Summary
We have used protein electrophoresis through polyacrylamide gels derivatized with the proprietary ligand Phos-tag™ to separate the response regulator BvgA from its phosphorylated counterpart BvgA~P. This approach has allowed us to readily ascertain the degree of phosphorylation of BvgA in in vitro reactions, or in crude lysates of Bordetella pertussis grown under varying laboratory conditions. We have used this technique to examine the kinetics of BvgA phosphorylation after shift of B. pertussis cultures from non-permissive to permissive conditions, or of its dephosphorylation following a shift from permissive to non-permissive conditions. Our results provide the first direct evidence that levels of BvgA~P in vivo correspond temporally to the expression of early and late BvgA-regulated virulence genes. We have also examined a number of other aspects of BvgA function predicted from previous studies and by analogy with other two component response regulators. These include the site of BvgA phosphorylation, the exclusive role of the cognate BvgS sensor kinase in its phosphorylation in Bordetella pertussis, and the effect of the T194M mutation on phosphorylation. We also detected the phosphorylation of a small but consistent fraction of BvgA purified after expression in Escherichia coli.
Keywords: Bordetella pertussis, transcriptional activation, RNA polymerase, two-component systems, phosphorylation
Introduction
Throughout their evolution, bacteria faced a number of puzzles that had to be solved to enable their adaptation to multiple, variable environments. One of these was how changes in external conditions could be transduced into relevant adaptive changes taking place in the cytoplasm, particularly changes in gene expression. A highly successful solution to this problem is represented by two-component systems. These systems typically comprise a membrane-spanning sensor kinase protein (SK), the activity of which is responsive to relevant environmental or physiological signals, together with a cognate target response regulator (RR) protein. The degree of phosphorylation of a given RR is generally an indicator of its activity as a transcriptional activator of relevant genes. Two-component systems are found in virtually every species of bacteria and have been found to regulate virtually all cellular processes. These processes include essential metabolic and physiologic activities but also secondary characteristics that confer highly adaptive traits for specialized environments, such as virulence factors of bacterial pathogens.
Given their importance, it is not surprising that two-component systems have been the subjects of intensive study. In vitro biochemical methods have been crucial in describing basic functional aspects, notably the flow of phosphate (Uhl & Miller, 1994). Genetic approaches, while contributing to these in vitro studies, have been particularly useful in vivo. For example, while the phosphorylation of response regulators by sensor kinases was first demonstrated in vitro, the importance of this in vivo can be investigated by altering the conserved aspartate residue that is the site of phosphorylation. Experiments like this demonstrate the importance of phosphorylation in a qualitative sense and indicate the extreme phenotype resulting from a complete lack of phosphorylation (Lukat et al., 1991). But quantifying the level of a phosphorylated protein relative to its unphosphorylated form has been difficult. Response regulators, which are phosphorylated at an aspartate residue, have been particularly challenging, in part because the phospho-Asp acylphosphate bond is easily hydrolysed (Lukat et al., 1991). Although a number of approaches have been successfully used to assess in vivo phosphorylation, these have generally been cumbersome or have involved radioactive labeling of response regulators (Head et al., 1998, Idelson & Amster-Choder, 1998, Ladds et al., 2003).
Recently a simpler, less expensive, and more direct procedure has been employed (Barbieri & Stock, 2008). This approach uses electrophoresis of total cellular proteins through a polyacrylamide gel cast from monomers derivatized with a proprietary adduct termed Phos-tag™. Phos-tag™ is a dinuclear metal complex, which together with Zn2+ or Mn2+, forms a complex with a phosphomonoester dianion, such as the phosphorylated aspartate of a RR. During SDS-PAGE through this matrix, the phosphorylated protein migrates more slowly than its non-phosphorylated form (Barbieri & Stock, 2008). As a result, a Western blot of total cellular proteins separated on a Phos-tag™ gel can be interrogated using antibody specific to a protein of interest. The relative amounts of the two species can then be estimated.
We report here our application of this method to the BvgAS two-component system involved in regulation of virulence in Bordetella pertussis. In this human respiratory pathogen, the BvgAS two-component system is a global regulator that controls the expression of over 100 virulence genes in Bordetella species (Cummings et al., 2006, Hot et al., 2003). Different laboratory conditions are known to modulate activity of this system and consequently, the level of virulence gene expression. At least three different modes (Bvg+, Bvgi, Bvg−) correspond to three different expression patterns for virulence genes. In the Bvg+ mode (non-modulating condition) characterized by growth in the absence of modulating factors, all of the virulence genes are expressed. In the Bvg− mode, i.e. under “modulating” conditions such as a temperature of 25°C or presence of nicotinic acid or MgSO4, virulence gene expression is off [for review,(Cotter & Jones, 2003, Decker et al., 2012)]. The Bvgi mode is manifested under growth conditions between these two extremes. Some virulence genes, so called “early genes”, are expressed but others, termed “late genes”, are not. This difference has been attributed to a requirement by late genes for higher levels of BvgA~P. Another feature of the intermediate mode is the maximal expression of another class of genes, most notably bipA. This pattern of expression has been shown to be due to the presence of an upstream high-affinity BvgA-binding site that activates expression, even at low BvgA~P levels, together with low-affinity BvgA-binding sites downstream that can repress bipA transcription at high levels of BvgA~P (Williams et al., 2005). These features have led to the characterization of the BvgAS system as a “rheostat” rather than an on/off switch. As such, the ability to determine in vivo levels of BvgA phosphorylation is especially relevant.
Previous work has identified mutations in bvgS and bvgA that affect their behavior in ways that have been interpreted in terms of their hypothetical effects on BvgA phosphorylation. Some in vitro work supports these interpretations, but the ability to test their validity by monitoring effects on phosphorylation in vivo has heretofore been lacking. Similarly, modulation has been predicted to control the level of BvgA phosphorylation in vivo, but this has not been testable until now. In this report we demonstrate the use of the Phos-tag™ gel system to address these and related issues.
Results
BvgA and BvgA~P can be separated by Phos-tag™ SDS-PAGE
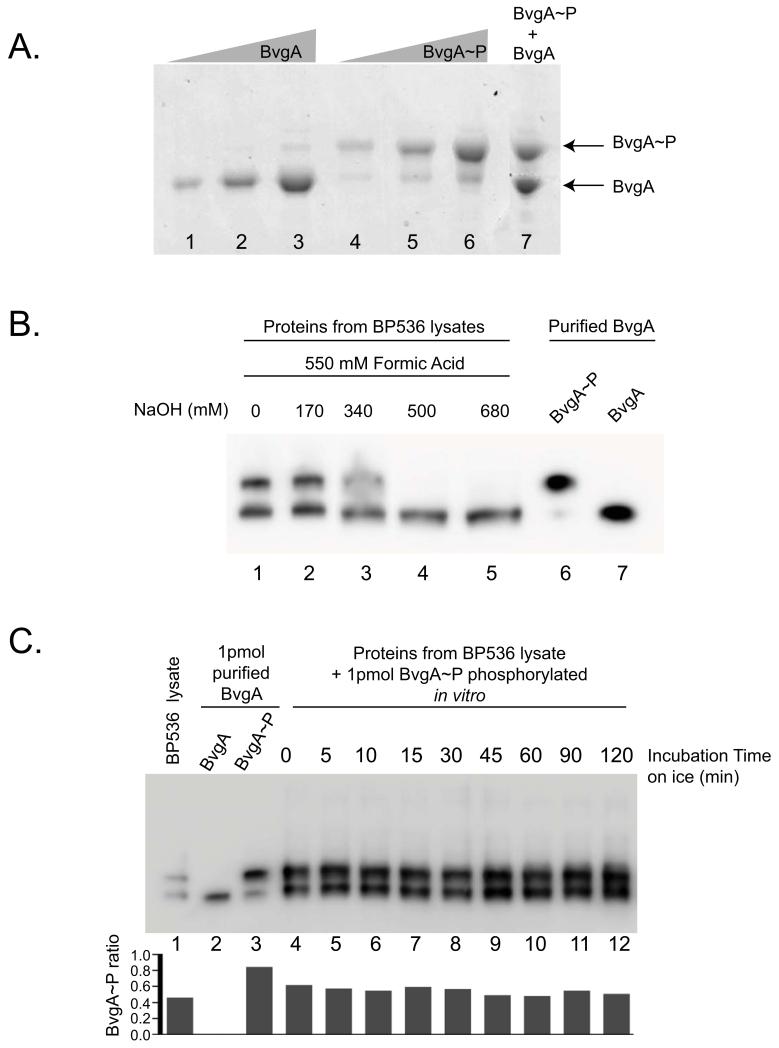
To determine if SDS-PAGE with Phos-tag™ could be used to separate BvgA~P from BvgA, we initially followed the protocol reported by Barbieri et al. for the analysis of E. coli PhoB and PhoB~P (Barbieri & Stock, 2008). PhoB and BvgA have similar molecular weights; 26kDa and 23 kDa, respectively. Initially unsuccessful, we tried recently described modifications including performing electrophoresis at neutral pH and substituting Zn2+ in place of Mn2+. Both have been reported to increase the separation between phosphorylated and non-phosphorylated species (Kinoshita & Kinoshita-Kikuta, 2011). We phosphorylated BvgA in vitro with acetylphosphate (Ac~P) or treated with buffer as a control, and performed SDS-PAGE in a gel containing Zn2+ and the Phos-tag™ ligand. Different amounts of BvgA (Fig. 1A, lanes 1-3), BvgA~P (lanes 4-6) or an equal mix of both forms (lane 7) were electrophoresed on a Phos-tag™ polyacrylamide gel.
Figure 1. BvgA and BvgA~P can be separated by Phos-tag™ polyacrylamide gel electrophoresis and detected in vivo by Western analysis.
A. Phosphorylation of BvgA in vitro. A Coomassie-stained gel shows different amounts [10 pmol (lane 1 and 4), 25 pmol (lane 2 and 5), 50 pmol (lane 3 and 6)] of BvgA after treatment with buffer alone (lanes 1-3) or buffer plus 20 mM Ac~P (lanes 4-6), followed by SDS-PAGE with Phos-tag™. Lane 7 contains a mix of 40 pmol of BvgA and 40 pmol of BvgA~P.
