Camalexin (3-thiazol-2′-yl-indole) is the major phytoalexin in Arabidopsis thaliana (Glawischnig, 2007) and is involved in defense against a wide range of pathogens, such as Botrytis cinerea and Alternaria brassicicola (Kagan and Hammerschmidt, 2002; Denby et al., 2004). The pathway leading to this model phytoalexin has been almost fully elucidated, and recent focus has been on the biosynthetic origin of the thiazole ring. Several publications have shown that glutathione, and not Cys, is the direct source of the heterocycle and have demonstrated that both glutathione and Cys conjugates of indole-3-acetonitrile (IAN) are intermediates in the pathway (Böttcher et al., 2009; Geu-Flores et al., 2011; Su et al., 2011). However, two recent studies published in this journal arrived at different conclusions regarding the conversion of the glutathione conjugate (γ-Glu-Cys[IAN]-Gly) to the Cys conjugate (Cys[IAN]), particularly regarding the family of enzymes cleaving off the γ-Glu residue. Su et al. (2011) reported that known members of the γ-glutamyl transpeptidase (GGT) family conducted this reaction, whereas we (Geu-Flores et al., 2011) found that members of the newly found γ-glutamyl peptidase (GGP) family performed the same reaction. This has created some confusion in the literature, with several subsequent depictions of the pathway containing both GGTs and GGPs (Ahuja et al., 2012; Saga et al., 2012), although some have made a clear distinction (Bednarek, 2012). Based on the results from these two reports, together with other published data and additional experimental results included here, we argue that GGPs and not GGTs are the γ-glutamyl cleaving enzymes in the camalexin pathway in Arabidopsis. Our line of argumentation leads to a discussion regarding the prerequisites for identification of specific genes as responsible for particular enzymatic steps in biosynthetic pathways.
THE CAMALEXIN BIOSYNTHETIC PATHWAY
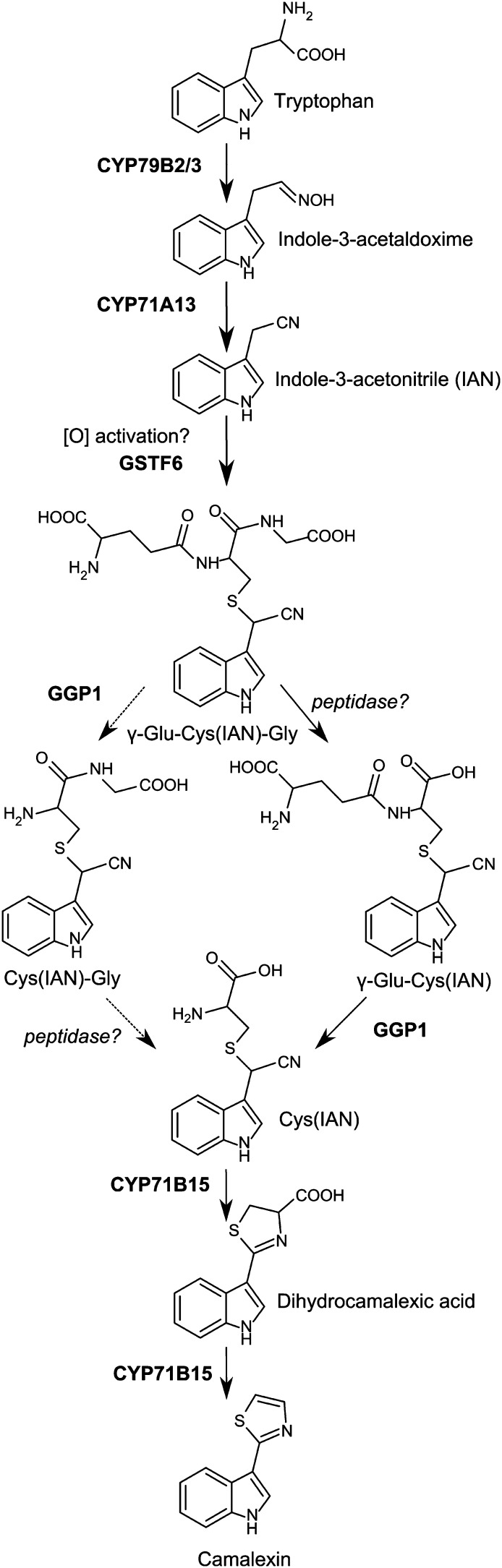
Camalexin is made from Trp, which is converted to indole-3-acetaldoxime by the cytochrome P450 CYP79B2 or by its close homolog CYP79B3 (Hull et al., 2000; Mikkelsen et al., 2000; Glawischnig et al., 2004). Subsequently, CYP71A13 converts the aldoxime to IAN (Nafisi et al., 2007), which, after an unknown activation step, is conjugated to the tripeptide glutathione (γ-Glu-Cys-Gly) by glutathione S-transferases, including GSTF6 (Böttcher et al., 2009; Su et al., 2011). The glutathione conjugate γ-Glu-Cys(IAN)-Gly is then hydrolyzed by two consecutive peptidase reactions (discussed in detail below) to Cys(IAN), which is finally converted to camalexin by a two-step reaction catalyzed by CYP71B15 (PAD3) (Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009) (Figure 1).
