Abstract
BACKGROUND & AIMS
Heme oxygenase-1 (HO-1) and its metabolic by-product, carbon monoxide (CO), protect against intestinal inflammation in experimental models of colitis, but little is known about their intestinal immune mechanisms. We investigated the interactions among CO, HO-1, and the enteric microbiota in mice and zebrafish.
METHODS
Germ-free, wild-type, and Il10−/− mice and germ free zebrafish embryos were colonized with pathogen-free (SPF). Germ-free or SPF-raised wild-type and Il10−/− mice were given intraperitoneal injections of cobalt protoporphyrin (CoPP), which upregulates HO-1, the CO releasing molecule ALF186, or saline (control). Colitis was induced in wild-type mice housed in SPF conditions by infection with S. typhimurium.
RESULTS
In colons of germ-free, wild-type mice, SPF microbiota induced production of HO-1 via activation of Nrf2–, IL-10–, and toll-like receptor–dependent pathways; similar observations were made in zebrafish. SPF microbiota did not induce HO-1 in colons of germ-free Il10−/− mice. Administration of CoPP to Il10−/− mice before transition from germ-free to SPF conditions reduced their development of colitis. In Il10−/− mice, CO and CoPP reduced levels of enteric bacterial genomic DNA in mesenteric lymph nodes (MLN). In mice with S. typhimurium-induced enterocolitis, CoPP reduced the numbers of live S. typhimurium recovered from the lamina propria, MLN, spleen, and liver. Knockdown of HO-1 in mouse macrophages impaired their bactericidal activity against E. coli, E. faecalis, and S. typhimurium, whereas exposure to CO or overexpression of HO-1 increased their bactericidal activity. HO-1 induction and CO increased acidification of phagolysosomes.
CONCLUSIONS
Colonic HO-1 prevents colonic inflammation in mice. HO-1 is induced by the enteric microbiota and its homeostatic function is mediated, in part, by promoting bactericidal activities of macrophages.
Keywords: mouse model, IBD, ulcerative colitis, anti-inflammatory agent
INTRODUCTION
The pathogenesis of the human inflammatory bowel diseases (IBD) involves complex interactions between genetic, immunologic and environmental factors1. The role of the environment in altering the course of IBD is exemplified by the epidemiological observation that cigarette smoking is protective against the development of ulcerative colitis2. However, factors that contribute to the protection from smoking remain unclear. Carbon monoxide (CO), a major component of cigarette smoke, has anti-inflammatory effects in numerous experimental models of inflammation3. Mammalian cells generate CO endogenously as a product of heme degradation by the heme oxygenase (HO) enzymes, of which, heme oxygenase-1 (HO-1) plays a homeostatic role in host defense and inflammation4. Our group previously demonstrated that CO ameliorates active inflammation in two models of chronic IBD, through mechanisms involving induction of HO-15, 6.
Consequently, we sought to further determine mechanisms through which CO and HO-1 maintain intestinal homeostasis. The human IBDs have been linked to genetic defects in innate immune responses to the enteric microbiota1. In this study, we show that the enteric microbiota induce colonic expression of HO-1 in WT but not colitis-prone Il10−/− mice. Pharmacologic induction of HO-1 protects Il10−/− mice from microbiota induced colitis when transitioned from germ free (GF) to conventional conditions. Induction of HO-1 by the microbiota represents a pathway conserved among diverse vertebrate hosts, as similar mechanisms were demonstrated in zebrafish. Moreover, HO-1 derived CO enhances macrophage bactericidal activity and bacterial clearance in vivo. Together, these experiments highlight the importance of HO-1 and CO in the maintenance of intestinal homeostasis through augmented bacterial clearance. HO-1, with pleiotropic anti-inflammatory effects, is a potential therapeutic molecule of interest in the human IBDs.
RESULTS
The enteric microbiota induce colonic HO-1 in vivo
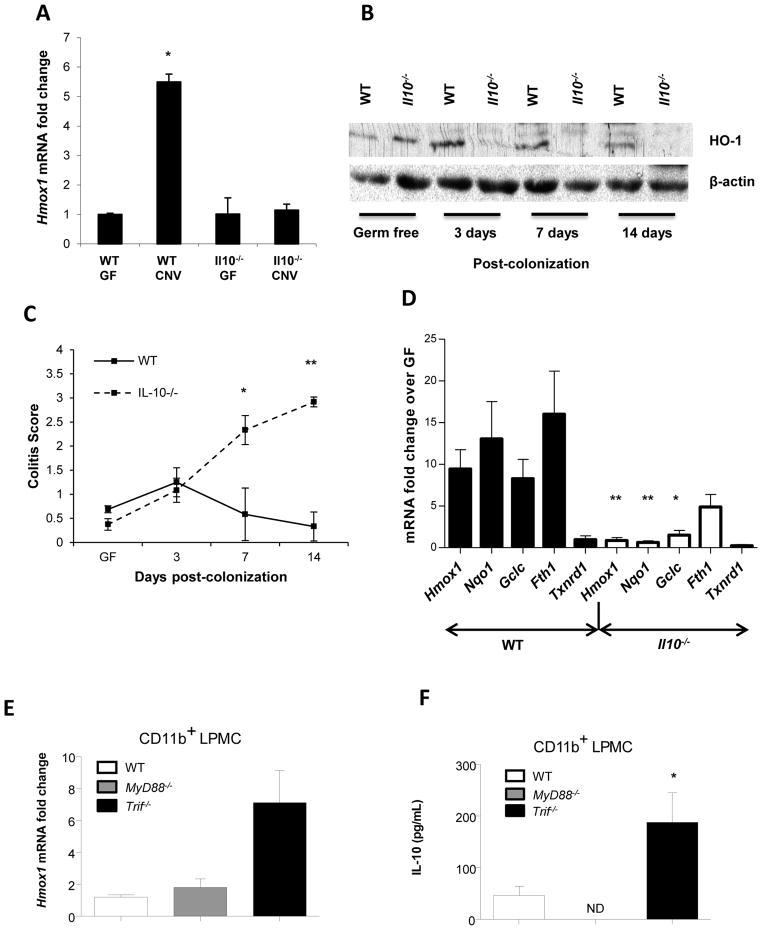
To determine whether the enteric microbiota regulate colonic HO-1 gene (Hmox1) expression, WT and colitis-prone Il10−/− mice raised germ-free (GF) were colonized with specific pathogen free (SPF) enteric microbiota (conventionalized, CNV). WT but not Il10−/− mice demonstrated increased colonic Hmox1 expression after colonization (Figure 1A). Increased colonic HO-1 protein by Western blot was detected in WT but not Il10−/− mice 3, 7 and 14 days after colonization (Figure 1B). This finding was confirmed by immunohistochemical staining for HO-1 in GF and CNV mice (Supplemental Figure 1). Attenuated colonic HO-1 correlated with the development of colonic inflammation in Il10−/− mice 14 days after colonization (Figure 1C). IL-12 p40 protein expression was augmented in colonic explant cultures from transitioned Il10−/− mice compared to WT mice (Supplemental Figure 2A). Likewise, Il12b and Tnf expression was increased in Il10−/− but not WT colonic tissue (Supplemental Figure 2B–C).