B. BvgA phosphorylation in B. pertussis. Cells were lysed and processed as described in Experimental Procedures using different amounts of NaOH (lanes 1-5) before electrophoresis on a Phos-tag™ gel and Western analysis. As a control, 1 pmol of purified BvgA~P (lane 6) and BvgA (lane 7) was loaded on the gel.
C. Stability of BvgA~P during cell lysate processing. One frozen aliquot of B. pertussis BP356 cells (harvested from 1 mL culture at OD600 0.15) was mixed with 15 pmol of in vitro phosphorylated BvgA. This combination of purified BvgA~P and B. pertussis cells was treated with 550 mM formic acid and 170 mM NaOH in a total volume of 60 μl for lysis (as described in Experimental Procedures) and incubated on ice for the indicated amount of time. From this mixture, 4 μl (containing 1 pmol of purified BvgA~P plus the in vivo B. pertussis BvgA~P) were electrophoresed on the Phos-tag™ gel. BvgA was detected by Western analysis. Lane 1 shows the pellet lysate with no purified BvgA~P added, to indicate the starting ratio of BvgA~P to BvgA. Lanes 2 and 3 correspond to 1 pmol of purified BvgA or BvgA~P, respectively. The graph represents the ratio of BvgA~P on the total amount of BvgA protein for each lane.
Under these conditions, BvgA and BvgA~P migrated at distinct positions and were easily separated on the gel. The amount of protein quantified in each band was proportional to the amount of protein loaded (data not shown). Treatment of BvgA with Ac~P (Fig. 1A, lanes 4-6) resulted in the majority of the protein being phosphorylated. Mixing BvgA and BvgA~P together prior to electrophoresis (lane 7) gave the expected ratio of nonphosphorylated to phosphorylated protein, indicating no significant loss of BvgA~P during the electrophoresis. Surprisingly, (as discussed further below), the purified, untreated BvgA appeared to contain a trace of phosphorylated protein (lane 3).
SDS-PAGE with Phos-tag™ can be used to determine the levels of BvgA and BvgA~P in vivo
To investigate whether the same approach could be used in vivo in B. pertussis we lysed cells with formic acid and then neutralized the acid with NaOH as described for E. coli (Barbieri & Stock, 2008). After electrophoresis, BvgA was visualized by Western analysis. This analysis demonstrated that BvgA and BvgA~P generated in vivo and in the context of a heterogenous sample migrated similarly to their in vitro counterparts (Fig. 1B, lanes 1-3 versus lanes 6 and 7). To determine whether the procedure for generating the cell lysate affected the level of BvgA~P observed we tested different conditions for processing the cells. The maximum level of BvgA~P was observed when either no NaOH or 170 mM NaOH was added to the lysate (Fig. 1B, lanes 1 and 2, respectively). BvgA~P was not detected when 500 mM or a higher concentration of NaOH was used (Fig. 1B, lanes 4 and 5). This is unlike the case of PhoB, for which a high level of NaOH could be used (Barbieri & Stock, 2008).
Because of the inherent instability of the phospho-aspartate bond, it was especially important to determine whether the treatments used for cell lysis and subsequent gel electrophoresis affected the level of BvgA~P we detected. To investigate this possibility, we added a constant amount of BvgA~P, formed by phosphorylation in vitro with Ac~P, to cell pellets, lysed the cells as above, and incubated the lysed samples on ice for different times. As shown in Fig. 1C, lanes 4-12, no significant difference in the amount of BvgA~P was observed, indicating that the extent of dephosphorylation of BvgA~P during the process was negligible.
These results demonstrate that the degree of BvgA phosphorylation can be quantitated easily and directly, both in vivo and in vitro, using Phos-tag™ technology. This experimental approach therefore allowed us to investigate the levels of BvgA~P in B. pertussis under a range of relevant growth conditions and genetic contexts.
Modulation of virulence gene expression in B. pertussis by MgSO4 occurs through a reduction in the level of BvgA~P
It is known that growth in the presence of 50 mM MgSO4 results in a Bvg− state, in which expression of the BvgA-activated genes is turned off. It has been assumed that the effect of MgSO4 and other modulating conditions is to reduce the level of BvgA~P in vivo. However, this assumption has not been tested directly. The Phos-tag™ system allowed us to directly observe the levels of BvgA and BvgA~P and to follow the kinetics of phosphorylation after cells were modulated or relieved from modulation.
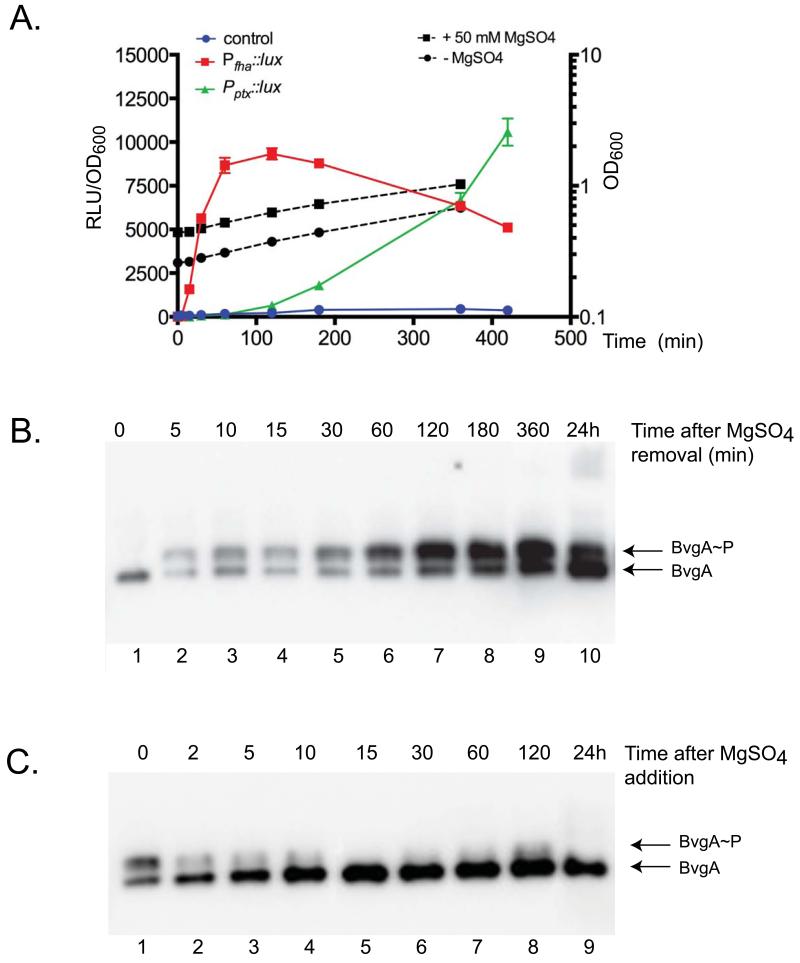
We determined levels of BvgA~P and BvgA in B. pertussis before, and at various times after, the addition of MgSO4. We also performed the complementary experiment in which cells grown in the presence of MgSO4 were shifted to a permissive medium lacking the modulator. As seen in Fig. 2A, in both cases, the cultures were growing exponentially over the time course of this experiment (with the exception of the 24 hour time point).
Figure 2. Kinetics of BvgA phosphorylation and gene expression in B. pertussis following modulation, or relief of modulation, of BvgAS kinase.
A. Level of gene expression and cell growth at different time points after removal of 50 mM MgSO4.
A vector carrying a transcriptional fusion of PfhaB, Pptx, or no promoter (control) with the luxCDABE operon was integrated in single copy in the BP536 chromosome. At the zero time point cells in liquid PLB medium were shifted from a non permissive (+MgSO4) to a permissive medium (−MgSO4) and OD600 as well as luminescence were monitored. The results shown here represent the assay repeated 4 times with triplicate sample reading at each time point. The left Y-axis corresponds to the relative luminescence in arbitrary units (RLU) normalized by OD600. Dashed black lines represent growth curves of both cultures monitored by OD600, with reference to the right Y-axis.
B., C. Levels of BvgA and BvgA~P at different time points after removal (B) or addition (C) of 50 mM MgSO4. At t=0, cells were shifted from a permissive (−MgSO4) to a non-permissive medium (+MgSO4) or vice versa. At the indicated times, cells were harvested. Cell lysates were subjected to Phos-tag™ gel electrophoresis and Western blot.
After many generations of growth in the presence of MgSO4, phosphorylated BvgA was not detected (Fig. 2B, lane 1) and the amount of BvgA protein was low. With the switch to permissive conditions (removal of MgSO4), BvgA~P could be detected within 5 minutes (lane 2), and the level of phosphorylation increased rapidly as incubation continued (lane 2-9) ultimately reaching a maximum level of 83% after two hours. In addition, the overall level of BvgA increased. This finding is reasonable since BvgA~P is known to activate expression of the bvgAS locus itself (Roy & Falkow, 1991, Scarlato et al., 1990). The reason for an apparent decrease in BvgA~P relative to BvgA at 24 hours is unknown at this time. Phosphorylation of all of the BvgA was never observed, even after continued incubation in the absence of MgSO4. Given that our control analyses suggested that BvgA~P is stable under our conditions (Fig. 1C), this result indicated a balance between the kinase activity of BvgS and mechanisms of BvgA dephosphorylation, either spontaneous or mediated by a hypothetical phosphatase activity of BvgS.