Figure 1.
The Camalexin Biosynthetic Pathway in Arabidopsis.
The carboxypeptidase cleaving off the Gly residue of γ-Glu-Cys(IAN)-Gly or Cys(IAN)-Gly is currently unknown, though PCS1 has been proposed to carry out the reaction (Böttcher et al., 2009; Su et al., 2011). The sequential order of the two consecutive reactions in the conversion of γ-Glu-Cys(IAN)-Gly to Cys(IAN) is not known. However, γ-Glu-Cys(IAN)-Gly, γ-Glu-Cys(IAN), and Cys(IAN), but not Cys(IAN)-Gly, have been found in Arabidopsis plants with induced camalexin biosynthesis (Böttcher et al., 2009; Figure 3).
In search for missing camalexin biosynthetic genes, Su et al. (2011) performed proteomics on Arabidopsis seedlings with camalexin production induced via a dexamethasone-stimulated mitogen-activated protein kinase 9 (MAPK9) signaling cascade. Protein profiling by two-dimensional gel electrophoresis identified 25 proteins significantly upregulated in the seedling producing camalexin, including GSTF6 and GGP1 (annotated as defense-related protein, At4g30530) (Su et al., 2011). In addition to this elegant proteomic study, which provided an important complementation to RNA-based coexpression studies, Su et al. (2011) provided support for the involvement of GSTF6 in the camalexin pathway by measuring camalexin levels in knockout and overexpression mutants and by demonstrating that extracts of GSTF6-expressing yeast could aid the formation of γ-Glu-Cys(IAN)-Gly from GSH and IAN.
The subsequent conversion of γ-Glu-Cys(IAN)-Gly to Cys(IAN) requires the action of both a GGP and a carboxypeptidase. In search for the GGP, Su et al. (2011) relied on pre-2009 literature, where GGTs were the only known plant enzymes capable of catalyzing GGP reactions (Martin et al., 2007; Ohkama-Ohtsu et al., 2007a, 2007b). They analyzed GGT expression patterns and ggt knockout mutants and assigned GGTs a role in the camalexin pathway (Su et al., 2011). However, we had shown in 2009 that GGP1 could hydrolyze the γ-glutamyl peptide bond of a glucosinolate-related glutathione conjugate both in transgenic Nicotiana benthamiana and in in vitro assays (Geu-Flores et al., 2009). Based on this knowledge, we investigated the native role of GGPs in Arabidopsis and found that they were responsible for hydrolyzing γ-glutamyl bonds in both the camalexin and glucosinolate pathways (Geu-Flores et al., 2011). The contradictory nature of the conclusions about the GGP activity in our report and in the report by Su et al. (2011) has created confusion for those working in the field. In the following sections, we evaluate the experimental evidence in favor of GGPs and GGTs as GGPs operating in the camalexin pathway using four criteria: end-product phenotype, accumulation of pathway intermediates, enzyme activity, and temporal/spatial localization. These criteria provide benchmarks that can be used to establish the direct involvement of any gene at a defined step in a biosynthetic pathway.
EVIDENCE FOR GENE FUNCTION
End-Product Phenotype
The term end-product is often used to describe the main metabolic outcome of a given pathway, and we adopt this definition here even though the term can be misleading, as many such products, including camalexin, can be further modified, for example, by hydroxylation or glucosylation (Böttcher et al., 2009). To provide experimental evidence for the involvement of GGTs, Su et al. (2011) analyzed camalexin levels (end-product levels) in media of liquid-cultured single T-DNA knockout mutants of ggt1 and ggt2 and found a decrease of 40 to 60% in both mutants. This is comparable with the decrease that we observed in leaves of knockdown mutants of GGP1. The double mutant ggp1 ggp3, with reduced transcripts of both GGP1 and GGP3, showed a 90% reduction in camalexin levels (Geu-Flores et al., 2011). This reduction is in turn comparable to the one observed in the media of MAPK9-induced seedlings upon treatment with the GGT inhibitor acivicin (Su et al., 2011). Based solely on end-product levels in mutants of candidate biosynthetic genes, both GGTs and GGPs appear to be involved in the camalexin pathway.
However, perturbations in the levels of end-product do not necessarily reflect direct involvement of a gene in a pathway; conversely, mutations in a gene in a biosynthetic pathway do not always result in measurable perturbations in end-product levels. End-products are often subject to tight feedback regulation, which is why perturbations in their levels may lead to the rapid activation of compensatory mechanisms. In order for a knockout mutation to result in an end-product phenotype, the pathway of interest must be the only one leading to the desired end-product, and there must be no compensation at the reaction level by other enzymes or nonenzymatic chemical reactions. In addition, mutations in genes not directly involved in a given pathway can have a big impact on the levels of end-product. Such mutations may include not only biosynthetic genes in other pathways but also genes involved in general plant fitness or regulatory networks. One example of a biosynthetic gene not directly involved in the camalexin pathway but with a considerable effect on end-product levels is phytoalexin-deficient2 (PAD2). The pad2-1 mutant was identified by its camalexin deficiency (Glazebrook and Ausubel, 1994) and was later shown to encode for γ-glutamylcysteine synthetase, which catalyzes the first committed step of glutathione biosynthesis (Parisy et al., 2007). Accordingly, pad2 is not directly part of the camalexin pathway but is involved in the synthesis of one of the cosubstrates. In order to investigate whether an end-product phenotype can be directly ascribed to the decreased catalysis of a particular enzymatic step, feeding experiments in which pathway intermediates are provided externally can be a powerful approach. Cases in which the putative product, but not the putative substrate, is able to complement the phenotype speak in favor of the proposed gene-to-reaction link. However, care must be taken when interpreting the results of such feeding experiments, as the results are affected by many factors, including differences in uptake rates of substrate and product as well as unwanted enzymatic conversion or chemical degradation of the applied pathway intermediates.