Figure 1. The enteric microbiota regulate colonic Hmox1 expression through Nrf2 and TLR dependent pathways.
WT (n = 3) and Il10−/− mice (n = 3) in GF conditions were conventionalized (CNV) with SPF enteric microbiota at 8 weeks of age. (A) Total RNA was isolated 14 days post-CNV and Hmox1 mRNA detected by real-time RT-PCR. Error bars represent mean ± SEM of three mouse colons analyzed independently. *p < .05 versus WT GF mice. (B) Total colonic protein was isolated and analyzed for HO-1 expression by Western blot. Data are representative of three independent blots on individual mouse colons. (C) Colitis scores of 8-week-old GF and CNV WT and Il10−/− mice represent mean ± SEM from three individual mouse colons. *p < .05, **p < .01 versus WT GF mice. (D) Total colonic RNA was isolated from WT (7 mice/group) and Il10−/− (7 mice/group) mice raised either GF or 14 days post-CNV. Expression of Nrf2 inducible genes (Hmox1, Nqo1, Gclc, Fth1, Txnrd1) were detected by real-time RT-PCR (normalized to β-actin mRNA). Results are from two independent experiments, presented as fold change of CNV mice relative to GF baseline controls. *p < .05, **p < .005. (E) Hmox1 expression was detected by real-time RT-PCR. Data represent mean ± SEM from 2 independent experiments with 3 mice per group, presented as fold change of CNV mice relative to GF baseline controls (normalized to β-actin mRNA). (F) Colonic CD11b+ LPMCs were isolated from WT, MyD88−/−, and Trif−/− mice. IL-10 secretion was determined by ELISA. Data represents mean ± SEM from 2 independent experiments with 3 mice per group. *p < .05.
Nuclear factor, erythroid 2 related factor 2 (Nrf2), a basic region leucine zipper transcription factor, is activated during cellular stress and inflammation, and is largely responsible for activation of Hmox1 transcription7. To determine whether Nrf2 is involved in transcriptional regulation of colonic stress response genes induced by the microbiota, expression of Nrf2 regulated genes (Hmox1, Nqo1, Gclc, Fth1, Txnrd1)8 was determined in GF and conventionally raised WT and Il10−/− mice. Significantly higher induction of Nrf2 regulated genes was observed in WT CNV mice compared to GF mice (Figure 1D), implicating Nrf2 as a homeostatic regulator of colonic Hmox1 induction by the microbiota. Consequently, attenuated induction of Nrf2 regulated genes was found in Il10−/− CNV compared to GF mice (Figure 1D).
We have previously shown that TLR ligands induce Hmox1 expression in bone marrow-derived macrophages (BMDMs) in a MyD88 dependent manner6. Moreover, IL-10 is a cofactor for HO-1 induction by TLR ligands through MyD88 independent mechanisms6, consistent with attenuated colonic Hmox1 induction by the microbiota in Il10−/− mice (Figure 1A, B, D). To advance these observations, Hmox1 and IL-10 expression was evaluated in colonic CD11b+ lamina propria mononuclear cells (LPMC) from WT mice and mice deficient in the downstream TLR signaling molecules MyD88 and TRIF. Interestingly, Hmox1 expression was similar in CD11b+ LPMC from MyD88−/− mice and WT mice (Figure 1E), demonstrating MyD88 independent mechanisms of Hmox1 regulation. Notably, in Trif−/− CD11b+ LPMC, there was increased expression of Hmox1 (Figure 1E) and IL-10 (Figure 1F), compared to CD11b+ cells from MyD88−/− and WT mice, suggesting that TRIF is a negative regulator of the mutually dependent homeostatic IL-10 and Hmox1 pathways6.
HO-1 induction by the microbiota is conserved in vertebrates
If HO-1 plays a prominent homeostatic role in the intestinal immune system, we reasoned that Hmox1 induction by the enteric microbiota would be conserved among vertebrate hosts. To test this hypothesis, hmox1 transcript levels were measured in gnotobiotic zebrafish larvae at 6 days post-fertilization (dpf) after rearing them under GF conditions or colonizing with a conventional microbiota at 3 dpf (conventionalized, CNV). Significantly increased expression of hmox1 was observed in CNV zebrafish relative to GF controls (Figure 2A). Next, zebrafish embryos were injected with nrf2 morpholino (MO) oligonucleotides to inhibit Nrf2 translation or a standard control MO, and then reared under GF or CNV conditions until 6 dpf. The efficiency of nrf2 MO knockdown was confirmed by reduced transcript levels of the known Nrf2 target gene, glutathione S-transferase pi (gstp1)9 (Figure 2B). Induction of hmox1 expression in CNV zebrafish injected with nrf2 MO was significantly diminished relative to those injected with standard control MO, indicating that hmox1 induction by the microbiota is nrf2 dependent in zebrafish, (Figure 2A).
Figure 2. HO-1 induction by the microbiota is conserved in vertebrates and regulated by nrf2.
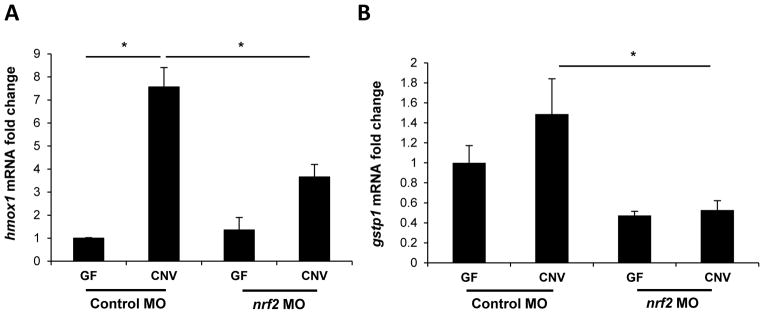
Zebrafish embryos derived in GF conditions were either transitioned to conventional microbiota (CNV) at 3 days post fertilization (dpf) or maintained GF. Total RNA isolated from zebrafish larvae at 6 dpf was analyzed for hmox1 expression by real-time RT-PCR. (A) Zebrafish embryos were injected with nrf2 morpholino (MO) to knockdown nrf2 mRNA translation, or standard control MO, then derived into GF conditions. At 3 dpf, zebrafish were transitioned to conventional microbiota or maintained GF. At 6 dpf, hmox1 expression was again analyzed by real-time RT-PCR. (B) Results of gstp1 expression in nrf2 MO and control MO treated zebrafish are shown as a positive control. Data represent mean ± SEM of biological duplicate pools (5–10 larvae/pool, 2 pools/condition) normalized to rpl32 mRNA levels.
