This work identifies direct binding targets of the master ripening regulator RIN, a MADS box transcription factor, and examines RIN-based transcriptional regulation, finding that RIN has positive and negative regulatory roles throughout tomato fruit ripening, including in lycopene accumulation, ethylene production, chlorophyll degradation, and many other physiological processes.
Abstract
The fruit ripening developmental program is specific to plants bearing fleshy fruits and dramatically changes fruit characteristics, including color, aroma, and texture. The tomato (Solanum lycopersicum) MADS box transcription factor RIPENING INHIBITOR (RIN), one of the earliest acting ripening regulators, is required for both ethylene-dependent and -independent ripening regulatory pathways. Recent studies have identified two dozen direct RIN targets, but many more RIN targets remain to be identified. Here, we report the large-scale identification of direct RIN targets by chromatin immunoprecipitation coupled with DNA microarray analysis (ChIP-chip) targeting the predicted promoters of tomato genes. Our combined ChIP-chip and transcriptome analysis identified 241 direct RIN target genes that contain a RIN binding site and exhibit RIN-dependent positive or negative regulation during fruit ripening, suggesting that RIN has both activator and repressor roles. Examination of the predicted functions of RIN targets revealed that RIN participates in the regulation of lycopene accumulation, ethylene production, chlorophyll degradation, and many other physiological processes. Analysis of the effect of ethylene using 1-methylcyclopropene revealed that the positively regulated subset of RIN targets includes ethylene-sensitive and -insensitive transcription factors. Intriguingly, ethylene is involved in the upregulation of RIN expression during ripening. These results suggest that tomato fruit ripening is regulated by the interaction between RIN and ethylene signaling.
INTRODUCTION
The fruit ripening developmental program dramatically changes the characteristics of fleshy fruits, including color, texture, flavor, and aroma. In general, fruit color changes during ripening because of the accumulation of pigments such as carotenoids and anthocyanins and by degradation of chlorophylls. Also, fruit firmness decreases because of cell wall degradation. The constituents of fruits, such as vitamins, sugars, and aroma and flavor components, also develop during ripening. Fully ripe fruits become edible and attractive (i.e., sweet, soft, juicy, and nourishing) to humans and animals that help to disseminate seeds. Moreover, ripening is crucial for the quality of cultivated fruits. Therefore, elucidating the regulatory mechanisms of fruit ripening may provide beneficial information for improving breeding strategies, evaluating cultivation conditions and optimizing postharvest storage methods, and ultimately for improving and maintaining fruit quality.
Fruit ripening is controlled by an intricate genetically programmed mechanism that is driven by the action of phytohormones and developmental factors and affected by environmental stimuli, such as temperature and light (Dumas et al., 2003). Ethylene is one of the most important ripening-promoting factors, especially for climacteric-type fruits, which show a rapid increase in ethylene levels followed by a rise in respiration rate at the onset of the ripening. Climacteric ethylene induces and promotes many physical and physiological changes, such as pigment accumulation and fruit softening. Furthermore, recent studies have identified a number of transcription factors (TFs) required for the initiation and promotion of fruit ripening. For example, in tomato (Solanum lycopersicum), RIPENING INHIBITOR (RIN; also called MADS-RIN), NON-RIPENING (NOR; also called NAC-NOR), and COLORLESS NON-RIPENING (CNR; also called SPL-CNR) have been identified as key TF genes for the regulation of fruit ripening (Vrebalov et al., 2002; Manning et al., 2006; Giovannoni, 2007). Mutations in these TF genes inhibit fruit ripening steps, such as softening, color development, and the climacteric rise in respiration rate and ethylene (Tigchelaar et al., 1978; Thompson et al., 1999; Vrebalov et al., 2002; Eriksson et al., 2004; Manning et al., 2006; Giovannoni, 2007). These TFs are thus believed to lie upstream of ethylene-mediated and -independent ripening regulatory pathways and to regulate, directly and indirectly, the expression of numerous ripening-related genes (Giovannoni, 2007). In addition, several TF genes, including AGAMOUS-LIKE1 (TAGL1), HD-ZIP HOMEOBOX PROTEIN-1 (HB-1), APETALA2a (AP2a), ETHYLENE RESPONSE FACTOR6 (ERF6), and two FRUITFULL homologs (TDR4 and MBP7), have been recently found to play significant roles in fruit ripening in tomato (Lin et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009; Chung et al., 2010; Giménez et al., 2010; Pan et al., 2010; Karlova et al., 2011; Bemer et al., 2012; Lee et al., 2012; Zhou et al., 2012).
We focused our studies on understanding how RIN regulates tomato fruit ripening. RIN belongs to the SEPALLATA subfamily of MADS box TFs and is strongly expressed in a ripening-specific manner (Vrebalov et al., 2002). The rin mutation was identified as a deletion stretching over a part of the RIN gene and the intergenic region between RIN and the adjacent MADS box gene MACROCALYX (also called MADS-MC) (Vrebalov et al., 2002). The rin mutant bears unripe fruit in which expression of numerous ripening-related genes is inhibited (Osorio et al., 2011; Fujisawa et al., 2012). Recent studies using chromatin immunoprecipitation (ChIP) revealed that direct RIN target genes are involved in ethylene synthesis and signaling, cell wall modification, carotenoid accumulation, aroma formation, and transcriptional regulation of ripening-related TF genes in tomato, including NOR, CNR, TDR4, and HB-1 (Ito et al., 2008; Fujisawa et al., 2011, 2012; Martel et al., 2011; Qin et al., 2012). Based on these findings, RIN is considered to be a master regulator of fruit ripening. However, targets of RIN have not been comprehensively identified; therefore, our current understanding does not provide a complete picture of RIN-based regulation of ripening in tomato. To examine RIN regulation of ripening, we recently performed a microarray analysis of wild-type and rin mutant tomatoes, identifying hundreds of genes whose expression was positively or negatively regulated by RIN (Fujisawa et al., 2012). These genes may be direct targets of RIN, but although several candidates were confirmed to be direct RIN targets, the relationship of most of these genes to RIN remains to be elucidated.
Here, we report the genome-wide identification of direct RIN target genes by a ChIP assay coupled with DNA microarray analysis (ChIP-chip) for putative promoter regions of all tomato predicted genes. Our combined ChIP-chip and transcriptome analysis identified 241 direct RIN targets that exhibit RIN-dependent positive or negative regulation during fruit ripening. Annotation of the functions of these targets revealed that RIN functions in a wide range of ripening processes, especially in lycopene accumulation. We also demonstrated that ethylene participates in the regulation of the expression of RIN and its target TFs during fruit ripening. Based on these results, we propose a revised regulatory model for tomato fruit ripening; this model describes the regulation of ripening in tomato by the robust interaction between RIN and ethylene signaling.
RESULTS
Large-Scale Identification of RIN Binding Sites and Potential Direct RIN Target Genes by ChIP-chip
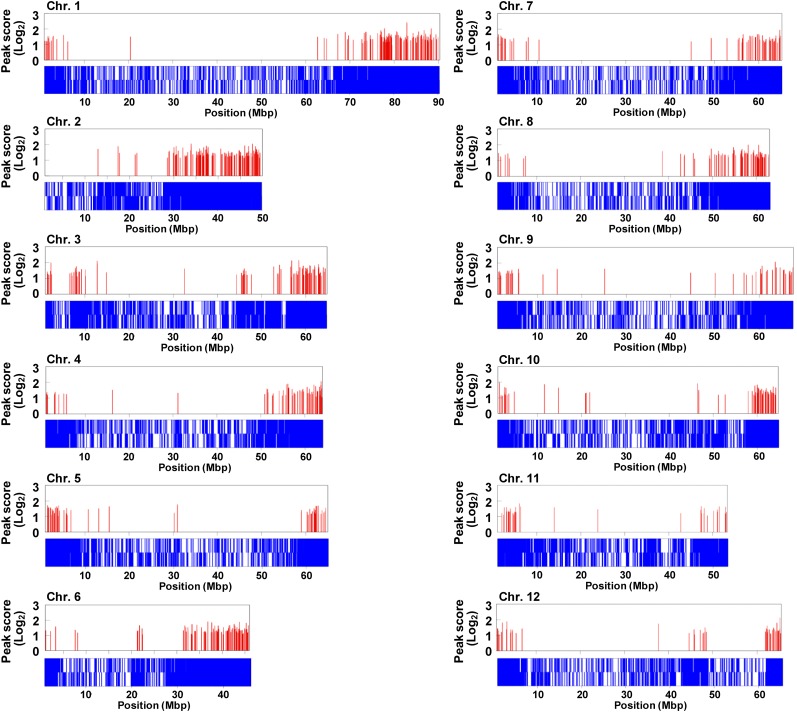
To identify, at the large scale, direct RIN target genes whose promoters are bound by RIN, we performed a ChIP-chip analysis using anti-RIN antibodies and a Roche NimbleGen custom microarray carrying probes designed for the putative promoters (2-kb upstream regions from the translation initiation sites) of the 35,802 predicted genes provided by the International Tomato Annotation Group (ITAG2). The chromatin for ChIP was prepared from wild-type tomato fruits at the ripening stage and immunoprecipitated with anti-RIN antibodies. The ChIPed and input (no ChIP) DNAs from three biologically independent preparations were amplified by random priming to prepare enough DNA for the microarray analysis. The quality of the ChIPed DNAs was confirmed by quantitative ChIP-PCR (qChIP-PCR) testing for enrichment of a known RIN binding sequence, the promoter region of the gene for 1-aminocyclopropane-1-carboxylic acid synthase (ACS2; Ito et al., 2008; Fujisawa et al., 2011) (see Supplemental Figure 1 online). We then hybridized the ChIPed and input DNAs to the NimbleGen microarrays. In each experiment, we calculated the log2-scale fold change (FC) values of the ChIPed DNA relative to the input DNA for each probe and detected ChIP-chip peak regions, which were defined by consecutively arranged probes with high FC at statistically significant levels (false discovery rate [FDR] ≤ 0.05) (for more details, see Methods). We regarded a ChIP-chip peak region that was detected in at least two of the three independent experiments as a RIN binding site. Using these criteria, we identified 1046 RIN binding sites, each of which was assigned the average FC value, ranging from 0.9 to 3.1 (corresponding to 1.9- to 8.6-fold enrichment), as a peak score (see Supplemental Table 1 online). The average width of these sites was 469 bp. Of the sites, 1002 were mapped in silico onto the 12 tomato chromosomes (Figure 1), whereas the remaining sites were mapped onto chromosome 0 (the Solanaceae Genomics Network [http://solgenomics.net] identifier Solyc00), which is an artificial pseudomolecule composed of scaffolds that could not be placed on either of the 12 tomato chromosomes (Tomato Genome Consortium, 2012). Using the information about genomic positions of the RIN binding sites, we identified 1200 genes as potential direct RIN targets that carried one or more RIN binding sites in the transcriptional regulatory region (2-kb upstream putative promoter) or in other gene regions, such as exons or introns, or downstream from the translation termination site (1 kb), in the region of overlap with the promoter region of a neighboring gene (Table 1; see Supplemental Data Set 1 online).