The kinetics of virulence gene expression following a shift from modulating to non-modulating conditions have been demonstrated previously, most notably by Scarlatto et al. (1991). Experiments such as this led to the concept of “early” and “late” genes, typified by fha and ptx, respectively. It has been assumed that the differential expression patterns observed between these two classes were orchestrated by rising levels of BvgA~P following relief of modulation. Indeed, Scarlatto et al. showed, as we have here, that overall levels of BvgA protein were increasing over the time course of their experiment. We have been able, through the use of the electrophoresis system described here, to extend this observation and to demonstrate that, as predicted, the level of BvgA~P correlates with gene expression. The early fha promoter can be activated at relatively low levels of BvgA~P, while activation of the late ptx promoter does not occur until higher levels have been reached. Since our study was performed somewhat differently from that of Scarlatto et al., we verified that differential expression of fha and ptx was seen in our hands as well. As shown in Fig. 2A, generation of light resulting from expression of lux fusions to these two promoters followed the same general patterns of early and late expression under our conditions of growth in PLB and induction by removal of MgSO4 as had been observed previously after growth in Stainer-Scholte medium and temperature shift (Scarlatto et al., 1991).
Lysates of cells obtained after long-term growth in permissive conditions contained approximately equal levels of BvgA and BvgA~P (Fig. 2C, lane 1). Upon the addition of MgSO4, there was a rapid decrease in the level of BvgA~P (lane 2-9) with no detectable BvgA~P remaining after 15 minutes of incubation (lane 5). However, the total amount of BvgA protein actually appeared to increase with continued incubation, until 15 minutes after addition of MgSO4, and then remained stable for hours. This finding is consistent with the idea that the BvgA protein itself is stable in vivo, but may also indicate a hitherto unappreciated regulatory process that stimulates BvgA synthesis immediately upon modulation. After 24 hours, the level of BvgA had decreased somewhat. This may indicate a very slow return to the low level observed at the zero time point in the panel above, which represents long term (2 days) growth on solid media under modulating conditions.
BvgS is the only kinase that phosphorylates BvgA in B. pertussis
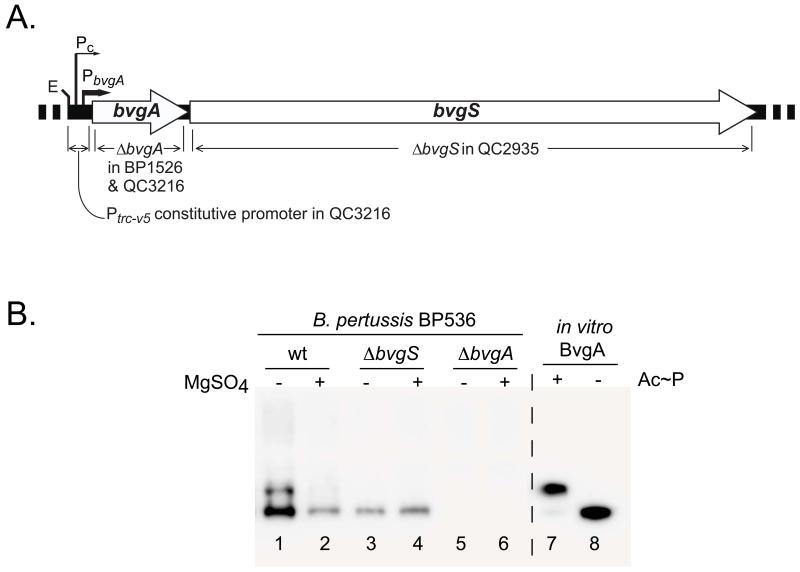
Genetic evidence supports the idea that BvgS is the major effector of BvgA phosphorylation. The bvgS gene is the locus affected by mutations that result in a variety of phenotypes, including a null phenotype, resistance to modulation, and the intermediate phase phenotype (Cotter & Miller, 1997, Goyard et al., 1994, Manetti et al., 1994, Miller et al., 1992). However it is not known if any other B. pertussis factors can phosphorylate BvgA. To address this directly, we determined the level of BvgA phosphorylation in wild-type and mutant B. pertussis (Fig. 3). As observed previously, BvgA~P was present when wild-type cells were grown in permissive conditions (Fig. 3B, lane 1), but only BvgA was observed under non-permissive growth conditions (lane 2). Growth under permissive or non-permissive conditions of a strain in which bvgS had been deleted (strain QC2935, depicted in Fig. 3A) also resulted in no BvgA phosphorylation, and the overall amount of BvgA protein was low (lanes 3 and 4), indicating that BvgS is the only kinase that phosphorylates BvgA in B. pertussis under these, typical, growth conditions. As expected, no BvgA was detected when bvgA was deleted (Fig. 3B, lanes 5 and 6; strain BP1526 depicted in Fig. 3A). The lack of any detectable species in lanes 5 and 6 also demonstrated the high specificity of the BvgA antibody used in these experiments.
Figure 3. BvgS is the only kinase that phosphorylates BvgA in B. pertussis under typical growth conditions.
A. Schematic illustration of the B. pertussis bvgAS locus showing specific deletions of bvgA and bvgS used in this study. Also shown are the locations of the native BvgA-inducible and constitutive promoters and the Ptrc-v5 constitutive promoter added to the bvgA deletion strain QC3216 to drive bvgS expression.
B. Effect of specific chromosomal deletions on BvgA phosphorylation. B. pertussis strains BP536 (wild-type, lanes 1 and 2), QC2935 (ΔbvgS, lanes 3 and 4) and BP1526 (ΔbvgA lanes 5 and 6) grown in the absence (lanes 1, 3, and 5) and presence (lanes 2, 4, and 6) of MgSO4 were analyzed by Phos-tag™ gel-electrophoresis and Western blot. Control lanes contained purified wild-type BvgA incubated with (lane 7), or without (lane 8) Ac~P.
Mutation of BvgA residue D54 eliminates BvgA phosphorylation in B. pertussis
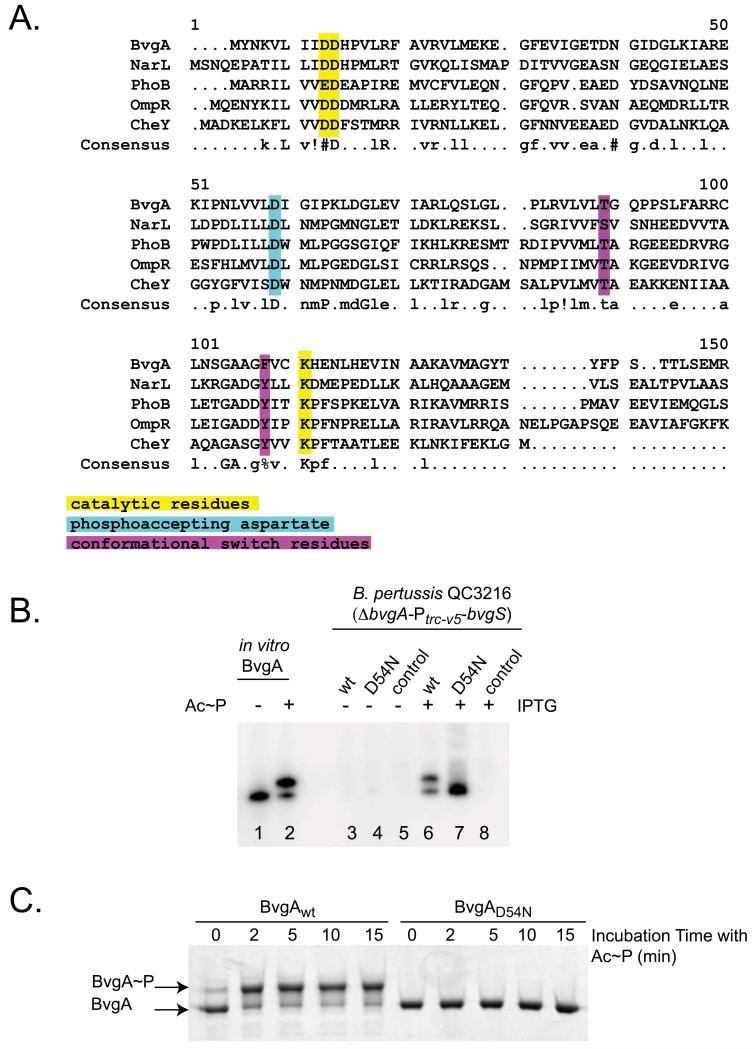
Key amino acids involved in BvgA function can be described as belonging to three groups (Dyer & Dahlquist, 2006, Ruiz et al., 2008): a phosphoaccepting aspartate, catalytic residues, and conformational switch residues. These three groups of amino acids have been identified for CheY (Dyer & Dahlquist, 2006). Although BvgA contains 11 aspartic acid residues, D54 has been assumed to be the phospho-accepting aspartate based on protein sequence alignments of BvgA with other well-characterized response regulators, such as CheY, PhoB, and OmpR, as well as NarL, on which the structure of BvgA has been modeled (Fig. 4A, residue shaded in cyan). In addition, this mutation confers a Bvg− colonial phenotype (flat, non-hemolytic) and renders BvgA incapable of activating fha-lux or ptx-lux transcriptional fusions in vivo (data not shown). Consequently, we introduced D54N substitution into the cloned bvgA gene driven by an IPTG inducible promoter on the plasmid pSS4983. This expression plasmid, together with those containing wild-type bvgA, or no insert, were transferred to B. pertussis strain QC3216, in which bvgA together with the bvgAS promoter were deleted and replaced with the constitutive promoter Ptrc-v5 (depicted in Fig. 3A) to drive bvgS expression in a constant, non-Bvg-dependent fashion.
Figure 4. Mutation of the aspartic residue at position 54 eliminates BvgA phosphorylation in B. pertussis.
A. Alignment of different response regulator protein sequences using the Multalin web program. (http://multalin.toulouse.inra.fr/multalin/). The parameters used correspond to the default parameters of the program. Consensus symbols are:! for L, M; % for F, Y; and # for N, D, Q, E, B, Z. Catalytic residues are highlighted in yellow, the phosphoaccepting aspartate in teal, and conformational switch residues in purple, with reference to Dyer & Dahlquist (2006) and Ruiz et al. (2008).