The observation that treatment with the Gln analog acivicin reduced camalexin production is important; however, acivicin binds to the substrate binding pocket of all GGTs (Wada et al., 2008), and it may also bind to and inhibit GGPs, which, apart from carrying out similar reactions as GGTs, are evolutionarily derived from Gln-metabolizing enzymes (Geu-Flores et al., 2009; Geu-Flores et al., 2011). An alternative explanation for the reduced camalexin production following acivicin treatment is that termination of all GGT activity could arrest glutathione and glutathione conjugates in the vacuole and extracellular space (Ferretti et al., 2009), thereby affecting glutathione availability in the cytosol. A source of uncertainty regarding the end-product phenotypes observed by Su et al. (2011) is the fact that the end-product measurements were performed in the liquid media used to grow the seedlings in, and not directly in the plant tissue. This adds the possibility that GGTs are involved in the secretion of camalexin rather than in its biosynthesis, a fact that has been noted by Bednarek (2012).
In summary, end-product phenotypes do not provide conclusive evidence of involvement in a pathway, and additional experiments are needed to assign the in planta function of a gene.
Accumulation of Pathway Intermediates
Mutations in a biosynthetic gene and the corresponding reduction or elimination of enzyme activity are likely to cause metabolic bottlenecks and can therefore lead to the accumulation of pathway intermediates as well as to a decrease in end-product. However, compromised enzyme functions can lead to accumulation of intermediates without having effects on the end-products. Such was the case of GGP1 in the glucosinolate pathway, where a T-DNA insertion in GGP1 resulted in accumulation of the expected substrate for GGP1, the corresponding glutathione conjugates, without affecting glucosinolate levels significantly (Geu-Flores et al., 2011). These situations are especially common in cases of partial genetic redundancy (like that of GGP1 and GGP3) in which the final products accumulate to high and variable levels (making small variations difficult to detect) and/or in which the relevant intermediates do not accumulate at all (making their accumulation more easily detectable). Furthermore, the recruitment of functionally homologous enzymes could result in such phenotypes as a result of a lack of channeling with respect to the previous enzyme in the pathway (arising from lack of proper protein–protein interactions), due to localization in a different subcellular compartment (which would require the transport of the intermediate to another compartment), or due to less favorable kinetics (which would require a buildup of substrate concentrations before reaching compensatory reaction rates). All these can potentially lead to the accumulation of intermediates with no changes in end-product levels. Situations where loss of enzyme function does not lead to detectable accumulation of intermediates are also foreseeable. Likely scenarios include cases in which the substrate is a central intermediate used in other reactions, which is the case if the enzyme of interest carries out the first committed step in a pathway or acts at the intersection between pathways. However, when accumulation of a particular pathway intermediate (the putative substrate) can be proven, this is a more specific indicator of enzyme function than the reduction in end-product levels, as fewer factors can influence it.
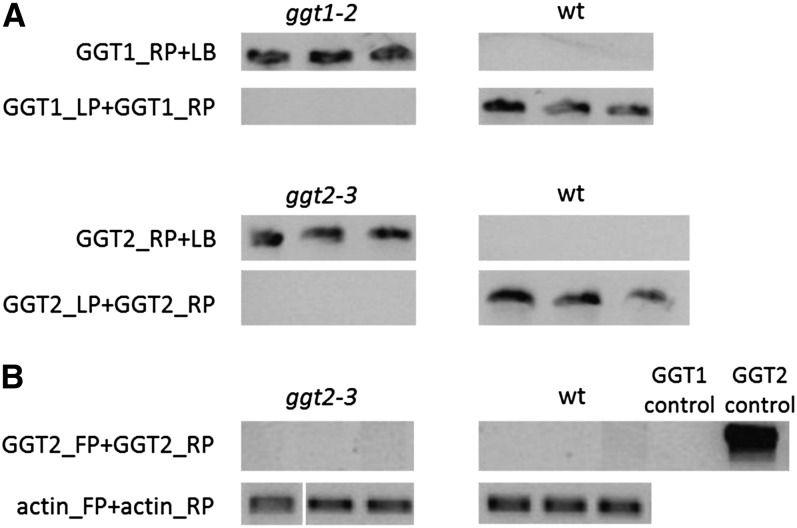
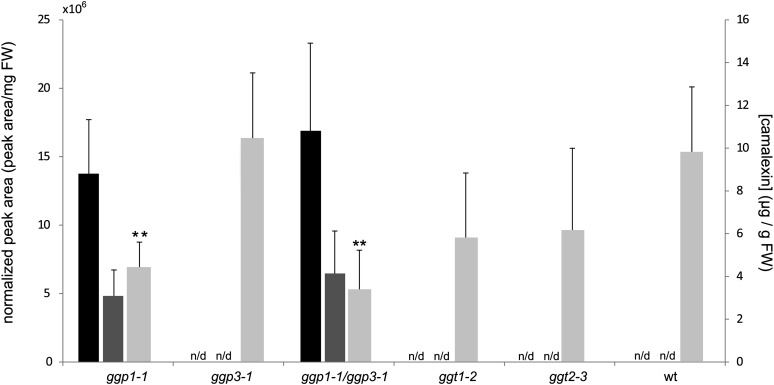
Dependent on the sequential order of the two consecutive reactions in the conversion of γ-Glu-Cys(IAN)-Gly to Cys(IAN), either γ-Glu-Cys(IAN)-Gly or γ-Glu-Cys(IAN) is the substrate of both the GGTs and the GGPs in vivo. Blinded by our history of research within the glucosinolate pathway, where only a GGP reaction appears to be needed to generate the substrate for the ensuing enzyme (Geu-Flores et al., 2009, 2011), we previously analyzed only for accumulation of the γ-Glu-Cys(IAN)-Gly intermediate, which we found accumulating in ggp1 knockdown mutants (Geu-Flores et al., 2011). Su et al. (2011) did not mention whether γ-Glu-Cys(IAN)-Gly or γ-Glu-Cys(IAN) accumulated in the ggt1 and ggt2 