Figure 1.
Distribution of the RIN Binding Sites Detected by ChIP-chip on the Tomato Chromosomes.
Genomic positions of the RIN binding sites on the 12 tomato chromosomes are indicated by red bars with the log2 scale peak score. Positions of the promoters (top, forward strand; bottom, complementary strand) where ChIP-chip probes were designed are indicated by blue bars.
Table 1. Distribution of the RIN Binding Sites and the Number of Potential Direct RIN Target Genes.
| The No. of RIN Binding Sites | (%) | The No. of Genes Containing a RIN Binding Site | (%) | |
|---|---|---|---|---|
| Total | 1046 | (100) | 1200 | (100) |
| Region | ||||
| 2-kb upstream | 1046 | (100) | 1090 | (91) |
| > Exon | 55 | (5) | 51 | (4) |
| > Intron | 5 | (0.5) | 5 | (0.4) |
| > 1-kb downstream | 88 | (8) | 87 | (7) |
| RIN-binding sites | ||||
| > With CArG box | 620 | (59) | 718 | (60) |
| > C(C/T)(A/T)6(A/G)G | 380 | (36) | 437 | (36) |
| > C(A/T)8G | 423 | (40) | 489 | (41) |
| > C(C/T)(A/T)G(A/T)4(A/G)G | 130 | (12) | 174 | (15) |
| > Without CArG box | 426 | (41) | 539 | (45) |
Detection of Conserved DNA Sequences in the RIN Binding Sites
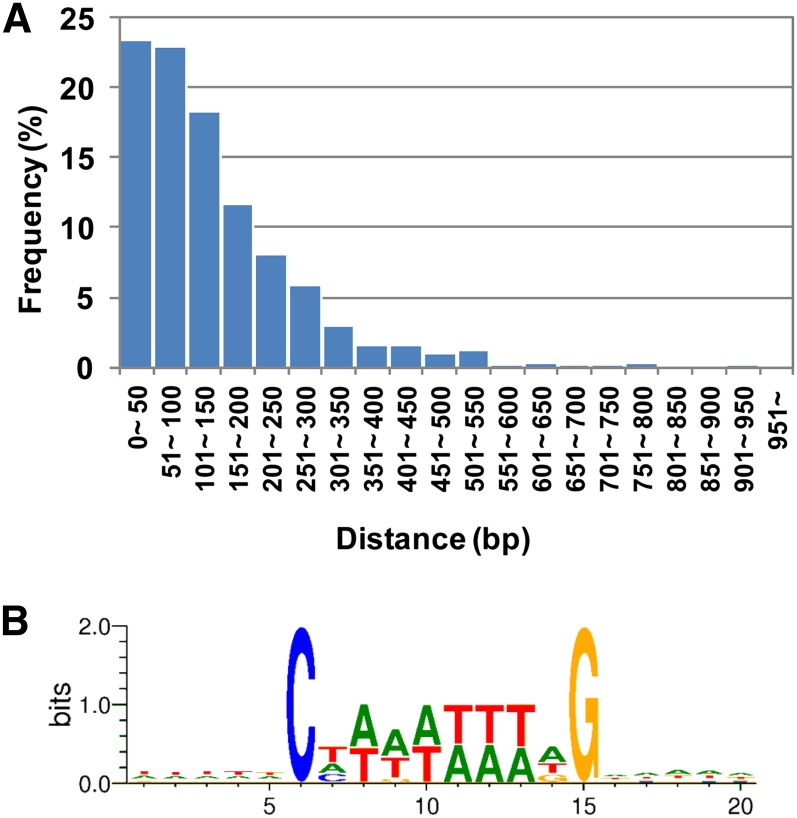
Previously, our in vitro analysis revealed that RIN binds to DNA sequences known as the C-A/T-rich-G (CArG) box, which is the general target of MADS box proteins (Ito et al., 2008). To detect conserved DNA sequences in the RIN binding sites in vivo, we first searched the RIN binding sites for CArG-box sequences. As summarized in Table 1, we found that a total of 620 (59%) RIN binding sites contained a DNA sequence that met the patterns for any of the possible three RIN biding CArG-box motifs, C(C/T)(A/T)6(A/G)G, C(A/T)8G, or C(C/T)(A/T)G(A/T)4(A/G)G (Ito et al., 2008; Fujisawa et al., 2011). The 620 sites were located in the 2-kb promoters or other gene regions of 718 potential target genes (see Supplemental Data Set 1 online). The frequency of occurrence of the motif, C(C/T)(A/T)6(A/G)G, which seems to be preferentially bound by RIN (Ito et al., 2008), was significantly higher in the RIN binding sites than in the whole set of 2-kb promoters (P < 0.001 by χ2 test), whereas the frequency of the other two motifs was not (see Supplemental Table 2 online). In the 620 RIN binding sites, the CArG boxes tended to be located closer to the centers (Figure 2A). To predict the in vivo RIN binding CArG-box motif, we cataloged and analyzed CArG-box sequences that lie in the 2-kb promoters along with the strands encoding the genes. The analysis of the collected 1063 strand-oriented CArG-box sequences yielded a 10-bp motif consensus sequence, C(T/A/C)(A/T)6(A/T/G)G (Figure 2B). Unlike a previous in vitro assay for RIN binding (Ito et al., 2008), no conservation was observed in the flanking regions upstream and downstream of the 10-bp core.
Figure 2.
Position and Conservation of the CArG Boxes in the RIN Binding Sites.
(A) Histogram of the distance between the center of the RIN binding sites and CArG boxes found in the sites.
(B) The consensus sequence of the CArG boxes in the RIN binding sites of the gene promoters.
Furthermore, we searched the RIN binding sites for known plant TF binding sequences, based on the motifs deposited in the Arabidopsis Gene Regulatory Information Server database (Davuluri et al., 2003). Sixteen out of the 87 examined motifs exhibited a significantly (P < 0.001) higher frequency of occurrence in the RIN binding sites than that expected from the 2-kb promoters of the set of all predicted genes (see Supplemental Table 2 online). In addition to the CArG-box–type motifs, a remarkably high frequency of occurrence was observed for the GCC-box motif (GCCGCC), although the numbers of these motifs in the RIN binding sites were much fewer than the CArG boxes. The GCC-box motif is a target of the AP2/ERF TF family proteins (Stockinger et al., 1997; Shinozaki and Yamaguchi-Shinozaki, 2000; Hao et al., 2002). Unfortunately, we could not detect any significantly conserved DNA motifs, even the CArG-box, in the RIN binding sites using authentic programs for de novo motif discovery.
Combined ChIP-chip and Transcriptome Analysis for Identification of RIN Direct Target Genes Whose Expression Was Positively or Negatively Regulated by RIN during Ripening
In our earlier study, to select genes with RIN-dependent expression, we calculated the gene expression FC ratio of the ripening (pink coloring [P]) stage relative to the preripening (mature green [G]) stage (FCWT for the wild type and FCrin for the rin mutant; for more details, see Methods) from a microarray data set for tomato fruit ripening (Fujisawa et al., 2012). We also calculated the expression change score (ECS), defined as the ratio of FCWT relative to FCrin, and used the ECS to evaluate the dependency of gene expression on the wild-type RIN gene.
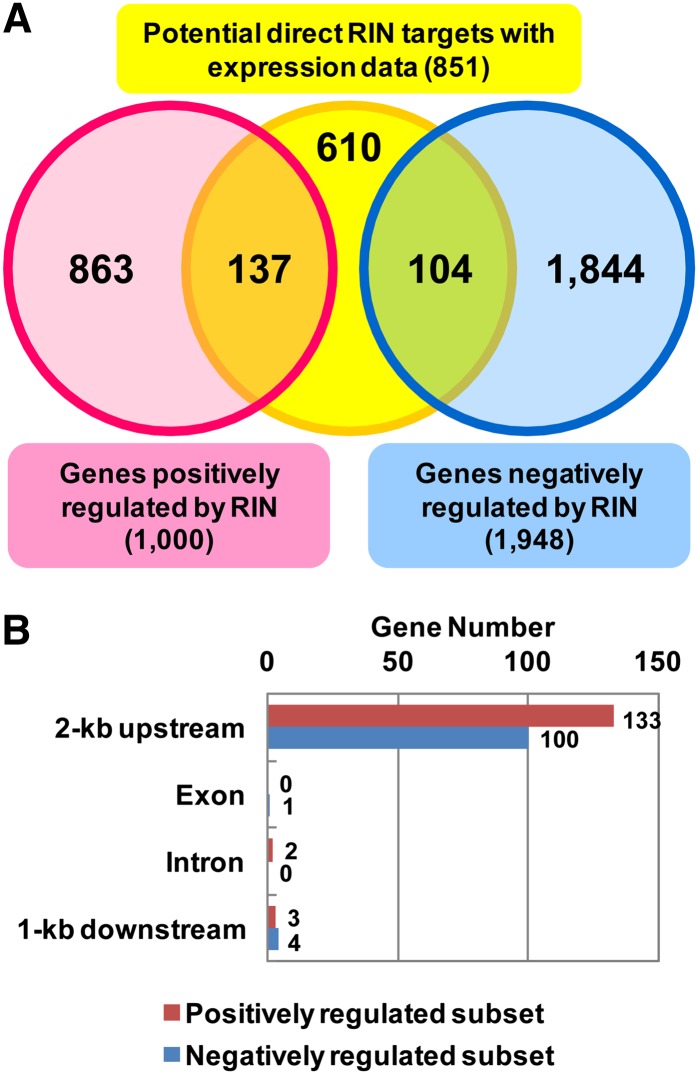
In this study, we reevaluated our microarray expression data, using lower FC and ECS stringency thresholds, to identify RIN-dependent targets more broadly. Using a threshold of FCWT > 2 (q-value < 0.05) and ECS > 2, we selected 1000 of the 17,307 genes with valid expression data as genes positively regulated by RIN. We next examined whether the set of genes positively regulated by RIN overlapped with the identified RIN direct binding targets. Of the 1200 potential targets, 851 were covered in the microarray expression data, but the remaining 349 were not because there were no probes specific to these loci in the microarray. Of the potential targets, 137 were included in the set of positively regulated genes (Figure 3A; see Supplemental Data Set 2 online). Similarly, we selected 1948 genes negatively regulated by RIN using a threshold of FCWT < 0.5 (q-value < 0.05) and ECS < 0.5. Of the potential targets, 104 were included in the negatively regulated gene set and then identified as negatively regulated direct targets of RIN (Figures 3A and 3B; see Supplemental Data Set 2 online). The remaining 863 positively regulated and 1844 negatively regulated genes were not selected as direct RIN targets and may be regulated by an indirect effect of RIN. Furthermore, 610 of the 851 potential targets were also not selected despite RIN binding to a site close to them because their expression changes did not meet the criteria for selection and thus are likely not transcriptionally regulated by RIN.
