This work reveals the function of the TIE1 transcriptional repressor in leaf development. TIE1 interacts with TPL/TPR corepressors through its C-terminal EAR motif and with CIN-like TCP transcription factors through its N-terminal region. TIE1 regulates leaf development by bridging the interactions between TPL/TPRs and CIN-TCPs.
Abstract
Leaf size and shape are mainly determined by coordinated cell division and differentiation in lamina. The CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors are key regulators of leaf development. However, the mechanisms that control TCP activities during leaf development are largely unknown. We identified the TCP Interactor containing EAR motif protein1 (TIE1), a novel transcriptional repressor, as a major modulator of TCP activities during leaf development. Overexpression of TIE1 leads to hyponastic and serrated leaves, whereas disruption of TIE1 causes epinastic leaves. TIE1 is expressed in young leaves and encodes a transcriptional repressor containing a C-terminal EAR motif, which mediates interactions with the TOPLESS (TPL)/TOPLESS-RELATED (TPR) corepressors. In addition, TIE1 physically interacts with CIN-like TCPs. We propose that TIE1 regulates leaf size and morphology by inhibiting the activities of TCPs through recruiting the TPL/TPR corepressors to form a tertiary complex at early stages of leaf development.
INTRODUCTION
Leaf size and morphology are determined by fine-tuning of cell division, differentiation, and expansion during leaf development (Nath et al., 2003; Kuchen et al., 2012). The TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) family transcription factors are among the best-characterized regulators of leaf development and play an essential role in the determination of leaf size and shape (Nath et al., 2003; Palatnik et al., 2003). The TCP genes encode plant-specific transcription factors with a conserved noncanonical basic helix-loop-helix domain, which mediates DNA binding or interactions with other proteins (Cubas et al., 1999). TCP proteins have been grouped into two subclasses on the basis of sequence similarity (Navaud et al., 2007; Martín-Trillo and Cubas, 2010). In Arabidopsis thaliana, 13 class I TCPs and 11 class II TCPs have been identified (Martín-Trillo and Cubas, 2010). The TCP genes are implicated in various aspects of plant development by regulating cell proliferation and differentiation, and Arabidopsis TCPs appear to have overlapping functions. Inactivation of a single TCP gene does not lead to dramatic developmental changes. However, simultaneous disruption of multiple TCP genes greatly affects leaf development (Schommer et al., 2008; Koyama et al., 2010).
TCP transcription factors are found in lycophytes, ferns, mosses, and green algae (Navaud et al., 2007). Their functions have been characterized in several species. Disruption of CINCINNATA (CIN), which encodes a TCP transcription factor in Antirrhinum majus, leads to delayed cell differentiation and results in abnormal leaf size and curvature (Nath et al., 2003). Recently, it has been shown that the diverse shapes of leaves at different developmental stages of tomato (Solanum lycopersicum) and other Solanaceae species are correlated with the temporal and spatial expression of the TCP genes. Genetic alteration of TCP activities causes changes of leaf maturation schedule and final leaf shape (Efroni et al., 2008; Shleizer-Burko et al., 2011). It is suggested that TCPs control leaf shape by promoting leaf maturation in a threshold activity manner (Shleizer-Burko et al., 2011). The functions of CIN-like TCPs during leaf development appear to be conserved across plant species.
The biological activities of TCPs require strict regulation for control of leaf size and shape. One key regulatory mechanism involves microRNA319 (miR319), which determines the abundance of some class II TCP genes at the posttranscriptional level (Palatnik et al., 2003; Ori et al., 2007). Overexpression of miR319 causes leaf curvature and wavy margins in the jaw-D mutants by downregulation of five class II TCP genes (Palatnik et al., 2003). Mutations in the miR319 binding site that result in TCP mRNAs resistant to microRNA degradation lead to a severe leaf phenotype or seedling lethality in Arabidopsis and the alteration of compound leaves to simple leaves in tomato (Palatnik et al., 2003, 2007; Ori et al., 2007). We previously linked the plant hormone auxin to TCP regulation during leaf development. Overexpression of IAMT1, which encodes an indole-3-acetic acid (IAA) carboxyl methyltransferase, presumably decreases free IAA levels and leads to hyponastic leaves. Some of the leaf phenotypes in IAMT1 overexpression lines are caused by decreased expression of several class II TCP genes (Qin et al., 2005). The activities of TCP transcription factors are also modulated by interacting with other proteins. TCP24 forms a complex with the Armadillo BTB Arabidopsis protein1 (ABAP1) in the regulation of cell proliferation in leaves (Masuda et al., 2008). The CCA1 HIKING EXPEDITION (CHE) encoding a TCP transcription factor which interacts with the core clock element TIMING OF CAB EXPRESSION1 (TOC1) and binds to the promoter region of CIRCADIAN CLOCK ASSOCIATED1 (CCA1) to form transcriptional feedback loops in the plant circadian clock (Pruneda-Paz et al., 2009). More recently, the class I TCP proteins TCP14 and TCP15 have been found to act with the O-linked N-acetylglucosamine transferase SPINDLY and may be modified by SPINDLY (Steiner et al., 2012). However, the regulatory mechanisms that control TCP transcriptional activities are still largely unknown.
We conducted a large-scale genetic screen for mutants with defects in leaf curvature by activation tagging. We identified a TCP-interacting protein, TCP Interactor containing EAR motif protein1 (TIE1), which plays an essential role in leaf development. Overexpression of TIE1 causes curly leaves. TIE1 encodes a novel transcriptional regulator. We show that TIE1 is located in the nucleus and behaves like a transcriptional repressor. The phenotypes of TIE1 overexpression lines resemble those displayed in some of the TCP loss-of-function mutants and in the miR319 overexpression lines, which suggests that TIE1 may affect leaf development by directly regulating TCP activities. TIE1 physically interacts with TCPs both in vitro and in vivo. In addition, we show that TIE1 also interacts with TOPLESS (TPL)/TOPLESS-RELATED (TPR) proteins, which are known transcriptional corepressors. Our data suggest that TIE1 brings the TCPs and TPL/TPRs together to repress the transcriptional activities of TCPs during leaf development.