mutants. We analyzed Arabidopsis mutants of both GGPs and GGTs for accumulation of either one of the possible pathway intermediates. In addition to ggt1-2 (SALK_133807C) (Su et al., 2011), we used an alternative GGT2 knockout line (ggt2-3, SALK_147881.52.00; Figure 2) (kindly provided by Erich Glawischnig, Technische Universität Münich, Germany). We analyzed these lines in parallel with our ggp mutants and wild-type plants. In this analysis, we found γ-Glu-Cys(IAN)-Gly and γ-Glu-Cys(IAN) only in AgNO3-induced ggp1-1 and ggp1-1/ggp3-1 plants and not in ggt1-2, ggt2-3, ggp3-1, or wild type plants (Figure 3). With respect to camalexin levels, the ggp1-1 and ggp1-1/ggp3-1 mutants showed a significant reduction (Figure 3), whereas the ggt mutants did not, showing that mutations in GGT1 and GGT2 only affect camalexin levels significantly in media of liquid-grown seedlings and not in leaves. We did not observe any effect on intermediate or camalexin levels for the single knockdown line of ggp3 or significant differences between ggp1-1 plants and ggp1-1/ggp3-1 plants, which leads us to suggest that only GGP1 is part of the camalexin pathway under normal conditions. We find that the accumulation of pathway intermediates is a strong single piece of evidence in favor of the involvement of GGP1 and argues against the involvement of GGT1 and GGT2 in the camalexin pathway in planta.
Figure 2.
Genotyping and RT-PCR.
The primer pairs used are listed to the left of the gel pictures. The genotypes of the individual leaves whose DNA was used as template are listed above the gel pictures.
(A) Genotyping of ggt1-2, ggt2-3, and wild-type (wt) plants.
(B) RT-PCR searching for GGT2 transcript in ggt2-3 and wild-type plants. “GGT1 control” and “GGT2 control” represent specificity control reactions, where plasmid carrying the respective coding sequences of GGT1 and GGT2 were used as templates. All PCR reactions were performed on material extracted from independent plants.
Figure 3.
Accumulation of γ-Glu-Cys(IAN)-Gly, γ-Glu-Cys(IAN), and Camalexin in ggp and ggt Mutants.
Rosette leaves of 3-week-old Arabidopsis plants were induced with AgNO3 and analyzed by liquid chromatography–tandem mass spectrometry 24 h after induction. Left axis, normalized peak areas for γ-Glu-Cys(IAN)-Gly (mass-to-charge ratio [m/z] [M+H]+ = 462; black bars) and γ-Glu-Cys(IAN) (m/z [M+H]+ = 405; dark-gray bars). Right axis, camalexin levels (m/z [M+H]+ = 201; light-gray bars). FW, fresh weight. Error bars represent sd; stars indicate data points with significant differences with respect to the wild type (wt) (Student’s t test, n = 6, P < 0.05). n/d, not detected.
Enzymatic Activity
With the power of today’s sequencing technologies and the corresponding rapid expansion of genome and transcriptome databases, in silico searches and coexpression analyses have become essential tools for gene discovery and pathway elucidation (Saito et al., 2008). With these methods, it is often possible to find candidate genes that have been annotated to encode enzymes that are likely to catalyze the reaction of interest. However, an inherent risk exists that a candidate enzyme catalyzes the desired reaction with physiologically relevant Km and Vmax parameters in vitro but is still unrelated to the reaction in vivo. This risk becomes even higher when working with pathways producing specialized plant metabolites, where many enzymes have been recruited from primary metabolism or general detoxification pathways (for example, glycosyltransferases or glutathione S-transferases) and might thus have functional homologs or possess vestigial substrate promiscuity not reflecting their role in planta (Weng et al., 2012). In forward genetic studies, where genes are identified by an unbiased approach, enzyme activity provides an independent proof of function, as exemplified by the discovery of PAD3 (Schuhegger et al., 2006). However, enzyme activity per se cannot be regarded as independent evidence when using reverse genetic in silico approaches, as the predicted function is usually among the criteria for choosing the gene; thus, enzyme activity can become a self-fulfilling prophecy. Nevertheless, confirmation of the predicted enzyme activity, preferably with the native substrate, is a prerequisite for determining its function.
