1. Introduction
Mesenchymal cells’ physiologic healing response to injury entails the formation of scar tissue. If the injury persists, this wound healing response leads to fibrogenesis and extracellular matrix accumulation. Although there are organ-specific differences in fibrogenic pathways, certain core elements characterize the fibrogenic response in almost all tissues. Damage to the epithelium or endothelium, instigated by infection, autoimmune processes, toxins, or mechanical disturbance, triggers the release of inflammatory mediators and initiation of fibrosis response.
Despite significant progress made in elucidating the mechanisms of fibrogenesis, new components of the fibrotic machinery continue to be unearthed, and effective antifibrotic therapies remain elusive [1], [2].
Cells respond to changes in the microenvironment through alteration of their anabolic and catabolic pathways. Autophagy is a catabolic pathway essential for cellular homeostasis that involves the self-degradation of intracellular components in lysosomes as part of cytoplasmic turnover. Autophagy has been implicated in the pathophysiology of many human disorders including fibrotic diseases. Here we review its role in the activation of mesenchymal cells in the context of fibrosis.
There are three types of autophagy which differ in the mechanism by which they deliver materials to the lysosomal lumen: microautophagy, chaperone-mediated autophagy and macroautophagy. Macroautophagy is the most prevalent form and unless otherwise specified, the term autophagy used in this paper refers to it.
Microautophagy is a poorly understood process in mammalian cells in which the material to be degraded is engulfed by direct invagination of the lysosomal membrane.
Chaperone-mediated autophagy is a more selective process by which cytosolic proteins marked by the pentapeptide motif KFERQ bind the lysosomal membrane through the lysosomal membrane receptor LAMP-2a (lysosomal-assocated membrane protein 2a). Once translocated to the lumen, proteins are degraded by lysosomal hydrolases.
In macroautophagy, hereafter referred to as ‘autophagy’, the cytoplasmic component is surrounded by an expanding membrane sac, termed the phagophore, which elongates until its two edges fuse, forming a double membrane called autophagosome. The outer membrane of the autophagosome then fuses with the lysosome generating the autolysosome or autophagolysosome and the lysosomal enzymes degrade the inner membrane together with sequestered material [3].
2. Autophagy machinery
Autophagy is a conserved and ubiquitous pathway essential for maintaining cellular energy homeostasis. Under normal conditions autophagic activity proceeds at a very low level, and is tightly regulated following induction in many cellular stress conditions. There have been tremendous advances in our knowledge of the molecular mechanism of autophagy in recent years, in particular the discovery of the autophagy-related genes (Atg). These 14 genes, conserved between yeast and humans, are required for autophagosome formation and regulate the formation of the core autophagic machinery [4]. The autophagy process is divided into several steps: induction, autophagosome formation, cargo recognition and selection, vesicle formation, autophagosome formation, fusion and breakdown.
In unstressed conditions, autophagy levels are low and are efficiently inhibited by the serine/threonine protein kinase mTOR (mammalian target of rapamycin). In mammals, mTOR inactivates the complex formed by the Unc-51-like kinase 1 (ULK1), 2 (ULK2), the focal adhesion kinase family-interacting protein of 200kD (FIP200) and Atg13 (ULKs-Atg13-FIP200 complex). However upon mTOR inhibition, ULK1 and ULK2 are activated and phosphorylate Atg13-FIP200, leading to a conformational change essential to autophagy induction[5]
Induction of autophagy provokes formation of the autophagosome. The origin of the autophagosome membrane is a matter of debate and available data supports three potential sources: endoplasmic reticulum (ER) [6], mitochondrial membrane [7] and plasma membrane [8].
