Abstract
Deletion of single genes from expanded gene families in bacterial genomes often does not elicit a phenotype thus implying redundancy or functional non-essentiality of paralogous genes. The molecular mechanisms that facilitate evolutionary maintenance of such paralogs despite selective pressures against redundancy remain mostly unexplored. Here, we investigate the evolutionary, genetic, and functional interaction between the Helicobacter pylori cysteine-rich paralogs hcpG and hcpC in the context of H. pylori infection of cultured mammalian cells. We find that in natural H. pylori populations both hcpG and hcpC are maintained by positive selection in a dual genetic relationship that switches from complete redundancy during early infection, whereby ΔhcpC or ΔhcpG mutants themselves show no growth defect but a significant growth defect is seen in the ΔhcpC,ΔhcpG double mutant, to quantitative redundancy during late infection wherein the growth defect of the ΔhcpC mutant is exacerbated in the ΔhcpC,ΔhcpG double mutant although the ΔhcpG mutant itself shows no defect. Moreover, during early infection both hcpG and hcpC are essential for optimal translocation of the H. pylori HspB/GroEL chaperone, but during middle-to-late infection hcpC alone is necessary and sufficient for HspB/GroEL translocation thereby revealing the lack of functional compensation among paralogs. We propose that evolution of context-dependent differences in the nature of genetic redundancy, and function, between hcpG and hcpC may facilitate their maintenance in H. pylori genomes, and confer robustness to H. pylori growth during infection of cultured mammalian cells.
Introduction
Gene duplication provides the raw material for functional innovation, and is a source of genetic redundancy and phenotype robustness [1], [2], [3]. Evolutionary theories on the fate of duplicate genes are based on the premise that gene duplication creates functional redundancy thereby relieving selection pressure on one or both gene copies [1], [4], [5]. Thus, mutations normally deleterious to gene function escape purifying selection and, over time, the mutation-containing gene is pseudogenized and lost from the population owing to genetic drift. However, duplicate genes are retained when accumulating mutations cause complementary loss of functional attributes in each copy such that both are required for full functionality [5]. Rarely, some duplicate genes are retained because accumulated mutations confer a new advantageous function [1], [6]. Also, a duplicate gene may be retained because it provides a buffer against deleterious mutations in the ancestral gene [2], [3], [7], [8], [9], [10], [11]. However, because true genetic redundancy is evolutionarily unstable [12], at best transient [8], [13] and only theoretically sustainable on an evolutionary time-scale [3], its contribution to maintenance of duplicate genes remains a subject of intense debate [2], [3], [7], [9], [13].
Sequence-related gene families produced by gene duplications are frequently observed in the genomes of bacteria that maintain long evolutionary associations with their eukaryotic hosts [14], [15]. In such bacteria, gene family expansions are likely associated with host-specific adaptations [14], [15], [16], [17], [18]. Paradoxically, deletion of single genes from expanded gene families often has little or no phenotypic consequence (e.g., on fitness) implying redundancy among paralogs [19], [20]. Conceptually, a lack of a notable phenotype is generally equated with the ability of a system to continue functioning after genetic change, although underlying molecular mechanisms for this mutational robustness remain largely unidentified. In the context of bacterial infection and virulence, whether gene dispensability in expanded gene families reflects the ability of paralogs to functionally compensate each other or a lack of essentiality of ancestral function is not known. Moreover, bacterial genes that are redundant and not under sufficient selection should be rapidly deleted [21], and the evolutionary drive toward small specialist genomes in host-adapted bacteria [16] should exacerbate loss of redundant genes. The mechanisms that facilitate evolutionary maintenance of expanded gene families in bacterial genomes remain largely unexplored.
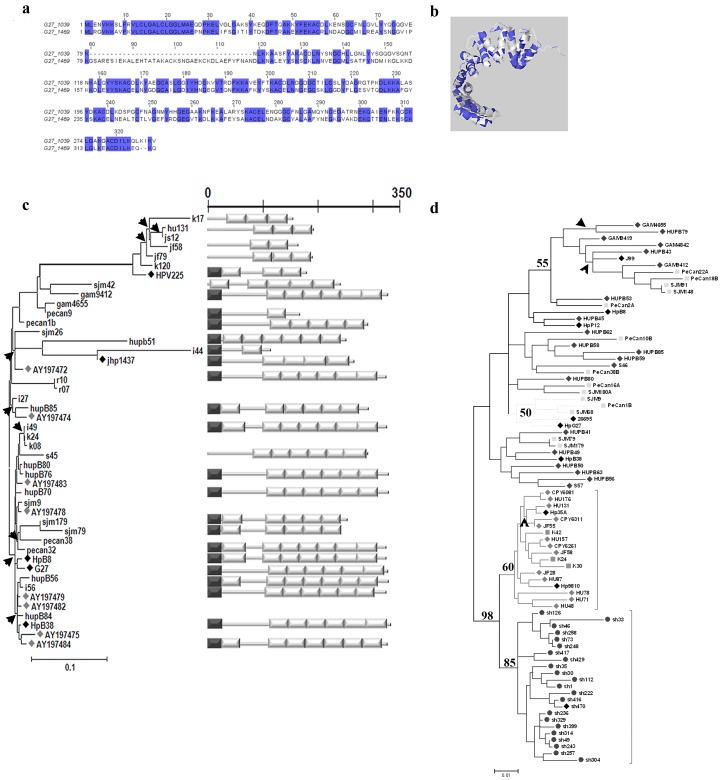
Helicobacter pylori is an important human pathogen in that it can establish decades-long infections and is the main cause of serious gastric diseases, including ulcers and cancer [22]. Nearly 17% of the H. pylori genome is composed of duplicate genes [23], [24], which are categorized into several gene families. Prominent among these is the Sel1-like gene family, which arose from H. pylori genome-specific expansion and contains eight diverse, rapidly evolving genes [18]. This gene family is characterized by the presence of modular Sel1-like repeat (Slr; PFAM entry, PF08238), which is eukaryotic in origin. Many of the Slr-containing genes encode Helicobacter cysteine-rich proteins (Hcp), which are highly immunogenic secreted proteins [18], [25], [26], [27], [28], [29], thought to contribute to H. pylori infection and pathogenesis. However, little is known about the genetic relationships among Slr-containing paralogs or about their functional relevance in the context of H. pylori infection or pathogenesis. For example, in H. pylori strain HpG27MA HcpG (HpG27_1469) is 53% similar to HcpC (HpG27_1039) ( Fig. 1a ; Table S6 in File S1) and predicted to adopt a helical conformation similar to that of HcpC ( Fig. 1b ). However, whereas 1) the Slr-gene hcpG (also called hsp12) is strain-specific, highly polymorphic, and apparently expressed under stress [30], and 2) the divergence of hcpG from its closest paralog in the H. pylori genome, hcpC, is driven by positive selection, indicating functional divergence between the two paralogs [18], hcpG appears dispensable to H. pylori growth in vitro under both normal and stress conditions [30]. Thus, does the dispensability of hcpG to H. pylori growth reflect that hcpG is genetically redundant with hcpC? And, does hcpC functionally compensate for the lack of hcpG? In the present study, we investigated the molecular evolutionary, genetic, and functional relationship between hcpG and hcpC to explore their relevance to H. pylori pathogenesis, and to gain general insight into the mechanisms that maintain duplicate genes in expanded bacterial gene families.
Figure 1. hcpG alleles are highly polymorphic whereas hcpC alleles are relatively conserved.
(a) Amino acid alignment of HcpC (G27_1039) and HcpG (G27_1469) from H. pylori strain G27MA. Identical amino acids are shaded in blue. (b) Mapping of amino acids shared by HcpC and HcpG onto the HcpC crystal structure. The alignment and amino acid mapping was performed using the Jalview multiple sequence editor (version 2.5.1). (c) Phylogenetic and corresponding domain architecture analyses of hcpG alleles (N = 46). Left, The ML phylogeny reconstructed using the TVM+Γ substitution model (Table S3 in File S1). Bar = 0.1 nucleotide substitutions per site and the arrowheads indicate hcpG lineages that experienced positive selection. Black diamonds, hcpG aleles obtained from sequenced H. pylori genomes; and grey diamonds, hcpG alleles obtained from GenBank. Right, domain architectures of representative hcpG alleles drawn to scale. The white rectangles are Slrs, and the black rectangles are predicted secretion signal sequences. (d) ML phylogeny of hcpC alleles (N = 81) reconstructed using the TrN+I+Γ substitution model (Table S4 in File S1). The geographic origins of the strains included are listed in Table S1 in File S1: black diamonds indicate hcpC alleles obtained from sequenced H. pylori genomes. Bar = 0.1 nucleotide substitutions per site. Arrowheads indicate hcpC lineages that experienced positive selection. Phylogenetic datasets used in generating panels (c) and (d) have been submitted to GenBank® with the following accession numbers: 1) hcpC dataset, KC007946–KC008026 and 2) hcpG dataset KC008027–KC008064).