B. BvgAD54N is not phosphorylated in B. pertussis. Strains of B. pertussis strain QC3216 (ΔbvgA-Ptrc-v5-bvgS) harboring plasmids directing the synthesis of wild-type BvgA (pSS4983, lanes 3 and 6), BvgAD54N (pSS5027, lanes 4 and 7), or the empty pQC1883 vector control (lanes 5 and 8) were analyzed for BvgA synthesis and phosphorylation without (lanes 3-5) and with (lanes 6-8) IPTG-induction. Control lanes contained 1 pmol of purified BvgA (lane 1) or BvgA~P, (lane 2)
C. Kinetics of in vitro phosphorylation of wild-type BvgA and BvgAD54N. Purified wild-type BvgA and BvgAD54N (25 pmol each) were phosphorylated by treatment with Ac~P for the indicated amount of time prior to sample preparation and Phos-Tag™ gel electrophoresis. Proteins were visualized by Coomassie staining.
Cells were grown in permissive conditions either in the absence of IPTG, which yielded only a very low level of the plasmid-encoded protein (Fig. 4B, lanes 3-5) or in the presence of IPTG, which yielded high levels of protein (lanes 6-8). The wild-type BvgA that was thereby produced in trans was phosphorylated (lane 6), but as expected, no phosphorylation was observed of the D54N (lane 7) mutant protein. In addition, no BvgA protein was detected when the empty vector-containing strain was induced (lane 8). A parallel experiment was performed in vitro, using Ac~P to phosphorylate BvgA or BvgAD54N. As shown in Fig. 4C, phosphorylation of wild-type BvgA was very rapid, with 75% of BvgA phosphorylated in less than 2 minutes. However, no phosphorylation of BvgAD54N was detected, even at the longest time point examined (15 minutes). These results demonstrate directly that BvgA phosphorylation requires D54. Given these results and the sequence homologies among response regulators (Fig. 4A), it is highly likely D54 is the target of phosphorylation.
Effects of mutation at BvgA T194 on phosphorylation by Ac~P
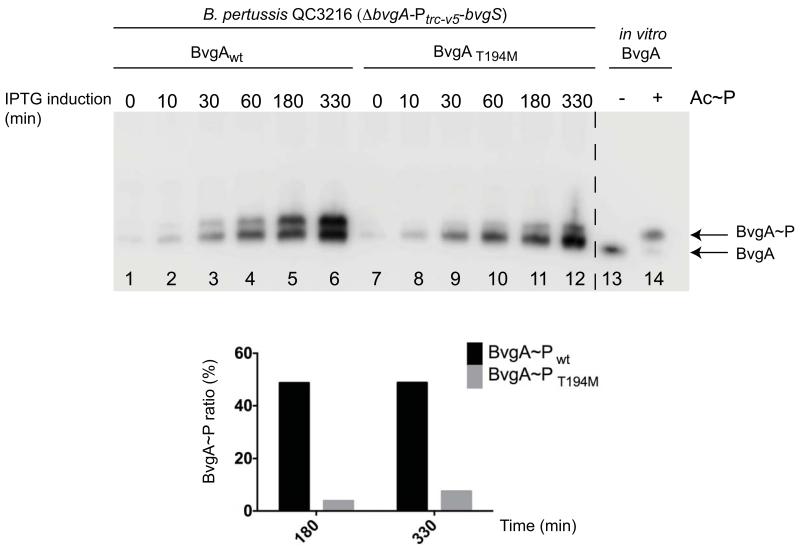
Two different substitutions of the threonine residue at position 194 have been examined in vivo in at least two different experimental contexts, with different results. Jones et al. (2005) observed an effect of the T194M mutation on late gene (cya) expression, while Boucher et al. (2003) found no impact of the T194C mutation (in the context of C93A and C103A) on fha or ptx expression. As shown in Fig. 5, in the same genetic context used above for BvgAD54N, following IPTG-induction, BvgAT194M was phosphorylated less efficiently than wild-type BvgA. This finding was consistent with our observation that, on BG agar containing IPTG, the BvgAT194M-containing strain demonstrated markedly less hemolysis (data not shown).
Figure 5. Kinetics of in vivo phosphorylation of wild-type and mutant BvgA.
Liquid PLB cultures of B. pertussis strain QC3216 (ΔbvgA-Ptrc-v5-bvgS) harboring plasmids directing the synthesis of wild-type BvgA (pSS4983, lanes 1-6), or BvgAT194M (pBvgAT194M, lanes 7-12), were induced with 1 mM IPTG at t=0, and sampled at various times post induction. Cells were harvested by centrifugation, samples were prepared, and Phos-tag™ gel electrophoresis followed by Western blot analysis was performed as described above. Control lanes contained 1 pmol of purified BvgA (lane 13) or BvgA~P, (lane 14). The graph represents the ratio of BvgA~P relative to the total amount of BvgA protein for lanes 5, 6, 11 and 12.
When expressed in E. coli, BvgA is phosphorylated by a BvgS-independent process
A small amount of BvgA~P was detected in our purified BvgA, which was isolated after high level expression from a plasmid in E. coli BL21(DE3) pLysE cells (Fig. 1A, lane 3). Furthermore, during in vitro transcription assays, a trace of BvgA~P-dependent transcription has been observed even when the purified BvgA has not been phosphorylated by Ac~P, when high concentrations of BvgA have been used (data not shown). These results suggested that BvgA might be phosphorylated by an E. coli kinase or by some other BvgS-independent process.
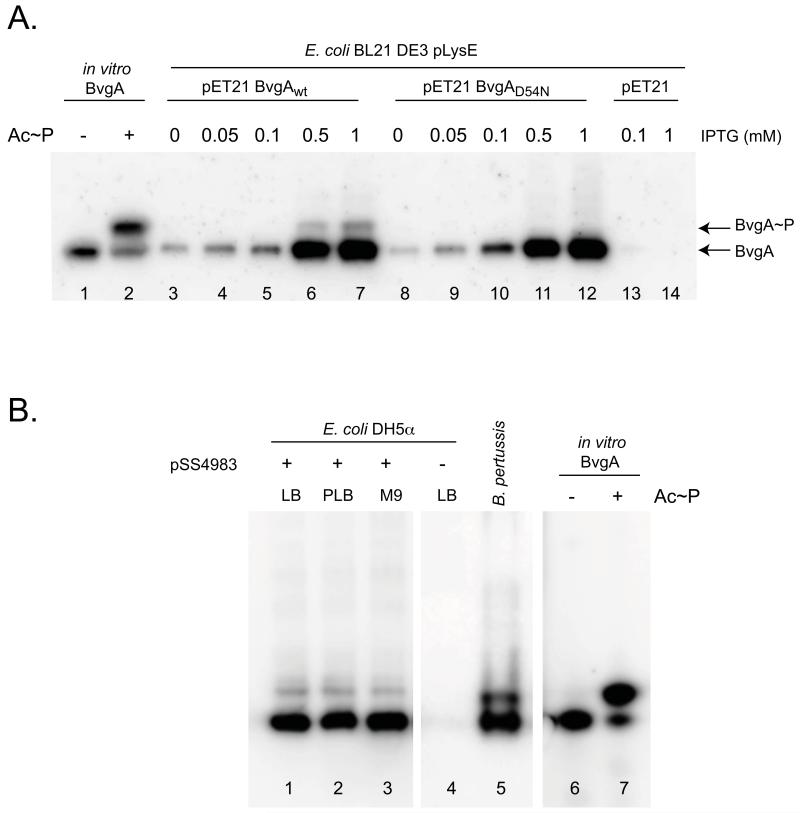
To test this possibility, we compared the levels of BvgA~P present in E. coli containing IPTG-inducible pET21a(+) expression plasmids for wild-type BvgA or BvgAD54N (Fig. 6A). When using low concentrations of IPTG, the level of either wild-type or mutant BvgA protein was low, and no BvgA~P was detected in either case (lanes 3-5 and 8-10). However, at higher levels of IPTG (0.5 or 1 mM), the level of BvgA increased sharply (lanes 6, 7, 11 and 12) and a species that co-migrated with BvgA~P was detected when using the wild-type BvgA plasmid (lanes 6 and 7). This species was not present in the D54N extracts (compare lane 6 to 11 and 7 to 12). Neither BvgA nor BvgA~P was seen when E. coli containing the empty vector was analyzed (lanes 13 and 14). We conclude that BvgA can be phosphorylated in E. coli and that this phosphorylation occurs at the D54 residue. Under these conditions, BvgA~P constitutes about 5% of the total amount of BvgA.
Figure 6. BvgA is phosphorylated in E. coli in the absence of BvgS.
A. Phosphorylation of BvgA in E. coli is dependent on D54. Cultures of E. coli BL21(DE3)/pLysE cells containing pET21a(+) alone (lanes 13-14), or pET21a(+) expressing wild-type bvgA (lanes 3-7) or bvgAD54N (lanes 8-12) were incubated for one hour with increasing concentrations of IPTG and analyzed by Phos-tag™ gel electrophoresis and Western blot as described above. Control lanes contained 1 pmol of purified BvgA (lane 1) or BvgA~P (lane 2).
B. Effect of growth medium on BvgS-independent phosphorylation of BvgA in E. coli. Cell lysates of E. coli DH5α with (lanes 1-3) or without (lane 4) pSS4983 (expressing wild-type BvgA under IPTG induction) were assessed for phosphorylation of BvgA in vivo after growth for three hours in LB (lanes 1 and 4), PLB (lane 2), or M9-glucose (lane 3) media, each supplemented with 2 mM IPTG. A B. pertussis culture (lane 5), purified BvgA (lane 6), and in vitro phosphorylated BvgA (lane 7) are shown for comparison.
Uhl and Miller (1995) previously reported that BvgAS-mediated activation of the ptx promoter in E. coli was dependent on growth medium, being higher when grown in M9-glucose minimal medium than when grown in LB. It was possible that, in those experiments, these different growth conditions might have also affected the background, non-BvgS-mediated phosphorylation we have demonstrated here. To determine if this was the case we repeated our analysis with BvgA-expressing E. coli strains grown in LB, PLB, or M9-glucose broth. As shown in Fig. 6B, lanes 1-3, no effect of growth medium was observed. As shown in lane 4, in the absence of the BvgA expression plasmid, no BvgA or BvgA~P was detected.