Pharmacologic induction of HO-1 attenuates microbiota-induced colitis in Il10−/− mice
Induction of colonic HO-1 by the microbiota correlated with protection from colitis in GF to SPF transitioned WT mice. If protection from colitis development is secondary to HO-1 activity, then pharmacologic induction of HO-1 in GF Il10−/− mice should attenuate colitis development induced by microbiota conventionalization. To test this hypothesis, GF Il10−/− mice were administered cobalt protoporphyrin (CoPP), an inducer of HO-1, or vehicle (DMSO) intraperitoneally (IP), every other day for one week prior to SPF transition (Figure 3A). CoPP treated GF Il10−/− mice demonstrated a robust increase in colonic HO-1 protein compared with vehicle treated mice (Figure 3B). Moreover, CoPP treated Il10−/− mice demonstrated attenuated colitis (Figure 3C) and decreased IL-12 p40 secretion in colonic explant cultures (Figure 3D) compared to the vehicle treated group.
Figure 3. Pharmacologic HO-1 induction prevents enteric microbiota induced colitis in Il10−/− mice.
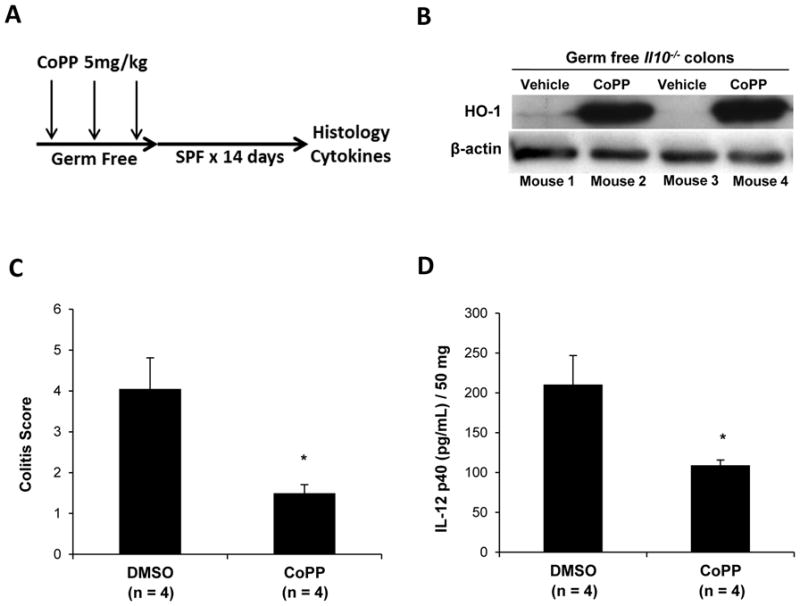
(A) GF Il10−/− mice were injected IP with 5 mg/kg CoPP (n = 4) or DMSO (vehicle, n = 4) every 48 h for 7 days, then conventionalized for 14 days. (B) Western blot on colonic tissue from 2 GF mice after 7 days of CoPP administration. (C) At 14 days after CNV, colitis scores were determined in CoPP injected mice and compared with DMSO injected mice. Results are presented as the mean of four averaged scores from five regions of the large intestine. Bars represent mean ± SEM (4 mice/group). (D) Spontaneous IL-12 p40 secretion was determined by ELISA in 24 hour supernatants from colonic explants from CoPP and DMSO treated Il10−/− mice. Results represent mean ± SEM from three independent colonic explant cultures (n = 3 mice/group). *p < .05, versus DMSO (vehicle) injected mice.
CoPP and ALF186 increase intestinal bacterial clearance in Il10−/− mice
We have previously shown that inhaled CO ameliorates chronic colitis in Il10−/− mice with induction of HO-1 necessary for protection5. The CO releasing molecule (CO-RM) ALF186 is a member of a family of related compounds where CO is covalently bound to a transition metal and released in aqueous solution10. To validate that ALF186 has similar effects as inhaled CO in Il10−/− mice, SPF-raised Il10−/− mice (5–6 weeks of age) were given the cyclooxygenase inhibitor piroxicam for 14 days in the diet to accelerate the development and severity of chronic colitis11. Following piroxicam treatment, ALF186 or inactive (i)ALF186 was administered IP twice daily for 2 weeks. ALF186 treated mice displayed a significant decrease in colitis severity scores (Supplemental Figure 3A) and less colonic secretion of IL-12 p40 in colon explant cultures (Supplemental Figure 3B) compared with mice treated with inactivated (i)ALF186 mice.
CO augments clearance of gram positive bacteria in polymicrobial sepsis, while blockade of HO-1 inhibited bactericidal activity in a S. typhimurium peritonitis model12, 13. We therefore hypothesized that HO-1 and CO may ameliorate colitis in part by augmenting clearance of enteric microbes that breach the epithelial barrier. The abundance of bacterial DNA in mesenteric lymph nodes (MLNs) of Il10−/− mice was measured as a marker of intestinal bacterial clearance. Eight week old Il10−/− mice were treated with CoPP, ALF186 or PBS by IP injection every 48 hours for two weeks and bacterial DNA abundance in MLNs quantitated by RT-PCR using universal bacterial 16S rRNA primers. There was significantly less bacterial DNA in CoPP and ALF186 treated mice compared to PBS controls (Figure 4A). The integrity of the intestinal epithelial barrier affects bacterial translocation, so proximal colon epithelial permeability from in vivo experiments was evaluated by measuring FITC-dextran permeability in colonic tissues mounted on Ussing chambers. There was an increase in the permeability of FITC-dextran in ALF186 and CoPP treated groups (Figure 4B). This result suggests that reduced bacterial detection in MLNs of ALF186 and CoPP treated Il10−/− mice was not secondary to increased intestinal epithelial barrier integrity. Additionally, CoPP and ALF186 did not inhibit enteric bacterial growth in culture (Supplemental Figure 4).
Figure 4. CoPP and ALF186 enhance intestinal bacterial clearance in Il10−/− mice.