Figure 3.
Identification of Direct RIN Targets Whose Expression Was Regulated by RIN.
(A) Venn diagram of potential direct RIN targets and genes positively and negatively regulated by RIN selected by microarray analysis.
(B) Distribution of RIN binding sites in the gene regulatory and transcribed regions of the positively and negatively regulated subsets.
Detection of Known and Novel Ripening-Related Direct RIN Targets
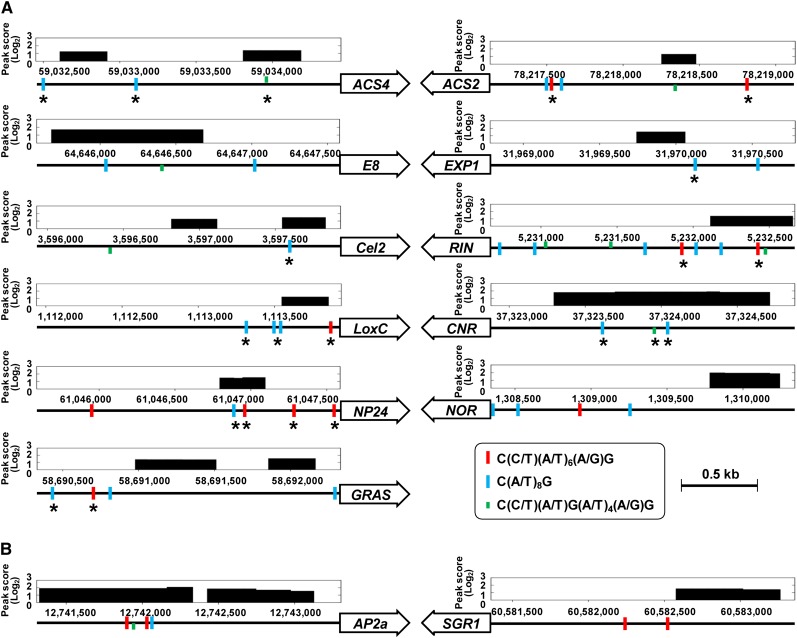
To test whether our combined ChIP-chip and expression analysis was capable of detecting RIN targets, we first determined whether known targets of RIN were identified in the analysis. The positively and negatively regulated subsets of direct RIN targets included 11 out of 24 known RIN target genes (Ito et al., 2008; Fujisawa et al., 2011, 2012; Martel et al., 2011; Qin et al., 2012) (see Supplemental Data Set 2 online). Five more known direct RIN targets, genes for β-d-xylosidase (XYL1), alcohol dehydrogenase (ADH2), phytoene synthase (PSY1), TDR4, and HB-1, were also included in the set of identified genes when ChIP-chip peak detection was conducted at a lower stringency (FDR ≤ 0.2) (see Supplemental Figure 2 online). Of the 11 known targets, nine were analyzed by qChIP-PCR in our earlier studies (Fujisawa et al., 2011, 2012). We found overlaps between the RIN binding sites and the already confirmed RIN binding CArG boxes in five (genes for 1-aminocyclopropane-1-carboxylic acid synthase [ACS4], an endo-β-1,4-glucanase [Cel2], an osmotin-like protein [NP24], CNR, and RIN itself) of the nine targets (Figure 4A). In the remaining targets detected, the RIN binding sites did not overlap with but were located near (within 400 bp) the enriched CArG boxes already confirmed (Figure 4A). The results indicated that our ChIP-chip would identify successfully a large portion of the direct RIN targets in the tomato genome.
Figure 4.
RIN Binding Sites in the Promoters of Ripening-Associated Genes.
RIN binding sites detected by ChIP-chip in the promoters of known direct RIN targets (A) and previously unidentified direct RIN targets (B) involved in fruit ripening. Genomic position and log2 scale peak score of each RIN binding site is indicated above the 2-kb gene promoters (horizontal lines). Boxed arrows with gene identifier indicate the orientation of genes. Thin vertical lines indicate the positions of three types of CArG boxes in the promoter. Asterisks below the thin vertical lines in (A) indicate the position of CArG boxes that were confirmed to be enriched by the previous qChIP-PCR.
The set of genes identified as direct RIN targets in this study did not include more than half of known direct RIN targets due to the following reasons. A known target, a gene for hydroperoxide lyase (HPL), was identified as a potential direct RIN target by our ChIP-chip, but its expression levels were not altered enough in either the wild type or rin mutant fruits to meet the criteria for selection. In addition, another known RIN target, a gene for Never-ripe (NR; also called ETR3), was also not detected in this study because our ChIP-chip microarray did not cover the RIN binding site likely located beyond 2 kb upstream of the first exon of NR (Martel et al., 2011). Our failure to detect the other known targets might be caused by a difference in sensitivity between quantitative PCR–based assays and microarray-based ChIP assays (as mentioned in the next section), or by the ChIP-chip microarray, if it does not cover the entire functional promoter regions.
Based on gene annotations, on the other hand, we found that the positively regulated RIN targets included two important ripening-related genes for senescence-inducible chloroplast stay-green protein 1 (SGR1) and AP2a (Figure 4B). By contrast, many ripening-associated genes (e.g., genes for 1-aminocyclopropane-1-carboxylic acid oxidase [ACO1], 1-d-deoxyxylulose 5-phosphate synthase [DXS], and geranylgeranyl pyrophosphate synthase [GGPS2]; Fujisawa et al., 2012) were not identified as direct RIN targets, although their expression patterns are RIN dependent. It is likely that these genes are not directly regulated by RIN during ripening but are regulated by other factors, such as ethylene signaling or RIN-regulated TFs.
Verification of RIN Binding to the Sites Detected by ChIP-chip
We used diagnostic qChIP-PCR to verify RIN binding to the sites detected by ChIP-chip. As representatives, we monitored the enrichment of RIN binding sites that showed the 10 highest ChIP-chip peak scores among the sites mapped onto the 12 tomato chromosomes (excluding the sites of chromosome 0 [Solyc00]). The highest peak score among those of the sites mapped onto the 12 chromosomes was 2.56 (see Supplemental Data Set 3 online). The qChIP-PCR amplified the DNA fragments of nine of the 10 sites but not that of the remaining one, likely due to the difficulty of PCR amplification of the site. All of the amplified sites showed enrichment of close to or more than twofold by qChIP-PCR (see Supplemental Figure 3 online). A positive correlation between ChIP-chip peak scores and qChIP-PCR enrichments was confirmed (Pearson’s r = 0.78), indicating that our ChIP-chip results are reliable. The scores determined by ChIP-chip were lower than the enrichment levels measured by qChIP-PCR; this may be attributable to the differences in dynamic range between the two analyses. This lower sensitivity of ChIP-chip compared with qChIP-PCR might also cause the failure to identify some known targets in the set of direct RIN targets by ChIP-chip, as described above.
Furthermore, we verified RIN binding to the sites in the promoters of the negatively regulated subset. As described above, the combined ChIP-chip and transcriptome analysis detected RIN binding sites in the promoters or other gene regions of both positively and negatively regulated RIN targets, suggesting both activator and repressor roles of RIN. Whereas many positive targets have been reported previously (Ito et al., 2008; Fujisawa et al., 2011, 2012; Martel et al., 2011; Qin et al., 2012), HB-1 is the only reported negatively regulated target (Martel et al., 2011). To verify RIN binding to the sites of the negatively regulated subset, we used diagnostic qChIP-PCR to monitor the enrichment level of RIN binding sites in the promoters of three randomly selected negatively regulated direct RIN targets. All sites examined were indeed enriched in the ChIPed DNA relative to the input at similar levels to those detected by ChIP-chip (see Supplemental Figures 4A and 4B online). Taken together, our ChIP-chip results were generally reproducible, regardless of positive or negative regulation by RIN.
