Abstract
The transport of proteins between the cytoplasm and nucleus requires interactions between soluble transport receptors (karyopherins) and phenylalanine-glycine (FG) repeat domains on nuclear pore complex proteins (nucleoporins). However, the role of specific FG repeat-containing nucleoporins in nuclear protein export has not been carefully investigated. We have developed a novel kinetic assay to investigate the relative export kinetics mediated by the karyopherin Msn5/Kap142 in yeast containing specific FG-Nup mutations. Using the Msn5 substrate Crz1 as a marker for Msn5-mediated protein export, we observe that deletions of NUP100 or NUP2 result in decreased rates of Crz1 export, while nup60Δ and nup42Δ mutants do not vary significantly from wild type. The decreased Msn5 export rate in nup100Δ was confirmed using Mig1-GFP as a transport substrate. A nup100ΔGLFG mutant shows defects in nuclear export kinetics similar to a nup100Δ deletion. Removal of FG-repeats from Nsp1 also decreases export kinetics, while a loss of Nup1 FXFGs does not. To confirm that our export data reflected functional differences in protein localization, we performed Crz1 transcription activation assays using a CDRE::LacZ reporter gene that is upregulated upon increased transcription activation by Crz1 in vivo. We observe that expression from this reporter increases in nup100ΔGLFG and nsp1ΔFGΔFXFG strains that exhibit decreased Crz1 export kinetics but resembles wild-type levels in nup1ΔFXFG strains that do not exhibit export defects. These data provide evidence that the export of Msn5 is likely mediated by a specific subset of FG-Nups and that the GLFG repeat domain of Nup100 is important for Msn5-mediated nuclear protein export.
Keywords: nuclear pore complex, nuclear export, nucleoporin, karyopherin, exportin
1. Introduction
The nuclear envelope (NE) in eukaryotes separates the nucleoplasm from the cytoplasm. The ability to control the transport of proteins and RNAs across the nuclear envelope is essential for mediating eukaryotic cell processes such as gene expression and transcriptional regulation, cell division, and signal transduction. Nuclear pore complexes (NPCs) perforate the NE and are the only known conduit for transport between the nuclear and cytoplasmic compartments. NPCs are composed of 30 different proteins termed nucleoporins or “Nups” that are found in 8 – 64 copies per NPC [1, 2]. The Nups provide both structure and function to the nuclear pores, serving to perforate the NE, forming the proteinaceous channel between the cytoplasm and nucleus, and regulating the translocation of substrates through the NPCs (reviewed in [3, 4]). The Nups that line the channel within each NPC contain domains consisting of phenylalanine-glycine (FG) pairs separated by highly degenerate amino acid sequences. Nucleoporins containing one or more of these repeat motifs are termed “FG Nups” and have filamentous, unstructured regions that extend into the channel of the NPC, making up the permeability barrier and facilitating translocation [2, 5, 6]. In contrast, nucleoporins devoid of FG repeats have discrete structures and comprise the structural backbone of the NPC [7, 8]. In the thirteen FG-Nups expressed in yeast, three general types of these repeat sequences have been identified: FXFG-repeats, with small, charged spacer sequences; GLFG-repeats, which have non-acidic spacer sequences; and more degenerate FG-repeats found in several Nups, including some that also contain FXFG or GLFG-repeats (reviewed in [4, 9]). The functional differences between the different types of FG-repeats and between different Nups with similar repeat types remain unclear.
Protein transport through NPCs is mediated by karyopherins (Kaps). Specific Kaps function to either import or export specific cargo proteins (reviewed in [10, 11]). Kaps responsible for nuclear import (“importins”) bind a nuclear localization signal (NLS) on their transport substrate in the cytoplasm and traverse the NPC as a heterodimeric complex. Export Kaps (“exportins”) associate with a nuclear export signal (NES) in the nucleus in the presence of the GTP-bound form of the Ran GTPase within the nucleus. The Kap/NES-protein/Ran-GTP heterotrimer crosses the NPC and the exported protein substrate is released in the cytoplasm upon GTP hydrolysis. FG-Nups play an important role in Kap-mediated nucleocytoplasmic transport by providing sites for transient karyopherin binding (reviewed in [12]). Karyopherin mutants that fail to bind FG repeats exhibit significantly reduced import [13]. Although it is clear that nuclear transport requires a direct interaction between Kaps and FG domains, exactly how different karyopherins utilize FG Nups to move cargo through the complex is not well understood.
The karyopherin Msn5/Kap142 functions as an exportin for more than 15 protein substrates whose intracellular locations are tightly controlled, including the transcription factors Pho4, Msn2, Far1, Spo12, Maf1, Crz1 and Mig1 [14–19]. Interactions have been detected between Msn5 and at least eight of the thirteen yeast FG Nups, including Nup1, Nup2, Nup42, Nup49, Nup57, Nup100, Nup116, and Nup145 [20, 21]. These data suggest that Msn5 interactions with FG nucleoporins are integral to its function as a transport receptor.
The Crz1 transcription factor is exported from the yeast nucleus by Msn5 [17]. The intracellular localization of Crz1 is regulated by a calcium signaling pathway that begins with the activation of calcineurin, which dephosphorylates Crz1, thus inhibiting Crz1 nuclear export by Msn5 and inducing Crz1 import by the importin Nmd5 [17, 22]. Rapid nuclear export of Crz1 can be induced by the immunosuppressive drug FK506, which inhibits calcineurin phosphatase activity, allowing the phosphorylation of Crz1 and subsequent Msn5-mediated export. Thus, Crz1 accumulates in the nucleus in the presence of Ca2+ and undergoes Msn5-mediated export from the nucleus upon addition of FK506.
While kinetic assays have allowed nuclear protein import to be carefully examined in many yeast mutants [23–28], the export of proteins from the nucleus has not been as closely studied. Permeabilized cell assays have been developed in an effort to identify soluble factors important for protein export in mammalian cells [29, 30] and microinjection of photoactivatable proteins has recently allowed assessment of export rates for specific proteins in mouse embryonic cells [31]. But neither of these methods provide a mechanism for the large-scale analysis of gene mutations on the kinetics of nuclear export, nor do they allow investigation of the effects of specific Nup mutations on transport rate. Here we report the development of a kinetic nuclear export assay that uses Crz1-GFP localization to investigate the relative rate of Msn5-mediated export in yeast and the use of this assay to identify FG-Nups necessary for efficient protein export. Using this assay, we observe that the deletion of FG-Nups Nup100 or Nup2 decreases export rates, while the export rates in nup42Δ and nup60Δ are not significantly different from wild type. We observe a similar export defect in nup100Δ for Mig1-GFP, a second Msn5 export substrate. Export rates also vary in Nup mutants lacking FG repeats. nup100ΔGLFG and nsp1ΔFGΔFXFG cells exhibit reduced transport, while nup1ΔFXFG exports Crz1-GFP at a rate similar to wild type. Using a CDRE-LacZ reporter, we show that nup mutants with kinetic export defects retain higher levels of Crz1 transcription activation activity than mutants that export efficiently, providing additional evidence for increased nuclear accumulation of Msn5 export substrates in these mutants. These data provide in vivo evidence that the GLFG repeats of Nup100 are necessary for efficient Msn5-mediated transport through the NPC, suggesting that only a specific subset of FG repeats are essential for normal Msn5 transport kinetics and that this subset is different from Nups identified as being important for mRNA export and protein export by other Kaps [25, 32, 33]. Importantly, this new kinetic assay provides novel insights into protein export pathways and adds support to the hypothesis that translocation by different transport factors is dependent upon distinct subsets of nucleoporins.