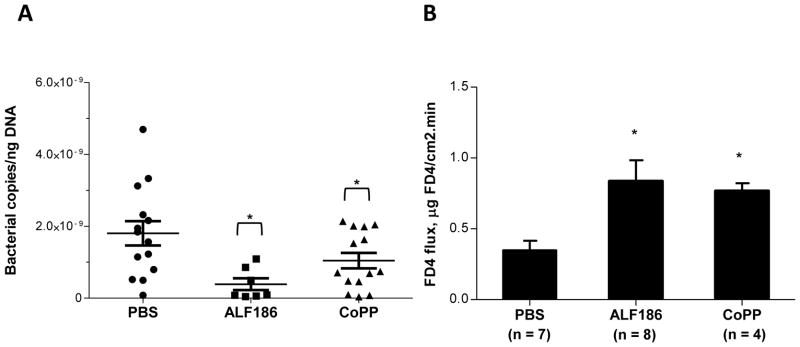
(A) Il10−/− mice (8–12 weeks old) were injected IP with 5mg/kg CoPP (n = 14), 30 mg/kg ALF186 (n = 14) or PBS control (n = 7), every 48 h for 14 days. Total DNA was isolated from MLNs and bacterial 16S rRNA gene copies were determined by real-time RT-PCR. Data are presented as combined mean ± SEM from three independent experiments. (B) Permeability was analyzed in proximal colon tissue in Ussing chambers by measuring mucosal-to-serosal FITC-dextran flux. Data represent mean ± SEM from duplicate experiments with n ≥ 4 per group. *p < .05 compared with PBS control.
Induction of HO-1 augments S. typhimurium clearance in an infectious colitis model
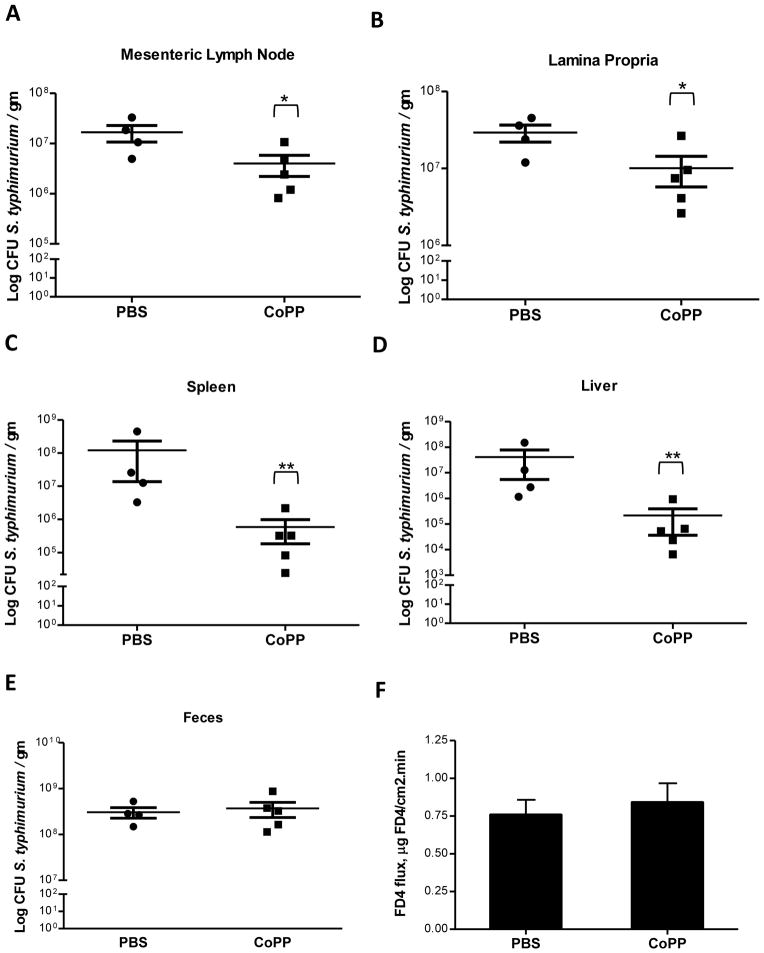
To further substantiate these observations, an enteric bacterial infection model (S. typhimurium) in WT mice that is characterized by invasion of mucosal and systemic lymphoid structures was utilized14. Significant reductions in live bacteria were noted in lamina propria, MLN, liver and spleen homogenates of CoPP treated mice relative to PBS controls (Figure 5A-D). S. typhimurium recovery from feces was not statistically different between groups (Figure 5E) and CoPP did not inhibit S. typhimurium growth in culture (Supplemental Figure 4C), suggesting that differences in mucosal and systemic bacterial recovery was not a consequence of differences in luminal bacterial abundance and/or growth. Moreover, there was no difference in FITC-dextran permeability between groups (Figure 5F) indicating that reduced systemic bacterial abundance in CoPP treated mice was not secondary to increased epithelial barrier integrity.
Figure 5. Pharmacologic HO-1 induction enhances S. typhimurium clearance in WT mice.
(A–E)Streptomycin treated WT mice were injected IP with CoPP (n = 5) or PBS (n = 4), 12 h prior to oral gavage with streptomycin resistant S. typhimurium. Treatment was repeated 48 h later and mice were euthanized 72 h after infection. Organs were recovered and CFUs were quantified by plating on LB agar with 50 μg/mL streptomycin. Data are presented as mean ± SEM and are representative of two independent experiments with similar results. (F) Permeability was analyzed in proximal colon tissue in Ussing chambers by measuring mucosal-to-serosal FITC-dextran flux. Data are presented as mean ± SEM. *p < .05, **p < .01.