Results
hcpG is Strain-specific, Highly Polymorphic, and Evolves Rapidly Subject to Positive Selection
This study began with our observation that hcpG was present in some fully sequenced H. pylori genomes but absent from others (Table S6 in File S1). Furthermore, in all genomes analyzed in this study hcpG appears to be a single copy gene whose chromosomal context is conserved (Table S6 in File S1). To confirm and extend these observations, we first sought to determine whether hcpG was present in 166 geographically diverse H. pylori isolates (Fig. S1 and Table S1 in File S1). Seventy-nine of the isolates were positive for hcpG with PCR primers located within genes flanking hcpG (Table S2 in File S1). PCR primers within hcpG-specific conserved internal regions confirmed the absence of hcpG among isolates that initially tested negative for hcpG. Thus, when present, the chromosomal context of hcpG appears conserved. We observed striking variation in the molecular size of hcpG in different H. pylori isolates (Fig. S3A in File S1). Thus, we next examined the molecular basis and effect of size variation on the modular domain architecture of hcpG alleles. The complete nucleotide sequences of the 79 PCR products described above revealed that 15 had a non-Slr containing gene in place of hcpG. Of the remaining 64 hcpG alleles, 28 encoded pseudogenes that contained premature stop codons in their nucleotide sequence introduced mostly via frameshift mutations (Fig. S3b in File S1). In hcpG nucleotide sequences encoding functional proteins (i.e., >100 amino acids), the number of Slr modules ranged from two to seven. Accordingly, the predicted proteins ranged from 107 to 330 amino acids with strikingly different domain architectures ( Fig. 1c ). To better understand hcpG evolution, we analyzed 46 unique alleles of hcpG phylogenetically. ML analysis revealed no significant geographic clustering of sequences or of different domain architectures ( Fig. 1c ). Next, we estimated selection pressures on individual hcpG codons and in branches of the phylogenetic tree using a combination of codon-based models of sequence evolution, ML, and Bayesian methods. Codon models that incorporated positive selection (ωS >>1) within the estimated parameters fit the data significantly better than those that did not ( Table 1 ), suggesting that functional hcpG alleles are subject to heterogeneous selective pressures. Moreover, Bayesian analysis confidently identified 11 sites under positive selection (Bayesian probability ≥0.99; ωS = 4.46) ( Table 1 ). Positive selection was also evident in several branches of the hcpG phylogenetic tree ( Fig. 1c and Table 1 ; Table S7 in File S1). Thus, we conclude that hcpG is highly polymorphic and present in only 38% of H. pylori strains, that some H. pylori strains only contain pseudogenized alleles while other strains only contain functional hcpG variants, and that positive selection maintains and drives the divergence of extant, functional hcpG alleles.
Table 1. Maximum-likelihood parameters of selection pressures acting on H. pylori hcpG codons.
| Model Code | -InL * | Tree length | κŦ | dN/dS | Parameter Estimates | D [d.f.]¶ | ?2 | P-value | Positively selected sites [Reference sequence: HpI49] |
| M0 (1-ratio) | 5242.663 | 3.2215 | 4.768 | 0.325 | ω = 0.325 | NA | NA | NA | NA |
| M1a (Nearly Neutral) | 5130.388 | 3.4698 | 4.867 | 0.341 | p0 = 0.742, ω0 = 0.111; p1 = 0.258, ω1 = 1 | NA | NA | NA | NA |
| M2a (Positive Selection) | 5108.087 | 3.5508 | 5.228 | 0.434 | p0 = 0.723, ω0 = 0.117; p1 = 0.256, ω1 = 1; [pS] = 0.021, [ωS] = 4.46 | M0 vs M2a [3] M1a vs M2a [2] | 269.152 44.602 | <0.0001<0.0001 | 28S, 30P, 36S, 58K, 66A, 79L, 262A |
| M3 (Discrete) | 5105.965 | 3.551 | 5.125 | 0.409 | p0 = 0.627, ω0 = 0.083; p1 = 0.334, ω1 = 0.674; [pS] = 0.039, [ωS] = 3.365 | M0 vs M3 [4] M1a vs M3 [4] | 273.396 44.846 | <0.0001<0.0001 | 21G. 28S, 30P, 36S, 58K, 66A, 79L, 221L, 224S, 262A, 313K |
| M7 (β) | 5135.398 | 3.4751 | 4.731 | 0.321 | p = 0.323, q = 0.684 | NA | NA | NA | NA |
| M8 (β,ω S >1) | 5106.781 | 3.556 | 5.096 | 0.405 | p = 0.465, q = 1.159; p0 = 0.961, [pS] = 0.393, [ωS] = 3.334 | M7 vs M8 [2] | 57.234 | <0.0001 | 21G. 28S, 30P, 36S, 58K, 66A, 79L, 221L, 224S, 262A, 272S, 313K |
| M8a (β, ωS = 1) | 5128.752 | 3.525 | 4.659 | 0.362 | p = 1.516, q = 9.983; p0 = 0.733, [pS] = 0.267, ωS = 1 | M8a vs M8 [1] | 43.942 | <0.0001 | |
| M1bra | 5177.115 | 3.2376 | 4.649 | NA | Free ω for each lineage | M0 vs M1bra | 131.1 | <0.002 | NA |
-InL, Log Likelihood Score.
∓, κ, kappa, ratio of transition to transversions.
, D, hierarchical Likelihood Ratio Test statistic and d.f., degrees of freedom.
hcpC Evolution is Characterized by Genomic Conservation and Relatively Stronger Functional Constraint
To better understand the divergence mechanisms of duplicate genes, we next characterized the evolutionary dynamics of hcpC. Although there are several slr genes encoded in H. pylori genome we focus here on hcpC because it is the closest paralog of hcpG [18]. We found that in contrast with hcpG, which was present in only a subset of H. pylori strains, hcpC was present in all H. pylori isolates screened. Complete nucleotide sequence analysis of the hcpC alleles from 100 H. pylori strains revealed that, unlike hcpG, their modular domain architecture was conserved. Also, hcpC alleles exhibited less overall nucleotide diversity than did hcpG alleles (Table S8 in File S1). Population genetic and ML phylogenetic analyses of 81 unique sequences revealed that unlike hcpG alleles, hcpC alleles clustered according to their geographic origins (Fig. 1d; Table S9 in File S1). Such geographic clustering is typical of H. pylori gene sequences. Because of striking differences in overall genomic conservation and evolution of hcpG and hcpC, we next examined the selective pressures on individual hcpC codons and branches of the hcpC phylogenetic tree. We found that hcpC codons experienced heterogeneous selective pressures similar to those on hcpG codons (Table 2). Corresponding Bayesian analysis confidently (Bayesian probability >0.99) identified 23 hcpC codons under positive selection (ωs = 2.73). All 23 sites mapped to the molecular surface of HcpC, some in close proximity to the experimentally identified peptide binding site [31] (Table 2; Fig. S4 in File S1). Thus, unlike hcpG, in which large-scale domain architecture changes are positively selected (Fig. 1c) and likely drive gain or loss of protein function [32], we predict that positive selection may simply fine tune hcpC functions by modulating its interaction with other bacterial or host proteins [18]. Most notably, although evolutionary rates vary significantly in the hcpC phylogenetic tree, few hcpC lineages preferentially accumulate non-synonymous substitutions (Fig. 1d, Table S10 in File S1). Moreover, the MacDonald-Kreitman test revealed that hcpC evolution was predominantly neutral in different populations (Table S9 in File S1). Note that this test was not useful on hcpG sequences because of lack of any fixed differences and geographic partitioning among hcpG alleles (Fig. 1C). Thus, we conclude that although divergence of hcpG and hcpC alleles is driven by positive selection, the intensity of positive selection is stronger on hcpG (ωs hcpG = 4.46 versus ωs hcpC = 2.73). Consequently, functional constraints on hcpC are likely stronger and manifest in its overall structural and genomic conservation.
Table 2. Maximum-likelihood parameters of selection pressures acting on H. pylori hcpC codons.
| Model Code | -InL * | Tree length | κŦ | dN/dS | Parameter Estimates | D [d.f.]¶ | ?2 | P-value | Positively selected sites [Reference sequence: HpSh30] | |
| M0 (1-ratio) | 6431.304 | 3.358 | 3.448 | 0.1311 | ω = 0.1311 | NA | NA | NA | NA | |
| M1a (Nearly Neutral) | 6137.837 | 3.447 | 3.478 | 0.135 | p0 = 0.881, ω0 = 0.018; p1 = 0.119, ω1 = 1 | NA | NA | NA | NA | |
| M2a (Positive Selection) | 6127.888 | 3.499 | 3.82 | 0.171 | p0 = 0.878, ω0 = 0.018; p1 = 0.102, ω1 = 1; [pS] = 0.019, [ωS] = 2.73 | M0 vs M2a [3] M1a vs M2a [2] | 606.832 19.898 | <0.0001<0.0001 | 9L, 43Q, 75Q, 161T, 229M, 248V | |
| M3 (Discrete) | 6124.796 | 3.483 | 3.695 | 0.155 | p0 = 0.794, ω0 = 0.004; p1 = 0.119, ω1 = 0.258; [pS] = 0.085, [ωS] = 1.4 | M0 vs M3 [4] M1a vs M3 [4] | 613.016 26.082 | <0.0001<0.0001 | 3E. 8S, 9L, 17A, 32L, 43Q, 46T, 75Q. 86S, 89A, 95D, 161T, 176A, 192G, 210D, 212V. 213A, 229M, 230Q, 248V, 249T, 273L, 281A | |
| M7 (β) | 6131.577 | 3.389 | 3.452 | 0.117 | p = 0.021, q = 0.145 | NA | NA | NA | NA | |
| M8 (β,ω S >1) | 6120.872 | 3.547 | 3.742 | 0.162 | p = 0.045, q = 0.375; p0 = 0.978 [pS] = 0.022, [ωS] = 2.51 | M7 vs M8 [2] | 21.41 | <0.0001 | 8S, 9L, 17A, 43Q, 46T, 75Q, 86S, 89A, 95D, 161T, 176A, 192G, 210D, 212V, 213A, 229M, 230Q, 248V, 249T, 281A | |
| M8a (β, ωS = 1) | 6129.478 | 3.464 | 3.426 | 0.127 | p = 0.102, q = 3.291; p0 = 0.895, [pS] = 0.104, ωS = 1 | M8a vs M8 [1] | 17.212 | <0.0001 | NA | |
| M1bra | 6324.72 | 3.362 | 3.458 | NA | Free ω for each lineage | M0 vs M1bra | 213.168 | <0.004 | NA | |
-InL, Log Likelihood Score.