Predicted Functions of the Direct RIN Target Genes
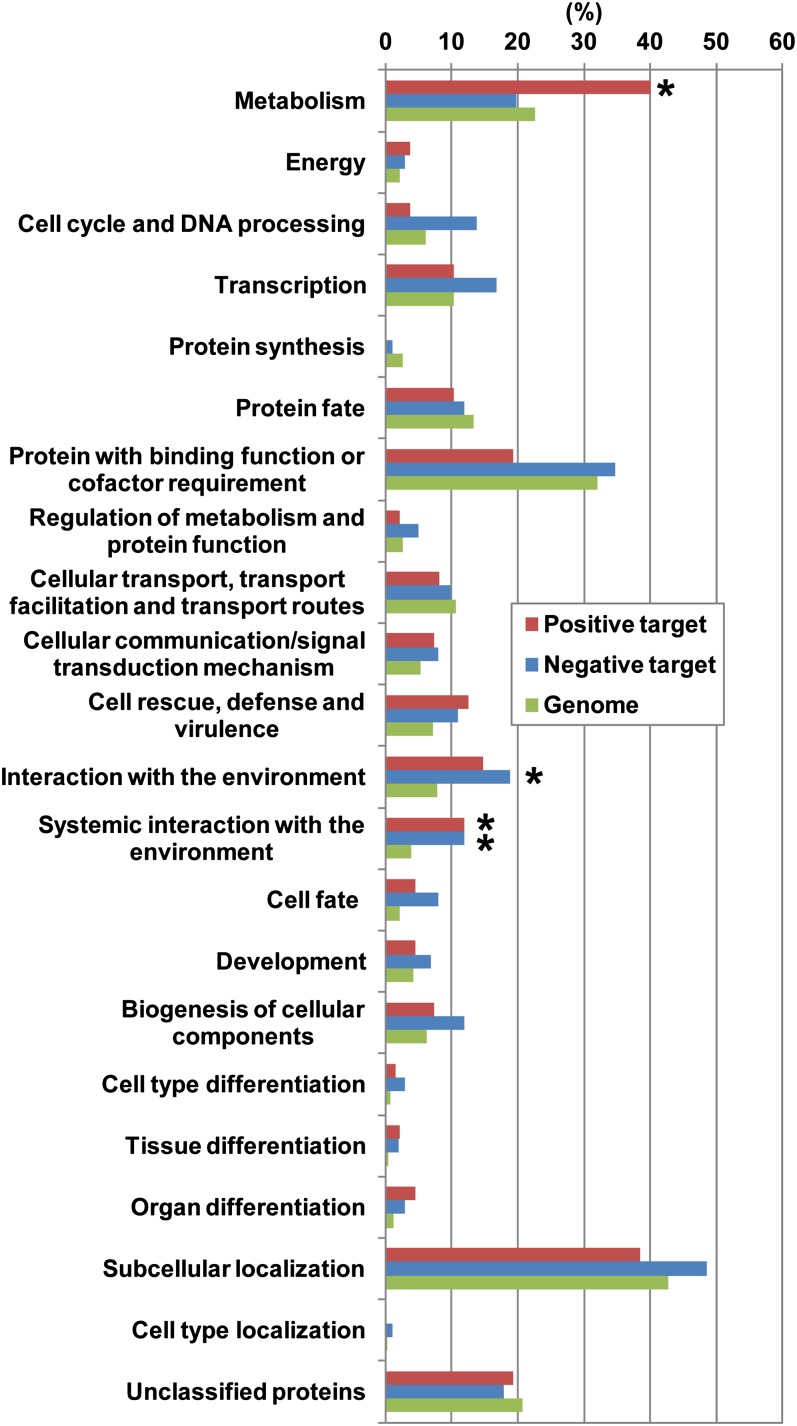
To provide an overview of the functional roles of RIN, we next estimated the functions of the direct RIN targets by searching for similarity to Arabidopsis thaliana proteins and then classified these proteins based on their predicted biological pathways. We found that the products of the direct RIN targets were involved in a total of 67 biological pathways, especially rich in “metabolic pathways,” including “biosynthesis of secondary metabolites” (see Supplemental Data Set 4 online). We also classified the direct RIN targets using the Munich Information Center for Protein Sequences (MIPS) functional catalog database based on the similarity to the Arabidopsis genes (see Supplemental Data Set 5 online). As found for the pathway search described above, the MIPS classification also showed that RIN targets were implicated in a wide range of fruit ripening processes. In particular, the classification of the positively regulated subset showed a significantly (P < 0.001 by Fisher’s exact test) higher frequency of genes involved in two functional categories, “metabolism” and “systemic interaction with the environment,” than the frequency in the whole genome (Figure 5). In the “metabolism” category, genes involved in the subcategories related to Met metabolism were significantly enriched in the positively regulated subset (see Supplemental Data Set 6 online). Because Met is a precursor of ethylene, this enrichment is likely associated with ethylene production during ripening. In addition to known direct RIN targets involved in ethylene synthesis (ACS2, ACS4, and E8), the positively regulated subset included two genes for homocysteine S-methyltransferase (HMTs), which belong to the S-adenosyl-Met-homocysteine cycle upstream of an ethylene synthesis pathway, and a gene for 1-aminocyclopropane-1-carboxylic acid oxidase (ACO6) catalyzing the ethylene synthesis reaction (annotated as the categories related to the Met metabolism; see Supplemental Data Sets 5 and 6 online). Furthermore, in the category of “systemic interaction with the environment,” genes belonging to such subcategories as “plant defense response” and “response to wounding” were also significantly enriched in the positively regulated subset (see Supplemental Data Set 6 online). These subcategories, which were associated with various stress responses, are frequently observed in tomato fruit ripening (Fujisawa et al., 2012). By contrast, the negatively regulated subset showed a significantly higher frequency of genes classified into the categories “interaction with the environment” and “systemic interaction with the environment” than the frequency in the whole genome (Figure 5; see Supplemental Data Set 6 online). Both categories shared the negatively regulated genes involved in response to plant hormones such as auxin and gibberellin (see Supplemental Data Sets 5 and 6 online).
Figure 5.
Functional Classification of the Direct RIN Targets.
For the analysis, we used genes that were assigned MIPS information based on similarity to Arabidopsis genes (135 positively regulated targets, 101 negatively regulated targets, and 28,440 genes as whole genome). Bars represent the ratio of the genes included in the categories. Asterisks indicate a significant enrichment (P < 0.001 by Fisher's exact test) compared with the genome.
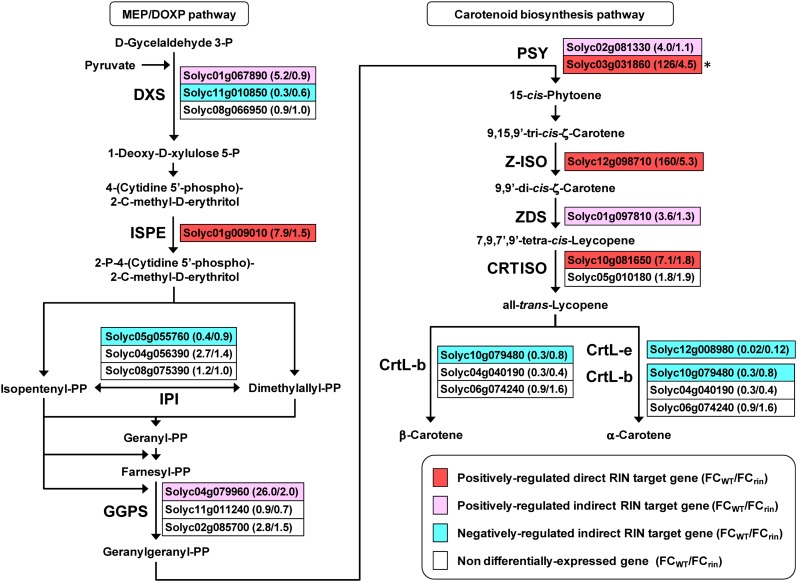
Pathway Analysis for Carotenoid Synthesis Based on the Expression Profiles of RIN Targets
Our functional estimation found that a number of the direct RIN target genes were implicated in lycopene accumulation, a process characteristic of tomato fruit ripening. To analyze how RIN participates in lycopene accumulation during ripening, we mapped the direct RIN targets and their isozyme genes with their expression profiles on the carotenoid pathway and the 2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate (MEP/DOXP) pathway (also called the nonmevalonate pathway). The MEP/DOXP pathway is one of the terpenoid backbone biosynthesis pathways and is considered to be the main route for plastidic carotenoid production during tomato fruit ripening (Rodriguez-Concepcion and Gruissem, 1999). We found that in the MEP/DOXP pathway, a gene for 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (ISPE; also called CDPMEK) was positively regulated by RIN in a direct manner (Figure 6). By contrast, DXS and GGPS2 were not included in the targets of RIN but were positively regulated in a RIN-dependent manner, suggesting that DXS and GGPS2 expressions are regulated by an indirect effect of RIN (Figure 6). Similarly, another DXS and a gene for isopentenyl pyrophosphate isomerase (IPI) were negatively regulated by an indirect effect of RIN (Figure 6). In the carotenoid pathway from phytoene to α- and β-carotenes, RIN positively regulates a known target, PSY1 (Martel et al., 2011), which encodes a rate-limiting enzyme of carotenoid synthesis (Fraser et al., 2002), although our ChIP-chip analysis surveying the 2-kb promoter regions could not detect any significantly enriched peaks (FDR ≤ 0.05) in the PSY1 promoter. Also, RIN positively regulated genes for ζ-carotene isomerase (Z-ISO) and carotenoid isomerase (CRTISO) in a direct manner. In addition, RIN positively regulated genes for another phytoene synthase (PSY2) and ζ-carotene desaturase (ZDS) and negatively regulated genes for lycopene β-cyclase (CrtL-b) and lycopene ϵ-cyclase (CrtL-e) by an indirect effect (Figure 6).
Figure 6.
A Diagram of the Terpenoid Backbone Synthesis and Carotenoid Biosynthesis Pathways with Expression Profiles of Direct and Indirect RIN Target Genes during Ripening.
Arrows indicate reaction flows in the pathways. The names of enzymes that catalyze the reaction are indicated on the left or below the arrows when the enzymes are encoded by direct or indirect RIN target genes. Boxes represent the predicted gene identifiers encoding the enzymes. FC value of the genes in the wild-type (FCWT) and rin mutant (FCrin) fruits during ripening are shown in parentheses. A known direct RIN target, PSY1 (Solyc03g031860; indicated by an asterisk), is also shown in red and was not detected by our ChIP-chip analysis. CRTISO, carotenoid isomerase; CrtL-b, lycopene β-cyclase; CrtL-e, lycopene ϵ-cyclase; DXS, 1-deoxy-d-xylulose 5-phosphate synthase; IPI, isopentenyl pyrophosphate isomerase; ISPE, 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase; P and PP, phosphate and pyrophosphate, respectively; PSY, phytoene synthase; ZDS, ζ-carotene desaturase; Z-ISO, ζ-carotene isomerase.