RESULTS
The Mutant tie1-D Displays Defects in Leaf Development
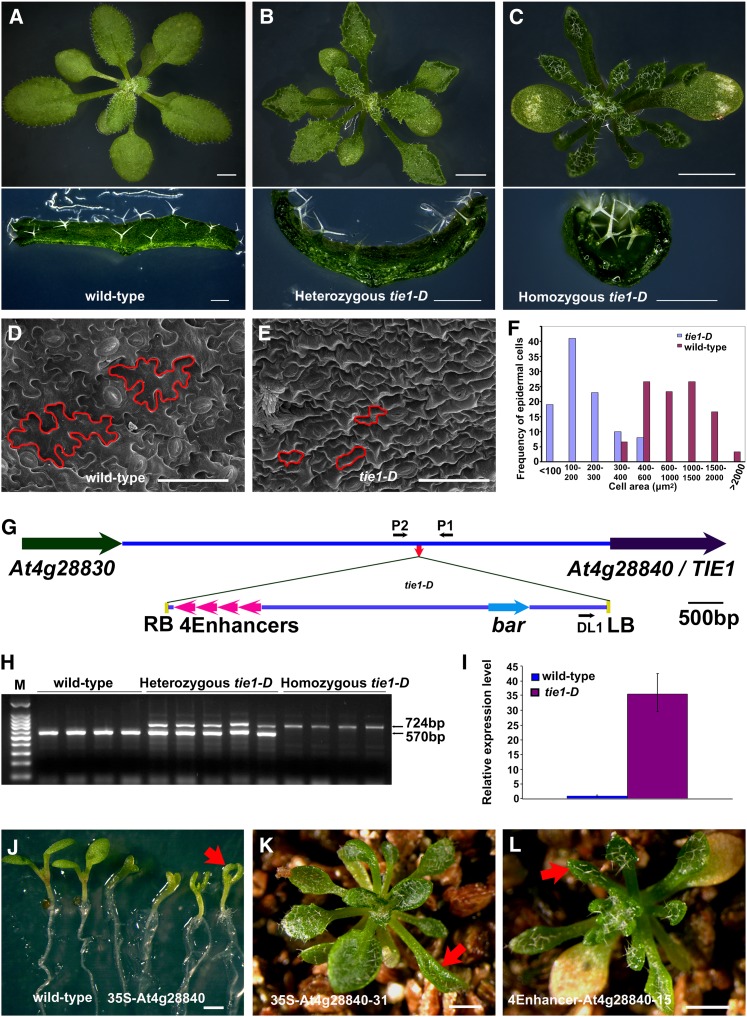
We previously identified and characterized mutants with curly leaves from a collection of activation tagged Arabidopsis mutants (Qin et al., 2003; Qin et al., 2005). In the present study, an additional curly leaf mutant, tie1-D, was characterized. Unlike wild-type leaves, which were flat, the tie1-D leaves were curled upwards (Figures 1A to 1C). The leaves of tie1-D were also smaller and narrower than those of wild-type plants. The leaf margins of the mutant were wavy and more serrated (Figures 1A to 1C, and see Supplemental Figure 1 online). Epidermal cells from mature leaves of tie1-D mutants were polygonal, whereas wild-type cells had a typical jigsaw shape (Figures 1D and 1E). The tie1-D mutant had much smaller leaf cells than the wild-type (Figure 1F), suggesting that TIE1 may affect leaf cell differentiation.
Figure 1.
Identification of the tie1-D Mutant.
(A) to (C) Top, from left to right, the 21-d-old wild-type plants and heterozygous and homozygous tie1-D mutants. The leaves of tie1-D were serrated and up-curled. Bars = 2 mm. Bottom, transverse sections of leaves from wild-type plants and heterozygous and homozygous tie1-D mutants. The leaves from homozygous tie1-D curled more than those of heterozygous tie1-D. Bars = 500 µm.
(D) and (E) Scanning electron micrographs of the leaf epidermal cells at the base of abaxial sides of mature leaves from the wild type (D) and tie1-D mutants (E). Bars = 50 µm.
(F) Distribution of cell size of the leaf epidermal cells from tie1-D mutants and wild-type control.
(G) Schematic representation of T-DNA insertion site in tie1-D mutant. The colored arrows represent genes and lines indicate the intergenic DNA region. The small red arrow in the intergenic region indicates the T-DNA insertion site in tie1-D. The four magenta arrowheads represent the four CaMV 35S enhancers of pSKI015 (Weigel et al., 2000). The small black arrows represent the DL1, P1, and P2 primers used in the cosegregation analysis. LB, T-DNA left border; RB, T-DNA right border; 4Enhancers, four CaMV 35S enhancers; bar, Basta resistance gene.
(H) Cosegregation analysis of T-DNA insertion with the hyponastic leaves. The 570-bp DNA bands were amplified from the wild-type genomic DNA and the 724-bp bands from tie1-D genomic DNA.
(I) The relative expression level of At4g28840 (TIE1) in the wild type and the homozygous tie1-D. The expression level of TIE1 in the wild type was set to 1.0. The error bars represent the sd of three biological replicates.