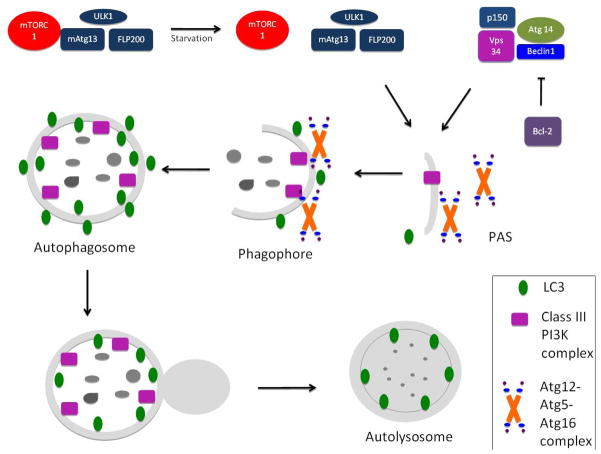
The cargo to be degraded has to be recognized by the autophagy machinery. In mammals this selection process is mediated by the cytosolic adaptor protein P62/sequestosome 1. P62 binds by its ubiquitin-associated domain to the microtubule-associated protein 1 light chain 3 (LC3) and delivers the ubiquitinated cargo for autophagic degradation [9]. The formation of the autophagosome requires the class III phosphatidylinositol 3-kinase (PI3K) macromolecular complex, composed of the PI3K vacuolar protein sorting 34 (Vps34), beclin1, Atg 14 and p150. Beclin 1 can enhance Vps34 activity; however, under homeostatic conditions it is inactivated through its binding to the anti-apoptotic protein Bcl2 (B-cell lymphoma/leukemia-2) [10]. The PI3K complex recruits two interconnected ubiquitin-like (Ubl) conjugation complexes that share a single E1-like activating enzyme, Atg7 [3], These Ubl complexes are essential for autophagosome formation (Atg12-Atg5-Atg16L1) and elongation (Atg8-LC3) respectively. Atg7 activates Atg12 and is then transferred to Atg10 and attached covalently to Atg5. The Atg12-Atg5 complex then conjugates with Atg16L1 and attaches to the phagophore (Figure 1).
Figure 1.
Autophagosome formation
Autophagosome formation requires the ULK1/2-mAtg13-FIP200 complex as well as the class III phosphatidylinositol 3-kinase (PI3K) macromolecular complex, which is composed of the PI3K vacuolar protein sorting 34 (Vps34), beclin1, Atg 14 and p150. This complex recruits two interconnected ubiquitin-like (Ubl) conjugation complexes that are essential for autophagosome formation (Atg12-Atg5-Atg16L1) and elongation (Atg8-LC3) respectively.
Although it is eventually delipidated by Atg4 and recycled, LC3-II is a useful marker of the autophagy membrane because it remains on the inner and outer membrane of the autophagosome until its fusion with the lysosomes. After fusion, the acidic lysosomal hydrolases degrade the engulfed materials. The resultant small molecules are finally transported back to the cytosol for maintenance of cellular homeostasis. The origin of the autophosome membrane remains unknown, although the plasma membrane, endoplasmic reticulum, and mitochondrial membrane have all been implicated.
In the second Ubl conjugation, the LC3 precursor is cleaved by Atg4 and converted to LC3-I. Atg7 and Atg3 promote the conjugation of LC3-I to phosphatidylethanolamine to form LC3-II, a process that is facilitated by the Atg12-Atg5-Atg16L1 complex. Although it is eventually delipidated by Atg4 and recycled, LC3-II is a useful marker of the autophagy membrane as it remains on the inner and outer membrane of the autophagosome until its fusion with the lysosomes[11]. After fusion, the acidic lysosomal hydrolases degrade the sequestered materials. [12] Finally, the resultant small molecules are transported back to the cytosol for maintenance of cellular homeostasis.
3. Autophagy regulation
3.1. Signaling pathways regulating autophagy
3.1.1 mTOR dependent regulation
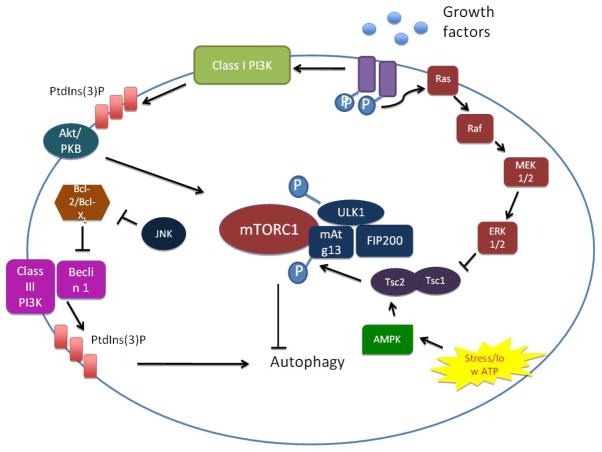
Autophagy is strongly induced under stress conditions such as amino acid starvation and ATP depletion. This nutrient-sensitive induction is mediated in part through the mTOR pathway. The mTOR pathway is composed of the rapamycin-sensitive mTOR complex (mTORC) 1 and 2, although only mTORC1 is a direct regulator of autophagy [13]. Under nutrient-rich conditions, mTORC1 phosphorylates and inhibits the ULKs-Atg13-FIP200 complex and autophagy is kept at very low levels. In situations of metabolic stress, mTORC dissociates from the complex, leading to dephosphorylation-dependent activation of ULKs and subsequent induction of autophagy[5]. A major signaling pathway regulating mTORC1 is the PI3K complex. Different members of the PI3K complex have discrete roles in autophagy modulation. The class I PI3K acts through Akt activation and suppresses autophagy [14]; however the class III PI3K, Vps34, stimulates autophagy by increasing local concentrations of phosphatidylinositol 3-phosphate (PI3P) [15]. Besides PI3K, other kinases can modulate autophagy through mTOR, including Adenosine Monophosphate-Activated Protein Kinase (AMPK) [16], IκB kinase [17] and the members of the mitogen-activated proteinase kinase (MAPK) pathway (ERK [18], p38 [19], JNK [20]) (Figure 2).