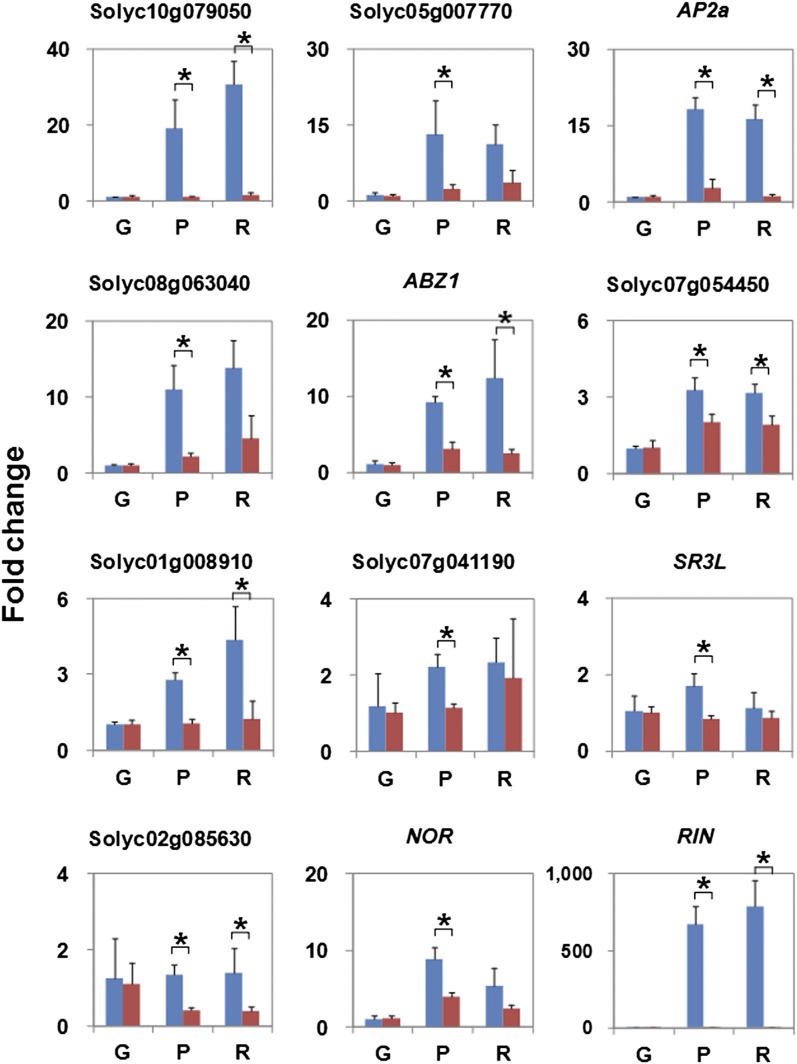
Expression Analysis of the TF Genes Directly Regulated by RIN
The positively and negatively regulated subsets of the direct RIN targets contained 14 and 17 TF genes, respectively, which were assigned a MIPS subcategory of “transcription” (see Supplemental Data Set 7 online). MADS box, NAC, SBP, and bZIP family genes were found in the positively regulated subset but not in the negatively regulated subset. By contrast, IAA and WRKY family genes were found in the negatively regulated subset but not in the positively regulated subset. Genes belonging to the AP2/ERF, bHLH, GRAS, HB, and zinc-finger families were found in both subsets, but the numbers of AP2/ERF family genes in the negatively regulated subset were relatively larger than those in the positively regulated subset. Apart from the known ripening regulator genes, such as RIN, NOR, CNR, and AP2a, there were no TF genes whose roles in fruit ripening have been characterized. In this study, we focused on the 14 positively regulated TF genes (Table 2) because they were expected to be associated with phenomena activated during ripening. Of the 14 TF genes, only four (CNR, NOR, a SCL32-like GRAS gene [Solyc07g052960], and RIN itself) were identified previously as direct RIN targets; the remaining 10 genes were newly identified as direct RIN targets (Table 2). Expression analysis by quantitative RT-PCR (qRT-PCR) revealed that nine targets (the RIN binding sites by ChIP-chip for the AP2a promoter are shown in Figure 4; the binding sites for the other eight targets are shown in Supplemental Figure 5 online) showed increased expression levels in the wild-type fruits for at least one of the ripening (P and red ripe [R]) stages relative to the levels at the preripening (G) stage. The increases were statistically significant (P value < 0.05) compared with the changes in the rin mutant fruits at the same age as observed in RIN and NOR (Figure 7). By contrast, the expression level of Solyc02g085630 during ripening was not increased in the wild-type fruits but decreased gradually in the rin mutant (Figure 7).
Table 2. TF Genes That Are Directly Targeted and Positively Regulated by RIN Found by ChIP-chip and Transcriptome Analysis in This Study.
| Microarray Gene Expression |
ChIP-chipb | ||||
|---|---|---|---|---|---|
| ITAG2 Gene ID | Description | FCWTc | FCrind | ECSe | Peak Score (Log2) |
| Solyc05g012020 | MADS box TF (RIN) | 847.2 | 70f | 12.1 | 1.40 |
| Solyc07g052960 | GRAS family TF (SCL32-like) | 683.5 | 6.8 | 100.6 | 1.43, 1.57 |
| Solyc10g079050a | BHLH TF (bHLH130 homolog) | 23.8 | 1.3 | 17.7 | 1.84 |
| Solyc05g007770a | NAC domain TF (NAP homolog) | 17.8 | 2.5 | 7.2 | 1.11 |
| Solyc08g063040a | Zinc-finger protein (IDD2 homolog) | 17.4 | 1.7 | 10.4 | 1.75, 1.65 |
| Solyc03g044300a | AP2-like ethylene-responsive TF (AP2a) | 15.8 | 1.7 | 9.0 | 1.96, 1.76 |
| Solyc01g100460a | BZIP TF (ABZ1) | 12.1 | 3.0 | 4.0 | 1.31 |
| Solyc10g006880 | NAC domain protein (NOR) | 9.4 | 4.6 | 2.1 | 1.97 |
| Solyc02g077920 | Squamosa promoter binding-like protein (C) | 4.8 | 1.7 | 2.8 | 1.87 |
| Solyc07g054450a | TF (fragment) ULTRAPETALA1 (ULT1 homolog) | 4.1 | 1.9 | 2.2 | 1.53, 1.57 |
| Solyc01g008910a | Scarecrow TF family protein (SCL3 homolog) | 4.0 | 1.1 | 3.7 | 1.50 |
| Solyc02g085630g | Homeobox-Leucine-zipper-like protein (HB40 homolog) | 2.9 | 0.3 | 9.1 | 1.52 |
| Solyc07g041190a |
Alfin-like 5 PHD finger family protein (AL5 homolog) | 2.5 | 1.2 | 2.1 | 1.44 |
| Solyc12g099340a | Calmodulin binding transcription activator 5 (SR3L) | 2.3 | 0.9 | 2.4 | 1.28 |
Direct RIN target genes newly identified.
All RIN binding sites presented here were located in the gene promoters. When there were two sites in a gene regulatory and transcribed region, both site values are listed.
FC value of a gene in the wild-type tomato fruits at the ripening (P) stage relative to that at the preripening (G) stage.
FC value of a gene in the rin mutant tomato fruits at the same age as the wild-type P stage relative to that at the same age as the wild-type G stage.
ECS is the ratio of FCWT to FCrin, representing the dependency of gene expression on RIN.
The apparent RIN expression in the rin mutant due to the expression of mutated rin gene.
Solyc02g085630 was eliminated later from the subset because qRT-PCR did not reproduce the ripening-induced expression obtained by the microarray analysis.
Figure 7.
Expression Targets during Ripening of Wild-Type and rin Mutant Fruits of TF Genes That Are Direct RIN Targets.
The change in expression level of the genes in the fruits at the ripening (P and R) stages is shown as FC relative to that in the G stage in the wild type and rin mutant. Bars represent the mean of three biological replicates. Error bars represent sd of the mean. Asterisks indicate a statistically significant (P < 0.05) difference in FC between the wild-type and rin mutant fruits at each P or R and G stage.
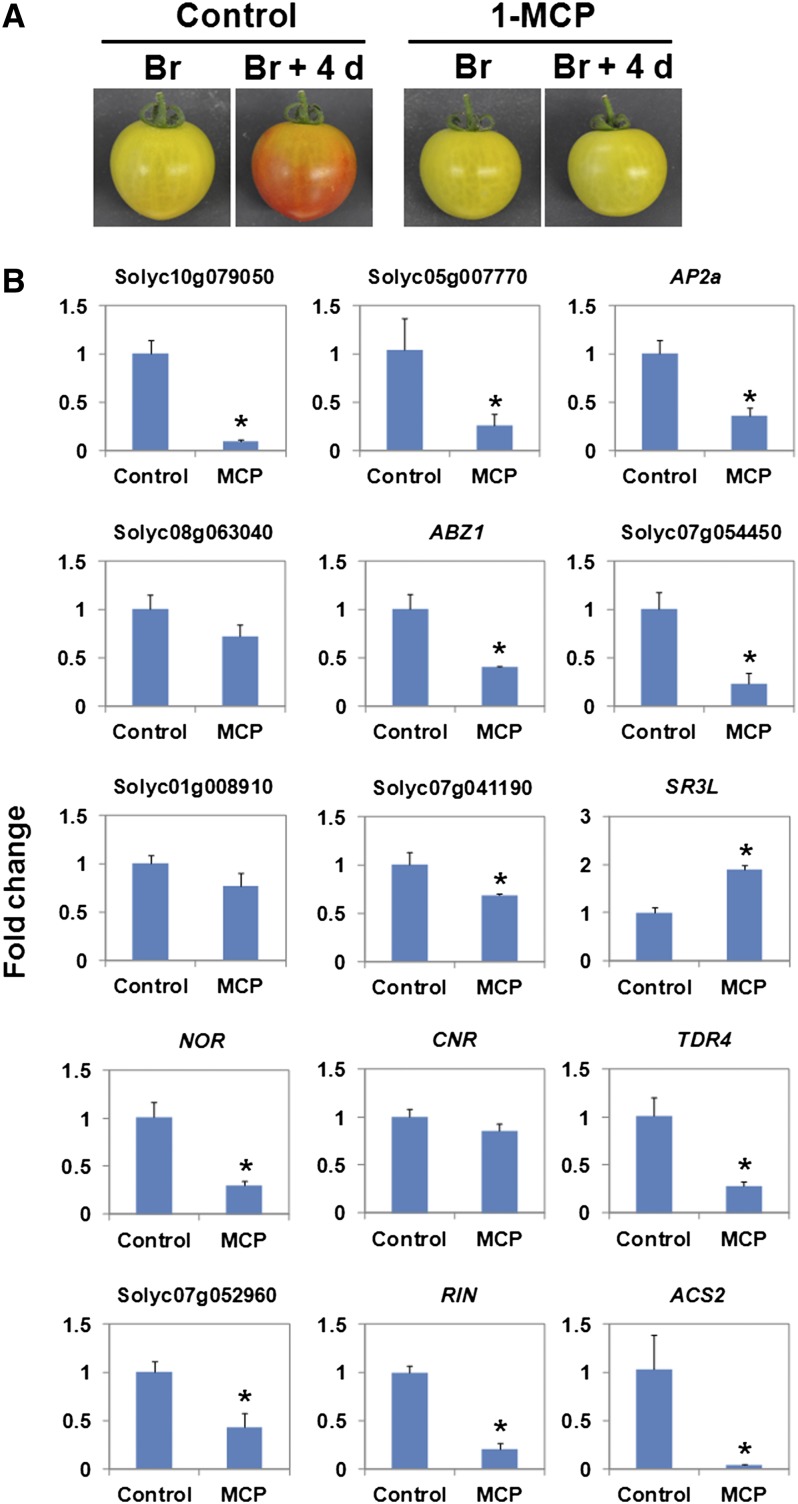
Ethylene Responsiveness of Positively Regulated Direct RIN Target TFs
To examine ethylene responsiveness of the TF genes that were identified as positively regulated direct targets of RIN in the previous and current studies, we monitored their expression levels in tomato fruits treated with an ethylene receptor inhibitor, 1-methylcyclopropene (1-MCP). Tomato fruits harvested at the breaker stage were treated with 1-MCP for 4 d and subjected to a qRT-PCR analysis of the nine novel and five known target TF genes. The treatment severely inhibited color development of tomato fruits during ripening (Figure 8A). The qRT-PCR confirmed that expression of an ethylene-inducible gene, ACS2, was suppressed in the 1-MCP–treated fruits compared with the control (Figure 8B), indicating the successful inhibition of ethylene signaling by the treatment. The qRT-PCR also showed that the treatment significantly decreased the expression of six novel target TFs and NOR, TDR4, Solyc07g052960, and RIN itself in the 1-MCP–treated tomato fruits (Figure 8B). In particular, the treatment substantially decreased the expression levels of nine TFs to 50% or less of the control. As for RIN, its expression level was decreased to 20% of the control (Figure 8B). By contrast, the treatment did not significantly change the expression levels of five target TFs, including CNR, whereas the level of a gene for calcium/calmodulin-regulated SR/CAMTA TF, SR3L, was increased (Figure 8B).