2. Materials and methods
2.1. Reagents, Strains, Plasmids, and Media
Yeast strains and bacterial plasmids used in this work are shown in Table 1 and Table 2, respectively. Yeast transformations were performed as described [34]. Cell cultures and media preparations were also carried out as described [35]. Enzymes for molecular biology were purchased from New England Biolabs (Beverly, MA) and used per the manufacturer’s instructions. Plasmid DNA was purified using the QIAprep Spin Miniprep Kit (QIAGEN Sciences, Germantown, MD).
Table 1.
Yeast strains used.
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATα his3 leu2 lys2 ura3 | OpenBiosystems |
| BY4742 | MATa his3 leu2 met15 ura3 | OpenBiosystems |
| KBY643 | MATα nup60ΔKANR his3 leu2 lys2 ura3 | OpenBiosystems |
| KBY793 | MATα nup100ΔKANR his3 leu2 lys2 ura3 | OpenBiosystems |
| KBY795 | MATa nup170ΔKANR his3-11 leu2-3,112 ura3-1 lys2 | OpenBiosystems |
| KBY893 | MATa msn5Δ ura3 leu2 his3 trp1 ade2 | OpenBiosystems |
| KBY1350 | MATα nup42ΔKANR his3 leu2 lys2 ura3 | OpenBiosystems |
| KBY1352 | MATα nup2ΔKANR his3 leu2 lys2 ura3 | OpenBiosystems |
| SWY2283 | MATa ura3-1 his3-11, 15 leu2-3, 112 lys2 | 25 |
| SWY2762 | MATα HA-LoxP-nup100ΔGLFG trp1 ura3 leu2 his3 | 25 |
| SWY2801 | MATα T7-nup1ΔFxFG trp1 ura3-52 leu2 his3 | 25 |
| SWY2811 | MATa lys2 ura3 leu2 his3 HA-LoxP-nsp1 ΔFG | 25 |
| SWY2825 | MATα T7-LoxP-nup49ΔGLFG trp1 ura3 leu2 his3 | 25 |
| SWY2919 | MATa lys2 ura3 leu2 his3 Flag-LoxP-nsp1 ΔFxFG- FG | 25 |
| SWY3030 | MATa lys2 ura3 leu2 his3 HA-LoxP-nsp1Δ FxFG | 25 |
Table 2.
Plasmids used.
| Strain | Genotype | Source |
|---|---|---|
| pRS314 | CEN TRP1 ampR | 50 |
| pRS315 | CEN LEU2 ampR | 50 |
| pLMB127 | CEN URA3 3xGFP-Crz1 ampR | 17 |
| pAMS367 | URA3-4xCDRE-LacZ ampR | 17 |
| LDB107 | CEN TRP1 NUP1 ampR | L. Davis |
| pUN100-NSP1 | CEN LEU2 NSP1 ampR | 51 |
| pBM3495 | 2μ URA3 Mig1(217-400)-GFP-β-gal | M. Johnston |
2.2. Fluorescence Microscopy
To visualize Crz1 localization using fluorescence microscopy, cells were transformed with a plasmid expressing Crz1 fused to GFP (pLMB127) and selected for on SD -Ura media. For fluorescence microscopy, the transformants were grown overnight at 24°C in SD –Ura media with NH4Cl (1.5 mg/ml yeast nitrogen base, 1 mg/ml NH4Cl, 20 mg/ml dextrose, 0.75 mg/ml CSM-URA). Cells were grown to early log phase (A600 = 0.05–0.2) and visualized under direct fluorescence using a Nikon E400 microscope. Images were captured using SPOT RTKE (Diagnostic Instruments, Inc., Sterling Heights, MI) cameras.
2.3. Crz1 Export Assays
To measure the relative rate of Crz1-GFP export, cells were prepared as described above. CaCl2 was added to a final concentration of 170mM and cells incubated at 24°C for 10 min. To initiate Crz1-GFP export, cells were incubated in media containing 170 mM CaCl2 and 1.4 μg/ml FK506. The percent of cells with nuclear Crz1-GFP was determined by direct fluorescence microscopy prior to FK506 addition (t=0) and at regular intervals after addition. At least 100 cells were counted for nuclear vs. cytoplasmic fluorescence at each timepoint. Each experiment was replicated three or more times and all assays were performed with the microscopist blind to the yeast strain being assayed. The percent of cells with nuclear Crz1 fluorescence (%N) was plotted vs. time for each strain to generate curves representative of the relative rate that Crz1 is exported out of the nucleus. To determine whether variation in export rate between strains was statistically significant, a natural logarithm transformation (ln [%N] +1) was used to generate a linear relationship between %N and time for each strain. ANCOVA was then used to determine the main effects of strain, time, and the strain*time interaction. If the interaction term explains a significant amount of the total variation in %N, there is significant among-strain variation in export rate. Following this analysis, the estimated marginal means of %N for each strain were calculated to determine the value of %N for each strain averaged over all time points in the study. To determine whether there was significant among-strain variation for %N, one-way ANOVA and post-hoc LSD tests were used for these estimated marginal mean data. The results of these analyses show whether there is significant among-strain variation for %N when time is held constant, and show whether there are significant differences in %N among all pair-wise strain comparisons.
2.4. Mig1 Export Assays
Yeast cells expressing Mig1 residues 217–400 fused to GFP and β-galactosidase (pBM3495) were grown overnight at 24°C in SD -Ura. Prior to assaying, cells were incubated for 30 min in fresh SD –Ura, then observed for Mig1-GFP localization by direct fluorescence microscopy. Cells with nuclear and cytoplasmic localization were counted and the % cells with nuclear fluorescence (%N) determined (t=0). Cells were then pelleted and resuspended in SD –Ura lacking dextrose and containing 5% glycerol. Cells were observed under direct fluorescence microscopy and %N determined at regular timepoints up to 1 hour after glycerol addition. Export assays were performed blind with each yeast strain assayed at least two times.