Figure 2.
mTORC-dependent regulation of autophagy
Autophagy is induced under stress conditions such as nutrient or ATP depletion. The rapamycin-sensitive serine/threonine protein kinase mTORC1 mediates the nutrient-sensitive regulation of autophagy. In unstressed conditions, or in the presence of ample nutrients, autophagy levels are maintained at a low level through activation of mTORC1, which inactivates the ULK1/2-Atg13-FIP200 complex. Nutrient starvation, on the other hand, instigates dissociation of mTORC1 and partial dephosphorylation of ULK1/2. Activated ULK1/2 phosphorylates the Atg13-FIP200 complex, which instigates conformational changes essential for autophagy. The PI3K pathway mediates upstream regulation of the mTOR pathway. Growth factor binding to receptor tyrosine kinases instigates their autophosphorylation and activation, which leads to the activation of the class I PI3K and the generation of phosphatidyinositol-3-phosphate (PI3P). These changes provoke the recruitment of Akt, which activates mTORC1 and suppresses autophagy. However, the class III PI3K (Vps34) stimulates autophagy independently of mTORC1, through the generation of PI3P. Certain members of the MAPK pathway, including ERK1/2 and JNK, as well as AMPK, also modulate autophagy through mTORC1.
3.1.2 mTOR independent regulation
Although the mTOR pathway is the most studied and best-characterized autophagy regulatory pathway, mTOR-independent regulation has also been described.
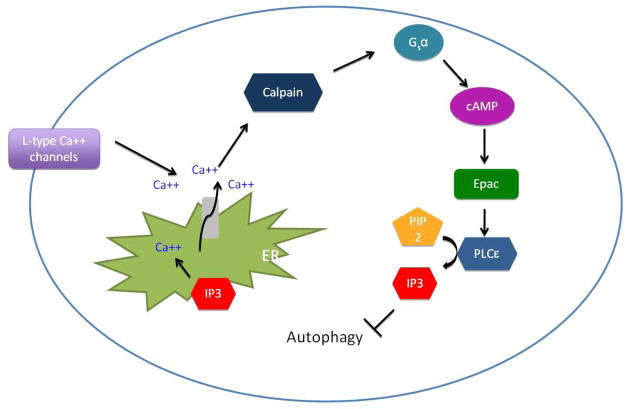
Autophagy is negatively regulated by intracellular inositol and inositol 1,4,5-triphosphate (IP3). IP3 binds to its receptor on the ER and release the stored Ca2+ into the cytoplasm, which provokes several cellular responses and inhibits autophagy [21, 22]. cAMP and cytosolic Ca2 downregulates autophagy trough the cAMP-Epac-PLC-ε-IP3 [22] and Ca2+ -calpain-Gsα[23] pathways, respectively (Figure 3).
Figure 3.
mTORC-independent regulation of autophagy
Inositol 1,4,5-triphosphate (IP3) mediates mTORC-independent regulation of autophagy. IP3 binding to the endoplasmic reticulum (ER) instigates the release of calcium into the cytoplasm. Binding of agonists to L-type calcium channels also increases the cytoplasmic levels of calcium. The increase in cytoplasmic calcium induces the activation of a family of calcium-dependent cysteine proteases called calpains. Activated calpains cleave and activate Gsα, which induces adenylyl cyclase activity to increase the concentration of cAMP. Elevated cAMP activates the Epac molecule, which activates phospholipase C (PLC)-ε and increases IP3, thereby completing this regulatory loop.
3.2 Transcriptional regulation
Although not much is known about the transcriptional regulation of autophagy, some autophagy genes such as lc3 are rapidly up-regulated. The transcription factor FoxO3 induces transcription of several autophagy genes including lc3, atg12, atg4 and beclin 1[10]. In yeast, atg1 and atg13 have been recognized as targets of the transcriptional factor GCN4 [24].