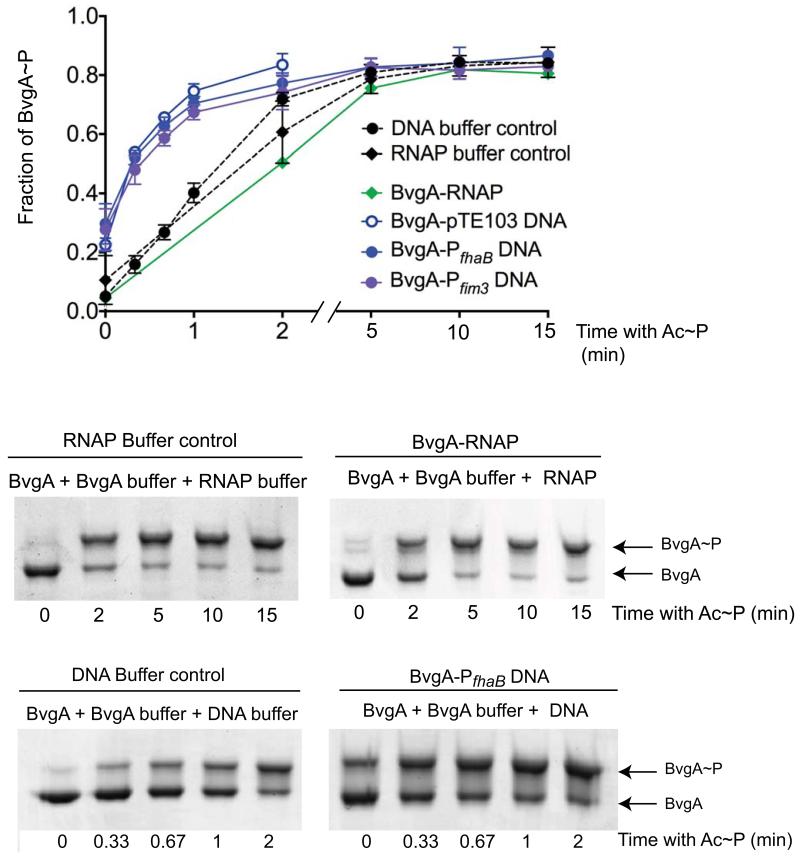
The presence of DNA but not RNAP stimulates in vitro phosphorylation of BvgA by Ac~P
Non-phosphorylated BvgA has been shown to bind the promoter regions of some B. pertussis virulence genes, including fhaB (Boucher et al., 1997, Roy & Falkow, 1991), and fim3 (data not shown). Previous work has also demonstrated that DNA binding stimulates phosphorylation of the RR OmpR (Ames et al., 1999, Head et al., 1998). We wondered if DNA binding could either accelerate or impair phosphorylation of BvgA or whether the presence of BvgA’s transcriptional partner, RNA polymerase (RNAP), might affect phosphorylation. We used E. coli RNAP for this study since previous work has documented that it behaves similarly to B. pertussis RNAP in terms of promoter binding and transcriptional activation by BvgA~P (Baxter et al., 2006, Boucher et al., 1997, Decker et al., 2011, Steffen & Ullmann, 1998), We found that the kinetics of BvgA phosphorylation by Ac~P in the presence of RNAP were the same as those observed with BvgA alone. However, as shown in Fig. 7, in the presence of plasmid DNA consisting of either the BvgA-regulated promoter Pfim3-15C or PfhaB cloned into pTE103 (Chen et al., 2010, Decker et al., 2011) BvgA phosphorylation was stimulated as previously observed for OmpR (Ames et al., 1999). Approximately 50% of the BvgA was phosphorylated 20 seconds after addition of Ac~P in the presence of Pfim3-15C or PfhaB while only 16% was phosphorylated in absence of DNA (Fig. 7). Interestingly, stimulation of BvgA phosphorylation was observed to the same degree even when empty pTE103 vector was added, indicating that the stimulation did not depend on the presence of sequences known to interact with BvgA.
Figure 7. The presence of DNA but not RNAP stimulates in vitro phosphorylation of BvgA by Ac~P.
BvgA phosphorylation in vitro was followed by Phos-tag™ gel electrophoresis and Coomassie staining. Shown below are gels from typical experiments in which BvgA was supplemented with either buffer alone, as control, or buffer plus RNAP or pPfhaB plasmid DNA. Since the two buffers differed, a separate buffer control for each is presented. The graph represents the combined data (mean and standard error) from three independent experiments for each set of conditions and also includes the results of using a Pfim3 clone and the pTE103 vector alone.
Discussion
It has been nearly three decades since the recognition of two-component regulatory systems as a distinct paradigm for environmentally responsive gene regulation (Ninfa & Magasanik, 1986). Since that time a great deal of research has given us a significantly more detailed and comprehensive understanding of the structure/function relationships involved in their action. Much of the more precise physicochemical description of these mechanisms has come from structural determination and from biochemical studies performed in vitro. But there has also been a synergy between these and genetic and functional studies performed in vivo. The latter have helped to put mechanistic knowledge into a relevant context and have provided a “reality check” to test the implications and conclusions of in vitro studies. In this regard, it has been frustrating that arguably the most salient feature of two-component systems, i.e. the activation of the response regulator component by transfer of phosphate from the sensor histidine component, has been difficult to monitor in in vivo systems. This is perhaps particularly true with two-component systems such as BvgAS, where phosphorylation is thought to be not simply an on/off switch, but rather to produce different effects at different promoters at different levels of BvgA~P. In this regard, at least three different classes of B. pertussis Bvg-activated genes (early, late, and intermediate) have been described.
Studies of protein kinases that have tyrosine, or serine as targets, i.e. mostly eukaryotic systems, have benefited greatly from the availability of specific antibody reagents to detect and measure these modifications in vivo. However analogous tools have not been developed for two-component systems. A number of other approaches to this issue have been employed. Head et al (1998) separated purified OmpR from OmpR~P using HPLC and were able to quantify the extent of phosphorylation in vitro. Ladds et al (2003) demonstrated that Spo0A of B. subtilis was phosphorylated in vivo when produced in E. coli. In this case, Spo0A and Spo0A~P were separable by gel filtration chromatography or native gel electrophoresis of trypsin-digested purified protein. SacY of B. subtilis was shown to be reversibly phosphorylated in vivo by 35S-labeling following specific induction of a cloned sacY gene and subsequent separation of SacY and SacY~P by 2-D gel electrophoresis (Idelson & Amster-Choder, 1998). In all of these examples, demonstration of RR phosphorylation did not readily lend itself to routine in vivo examination of multiple conditions or time points.
In contrast, using the recently available Phos-tag™ ligand in gel electrophoresis, we were able to readily ascertain the level of BvgA~P in vivo and in vitro, under multiple conditions and at varying time points. As we began these studies, an initial overriding concern, given the intrinsic lability of the aspartyl-phosphate bond, was the stability of BvgA~P. Failure to maintain the modification during the time and under the conditions required for sample processing could confound a reliable assessment of in vivo BvgA~P levels. Gratifyingly, simple control experiments have indicated that, under the conditions we employed, the amount BvgA~P in our samples did not significantly decline over the time course of a typical experiment. In addition, in some samples (see Fig. 2B, lane 6) a majority of the BvgA was found to be phosphorylated, again indicating that significant degradation did not occur.
As a dynamic application of this technique, we monitored the kinetics of BvgA phosphorylation following a shift of B. pertussis from permissive (i.e. non-modulating) to non-permissive (i.e. modulating) growth conditions, or the reverse. It has previously been observed that after a shift from non-permissive to permissive conditions, so-called “early genes” such as fha were expressed within minutes, while other, “late genes”, such as ptx and cya, were expressed only after several hours (Scarlato et al., 1991). A presumably related observation is the separation of early and late gene expression at different concentrations of modulating agents (Stibitz, 1998). Scarlato et al. interpreted the lag in late gene expression in their time course experiment to be the time required for BvgA~P levels to reach a higher threshold required for ptx and cya activation, compared to that required for fha activation. Indeed these researchers observed an increase in total BvgA concentration with time following induction, consistent with the known autoregulation of the bvgAS locus. In this study we have been able to recreate this scenario, but with the added ability to monitor BvgA phosphorylation. Although none was observed at the zero time point, at 5 minutes after a shift to permissive conditions, a significant amount of BvgA~P was present. This corresponds to a minimal level sufficient to activate the “early” fha promoter. Both the total amount of BvgA, as well as the level of BvgA~P increased dramatically over the following minutes and hours, reaching a maximum at 2-6 hours. These results are in marked agreement with prediction, and offer the first direct demonstration that the timing and degree of BvgA phosphorylation coincides with early and late promoter activity.
In the reverse experiment, involving a shift to non-permissive conditions, a number of unexpected observations were made. One was that the initial steady state level of total BvgA, after long-term growth under permissive conditions, was lower than expected (Fig. 2C, lane 1) and was in fact comparable to the initial amount after long term growth under non-permissive conditions. As expected, BvgA~P declined rapidly and was absent by 15 minutes. Unexpectedly, the total amount of BvgA appeared to increase to a maximum at the same time. This higher level appeared quite stable, remaining elevated at 2 hours, and declining somewhat by 24 hours. Both the mechanism leading to, and the utility of, this increase are unknown at this time. However, we can speculate that it could represent an adaptation involved in inter-host transmission. Transmission of B. pertussis is likely to involve a brief sojourn in aerosolized particles; assuming non-permissive conditions in these droplets, the total level of BvgA could increase during this time. Upon subsequent restoration of permissive conditions after redeposition in a human respiratory tract, rapid phosphorylation would lead to an initially high level of BvgA~P, thus favoring early synthesis of crucial virulence factors.