2.5. β-Galactosidase assays
To quantify the relative activity of the calcineurin-dependent response element in strains with mutated FG nucleoporins, CDRE::LacZ (pAMS367) was integrated into the genome at URA3 locus of nup100ΔGLFG (SWY2762), nup49ΔGLFG (SWY2825) nup1ΔFXFG (SWY2919), nup1ΔFXFG + NUP1 (SWY2801 + pLDB107), nsp1ΔFGΔFXFG (SWY2919), nsp1ΔFXFG (SWY3030), nsp1ΔFG (SWY2811), and wild-type (SWY2283) yeast strains after being linearized using the restriction enzyme Stu1. Yeast cells were grown overnight in selective media to exponential growth phase. Cells were resuspended in 500μl Z-buffer (100 mM Na2HPO4, 40 mM NaH2PO4, 10 Mm KCl, 1 mM MgSO4, 0.027% β-mercaptoethanol, 0.1% SDS) and vortexed for 5 minutes. Chloroform (5%) was added to the Z-buffer suspension and the cells were lysed by vortexing for 1 minute. Chlorophenol red-β-D-galactopyranoside (3.84mM) was added and reactions were incubated at room temperature for 1–2 hours after which each sample was measured for absorbance at A595. Miller units of β-galactosidase activity were calculated as per Miller [36]. Because activity levels varied among the days that the assays were performed, β-galactosidase activity was converted into units of standard deviation away from the mean for each day by subtracting the activity level of each sample from the average activity level in all samples for that day and then dividing by the standard deviation in activity level for that day. The average units of deviation away from the day mean for each mutant were then calculated and plotted relative to wild type. Statistical analyses were performed by one-way ANOVA to determine if variation among the strains for β-gal activity was greater than the variation within the strains, and post-hoc Tukey tests carried out to perform all pair-wise strain comparisons in activity level. Comparisons between the nup1ΔFXFG strain and the corresponding wild-type is expressed in Miller units because standard deviations from day mean cannot be calculated for only two strains.
3. Results
Nuclear protein import through the NPC has been studied extensively using biochemical and genetic approaches, steady-state microscopic analysis, and kinetic in vivo import assays (reviewed in [4, 37]). However, the function of specific Nups in the export of nuclear proteins to the cytoplasm has not been as carefully examined and thus far has not included investigation of the relative kinetics of protein export through the NPCs. Impaired nuclear protein export should result in nuclear accumulation of the transporter, and thus of the cargo that would normally be exported. When the rate of import is greater than the rate of export, the cargo will appear to accumulate in the nucleus. Conversely, when the rate of export out of the nucleus surpasses that of import, the cargo appears to be cytoplasmically localized when examined under steady state conditions. However, defects in the rate of transport under particular conditions will not be readily apparent using steady state localization if those conditions do not alter the overall direction of transport. An assay that examines changes in localization of a marker protein over time is required to identify alterations in the kinetics of nucleocytoplasmic transport. While such assays exist for investigating nuclear protein import [23], we are not aware of any such kinetic assays investigating variations in the rate of nuclear protein export.
3.1. Assay to examine nuclear protein export kinetics
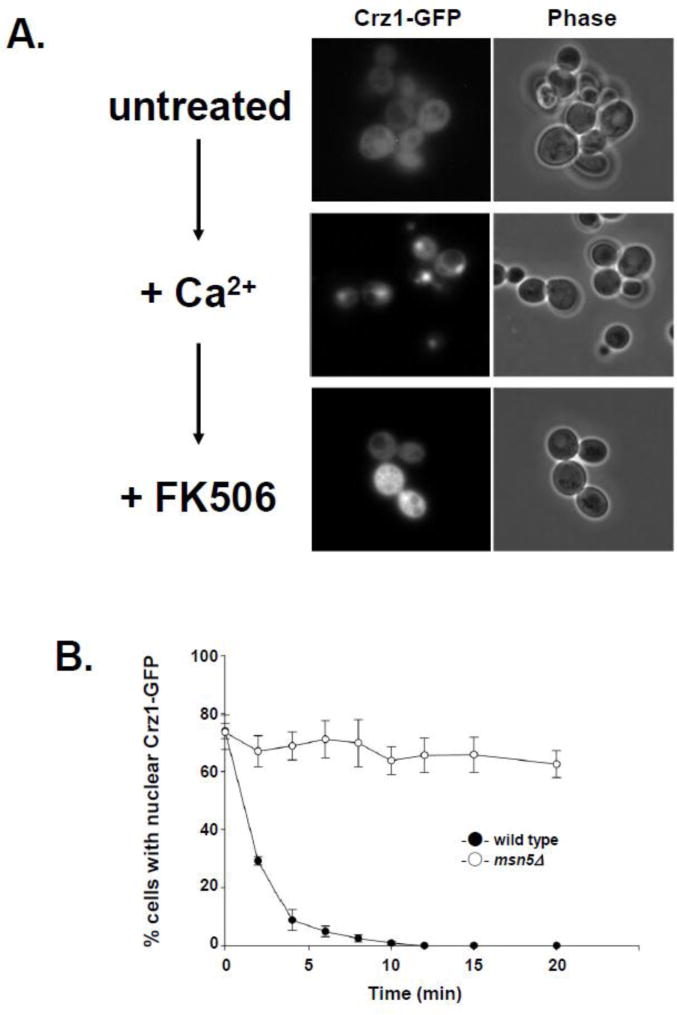
Using Crz1-GFP as a marker for inducible Msn5-mediated protein export, we have developed a method for detecting changes in the rate of nuclear protein export in nup mutant cells (Fig. 1). Crz1 is primarily cytoplasmic in cells grown in synthetic complete (SC) media, but rapidly accumulates in the nucleus upon addition of Ca2+ ([22]; Fig. 1A). Upon exposure to the calcineurin inhibitor FK506, Nmd5-mediated Crz1 import is inhibited and Msn5-mediated Crz1 export is activated, resulting in a rapid reaccumulation of Crz1 in the cytoplasm [17, 22]. We have developed an assay to follow the progression of this movement by counting the number of cells with nuclear Crz1-GFP accumulation and the number with cytoplasmic signal and using those values to determine the ratio of nuclear vs. cytoplasmically fluorescent cells. This ratio was measured at regular timepoints from FK506 addition through 25 minutes after addition and plotted to yield a curve that provides information about the kinetics of Crz1 export in the population of cells assayed (Fig. 1B). This shift from nuclear to cytoplasmic Crz1 localization is rapid, occurring in less than 10 minutes in wild type yeast. As expected, cells lacking Msn5 fail to export Crz1-GFP from the nucleus upon FK506 addition. Thus, this in vivo nuclear export assay can be used to investigate the kinetics of Msn5-mediated Crz1-GFP export and provide a novel mechanism to gain insight into the kinetics of nuclear protein export.
Figure 1.