(J) Overexpression of At4g28840 by 35S promoter caused small and up-curled cotyledons. From left to right, two wild-type 7-d-old seedlings and four independent 35S-At4g28840 transgenic lines. Red arrow indicates a small and curly cotyledon. Bar = 1 mm.
(K) Recapitulation of tie1-D hyponastic leaf phenotype by overexpressing At4g28840 using a CaMV 35S promoter. Red arrow indicates a curly leaf. Bar = 1 mm.
(L) Recapitulation of tie1-D curly leaf phenotype by the overexpression of At4g28840 using the four CaMV 35S promoter enhancers and its own promoter. Red arrow indicates a curly leaf. Bar = 1 mm.
The Defects in tie1-D Are Caused by the Overexpression of At4g28840
We determined that tie1-D was a gain-of-function mutant. The tie1-D phenotypes were caused by a single T-DNA insertion located in the intergenic region between At4g28830 and At4g28840 (Figure 1G). The T-DNA insertion cosegregated with the tie1-D phenotypes (Figure 1H) and led to a significant increase in the expression level of At4g28840 (Figure 1I). We generated the construct 35S-At4g28840 in which At4g28840 was driven by a cauliflower mosaic virus (CaMV) 35S promoter and transformed it into wild-type Arabidopsis; 251 independent transgenic lines displayed hyponastic leaves (Figures 1J and 1K), indicating that overexpression of At4g28840 phenocopied tie1-D. Most transgenic lines also produced hyponastic cotyledons (Figure 1J). At4g28840 was also overexpressed under the control of its own promoter and four copies of CaMV 35S enhancers, a construct that mimics the T-DNA insertion in the tie1-D mutants. Seventy-six independent transgenic lines displayed curly leaf phenotype (Figure 1L). These results indicated that tie1-D phenotypes were caused by the increased expression levels of At4g28840. Thus, At4g28840 was designated as TIE1.
TIE1 Encodes a Nuclear Transcriptional Repressor Containing an EAR Motif
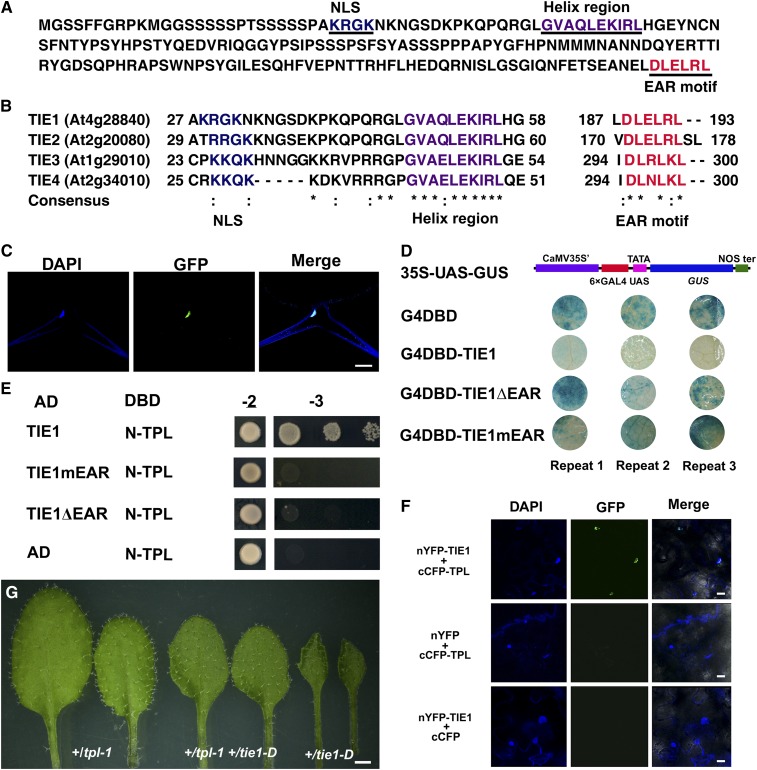
TIE1 encodes a protein of 193 amino acids that contains a basic region (between residue 29 and 45) followed by a helix region (between residue 47 and 57) in the N-terminal region (Figure 2A). Interestingly, a typical EAR motif (DLELRL) is located at the C-terminal end of the TIE1 protein (Figure 2A), suggesting that TIE1 may function as a transcriptional repressor. TIE1 also contains a putative monopartite nuclear localization signal (KRGK) located in the N terminus of the TIE1 protein (Figure 2A) (Dingwall et al., 1988). We found three genes in the Arabidopsis genome, At2g20080, At1g29010, and At2g34010, encoding proteins with high amino acid sequence similarity to TIE1, which were thus designated as TIE2, TIE3, and TIE4, respectively (Figure 2B).
Figure 2.
TIE1 Is a Novel Nuclear Transcription Repressor.
(A) The amino acid sequence of TIE1 (At4g28840). TIE1 contains a monopartite nuclear localization signal (NLS) (underlined, blue), a helix region (underlined, purple), and an EAR motif (underlined, red).
(B) Partial amino acid sequence alignment of TIE1, TIE2 (At2g20080), TIE3 (At1g29010), and TIE4 (At2g34010). They all contain a nuclear localization signal, a helix region, and an EAR motif. Asterisks indicate identical residues and colons indicate similar residues.
(C) TIE1 is localized in the nucleus. Trichome cell from a leaf of 35S-TIE1-GFP-3 transgenic line expressing TIE1-GFP fusion protein. From left to right, the staining of nucleus by DAPI, fluorescence of GFP, and merge of DAPI and GFP. Bar = 25 µm.