In the camalexin case, purified recombinant GGP1 expressed in Escherichia coli was shown to convert γ-Glu-Cys(IAN)-Gly to Cys(IAN)-Gly in vitro, supporting its proposed role in vivo. However, we have not yet assessed whether GGP1 can also metabolize γ-Glu-Cys(IAN) because of the lack of availability of this alternative substrate. To the best of our knowledge, no such assays have been performed with any of the GGTs from Arabidopsis. Because the GGTs are characterized by their ability to hydrolyze a broad range of glutathione conjugates within detoxification of xenobiotics and degradation of oxidized glutathione, it is highly likely that they can hydrolyze γ-Glu-Cys(IAN)-Gly and Cys(IAN)-Gly (Ohkama-Ohtsu et al., 2009). If that turns out to be the case, it cannot be interpreted as evidence for their in planta role, but as supporting information that requires additional evidence for the assignment of gene function.
Temporal/Spatial Localization
For an enzyme to carry out a biochemical reaction, it must be in physical contact with its substrate and any potential cosubstrates. In other words, it needs to be expressed in the right place at the right time. Therefore, for pathways with known temporal and spatial localization, investigating the cellular and subcellular localization of a candidate enzyme is of considerable importance. Examples do exist of pathways where the biosynthetic machinery is allocated to different subcellular compartments and even different types of cells, demonstrating the possibility of transport of intermediates (Ziegler and Facchini, 2008). But even in such pathways, stretches of consecutive enzymatic reactions occur in single compartments, avoiding the transport of toxic or reactive intermediates. Thus, temporal and spatial localization are important clues for assessing the likelihood that a given enzyme is involved in a particular pathway in planta.
With regards to subcellular localization, the camalexin pathway is regarded as being cytosolic, as three of the four known enzymes, including the first and the last enzymes in the pathway, are endoplasmic reticulum–associated cytochrome P450s, having their catalytic domain facing the cytosol (Schuler and Werck-Reichhart, 2003). Green fluorescent protein gene fusions of GGP1 and GGP3 showed that both were localized in the cytosol (Geu-Flores et al., 2011), which is in accordance with their proposed role in the camalexin pathway. By contrast, enzymatic assays with Arabidopsis cell fractions demonstrated that both GGT1 and GGT2 are localized to the apoplastic space (Martin et al., 2007; Ohkama-Ohtsu et al., 2007b), where a homologous maize (Zea mays) GGT1 was also proven to be localized by immunocytochemistry (Ferretti et al., 2009). Accordingly, direct involvement of GGT1 and GGT2 in the camalexin pathway would require export of their γ-Glu–containing substrate and import of the corresponding product to the cytosol.
With respect to cellular localization, promoter-β-glucuronidase (GUS) fusions have shown that GGT1 is expressed in roots, leaves, and siliques and that GGT2 is expressed in only roots and siliques (Martin et al., 2007; Ohkama-Ohtsu et al., 2007b; Destro et al., 2011). In turn, promoter-GUS experiments with GGP1 and GGP3 have demonstrated expression in leaves (Geu-Flores et al., 2011). However, these experiments were all made with uninduced Arabidopsis plants. As camalexin is a phytoalexin and thus produced only upon induction, basal expression levels of camalexin-related genes are only marginally informative. Su et al. (2011) found increased transcript levels of both GGT1 and GGT2 in total liquid-grown seedlings after MAPK9-induced camalexin production, though they did not identify the mentioned GGTs among the upregulated proteins in their proteomic study. It should be noted, however, that seedlings include both roots and leaves; therefore, these GGT transcript measurements cannot be used to conclusively determine coherence between gene induction and camalexin accumulation on a tissue-specific level.