Msn5-mediated nuclear export kinetics can be followed in vivo using FK506 induced nuclear export of Crz1-GFP from the nucleus. (A) Crz1-GFP localization changes upon addition of Ca2+ and FK506. Yeast expressing Crz1-GFP were examined using direct fluorescence microscopy prior to calcium addition (untreated), after the addition of CaCl2 to 170 mM for 5 min (+Ca2+), and after addition of FK506 to 1.4 ug/ml for 10 min (+FK506). Fluorescence (Crz1-GFP) and phase-contrast (phase) photomicrographs are shown. (B) Wild type and msn5Δ cells export Crz1-GFP at different rates following addition of FK506. Data points represent the mean percentage of cells with nuclear Crz1-GFP fluorescence at each time point based on a minimum of three assays. Error bars represent standard error of the mean.
3.2. nup100Δ and nup2Δ cells have slowed Crz1 export
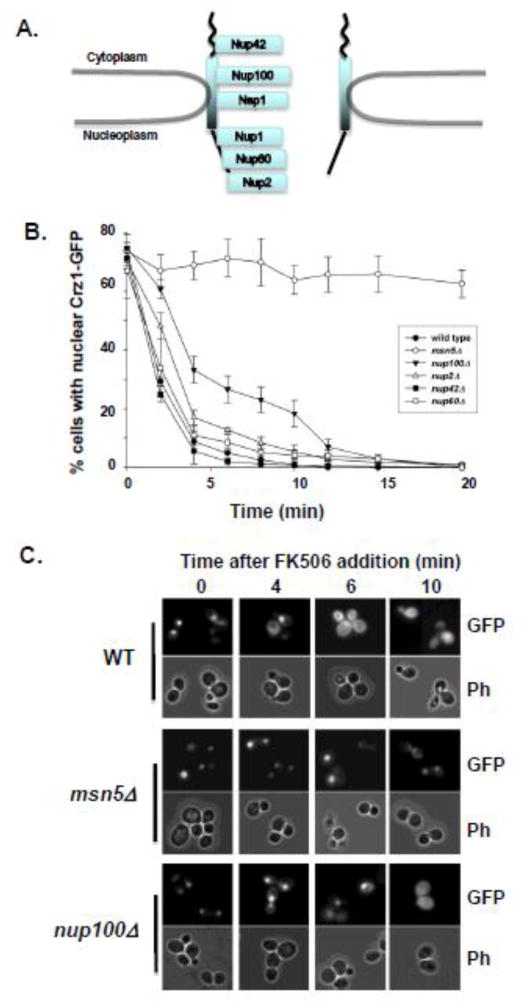
In order to investigate the function of specific Nups in Msn5-mediated nuclear protein export, we repeated this kinetic assay on several yeast strains containing complete deletions of specific nup genes. We selected deletions of four FG-containing Nups for comparison with an isogenic wild type strain (Fig. 2A): Nup42, a cytoplasmically oriented Nup with FG repeats; Nup100, a GLFG Nup from the NPC channel that exhibits a slight bias in abundance toward the cytoplasmic side of the NPC; Nup60, an FXFG Nup from the nucleoplasmic face of the NPC; and Nup2, a nucleoplasmically oriented FXFG Nup that is only transiently associated with the NPC [1, 38]. These nup mutants were selected to investigate the importance of several different types of FG repeats on Crz1 export and because complete deletions of each of these Nups remain viable. In each strain, Crz1-GFP was induced to accumulate in the nucleus, then cells were treated with FK506 to stimulate Crz1 export. The percent of cells retaining strong nuclear fluorescence was measured at two-minute intervals following FK506 addition (Fig. 2A). The export rate of Crz1 varied among the five strains tested over the first six minutes (ANCOVA strain*time: F(1,4) = 4.522, p<0.005) with nup100Δ exhibiting the slowest Crz1-GFP export kinetics. However, by 20 minutes after FK506 addition all four nup mutants and the wild type cells showed nearly complete cytoplasmic localization of Crz1. Only an msn5Δ mutant, which lacks the karyopherin required for Crz1 export, failed to eventually export Crz1-GFP.
Figure 2.
Crz1-GFP is exported from the nucleus at different rates in different nup mutants. (A) Schematic cartoon of the location within the NPC of Nups investigated using the in vivo nuclear export kinetics assay. (B) Yeast cells expressing Crz1-GFP were examined for Crz1 export kinetics after addition of FK506 as described in figure 1 and Materials and Methods. Wild type cells (-●-) and mutants lacking Msn5 (msn5Δ -○-), Nup100 (nup100Δ -▼-), Nup2 (nup2Δ -△-), Nup42 (nup42Δ -■-), and Nup60 (nup60Δ -□-) were assayed. Data points represent mean percentage of cells with nuclear Crz1-GFP and error bars represent standard error of the mean. (C) Direct fluorescence microscopy was performed on wild type (WT), msn5Δ, and nup100Δ cells expressing Crz1-GFP. Cells were treated with Ca2+ and photomicrographs were taken prior to FK506 addition (0), and at 4, 6, and 10 minutes after the introduction of FK506.
We next carefully analyzed the Crz1-GFP export kinetics of each nup mutant compared to wild type cells. A deletion of the GLFG nucleoporin Nup100 results in a significantly reduced rate of Crz1-GFP nuclear export following FK506 treatment compared to wild type cells (Fig. 2B, C). Two minutes after FK506 addition over 60% of nup100Δ cells retain strong nuclear Crz1-GFP, while less than 30% of wild type cells show nuclear fluorescence. By 6 min after inducing export, approximately 30% of nup100Δ cells show nuclear fluorescence, as compared to 3% of wild type cells. A loss of the FXFG-Nup, Nup2, also significantly slows Crz1-GFP export, with 49% of cells displaying nuclear fluorescence at 2 min and 14% nuclear at 6 min following FK506 addition. Neither a complete deletion of Nup42 nor Nup60 results in a significant change in Crz1-GFP export kinetics. These data indicate that a subset of FG-Nups are essential for efficient Msn5-mediated export of Crz1 from the nucleus, while the absence of other tested FG-Nups does not affect the kinetics of Crz1 export.
3.3. Deletion of NUP100 reduces Mig1 protein export kinetics
To determine if the differences in transport observed between nup100Δ and wild type cells are specific to Crz1-GFP or are an indicator of a more general affect on Msn5-mediated protein export, we investigated the export kinetics of Mig1-GFP. Mig1 is a transcription factor that regulates glucose repression and shuttles in and out of the nucleus in response to changes in glucose concentration [39]. We grew wild-type (BY4742), msn5Δ, nup100Δ, nup42Δ, and nup170Δ mutant cells expressing Mig1-GFP in media containing glucose, allowing Mig1 to accumulate in the nucleus. We then induced Mig1 export by shifting the cells to media containing glycerol as the primary carbon source and examined the intracellular location of Mig1-GFP over time (Fig. 3). Wild-type cells export Mig1-GFP rapidly so that fewer than 25% of cells show nuclear Mig1-GFP within 3 minutes following the shift to glycerol. The percent of wild-type cells with nuclear Mig1-GFP fluorescence then remains steady at about 20%. Populations of cells lacking Msn5 undergo a decrease in nuclear fluorescence from 89% to 64% in the first 3 min after shift to glycerol, but then retain nuclear fluorescence at levels above 60% for the remainder of the assay, confirming previous reports that Msn5 is a karyopherin mediating Mig1 protein export [39]. For the nup100Δ mutant, 55% of cells show nuclear Mig1-GFP at 3 min and over 40% retain nuclear Mig1-GFP throughout the assay. Deletion of either Nup170, a non-FG Nup important for efficient NPC assembly and organization, or Nup42 result in import kinetics similar to those seen for wild type cells, although nup170Δ mutants show a slight decrease in cells with nuclear Mig1 and nup42Δ mutants show a slight increase at 3 min. By 6 min post-glycerol addition, the percent of nup42Δ or nup170Δ cells with nuclear Mig1-GFP is nearly identical to wild type cells. The reduced export kinetics of both Crz1 and Mig1 in nup100Δ cells indicate that Nup100 is important for general Msn5-mediated nuclear protein export.