(D) TIE1 is a transcription repressor. The transcription activity of TIE1 was tested in tobacco leaves using a GAL4/UAS-based system. CaMV 35S', 35S promoter without TATA box; 6×GAL4 UAS, six copies of GAL4 binding site (UAS); NOS ter, terminator of nopaline synthase gene; G4DBD, GAL4 DNA binding domain; G4DBD-TIE1, G4DBD fused with TIE1; G4DBD-TIE1ΔEAR, G4DBD fused with TIE1ΔEAR in which the EAR motif was deleted; G4DBD-TIE1mEAR, G4DBD fused with TIE1mEAR in which the three conserved Leu residues of the EAR motif was mutated into Ser residues.
(E) TIE1 interacted with TPL protein through the EAR motif in yeast two-hybrid assays. AD, activation domain; DBD, DNA binding domain; TIE1mEAR, mutated TIE1 in which the three conserved Leu residues of the EAR motif was mutated into Ser residues. TIE1ΔEAR, deleted TIE1 in which the EAR motif was deleted; N-TPL, the TPL N terminus including residues from 1 to 188. Transformed yeasts were spotted on control medium (-2) or selective medium (-3) in 10-, 100-, and 1000-fold dilutions. The empty vectors were used as controls.
(F) BiFC analysis of the interaction between TIE1 and TPL. From left to right, DAPI staining showing the nuclei, GFP fluorescence, and merge of DAPI and GFP. Bars = 20 µm.
(G) Genetic interaction between tpl-1 and tie1-D. From left to right, the 4th and 5th leaves from 20-d-old +/tpl-1, +/tpl-1 +/tie1-D double mutant, and +/tie1-D. Bar = 1 mm.
To test whether TIE1 is a transcriptional repressor, we first expressed TIE1 with a C-terminal green fluorescent protein (GFP) tag in Arabidopsis under the control of a CaMV 35S promoter. Twenty-one independent transgenic plants displayed small, hyponastic leaves that were similar to those of tie1-D mutants, demonstrating that the TIE1-GFP fusion protein was functional. The TIE1-GFP protein was clearly localized in the nucleus (Figure 2C), indicating that TIE1 is a nuclear protein. Then we generated a reporter construct 35S-UAS-GUS in which the β-glucuronidase (GUS) gene was under the control of a synthetic promoter that contained six copies of GAL4 binding site (UAS) fused with a CaMV 35S promoter (Figure 2D). We fused TIE1 to the GAL4 DNA binding domain to generate G4DBD-TIE1 and cotransformed 35S-UAS-GUS with G4DBD or G4DBD-TIE1 into tobacco (Nicotiana tabacum) leaves to determine TIE1 transcriptional activities. As expected, GUS staining was strong in leaves cotransformed with 35S-UAS-GUS and G4DBD combination (Figure 2D). However, GUS expression was severely attenuated when 35S-UAS-GUS was cotransformed with G4DBD-TIE1, suggesting that TIE1 is probably a transcriptional repressor. We then tested whether the EAR motif in TIE1 was responsible for the repression activity of TIE1. We fused the GAL4 DNA binding domain either with TIE1ΔEAR in which EAR motif was deleted or with TIE1mEAR in which the conserved Leu residues in the EAR motif were changed to Ser residues. Unlike G4DBD-TIE1, G4DBD-TIE1ΔEAR and G4DBD-TIE1mEAR did not repress GUS expression in tobacco leaves (Figure 2D). These results indicate that the EAR motif is required for the repression activities of TIE1.
TIE1 Interacts with TPL Family Proteins
Recent studies revealed that TPL and TPR proteins interacted with the EAR motif to function as corepressors in auxin and jasmonic acid (JA) signaling pathways (Szemenyei et al., 2008; Pauwels et al., 2010). TIE1 interacted with the TPL N-terminal region (Figure 2E), which is reported to be sufficient for TPL to interact with EAR-containing proteins (Szemenyei et al., 2008; Causier et al., 2012). The interaction between TIE1 and TPL was dependent on the EAR motif, as deletion of the EAR motif (TIE1ΔEAR) or mutation of the three conserved Leu residues of the EAR motif (TIE1mEAR) abolished the interaction between TIE1 and TPL (Figure 2E). The interaction between TIE1 and TPL was confirmed in a bimolecular fluorescence complementation (BiFC) assay (Figure 2F). Our results also showed that TIE1 interacted with all of the TPR proteins, including TPR1, TPR2, TPR3, and TPR4, but not with SAP18, which interacts with some EAR-containing transcription factors (Song and Galbraith, 2006; Hill et al., 2008; Liu et al., 2009) (see Supplemental Figure 2A and Supplemental Methods 1 online).
Our data suggest that TIE1 probably recruits TPL, rather than SAP18, to form a transcriptional repressor complex. To test this hypothesis, we fused TIE1ΔEAR with the C-terminal portion of TPL (TPLC) (see Supplemental Figure 2B online). We expected that TIE1ΔEAR is not functional as a repressor (Figure 2D), but TIE1ΔEAR-TPLC and TIE1 should behave similarly in plants. Indeed, four independent transgenic lines in which the expression of TIE1ΔEAR-TPLC was driven by a CaMV 35S promoter led to curly leaf phenotypes similar to those observed in tie1-D (see Supplemental Figure 2B online). However, the curly leaf phenotypes may also be caused by expressing TIE1ΔEAR fused with other corepressors, such as SAP18. To further clarify the relationship between TIE1 and TPL, we crossed tie1-D to the tpl-1 mutant (Szemenyei et al., 2008). The small and up-curled leaf phenotype of +/tie1-D was largely rescued in the double mutant +/tie1-D +/tpl-1, suggesting that TPL is required for the function of TIE1 (Figure 2G). These results indicate that TIE1 functions as a transcriptional repressor during leaf development by interacting with TPL/TPRs through the EAR motif.