3.3 Post-translational regulation
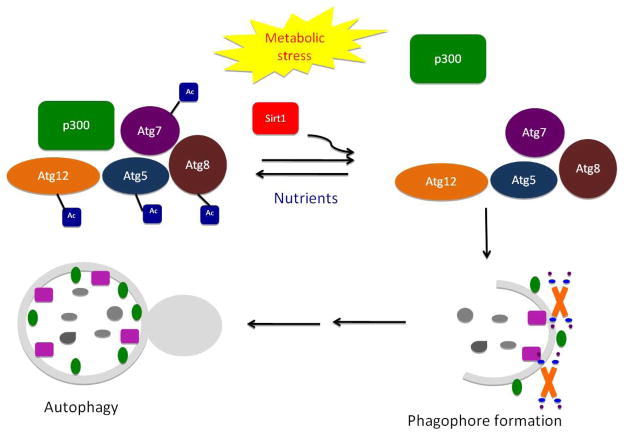
Recent studies have shown that post-translational modifications modulate the autophagic flow[25]. As mentioned before ubiquitylation of the autophagic cargo and ubiquitylation-like conjugation of the Atg12-Atg5-Atg16L1 and Atg8-LC3 complexes are essential autophagy triggers. Postranscriptional lipidation of LC3 also induces autophagosome formation. Phosphorylation also plays an important role in autophagy regulation. Phosphorylation of either Bcl-2 or Beclin 1 reduces Bcl2-Beclin1 interaction and induces autophagy [26]. Phosphorylation of ULK1 and dephosphorylation of Atg13 are also essential for autophagy initiation [25]. Acetylation of Atg proteins by acetyltransferases also modifies autophagy. Atg7 interaction with p300 maintains the acetylation of Atg5, Atg7, Atg8 and Atg12 proteins, thereby suppressing autophagy [27]. Upon metabolic stress conditions, p300 dissociates from Atg7 and the deacetylase Sirt1 removes the acetyl groups from Atg7, Atg5, Atg8 and Atg12 and triggers autophagy [28] (Figure 4). TIP60 is another acetyltransferase that has been recently identified as a positive regulator of autophagy through its interaction with ULK[29].
Figure 4.
Post-translational modification of autophagy
Autophagy is subject to post-translational modification. As depicted in Figure 2, activated mTORC1 suppresses autophagy through phosphorylation of of ULK1/2 and of the Atg13-FIP200 complex. Acetylation of Atg proteins by acetyltransferases also modifies autophagy. Atg7 interaction with p300 maintains the acetylation of Atg5, Atg7, Atg8 and Atg12 proteins, thereby suppressing autophagy. Upon metabolic stress conditions, p300 dissociates from Atg7 and the deacetylase Sirt1 removes the acetyl groups from Atg7, Atg5, Atg8 and Atg12 and triggers autophagy
3. Autophagy measurement
The accurate measurement of autophagy is a critical and controversial issue, especially because autophagy is a dynamic process with several steps that can be individually regulated. The main issue is the differentiation between formation and accumulation of autophagosomes. Measurement of the autophagic flux constitutes the most accurate quantification of the complete process, including the delivery of cargo to lysosomes and its breakdown and recycling [30]. Here we review some of the recommended methods for monitoring autophagy. However, the absence of an approach with high sensitivity and specificity necessitates the combination of at least two of the methods discussed here.
Electron microscopy (EM) is one of the most sensitive techniques to detect the presence of autophagic vacuoles (AV). EM permits the visualization of the entire maturation process from the formation of the phagophore to the assembly of the autophagolysosome [31]. Quantification of the AV is required; the preferred method is to quantify autophagosome volume as the percent of cytoplasmic volume. Immuno-EM with gold-labeling using antibodies to cargo proteins increases specificity and facilitates quantification [32].
LC3 western blotting is another widely used steady state method. Detection of LC3II is a reliable marker of autophagy, as it is localized in the AV membrane from the very early state and is eventually degraded via lysosomal enzymes. Remarkably, in mammalian cells, total levels of LC3 do not necessarily fluctuate with changes in autophagy; therefore, the conversion of LC3I to LC3II must be monitored[33]. The main limitation of LC3 western blotting is its interpretation. An increase in LC3 levels can be due either to an increase in autophagosome synthesis or to reduced turnover. To better interpret the changes, autophagy flux should be measure after preventing lysosomal degradation with protease inhibitors (leupeptin), drugs that alter the lysosomal pH (bafilomycin A, chloroquine), or agents that block the fusion of autophagosomes with lysosomes. In the presence of such inhibitors, an accumulation of LC3II would indicate a deficient autophagic flux, whereas a failure of LC3 to increase would indicate a defect earlier in the process [34].