HO-1 derived CO enhances macrophage bactericidal activity
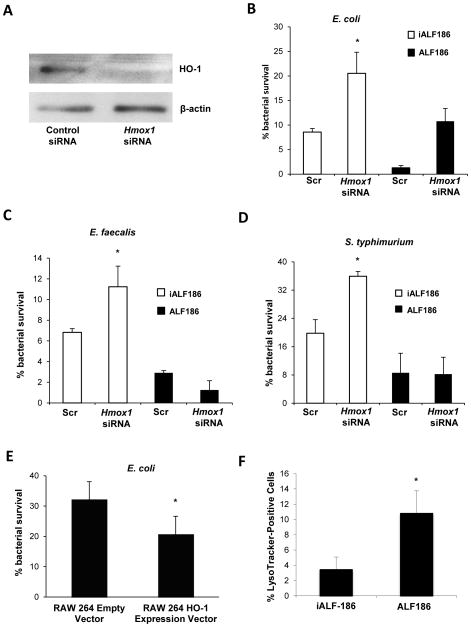
Based on our in vivo findings, we speculated that CO and HO-1 induction may enhance the ability of macrophages to eradicate enteric bacteria. WT BMDMs were transfected with Hmox1 siRNA and demonstrated significant knockdown of HO-1 compared with control scrambled siRNA transfected BMDMs (Figure 6A). Hmox1 siRNA transfected BMDMs showed impaired killing of intracellular K12 E. coli, E. faecalis and S. typhimurium (Figure 6B, C and D) compared with scrambled siRNA transfected BMDMs. The RAW 264.7 macrophage cell line stably transfected to over-express HO-1 demonstrated enhanced E. coli clearance compared with control plasmid transfected cells (Figure 6E). To demonstrate whether HO-1 derived CO is a downstream mediator of bactericidal activity, Hmox1 siRNA and control transfected BMDMs were incubated with ALF186 or iALF186. Incubation with ALF186 enhanced macrophage bactericidal activity in control and Hmox1 siRNA transfected cells compared to iALF186 treated BMDMs (Figure 6B–D, black bars). ALF186 but not iALF186 resulted in restitution of bactericidal activity in Hmox1 siRNA transfected macrophages exposed to E. coli, E. faecalis or S. typhimurium. ALF186 and iALF186 had no effect on BMDM viability (Supplemental Figure 5A). Additionally, similar levels of phagocytosis were observed in Hmox1 siRNA transfected BMDMs versus scrambled siRNA BMDMs and ALF186 versus iALF186 treated BMDMs (Supplemental Figure 5B–C).
Figure 6. HO-1 and CO enhance macrophage bactericidal activity.
(A) siRNA was used to knock down Hmox1 in WT BMDMs. Total protein was isolated and analyzed for HO-1 expression by Western blot to assess gene silencing. BMDMs were analyzed for macrophage bactericidal activity against (B) E. coli (C) E. faecalis and (D) S. typhimurium using gentamicin protection assay. siRNA transfected cells were also cultured in the presence of ALF186 (black bars) or inactive iALF186 (white bars) to study the effect of CO on bactericidal activity. Data are presented as mean ± SEM of 3 independent experiments. (E) RAW 264.7 macrophages that stably overexpress Hmox1 were used in gentamicin protection assays against E. coli K12. Results are expressed as mean ± SEM from 3 independent experiments, *p < .05 vs. scrambled siRNA transfected BMDMs. (F–G) BMDMs were incubated with ALF186, iALF186 (100 μM), CoPP (10 μM) or DMSO, then cells were treated with LysoTracker® (100 nM), and exposed to K12 E. coli. Cells were then fixed and nuclei stained with Topro-3, 5 nM. LysoTracker® positive cells were calculated as a percentage of total cells. At least 10 high powered fields and at least 100 cells were counted. Data are presented as mean ± SEM of 3 independent experiments. *p < .05.
We next sought to determine if treatment with ALF186 affects phagolysosomal maturation in macrophages as a mechanism for enhanced bactericidal activity. ALF186 and iALF186 treated WT BMDMs were incubated with medium containing LysoTracker® Red DND-99, a weak base that permeates cell membranes and fluoresces upon protonation in low-pH environments. ALF186-treated BMDMs demonstrated a significantly increased percentage of LysoTracker®-positive cells when cultured with E. coli (Figure 6F), suggesting that HO-1 derived CO may enhance phagolysosomal formation/acidification. E. coli labeled with eGFP was ultilized to localize bacteria to phagolysosomes (representative photomicrograph shown with LysoTracker staining in Supplemental Figure 5D).
DISCUSSION
The dynamic balance between the enteric microbiota and host innate responses in the intestine plays a pivotal role in the pathogenesis of IBD. Disordered homeostatic mechanisms such as autophagy, intracellular microbial killing, and cytokine expression have been implicated in disrupting this balance15. Utilizing GF WT and Il10−/− mice, this study demonstrates that the enteric microbiota regulate expression of HO-1 in mice and zebrafish, and that HO-1 induction inversely correlates with colonic inflammation in Il10−/− mice. Induction of colonic IL-10 has previously been shown to be dependent on the presence of enteric microbiota, with macrophages and regulatory T cells as primary sources16. We have previously demonstrated that HO-1 is induced by TLR ligands in BMDMs in an IL-10 and MyD88 dependent manner6. However, the present findings describe unique mechanisms of Hmox1 regulation in colonic macrophages. Increased levels of Hmox1 and IL-10 were found in Trif−/− mice compared with WT and MyD88−/− mice (Figure 1E, F). This result is consistent with our previous report where Trif was identified as a negative regulator of IL-10 in CD11b+ LPMC17. However, unlike BMDMs where Hmox1 induction is MyD88 dependent6, in CD11b+ LPMC, Hmox1 is induced in vivo through MyD88 independent and IL-10 dependent pathways.
Consequently, we used Il10−/− mice to elucidate functional effects of HO-1 and CO on clearance of commensal bacteria. Treatment of Il10−/− mice with ALF186 or CoPP was associated with increased enteric bacterial clearance in intestinal secondary lymphoid tissue. Increased intestinal permeability defects, modulated by the enteric microbiota, precede the development of colitis in Il10−/− mice18. In this study, enhanced intestinal barrier function was not responsible for reduced extraluminal bacterial detection, which instead implicates host clearance mechanisms. Moreover, CoPP and ALF186 did not exhibit direct antibacterial properties.
Differences in bacterial recovery at extraluminal sites between experimental groups may be a secondary consequence of differences in the intestinal inflammatory state. To mitigate this confounding issue in the assessment of bacterial clearance, we studied Il10−/− mice at a young age (eight weeks) when colitis is mild to assess bacterial clearance in a model that wouldn’t be biased by major differences in histologic inflammation between treatment groups. Indeed, significant differences in colonic inflammation were not detected between treatment groups (Supplemental Figure 6A). Likewise, an early time-point was chosen to evaluate bacterial clearance in the S. typhimurium model to exclude differences in bacterial recovery as a consequence of differences in inflammation and cellular infiltration (Supplemental Figure 6B). Early control of intracellular S. typhimurium infection is dependent on professional phagocytes. Macrophages are the main harbor of this intracellular pathogen in vivo19. S. typhimurium has also been shown to disrupt epithelial barrier integrity20. Consequently, this infectious model of colitis presents a more direct measure of bacterial clearance in vivo and allows correlation with our cell-based findings of HO-1 augmented bactericidal activity in macrophages. In summary, our studies show that induction of HO-1 leads to augmented clearance of bacteria from secondary mucosal lymphoid and systemic sites in two colitis models.