Figure 8.
Expression of the TF Genes That Are Direct RIN Targets in Tomato Fruits Treated with 1-MCP.
(A) Tomato fruits at the breaker (Br) stage and after 4 d of treatment (Br + 4 d) with 1-MCP. Fruits harvested at the Br stage and treated with water for 4 d were used as a control.
(B) Expression analysis of direct RIN target TF genes in the control and 1-MCP–treated tomato fruits. The change in expression levels of the genes in the fruits treated with 1-MCP is shown as FC relative to the control. Bars represent the mean of three biological replicates. Error bars represent sd of the mean. Asterisks indicate a statistically significant (P < 0.05) difference in FC between the 1-MCP–treated fruits and control.
DISCUSSION
Because the rin mutation causes a severe inhibition of tomato fruit ripening, RIN has been considered to be a pleiotropic regulator of diverse ripening processes. By combined ChIP-chip and transcriptome analysis, we revealed here that RIN possibly contributes to at least 67 metabolic pathways by direct transcriptional regulation of 241 genes, which explains the pleiotropic role of RIN. However, a large portion of the potential target genes does not apparently show a RIN-dependent expression change, at least under our experimental conditions. The presence of these unaffected targets has also been found in previous ChIP-chip experiments (Lee et al., 2007; Kaufmann et al., 2009; Oh et al., 2009; Zheng et al., 2009; Fujisawa et al., 2011; Tao et al., 2012). This result suggests that association with an additional factor may be necessary for RIN to induce transcription of direct RIN targets. It remains unclear what kind of factor and cis-element may assist the function of RIN, and a higher resolution survey of RIN binding sites will provide a clue to clarify this issue. In addition, the set of direct RIN targets in this study did not include more than half of the previously identified RIN targets, including some well-known, authenticated ripening-related genes, such as those encoding NR, E4, PSY1, TDR4, HB-1, and polygalacturonase (PG2A) or other cell wall–modifying proteins (Fujisawa et al., 2011, 2012; Martel et al., 2011). These genes were not identified in this study, possibly due to either the limitations of the genomic (2-kb upstream promoter) regions covered by the ChIP-chip microarray, the lower sensitivity of ChIP-chip compared with qChIP-PCR, or both. Five more known targets are included in the set of RIN targets identified at a lower stringency for ChIP-chip peak selection (FDR ≤ 0.2), suggesting that some direct RIN targets might have remained unidentified. For example, a TDR4 homolog, MBP7, might be an unidentified direct RIN target because a RIN binding site was detected in the MBP7 promoter at a lower stringency (FDR ≤ 0.2) and also because it has been suggested that the ripening-induced upregulation of MBP7 depends on RIN (Bemer et al., 2012). Nevertheless, our findings provide meaningful information about the RIN-mediated ripening regulatory mechanisms, as described below.
Structural Features of the RIN Binding Sites Detected by ChIP-chip
Our ChIP-chip analysis showed that 59% of the RIN binding sites contained CArG-box motifs. These CArG boxes were frequently observed close to the center of the CArG box–containing sites (Figure 2A), suggesting that these CArG boxes are possibly recognized by RIN. However, RIN binding to sites without a CArG box was also confirmed. Similar cases have been described in previous reports on ChIP-chip analyses for several types of Arabidopsis TFs (Lee et al., 2007; Kaufmann et al., 2009; Oh et al., 2009; Zheng et al., 2009; Tao et al., 2012), but the reason for detecting such sites remains unclear. We assume that RIN may bind to non-CArG-box motif sites via an additional DNA binding factor that forms a complex with RIN. Indeed, Arabidopsis MADS box proteins interact with various types of TFs and chromatin remodeling proteins in vivo (Smaczniak et al., 2012). Our results also showed that the RIN binding sites overrepresented a number of TF binding motifs, especially the GCC-box motif. The GCC box is recognized by ERFs (Fujimoto et al., 2000). The overrepresentation indicates the possibility that DNA binding factors, such as ERF family proteins, may form a complex with RIN and bring RIN to the genome sites with the DNA binding factor–specific motifs.
RIN Affects a Wide Range of Fruit Ripening Processes through Transcriptional Regulation of Key Genes
Our results demonstrated that direct targets of RIN are involved in various pathways for biosynthesis, metabolism, and signaling, such as carotenoid accumulation, ethylene synthesis, and chlorophyll degradation, suggesting the contribution of RIN to a wider range of fruit ripening processes than previously known. In particular, RIN appears to regulate carotenoid biosynthesis and its upstream MEP/DOXP pathway through the direct regulation of many target genes. The role of several carotenogenesis genes targeted by RIN has been shown in the reports on their mutants (Fray and Grierson, 1993; Isaacson et al., 2002). Lycopene accumulation in the ripening tomato fruit seems to be explained by our result (Figure 6), demonstrating both the upregulation of the genes involved in the upstream pathway of lycopene and downregulation of the genes in the downstream lycopene cyclization to generate α- or β-carotenes, as suggested previously (Alba et al., 2005). We demonstrate that RIN regulates these up- and downregulations directly and indirectly and thus consider that RIN acts to enhance the levels of carotenoid (lycopene in common cultivars) in the ripening tomato fruit.
Among the identified direct RIN targets, SGR1 is responsible for the tomato green-flesh mutation, which causes the production of brown fruits as the result of carotenoid accumulation and lack of chlorophyll degradation occurring simultaneously (Barry et al., 2008). SGR1 is a tomato homolog of SGR family proteins that destabilize the light-harvesting chlorophyll binding protein complexes of the thylakoid membranes (Park et al., 2007). Our finding that SGR1 is a direct RIN target agrees with the ripening-related expression of SGR1, as described previously (Barry et al., 2008), suggesting that RIN regulates chlorophyll degradation during ripening via transcription of SGR1.
TF Genes Directly Targeted and Positively Regulated by RIN during Fruit Ripening
At the onset of ripening, the increase and decrease in expression of numerous ripening-associated genes occurs after the rise in RIN expression. This implies that, in addition to the direct regulation of RIN, other TFs under the control of RIN participate in transcriptional regulation. Here, we identified at least nine additional TF genes that are positively regulated during ripening as direct targets of RIN. The target TFs are expected to regulate the expression of their own targets downstream of RIN. We observed slight increases in expression levels of several target TF genes also in the rin mutant fruits, suggesting that these targets are affected by other developmental factors in addition to RIN. Several of the RIN targets have been reported to be associated with fruit ripening. For example, AP2a, a member of the AP2/ERF family, is a negative regulator of ethylene synthesis during ripening (Chung et al., 2010; Karlova et al., 2011). Our finding that AP2a is a direct RIN target suggests that RIN plays a role in its regulation, but ethylene and other factors are also involved (Karlova et al., 2011). Interestingly, the subsets of direct RIN targets share several genes, such as CRTISO, SGR1, and a lipoxygenase gene (TomLoxC), with the set of AP2a-affected genes (Chung et al., 2010; Karlova et al., 2011). This suggests that AP2a activity is required to express a subset of the direct RIN targets. Another RIN target, SR3L, which is a homolog of calcium-signaling TF genes, has been recently reported to be affected in expression by RIN and ethylene during ripening (Yang et al., 2012). Calcium is associated with various ripening processes, such as ripening rate, respiration rate, ethylene production, and fruit softening (Ferguson, 1984). Although the exact role of SR3L in fruit ripening remains uncertain, RIN may contribute to calcium-mediated ripening processes by regulating SR3L expression. The roles of the remaining novel RIN target TF genes, and also most of the negatively regulated TF genes, in tomato fruit ripening remain unclear. Functional analysis of the target TFs will lead to a more comprehensive understanding of the ripening regulatory mechanism.
The Interplay between RIN and Ethylene May Have a Crucial Role in the Regulation of Fruit Ripening
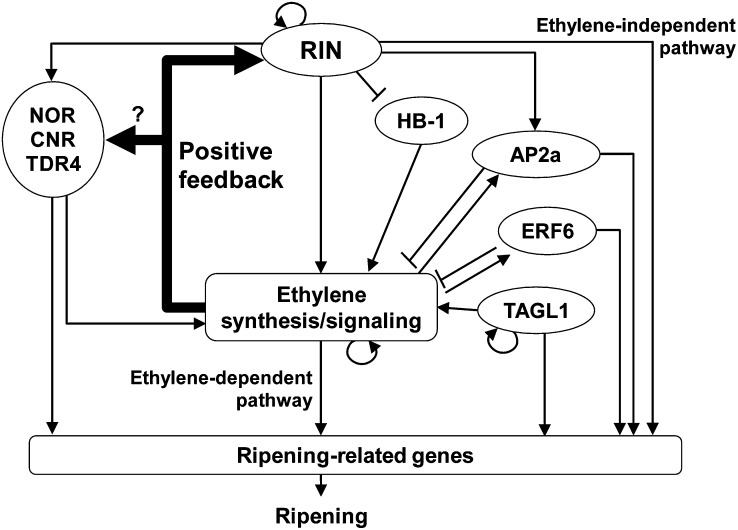
The analysis of ethylene responsiveness using the ethylene receptor inhibitor 1-MCP revealed that ethylene is implicated in the upregulation during ripening of at least 10 TF genes that are direct RIN targets and is especially critical for the upregulation of nine TFs, including RIN; more than half of their expression levels during ripening depend on ethylene action. This observation indicates that ethylene is required for activating RIN-mediated ripening regulation. The ethylene effect on the expression of TDR4 and AP2a is consistent with results reported previously (Karlova et al., 2011; Osorio et al., 2011). By contrast, it is surprising that ethylene is involved in the upregulation of main ripening regulators, such as RIN and NOR, because they are believed to lie upstream of both the ethylene-dependent and -independent pathways in the fruit ripening regulatory mechanism (Giovannoni, 2007; Lozano et al., 2009). The induction of RIN at the onset of ripening is not influenced by exogenous ethylene (Vrebalov et al., 2002), indicating that ethylene likely affects the enhancement but not the initial induction of RIN expression. Concerning ethylene production during ripening, Yokotani et al. (2009) demonstrated that ethylene production for tomato fruit ripening is initially induced by a developmental factor and then massively increased by ethylene autocatalytic regulation. They also suggest that RIN is a possible inducer of the initial ethylene production. Other ripening regulators, such as NOR, CNR, TAGL1, and HB-1, are also involved in the regulation of ethylene levels during ripening (Thompson et al., 1999; Giovannoni, 2004; Lin et al., 2008; Itkin et al., 2009; Vrebalov et al., 2009). In addition to this study, recent molecular and biochemical studies suggest that RIN participates in ethylene production by inducing many of the ethylene synthesis/signaling genes (e.g., ACS2 and ACS4), by upregulating NOR and CNR and by downregulating HB-1 (Ito et al., 2008; Fujisawa et al., 2011, 2012; Martel et al., 2011; Qin et al., 2012; Zhou et al., 2012). Furthermore, TAGL1 and HB-1 may directly regulate the expression of ACS2 (Itkin et al., 2009) and ACO1 (Lin et al., 2008), respectively. Furthermore, ethylene production is negatively regulated by AP2a and ERF6 (Chung et al., 2010; Karlova et al., 2011; Lee et al., 2012). Taking the observations and the existing ripening regulatory model (Giovannoni, 2007; Lozano et al., 2009) into account, we propose a revised model for tomato fruit ripening (Figure 9). In this model, RIN induced at the onset of ripening initiates both the production of climacteric ethylene and the ethylene-independent transcriptional regulatory pathway. The RIN-induced and subsequent autocatalytic ethylene further induces the ethylene responsive genes via the ethylene signal transduction pathway. The signaling pathway also enhances RIN expression in a positive feedback loop. We previously proposed that the expression of RIN is autoregulated (Fujisawa et al., 2011). The two enhancement systems, the autoregulation and the ethylene-mediated positive feedback loop, may bring about a rapid and remarkable upregulation (up to a several hundred-fold increase) of RIN, leading to the subsequent rise in ethylene level and a rapid up- and downregulation of the vast number of genes associated with fruit ripening. The proper ethylene level for each ripening stage may be fine-tuned by both the positive loop regulated by RIN and the negative regulation of ethylene production by AP2a and ERF6. Thus, a robust linkage between RIN and ethylene signaling is central to the regulation of fruit ripening. The positive feedback loop requires an additional factor that is implicated in the ethylene signal transduction pathway and directly regulates RIN expression during ripening. Identifying the factor implicated directly or indirectly in the ethylene signal transduction pathway will substantially unveil the regulatory mechanism acting between RIN and ethylene during fruit ripening.