LC3 can also detected by direct fluorescence microscope by tagging it to a fluorescence protein such as GFP (GFP-LC3). Quantification of the number of fluorescence punctae per cell is an accurate and useful approach[30]. As the mTOR pathway is one of the most important autophagy regulators, measurement of mTORC1 activity via quantification of the phosphorylation of its downstream targets (p70S6 and S6 protein) has been proposed as a potential method to monitor autophagy [35]. Nonetheless, it should be taken into account that there are mTOR-independent mechanisms that also induce autophagy.
As most of the Atg genes do not show significant changes in mRNA levels upon autophagy induction [36], protein quantification is a better readout.
The p62 protein binds strongly to LC3 as well as to ubiquitinated substrates and is mainly degraded through autophagy; therefore, inhibition of autophagy causes its accumulation. Detection of endogenous p62 by Western blot is useful to monitor autophagy flux. However, p62 detection is recommended in conjunction with another method as inhibition of the proteasome also induces its accumulation [37].
The autophagosome maturation process can be also monitored using a tandem RFP-GFP- tagged LC3 [38]. The RFP signal is resistant to the acidic/proteolytic conditions of the lysosome lumen, whereas GFP is sensitive. Therefore, the co-localization of both GFP and RFP indicates the absence of contact with the lysosomal compartment. On the other hand, expression of RFP without GFP signals the presence of an autolysosome.
Generation of transgenic GFP-LC3 mice with basal GFP-LC3 expression that can be further induced under conditions that activate autophagy has led to the capability to monitor autophagy in vivo[39]. For a more extensive review in the use and interpretation of assays for monitoring autophagy see reference [28].
4. Autophagy and mesenchymal cells
Recent studies implicate autophagy in the pathogenesis of multiple diseases, including infection, cancer, and neurodegenerative disease. Autophagy is increasingly recognized as a mediator of survival and proliferation, although its role in fibrogenesis varies greatly in different tissues and settings. Little is known about the role of autophagy in the regulation of mesenchymal cell differentiation into myofibroblast like cells, and there is evidence suggesting that this role may differ from its role in epithelial cells [40, 41] (Table 1).
Table 1.
The role of autophagy according to mesenchymal cell type.
| MESENCHYMAL CELL TYPE | ROLE OF AUTOPHAGY |
|---|---|
| Hepatic stellate cells [40, 43], | Fuels catabolic pathways of cellular activation and thereby promotes acquisition of a fibrogenic phenotype |
| Scleroderma fibroblasts [46] | Provides energy to fuel acquisition of a fibrotic phenotype |
| Synovial fibroblasts [47] | Pro-survival pathway that perpetuates injury |
| Cancer-associated fibroblasts [50, 51] | Generates high energy nutrients that feed adjacent cancer cells and promotes tumor growth, proliferation, and metastasis |
| Cardiac fibroblasts [41, 44] | Ameliorates fibrosis by promoting collagen degradation |
| Kidney mesangial cells [45] | Ameliorates fibrosis by promoting collagen degradation |
Here we review the still scarce knowledge on the role of autophagy in mesenchymal cells:
Autophagy has been shown to play a crucial role in survival of primary human fibroblasts [42] and high levels of autophagy have been reported in fibrogenic cells in almost every tissue: stellate cells[40, 43], cardiac fibroblasts[41, 44], mesangial cells[45], dermal fibroblasts[46], and synovial fibroblasts[47].
In the liver, injury of any kind leads to the transdifferentation of quiescent stellate cells into myofibroblast-like cells that are key players in the wound-healing response. Activation of stellate cells leads to the deposition of massive quantities of extracellular matrix (ECM) that over time culminates in hepatic fibrosis. Our group recently reported that autophagy provides cellular energy to fuel the catabolic pathways of cellular activation[40]. Indeed, autophagy inhibition in the liver fibrogenic cells leads to an impairment of activation and subsequent reduction in ECM deposition[40] We reproduced this effect on the fibrogenic response by blocking autophagy in fibrogenic cells from kidney and lung, suggesting that autophagy may comprise a core pathway of fibrogenesis and an attractive targetable antifibrotic candidate.
Similar data have been reported in skin fibrosis[46, 48]. Scleroderma fibroblasts display increased autophagy levels that provide the energy required for the acquisition of the profibrotic phenotype[46].
In rheumatoid arthritis, synovial fibroblasts synthesize excessive amounts of ECM and are resistant to apoptosis. In this setting, autophagy promotes synovial fibroblasts survival [47, 49] and therefore perpetuation of the injury.
ECM deposition by cancer-associated fibroblasts in the tumor microenvironment drives tumor recurrence and metastasis. Stromal cells up-regulate autophagy and produce high energy nutrients that fuel the adjacent cancer cells, thereby promoting tumor growth, proliferation and metastasis[50, 51]. Autophagy inhibition in the tumor microenvironment has been proposed as a promising new anticancer therapy[52].