κ, kappa, ratio of transition to transversions.
D, hierarchical Likelihood Ratio Test statistic and d.f., degrees of freedom.
hcpG is Expressed during H. pylori Growth and Infection, but Deletion of hcpG does not Cause Defects in H. pylori Growth or Fitness
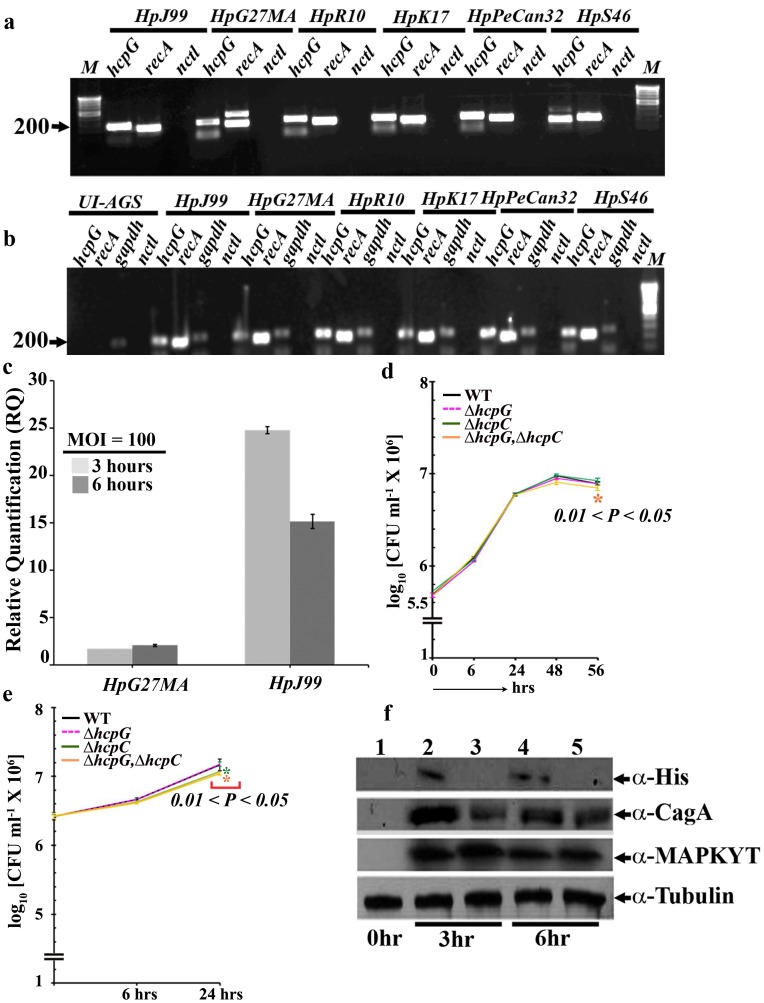
To test the functional essentiality of hcpG, we determined whether it was expressed in broth cultures during H. pylori growth and cultured AGS cells during H. pylori infection. Under both conditions, we detected robust hcpG mRNA transcripts from diverse strains ( Fig. 2a and 2b ). We next quantified the hcpG mRNA transcripts during infection of cultured AGS cells with the H. pylori strains HpG27MA and JHp99, both of which have been extensively characterized. This analysis revealed that 3 and 6 h after infection with HpG27MA, hcpG expression was up-regulated 2.0 and 2.5 fold, respectively. However, following infection with JHp99, hcpG expression was up-regulated 25-fold at 3 h but then down-regulated to 16-fold at 6 h ( Fig. 2c ). These data suggested that hcpG is expressed in vitro and during infection and that its expression is likely to be differentially regulated in distinct H. pylori strains.
Figure 2. Biological relevance of hcpG during H. pylori growth and infection.
(a) RT-PCR amplification of hcpG transcripts from 24 h growth in pure broth cultures. recA, positive control; nctl, negative control, in which extracted RNA was used as a template for RT-PCR. The geographic origins of the strains used were as follows: HpJ99, United States; HpG27MA, Italy; HpR10, South Africa; HpK17, South Korea; HpPeCan32, Lima, Peru; and HpS46, Spain. (b) RT-PCR amplification of hcpG transcripts in cultured AGS cells 6 h after infection (multiplicity of infection [MOI] = 100). gapdh was included as a positive control for AGS cell transcription in addition to the controls listed in Fig. 3a. UI, uninfected. (c). Quantitative RT-PCR analysis of hcpG transcripts in cultured AGS cells 3 and 6 h after infection with HpG27MA and HpJ99, respectively. The results show the averages from three experiments (± standard error of the mean). (d) Growth curves of WT HpG27MA and hcpG and/or hcpC derivatives in pure broth cultures; the results show averages from three experiments (± standard deviation [SD]). (e). Growth curves of WT HpG27MA strain and hcpG and/or hcpC deletion derivatives during infection of cultured AGS cells. The results show the averages from five experiments (± SD). (f). Verification of HcpG synthesis during infection of cultured AGS cells with the HpG27MA derivative expressing the HcpG::6xHis fusion protein; Lane 1, uninfected AGS cells (control); lanes 2 and 3, 3 h after infection with HpG27MA::HcpG-6XHis and HpG27MAΔhcpG, respectively; lanes 4 and 5∶6 h after infection with HpG27MA::HcpG-6XHis and HpG27MAΔhcpG, respectively. α-Tubulin was used as a measure for equal gel loading. CFU, colony-forming unit.
To assess the contribution of hcpG to H. pylori growth and infection, the growth of the ΔhcpG mutant strain in broth culture, and then during infection of cultured AGS cells was compared with that of the WTHpG27MA strain. This comparison revealed that the ΔhcpG mutant had no significant growth defect in either broth culture or during infection of cultured AGS cells ( Fig. 2d and 2e ).
We next determined whether the mild up-regulation of hcpG transcript expression seen in HpG27MA resulted in protein production. For this purpose we reverse engineered the ΔhcpG mutant strain by replacing the rpsL,catR cassette with a hcpG::6xHis fusion assembly (Fig. S2 in File S1). We detected the HcpG::6xHis fusion protein following infection of AGS cells with the HpG27MA::hcp-6xhis strain ( Fig. 2f ). Furthermore, the ΔhcpG mutant and HpG27MA::hcp-6xhis strains both triggered bacterial CagA translocation and subsequent activation of cellular MAPK (ERK2) in AGS cells suggesting that both strains initiated normal infection-induced signaling events [33] ( Fig. 2f ).
We next assayed the growth and fitness of the ΔhcpG mutant strain relative to that of the WT strain during competition in broth culture or during infection in cultured AGS cells. These competition assays revealed no significant reduction in growth or fitness of the ΔhcpG mutant strain in broth culture or during infection of cultured AGS cells ( Figs. 3a and 3d ; Figs. S5a and S5d in File S1). Thus, data from absolute and relative measures of fitness suggest that hcpG is dispensable to H. pylori growth and infection even though hcpG is expressed in vitro and during infection in cultured AGS cells.