Both TIE1 and TPL Are Expressed in Young Leaves
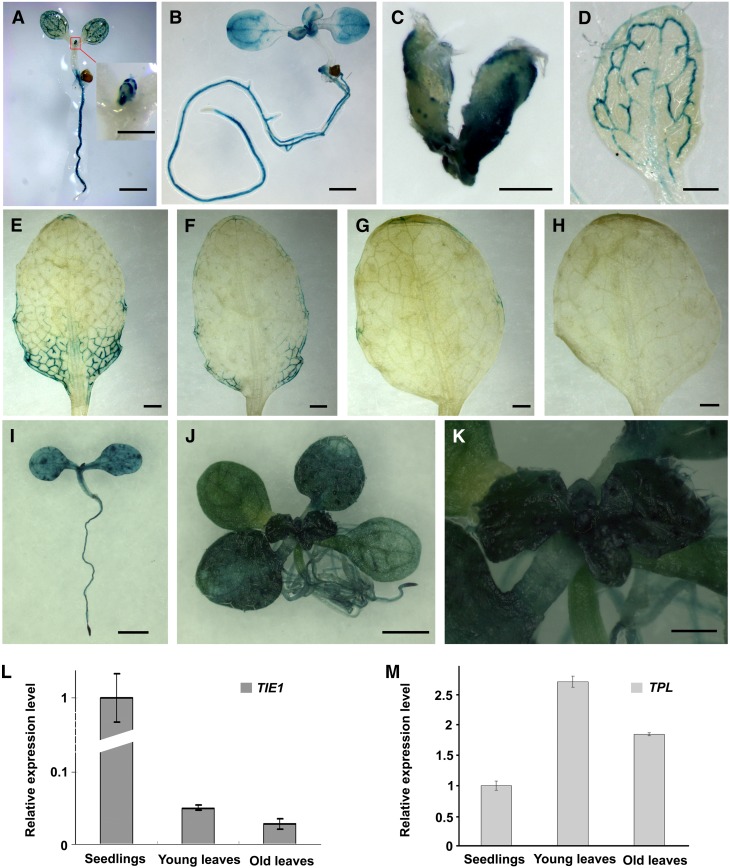
To investigate the spatial and temporal expression pattern of TIE1, we cloned a 2790-bp-long promoter fragment of TIE1 and fused it to the GUS reporter gene to generate the construct TIE1P-GUS. Ten TIE1P-GUS transgenic lines showed similar staining patterns. GUS activities were observed in shoot apical meristem, cotyledons, and leaves (Figures 3A and 3B). In young leaves, GUS staining was strong at the leaf margins and at the base of the leaves (Figures 3A to 3C). As the leaves matured, GUS staining became weaker (Figures 3D to 3H). Initially, GUS staining was restricted to the leaf margin and the developing vascular bundles of the complete leaf blade (Figure 3D). Subsequently, staining gradually concentrated in the vascular bundles at the base of the leaves as the leaves developed (Figures 3E to 3G). No GUS staining was observed in mature leaves (Figure 3H). The expression of TIE1 in young leaves and at the leaf margins was consistent with the observed phenotypes of the tie1-D mutants in which the leaf marginal regions curled upward.
Figure 3.
TIE1 and TPL Are Expressed in Young Leaves.
(A) and (B) The 7-d-old TIE1P-GUS-10 seedling (A) and 9-d-old TIE1P-GUS-10 seedling (B). Inset in (A) is the close-up view of young leaves and shoot apical meristem. TIE1 was strongly expressed at the leaf margin and the base of the young leaves. GUS staining was also observed in the cotyledons, shoot apical meristem, and roots. Bars = 1 mm in (A) and (B) and 250 µm in the inset in (A).
(C) to (H) The young to old leaves from a 24-d-old TIE1P-GUS-10 plant. The GUS staining faded as the leaves grew old. No GUS staining was observed in the first leaf of the 24-d-old TIE1P-GUS-10 plant. Bars = 250 µm in (C) and 1 mm in (D) to (H).
(I) to (K) TPL expression overlapped with TIE1. Bars = 1 mm in (I) and (J) and 300 µm in (K).
(I) The 7-d-old TPLP-GUS-3 seedling.
(J) The 12-d-old TPLP-GUS-8 seedling.
(K) The close-up view of shoot apical meristem from the 12-d-old TPLP-GUS-8 seedling.
(L) and (M) The expression level of TIE1 (L) and TPL (M) in seedlings and leaves. The expression levels in seedlings were set to 1.0. The error bars represent the sd of three biological replicates.
TPL was reported to be expressed in the embryo (Long et al., 2006), but it is not clear whether TPL is expressed in other tissues. To reveal the expression pattern of TPL, we cloned a 2.8-kb-long promoter fragment of TPL and fused to GUS reporter gene to generate the TPLP-GUS construct. Five of six TPLP-GUS transgenic lines showed a similar GUS staining pattern. Strong GUS activities were observed in the vascular tissues, shoot apical meristem, and cotyledons (Figure 3I). At late seedling stage, TPL was strongly expressed in young leaves (Figures 3J and 3K).
We then used quantitative RT-PCR to further confirm whether TIE1 and TPL are expressed in leaves and found that TIE1 was expressed at higher levels in seedlings than in older plants or old leaves (Figure 3L). The results are consistent with those observed from the GUS staining of TIE1P-GUS transgenic lines. Meanwhile, TPL was also expressed in leaves (Figure 3M). These results indicate that TPL shows an overlapping expression pattern with TIE1 in leaves, providing spatial evidence for the interaction between these two proteins.