Figure 3.
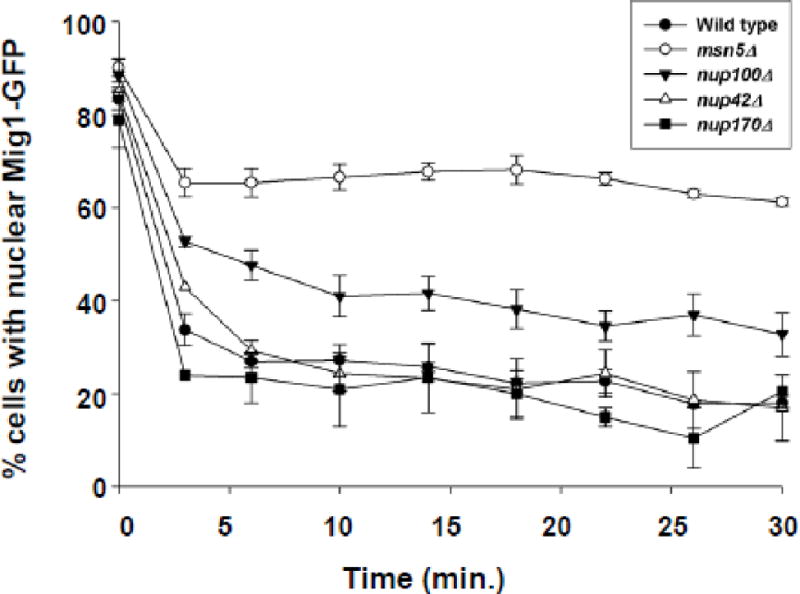
Mig1-GFP is exported less efficiently in cells lacking Nup100 than control wild type cells. Mig1-GFP was expressed in wild type (-●-), msn5Δ (-○-), nup100Δ (-▼-), nup42Δ (-△-), and nup170Δ (-■-) cells in the presence of 2% dextrose. After confirming Mig1-GFP localization in the nucleus (t = 0 min), cells were pelleted, rinsed in media lacking a carbon source, resuspended in media containing 5% glycerol, and assayed for the percent of cells exhibiting nuclear fluorescence every 3–4 min. Error bars represent standard error of the mean.
3.4. Nup100 GLFG repeats are necessary for efficient Crz1 export
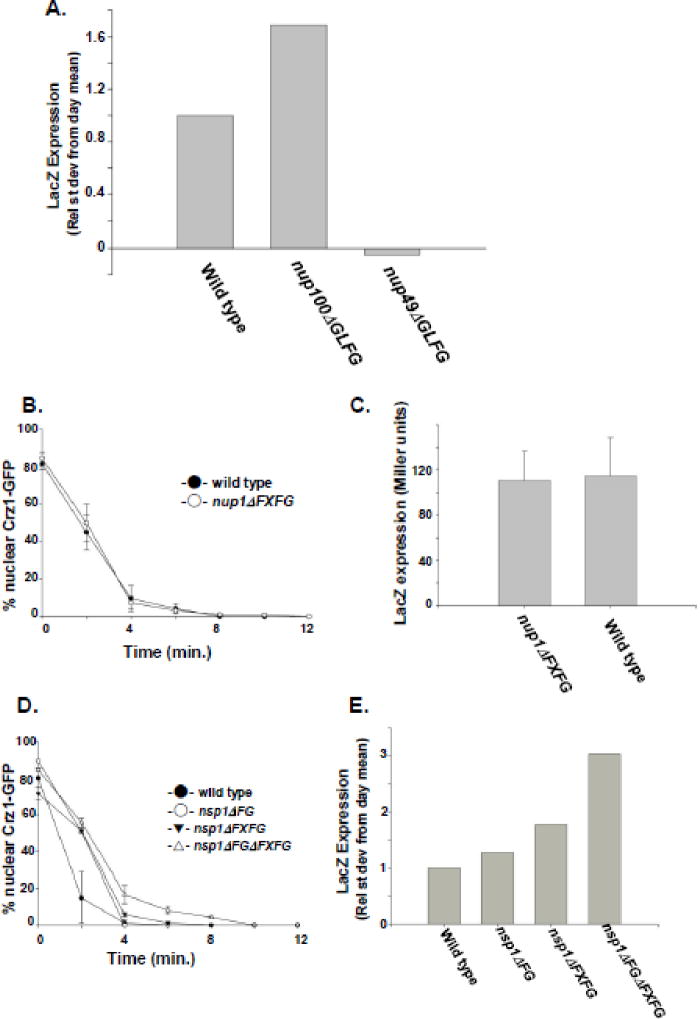
Nup100 associates with Msn5 and other karyopherins via its GLFG repeats [21, 40–42]. To determine if the GLFG repeats of Nup100 are necessary for efficient Msn5-mediated protein export, we performed kinetic Crz1-GFP export assays on nup100ΔGLFG yeast [25] that completely lack their GLFG repeat domains (Fig. 4). The nup100ΔGLFG yeast showed a decreased rate of export when compared to wild type cells (70% and 50% nuclear Crz1-GFP, respectively, after 2 min; 59% and 39% nuclear Crz1-GFP after 4 min), but nup100ΔGLFG and nup100Δ mutants exhibited export kinetics that are not significantly different from each other. Thus, the Nup100 GLFG domain is necessary for efficient Msn5-mediated export and this domain appears to be the primary reason that nup100Δ mutants have reduced export kinetics.
Figure 4.
