar]
- 68. Zverlov VV, Schantz N, Schmitt-Kopplin P, Schwarz WH (2005) Two new major subunits in the cellulosome of Clostridium thermocellum: xyloglucanase Xgh74A and endoxylanase Xyn10D. Microbiology 151: 3395–3401. [DOI] [PubMed] [Google Scholar]
- 69. Izquierdo JA, Goodwin L, Davenport KW, Teshima H, Bruce D, et al. (2012) Complete genome sequence of Clostridium clariflavum DSM 19732. Stand Genomic Sci 6: 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Erbeznik M, Ray M, Dawson KA, Strobel HJ (1998) Xylose transport by the anaerobic thermophile Thermoanaerobacter ethanolicus and the characterization of a D-xylose binding protein. Curr Microbiol 37: 295–300. [DOI] [PubMed] [Google Scholar]
- 71. Erbeznik M, Hudson SE, Herman AB, Strobel HJ (2004) Molecular analysis of the xylFGH Operon, coding for xylose ABC transport, in Thermoanaerobacter ethanolicus . Curr Microbiol 48: 295–299. [DOI] [PubMed] [Google Scholar]
- 72. Lin L, Song H, Tu Q, Qin Y, Zhou A, et al. (2011) The Thermoanaerobacter glycobiome reveals mechanisms of pentose and hexose co-utilization in bacteria. PLoS Genet 7: e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ng TK, Zeikus JG (1982) Differential metabolism of cellobiose and glucose by Clostridium thermocellum and Clostridium thermohydrosulfuricum . J Bacteriol 150: 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cook GM, Janssen PH, Morgan HW (1993) Uncoupler-resistant glucose uptake by the thermophilic glycolytic anaerobe Thermoanaerobacter thermosulfuricus (Clostridium thermohydrosulfuricum). Appl Environ Microbiol 59: 2984–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30: 279–291. [DOI] [PubMed] [Google Scholar]
- 77. Shallom D, Shoham Y (2003) Microbial hemicellulases. Curr Opion Microbiol 6: 219–228. [DOI] [PubMed] [Google Scholar]
- 78.Moreira LRS, Milanezi NvG, Filho EXF (2011) Enzymology of plant cell wall breakdown: an update. In: Buckeridge MS, Goldman GH, editors. Routes to Cellulosic Ethanol. New York: Springer. 73–96.
- 79. Demirbaş A (2005) Bioethanol from cellulosic materials: a renewable motor fuel from biomass. Energy Sources 27: 327–337. [Google Scholar]
- 80. Galbe M, Sassner P, Wingren A, Zacchi G (2007) Process engineering economics of bioethanol production. Adv Biochem Eng Biotechnol 108: 303–327. [DOI] [PubMed] [Google Scholar]
- 81. Feng X, Mouttaki H, Lin L, Huang R, Wu B, et al. (2009) Characterization of the central metabolic pathways in Thermoanaerobacter sp. strain X514 via isotopomer-assisted metabolite analysis. Appl Environ Microbiol 75: 5001–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qian Z, Meng B, Wang Q, Wang Z, Zhou C, et al. (2009) Systematic characterization of a novel gal operon in Thermoanaerobacter tengcongensis. . Microbiology 155: 1717–1725. [DOI] [PubMed] [Google Scholar]
- 83. Cayol JL, Ollivier B, Patel BKC, Ravot G, Magot M, et al. (1995) Description of Thermoanaerobacter brockii subsp. lactiethylicus susbp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii . Int J Syst Bacteriol 45: 783–789. [DOI] [PubMed] [Google Scholar]
- 84. Cook GM, Rainey FA, Patel BKC, Morgan HW (1996) Characterization of a new obligately anaerobic thermophile, Thermoanaerobacter wiegelii sp. nov. Int J Syst Bacteriol 46: 123–127. [DOI] [PubMed] [Google Scholar]
- 85. Roh Y, Liu SV, Li G, Huang H, Phelps TJ, et al. (2002) Isolation and characterization of metal-reducing Thermoanaerobacter strains from deep subsurface environments of the Piceance Basin, Colorado. Appl Environ Microbiol 68: 6013–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Furdui C, Ragsdale SW (2002) The roles of coenzyme A in the pyruvate:ferredoxin oxidoreductase reaction mechanism: rate enhancement of electron transfer from a radical intermediate to an iron-sulfur cluster. Biochemistry 41: 9921–9937. [DOI] [PubMed] [Google Scholar]