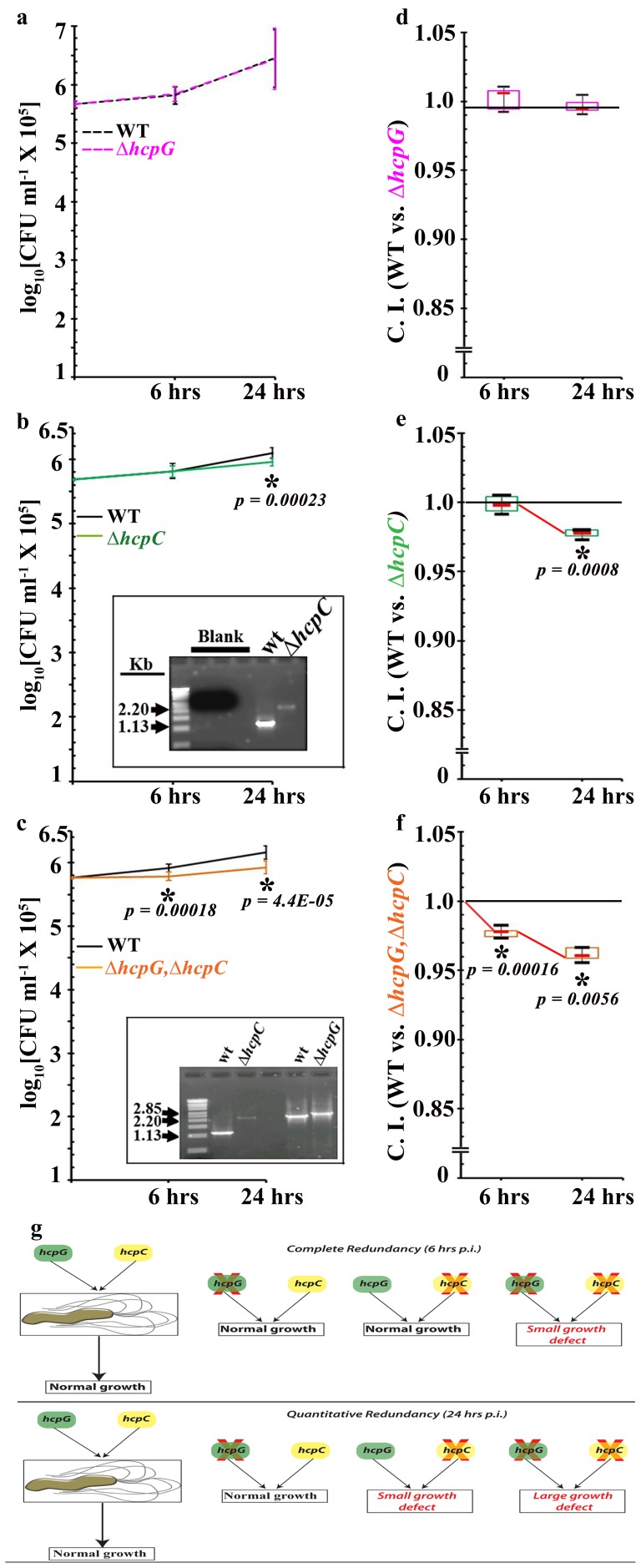
Figure 3. hcpC and hcpG can be completely or quantitatively redundant.
(a) Growth curves for the ΔhcpG mutant derivative and HpG27MA WT strain. (b) Growth curves for the ΔhcpC mutant and HpG27MA WT strains. Inset: PCR performed to verify the presence of both strains in the infection mix. (c) Growth curves for the ΔhcpG,ΔhcpC double mutant and HpG27MA WT strains. Inset, PCR performed to verify the presence of both strains in the infection mix. (d) Unbiased box and whisker plot representation of the CI for the ΔhcpG mutant. (e) CI measurements for the ΔhcpC mutant. (f) CI measurements for the ΔhcpC,ΔhcpG double mutant. The median reduction in the CI from 6 to 24 h after infection is indicated by the red connector. The asterisks indicate significant P-values. The results show the averages from five experiments (± SD). (g) Interpretation and representation of complete and quantitative redundancy of hcpG and hcpC in competition assays.
hcpG and hcpC are Completely and Quantitatively Redundant Specifically during Infection
We next asked whether the dispensability of hcpG indicated lack of essentiality of its ancestral function or some form of genetic buffering by its paralog, hcpC. To answer this question, we engineered two mutant H. pylori strains for use in growth and fitness assays: 1) HpG27MAΔhcpC, and 2) the HpG27MAΔhcpG,ΔhcpC double mutant [See supplementary methods in File S1]. We found that compared to the WT HpG27MA strain, the ΔhcpC mutant had no growth defects in pure broth culture ( Fig. 2d ). However, we observed a small but significant growth defect in the ΔhcpC mutant in cultured AGS cells 24 h after infection (0.01<P<0.05; Fig. 2e ). The ΔhcpG,ΔhcpC double mutant exhibited no defects for up to 48 h of growth in pure broth culture, but measurements at 56 h revealed a small but significant growth defect (0.01<P<0.05; Fig. 2d ). Similar to the ΔhcpC mutant, the ΔhcpG,ΔhcpC double mutant had a small growth defect in cultured AGS cells 24 h after infection (0.01<P<0.05; Fig. 2e ). These data suggest that hcpC may be required for optimal H. pylori growth late during infection of cultured AGS cells and that deletion of both hcpG and hcpC is mildly deleterious for growth in late broth culture and during late infection. Thus, collectively these results indicate a possible genetic interaction between hcpG and hcpC.
To confirm and clarify the nature of the genetic interaction between hcpG and hcpC, we co-cultured the ΔhcpC and ΔhcpG,ΔhcpC mutants, respectively, with the WT strain in broth cultures and during infection in cultured AGS cells. The ΔhcpC mutant exhibited no growth defects in competition assays in broth culture (Fig. S5b and S5e in File S1) or in cultured AGS cells 6 h after infection. However, unlike the ΔhcpG mutant, the ΔhcpC mutant had a significant growth defect and reduced fitness relative to that of the WT strain in cultured AGS cells 24 h after infection ( Fig. 3b and 3e ). Strikingly, the ΔhcpG,ΔhcpC double mutant, unlike the ΔhcpC and ΔhcpG single mutants, had significant fitness reduction relative to that of the WT strain in cultured AGS cells 6 h after infection ( Fig. 3c and 3f ). Moreover, the ΔhcpG,ΔhcpC double mutant showed significant fitness reduction, even more than that observed for the ΔhcpC mutant, in cultured AGS cells 24 h after infection ( Fig. 3c and 3f ). Similar to the ΔhcpG and ΔhcpC mutants, the ΔhcpG,ΔhcpC double mutant experienced no fitness reduction when co-cultured with the WT strain in broth culture (Fig. S5c and S5f in File S1). Thus, we identify two categories of genetic interactions between hcpG and hcpC depending on the temporal context of H. pylori infection ( Fig. 3g ). First, during early infection, hcpG and hcpC are completely redundant in that disrupting either gene alone has no effect on H. pylori growth or fitness, but disrupting both genes causes significant reduction in H. pylori growth fitness. Second, during late infection hcpG and hcpC are quantitatively redundant in that the fitness phenotype of the ΔhcpC mutant is exacerbated in the ΔhcpG,ΔhcpC double mutant. Thus, hcpG and hcpC are coupled in a redundant relationship that differs depending on the temporal context of the infection. We conclude that context-dependent redundant relationships between hcpG and hcpC contribute significantly to the mutational robustness of H. pylori growth during infection and likely contribute to the retention of hcpG along with hcpC in select H. pylori genomes.
Molecular Role of hcpG and hcpC: Infection-induced Signaling-dependent Regulation of H. pylori HspB/Hsp60/GroEL Export
We next considered molecular mechanisms underlying the genetic redundancy of hcpG and hcpC. Using a combination of co-immunoprecipitation and mass-spectrometry the Helicobacter HspB/Hsp60/GroEL chaperone was identified as a potential interacting partner of HcpC [34]. Using ELISA we confirmed that HcpC can bind directly to HspB ( Fig. 4a ). HspB/Hsp60/GroEL is an essential chaperone that is cytoplasmic in most bacteria except H. pylori in which it can also be translocated to the bacterial surface or extracellular milieu [35], [36]. The translocated HspB protein then associates with the UreB subunit of the H. pylori urease complex and contributes to H. pylori pathogenesis via multiple pathways [37], [38]. The mechanism of HspB translocation is controversial, although it is likely translocated actively [36]. Because Slr-containing proteins facilitate protein-protein interactions, we hypothesized that the redundant partners HcpC and HcpG may mediate or modulate HspB translocation.
Figure 4. Modulation of HspB translocation by HcpG and HcpC during H. pylori infection.
(a) ELISA performed to determine the specificity of HcpC-HspB interaction, in vitro. (b–e) FACS analyses of HspB translocation in single- and double hcp gene mutants and in the parent HpG27MA WT strain (b) 3, (c) 6, (d) 12, and (e) 24 h after infection of serum-starved confluent AGS cell cultures. Overlapping histograms of mean geometric intensity from individual infections were generated using the WINMDI software program (version 2.9; Joseph Trotter, Scripps Research Institute, LA Jolla, CA). x-axis, α-HspB fluorescence; y-axis, number of events measured for each sample (104 cells/sample). A shift of fluorescence to the left indicates reduced intensity. The FACS parameters are listed in supplementary methods available in File S1. Inset, bar graphs showing the geometric mean intensity of α-HspB fluorescence at each time point after infection with mutant and WT strains, respectively. The strains are color-coded as follows: Black, WT; blue, ΔhcpC; red, ΔhcpG; and green, ΔhcpG,ΔhcpC. The results show the averages from three experiments (± SD). (f). Model of observed functional interactions of HcpC, HcpG, and HspB translocation depending on the temporal context of the infection.
To determine whether HspB translocation is affected by hcpG or hcpC, we analyzed HspB expression in unpermeabilized WT and mutant H. pylori strains in broth cultures and during infection of AGS cells using fluorescence-activated cell sorter (FACS) analysis. We found that 3 h after infection, HspB fluorescence was significantly reduced in the ΔhcpG and ΔhcpC single mutants and in the ΔhcpG,ΔhcpC double mutant ( Fig. 4b ). Strikingly, 6 h after infection with the, HspB fluorescence in the ΔhcpG mutant strain recovered nearly to the same level as seen with WT strain ( Fig. 4c ); by 12 and 24 h after infection, the ΔhcpG mutant and WT strain demonstrated similar HspB fluorescence levels ( Fig. 4d and 4e ). In contrast, HspB fluorescence remained significantly reduced throughout the infection in the ΔhcpC mutant and ΔhcpG,ΔhcpC double mutant ( Figure 4b–4e ). Importantly, parallel experiments with pure broth cultures maintained for up to 56 h revealed similar HspB fluorescence levels in the WT and mutant strains (Fig. S6 a–c in File S1). Thus, these data demonstrate apparent modulation of HspB translocation specifically in response to infection-induced signals.