Rosette leaves have been shown to express the camalexin biosynthetic machinery after treatment with AgNO3 or infection with B. cinerea and are accordingly the most used tissue for camalexin studies (Glawischnig et al., 2004; Schuhegger et al., 2006; Böttcher et al., 2009), though other tissues, such as roots, are also known to accumulate camalexin upon induction (Bednarek et al., 2005). To complement the existing expression data for both GGP1 and the GGTs, we investigated the expression of GGT1, GGT2, and GGP1 transcripts in rosette leaves upon camalexin induction using an in silico approach. We used the Arabidopsis eFP browser (Winter et al., 2007) to extract data from existing microarray experiments made on leaves 18 h after treatment with B. cinerea. The total expression value of GGP1 was 1596 units compared with that of the mock-treated control, which was 700 units. The same values for GGT1 and GGT2 were 208 (control) and 174 (treated), and 2 (control) and 8 (treatment), respectively. This demonstrated that GGP1 was expressed in leaves and upregulated upon camalexin induction, whereas GGT1 was expressed in leaves but not upregulated upon camalexin induction. Furthermore, it demonstrated that GGT2 was not expressed in leaves at any point, which is in agreement with the literature (Martin et al., 2007; Ohkama-Ohtsu et al., 2007b; Destro et al., 2011). Finally, based on seedling microarray data extracted via the Arabidopsis eFP browser, basal levels of GGT1 in seedlings were found to be ∼60 times higher than basal levels of GGT2. Since Su et al. (2011) demonstrated that the expression of both GGT1 and GGT2 was increased ∼20-fold in the MAPK9-induced seedlings, we find it peculiar that knockout mutants of two genes with such dissimilar expression levels can result in the same reduction of camalexin.
SOLVING THE PUZZLE OF CAMALEXIN BIOSYNTHESIS
Involvement of Carboxypeptidases
It is apparent that a carboxypeptidase reaction is needed to remove Gly from the glutathione backbone in the process of converting γ-Glu-Cys(IAN)-Gly into Cys(IAN), as high levels of both γ-Glu-Cys(IAN)-Gly and γ-Glu-Cys(IAN) accumulate in ggp1 knockdown plants (Figure 3). An Arabidopsis enzyme well known for hydrolyzing the Cys-Gly bond in glutathione conjugates is phytochelatin synthase (PCS), also known as glutathione γ-glutamylcysteinyltransferase (Blum et al., 2007). Accordingly, PCS1 has been suggested to be part of the camalexin pathway (Böttcher et al., 2009), but the hypothesis has not been experimentally supported by in planta evidence. Su et al. (2011) showed a tendency to a reduction in camalexin levels in psc1 mutants using the MAPK9-induced seedling system, but the reduction was not reported as statistically significant. Only proper identification of the carboxypeptidase will allow complete elucidation of the camalexin biosynthesis puzzle. Böttcher et al. (2009) demonstrated the presence of γ-Glu-Cys(IAN)-Gly, γ-Glu-Cys(IAN), Cys(IAN), and dihydrocamalexic acid in AgNO3-induced Arabidopsis plants. This set of compounds corresponds to all possible intermediates in the pathway from IAN to camalexin except for Cys(IAN)-Gly. Feeding with both Cys(IAN)-Gly and γ-Glu-Cys(IAN) was able to restore camalexin production in pad2 plants (Su et al., 2011). However, as chemical compounds can undergo many reactions during feeding experiments, this does not necessarily mean that both chemicals are pathway intermediates. In agreement with Böttcher et al. (2009), we did not detect any Cys(IAN)-Gly in the ggp mutants, which might indicate that this compound is not part of the pathway and that the in vivo pathway follows the route depicted to the right in Figure 1. At present, however, we do not have enough experimental evidence to determine which of the two possible hydrolysis routes predominates in planta.
Alternative Camalexin Pathways?
Recently, it was reported that the dwarfed activation-tagged acetyl-amido synthetase Arabidopsis mutant gh3.5-1D had highly increased levels of camalexin (Wang et al., 2012). The authors showed that the acetyl-amido synthetase was able to conjugate indole-3-carboxylic acid to Cys in vitro, leading them to suggest a new parallel route within the camalexin biosynthetic pathway. However, the increased camalexin content of the mutant can be explained with the existing pathway as the mutant contained elevated levels of salicylic acid and accordingly was primed in defense reactions, including a strong induction of camalexin-related transcripts. Furthermore, the in vitro activity of the acetyl-amido synthetase is not supported by in planta evidence, such as accumulation of intermediates or reduction of camalexin in knockout mutants. This example shows the importance of using a combination of criteria to establish the role of genes in metabolic pathways and emphasizes the need for general discussion of those criteria.
CONCLUSION
In this letter, we described four central criteria with which to evaluate the involvement of a gene in a biosynthetic pathway in planta: (1) end-product phenotype, defined as the alteration of end-product levels in planta upon changes of enzyme levels or activity and commonly evaluated using knockout mutants, RNA interference, overexpression lines, or feeding of specific enzyme inhibitors; (2) phenotype of biosynthetic intermediates, defined as the alteration of levels of pathway intermediates or their derivatives in planta upon changes in enzyme levels or activity and commonly evaluated using similar means as in the previous criterion; (3) enzymatic activity, defined as the ability of the enzymes of interest to metabolize the substrate of interest and commonly evaluated by in vitro assays with heterologously expressed proteins or functional characterization in heterologous hosts; and, finally, (4) cellular and subcellular localization, demonstrating the likelihood that an enzyme encounters its presumed substrate in planta and commonly evaluated using green fluorescent protein fusions, immunocytochemistry, promoter-GUS fusions, and tissue-specific microarray-based expression analysis.