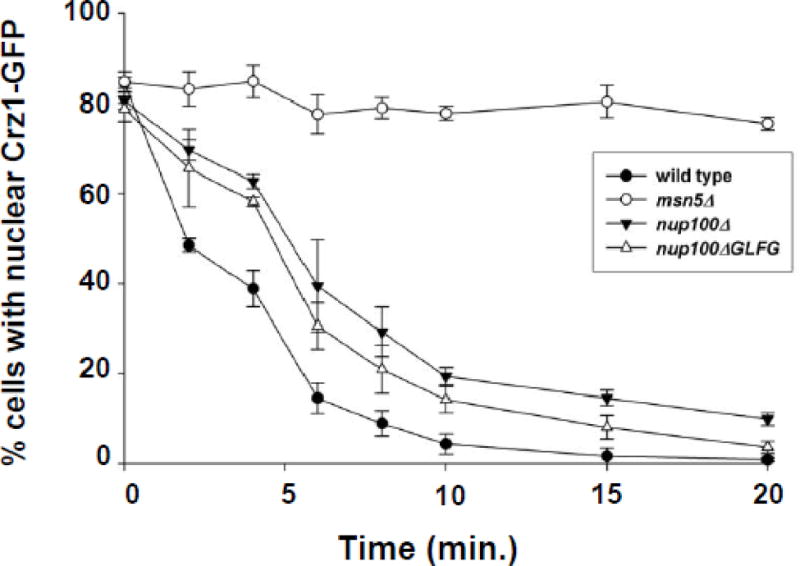
Cells lacking Nup100 GLFG domains have reduced Crz1-GFP export kinetics as compared to wild type control cells. Export assays were performed on wild type (-●-), msn5Δ (-○-), nup100Δ (-▼-), and nup100ΔGLFG (-△-) cells as described for Figure 2A. Data points represent the mean percent of cells with nuclear Crz1-GFP localization following incubation in FK506 for the amount of time indicated, based on a minimum of three assays. Error bars represent the standard error of the mean.
3.5. Deletion of Nup100 GLFGs reduces Crz1-dependent transcription
Crz1 functions as a transcription factor for multiple genes by associating with a calcineurin-dependent response element (CDRE) in the promoter of responding genes [43]. The nuclear localization of Crz1 is necessary for efficient binding and activation of the CDRE and activation of a CDRE-LacZ reporter has been used previously as a reporter for the accumulation of Crz1 within the nucleus [17]. To further characterize the role of specific nup mutations in the nucleocytoplasmic transport of Crz1, we have used a CDRE-LacZ reporter to investigate the steady-state transcriptional activation activity of Crz1-GFP in several isogenic yeast strains containing specific nup mutations (Fig. 5). Given the results of our export kinetics assay indicating that Nup100 GLFG repeats are important for efficient Crz1 nuclear export, we tested whether removal of Nup100 GLFGs altered Crz1-dependent activation of the CDRE-LacZ reporter. A comparison of the relative standard deviation from the mean between wild type, nup100ΔGLFG, and nup49ΔGLFG cells reveals that nup100ΔGLFG cells have a significantly higher level of steady state CDRE-LacZ activation than do wild-type cells (Fig. 5A). Thus, nup100ΔGLFG mutants have increased Crz1 transcriptional activation activity, consistent with a nuclear accumulation of Crz1 resulting from a decrease in Crz1 nuclear export. Conversely, nup49ΔGLFG mutants have a lower level of CDRE activation, in agreement with our observation that a yeast strain lacking Nup49 GLFG repeats accumulate Crz1-GFP in the nucleus after Ca2+ addition in fewer than 50% of cells (E. DeRoo, G. Clement, and K. Belanger, unpublished). These data suggest that the CDRE-LacZ assay can be used to assess the Crz1 nuclear export efficiency in yeast that undergo Crz1 nuclear import.
Figure 5.
Deletion of different FG-Nup domains affects Crz1 export in different ways. (A) Wild type, nup100ΔGLFG, and nup49ΔGLFG cells expressing CDRE-LacZ were assayed for β-galactosidase activity. Activity is represented as relative standard deviation from day mean, as described in Materials and Methods. (B) Export assay comparing Crz1-GFP export kinetics in wild type (-●-) and nup1ΔFXFG (-○-) cells. (C) Comparison of CDRE-LacZ expression in wild type and nup1ΔFXFG cells. Expression is measured in Miller units of β-galactosidase activity and represents the mean values from at least four independent assays. Error bars represent standard error of the mean. (D) Export assay comparing export kinetics of Crz1-GFP between wild type (-●-), nsp1ΔFG (-○-), nsp1ΔFXFG (-▼-), and nsp1ΔFGΔFXFG (-△-) cells after addition of FK506. (E) CDRE-LacZ reporter assays comparing nuclear Crz1 transcription activity between wild type cells and nsp1ΔFG, nsp1ΔFXFG, and nsp1ΔFGΔFXFG mutants. LacZ expression is represented as the relative standard deviation from the day mean.
3.6. Nsp1 but not Nup1 FG mutations affect Crz1 nuclear export and Crz1-dependent transcription
Given the potential utility of the CDRE-LacZ assay for investigating Msn5-mediated export, we compared Crz1-GFP export rates and CDRE activation in several additional nup mutant strains. First, we compared export rates between mutant cells containing a mutation in FXFG-repeat containing Nup1 and an isogenic wild type strain. The FXFG repeat domain of Nup1 associates with the karyopherin Kap95 [44, 45] but a physical interaction with Msn5 has not been detected by biochemical methods. Using our export assay, we determined that the deletion of Nup1 FXFG repeats does not alter the kinetics of Crz1-GFP export (Fig. 5B). Furthermore, we find that CDRE activation assayed using the CDRE-LacZ reporter does not differ between nup1ΔFXFG and wild type cells (ANOVA: F(1,126)=1.185, p>0.27; Fig. 5C). Together these data provide evidence that the FXFG repeats of Nup1 are not important for Msn5-mediated Crz1 export.
For comparison, we investigated the Crz1 export rate and CDRE activation in several mutants of Nsp1, an FG-Nup that contains a series of FXFG repeats as well as some degenerate ‘FG’ repeats. A complete deletion of Nsp1 is lethal, so we focused our assays on three nsp1 mutants containing deletions of just the FXFG region (nsp1ΔFXFG), just the FG region (nsp1ΔFG), or both (nsp1ΔFGΔFXFG). The export kinetics of all three mutants are slower than a wild type control (Fig. 5D), with the export rates of nsp1ΔFGΔFXFG significantly slower than those with only FG or FXFG repeat sequences removed. These differences in Crz1 transport are supported by the level of CDRE-LacZ activity in each nsp1 mutant strain (Fig. 5E). Comparing the relative standardized CDRE-LacZ expression between nsp1ΔFGΔFXFG, nsp1ΔFG, nsp1ΔFXFG, and wild type cells, nsp1ΔFGΔFXFG cells have significantly greater Crz1 transcription activation activity than either of the less severely impacted nsp1ΔFG or nsp1ΔFXFG deletions. Variation among the strains for β-galactosidase activity was greater than the variation within the strains (ANOVA: F(3,15) = 16.854, p<0.001). Thus, while the FG- and FXFG-repeats each contribute to optimal transport efficiency, the absence of both severely alters Crz1 nuclear export. Together, the Nup1 and Nsp1 data provide strong evidence that different FG domains on different Nups have different influences on Msn5-mediated nuclear protein export and that measuring CDRE activity provides an additional assay for investigating Crz1 nucleocytoplasmic transport.