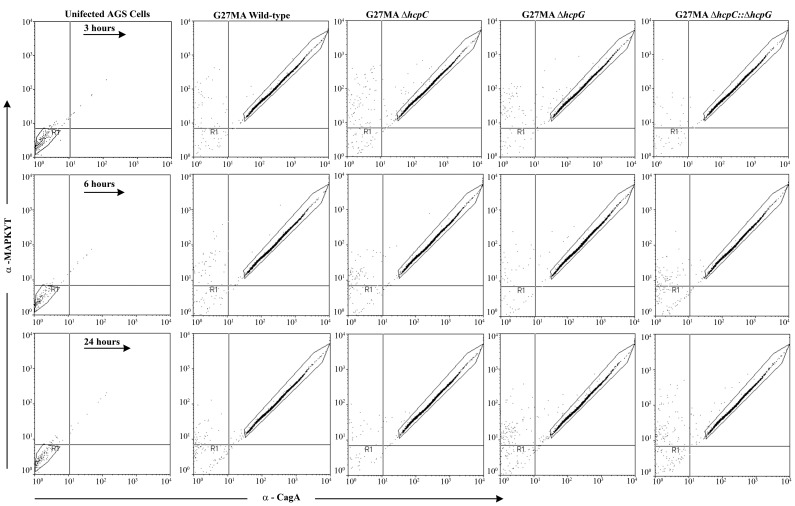
To confirm that the modulation of HspB expression did not reflect generalized disruption of infection-induced signaling events in response to deletion of hcpG and/or hcpC, we monitored CagA and MAPK (ERK2) expression levels in infected permeabilized AGS cells using FACS analysis. We found that the mutant and WT strains described above all similarly triggered the release of bacterial CagA accompanied by activation of cellular MAPK ( Fig. 5 ). Thus, hcpG and hcpC specifically modulate HspB expression whereas independent H. pylori infection-induced signaling events remain unaffected.
Figure 5. Deletion of hcpG and/or hcpC does not alter independent infection-induced signaling events.
CagA and activated MAPK (ERK2) expression in hcp mutants, WT G27MA and uninfected AGS cells over the course of infection. A shift in the fluorescence to the upper right quadrant in AGS cells infected with either WT or hcpC or hcpG mutants compared with uninfected AGS cells indicates increased CagA and activated-MAPK expression after infection. Results are representative of three experiments.
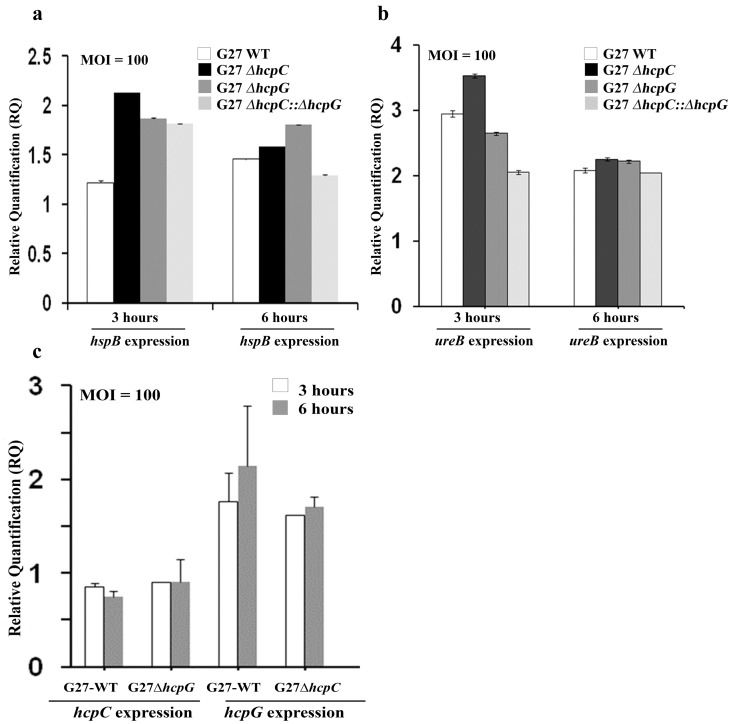
We next asked whether hcpG and hcpC affected the transcriptional regulation of hspB, which in turn may alter HspB expression. Real-time (RT) PCR analysis revealed no significant alterations in hspB expression levels in the mutant and WT strains following infection of AGS cells ( Fig. 6a ). Because HspB is known to associate with the UreB subunit of the urease complex we also measured the ureB expression levels to rule out indirect causes of altered HspB expression. We found no significant alteration in ureB expression in the mutant and WT strains at equivalent intervals during infection ( Fig. 6b ). Thus, these data suggest that altered HspB fluorescence does not result from modulation of hspB expression in hcpG or hcpC mutants.
Figure 6. hcpC and hcpG are transcriptionally uncoupled, and do not affect hspB or ureB expression during H. pylori infection of AGS cells.
(a) Fold change in hspB expression levels 3 and 6 h after infection with WT and hcp mutant bacteria. (b) Fold change in ureB expression levels 3 h and 6 h after infection with WT and hcp mutant bacteria. A P>0.1 (not significant) was observed using a one-way ANOVA test in comparisons of observed differences. (c) Fold change in hcpC expression levels in the ΔhcpG mutant of G27MA and in hcpG expression levels in the ΔhcpC mutant 3 and 6 h after infection. A P>0.1 (not significant) was observed using a one-way ANOVA test in comparisons of observed differences.
We next determined whether the deletion of hcpG resulted in the upregulation of hcpC expression, which, if true, could explain the apparent normalization of GroEL/HspB fluorescence to WT levels in the ΔhcpG mutant ( Fig. 4c–4e ). We found that hcpC expression in the ΔhcpG mutant was not significantly affected ( Fig. 6c ). Similarly, deletion of hcpC had no significant effect on hcpG expression. Together these data suggest that hcpC and hcpG are transcriptionally uncoupled.
Taken together, our results reveal two categories of functional interactions between hcpG and hcpC depending on the temporal context of H. pylori infection ( Fig. 4f ). First, during early infection, hcpG and hcpC are both essential for optimal HspB translocation and that neither of them can functionally compensate for deletion of the other gene. Thus, hcpG and hcpC are selected independently to perform HspB translocation. Second, during middle to late infection, hcpC alone is necessary and sufficient for optimal HspB translocation whereas hcpG is not required for it. Given the quantitatively redundant fitness phenotypes exhibited by hcpG and hcpC, our results suggest that these two genes are likely important because of their capacity to perform distinct functions. We conclude that hcpG and hcpC partially overlap in their function but lack the generic functional backup capacity expected among genetically redundant paralogs.
Discussion
Simultaneous Occurrence of Different Types of Genetic Interactions between hcpG and hcpC in H. pylori Infection
Genetic buffering interactions are most commonly studied by measuring the fitness of an organism under standard laboratory growth conditions, in which the spatial and temporal flux in the organism’s interactions with its environment is inherently underestimated. Moreover, in most cases, the molecular functions underlying such genetic interactions remain relatively unexplored. In our study, by taking into account the temporal context of H. pylori infection modeled in cultured mammalian cells, we uncovered the simultaneous occurrence of different types of genetic buffering interactions between the H. pylori paralogs hcpG and hcpC. A recent study reported that multiple genetic interactions in yeast paralogs conferred robustness to yeast signaling and regulatory networks [11]. In bacteria, studies of duplicate genes have historically focused on tandem gene duplications (gene amplification) but rarely on expanded gene families [39]. To the best of our knowledge, no previous reports have generally described multiple genetic buffering interactions among duplicate genes from expanded bacterial gene families in the context of pathogen-host interaction. Moreover, our data caution against the prevalent notion that absence of phenotypes upon deletion of single genes from expanded gene families reflects either compensation of function by other paralogs or lack of essential function of paralogs in mediating pathogen-host interactions.
Selective Advantage of hcpG and hcpC in H. pylori Infection
Previous studies have proposed that maintenance of redundant paralogs can have several selective advantages, including mutational robustness [2], [3], [7], robustness against random fluctuations in gene expression [8], [9] and robustness of regulatory signaling networks [11]. In bacteria, tandem gene duplications are known to contribute to specific environmental adaptations [39]. Our present findings clearly show that genetic redundancy of hcpG and hcpC contributes significantly to the mutational robustness of H. pylori growth specifically during infection of cultured AGS cells ( Figs. 3a–3f ). Genetic redundancy among paralogs generally tends to be condition-dependent [40]; thus, that effects of hcpG and hcpC deletions on H. pylori growth and fitness were more apparent specifically during infection, a physiologically more relevant condition, and not in pure broth culture is not surprising (Figs. S5a–S5f in File S1). What is surprising, however, is that the nature of this genetic redundancy switches from complete to quantitative depending on the temporal context of the infection ( Fig. 3g ). This suggests that hcpG and hcpC are also coupled via infection-induced regulatory links that mediate such switches. Constituting such a regulatory module can be advantageous for H. pylori because depending on when hcpG and hcpC are active, regulation of distinct processes mediated by them (see discussion below) can be coupled or uncoupled from each other in response to temporal or spatial context of the infection.