Disruption of TIE Led to Epinastic Leaves
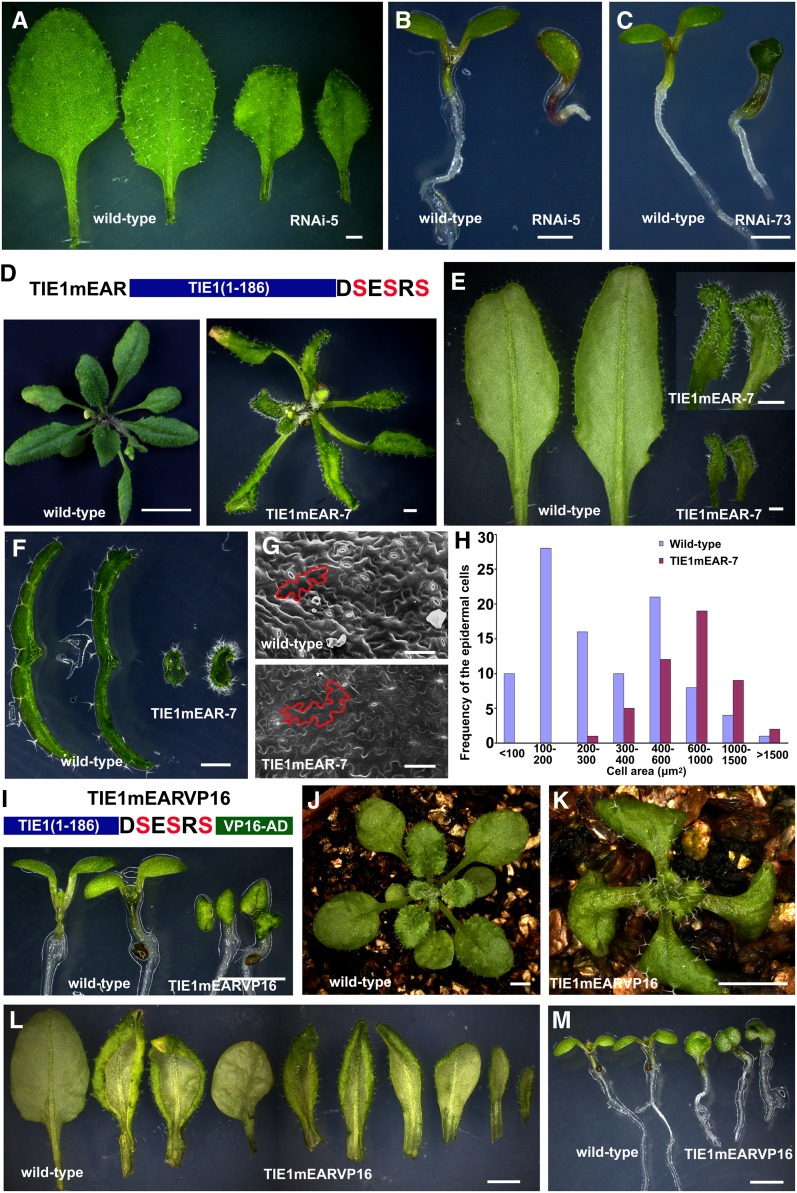
We identified one T-DNA insertion mutant for TIE1 from the available public mutant collections (see Supplemental Figure 3A online). However, it was not a null mutant (see Supplemental Figure 3B online), and no obvious phenotypes were observed in this mutant. We therefore used the inducible insertional mutagenesis system based on Ac-Ds induction by heat shock to generate knockout mutants for TIE1 (Nishal et al., 2005). We first identified one T-DNA insertion mutant, WiscDsLoxHs225_11A, in which the T-DNA insertion was located at a position ∼13 kb away from TIE1 (see Supplemental Figure 3A online) and then performed the heat shock treatment on the homozygous WiscDsLoxHs225_11A mutants (see Supplemental Methods 1 online). From ∼5000 HS2 (second generation after heat shock) plants, we identified one mutant plant, tie1-455, in which the Ds jumped into the splice site of the second intron of TIE1 (see Supplemental Figure 3A online), and the expression of TIE1 was completely knocked out (see Supplemental Figure 3C online). Although tie1-455 is a TIE1 null mutant, it displayed no obvious leaf phenotype. We speculate that TIE2, TIE3, and TIE4 are functionally redundant with TIE1, since they have similar domains with TIE1 (Figures 2A and 2B) and interact with TPL and the other four TPR proteins in yeast two-hybrid assays (see Supplemental Figure 3D online). Moreover, overexpression of TIE2, TIE3, or TIE4 in Arabidopsis led to phenotypes similar to those of tie1-D (see Supplemental Figures 3E to 3H and Supplemental Methods 1 online). In order to clarify the expression pattern of these three genes, we conducted RT-PCR analysis and found that TIE3 and TIE4 were expressed in leaves, whereas TIE2 was not expressed in leaves (see Supplemental Figure 3I online), suggesting that TIE3 and TIE4 may also participate in leaf development. Because the DNA sequence of TIE3 is highly similar to TIE4 (see Supplemental Figure 3J online), we generated an RNA interference (RNAi) construct, 35S-TIE3-RNAi, to knock down the expression of both TIE3 and TIE4 (see Supplemental Figure 3K online). We transformed 35S-TIE3-RNAi into tie1-455 and found that nine independent transgenic lines displayed epinastic leaves, a phenotype that is opposite to that observed in tie1-D mutants (Figure 4A). Furthermore, three independent transgenic plants exhibited even more severe phenotypes, for instance, production of deformed leaves (see Supplemental Figures 3L and 3M online), single cotyledons, or cup-shaped cotyledons (Figures 4B and 4C), which is similar to tpl-1 mutants (Long et al., 2006). These results indicate that TIEs play pivotal roles in leaf development.