We previously demonstrated that CO ameliorates chronic inflammation through HO-1 dependent mechanisms in Il10−/− mice5. We have more recently shown different anti-inflammatory mechanisms of CO and HO-1 in TCRα−/− mice, where amelioration of colitis by CO and HO-1 induction was associated with increased IL-10 and IL-22 production, and decreased Th2 cytokine expression6. These studies demonstrate pleiotropic effects of the HO-1 pathway in different models of microbiota dependent colitis and combined with our current findings, suggest that the HO-1 pathway affects multiple central homeostatic processes. Moreover, we observed a similar phenomenon in GF zebrafish transitioned to a conventional microbiota, suggesting that HO-1 is important across diverse vertebrate lineages. Nrf2 is a well described transcriptional factor responsible for HO-1 induction in mammals and zebrafish in response to oxidative stress7. Here, we show that Nrf2 is responsible for HO-1 induction by the microbiota in mice and zebrafish. Moreover, decreased induction of Nrf2 inducible genes in Il10−/− GF mice colonized with microbiota, compared to WT mice suggests a key homeostatic interrelated regulatory pathway involving IL-10, Nrf2, and HO-1 in the intestine (Figure 1D).
This study also reveals mechanisms through which HO-1 and CO affect macrophage bactericidal activity. The importance of microbicidal pathways in the pathogenesis of IBD is highlighted by the discovery that synonymous SNPs in the autophagocytic genes ATG16L1 and IRGM; and a SNP in the phagosomal gene NCF4 are associated with enhanced risk for IBD21, 22. Incubation with ALF186 resulted in significantly increased phagolysosomal formation/activation in macrophages associated with enhanced killing of intracellular E. coli, E. faecalis and S. typhimurium. Chung and colleagues recently demonstrated that a CO-RM increases phagocytosis, decreases circulating bacterial counts and rescues Hmox1−/− mice from mortality in polymicrobial sepsis, interestingly via mechanisms that involve the Crohn’s disease susceptibility gene product NOD212. Our studies show similar effects between HO-1 induction and CO release implicating CO as a downstream effector of HO-1. Similarly, Otterbein and colleagues have shown in vitro that CO increased macrophage phagocytosis of E. coli23. However, we did not observe increased phagocytosis in cells, possibly due to differences in cell types and CO-RMs used in the respective experiments. Other interrelated mechanisms through which HO-1 promotes homeostasis are possible. Induction of HO-1 may foster the development of a microbiota that sustains colonic HO-1 expression and ameliorates inflammation. Likewise, HO-1 may induce the expression of unidentified cytokines/growth factors that directly mediate homeostasis.
In conclusion, the enteric microbiota induce colonic HO-1 in WT but not colitis-prone Il10−/− mice. HO-1 and its metabolic product CO exert their anti-inflammatory effects in part through enhancement of macrophage bactericidal activity. Consequently, defective HO-1 induction in macrophages may lead to defective clearance of enteric bacteria, contributing to the pathogenesis of IBD.
MATERIALS AND METHODS
Reagents
Cobalt(III) protoporphyrin IX chloride (CoPP) was from Frontier Scientific. ALF186, a CO-RM with structure [Mo(CO)3(histidinato)]Na, was obtained from Alfama, Inc (Lisbon, Portugal). ALF186 is water soluble and stable under anaerobic conditions but releases CO in a pH dependent fashion under aqueous aerobic conditions with a half-life of ~30 minutes. In vivo, peak carboxyhemoglobin levels are seen in 5–10 minutes after intraperitoneal, intravenous and oral administration (data not shown). On completion of in vivo experiments, blood carboxyhemoglobin levels of 10% to 12% were detected in individual mice, consistent with previously reported results in mice exposed to CO in ambient air5. A RAW 264.7 cell line stably transfected to overexpress HO-1 was generated by L. Otterbein. Bacterial strains used were E. coli K12 strain ATCC 29181, S. typhimurium strain ATCC 14028, E. faecalis OG1RF and streptomycin resistant S. typhimurium strain CA32.
Mice
All animals were housed in accordance with guidelines from the American Association for Laboratory Animal Care and Research Protocols and experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina. WT, Il10−/− MyD88−/−, and Trif−/− mice in SPF housing were on the C57BL/6 background and matched for age in all experiments. 129S6/SvEv germ free (GF) mice (WT and Il10−/−) were derived and maintained by standard techniques24 at the University of North Carolina Gnotobiotic Facility.
Il10−/− colitis model
GF to CNV transition: 129S6/SvEv GF Il10−/− mice were colonized at 10–12 weeks of age with an enteric microbiota isolated from 129S6/SvEv WT mice raised in SPF conditions25. In other experiments, GF Il10−/− mice were injected IP with 5 mg/kg of CoPP every 48 hours for 7 days then transitioned to SPF microbiota. Bacterial translocation: SPF raised C57Bl/6 Il10−/− mice were injected IP with either PBS or 5 mg/kg CoPP every 48 hours for 14 days. Mice were euthanized at day 14, MLNs were harvested and total DNA recovered for measurement of bacterial 16S rRNA gene copies by real-time PCR. Piroxicam model: 5–6 week old C57Bl/6 Il10−/− mice raised in SPF conditions were fed piroxicam chow for 2 weeks as described5. Mice were subsequently injected IP with 30 mg/kg of ALF186 or inactive ALF186, every 48 hours for two weeks.
S. typhimurium colitis model
WT C57Bl/6 mice housed in SPF conditions were infected with S. typhimurium as described with modifications14. Mice were pretreated with streptomycin by oral gavage and 12 hours later were treated with 5 mg/kg CoPP via IP injection or 100 μl of PBS (vehicle). 12 hours following, streptomycin resistant S. typhimurium (108 CFU) was introduced by oral gavage. CoPP or PBS treatment was repeated 48 hours later. Mice were euthanized 72 hours after infection. Organs were recovered under sterile conditions and homogenized using a rotor homogenizer. Cecal tissue was thoroughly washed in sterile PBS prior to homogenization. Dilutions in sterile PBS were plated on LB agar with 50 μg/ml streptomycin to determine CFUs.
Permeability Assay
Segments of proximal colon were harvested, placed in oxygenated (95% O2-5% CO2) Ringer solution and tissues were then mounted in 0.3 cm2 aperture Ussing chambers (Physiologic Instruments, San Diego, CA) as previously described 26, 27. Mucosal-to-serosal flux of (FITC)-dextran (4 kDa; Sigma-Aldrich, St. Louis, MO) (FD4) was performed as described28. The presence of FD4 fluorescence intensity was measured by fMax Fluorescence Microplate Reader (Molecular Devices, Sunnyvale, CA) and concentrations were determined from standard curves generated by serial dilution of FD4. Data for FD4 flux are presented as μg FD4/cm2.min.