The dependence of paralogous redundancy on the context of H. pylori infection observed in the present study and in other earlier studies [40], [41] argues against a predominantly compensatory (backup) function of duplicate genes. During early infection, both hcpG and hcpC are essential for optimal HspB translocation, and neither of them functionally compensates for a lack of their redundant partner. This suggests that hcpG and hcpC are specialized in distinct manners to perform HspB translocation during early infection. This is intriguing, because from middle to late infection, HcpC alone appears to be necessary and sufficient for HspB translocation, whereas HcpG is dispensable and unable to functionally rescue HcpC deletion despite its apparent ability to mediate HspB translocation in early infection ( Fig. 4b–4e ). We ascribe the lack of generic backup functional capacity between hcpG and hcpC to two distinct factors: dosage amplification and functional or regulatory divergence.
Theoretical studies have suggested that duplicate genes whose products mediate stress responses or generally mediate organism-environment interactions can be retained in genomes of such organisms by positive selection for increased dosage [42], [43]. Specifically, surface-associated HspB appears to be important for gastric colonization early in H. pylori infection [44]. Thus, efficient, rapid HspB translocation early during infection should favor successful H. pylori colonization. We also observed that during early infection, both hcpG and hcpC were expressed at relatively low levels in WT HpG27 strain, and expression of both genes was not significantly altered when their redundant partner was deleted ( Fig. 6c ). Thus, given their relatively low expression levels, HcpG and HcpC appear independently selected because of their combined contribution to efficient and rapid HspB translocation in early H. pylori infection.
Temporal variation in the relative necessity of H. pylori paralogs for HspB translocation indicates the potential regulatory influence of infection-induced cellular signals on HcpG and HcpC activity and function. This regulatory influence is also suggested by our observation that hcpG could not functionally rescue hcpC deletion ( Figure 4b–4e ). Thus, we speculate that whereas early infection-induced signals likely activate both HcpG and HcpC the transition from early infection to middle to late infection phase predominantly elicits HcpC-activating signals ( Fig. 4f ). Two additional lines of evidence suggest functional divergence between hcpG and hcpC during late infection: (1) the additive effect of hcpG deletion on the ΔhcpC phenotype during late infection suggests that hcpG and hcpC likely perform distinct functions; and (2) positive (or diversifying) selection on extant hcpG and hcpC allele also suggests potential functional divergence between HcpG and HcpC. Collectively, these data suggest that during late infection hcpG and hcpC appear to be selected primarily for their divergent functions that are likely regulated by infection-induced signals.
Evolution of hcpG and hcpC Duplication in H. pylori Populations
The context-variable redundancy of hcpG and hcpC described here may also have broader implications on understanding the evolution of gene duplications. Models explaining the maintenance of paralogs typically invoke functional subdivision and/or novelty in duplicate copies and are classified into multiple categories [42]. Although, our present data best fit the positive dosage model [42], [43], in which paralogs are selected independently because of their cumulative contribution to the same function, additional models may be required because of different redundancy dynamics during late infection. The positive dosage model predicts two possible outcomes based on the strength of selection on cumulative hcpG and hcpC action: under strong selection, the duplicate copy (hcpG) may be quickly fixed whereas under weak selection, a null mutation may become fixed by random genetic drift resulting in hcpG pseudogenization, and eventually loss of hcpG from H. pylori populations ( Fig. 7 ). In highly variable environments, such as those in H. pylori’s more than three billion human hosts, the strength of selection on increased dosage may periodically wax and wane, leading to cyclical gene duplication and gene loss [45]. Such variation in strength of selection may underlie the three distinct hcpG and hcpC genotype combinations we observed in H. pylori populations ( Fig. 7 ). Thus, each strain-specific hcpG-hcpC genotype may reflect an individual host-specific adaptation of that H. pylori strain. Alternatively, if cyclical bursts of gene duplication and pseudogenization are common then the presence of pseudogenized hcpG alleles in H. pylori populations could be re-interpreted to reflect inferior hcpG variants outcompeted by hcpG alleles better adapted to a specific biochemical niche [45]. Such cyclical duplication-pseudogenization may permit selection to explore a wide mutational landscape, likely fixing only those hcpG variants that provide additional functional capability ( Fig. 7 ). The extreme genetic heterogeneity among functional hcpG alleles driven by positive selection further supports the possibility that hcpG alleles may encode functionally divergent proteins. Taken together, we propose that context-variable redundant behavior and coupling of paralogs via regulatory links generated by infection-induced signals may have wide-ranging implications on understanding of the evolution of gene duplications, and may require additional sub classification of existing models.
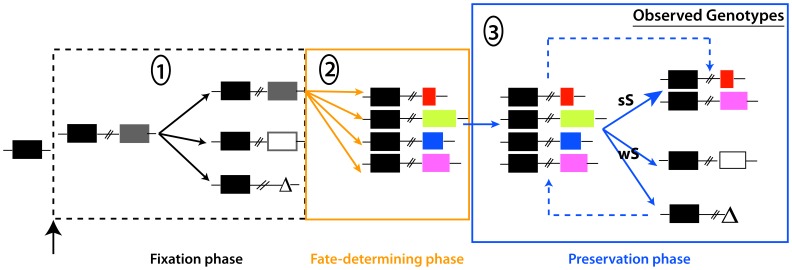
Figure 7. Simplified model of stable maintenance of hcpC and hcpG in H. pylori populations.
The fate of a duplicate gene can be studied in three distinct phases beginning with fixation of the duplicate in the H. pylori population [42]. We propose that in the preduplication phase, hcpC is fixed in H. pylori populations; the fixation phase begins with the origin of hcpG. During the fixation phase, the duplicate copy escapes loss because of genetic drift and is fixed. This initiates the second phase, in which the duplicate accumulates fate-determining mutations. We suggest that multiple fate-determining mutations give rise to polymorphic hcpG variants. In the preservation phase, hcpC and hcpG are maintained subject to selection intensity for 1) dosage amplification in early infection or 2) functional divergence during late infection. Strains harboring pseudogenized hcpG or lacking hcpG altogether are postulated to reflect weak selection or competition among variants for occupation of specific biochemical niches, respectively. The dashed line in the preservation phase indicates cyclical duplication and loss events that may periodically give rise to hcpG alleles with new functional capacities; only well adapted hcpG variants survive whereas the others are pseudogenized. Black rectangles, hcpC; grey rectangle, duplicate copy; white rectangle, pseudogenized hcpG; Δ, hcpG deletion; colored rectangles, hcpG variants; sS, strong selection; wS, weak selection.
Implications for H. pylori Pathogenesis
Our study has identified two new potential bacterial determinants, HcpG and HcpC, which may contribute to H. pylori pathogenesis via regulation of HspB/GroEL/Hsp60 translocation or export to the bacterial surface. The H. pylori HspB/GroEL/Hsp60 appears essential for H. pylori colonization during early infection [44], for induction of innate immune responses [37], and can enhance angiogenesis [38] and tumorigenesis [46]. In most bacteria the HspB chaperone protein is cytoplasmic but in H. pylori this protein is often found on bacterial surface and in the extracellular milieu. The mechanism of HspB export is somewhat controversial. While Phadnis et al [35] argued that HspB is reabsorbed to intact cell membranes following its release into the extracellular milieu via autolysis, Vanet and Labigne [36] showed that HspB/GroEL/Hsp60 more likely underwent active secretion rather than autolysis. Because of the apparent HcpG- and HcpC-dependent modulation of HspB translocation in intact unpermeabilized H. pylori cells, and the demonstration that HcpC can directly interact with HspB, we favor the idea that HspB may be actively secreted rather than exported via autolysis. The precise mechanism of how HcpC and HcpG might mediate HspB export, however, remains to be determined. It will be important to determine whether HcpG can also interact directly with HspB, whether HcpG and HcpC co-localize with HspB to the bacterial surface, and identify the infection-induced signals that seem to temporally regulate the functions and/or activity of HcpC and HcpG in the context of infection.
Materials and Methods
H. Pylori Strains
The 166 H. pylori isolates included in this study were obtained from phylogenetically distinct European (Spain), African (The Gambia and South Africa), East Asian (Japan, South Korea), South Asian (India), and South American (Lima and Shimaa, Peru) populations (Figure S1 and Table S1 in File S1). All of these strains were obtained from patients who had sought medical attention and undergone endoscopic biopsies after giving their informed consent at their respective institutions. Detailed listing of strains, along with their culture, growth and maintenance conditions is described in supplementary methods available in File S1.
DNA Extractions, PCRs and Nucleotide Sequencing
Standard procedures were used for extracting H. pylori genomic DNA [47], PCR and nucleotide sequencing. Briefly, specific PCRs were carried out in 25 µl reaction mixtures in a PCR buffer supplied by the manufacturer (Biolase; MidSci, St. Louis, MO) and containing 5–10 ng of genomic DNA, 1 U of Taq polymerase (Biolase; MidSci), 1.5 mM MgCl2, 0.8 pmol each of forward and reverse primers, and 100 µM dNTP mix for 30–35 cycles of denaturation (94°C), annealing (55°–64°C, as required), and elongation (68°–72°C, as required). PCR products were purified (QIAquick PCR purification kit; Qiagen, Valencia, CA) and then quantified using the Biophotometer (Eppendorf, Hauppauge, NY). Nucleotide sequencing was performed using both DNA strands at the high-throughput genomics unit at the University of Washington, Seattle, WA. The primers used in PCR, and nucleotide sequencing are listed in Table S2 in File S1.