Figure 4.
Disruption of TIE Genes Led to Epinastic Leaves.
(A) to (C) Phenotypes of 35S-TIE3-RNAi plants in the TIE1 null mutant tie1-455 background: epinastic true leaves (A), single cotyledon (B), or cup-shaped cotyledon (C). Bar = 1 mm.
(D) to (F) Expression of TIE1mEAR in wild-type Arabidopsis led to epinastic leaves.
(D) Top, schematic representation of TIE1mEAR protein. The three conserved Leu residues of EAR motif were mutated into Ser residues in TIE1mEAR. Bottom, left, 28-d-old wild-type plant. Bar = 1 cm. Right, 28-d-old TIE1mEAR-7 transgenic line. Bar = 2 mm.
(E) The close-up views of the 6th and 7th leaves from 28-d-old wild-type (left) and TIE1mEAR-7 plants (right). Bar = 1 mm.
(F) Transverse sections of leaves from 28-d-old wild-type (left) and TIE1mEAR-7 plants (right). Bar = 1 mm.
(G) Scanning electron micrographs of the leaf epidermal cells at the base of abaxial sides of the 5th leaves from 27-d-old wild-type (top) and TIE1mEAR-7 plants (bottom). Bars = 50 µm.
(H) Distribution of cell size of the leaf epidermal cells at the base of abaxial sides of the 5th leaves from 27-d-old wild-type and TIE1mEAR-7 plants.
(I) to (M) Expression of TIE1 chimeric protein of TIE1mEARVP16 caused severe cotyledon and leaf phenotypes.
(I) Top, schematic representation of TIE1mEARVP16 fusion protein. Bottom, 9-d-old seedlings from wild-type and TIE1mEARVP16 plants.
(J) and (K) Twenty-one-day-old plants from wild-type (J) and TIE1mEARVP16 (K) plants. Bar = 2 mm.
(L) Close-up views of leaves from 28-d-old wild-type and TIE1mEARVP16 plants. From left to right, the 5th leaf from the wild type and nine 5th leaves from nine independent TIE1mEARVP16 transgenic lines. Bar = 2 mm.
(M) The 8-d-old seedlings from wild-type and TIE1mEARVP16 plants. From left to right, two wild-type seedlings and three seedlings from three independent TIE1mEARVP16 transgenic lines. The first one displayed one cotyledon, the second one had two cotyledons fused, and the third one had three cotyledons fused. Bar = 2 mm.
To further investigate the role of TIE1 in leaf development, we expressed TIE1mEAR, which may elicit a dominant-negative effect in wild-type Arabidopsis in an attempt to determine the consequences of inactivation of TIEs. TIE1mEAR does not interact with TPL/TPRs and is no longer a transcriptional repressor (Figures 2D and 2E). However, TIE1mEAR may compete with and disrupt the endogenous TIEs. This strategy was used successfully in the study of EAR motif–containing proteins (Ciftci-Yilmaz et al., 2007; Li et al., 2011). Expression of TIE1mEAR by a CaMV 35S promoter or TIE1 promoter caused interesting phenotypes. Eleven independent 35S-TIE1mEAR transgenic lines displayed epinastic leaves, opposite to the leaf phenotypes observed in tie1-D mutants (Figures 4D to 4F; see Supplemental Figures 4A and 4B online). Fifteen independent TIE1P-TIE1mEAR transgenic lines also exhibited epinastic leaves (see Supplemental Figures 4C and 4D online). The leaf epidermal cells of TIE1mEAR-7 transgenic lines differentiated earlier (Figures 4G), and the cell sizes were significantly larger than those of the wild-type control (Figure 4H). These results further confirm that TIE1 and other TIEs regulate leaf development, possibly by affecting cell differentiation.
Expression of the TIE1 Activation Fusion Proteins TIE1mEARVP16 or TIE1ΔEARVP16 Caused Severe Epinastic Leaves
To further investigate the TIE1 function, we fused the transcriptional activation domain of the VP16 protein to mutated TIE1 in which the EAR motif was either deleted or mutated. We hypothesized that overexpression of TIE1mEARVP16 or TIE1ΔEARVP16 driven by a CaMV 35S promoter in transgenic plants was not only able to play a dominant-negative role by competing with the endogenous TIEs, but also might activate the target genes of TIEs that are normally repressed. We found that fifty independent TIE1mEARVP16 transgenic lines and thirty-nine independent TIE1ΔEARVP16 transgenic lines produced epinastic leaves, a phenotype observed in TIE1mEAR transgenic lines (Figures 4I to 4L; see Supplemental Figures 4E to 4I online). Furthermore, the TIE1mEARVP16 and TIE1ΔEARVP16 lines displayed more extreme curvature in both cotyledons and leaves than those observed in TIE1mEAR lines (Figures 4I to 4K; see Supplemental Figures 4F to 4I online). Six TIE1mEARVP16 as well as four TIE1ΔEARVP16 independent transgenic lines produced single cotyledons or fused cotyledons (Figure 4M; see Supplemental Figure 4F online), which were similar to those observed in tpl-1 mutants (see Supplemental Figure 4J online) (Long et al., 2006). Notably, these epinastic and deformed leaf phenotypes were also observed in tpl-1 (see Supplemental Figures 4J and 4K online). These findings are consistent with our observation that TIE1 interacts with TPL proteins (Figure 2), again indicating that TIE1 plays an important role in leaf development.