- 87. Pierce E, Xie G, Barabote RD, Saunders E, Han CS, et al. (2008) The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ Microbiol 10: 2550–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lamed R, Zeikus JG (1980) Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii . J Bacteriol 144: 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ben-Bassat A, Lamed R, Zeikus JG (1981) Ethanol production by thermophilic bacteria: metabolic control of end product formation in Thermoanaerobium brockii . J Bacteriol 146: 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lovitt RW, Shen GJ, Zeikus JG (1988) Ethanol production by thermophilic bacteria: biochemical basis for ethanol and hydrogen tolerance in Clostridium thermohydrosulfuricum . J Bacteriol 170: 2809–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shaw AJ, Covalla SF, Hogsett DA, Herring CD (2011) Marker removal system for Thermoanaerobacterium saccharolyticum and development of a markerless ethanologen. Appl Environ Microbiol 77: 2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mayer MAG, Bronnenmeier K, Schwarz WH, Schertler C, Staudenbauer WL (1995) Isolation and properties of acetate kinase- and phosphotransacetylase-negative mutants of Thermoanaerobacter thermohydrosulfuricus . Microbiol 141: 2891–2896. [Google Scholar]
- 93. Pei J, Zhou Q, Jian Y, Le Y, Li H, et al. (2010) Thermoanaerobacter spp. control ethanol pathway via transcriptional regulation and versatility of key enzymes. Metab Eng 12: 420–428. [DOI] [PubMed] [Google Scholar]
- 94. Bryant FO, Wiegel J, Ljungdahl LG (1988) Purification and properties of primary and secondary alcohol dehydrogenases from Thermoanaerobacter ethanolicus . Appl Environ Microbiol 54: 460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Holt PJ, Williams RE, Jordan KN, Lowe CR, Bruce NC (2000) Cloning, sequencing and expression in Escherichia coli of the primary alcohol dehydrogenase gene from Thermoanaerobacter ethanolicus JW200. FEMS Microbiol Lett 190: 57–62. [DOI] [PubMed] [Google Scholar]
- 96. Burdette D, Zeikus JG (1994) Purification of acetaldehyde dehydrogenase and alcohol dehydrogenases from Thermoanaerobacter ethanolicus 39E and characterization of the secondary-alcohol dehydrogenase (2° Adh) as a bifunctional alcohol dehydrogenase–acetyl-CoA reductive thioesterase. Biochem J 302: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Burdette DS, Vieille C, Zeikus JG (1996) Cloning and expression of the gene encoding the Thermoanaerobacter ethanolicus 39E secondary-alcohol dehydrogenase and biochemical characterization of the enzyme. Biochem J 316: 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pei J, Zhou Q, Jing Q, Li L, Dai C, et al. (2011) The mechanism for regulating ethanol fermentation by redox levels in Thermoanaerobacter ethanolicus . Metab Eng 13: 186–193. [DOI] [PubMed] [Google Scholar]
- 99. Soboh B, Linder D, Hedderich R (2004) A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. . Microbiol 150: 2451–2463. [DOI] [PubMed] [Google Scholar]
- 100. Schut GJ, Adams MWW (2009) The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol 191: 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Taylor BL, Zhulin IB (1999) PAS Domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63: 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Schut GJ, Bridger SL, Adams MWW (2007) Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J Bacteriol 189: 4431–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Biegel E, Schmidt S, Müller V (2009) Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii . Environ Microbiol 11: 1438–1443. [DOI] [PubMed] [Google Scholar]
- 104. Biegel E, Schmidt S, González JM, Müller V (2011) Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci 68: 613–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mulkidjanian AY, Dibrov P, Galperin MY (2008) The past and present of sodium energetics: may the sodium-motive force be with you. Biochim Biophys Acta 1777: 985–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. van de Werken HJG, Verhaart MRA, VanFossen AL, Willquist K, Lewis DL, et al. (2008) Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus . Appl Environ Microbiol 74: 6720–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Cheng J, Guffanti AA, Krulwich TA (1997) A two-gene ABC-type transport system that extrudes Na+ in Bacillus subtilis is induced by ethanol or protonophore. Mol Microbiol 23: 1107–1120. [DOI] [PubMed] [Google Scholar]
- 108. Luoto HH, Belogurov GA, Baykov AA, Lahti R, Malinen AM (2011) Na+-translocating membrane pyrophosphatases are widespread in the microbial world and evolutionarily precede H+-translocating pyrophosphatases. J Biol Chem 286: 21633–21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Cook GM (2000) The intracellular pH of the thermophilic bacterium Thermoanaerobacter wiegelii during growth and production of fermentation acids. Extremophiles 4: 279–284. [DOI] [PubMed] [Google Scholar]