Zebrafish
Zebrafish (Danio rerio) embryos were derived into GF conditions using published methods 29. GF embryos were either maintained under GF conditions until 6 dpf or inoculated with a conventional zebrafish microbiota at 3 dpf. GF and CNV zebrafish were provided an autoclaved diet (ZM-000, ZM Ltd) beginning 3 dpf. Zebrafish embryos were injected with nrf2/nfe2l2 morpholino (nfe2l2.e1.ATG, 2.5 pmol/embryo; CATTTCAATCTCCATCATGTCTCAG; Entrez Gene ID 360149) or standard control morpholino oligonucleotides (Gene Tools LLC) as previously described30. At 6 dpf, total RNA from whole zebrafish was harvested from pools of GF and CNV larvae (5–10 larvae/pool, 2 biological replicate pools/condition), and transcript levels for hmox1 and gstp1 were evaluated by real-time RT-PCR and normalized to rpl32 levels as described30.
Bone marrow-derived macrophages
BMDMs were cultured as described31.
Cytokine ELISAs
Murine IL-12 p40 and TNF ELISA kits (R&D Systems) was used according to manufacturers’ instructions.
Western immunoblot
Western immunoblot analyses were performed on whole cell extracts as described31. Anti–HO-1 antibodies were from Stressgen and β-actin antibodies were purchased from Abcam, MA.
Real time RT-PCR
Real time RT-PCR was performed as described6. Primer sequences are listed in Supplemental Table 1.
siRNA transfection
BMDMs were transfected with ON-TARGETplus Hmox1 siRNA, Chicago, IL using AMAXA Nucleofector Technology (AMAXA) by the protocol for murine macrophages. Control scrambled siRNA was purchased from Dharmacon, IL.
Colonic tissue explant cultures
Sections of colon were processed as described5.
Histology
Colitis scoring from colonic tissue sections from Il10−/− mice was performed as described5. In the S. typhimurium model, histological inflammation was assessed using a previously reported scoring system14. All histological scores were determined by a veterinary pathologist (LBB) blinded to the experimental treatment groups.
Bacterial DNA Isolation
Total DNA was extracted from mesenteric lymph nodes as described32. Bacterial 16S rRNA gene copies were determined by real-time RT-PCR using universal bacterial primer sequences (Supplemental Table 1) and normalized to total DNA33.
Gentamicin protection assay
1×106 WT and Il10−/− BMDMs were cultured in 12-well plates in triplicate and incubated with E. coli, E. faecalis or S. typhimurium at 10:1 ratio in antibiotic-free medium for 1 hour. Cells were then washed with PBS plus gentamicin (200 μg/ml) and incubated in medium supplemented with gentamicin (200 μg/ml) at 37°C for an additional hour to eliminate extracellular bacteria. New medium supplemented with gentamicin (100 μg/ml) was added to cells. ALF186 (100 μg/ml) and iALF186 (100 μg/ml) were added to the cells after elimination of extracellular bacteria. Cells were lysed with 1% Triton X-100, diluted, plated on BHI agar plates, and incubated at 37°C. CFUs calculated at 1 hour reflected the total uptake of bacteria (phagocytosis). CFUs recovered at 12 hours are represented as the percent survival of total bacteria phagocytosed at the 1 hour time point.
Phagolysosomal acidification assay
BMDMs from WT mice were incubated with ALF186 (100 μM) or iALF186 (100 μM), for 1 hour. Cells were then treated with LysoTracker® Red DND-99 (Invitrogen) 100 nM for 30 mins and E. coli K12 for 1h, fixed in 4% paraformaldehyde and stained with nuclear stain Topro-3, 5 nM (Invitrogen). E. coli labeled with eGFP (to localize bacteria to phagolysosomes) was used as a positive control. Cells were visualized using confocal microscopy with a Carl Zeiss LSM 710 laser scanning microscope. At least 10 high powered fields and 100 cells were counted.
Statistical Analysis
Statistical significance for data subsets from experiments performed in cells was assessed by the two-tailed Student’s t-test. Statistical significance for in vivo data subsets was assessed by the Mann-Whitney U test (SPSS, Chicago, IL, USA) with Bonferroni correction or one-way ANOVA where appropriate. A value of p < .05 was considered statistically significant.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by National Institutes of Health grants R01 DK54452 (to S.E.P.), R01 DK081426 (to J.F.R.), and K08 DK084313 (to A.J.M); Gastroenterology Research Training Grant T32 DK007737 (to S.Z.S. and J.C.O.); National Research Service Award F32 DK083186 (to S.Z.S.); UNC Center for Gastrointestinal Biology and Disease P30 DK034987 (Immunotechnologies, Gnotobiotic and Histology Cores), and a Crohn’s and Colitis Foundation of America Research Fellowship Award (N.M. and T.K.).
Abbreviations
- ALF186
Alfama CO-releasing molecule 186
- BMDMs
bone marrow derived macrophages
- CO
carbon monoxide
- CoPP
cobalt protoporphyrin
- CFU
colony forming unit
- ELISA
enzyme-linked immunosorbant assay
- Fth1
ferritin, heavy polypeptide 1
- GF
germ-free
- Gclc
glutamate-cysteine ligase, catalytic subunit
- Hmox1
murine heme oxygenase 1 gene
- HO-1
heme oxygenase-1
- iALF186
inactivated ALF186
- IFN
interferon
- IL
interleukin
- IL-10
deficient
- Il10
murine IL-10 gene
- Nqo1
NAD(P)H:quinone oxidoreductase
- Nrf2
NF-E2-related factor 2
- PCR
polymerase chain reaction
- SPF
specific pathogen freeIl10−/−
- TCRα−/−
T cell receptor- alpha deficient mice
- Th
T-helper cell
- Txnrd1
thioredoxin reductase 1
- WT
wild-type
Footnotes
Disclosures: L.E.O. is a scientific consultant for Alfama, Inc., and provided CO-releasing molecules (ALF186) for some experiments. The other authors have no financial conflicts of interest.