Cardiac fibroblasts regulate the structural, biochemical, mechanical and electrical properties of the heart by regulating the homeostasis of the ECM [53]. However, up-regulation of autophagy in cardiac fibroblasts has a beneficial effect on cardiac fibrosis. Beta-2 adrenergic receptor stimulation in cardiac myofibroblasts is associated with increased autophagy levels and increased collagen degradation[41]
In renal fibrosis, autophagy induction during renal injury protects mesangial cells (MC), the fibrogenic cell type in the kidney, and increases their survival[54, 55]. Disruption of the essential autophagy gene Atg7 with specific siRNA or treatment with the autophagy inhibitor 3MA in cultured MC has been associated with decreased levels of collagen type I (COL 1) [40]. Conversely, Beclin 1-deficient MCs (siBeclin1) exhibit accumulation of COL 1 when compared with autophagy competent MC [56]. Therefore, this data suggest a potential dual role of autophagy during kidney injury as both inducer of COL 1 synthesis and degradation.
5. Chemical modulators of autophagy
The recent progress made in elucidating autophagy regulation has led to its implication in a wide range of human diseases. As a result, pharmacological manipulation of the autophagy pathway has become an attractive and promising target. Development of specific compounds is necessary in order to better understand the molecular regulation of the autophagic pathway and to identify new treatment strategies for human diseases associated with autophagy deregulation.
Small molecules that enhance or inhibit autophagy may constitute a useful pharmacological approach depending on the disease context. Here we briefly describe some of them.
Inhibitors of autophagy
Inhibition of the class III PI3K (Vps34) pathway can be achieved with drugs such as 3-MA, wortmannin, and LY294002 [57, 58] (Table 2).
Table 2.
Inhibitors of autophagy and their current clinical applications.
| MECHANISM | COMPOUNDS | CURRENT CLINICAL USE? |
|---|---|---|
| Inhibition of the class III PI3K (Vps34) pathway | • 3-methyladenine | |
| • Wortmannin | ||
| • LY294002 | ||
| Impairment of AV fusion/alteration of lysosomal pH | • Chloroquine | • Antimalarial |
| • Bafilomycin A1 | • Macrolide antibiotic | |
| Increase of cytosolic Ca++ levels | • Thapsigargin | |
| • Bay K8644 | ||
| Inhibition of histone deacetylase | • Trichostatin A | |
| Interaction with AMPK | • Dorsomorphin | |
| • AICAR (5-aminoimidazole-4-carboxyamide ribonucleoside) |
Impairment of fusion of the AV with the lysosome, or modification of the pH in the lysosome with drugs such as the antimalarial chloroquine and the macrolide antibiotic bafilomycin A1 are also effective autophagy inhibitors[59, 60].
Drugs such as thapsigarin and Bay K8644 that increase cytosolic Ca2+ levels have been described as mTOR independent-inhibitors of autophagy [61]
Trichostain A is a histone deacetylase inhibitor that has been identified as a promising compound able to reduce excessive levels of autophagy without altering basal levels[62].
Dosomorphin blocks autophagy via inhibition of AMPK[63], however AICAR (5-aminoimidazole-4-carboxamide ribonucleoside), an activator of AMPK, can also inhibit autophagy in hepatocytes[64]. This demonstrates the complexity of the autophagy process and the need to develop more selective targets.
Inducers of autophagy
Manipulation of the mTOR pathway is the most widely used strategy to enhance autophagy. Although rapamycin is the classic example, perhexilene, niclosamide, amoidarone and rottlerin inhibit mTORC1 but not mTORC2 [65, 66]. PP242 and Torin1 inhibit both mTORC1 and mTORC2 complexes in a selective ATP- competitive way[67] (Table 3).
Table 3.
Chemical inducers of autophagy and their current clinical applications.
| MECHANISM | COMPOUND | CURRENT CLINICAL USE |
|---|---|---|
| mTORC-dependent | ||
| mTORC1-specific inhibition | • Rapamycin | • Immunosuppressant |
| • Perhexilene | ||
| • Niclosamide | • Antihelminthic | |
| • Amiodarone | • Antiarrhythmic | |
| • Rottlerin | ||
| Inhibition of both mTORC1 and mTORC2 | • PP242 | |
| • Torin1 | ||
| Inhibition of both class I PI3K and mTORC1 | • PI103 | |
| mTORC-independent | ||
| Reduction of intracellular levels of IP3 | • Lithium | Mood stabilizers |
| • Carbamazepine | ||
| • Valproic acid | ||
| Regulation of camp-Epac-PLC-ε-IP3 and Ca2+-calpain-Gsα pathways | • Verapamil | • Antihypertensive |
| • Loperamide | • Antidiarrheal | |
| • Nimodipine | • Antihypertensive | |
| • Pimozide | • Antipsychotic | |
| • Nitrendipine | • Antihypertensive | |
| • Clonidine | • Antihypertensive, pain management, anxiolytic | |
| • Rilmenidine | • Antihypertensive | |
Although PI3K plays an important role in controlling the mTOR pathway, its pharmacological manipulation has not been extensively explored. PI103 is a potent autophagy inducer that blocks the class I PI3K and also mTORC1 in an ATP-competitive manner[68].