We have been able to directly test several other prevailing assumptions regarding BvgAS function predicted by analogy with other two-component systems. For example, we have shown that BvgS is necessary and sufficient for phosphorylation of BvgA in vivo. Crosstalk between two-component systems, i.e. the ability of a sensor kinase to phosphorylate a non-cognate response regulator, although found infrequently, is an important issue to address. Our finding that deletion of the bvgS gene abolished BvgA phosphorylation indicates that, under standard laboratory growth conditions, BvgA does not receive inputs from other sensor kinases. This experiment also provides evidence against in vivo phosphorylation of BvgA by small molecule phospho-donors such as Ac~P, as was reported for the E. coli RR CpxR (Lima et al., 2012). Based on protein sequence alignment of BvgA with more fully characterized response regulators, it is predicted that aspartic acid residue number 54 is the site of phosphorylation. In support of this prediction, we have shown that substitution with asparagine abolished phosphorylation of BvgA in vivo and in vitro. Another property of interest is one that has been reported for OmpR of E. coli, but is not necessarily predicted for BvgA. This is the stimulation of phosphorylation by interaction with a cognate DNA binding site. In cases where this has been shown, it has been posited that binding to a high affinity site might shift the equilibrium of the phosphorylation reaction by shifting a conformational equilibrium from a non-activated toward an activated structure. We used the Phos-tag™ gel system to monitor phosphorylation of BvgA in vitro in the absence and presence of plasmid DNA containing either the fim3 or the fhaB promoter, each of which contains a high affinity BvgA binding site. Phosphorylation was significantly stimulated by the presence of the DNA. Even at the zero time point, corresponding to simply addition of Ac~P, mixing and freezing, phosphorylation was approximately 20% complete. Interestingly, the empty vector DNA had the same effect, suggesting that this stimulation was not the result of BvgA interacting with specific binding sites, but rather to interactions with the DNA backbone, i.e. in a non-sequence-specific way. This is not altogether surprising when one recalls that the crystal structure of the C-terminal domain of the homologous NarL RR bound to DNA indicated the presence of extensive backbone contacts (Maris et al., 2002).
Similarly, we looked for stimulation by the presence of RNAP in the phosphorylation reaction. In this case no stimulation of phosphorylation with Ac~P was observed. However, the possibility remains that phosphorylation by BvgS might respond to such conditions.
Mutations at the threonine residue at position 194 have been examined in at least two different experimental contexts. Jones et al. (2005) isolated the BvgA T194M mutation after an in vivo screen for mutants that failed to express Bvg+ mode genes but did express Bvg− and Bvgi genes. When kinetics of expression were examined, it was found that the early gene fha and the intermediate gene bipA were somewhat delayed but eventually reached wild type levels. These observations were suggestive of a defect in phosphorylation. In vitro phosphorylation studies using either Ac~P or truncated BvgS supported this. In contrast, the T194C mutation, in the context of the C93A and C103A mutations did not adversely affect BvgAS-mediated activation of fha or ptx in vivo, and when conjugated with Fe-BABE, this protein successfully formed productive transcription complexes with RNAP at BvgA-regulated promoters in vitro (Boucher et al., 2003, Chen et al., 2010). In this study, we found that, while BvgAT194M phosphorylation in vitro using Ac~P (probably at higher concentrations than used by Jones et al.) was not impaired (data not shown), the kinetics of BvgAT194M in vivo were indeed somewhat impaired. Taken together, these results support a role for the T194 residue in efficient phosphorylation at D54, perhaps by a conformational effect.
In vitro experimentation with BvgA has invariably utilized recombinant protein produced in E. coli. A working assumption has been that this protein, produced in the absence of BvgS, is not phosphorylated. However, we have shown in this study that a low, but constant, fraction of BvgA purified from E. coli is in fact phosphorylated by a BvgS-independent process. It is quite possible that this represents two-component crosstalk, i.e. phosphorylation of BvgA by a non-cognate sensor kinase protein. The observation that this phosphorylation requires the aspartate 54 residue is consistent with this, but not dispositive. We are currently working to identify the E. coli factor responsible. However, whatever the source, the presence of this low level of BvgA~P requires us to examine more closely activities ascribed to BvgA preparations heretofore assumed to be devoid of phosphorylation. An example of this is the ability of BvgA to bind to the high-affinity binding site in the fha promoter, first shown by Roy and co-workers (Roy & Falkow, 1991). Similarly, in in vitro transcription assays, we have consistently observed low basal levels of activation of the fhaB and fim3 promoters, even in the absence of acetyl phosphate (Chen et al., 2010). We consider it quite possible that these activities are due to the low level of BvgA~P in such preparations. Once the source of this E. coli kinase activity has been identified and inactivated, we will be able to produce truly unphosphorylated BvgA to test this hypothesis.
Experimental Procedures
Bacterial strains and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in LB broth, on LB agar, or in M9 medium supplemented with 2% glucose (Miller, 1992). B. pertussis strains were grown on Bordet Gengou (BG) agar described in Chen, et al., (2010). For the BvgAS modulation and induction kinetics assays, B. pertussis strains were grown in liquid Pertussis LB (PLB) (Vanderpool & Armstrong, 2001), in which the LB broth was supplied with 0.12% Dimethyl β cyclodextrin (Cyclodextrin Technologies Development Inc.) and 0.2% bovine serum albumin (BSA, Sigma). Unless indicated elsewhere, the antibiotics in the culture media were used in following concentrations in LB for E. coli strains: ampicillin, 100 μg/mL; gentamicin, 5 μg/mL; kanamycin, 50 μg/mL. Antibiotic concentrations used in BG agar or PLB for Bordetella strains were: streptomycin, 50μg/mL; gentamicin, 5 μg/mL; kanamycin, 20 μg/mL.
Table 1. Strains and plasmids used in this work.
| Strain or plasmid | Relevant features | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | High-efficiency transformation | Bethesda Research Laboratories |
| BΛ21(ΔE3)[πΛψσE] | Expression strain for pET vectors, CamR | Studier et al., 1990 |
| BΛ21(ΔE3)[πΛψσS] | Expression strain for pET vectors, CamR | Studier et al., 1990 |
| SM10 | Mobilization of RK2 oriT plasmids | Simon et al., 1983 |
| B. pertussis | ||
| Tohama I | Patient isolate | Kasuga, et al. (1954) |
| BP536 | Tohama I, StrR, NalR | Stibitz and Yang (1991) |
| QC2935 | BP536, ΔbvgS | This study |
| BP1526 | BP536, ΔbvgA | Chen et al. (2010) |
| QC1416 | BP536, pSS3967::Pfim3-i5c | Chen et al. (2010) |
| QC3216 | BP536, ΔbvgA-Ptrc-v5-bvgS | This study |
| Plasmids | ||
| pSS1827 | Helper plasmid,for triparental mating, RP4 tra genes, AmpR | Stibitz and Carbonetti (1994) |
| pQC1883 | B. pertussis expression vector, KanR | This study |
| pSS4983 | pQC1883::bvgA-wild-type, KanR | This study |
| pBvgAD54N | pQC1883::bvgAD54N, KanR | This study |
| pBvgAT194M | pQC1883::bvgAT194M, KanR | This study |
| pSS4661 | Vector for allelic exchange in Bordetella, KanR | This study |
| pQC1805 | pSS4661::ΔbvgS, kanR | This study |
| pSS4871 | pSS4661::ΔbvgA-Pc-bvgS, KanR | This study |
| pET21a(+) | Expression vector for BvgA in E. coli | EMD Millipore/Novagen |
| pET21a(+)-BvgAwt | pET21a(+)::bvgA | This study |
| pET21a(+)-BvgAD54N | pET21a(+)::bvgAD54N | This study |
| pTE103 | In vitro transcription vector | Elliot and Geiduscheck (1984) |
| pPfim3-15C | pTE103::Pfim3-15C | Chen et al. (2010) |
| pPfhaB | pTE103::PfhaB | Boucher et al. (1997) |
| pSS3967 | luxCDABE promoter assay vector | Chen et al. (2010) |
| pQC1552 | pSS3967::Pptx | This study |
| pQC1557 | pSS3967::PfhaB | This study |
Plasmid and strain constructions
The allelic exchange plasmid pSS4661 was used for the introduction of defined deletions into the B. pertussis BP536 chromosome. This vector can be transferred to B. pertussis strains by conjugation but is incapable of replication. Thus, integration by recombination can be forced after transfer by selection for kanamycin resistance. A second recombination event leading to allelic exchange can then be forced by initiating cleavage of an I-SceI site within the vector, in a manner similar to that described by Posfai et al. (1999). The I-SceI restriction enzyme catalyzing this cleavage is encoded within the vector but is not expressed until the ptx promoter driving its expression is activated by removal of MgSO4 from the growth medium. Details of the construction of pSS4661 and related plasmids will be published separately. Plasmids constructed to introduce specific deletions in the B. pertussis chromosome consisted of sequences flanking and defining the desired deletion endpoints, cloned between the NotI and BamHI sites of pSS4661. To create the bvgS deletion strain QC2935, the plasmid pQC1805 was used. The deletion in this construct removed all but the first five and last five codons of the bvgS orf and was flanked on both sides by segments of approximately 800 bp (Fig. 3A). In the construct pSS4871, similarly sized segments flanked a similarly designed inframe deletion of the bvgA gene. In addition, a segment that extended from 61 bp upstream of the bvgA start codon to the nearby EcoRI site and which included the native bvgAS promoter was replaced with the sequence GAATTCagctgTTGACAattaatcatccggctcgTATAAGgtgtggaattgtgaGTCGAC, which contains a variant of the trc promoter (Ptrc-v5). This plasmid was used to perform allelic exchange on BP536 resulting in strain QC3216 (Fig. 3A).
The Bordetella expression plasmid pQC1883 was derived by the addition of a PCR fragment containing the lac promoter and the lacIq gene from the relA expression plasmid pALS10 (Svitil et al., 1993) to pTM203, itself derived from the broad host-range vector pBBR1 (Antoine & Locht, 1992) by the addition of a kanamycin resistance gene and transcription terminators flanking a multiple cloning site. Plasmid pSS4983 consists of pQC1883 plus a PCR-generated fragment comprising the wild-type bvgA orf together with an AAGGAG ribosome binding site, downstream of, and driven by, the IPTG-inducible lac promoter of pQC1883. The D54N and T194M substitutions were introduced into pSS4983 to create pSS5027 and pBvgAT194M, respectively, by a variation on the method of Stemmer and Morris (1992) or through the use of the QuickChange kit (Agilent). These pQC1833-based expression plasmids were introduced into B. pertussis strain QC2316 by conjugation using E. coli strain SM10 as a host donor strain. For the expression of wild-type BvgA and mutant derivatives in E. coli, preparatory to purification, derivatives based on the pET21a+ vector (Novagen) were constructed.
