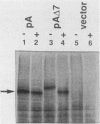
Abstract
Both 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase isozymes of the yeast Saccharomyces cerevisiae are predicted to contain seven membrane-spanning domains. Previous work had established the utility of the histidinol dehydrogenase protein domain, encoded by HIS4C, as a topologically sensitive monitor that can be used to distinguish between the lumen of the endoplasmic reticulum and the cytoplasm. This study directly tested the structural predictions for HMG-CoA reductase by fusing the HIS4C domain to specific sites in the HMG-CoA reductase isozymes. Yeast cells containing the HMG-CoA reductase-histidinol dehydrogenase fusion proteins grew on histidinol-containing medium if the HIS4C domain was present on the cytoplasmic side of the endoplasmic reticulum membrane but not if the HIS4C domain was targeted to the endoplasmic reticulum lumen. Systematic exchanges of transmembrane domains between the isozymes confirmed that both isozymes had equivalent membrane topologies. In general, deletion of an even number of putative transmembrane domains did not interfere with the topology of the protein, but deletion or duplication of an odd number of transmembrane domains inverted the orientation of the protein. The data confirmed the earlier proposed topology for yeast HMG-CoA reductase, demonstrated that the yeast enzymes are core glycosylated, and provided in vivo evidence that the properties of transmembrane domains were, in part, dependent upon their context within the protein.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama Y., Ito K. Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli. EMBO J. 1987 Nov;6(11):3465–3470. doi: 10.1002/j.1460-2075.1987.tb02670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. G., Orci L., Brown M. S., Garcia-Segura L. M., Goldstein J. L. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J Cell Sci. 1983 Sep;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]
- Audigier Y., Friedlander M., Blobel G. Multiple topogenic sequences in bovine opsin. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5783–5787. doi: 10.1073/pnas.84.16.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Finer-Moore J., Stroud R. M., Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988 Sep;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. A., Simoni R. D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, an integral glycoprotein of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1674–1678. doi: 10.1073/pnas.81.6.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Faust J. R., Goldstein J. L., Brown M. S., Luskey K. L. Sterols accelerate degradation of hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase encoded by a constitutively expressed cDNA. Mol Cell Biol. 1985 Apr;5(4):634–641. doi: 10.1128/mcb.5.4.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. F., Fogelman A. M., Edwards P. A. Transcriptional regulation of the 3-hydroxy-3-methylglutaryl coenzyme A reductase gene in rat liver. J Biol Chem. 1985 Nov 15;260(26):14363–14367. [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into the endoplasmic reticulum. J Cell Biol. 1987 Aug;105(2):633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Bouvier M., Benovic J. L., Caron M. G., Lefkowitz R. J. The multiple membrane spanning topography of the beta 2-adrenergic receptor. Localization of the sites of binding, glycosylation, and regulatory phosphorylation by limited proteolysis. J Biol Chem. 1987 Oct 15;262(29):14282–14288. [PubMed] [Google Scholar]
- Edwards P. A., Lan S. F., Fogelman A. M. Alterations in the rates of synthesis and degradation of rat liver 3-hydroxy-3-methylglutaryl coenzyme A reductase produced by cholestyramine and mevinolin. J Biol Chem. 1983 Sep 10;258(17):10219–10222. [PubMed] [Google Scholar]
- Finidori J., Rizzolo L., Gonzalez A., Kreibich G., Adesnik M., Sabatini D. D. The influenza hemagglutinin insertion signal is not cleaved and does not halt translocation when presented to the endoplasmic reticulum membrane as part of a translocating polypeptide. J Cell Biol. 1987 Jun;104(6):1705–1714. doi: 10.1083/jcb.104.6.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M., Blobel G. Bovine opsin has more than one signal sequence. 1985 Nov 28-Dec 4Nature. 318(6044):338–343. doi: 10.1038/318338a0. [DOI] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingami H., Brown M. S., Goldstein J. L., Anderson R. G., Luskey K. L. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J Cell Biol. 1987 Jun;104(6):1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesey J. K., Jr, Bigelis R., Fink G. R. The product of the his4 gene cluster in Saccharomyces cerevisiae. A trifunctional polypeptide. J Biol Chem. 1979 Aug 10;254(15):7427–7433. [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Learned R. M., Fink G. R. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase from Arabidopsis thaliana is structurally distinct from the yeast and animal enzymes. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2779–2783. doi: 10.1073/pnas.86.8.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. 3-Hydroxy-3-methylglutaryl-CoA reductase: a transmembrane glycoprotein of the endoplasmic reticulum with N-linked "high-mannose" oligosaccharides. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7165–7169. doi: 10.1073/pnas.80.23.7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985 Jan 10;260(1):522–530. [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Manoil C., Boyd D., Beckwith J. Molecular genetic analysis of membrane protein topology. Trends Genet. 1988 Aug;4(8):223–226. doi: 10.1016/0168-9525(88)90154-0. [DOI] [PubMed] [Google Scholar]
- Marsh L., Herskowitz I. STE2 protein of Saccharomyces kluyveri is a member of the rhodopsin/beta-adrenergic receptor family and is responsible for recognition of the peptide ligand alpha factor. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3855–3859. doi: 10.1073/pnas.85.11.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Parker R., Guthrie C. A point mutation in the conserved hexanucleotide at a yeast 5' splice junction uncouples recognition, cleavage, and ligation. Cell. 1985 May;41(1):107–118. doi: 10.1016/0092-8674(85)90065-0. [DOI] [PubMed] [Google Scholar]
- Schauer I., Emr S., Gross C., Schekman R. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985 May;100(5):1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro D. J., Rodwell V. W. Regulation of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and cholesterol synthesis. J Biol Chem. 1971 May 25;246(10):3210–3216. [PubMed] [Google Scholar]
- Wessels H. P., Spiess M. Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell. 1988 Oct 7;55(1):61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce "karmellae": a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988 Jul;107(1):101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., Huylebroeck D., Garoff H. Foreign transmembrane peptides replacing the internal signal sequence of transferrin receptor allow its translocation and membrane binding. Cell. 1987 Jan 16;48(1):147–155. doi: 10.1016/0092-8674(87)90365-5. [DOI] [PubMed] [Google Scholar]