Figure 9.
A Schematic Representation of the Proposed Model for a Regulatory Mechanism of Tomato Fruit Ripening, Including a Mechanism That Maintains Ethylene Levels via RIN and Other Factors.
Bold line arrows indicate an ethylene-mediated positive feedback loop that enhances RIN expression. It is unclear whether the loop regulates the expression of the other ripening regulators (such as NOR and TDR4) affected by ethylene during ripening directly or indirectly (via RIN). Arrows indicate the direction of the transcriptional regulatory pathways. Blunt-ended lines indicate repression. Circle arrows on RIN and TAGL1 indicate autoregulation and on ethylene indicate autocatalytic ethylene production.
METHODS
ChIP-chip
Chromatin immunoprecipitated (ChIPed) DNA was prepared using anti-RIN antibodies from input chromatin extracted from ripening tomato (Solanum lycopersicum) fruits basically as previously described (Ito et al., 2008; Fujisawa et al., 2011). For microarray hybridization, the ChIPed and input DNAs were amplified with the GenomePlex Complete Whole Genome Amplification kit 2 (Sigma-Aldrich) according to the manufacturer’s instructions. Each of the amplified ChIPed DNA samples was labeled with Cy3 and competitively hybridized with the amplified input DNA labeled with Cy5 onto a NimbleGen custom-designed tomato gene promoter microarray (Roche NimbleGen). The 720K array had 694,802 probes (52- to 70-mers tiled throughout nonrepetitive sequences at a median spacing of 80 bp), designed from the 2-kb upstream regions (putative promoters; a total of ∼71.6 Mb) of all 35,802 tomato predicted genes provided by ITAG version 2 (ITAG2;http://solgenomics.net/genomes/Solanum_lycopersicum/index.pl) (Tomato Genome Consortium, 2012). Sequence source for probes was a draft genome sequence (∼781 Mb) of the tomato chromosomes released by the International Tomato Genome Sequencing Consortium (version 2.31; http://solgenomics.net/about/tomato_sequencing.pl) (Tomato Genome Consortium, 2012). Labeling of the ChIPed and input DNAs, two-color competitive hybridization, and peak detection were performed by Roche NimbleGen. Extraction of signal intensity data from scanned images of the ChIP-chip array and subsequent peak detection were performed with NimbleScan software (Roche NimbleGen). Three independent biological samples were used in separate hybridizations (including one dye swap).
Data Analysis
Normalization and peak detection were performed by Roche NimbleGen as per their protocols (http://www.nimblegen.com/products/chip/data_guide.html). In each ChIP-chip experiment, the FC ratio of the ChIP signal value to the input signal value for each probe was converted to a log2 scale. The log2 ratio was scaled to center the data around zero by subtracting the bi-weight mean for the log2 ratio values for all features on the array from each log2 ratio value. The details of the procedure for peak detection were described by Johnson et al. (2008).
Peaks were determined using a 500-bp sliding window with a probe height cutoff. A peak was recognized if the number of probes above a specific probe height cutoff value in the window was ≥4 (or two if there were fewer than four probes per window). As the window slid along the data track, the recognized peak could extend. The probe height cutoff values were defined as a percentage of a hypothetical maximum, which is the mean of the log2 ratio values of the probes + 6 sd. The default value of the probe height cutoff was 90%, and the peak detection scan was repeated. For each repetition, the probe height cutoff values were decreased by 1% per detection scan, down to 15% of the theoretical maximum. The probe height cutoff value at which a peak was first recognized was used as the score of the peak. Peaks were categorized based on their scores; peaks recognized during the first detection scan were in the category of cutoff = 90%, peaks recognized during the second scan but not during the first scan were in the category of cutoff = 89%, and so on, down to cutoff = 15%. The number of peaks in each category was used in the FDR calculation.
The FDR, which represents the probability of finding a peak of similar significance by chance (Johnson et al., 2008), was estimated for each peak using a permutation-based algorithm to find statistically significant peaks. For the FDR calculation, simulated noise data were made by the following method: The original data were scrambled (randomly permuted) and peaks were counted using the same peak detection procedure. We then computed the ratio of the average number of peaks recognized across 20 scrambled data tracks in a detection scan with a probe height cutoff to the number of peaks recognized in the original data track in the scan with the same cutoff and used the ratio as FDR for peaks that fall in the range of the cutoff.
Next, to identify reproducible RIN binding sites, the genomic start and end positions of the peaks (FDR ≤ 0.05) were compared among three experiments, and a region that was covered by the peak in at least two experiments was searched. The region was finally noted as a RIN binding site, and the width of the region was defined as the peak width.
In Silico Motif Analyses
Using the FUZZNUC program included in the EMBOSS package (Rice et al., 2000), we searched the nucleotide sequences of the RIN binding sites and all the 2-kb promoters of the tomato predicted genes for DNA motif sequences for possible RIN binding sites [CArG-box sequences: C(C/T)(A/T)6(A/G)G, C(A/T)8G and C(C/T)(A/T)G(A/T)4(A/G)G] (Ito et al., 2008; Fujisawa et al., 2011). We also searched the sequences for known TF binding sites in the Arabidopsis thaliana cis-regulatory database (AtcisDB) provided by the Arabidopsis Gene Regulatory Information Server of Ohio State University (http://Arabidopsis.med.ohio-state.edu/AtTFDB/) (Davuluri et al., 2003). Significance of the differences in the occurrence of the motifs between the RIN binding sites and the promoters was evaluated using a χ2 test. The consensus sequence for the CArG boxes was analyzed using the WebLogo 3 program (http://weblogo.threeplusone.com/) (Schneider and Stephens, 1990; Crooks et al., 2004).
qChIP-PCR
The enrichment of DNA fragments bound by RIN was measured by qChIP-PCR as described previously (Fujisawa et al., 2011, 2012). To confirm the ChIPed DNA quality, a pair of oligonucleotide primers for a known RIN binding site in the ACS2 promoter (Fujisawa et al., 2011) was used. The oligonucleotide sequences of primer pairs used for validation of the enrichment of RIN binding sites identified by ChIP-chip are listed in Supplemental Data Set 3 online. The measurements (cycle threshold [CT]) for the CArG-box sites were normalized with those for the Actin gene, which is free from RIN binding and was used for qChIP-PCR (Ito et al., 2008; Fujisawa et al., 2011). The quantitative PCR was conducted as described below in the gene expression analyses section.
Functional Gene Annotation
A similarity search of the tomato ITAG2 predicted gene products was conducted as described previously (Fujisawa et al., 2011) using the BLASTP program (Altschul et al., 1997) with an e-value cutoff of <0.01 against the Arabidopsis protein database provided by The Arabidopsis Information Resource (TAIR10; http://Arabidopsis.org/). Based on the similarity, the ITAG2 genes were functionally annotated using a functional catalog database provided by MIPS (Helmholtz Zentrum München) (http://www.helmholtz-muenchen.de/en/mips/projects/funcat/index.html) (Ruepp et al., 2004). For each MIPS functional category, significance of enrichment of the genes in the positively or negatively regulated subsets of direct RIN targets relative to those in the tomato genome (the whole set of ITAG2 genes) was calculated by Fisher’s exact test. A pathway search for direct RIN targets was conducted using the Kyoto Encyclopedia of Genes and Genomes pathway database for Arabidopsis (http://www.genome.jp/kegg/pathway.html).
Gene Expression Analyses
To select differentially expressed genes during ripening, we analyzed a microarray data set (accession number GSE28564 in the Gene Expression Omnibus database at the National Center for Biotechnology Information) (http://www.ncbi.nlm.nih.gov/geo/) for gene expression of the wild-type and rin mutant tomato fruits basically as reported earlier (Fujisawa et al., 2012). In brief, the FC of the expression level of each tomato gene was calculated as the relative signal intensity of the probes in the wild type at the ripening (P) stage to that at the preripening (G) stage (FCWT) or in the rin mutant fruits at the same age as the wild-type P stage relative to those at the G stage (FCrin). Note that the rin mutant tomatoes harvested at the same age as the wild-type P fruit expressed the mutated RIN gene and did not normally reach the pink stage. The significance of FCWT was tested using the P value from three independent experiments estimated by the t test with the FDR control (q-value) as previously described (Fujisawa et al., 2012). Furthermore, to select genes positively and negatively regulated by RIN, we analyzed the ECS (the ratio of FCWT relative to FCrin) (Fujisawa et al., 2012). We finally selected genes that met the criteria of FCWT > 2 (q-value < 0.05) and ECS > 2 as positively regulated genes by RIN and genes that met the criteria of FCWT < 0.5 (q-value < 0.05) and ECS < 0.5 as negatively regulated genes by RIN.