Phylogenetic Analyses and Computations
hcpC and hcpG nucleotide sequences were assembled and edited using the Seqman suite in the Lasergene software program (DNASTAR, Madison, WI). An hcpC multiple sequence alignment (MSA) was generated using MEGA version 5 [48] (www.megasoftware.net). The hcpG MSA was generated as follows: an initial MSA was generated by aligning HcpG sequences with crystal structures of H. pylori Slr proteins HcpB [49] and HcpC [31] using EXPRESSO [50] (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi). Because of high polymorphism levels in hcpG sequences, this alignment was further edited manually based on the biochemical features of amino-acids to correct for obvious mismatches. The resulting MSA was used to manually derive a corresponding nucleotide alignment using MEGA alignment editor. Phylogenetic reconstruction and analyses of selection pressures acting on hcpC and hcpG codons and lineages were performed using maximum likelihood (ML) methods implemented in PAUP* version 4b10 and PAML version 4.3b, respectively. Best-fit models of DNA sequence evolution used in phylogenetic reconstruction were selected using MODELTEST (Table S3 and Table S4 in File S1). Details of these analyses are described in supplementary methods available in File S1. Phylogenetic datasets generated in this study have been submitted to GenBank® (Accession numbers: 1) hcpC dataset, KC007946–KC008026 and 2) hcpG dataset KC008027–KC008064).
Population genetic computations
Estimates of total nucleotide diversity (π), Waterson’s θ, nucleotide diversity at synonymous and nonsynonymous sites (πS and πA), genetic differentiation among populations (FST) with accompanying permutation tests, and the McDonald-Kreitman tests and associated estimates of α, the proportion of amino acids under positive selection [51] were obtained using the DNASP software program (version 5.1) [52].
Structural and domain architecture analyses
Domain architecture analysis of hcpG and hcpC sequences was performed using the Simple Modular Architecture Research Tool (SMART) [53]. Positively selected residues of HcpC were mapped to the surface of the HcpC crystal structure (PDB code 1OUV [31]) using the PYMOL molecular visualization system (http://www.pymol.org).
H. pylori Genetic Engineering
hcpC and hcpG knockout derivatives (ΔhcpC and ΔhcpG single mutants and ΔhcpC,ΔhcpG double mutant) of H. pylori strain G27MA and the hcpG::6xHIS knock-in G27MA strain were generated using the streptomycin contraselection-based method described previously [54] while incorporating small modifications. Strategy for generating knockout and knock-in strains is shown in Fig. S2 in File S1 and is described in detail in supplementary material available in File S1.
Growth and Fitness Assays
In vitro growth and fitness assays
H. pylori G27MA WT and its hcpG and/or hcpC deletion derivatives were grown on BHI plates containing appropriate selective antibiotics for 3 days. Fifty milliliter tissue culture flasks were then inoculated with a bacterial suspension derived from plate cultures (OD600 = 0.05/mL for each strain). A liquid medium comprising BHI broth supplemented with 1% IsoVitaleX, 1% H. pylori-selective supplement, and 10% fetal bovine serum (FBS) (GIBCO, CA) was used to culture the H. pylori strains. Flasks were initially incubated at 37°C for 30 min in a 5% CO2 incubator and then transferred to GasPak jars and incubated at 37°C with shaking (120 rpm) for a maximum of 56 h [55]. At specific intervals, cell aliquots were from culture flasks were diluted serially and plated on selective BHI plates to enumerate the WT and mutant colony-forming units (CFUs). The log-transformed CFU mL−1 count was used to determine the competitive index (CI) in co-culture experiments. The CI was calculated according to the ratio of mutant to WT bacteria at each time point compared with the ratio of mutant to WT bacteria in the inoculum [56], [57]. A CI value greater than one indicated that the mutant out-competed the WT, whereas a value less than or equal to one indicated that the WT out-competed the mutant. Growth assays in pure cultures and fitness assays in broth co-cultures were repeated three times, and the statistical significance of observed differences in the growth or fitness of hcpC and/or hcpG mutants and WT G27MA was determined using a t-test with α = 0.05.
Growth and fitness assays during H. pylori infection of cultured AGS cells
Before each experiment in cultured AGS cells, bacteria were passaged once on BHI horse blood agar plates under standard microaerobic conditions as recommended [33]. AGS cells (ATCC CRL 1739) were normally cultured and maintained in antibiotic-free high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated FBS. Cells were allowed to grow and maintained in 50 mL tissue culture flasks at 37°C in a humidified atmosphere of 5% CO2. AGS cells were seeded at a density of 1×105 cells/mL into six-well plates and then allowed to grow to 80% confluence [58]. Just prior to infection of cultured AGS cells with H. pylori, the cell-medium mixture was removed and replaced with fresh DMEM containing 20% FBS. Bacteria were harvested from pure liquid cultures in BHI and washed in phosphate-buffered saline (PBS; pH 7.4); AGS cells were infected at an MOI of ∼ 100 H. pylori cells per AGS cell. Plates were centrifuged for 10 min at 1,000 g to ensure bacterial contact with the AGS cells and incubated at 37°C in a humidified atmosphere of 5% CO2. At specific intervals cells were gently scraped from a well, mixed, diluted serially, and plated on selective BHI plates to enumerate the WT and mutant CFUs. In co-cultures experiments the CI was calculated as described above. Each experiment was repeated five more times, and the statistical significance of observed differences in WT and mutant strains was calculated using a t-test with α = 0.05.
FACS, Immunoblot Analysis and ELISA
Antibodies
Antibodies used in this study are listed in supplementary methods available in File S1.
HspB expression dynamics in vitro and during infection
HspB expression was studied in unpermeabilized bacterial cells grown in pure cultures and during infection of cultured AGS cell using a FACS-Calibur™ flow cytometer (BD, Franklin Lakes, NJ). The data were analyzed using the WinMDI software program (version 2.9). HspB, CagA and activated-Mitogen Activated Protein Kinase (MAPK, ERK2) expression during infection was studied using serum-deprived AGS cells that were allowed to grow to 80% confluence. Parameters used in the FACS analyses are listed in Table S5 in File S1. Detailed methods for FACS analyses are presented in supplementary material available in File S1.
Protein expression, purification and immunoassays
HcpC fused to N-terminal MBP and C-terminal histidine tags was expressed and purified as described previously [26]. HspB (gene hp0010 from H. pylori strain 26695) was cloned into a pGEX-6P expression vector (GE Healthcare) and purified GST-HspB was concentrated using ultrafiltration and stored in PBS buffer supplemented with 10% (v/v) glycerol at −80°C. All ELISA experiments were performed using Nunc Maxisorp 96-well plates at volumes of 100 µL/well. Details of HspB expression, purification and subsequent use in ELISA are provided in supplementary methods available in File S1.
Reverse-transcription and Real-time PCRs
Standard methods were used and are detailed in supplementary material available in File S1.
Supporting Information
Detailed description of methods and supplementary figures (Fig. S1– Fig. S6) and tables (Table S1–Table S10).
(DOCX)
Acknowledgments
We thank Drs. Hideki Innan and Fyodor Kondrashov for discussions and comments on the manuscript. We thank Prof. Yousef Abu Kwaik and members of his laboratory for their help and advice with cell culture and RT-PCR experiments.
Funding Statement
S.A.M. and L.E.B. were supported by the university of Louisville undergraduate summer research fellowship. This study was not funded by any dedicated grants received from federal, state or private sources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ohno S (1970) Evolution by gene duplication.: Springer-Verlag.
- 2. Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, et al. (2003) Role of duplicate genes in genetic robustness against null mutations. Nature 421: 63–66. [DOI] [PubMed] [Google Scholar]
- 3. Nowak MA, Boerlijst MC, Cooke J, Smith JM (1997) Evolution of genetic redundancy. Nature 388: 167–171. [DOI] [PubMed] [Google Scholar]
- 4. Zhang J (2003) Evolution by gene duplication: an update. Trends in ecology & evolution (Personal edition) 18: 292–298. [Google Scholar]
- 5. Lynch M, Conery JS (2000) The Evolutionary Fate and Consequences of Duplicate Genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 6. Burki F, Kaessmann H (2004) Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet 36: 1061–1063. [DOI] [PubMed] [Google Scholar]
- 7. DeLuna A, Vetsigian K, Shoresh N, Hegreness M, Colon-Gonzalez M, et al. (2008) Exposing the fitness contribution of duplicated genes. Nat Genet 40: 676–681. [DOI] [PubMed] [Google Scholar]
- 8. Kafri R, Dahan O, Levy J, Pilpel Y (2008) Preferential protection of protein interaction network hubs in yeast: Evolved functionality of genetic redundancy. Proceedings of the National Academy of Sciences 105: 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kafri R, Pilpel T (2010) Evolutionary and Functional Aspects of Genetic Redundancy. In: Dittmar K, Liberles D, editors. Evolution After Gene Duplication. Hoboken, New Jersey: Wiley-Blackwell. 229–252.
- 10. Klein AM, Dioum EM, Cobb MH (2010) Exposing Contingency Plans for Kinase Networks. Cell 143: 867–869. [DOI] [PubMed] [Google Scholar]
- 11. van Wageningen S, Kemmeren P, Lijnzaad P, Margaritis T, Benschop JJ, et al. (2010) Functional Overlap and Regulatory Links Shape Genetic Interactions between Signaling Pathways. Cell 143: 991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brookfield J (1992) Can genes be truly redundant? Current Biology 2: 553–554. [DOI] [PubMed] [Google Scholar]
- 13.Lynch M (2007) The origins of genome architecture. Sunderland, MA: Sinauer Associates, Inc. Publishers.