4. Discussion
Nucleocytoplasmic transport requires transient interactions between soluble transport receptors and phenylalanine-glycine (FG) containing repeats of the nuclear pore complex. Here we report the investigation of FG-Nups important for efficient protein export from the nucleus using a novel assay for nuclear export kinetics. To our knowledge, this study provides the first in vivo examination of the effect of specific Nup mutations on the kinetics of protein export through the NPC. While permeabilized cell assays in metazoan cells [46] and in vivo import assays in yeast [23] have allowed for careful examination of the nucleoporins and soluble factors affecting the import of NLS-containing proteins into the nucleus, analysis of proteins required for export has been more difficult. Proteins that are exported from the nucleus are by definition “shuttling” proteins in that they must enter the nucleus prior to being exported. Thus these proteins must contain (or associate with a protein that contains) an NLS that mediated their nuclear import in addition to an NES that facilitates export. The apparent intracellular localization of a shuttling protein under steady state conditions is dependent upon the relative rate of protein import versus export. If the rate of import is greater, the protein will appear to be “nuclear.” If export occurs faster than import, the majority of the protein will be outside the nucleus and the localization will appear cytoplasmic. This shuttling leads to several experimental difficulties. In order for export to be studied, a protein must first be detectable in the nucleus, requiring faster import than export. Then a mechanism for following passage out of the nucleus must exist to provide a way to detect export, or at least to measure the relative rate of substrate export vs. import. A permeabilized cell assay has been used in efforts to identify soluble factors important for the export of specific substrates in mammalian cells [29] and the microinjection of photoactivatable fusion proteins has allowed investigation of nuclear export kinetics of the Oct4 transcription factor in cleavage-stage mouse embryos [31]. However, an assay that allows for investigation of export rates in large numbers of mutants, including nucleoporin mutants, has not been reported. The in vivo export kinetics assay we have developed in yeast provides the first mechanism to comprehensively investigate relative export rates in nucleoporin and transport factor mutants.
We have used the yeast nuclear export kinetics assay to investigate the transport of substrate proteins (Crz1 and Mig1) whose nuclear import is more rapid than export under one set of environmental conditions (the presence of Ca2+ or glucose, respectively) and whose export is more rapid than import under a different set of conditions (addition of FK506 or removal of glucose, respectively). By assessing substrate protein localization at intervals after conditional shift, we not only determine whether the substrate is exported, but also observe differences in the kinetics of export between mutants. By measuring changes in export kinetics we have been able to detect subtle but significant perturbations in export rate that are not detectable by examining localization of an NES-containing substrate in cells under steady-state conditions.
The sensitivity of our kinetic export assay has allowed us to identify a subset of FG-containing Nups that are necessary for efficient Msn5-mediated export, including Nup100, Nup2, and Nsp1, while also providing evidence that other FG-Nups and FG-repeats contribute less significantly, if at all, to Crz1 export. Nup100 is localized toward the cytoplasmic side of the central core of the NPC [1]. We observe that both a complete deletion of Nup100 and removal of the Nup100 GLFG repeats reduce the export kinetics of Crz1-GFP. This result is consisted with the finding that Msn5 biochemically associates with Nup100 [21]. We have confirmed the effects of a nup100Δ mutation on Msn5-mediated export using two different marker proteins, Crz1-GFP and Mig1-GFP, and have supported these data with a CDRE-LacZ assay that reports nuclear Crz1 transcription activity. While this study examines the export kinetics of Msn5 substrates Crz1 and Mig1, similar export assays could be developed for other karyopherin substrates that undergo regulated nucleocytoplasmic shuttling, including Far1 [15], Msn4 [18], Pho4 [14], Aft1 [47], Yap1 [48], and proteins whose shuttling is experimentally controlled by photoactivation or induced dimerization [31, 49].
A Nup2 deletion also results in a statistically significant decrease in export kinetics of Crz1-GFP, although the decrease is smaller than observed for nup100Δ or nup100ΔGLFG. FRET analysis has revealed a close juxtaposition of Msn5 and Nup2 in vivo [20] providing support for a functional role for this interaction. Interestingly, Msn5 also interacts with Nup42, Nup49, and Nup1 [20, 21], but we do not observe significant changes in Msn5-mediated export upon deletion of these Nups or their FG repeats. It is possible that although Msn5 may interact functionally with these Nups, other Nups compensate for their absence, resulting in an inability to detect even subtle perturbations of export kinetics. Alternatively, while biochemical and FRET assays reveal interactions under experimental conditions, the proteins still may not interact functionally in vivo. Interestingly, we observe a decrease in Crz1 export in Nsp1 mutants, despite the absence of published evidence for an Msn5-Nsp1 physical or functional interaction. Further experiments are necessary to determine if the export defect we observe is indicative of a direct functional relationship between Msn5 and Nsp1 or due to a more general perturbation of NPC function that indirectly influences Msn5-mediated translocation.
Our observations also reveal differences in Msn5-mediated transport kinetics between Nup mutants from which similar domains are deleted. Nup1, Nup2, and Nsp1 are all FXFG Nups, with Nup1 and Nup2 found in the NPC basket and Nsp1 in the central core of the NPC [1]. We did not observe a decrease in Crz1-GFP export kinetics in nup1ΔFXFG cells, while Crz1-GFP export does decrease in both nup2Δ and nsp1ΔFXFG mutants to different extents, suggesting that some functional differences exist between the FXFG repeats of these three proteins. Interestingly, export is also reduced in nsp1ΔFG cells and is significantly affected by removing both the FG and FXFG repeats of Nsp1, providing evidence that these dissimilar repeats may have a redundant or cooperative function in Msn5-mediated nuclear export. Future experiments that swap FG-repeats between different Nups and carefully investigate potential changes in export kinetics will provide insight into the specialized functions of similar FG-repeats found on distinct Nups.
Our observations also provide preliminary evidence for differing effects on nuclear import and export between related FG repeats. Nup100 and Nup49 are both GLFG-Nups found in the central core of the NPC, with Nup49 localized symmetrically and Nup100 biased slightly toward the cytoplasmic face. While nup49ΔGLFG cells do not accumulate Crz1-GFP in the nucleus efficiently enough to perform an export kinetics assay (E. DeRoo, G. Clement, K. Belanger, unpublished) and show low levels of Crz1 transcriptional activity (Fig. 5A). nup100ΔGLFG cells accumulate Crz1 in the nucleus but export it more slowly than wild-type and have increased Crz1 activity compared to wild-type. Differences in transport between GLFG mutants have been observed previously when examining Kap121-mediated import and Mex67-mediated mRNA export (29). The data we report here provide additional evidence for a functional distinction between some highly similar FG-repeat sequences.