We argued that end-product phenotypes are good indicators of the involvement of an enzyme in a pathway, but given the multiplicity of factors that can lead to such metabolic outcomes, phenotypes of accumulation of intermediates (or their derivatives) are more specific indicators. In addition, we discussed multiple scenarios leading to accumulation of intermediates in the absence of an end-product phenotype. Regarding the demonstration of enzymatic activity, we see it as a prerequisite for determining involvement in a pathway. However, given the possible promiscuity of many enzymes involved in specialized metabolism, enzymatic activity alone cannot stand as proof of a physiological role in that pathway. Finally, we have shown how information on cellular and subcellular localization can provide important evidence in favor or against a proposed enzymatic function in planta.
By applying these criteria to the camalexin pathway, we conclude that GGP1 and not GGT1 or GGT2 are part of the camalexin pathway. ggp1 mutants have reduced end-product (camalexin) levels and accumulate the expected intermediates [γ-Glu-Cys(IAN)-Gly and γ-Glu-Cys(IAN)]. GGP1 possesses the required enzymatic activity [at least for γ-Glu-Cys(IAN)-Gly] and is found in the cytosol, where it colocalizes with other enzymes of the camalexin pathway. By contrast, ggt1 and ggt2 mutants do not have reduced camalexin levels in leaves and neither do they accumulate the expected intermediates. GGT1 and GGT2 may or may not have the required enzyme activities, but both enzymes are found in the apoplast and only GGT1 seems to be expressed in the relevant tissues upon induction.
Given all the discussed considerations for each of the mentioned criteria, only a combination of the different pieces of evidence will make assignment of enzyme function robust. However, it is not always possible to obtain all the necessary evidence for conclusive assignment. It is of crucial importance, though, that experimental results within the avenue of functional gene assignment can be published without fulfilling all of the aforementioned criteria. Therefore, we invite all researchers involved in metabolism to make clear distinctions in their articles between (1) identification of genes coding for enzymes whose activities have only been determined in vitro, (2) genes that are proposed to be involved in a pathway based on a few of the mentioned criteria, and (3) genes that satisfy most or all of the mentioned criteria and can be assigned to a particular step of a metabolic pathway. We conclude our letter with this open invitation and with a genuine acknowledgment to The Plant Cell for its publication of excellent articles in our research field, both in the past and in the time to come.
METHODS
Plant Growth and Metabolite Analysis
Arabidopsis thaliana plants were grown in growth chambers at 20°C and 70% relative humidity with 16-h photoperiods as described by Geu-Flores et al. (2011). Metabolite analyses were performed using rosette leaves of 3-week-old plants. Camalexin production was induced by spraying leaves with 5 mM AgNO3 in 0.02% (v/v) Silwet L-77. Twenty-four hours after induction, entire leaves were harvested and crushed in 85% (v/v) methanol. This was followed by a 15,000g centrifugation for 10 min. Three microliters of supernatant was analyzed by liquid chromatography–tandem mass spectrometry by injecting into a Zorbax SB-C18 RRHT column (Agilent; 2.1 ⋅ 50 mm, 1.8 μm). Using a binary mobile phase consisting of A (0.1% [v/v) formic acid and 50 mM NaCl in water) and B (0.1% [v/v] formic acid in acetonitrile), the following gradient program was run at 0.2 mL min−1: 0 to 0.5 min, 6% B; 0.5 to 12.5 min, linear gradient 6 to 55% (v/v) B; 12.5 to 13.10 min, linear gradient 55 to 90% B; 13.10 to 15.5 min, 90% B. Camalexin was quantified using an external standard curve.
Genotyping and RT-PCR
The homozygosity of ggt2-3 (SALK_147881.52.00) and ggt1-2 (SALK_133807C) was confirmed by genotyping (Figure 2). ggt1-2 was genotyped using primers GGT1_LP, 5′-AAGCAATTTCTTTCCCACCAG-3′, and GGT1_RP, 5′-GTGTGTGGGCCAACTTTTATC-3′. ggt1-2 was similarly genotyped using primers GGT2_LP, 5′-CCCTCCGGCTTTTTGTATATC-3′, and GGT2_RP, 5′-CAGATGAGAGTTTGACCACAGG-3′. Both genotyping experiments were done together with the SALK left border primer 5′-ATTTTGCCGATTTCGGAAC-3′. RT-PCR was done on ggt2-3 as this has not been described before. Total RNA was extracted from 3-week-old rosette leaves using the RNeasy plant mini kit (Qiagen), including on-column DNase treatment. RNA concentration was estimated spectrophotometrically (Nano Drop ND-1000; Thermo Scientific). cDNA was synthesized from 2 μg RNA using the iScript cDNA synthesis kit (Bio-Rad). RT-PCR was performed using primers GGT2_FP, 5′-CGATGGACGGTGTTCTGCAATAGG-3′, and GGT2_RP, 5′-CCGTATTCAGCGATTTGGCTTAGTG-3′. No transcript could be found in leaves of either wild-type or ggt2-3 plants (Figure 2).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: GGP1, At4g30530; GGP3, At4g30550; GGT1, At4g39640; GGT2, At4g39650.
Acknowledgments
We thank Carl Erik Olsen for performing liquid chromatography–mass spectrometry and Erich Glawischnig for kindly providing seeds of ggt1 and ggt2 mutants. This study was supported by the Danish National Research Foundation (Grant DNRF99).
Acknowledgments
M.E.M. performed the experiments. All authors contributed to writing the Letter.