- 14. Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y (2010) Functional diversity of ankyrin repeats in microbial proteins. Trends in Microbiology 18: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgiades K, Raoult D (2011) Defining pathogenic bacterial species in the genomic era. Frontiers in Microbiology 2. [DOI] [PMC free article] [PubMed]
- 16. Toft C, Andersson SGE (2010) Evolutionary microbial genomics: insights into bacterial host adaptation. Nat Rev Genet 11: 465–475. [DOI] [PubMed] [Google Scholar]
- 17. Serres M, Kerr A, McCormack T, Riley M (2009) Evolution by leaps: gene duplication in bacteria. Biology Direct 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ogura M, Perez JC, Mittl PRE, Lee H-K, Dailide G, et al. (2007) Helicobacter pylori evolution: lineage- specific adaptations in homologs of eukaryotic Sel1-like genes. PLoS Comput Biol 3: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dumrese C, Slomianka L, Ziegler U, Choi SS, Kalia A, et al. (2009) The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Letters 583: 1637–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Habyarimana F, Al-khodor S, Kalia A, Graham JE, Price CT, et al. (2008) Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environmental Microbiology 10: 1460–1474. [DOI] [PubMed] [Google Scholar]
- 21. Mira A, Ochman H, Moran NA (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet 17: 589–596. [DOI] [PubMed] [Google Scholar]
- 22. Atherton JC (2006) The pathogenesis of Helicobacter pylori–induced gastro-duodenal diseases/. Annual Review of Pathology: Mechanisms of Disease 1: 63–96. [DOI] [PubMed] [Google Scholar]
- 23. Alm RA, Ling L-SL, Moir DT, King BL, Brown ED, et al. (1999) Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature 397: 176–180. [DOI] [PubMed] [Google Scholar]
- 24. Tomb J-F, White O, Kerlavage AR, Clayton RA, Sutton GG, et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori . Nature 388: 539–547. [DOI] [PubMed] [Google Scholar]
- 25. Deml L, Aigner M, Decker J, Eckhardt A, Schutz C, et al. (2005) Characterization of the Helicobacter pylori Cysteine-Rich Protein A as a T-Helper Cell Type 1 Polarizing Agent. Infect Immun 73: 4732–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roschitzki B, Schauer S, Mittl P (2011) Recognition of Host Proteins by Helicobacter Cysteine-Rich Protein C. Current Microbiology: 1–11. [DOI] [PubMed]
- 27. Mittl PRE, Luthy L, Reinhardt C, Joller H (2003) Detection of High Titers of Antibody against Helicobacter Cysteine-Rich Proteins A, B, C, and E in Helicobacter pylori-Infected Individuals. Clin Diagn Lab Immunol 10: 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabarth N, Lamer S, Zimny-Arndt U, Jungblut PR, Meyer TF, et al. (2002) Identification of Surface Proteins of Helicobacter pylori by Selective Biotinylation, Affinity Purification, and Two-dimensional Gel Electrophoresis. Journal of Biological Chemistry 277: 27896–27902. [DOI] [PubMed] [Google Scholar]
- 29. Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, et al. (2002) Proteome Analysis of Secreted Proteins of the Gastric Pathogen Helicobacter pylori . Infection and Immunity 70: 3396–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. De Vries N, van Ark EM, Stoof J, Kuipers EJ, van Vliet AHM, et al. (2003) The stress-induced hsp12 gene shows genetic variation among Helicobacter pylori strains. FEMS Immunology & Medical Microbiology 38: 45–51. [DOI] [PubMed] [Google Scholar]
- 31. Lüthy L, Grütter MG, Mittl PRE (2004) The Crystal Structure of Helicobacter Cysteine-rich Protein C at 2.0 Å Resolution: Similar Peptide-binding Sites in TPR and SEL1-like Repeat Proteins. Journal of Molecular Biology 340: 829–841. [DOI] [PubMed] [Google Scholar]
- 32. Todd AE, Orengo CA, Thornton JM (2001) Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol 307: 1113–1143. [DOI] [PubMed] [Google Scholar]
- 33. Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, et al. (2002) Grb2 Is a Key Mediator of Helicobacter pylori CagA Protein Activities. Molecular Cell 10: 745–755. [DOI] [PubMed] [Google Scholar]
- 34.Luthy L (2003) Structural and functional characterization of the Helicobacter pylori cysteine rich proteins. Zurich: Universitat Zurich.
- 35. Phadnis S, Parlow M, Levy M, Ilver D, Caulkins C, et al. (1996) Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun 64: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vanet A, Labigne A (1998) Evidence for Specific Secretion Rather than Autolysis in the Release of Some Helicobacter pylori Proteins. Infect Immun 66: 1023–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gobert AP, Bambou J-C, Werts C, Balloy V, Chignard M, et al. (2004) Helicobacter pylori Heat Shock Protein 60 Mediates Interleukin-6 Production by Macrophages via a Toll-like Receptor (TLR)-2-, TLR-4-, and Myeloid Differentiation Factor 88-independent Mechanism. Journal of Biological Chemistry 279: 245–250. [DOI] [PubMed] [Google Scholar]
- 38. Lin C-S, He P-J, Hsu W-T, Wu M-S, Wu C-J, et al. (2010) Helicobacter pylori-derived Heat shock protein 60 enhances angiogenesis via a CXCR2-mediated signaling pathway. Biochemical and Biophysical Research Communications 397: 283–289. [DOI] [PubMed] [Google Scholar]
- 39. Romero D, Palacios R (1997) Gene amplification and genomic plasticity in prokaryotes. Annu Rev Genet 31: 91–111. [DOI] [PubMed] [Google Scholar]
- 40. Musso G, Costanzo M, Huangfu M, Smith AM, Paw J, et al. (2008) The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res 18: 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS (2007) Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol 3. [DOI] [PMC free article] [PubMed]
- 42. Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11: 97–108. [DOI] [PubMed] [Google Scholar]
- 43. Kondrashov F, Rogozin I, Wolf Y, Koonin E (2002) Selection in the evolution of gene duplications. Genome Biology 3: research0008.0001–research0008.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huesca M, Borgia S, Hoffman P, Lingwood C (1996) Acidic pH changes receptor binding specificity of Helicobacter pylori: a binary adhesion model in which surface heat shock (stress) proteins mediate sulfatide recognition in gastric colonization. Infect Immun 64: 2643–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francino MP (2005) An adaptive radiation model for the origin of new gene functions. Nat Genet 37: 573–577. [DOI] [PubMed] [Google Scholar]
- 46. Lin C-S, He P-J, Tsai N-M, Li C-H, Yang S-C, et al. (2010) A potential role for Helicobacter pylori heat shock protein 60 in gastric tumorigenesis. Biochemical and Biophysical Research Communications 392: 183–189. [DOI] [PubMed] [Google Scholar]
- 47.Owen RJ, Bickely J (1997) Isolation of H. pylori genomic DNA and restriction Analysis Clayton CL, Mobley HLT, editors. Totowa, NJ: Human Press, Inc. [DOI] [PubMed]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al.. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. [DOI] [PMC free article] [PubMed]
- 49. Lüthy L, Grütter MG, Mittl PRE (2002) The Crystal Structure of Helicobacter pyloriCysteine-rich Protein B Reveals a Novel Fold for a Penicillin-binding Protein. Journal of Biological Chemistry 277: 10187–10193. [DOI] [PubMed] [Google Scholar]
- 50. O'Sullivan O, Suhre K, Abergel C, Higgins DG, Notredame C (2004) 3DCoffee: Combining Protein Sequences and Structures within Multiple Sequence Alignments. Journal of Molecular Biology 340: 385–395. [DOI] [PubMed] [Google Scholar]
- 51. Fay JC, Wu C-I (2001) The neutral theory in the genomic era. Current Opinion in Genetics & Development 11: 642–646. [DOI] [PubMed] [Google Scholar]
- 52. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 53. Letunic I, Doerks T, Bork P (2009) SMART 6: recent updates and new developments. Nucleic Acids Research 37: D229–D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dailidiene D, Dailide G, Kersulyte D, Berg DE (2006) Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori . Appl Environ Microbiol 72: 5908–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kitsos CM, Stadtlander CT (1998) Helicobacter pylori in liquid culture: evaluation of growth rates and ultrastructure. Curr Microbiol 37: 88–93. [DOI] [PubMed] [Google Scholar]
- 56. Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, et al. (2010) Peptidoglycan crosslinking relaxation promotes Helicobacter pylori's helical shape and stomach colonization. Cell 141: 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davis KM, Akinbi HT, Standish AJ, Weiser JN (2008) Resistance to mucosal lysozyme compensates for the fitness deficit of peptidoglycan modifications by Streptococcus pneumoniae. PLoS Pathog 4: e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shen F, Hobley L, Doherty N, Loh J, Cover T, et al. (2010) In Helicobacter pylori auto-inducer-2, but not LuxS/MccAB catalysed reverse transsulphuration, regulates motility through modulation of flagellar gene transcription. BMC Microbiology 10: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed description of methods and supplementary figures (Fig. S1– Fig. S6) and tables (Table S1–Table S10).
(DOCX)