FG-Nups are necessary for nucleocytoplasmic protein transport. However, the specificity of the contribution of any one FG-Nup to the transport of a specific karyopherin/cargo complex is unclear. We have developed a novel in vivo nuclear export assay that contributes kinetic evidence to a growing body of steady-state data [25, 32] indicating that specific Nups have distinct roles in nuclear export. Our data reveal that examining export kinetics will be critical dissecting the function of distinct Nups in nuclear protein export. Finally, we provide kinetic evidence that protein export – like protein import and RNA export – requires specific Nups and that the subset of Nups required for efficient Msn5-mediated protein export is distinct from those involved in protein import [25] or mRNA export [32, 33].
Highlights.
A novel approach to study nuclear protein export kinetics is described.
Cells lacking the nucleoporins Nup100 or Nup2 exhibit slowed export.
GLFG repeats of Nup100 are necessary for efficient protein export.
FXFG repeats in Nsp1 but not Nup1 affect nuclear export kinetics.
Nup mutations that affect export alter Crz1 transcription factor activity.
Acknowledgments
The authors are indebted to Mark Johnson, Ed Hurt, Laura Davis, Pamela Silver, and Martha Cyert, and especially to Lisa Strawn and Susan Wente, for very generous sharing of strains, plasmids, and reagents. We acknowledge Karyn Belanger and Susan Geier for expert technical assistance, Joy Commisso for preliminary export assays, and Anita Corbett for helpful comments on the export assay and critical reading of this manuscript. This work was supported by funding from Colgate University and NIH R15-GM065107 to K.D.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rout M, Aitchison J, Suprapto A, Hjertaas K, Zhao Y, Chait B. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 3.Lim RY, Ullman KS, Fahrenkrog B. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walde S, Kehlenbach RH. The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 2010;20:461–469. doi: 10.1016/j.tcb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Denning D, Patel S, Uversky V, Fink A, Rexach M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci USA. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 7.Antonin W, Ellenberg J, Dultz E. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Letters. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- 8.Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 9.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–27. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pemberton L, Paschal B. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 11.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran EJ, Wente SR. Dynamic nuclear pore complexes: Life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- 14.Kaffman A, Rank NM, O’Neill EM, Huang LS, O’Shea EK. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 15.Blondel M, Alepuz PM, Huang LS, Shaham S, Ammerer G, Peter M. Nuclear export of Far1p in response to pheromones requires the export receptor Msn5p/Ste21p. Genes Dev. 1999;13:2284–300. doi: 10.1101/gad.13.17.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeVit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boustany L, Cyert M. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Devel. 2002;16:608–618. doi: 10.1101/gad.967602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schuller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towpik J, Graczyk D, Gajda A, Lefebvre O, Boguta M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J Biol Chem. 2008;283:17168–74. doi: 10.1074/jbc.M709157200. [DOI] [PubMed] [Google Scholar]
- 20.Damelin M, Silver P. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–40. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- 21.Allen N, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 22.Polizotto RS, Cyert MS, Polizotto RS, Cyert MS. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J Cell Biol. 2001;154:951–60. doi: 10.1083/jcb.200104078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shulga N, Roberts P, Gu Z, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–39. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fahrenkrog B, Hubner W, Mandinova A, Pante N, Keller W, Aebi UU. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol Biol Cell. 2000;11:3885–3896. doi: 10.1091/mbc.11.11.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol. 2004;6:197–206. doi: 10.1038/ncb1097. [DOI] [PubMed] [Google Scholar]
- 26.Belanger KD, Gupta A, MacDonald KM, Ott CM, Hodge CA, Cole CM, Davis LI. Nuclear pore complex function in Saccharomyces cerevisiae is influenced by glycosylation of the transmembrane nucleoporin Pom152p. Genetics. 2005;171:935–947. doi: 10.1534/genetics.104.036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLane LM, Pulliam KF, Devine SE, Corbett AH. The Ty1 integrase protein can exploit the classical nuclear protein import machinery for entry into the nucleus. Nucleic Acids Res. 2008;36:4317–4326. doi: 10.1093/nar/gkn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holaska JM, Paschal BM. A cytosolic activity distinct from Crm1 mediates nuclear export of protein kinase inhibitor in permeabilized cells. Proc Natl Acad Sci USA. 1998;95:14739–14744. doi: 10.1073/pnas.95.25.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holaska JM, Black BE, Rastinejad F, Paschal BM. Ca2+-dependent nuclear export mediated by calreticulin. Mol Cell Biol. 2002;22:6286–6297. doi: 10.1128/MCB.22.17.6286-6297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plachta N, Bollenbach T, Pease S, Fraser SE, Pantazis P. Oct4 kinetics predict cell lineage patterning in the early mammalian embryo. Nat Cell Biol. 2011;13:117–123. doi: 10.1038/ncb2154. [DOI] [PubMed] [Google Scholar]
- 32.Terry L, Wente S. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol. 2007;178:1121–1132. doi: 10.1083/jcb.200704174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powrie EA, Zenklusen D, Singer RH. A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. RNA. 2011;17:134–144. doi: 10.1261/rna.1210411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agatep R, Kirkpatrick R, Parchaliuk DL, Woods RA, Gietz RD. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online. 1998;1:1525. [Google Scholar]
- 35.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Academic Press; San Diego, CA: 1991. [Google Scholar]
- 36.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 37.Fiserova J, Goldberg MW. Nucleocytoplasmic transport in yeast: a few roles for many actors. Biochem Soc Trans. 2010;38:273–277. doi: 10.1042/BST0380273. [DOI] [PubMed] [Google Scholar]
- 38.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–78. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeVit MJ, Johnston M. The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr Biol. 1999;9:1231–41. doi: 10.1016/s0960-9822(99)80503-x. [DOI] [PubMed] [Google Scholar]
- 40.Iovine MK, Wente SR. A nuclear export signal in Kap95p is required for both recycling the import factor and interaction with the nucleoporin GLFG repeat regions of Nup116p and Nup100p. J Cell Biol. 1997;137:797–811. doi: 10.1083/jcb.137.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2001;276:6445–52. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- 42.Patel SS, Rexach MF. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium. Mol Cell Proteomics. 2001;7:121–31. doi: 10.1074/mcp.M700407-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–44. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83(5):683–92. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 45.Liu SM, Stewart M. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-beta homologue, Kap95p. J Mol Biol. 2005;349:515–25. doi: 10.1016/j.jmb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Adam SA, Sterne Marr R, Gerace L. Nuclear import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2980–90. doi: 10.1091/mbc.E06-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–20. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geda P, Patury S, Ma J, Bharucha N, Dobry C, Lawson S, Gestwicki J, Kumar A. A small molecule-directed approach to control protein localization and function. Yeast. 2008;25:577–594. doi: 10.1002/yea.1610. [DOI] [PubMed] [Google Scholar]
- 50.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–71. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]