Plasmid pQC1552 contains a PCR-generated ptx promoter fragment cloned between the upstream EcoRI and SalI sites upstream of luxCDABE in the promoter assay vector pSS3967 (Chen et al., 2010). Similarly, plasmid pQC1557 contains a PCR-generated fhaB promoter fragment. The resulting pSS3967-based plasmids were integrated as single copies at a specific ectopic chromosomal location in B. pertussis BP536 following conjugation using E. coli SM10 strain as a host donor strain as described previously (Chen et al., 2010).
Proteins
E. coli RNAP core was purchased from Epicentre Technologies. Purification of σ70 was done using a modification of the method of Gribskov and Burgess (Gerber & Hinton, 1996, Hernandez et al., 1996) using E. coli BL21(DE3)/pLysS (Studier et al., 1990) containing the plasmid pLHN12 (Hernandez et al., 1996, Nguyen, 1996), which expresses rpoD.
Wild type, D54N, and T194M, BvgA were purified as previously described (Chen et al., 2010) except that the white precipitate formed after dialysis of the Hi-Prep 16/10 Q Sepharose flow-through fraction and centrifugation was resuspended in the inclusion body storage buffer [50 mM HEPES-OH (pH 7.5), 0.1 M NaCl, 1 mM EDTA, 1 mM DTT, 20% glycerol] and diluted to a final concentration of 0.15 mg/mL in 50 mM Na2HPO4 (pH 6.8), 6 M guanidinium-HCl, 2 mM MgCl2, 1 mM DTT. The protein was then dialyzed as previously described (Chen et al., 2010) and stored at −80°C. BvgA storage buffer contained 20 mM HEPES pH 7.4, 10 mM MgCl2, 50 mM KCl, 1 mM DTT, and 50 % glycerol.
Phosphorylation of BvgA
BvgA was phosphorylated by incubation in the presence of 20 mM acetyl phosphate, (lithium potassium acetyl phosphate from Sigma-Aldrich dissolved in 20 mM Tris-Cl, pH 8) for the indicated times at room temperature. Non-phosphorylated BvgA was incubated for the same period of time in the presence of 20 mM Tris-Cl (pH 8) only. Samples were collected on dry ice.
To investigate the effect of RNAP, Pfim3 or PfhaB DNA on BvgA phosphorylation, 25 pmol BvgA was mixed either with reconstituted RNAP [previously reconstituted using 3.8 pmol RNAP core and 9.4 pmol purified σ70 at 37°C for 10 min], RNAP buffer (33 mM Tris-Cl (pH 8), 0.1 mM EDTA, 355 mM NaCl, 0.6 mM DTT and 50% glycerol), and 1 pmol pPfim3-15C supercoiled DNA, 1 pmol pPfhaB supercoiled DNA, 1 pmol pTE103 (50 nM DNA) or the same volume of H2O. After 10 minutes incubation at 37°C, 20 mM Ac~P was added to start the reaction and samples were put back at 37°C for the indicated amount of time. To determine kinetics of phosphorylation of wild-type BvgA mutants, protein (25 pmol) was first incubated for 10 min at 37°C. Ac~P (20 mM) was then added and the solution incubated for the indicated amount of time at 37°C.
Sample preparation for in vivo detection of BvgA phosphorylation
B. pertussis strain BP536 and its derivatives were grown at 37° C for 2 days on BG agar supplemented with streptomycin (50 μg/mL) with or without MgSO4 (50 mM). To prepare cell lysates for the Phos-tag™ gel assay, cells were swabbed from the plate with a polyester-tipped applicator (Puritan Medical Products Company LLC.) and resuspended in 1.5 mL of Phosphate Buffered Saline (PBS, GIBCO) to an OD600 of 0.15. A 1 mL aliquot was centrifuged for 1 min at RT, the supernatant was removed, and the pellet was frozen in dry ice. Unless otherwise indicated, the frozen cell pellet was then lysed on ice by the addition of 33 μL of ice-cold 1M formic acid (0.55 M final concentration), immediately followed by the addition of 2 μL of 5 N NaOH (0.17 N final concentration) to neutralize the solution, 10 μL of H2O and 15 μL of 5X Loading Solution (1% SDS, 65 mM Tris-Cl (pH 8), 25% glycerol, 5% Bromophenol Blue). Resulting cell lysates (4 μl) were immediately loaded onto a Phos-tag™ gel for electrophoresis as described below. E. coli strains were grown overnight in LB with appropriate antibiotics before being diluted in fresh medium +/− IPTG, at an OD600 of 0.1. Cells were grown at 37°C for indicated amount of time, pelleted by centrifugation at 16,000 × g for 1 min, frozen on dry ice, and lysed as described above.
Phos-tag™ gel electrophoresis
BvgA and BvgA~P were separated on polyacrylamide gels containing acrylamide-Phos-tag™ ligand (Wako Pure Chemical) as previously described (Barbieri & Stock, 2008, Kinoshita & Kinoshita-Kikuta, 2011). Gels were composed of a 10% resolving solution [10% (w/v) 29:1 acrylamide:N,N’-methylene-bis-acrylamide (deionized 3 min with Ag 501-X8 Resin (Biorad) before filtration); 350 mM Tris-Cl (pH 6.8); 0.1 % SDS; 75 μM Phos-tag™ acrylamide; and 150 μM Zn(NO3)2] and a 4 % stacking solution [4% (w/v) 29:1 acrylamide:N, N’-methylene-bis-acrylamide (deionized 3 min with Ag 501-X8 Resin before filtration); 350 mM Tris-Cl (pH 6.8 @ 4°C); and 0.1 % SDS]. Protein samples were mixed with 5X loading solution (1% SDS, 65 mM Tris-Cl (pH 8), 25% glycerol, 5% Bromophenol Blue) to a final concentration of 1 × before loading on the gel. Electrophoresis was performed at 150 V at 4°C for 1 h 20 min in MOPS running buffer (pH 8) (0.1 M Tris-Cl, 0.1 M MOPS, 0.1% SDS and 5 mM sodium bisulfite). After being washed in water, gels were stained with Coomassie blue or used for Western blot analyses as described below. Stained gels were scanned using a Powerlook 2100XL densitometer and analyzed using Quantity One software from Bio-Rad, Inc.
Western Blot analyses
For Western Blot analyses, 4 μL of cell lysate (prepared as described above) and 1 pmol of purified BvgA (unphosphorylated or phosphorylated in vitro) were loaded onto the gel. After electrophoresis, the Phos-tag™ gel was washed 10 min at RT with Transfer Buffer (25 mM Tris base, 0.192 M glycine, 20% methanol) supplied with 1 mM EDTA to remove Zn+ from the gel, followed by a 20 min wash with the Transfer Buffer to remove the chelated metal. Transfer of the gel to the PVDF filter (Invitrogen) was carried out using MINI-PROTEIN II (Bio-Rad) at constant voltage 100V for 1 h at RT, using a cooling system at 4° C. The PVDF filter was blocked with 1% BSA in PBS, washed with PBS and then incubated with BvgA monoclonal antibody diluted 1:5000 in PBS containing 1% BSA at RT for 1 h, followed by 3 washes (15 min. each) with PBS + 0.05% Tween. The filter was then incubated with goat anti-mouse IgG-HRP conjugated (1:20,000, Santa Cruz) in PBS containing 1% milk at RT for 1 h. After 3 washes (15 min. each) with PBS + 0.05% Tween, the filter was developed using the Amersham ECL Plus Western Blotting Detection System (GE Healthcare), and the signal was detected using a FUJI LAS-3000 imaging system (Fuji).
Induction and modulation kinetics of BvgA~P in vivo
Strain QC1416, a derivative of BP536 (Chen et al., 2010) was used in these studies. Although not relevant to this work, it also harbors a chromosomal insertion of the vector pSS3967 containing an ectopic transcription fusion of Pfim3-15C.
For repression kinetics, strain QC1416 was grown on BG plates supplemented with streptomycin and gentamicin at 37°C in the absence of MgSO4 for 3 days to produce Bvg+ mode cells. After restreaking on the same medium and growth at 37°C overnight, cells were swabbed from plates and resuspended in PLB plus streptomycin and gentamicin. Cultures were incubated at 37°C with shaking until the OD600 had doubled (~3 h). An aliquot of cells, corresponding to time t=0 was removed, harvested and frozen on dry ice as described above. MgSO4 was added to the culture to a final concentration of 50 mM and growth was continued at 37°C.
For induction kinetics, strain QC1416 was grown on BG plates supplemented with streptomycin, gentamicin, and MgSO4 (50mM) at 37°C for 3 days, resulting in Bvg− mode cells. After restreaking on the same medium and growth at 37°C overnight, cells were swabbed from plates and resuspended in PLB plus streptomycin, gentamicin and 50 mM MgSO4. Cultures were incubated at 37°C with shaking until the OD600 had doubled (~3 h). An aliquot of cells, corresponding to time t=0 was removed, harvested and frozen on dry ice as described above. The remainder of the culture was harvested by centrifugation at 1200 × g for 10 min at RT, and resuspended in the same volume of pre-warmed PLB supplemented with streptomycin and gentamicin but no MgSO4. Cell growth was continued at 37°C.
In both cases aliquots were then taken at indicated time points, cell pellets were obtained by centrifugation, and held frozen on dry ice. Samples were normalized based on OD600 readings and processed together for electrophoresis as described above.
Induction kinetics of fhaB and ptx in vivo
B. pertussis BP536 harboring plasmids pQC1552, pQC1557, or pSS3967 vector integrated in the chromosome were cultured and induced as described above. A 200μl aliquot was removed at the zero time point before induction and at indicated times after. Aliquots were transferred to individual wells of a 96-well white microtiter plate with clear bottom. The OD600 reading and measurement of luminescence activity of each aliquot were carried out using Gen5 software on a Synergy 2 plate reader (BioTek Instruments, Inc.).