Autophagy can also be induced in an mTOR-independent manner. The mood-stabilizing drugs lithium, carbamazepine and valproic acid enhance autophagy by reducing the intracellular levels of IP3 [69]. Other drugs (verapamil, loperamide, nimodipine, pimozide, nitrendipine, clonidine, rilmenidine) [61] regulate the cAMP-Epac-PLC-ε-IP3 and Ca2+-calpain-Gsα pathways respectively and have recently been identified as autophagy inducers[66].
For a more extensive overview of the small molecules regulating autophagy we direct the reader to the outstanding review[64]
6. Conclusions
Mesenchymal cells are the key drivers of the fibrogenic process and share certain core properties and regulatory pathways among tissues. Despite recent advances in our understanding of fibrotic diseases, effective antifibrotic therapies remain elusive. A better understanding of mesenchymal cell biology and regulatory pathways is needed to promote the development of new antifibrotic drugs that exploit autophagic pathways.
Highlights.
Fibrotic diseases account for more than 45% of deaths in the industrialized world.
Despite the progress made in elucidating its regulation, there are many gaps in our understanding of autophagy.
Autophagy has recently been implicated in the pathophysiology of fibrosis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. The Journal of clinical investigation. 2007;117:524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA. Cellular and molecular mechanisms of fibrosis. The Journal of pathology. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 4.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 5.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 7.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 10.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]
- 14.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 15.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 16.Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, Tailler M, Delahaye N, Tesniere A, De Stefano D, Younes AB, Harper F, Pierron G, Lavandero S, Zitvogel L, Israel A, Baud V, Kroemer G. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcelle E, Nebout M, Bekri S, Gauthier N, Hofman P, Poujeol P, Fenichel P, Mograbi B. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006;66:6861–6870. doi: 10.1158/0008-5472.CAN-05-3557. [DOI] [PubMed] [Google Scholar]
- 19.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 22.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 23.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 24.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamai A, Codogno P. New targets for acetylation in autophagy. Sci Signal. 2012;5:pe29. doi: 10.1126/scisignal.2003187. [DOI] [PubMed] [Google Scholar]
- 26.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-X-L and induction of autophagy. Embo Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, Zhu L, Le Y, Gong X, Yan X, Hong B, Jiang FJ, Xie Z, Miao D, Deng H, Yu L. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 29.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Ruan K, Wang Z, Zhang CS, Chien KY, Wu J, Li Q, Han J, Lin SC. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- 32.Mayhew TM. Quantitative immunoelectron microscopy: alternative ways of assessing subcellular patterns of gold labeling. Methods Mol Biol. 2007;369:309–329. doi: 10.1007/978-1-59745-294-6_15. [DOI] [PubMed] [Google Scholar]
- 33.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth S, Glick D, Macleod KF. Autophagy: assays and artifacts. J Pathol. 2010;221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–8527. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 36.Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem. 2006;54:85–96. doi: 10.1369/jhc.5A6743.2005. [DOI] [PubMed] [Google Scholar]
- 37.Bardag-Gorce F, Francis T, Nan L, Li J, He Lue Y, French BA, French SW. Modifications in P62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci. 2005;77:2594–2602. doi: 10.1016/j.lfs.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 38.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 39.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aranguiz-Urroz P, Canales J, Copaja M, Troncoso R, Vicencio JM, Carrillo C, Lara H, Lavandero S, Diaz-Araya G. Beta(2)-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim Biophys Acta. 2011;1812:23–31. doi: 10.1016/j.bbadis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Kang HT, Lee KB, Kim SY, Choi HR, Park SC. Autophagy impairment induces premature senescence in primary human fibroblasts. PLoS One. 2011;6:e23367. doi: 10.1371/journal.pone.0023367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koesters R, Kaissling B, Lehir M, Picard N, Theilig F, Gebhardt R, Glick AB, Hahnel B, Hosser H, Grone HJ, Kriz W. Tubular overexpression of transforming growth factor-beta1 induces autophagy and fibrosis but not mesenchymal transition of renal epithelial cells. Am J Pathol. 