Gene expression levels in tomato fruits were confirmed by qRT-PCR as previously described (Fujisawa et al., 2011, 2012). Briefly, 1 μL of cDNA synthesis reaction mixture was used as template for analysis using Thunderbird SYBR qPCR mix (Toyobo). Three biological replicates were analyzed for each stage of the respective lines. A pair of primers specific to each gene was designed using the PRIMER3 program (http://frodo.wi.mit.edu/primer3/) with the following settings: 130 to 150 bp for product size range, 22-mer for optimal primer size, and 62°C for optimal annealing temperature. The oligonucleotide sequences of gene-specific primer pairs used in this study for RIN, CNR, TDR4, and a SCL32-like GRAS gene (Solyc07g052960) were described previously (Fujisawa et al., 2011, 2012), and for the other genes, the primer sequences are listed in Supplemental Data Set 3 online. The amplification efficiency of the primer pairs was checked using the Sequence Detection Software (version 1.3.1) included in the 7300 system (Applied Biosystems). The data were represented as FC relative to the G stage in each line calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001) using a primer pair for a gene encoding clathrin adaptor complexes medium subunit (CAC) as a reference (Expósito-Rodríguez et al., 2008). The significance of difference in FC (log2 scale) between the wild-type and rin mutant fruits at each P and G stage was evaluated by Student’s t test.
1-MCP Treatment
Tomato fruits at the breaker stage were placed into an airtight 1-liter plastic container with 5 ppm of 1-MCP that was generated by dissolving 24 mg of 1-MCP–releasing powder (SmartFresh, 0.14% of active ingredient by weight; Rohm and Haas) in 25 μL of water. The treatment was conducted for 4 d in an incubator under 16-h light at 26°C and 8-h dark at 20°C conditions. The 1-MCP solution was changed every other day. After the treatment, the fruits were sliced, frozen in liquid nitrogen, and subjected to RNA isolation followed by qRT-PCR as described above. The effect of 1-MCP was confirmed by qRT-PCR using a pair of primers for the protein-coding sequence of ACS2, whose expression is suppressed by 1-MCP treatment (Yokotani et al., 2009).
Accession Numbers
Sequence data from this article can be found in the Solanaceae Genomics Network or GenBank/EMBL databases under the following accession numbers: RIN, AF448522; CNR, DQ672601; TDR4, AY098732; Actin, U60482; and CAC, SGN-U314153. The accession numbers of the other data are shown in Supplemental Data Set 3 online. Sequence data of the ITAG2 identifiers for tomato genes from this article are shown in Supplemental Data Sets 1 to 3 online. The ChIP-chip data are MIAME compliant and have been deposited in a MIAME-compliant database (Gene Expression Omnibus accession number GSE40257) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Preparation and Validation of ChIPed DNA Samples Recovered with the Anti-RIN Antibodies.
Supplemental Figure 2. RIN Binding Sites Detected by ChIP-chip at a Lower Stringency in the Promoters of Five Known Direct RIN Targets.
Supplemental Figure 3. Validation of Enrichment of RIN Binding Sites by qChIP-PCR.
Supplemental Figure 4. Validation of Enrichment of RIN Binding Sites in the Promoters of Negatively Regulated Direct RIN Target Genes by qChIP-PCR.
Supplemental Figure 5. RIN Binding Sites Detected by ChIP-chip in the Promoters of Novel Positively Regulated Direct RIN Target Transcription Factors.
Supplemental Table 1. Statistics of the ChIP-chip Results.
Supplemental Table 2. Overrepresented Plant Transcription Factor Binding DNA Motif Sequences in the RIN Binding Sites.
Supplemental Data Set 1. Potential Direct RIN Target Genes Found by ChIP-chip and CArG Boxes in the RIN Binding Sites.
Supplemental Data Set 2. A List of Direct RIN Target Genes.
Supplemental Data Set 3. Oligonucleotide Primers Used in This Study.
Supplemental Data Set 4. KEGG Pathway Analysis for Direct RIN Targets Based on Similarity to Arabidopsis Genes.
Supplemental Data Set 5. Functional Annotation of Direct RIN Target Genes.
Supplemental Data Set 6. Details of the Overrepresented MIPS Categories in the Direct RIN Target Genes.
Supplemental Data Set 7. Classification of the Direct RIN Target Transcription Factors.
Acknowledgments
We thank Mamiko Kitagawa and Junji Kimbara (Research Institute, Kagome Co., Tochigi, Japan) for providing tomato fruits. We thank Rohm and Haas Japan for providing 1-MCP. This work was supported in part by the Program for Promotion of Basic and Applied Researches for Innovations in the Bio-oriented Technology Research Advancement Institution (BRAIN) of Japan to Y.I.
AUTHOR CONTRIBUTIONS
M.F. and Y.I. designed the experiments. M.F., T.N., Y.S., and Y.I. prepared samples. M.F., T.N., and Y.S. performed research. M.F. analyzed the data. M.F. and Y.I. wrote the article.
Glossary
- TF
transcription factor
- ChIP
chromatin immunoprecipitation
- ChIP-chip
ChIP assay coupled with DNA microarray analysis
- qChIP-PCR
quantitative ChIP-PCR
- FC
fold change
- FDR
false discovery rate
- ECS
expression change score
- MEP/DOXP
2-C-methyl-d-erythritol 4-phosphate/1-deoxy-d-xylulose 5-phosphate
- qRT-PCR
quantitative RT-PCR
- 1-MCP
1-methylcyclopropene
- MIPS
Munich Information Center for Protein Sequences
- P
pink coloring
- G
mature green
References
- Alba R., Payton P., Fei Z., McQuinn R., Debbie P., Martin G.B., Tanksley S.D., Giovannoni J.J. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C.S., McQuinn R.P., Chung M.Y., Besuden A., Giovannoni J.J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M., Karlova R., Ballester A.R., Tikunov Y.M., Bovy A.G., Wolters-Arts M., Rossetto Pde.B., Angenent G.C., de Maagd R.A. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24: 4437–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J.D., Klein P., Giovannoni J. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64: 936–947 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R.V., Sun H., Palaniswamy S.K., Matthews N., Molina C., Kurtz M., Grotewold E. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas Y., Dadomo M., Di Lucca G., Grolier P. (2003). Effects of environmental factors and agricultural techniques on antioxidant content of tomatoes. J. Sci. Food Agric. 83: 369–382 [Google Scholar]
- Eriksson E.M., Bovy A., Manning K., Harrison L., Andrews J., De Silva J., Tucker G.A., Seymour G.B. (2004). Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 136: 4184–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A.A., Borges-Pérez A., Pérez J.A. (2008). Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson I.B. (1984). Calcium in plant senescence and fruit ripening. Plant Cell Environ. 7: 477–489 [Google Scholar]
- Fraser P.D., Romer S., Shipton C.A., Mills P.B., Kiano J.W., Misawa N., Drake R.G., Schuch W., Bramley P.M. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. USA 99: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray R.G., Grierson D. (1993). Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 22: 589–602 [DOI] [PubMed] [Google Scholar]
- Fujimoto S.Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Ito Y. (2011). Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Shima Y., Higuchi N., Nakano T., Koyama Y., Kasumi T., Ito Y. (2012). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235: 1107–1122 [DOI] [PubMed] [Google Scholar]
- Giménez E., Pineda B., Capel J., Antón M.T., Atarés A., Pérez-Martín F., García-Sogo B., Angosto T., Moreno V., Lozano R. (2010). Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5: e14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J.J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16 (suppl.): S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J.J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Hao D., Yamasaki K., Sarai A., Ohme-Takagi M. (2002). Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41: 4202–4208 [DOI] [PubMed] [Google Scholar]
- Isaacson T., Ronen G., Zamir D., Hirschberg J. (2002). Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60: 1081–1095 [DOI] [PubMed] [Google Scholar]
- Ito Y., Kitagawa M., Ihashi N., Yabe K., Kimbara J., Yasuda J., Ito H., Inakuma T., Hiroi S., Kasumi T. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55: 212–223 [DOI] [PubMed] [Google Scholar]
- Johnson D.S., et al. (2008). Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 18: 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R., Rosin F.M., Busscher-Lange J., Parapunova V., Do P.T., Fernie A.R., Fraser P.D., Baxter C., Angenent G.C., de Maagd R.A. (2011). Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Joung J.G., McQuinn R., Chung M.Y., Fei Z., Tieman D., Klee H., Giovannoni J. (2012). Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 70: 191–204 [DOI] [PubMed] [Google Scholar]
- Lin Z., Hong Y., Yin M., Li C., Zhang K., Grierson D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55: 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lozano R., Giménez E., Cara B., Capel J., Angosto T. (2009). Genetic analysis of reproductive development in tomato. Int. J. Dev. Biol. 53: 1635–1648 [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38: 948–952 [DOI] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J.J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157: 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Alba R., Damasceno C.M., Lopez-Casado G., Lohse M., Zanor M.I., Tohge T., Usadel B., Rose J.K., Fei Z., Giovannoni J.J., Fernie A.R. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan I.L., McQuinn R., Giovannoni J.J., Irish V.F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61: 1795–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2007). The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Wang Y., Cao B., Wang W., Tian S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70: 243–255 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Concepcion M., Gruissem W. (1999). Arachidonic acid alters tomato HMG expression and fruit growth and induces 3-hydroxy-3-methylglutaryl coenzyme A reductase-independent lycopene accumulation. Plant Physiol. 119: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Güldener U., Mannhaupt G., Münsterkötter M., Mewes H.W. (2004). The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T.D., Stephens R.M. (1990). Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18: 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3: 217–223 [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70: 549–561 [DOI] [PubMed] [Google Scholar]
- Thompson A.J., Tor M., Barry C.S., Vrebalov J., Orfila C., Jarvis M.C., Giovannoni J.J., Grierson D., Seymour G.B. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigchelaar E.C., Mcglasson W.B., Franklin M.J. (1978). Natural and ethephon-stimulated ripening of F1 hybrids of ripening Inhibitor (rin) and non-ripening (nor) mutants of tomato (Lycopersicon esculentum Mill). Aust. J. Plant Physiol. 5: 449–456 [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Pan I.L., Arroyo A.J., McQuinn R., Chung M., Poole M., Rose J., Seymour G., Grandillo S., Giovannoni J., Irish V.F. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21: 3041–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., Schuch W., Giovannoni J. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Yang T., Peng H., Whitaker B.D., Conway W.S. (2012). Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 12: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N., Nakano R., Imanishi S., Nagata M., Inaba A., Kubo Y. (2009). Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. J. Exp. Bot. 60: 3433–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., et al. (2012). Virus-induced gene complementation reveals a transcription factor network in modulation of tomato fruit ripening. Sci. Rep. 2: 836. [DOI] [PMC free article] [PubMed] [Google Scholar]