Acknowledgements
The authors thank A. Battesti, L. Knipling, T. James, S. Jha, L. Abell, and J. Kassis for helpful discussions, Ilana Cohen for technical assistance, and J. P. Castaing for the construction of pET21a+ D54N. This research was supported in part by the Intramural Research Program of the NIH, NIDDK.
Abbreviations
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- RR
response regulator
- HK
histidine kinase
- HPLC
high pressure liquid chromatography
- BSA
bovine serum albumin
- BG
Bordet Gengou agar
- LB
Luria Broth
- orf
open reading frame
- Ac~P
acetyl phosphate
References
- Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine R, Locht C. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol Microbiol. 1992;6:1785–1799. doi: 10.1111/j.1365-2958.1992.tb01351.x. [DOI] [PubMed] [Google Scholar]
- Barbieri CM, Stock AM. Universally applicable methods for monitoring response regulator aspartate phosphorylation both in vitro and in vivo using Phos-tag-based reagents. Anal Biochem. 2008;376:73–82. doi: 10.1016/j.ab.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter K, Lee J, Minakhin L, Severinov K, Hinton DM. Mutational analysis of sigma70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA. J Mol Biol. 2006;363:931–944. doi: 10.1016/j.jmb.2006.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher PE, Maris AE, Yang MS, Stibitz S. The response regulator BvgA and RNA polymerase alpha subunit C-terminal domain bind simultaneously to different faces of the same segment of promoter DNA. Mol Cell. 2003;11:163–173. doi: 10.1016/s1097-2765(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Boucher PE, Murakami K, Ishihama A, Stibitz S. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J Bacteriol. 1997;179:1755–1763. doi: 10.1128/jb.179.5.1755-1763.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Decker KB, Boucher PE, Hinton D, Stibitz S. Novel architectural features of Bordetella pertussis fimbrial subunit promoters and their activation by the global virulence regulator BvgA. Mol Microbiol. 2010;77:1326–1340. doi: 10.1111/j.1365-2958.2010.07293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PA, Jones AM. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 2003;11:367–373. doi: 10.1016/s0966-842x(03)00156-2. [DOI] [PubMed] [Google Scholar]
- Cotter PA, Miller JF. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol. 1997;24:671–685. doi: 10.1046/j.1365-2958.1997.3821741.x. [DOI] [PubMed] [Google Scholar]
- Cummings CA, Bootsma HJ, Relman DA, Miller JF. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J Bacteriol. 2006;188:1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker KB, Chen Q, Hsieh ML, Boucher P, Stibitz S, Hinton DM. Different requirements for sigma Region 4 in BvgA activation of the Bordetella pertussis promoters P(fim3) and P(fhaB) J Mol Biol. 2011;409:692–709. doi: 10.1016/j.jmb.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker KB, James TD, Stibitz S, Hinton DM. The Bordetella pertussis model of exquisite gene control by the global transcription factor BvgA. Microbiology. 2012;158:1665–1676. doi: 10.1099/mic.0.058941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T, Geiduschek EP. Defining a bacteriophage T4 late promoter: absence of a “-35” region. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- Gerber JS, Hinton DM. An N-terminal mutation in the bacteriophage T4 motA gene yields a protein that binds DNA but is defective for activation of transcription. J Bacteriol. 1996;178:6133–6139. doi: 10.1128/jb.178.21.6133-6139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyard S, Bellalou J, Mireau H, Ullmann A. Mutations in the Bordetella pertussis bvgS gene that confer altered expression of the fhaB gene in Escherichia coli. J Bacteriol. 1994;176:5163–5166. doi: 10.1128/jb.176.16.5163-5166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head CG, Tardy A, Kenney LJ. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J Mol Biol. 1998;281:857–870. doi: 10.1006/jmbi.1998.1985. [DOI] [PubMed] [Google Scholar]
- Hernandez VJ, Hsu LM, Cashel M. Conserved region 3 of Escherichia coli final sigma70 is implicated in the process of abortive transcription. J Biol Chem. 1996;271:18775–18779. doi: 10.1074/jbc.271.31.18775. [DOI] [PubMed] [Google Scholar]
- Hot D, Antoine R, Renauld-Mongenie G, Caro V, Hennuy B, Levillain E, Huot L, Wittmann G, Poncet D, Jacob-Dubuisson F, Guyard C, Rimlinger F, Aujame L, Godfroid E, Guiso N, Quentin-Millet MJ, Lemoine Y, Locht C. Differential modulation of Bordetella pertussis virulence genes as evidenced by DNA microarray analysis. Mol Genet Genomics. 2003;269:475–486. doi: 10.1007/s00438-003-0851-1. [DOI] [PubMed] [Google Scholar]
- Idelson M, Amster-Choder O. SacY, a transcriptional antiterminator from Bacillus subtilis, is regulated by phosphorylation in vivo. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Boucher PE, Williams CL, Stibitz S, Cotter PA. Role of BvgA phosphorylation and DNA binding affinity in control of Bvg-mediated phenotypic phase transition in Bordetella pertussis. Mol Microbiol. 2005;58:700–713. doi: 10.1111/j.1365-2958.2005.04875.x. [DOI] [PubMed] [Google Scholar]
- Kasuga T, Nakase Y, Ukishima K, Takatsu K. Studies on Haemophilus pertussis. V. Relation between the phase of bacilli and the progress of the whooping-cough. Kitasato Arch Exp Med. 1954;27:57–62. [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 2011;11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- Ladds JC, Muchova K, Blaskovic D, Lewis RJ, Brannigan JA, Wilkinson AJ, Barak I. The response regulator Spo0A from Bacillus subtilis is efficiently phosphorylated in Escherichia coli. FEMS Microbiol Lett. 2003;223:153–157. doi: 10.1016/S0378-1097(03)00321-5. [DOI] [PubMed] [Google Scholar]
- Lima BP, Thanh Huyen TT, Basell K, Becher D, Antelmann H, Wolfe AJ. Inhibition of Acetyl Phosphate-dependent Transcription by an Acetylatable Lysine on RNA Polymerase. J Biol Chem. 2012;287 doi: 10.1074/jbc.M112.365502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- Manetti R, Arico B, Rappuoli R, Scarlato V. Mutations in the linker region of BvgS abolish response to environmental signals for the regulation of the virulence factors in Bordetella pertussis. Gene. 1994;150:123–127. doi: 10.1016/0378-1119(94)90870-2. [DOI] [PubMed] [Google Scholar]
- Maris AE, Sawaya MR, Kaczor-Grzeskowiak M, Jarvis MR, Bearson SM, Kopka ML, Schroder I, Gunsalas RP, Dickerson RE. Dimerization allows DNA target site recognition by the NarL response regulator. Nat Struct Biol. 2002;9:771–778. doi: 10.1038/nsb845. [DOI] [PubMed] [Google Scholar]
- Miller JF, Johnson SA, Black WJ, Beattie DT, Mekalanos JJ, Falkow S. Constitutive sensory transduction mutations in the Bordetella pertussis bvgS gene. J Bacteriol. 1992;174:970–979. doi: 10.1128/jb.174.3.970-979.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A short course in molecular genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 1992. [Google Scholar]
- Nguyen L. Ph.D. thesis. University of Wisconsin; Madison: 1996. [Google Scholar]
- Ninfa AJ, Magasanik B. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy CR, Falkow S. Identification of Bordetella pertussis regulatory sequences required for transcriptional activation of the fhaB gene and autoregulation of the bvgAS operon. J Bacteriol. 1991;173:2385–2392. doi: 10.1128/jb.173.7.2385-2392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz D, Salinas P, Lopez-Redondo ML, Cayuela ML, Marina A, Contreras A. Phosphorylation-independent activation of the atypical response regulator NblR. Microbiology. 2008;154:3002–3015. doi: 10.1099/mic.0.2008/020677-0. [DOI] [PubMed] [Google Scholar]
- Scarlato V, Arico B, Prugnola A, Rappuoli R. Sequential activation and environmental regulation of virulence genes in Bordetella pertussis. EMBO J. 1991;10:3971–3975. doi: 10.1002/j.1460-2075.1991.tb04967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlato V, Prugnola A, Arico B, Rappuoli R. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc Natl Acad Sci U S A. 1990;87:10067. doi: 10.1073/pnas.87.24.10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- Steffen P, Ullmann A. Hybrid Bordetella pertussis-Escherichia coli RNA polymerases: selectivity of promoter activation. J Bacteriol. 1998;180:1567–1569. doi: 10.1128/jb.180.6.1567-1569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP, Morris SK. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site-directed mutagenesis. Biotechniques. 1992;13:214–220. [PubMed] [Google Scholar]
- Stibitz S. Mutations affecting the alpha subunit of Bordetella pertussis RNA polymerase suppress growth inhibition conferred by short C-terminal deletions of the response regulator BvgA. J Bacteriol. 1998;180:2484–2492. doi: 10.1128/jb.180.9.2484-2492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibitz S, Carbonetti NH. Hfr mapping of mutations in Bordetella pertussis that define a genetic locus involved in virulence gene regulation. J Bacteriol. 1994;176:7260–7266. doi: 10.1128/jb.176.23.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibitz S, Yang MS. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J Bacteriol. 1991;173:4288–4296. doi: 10.1128/jb.173.14.4288-4296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- Uhl MA, Miller JF. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc Natl Acad Sci U S A. 1994;91:1163–1167. doi: 10.1073/pnas.91.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl MA, Miller JF. BvgAS is sufficient for activation of the Bordetella pertussis ptx locus in Escherichia coli. J Bacteriol. 1995;177:6477–6485. doi: 10.1128/jb.177.22.6477-6485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK, Armstrong SK. The Bordetella bhu locus is required for heme iron utilization. J Bacteriol. 2001;183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CL, Boucher PE, Stibitz S, Cotter PA. BvgA functions as both an activator and a repressor to control Bvg phase expression of bipA in Bordetella pertussis. Mol Microbiol. 2005;56:175–188. doi: 10.1111/j.1365-2958.2004.04526.x. [DOI] [PubMed] [Google Scholar]