2010;177:632–643. doi: 10.2353/ajpath.2010.091012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castello-Cros R, Whitaker-Menezes D, Molchansky A, Purkins G, Soslowsky LJ, Beason DP, Sotgia F, Iozzo RV, Lisanti MP. Scleroderma-like properties of skin from caveolin-1-deficient mice: implications for new treatment strategies in patients with fibrosis and systemic sclerosis. Cell Cycle. 2011;10:2140–2150. doi: 10.4161/cc.10.13.16227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Connor AM, Mahomed N, Gandhi R, Keystone EC, Berger SA. TNFalpha modulates protein degradation pathways in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther. 2012;14:R62. doi: 10.1186/ar3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oikarinen A. Hydroxychloroquine induces autophagic cell death of human dermal fibroblasts: implications for treating fibrotic skin diseases. J Invest Dermatol. 2009;129:2333–2335. doi: 10.1038/jid.2009.164. [DOI] [PubMed] [Google Scholar]
- 49.Shin YJ, Han SH, Kim DS, Lee GH, Yoo WH, Kang YM, Choi JY, Lee YC, Park SJ, Jeong SK, Kim HT, Chae SW, Jeong HJ, Kim HR, Chae HJ. Autophagy induction and CHOP under-expression promotes survival of fibroblasts from rheumatoid arthritis patients under endoplasmic reticulum stress. Arthritis Res Ther. 2010;12:R19. doi: 10.1186/ar2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castello-Cros R, Bonuccelli G, Molchansky A, Capozza F, Witkiewicz AK, Birbe RC, Howell A, Pestell RG, Whitaker-Menezes D, Sotgia F, Lisanti MP. Matrix remodeling stimulates stromal autophagy, “fueling” cancer cell mitochondrial metabolism and metastasis. Cell Cycle. 2011;10:2021–2034. doi: 10.4161/cc.10.12.16002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez CG, Penfornis P, Oskowitz AZ, Boonjindasup AG, Cai DZ, Dhule SS, Rowan BG, Kelekar A, Krause DS, Pochampally RR. Activation of autophagy in mesenchymal stem cells provides tumor stromal support. Carcinogenesis. 2011;32:964–972. doi: 10.1093/carcin/bgr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sotgia F, Martinez-Outschoorn UE, Howell A, Pestell RG, Pavlides S, Lisanti MP. Caveolin-1 and cancer metabolism in the tumor microenvironment: markers, models, and mechanisms. Annu Rev Pathol. 2012;7:423–467. doi: 10.1146/annurev-pathol-011811-120856. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Weber KT. Infarct scar: a dynamic tissue. Cardiovasc Res. 2000;46:250–256. doi: 10.1016/s0008-6363(00)00032-8. [DOI] [PubMed] [Google Scholar]
- 54.Ponnusamy M, Liu N, Sellamuthu R, Zhao TC, Mao H, Zhuang S. Autophagy protects against necrotic renal epithelial cell-induced death of renal interstitial fibroblasts. Am J Physiol Renal Physiol. 2012;303:F83–91. doi: 10.1152/ajprenal.00027.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Y, Kim JK, Kim SI, Na HJ, Jun SY, Lee SJ, Choi ME. TGF-{beta}1 protects against mesangial cell apoptosis via induction of autophagy. J Biol Chem. 2010;285:37909–37919. doi: 10.1074/jbc.M109.093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SI, Na HJ, Ding Y, Wang Z, Lee SJ, Choi ME. Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-beta1. J Biol Chem. 2012;287:11677–11688. doi: 10.1074/jbc.M111.308460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 59.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 61.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, Jahreiss L, Fleming A, Pask D, Goldsmith P, O’Kane CJ, Floto RA, Rubinsztein DC. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, Rothermel BA, Gillette TG, Hill JA. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 64.Baek KH, Park J, Shin I. Autophagy-regulating small molecules and their therapeutic applications. Chem Soc Rev. 2012;41:3245–3263. doi: 10.1039/c2cs15328a. [DOI] [PubMed] [Google Scholar]
- 65.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 67.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, Henley A, Di-Stefano F, Ahmad Z, Guillard S, Bjerke LM, Kelland L, Valenti M, Patterson L, Gowan S, de Haven Brandon A, Hayakawa M, Kaizawa H, Koizumi T, Ohishi T, Patel S, Saghir N, Parker P, Waterfield M, Workman P. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–5850. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 69.Williams RS, Cheng L, Mudge AW, Harwood AJ. A common mechanism of acton for three mood-stabilizing drugs. Nature. 2002;417:292–295. doi: 10.1038/417292a. [DOI] [PubMed] [Google Scholar]