- 110. Aich S, Delbaere LTJ (2007) Phylogenetic study of the evolution of PEP-carboxykinase. Evol Bioinform Online 3: 333–340. [PMC free article] [PubMed] [Google Scholar]
- 111. Heinonen JK, Drake HL (1988) Comparative assessment of inorganic pyrophosphate and pyrophosphatase levels of Escherichia coli, Clostridium pasteurianum, and Clostridium thermoaceticum . FEMS Microbiol Lett 52: 205–208. [Google Scholar]
- 112. Bielen AAM, Willquist K, Engman J, van der Oost J, van Niel EWJ, et al. (2010) Pyrophosphate as a central energy carrier in the hydrogen-producing extremely thermophilic Caldicellulosiruptor saccharolyticus . FEMS Microbiol Lett 307: 48–54. [DOI] [PubMed] [Google Scholar]
- 113. Bapteste E, Moreira D, Philippe H (2003) Rampant horizontal gene transfer and phospho-donor change in the evolution of the phosphofructokinase. Gene 318: 185–191. [DOI] [PubMed] [Google Scholar]
- 114. Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182. [DOI] [PubMed] [Google Scholar]
- 115. Gregg D, Saddler JN (1996) A techno-economic assessment of the pretreatment and fractionation steps of a biomass-to-ethanol process. Appl Biochem Biotechnol 57: 711–727. [Google Scholar]
- 116.Mikkelsen MJ, Ahring BK (2007) Biogasol. Thermoanaerobacter mathranii strain BG1, WO/2007/124607.
- 117. Lin L, Song H, Ji Y, He Z, Pu Y, et al. (2010) Ultrasound-mediated DNA transformation in thermophilic Gram-positive anaerobes. PLoS One 5: e12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Faudon C, Fardeau ML, Heim J, Patel B, Margot M, et al. (1995) Peptide and amino acid oxidation in the presence of thiosulfate by members of the genus Thermoanaerobacter . Curr Microbiol 31: 152–157. [Google Scholar]
- 119. Gavrilov SN, Bonch-Osmolovskaya EA, Slobodkin AI (2003) Physiology of organotrophic and lithotrophic growth of the thermophilic iron-reducing bacteria Thermoterrabacterium ferrireducens and Thermoanaerobacter siderophilus . Microbiol 72: 16–167. [PubMed] [Google Scholar]
- 120. de Hoon MJL, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 121. Schmid U, Giesel H, Schoberth SM, Sahm H (1986) Thermoanaerobacter finnii spec. nov., a new ethanologenic sporogeneous bacterium. Syst Appl Microbiol 8: 80–85. [Google Scholar]
- 122. Lovitt RW, Longin R, Zeikus JG (1984) Ethanol production by thermophilic bacteria: physiological comparison of solvent effects on parent and alcohol-tolerant strains of Clostridium thermohydrosulfuricum . Appl Environ Microbiol 48: 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. He Q, Lokken PM, Chen S, Zhou J (2009) Characterization of the impact of acetate and lactate on ethanolic fermentation by Thermoanaerobacter ethanolicus . Bioresour Technol 100: 5955–5965. [DOI] [PubMed] [Google Scholar]
- 124. Cook GM, Morgan HW (1994) Hyperbolic growth of Thermoanaerobacter thermohydrosulfuricus (Clostridium thermohydrosulfuricum) increases ethanol production in pH-controlled batch culture. Appl Microbiol Biotechnol 41: 84–89. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Partial sequence alignment of selected PFK genes in different bacteria.
(PDF)
Selected genome metadata for sequenced Thermoanaerobacter spp.
(XLS)
All CAZyme designated gene sequences within sequenced Thermoanaerobacter strains as are available within the CAZy database or identified through de novo analysis.
(XLS)
Identified genes associated with the utilization of the major carbohydrates produced through lignocellulose hydrolysis in sequenced Thermoanaerobacter strains.
(XLS)
Genes associated with pyruvate metabolism in sequenced Thermoanaerobacter strains.
(XLS)
Genes associated with transmembrane ion gradient generating and consuming reactions involved with cellular energetics in sequenced Thermoanaerobacter genomes.
(XLS)
Key amino acid residues responsible for imparting predicted substrate specificity and K+ dependence in annotated Thermoanaerobacter V-type pyrophosphatases.
(XLS)
Identification of key residues characteristic of ATP or PPi dependent 6-phosphofructokinase genes in Thermoanaerobacter .
(XLS)