Contribution of Authors: Study concept and design (J.C.O., S.Z.S., S.E.P.); Acquisition of data (J.C.O., S.Z.S., N.M., S.M.R., E.C.S., T.K., L.C.M., J.J.H, A.J.M., J.F.R., L.B.B., S.E.P.); Analysis and interpretation of data (J.C.O., S.Z.S., N.M., A.J.M., J.F.R., L.B.B., T.K., L.E.O., S.E.P.); Drafting of the manuscript (J.C.O., S.Z.S., S.E.P.); Critical revision of the manuscript for important intellectual content (J.C.O., S.Z.S., A.J.M., J.F.R., L.E.O., S.E.P.); statistical analysis (J.C.O., S.Z.S., S.E.P.); Obtained funding (J.C.O., S.Z.S., N.M., T.K., A.J.M., J.F.R., S.E.P.); Technical or material support (L.E.O.); study supervision (S.E.P.).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–34. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Jick H, Walker AM. Cigarette smoking and ulcerative colitis. N Engl J Med. 1983;308:261–3. doi: 10.1056/NEJM198302033080507. [DOI] [PubMed] [Google Scholar]
- 3.Ryter SW, Choi AM. Cytoprotective and anti-inflammatory actions of carbon monoxide in organ injury and sepsis models. Novartis Found Symp. 2007;280:165–75. discussion 175–81. [PubMed] [Google Scholar]
- 4.Otterbein LE, Soares MP, Yamashita K, et al. Heme oxygenase-1: unleashing the protective properties of heme. Trends Immunol. 2003;24:449–55. doi: 10.1016/s1471-4906(03)00181-9. [DOI] [PubMed] [Google Scholar]
- 5.Hegazi RA, Rao KN, Mayle A, et al. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 2005;202:1703–13. doi: 10.1084/jem.20051047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheikh SZ, Hegazi RA, Kobayashi T, et al. An anti-inflammatory role for carbon monoxide and heme oxygenase-1 in chronic Th2-mediated murine colitis. J Immunol. 2011;186:5506–13. doi: 10.4049/jimmunol.1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alam J, Stewart D, Touchard C, et al. Nrf2, a Cap‘n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–8. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 8.Thimmulappa RK, Mai KH, Srisuma S, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–203. [PubMed] [Google Scholar]
- 9.Suzuki T, Takagi Y, Osanai H, et al. Pi class glutathione S-transferase genes are regulated by Nrf 2 through an evolutionarily conserved regulatory element in zebrafish. Biochem J. 2005;388:65–73. doi: 10.1042/BJ20041860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motterlini R, Otterbein LE. The therapeutic potential of carbon monoxide. Nat Rev Drug Discov. 2010;9:728–43. doi: 10.1038/nrd3228. [DOI] [PubMed] [Google Scholar]
- 11.Berg DJ, Zhang J, Weinstock JV, et al. Rapid development of colitis in NSAID-treated IL-10-deficient mice. Gastroenterology. 2002;123:1527–42. doi: 10.1053/gast.2002.1231527. [DOI] [PubMed] [Google Scholar]
- 12.Chung SW, Liu X, Macias AA, et al. Heme oxygenase-1-derived carbon monoxide enhances the host defense response to microbial sepsis in mice. J Clin Invest. 2008;118:239–47. doi: 10.1172/JCI32730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaki MH, Fujii S, Okamoto T, et al. Cytoprotective function of heme oxygenase 1 induced by a nitrated cyclic nucleotide formed during murine salmonellosis. J Immunol. 2009;182:3746–56. doi: 10.4049/jimmunol.0803363. [DOI] [PubMed] [Google Scholar]
- 14.Barthel M, Hapfelmeier S, Quintanilla-Martinez L, et al. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–58. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–37. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda Y, Kayama H, Jeon SG, et al. Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. Int Immunol. 2010;22:953–62. doi: 10.1093/intimm/dxq449. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi T, Matsuoka K, Sheikh SZ, et al. IL-10 Regulates Il12b Expression via Histone Deacetylation: Implications for Intestinal Macrophage Homeostasis. J Immunol. 2012;189:1792–9. doi: 10.4049/jimmunol.1200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen KL, Malfair D, Gray D, et al. Interleukin-10 gene-deficient mice develop a primary intestinal permeability defect in response to enteric microflora. Inflamm Bowel Dis. 1999;5:262–70. doi: 10.1097/00054725-199911000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Mittrucker HW, Kaufmann SH. Immune response to infection with Salmonella typhimurium in mice. J Leukoc Biol. 2000;67:457–63. doi: 10.1002/jlb.67.4.457. [DOI] [PubMed] [Google Scholar]
- 20.Kohler H, Sakaguchi T, Hurley BP, et al. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am J Physiol Gastrointest Liver Physiol. 2007;293:G178–87. doi: 10.1152/ajpgi.00535.2006. [DOI] [PubMed] [Google Scholar]
- 21.Reinhard C, Rioux JD. Role of the IBD5 susceptibility locus in the inflammatory bowel diseases. Inflamm Bowel Dis. 2006;12:227–38. doi: 10.1097/01.MIB.0000202413.16360.a1. [DOI] [PubMed] [Google Scholar]
- 22.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–66. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 23.Otterbein LE, May A, Chin BY. Carbon monoxide increases macrophage bacterial clearance through Toll-like receptor (TLR)4 expression. Cell Mol Biol (Noisy-le-grand) 2005;51:433–40. [PubMed] [Google Scholar]
- 24.MacDonald TT, Carter PB. Contact sensitivity in the germ-free mouse. J Reticuloendothel Soc. 1978;24:287–93. [PubMed] [Google Scholar]
- 25.Sheikh SZ, Kobayashi T, Matsuoka K, et al. Characterization of an interferon-stimulated response element (ISRE) in the Il23a promoter. J Biol Chem. 2011;286:1174–80. doi: 10.1074/jbc.M110.147884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argenzio RA, Liacos JA. Endogenous prostanoids control ion transport across neonatal porcine ileum in vitro. Am J Vet Res. 1990;51:747–51. [PubMed] [Google Scholar]
- 27.Moeser AJ, Ryan KA, Nighot PK, et al. Gastrointestinal dysfunction induced by early weaning is attenuated by delayed weaning and mast cell blockade in pigs. Am J Physiol Gastrointest Liver Physiol. 2007;293:G413–21. doi: 10.1152/ajpgi.00304.2006. [DOI] [PubMed] [Google Scholar]
- 28.Moeser AJ, Borst LB, Overman BL, et al. Defects in small intestinal epithelial barrier function and morphology associated with peri-weaning failure to thrive syndrome (PFTS) in swine. Res Vet Sci. 2012 doi: 10.1016/j.rvsc.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Pham LN, Kanther M, Semova I, et al. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–75. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanther M, Sun X, Muhlbauer M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-kappaB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong H, Zhu C, Li F, et al. Inhibition of Interleukin-12 p40 Transcription and NF-{kappa}B Activation by Nitric Oxide in Murine Macrophages and Dendritic Cells. J Biol Chem. 2004;279:10776–10783. doi: 10.1074/jbc.M313416200. [DOI] [PubMed] [Google Scholar]
- 32.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horz HP, Vianna ME, Gomes BP, et al. Evaluation of universal probes and primer sets for assessing total bacterial load in clinical samples: general implications and practical use in endodontic antimicrobial therapy. J Clin Microbiol. 2005;43:5332–7. doi: 10.1128/JCM.43.10.5332-5337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.