References
- Ahuja I., Kissen R., Bones A.M. (2012). Phytoalexins in defense against pathogens. Trends Plant Sci. 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Bednarek P. (2012). Sulfur-containing secondary metabolites from Arabidopsis thaliana and other Brassicaceae with function in plant immunity. ChemBioChem 13: 1846–1859 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Schneider B., Svatos A., Oldham N.J., Hahlbrock K. (2005). Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 138: 1058–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum R., Beck A., Korte A., Stengel A., Letzel T., Lendzian K., Grill E. (2007). Function of phytochelatin synthase in catabolism of glutathione-conjugates. Plant J. 49: 740–749 [DOI] [PubMed] [Google Scholar]
- Böttcher C., Westphal L., Schmotz C., Prade E., Scheel D., Glawischnig E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby K.J., Kumar P., Kliebenstein D.J. (2004). Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 38: 473–486 [DOI] [PubMed] [Google Scholar]
- Destro T., Prasad D., Martignago D., Bernet I.L., Trentin A.R., Renu I.K., Ferretti M., Masi A. (2011). Compensatory expression and substrate inducibility of γ-glutamyl transferase GGT2 isoform in Arabidopsis thaliana. J. Exp. Bot. 62: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti M., Destro T., Tosatto S.C.E., La Rocca N., Rascio N., Masi A. (2009). γ-Glutamyl transferase in the cell wall participates in extracellular glutathione salvage from the root apoplast. New Phytol. 181: 115–126 [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Møldrup M.E., Böttcher C., Olsen C.E., Scheel D., Halkier B.A. (2011). Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23: 2456–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geu-Flores F., Nielsen M.T., Nafisi M., Møldrup M.E., Olsen C.E., Motawia M.S., Halkier B.A. (2009). Glucosinolate engineering identifies a γ-glutamyl peptidase. Nat. Chem. Biol. 5: 575–577 [DOI] [PubMed] [Google Scholar]
- Glawischnig E. (2007). Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Ausubel F.M. (1994). Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc. Natl. Acad. Sci. USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull A.K., Vij R., Celenza J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan I.A., Hammerschmidt R. (2002). Arabidopsis ecotype variability in camalexin production and reaction to infection by Alternaria brassicicola. J. Chem. Ecol. 28: 2121–2140 [DOI] [PubMed] [Google Scholar]
- Martin M.N., Saladores P.H., Lambert E., Hudson A.O., Leustek T. (2007). Localization of members of the γ-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 144: 1715–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275: 33712–33717 [DOI] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Fukuyama K., Oliver D.J. (2009). Roles of γ-glutamyl transpeptidase and γ-glutamyl cyclotransferase in glutathione and glutathione-conjugate metabolism in plants. In Advances in Botanical Research, J. Jean-Pierre, ed (London, United Kingdom: Academic Press), pp. 87–113
- Ohkama-Ohtsu N., Radwan S., Peterson A., Zhao P., Badr A.F., Xiang C., Oliver D.J. (2007b). Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 49: 865–877 [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N., Zhao P., Xiang C., Oliver D.J. (2007a). Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 49: 878–888 [DOI] [PubMed] [Google Scholar]
- Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. (2007). Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Saga H., Ogawa T., Kai K., Suzuki H., Ogata Y., Sakurai N., Shibata D., Ohta D. (2012). Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol. Plant Microbe Interact. 25: 684–696 [DOI] [PubMed] [Google Scholar]
- Saito K., Hirai M.Y., Yonekura-Sakakibara K. (2008). Decoding genes with coexpression networks and metabolomics - ‘Majority report by precogs’. Trends Plant Sci. 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Schuhegger R., Nafisi M., Mansourova M., Petersen B.L., Olsen C.E., Svatos A., Halkier B.A., Glawischnig E. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M.A., Werck-Reichhart D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Su T.B., Xu J.A., Li Y.A., Lei L., Zhao L., Yang H.L., Feng J.D., Liu G.Q., Ren D.T. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23: 364–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K., Hiratake J., Irie M., Okada T., Yamada C., Kumagai H., Suzuki H., Fukuyama K. (2008). Crystal structures of Escherichia coli γ-glutamyltranspeptidase in complex with azaserine and acivicin: novel mechanistic implication for inhibition by glutamine antagonists. J. Mol. Biol. 380: 361–372 [DOI] [PubMed] [Google Scholar]
- Wang M.-Y., Liu X.-T., Chen Y., Xu X.-J., Yu B., Zhang S.-Q., Li Q., He Z.-H. (2012). Arabidopsis acetyl-amido synthetase GH3.5 involvement in camalexin biosynthesis through conjugation of indole-3-carboxylic acid and cysteine and upregulation of camalexin biosynthesis genes. J. Integr. Plant Biol. 54: 471–485 [DOI] [PubMed] [Google Scholar]
- Weng J.-K., Philippe R.N., Noel J.P. (2012). The rise of chemodiversity in plants. Science 336: 1667–1670 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Glazebrook J. (1999). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J., Facchini P.J. (2008). Alkaloid biosynthesis: Metabolism and trafficking. Annu. Rev. Plant Biol. 59: 735–769 [DOI] [PubMed] [Google Scholar]