TIE1 Is Associated with TCP Transcription Factors
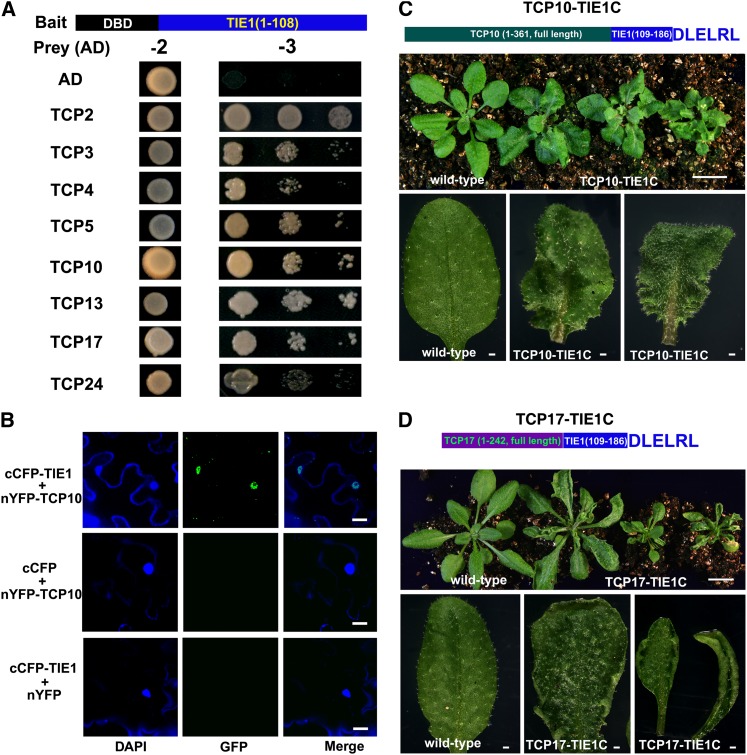
Because TIE1 does not have an apparent DNA binding domain, we hypothesized that TIE1 might bind other transcription factors to regulate gene expression. Therefore, we conducted a yeast two-hybrid screen to identify proteins interacting with TIE1 (Ou et al., 2011). Although TIE1 is a transcriptional repressor in plants, the full-length TIE1 is able to activate the reporter gene in yeasts (see Supplemental Figure 5 online). Activation of the reporter in yeast by a plant repressor has been reported previously (Hou et al., 2010; Sun et al., 2010). We then performed deletion analysis and found that the N terminus of TIE1, residues 1 to 108, showed no transcriptional activation activity in yeasts (see Supplemental Figure 5 and Supplemental Methods 1 online). We fused this TIE1 fragment with the GAL4 DNA binding domain (DBD) to generate the bait for the yeast two-hybrid screen and identified CIN-like TCP transcription factors. To confirm the interactions between TCPs and TIE1, we cloned all of the CIN-like TCPs in Arabidopsis and fused them with the AD domain. All of the CIN-like TCPs were able to interact with TIE1 in yeast (Figure 5A). We verified the association of TCP10 with TIE1 in vivo using BiFC analysis. Clear fluorescence signals were observed in the nuclei in the combinations cCFP-TIE1 and nYFP-TCP10, whereas the controls showed no fluorescence (Figure 5B), indicating that TIE1 physically interacts with TCP10 in vivo.
Figure 5.
TIE1 Interacts with CIN-Like TCP Transcription Factors.
(A) TIE1 interacted with CIN-like TCP transcription factors by yeast two-hybrid assays. Top, schematic representation of bait in which the 1 to 108 residues of TIE1 were fused with DBD. AD, activation domain; DBD, DNA binding domain. Transformed yeasts were spotted on control medium (-2) or selective medium (-3) in 10-, 100-, and 1000-fold dilutions. The empty vectors are used as controls.
(B) BiFC analysis of the interaction between TIE1 and TCP10. From left to right, DAPI staining showed the nuclei, GFP fluorescence, and merge of DAPI and GFP. Bars = 20 µm.
(C) Expression of chimeric protein TCP10-TIE1C in which TCP10 fused with C-terminal region (109 to 193 residues) of TIE1 leads to crinkled, serrated leaves with wavy margins. Top, schematic representation of chimeric protein TCP10-TIE1C. Middle, from left to right, 24-d-old wild-type plants and three independent TCP10-TIE1C transgenic lines. Bar = 1 cm. Bottom, from left to right, close-up views of 5th leaves from the wild type and two independent TCP10-TIE1C transgenic lines. Bars = 1 mm.
(D) Phenotypes of TCP17-TIE1C plants. Top, schematic representation of chimeric protein TCP17-TIE1C. Middle, 24-d-old wild-type plant and three independent TCP17-TIE1C transgenic lines (from left to right). Bar = 1 cm. Bottom, close-up views of the 5th leaves from the wild type and three independent TCP17-TIE1C transgenic lines (from left to right). Bars = 1 mm.
The fact that the N-terminal region of TIE1 (1 to 108 residues) interacts with TCP transcription factors and the EAR motif in the C-terminal region interacts with the corepressor TPL/TPRs suggests that TIE1 may serve as a bridge between TCPs and TPL/TPRs. To test this hypothesis, we fused TCP genes with the 3′-end region of TIE1 (corresponding to the protein region from residue 109 to 193 containing the EAR motif), and the fusion construct driven by a CaMV 35S promoter was transformed into wild-type Arabidopsis (Figures 5C and 5D). We expected that the transgenic plants expressing these chimeric proteins would mimic some phenotypes of tie1-D in which TIE1 was overexpressed and TIE1 had an increased chance to bind with TCP proteins in vivo. As expected, 16 independent TCP10-TIE1C transgenic plants displayed phenotypes similar to jaw-D (Figure 5C). Eleven independent TCP17-TIE1C transgenic plants showed hyponastic leaves similar to those observed in tie1-D (Figure 5D). These results suggest that TIE1 may regulate leaf development by interacting with TCP transcription factors.
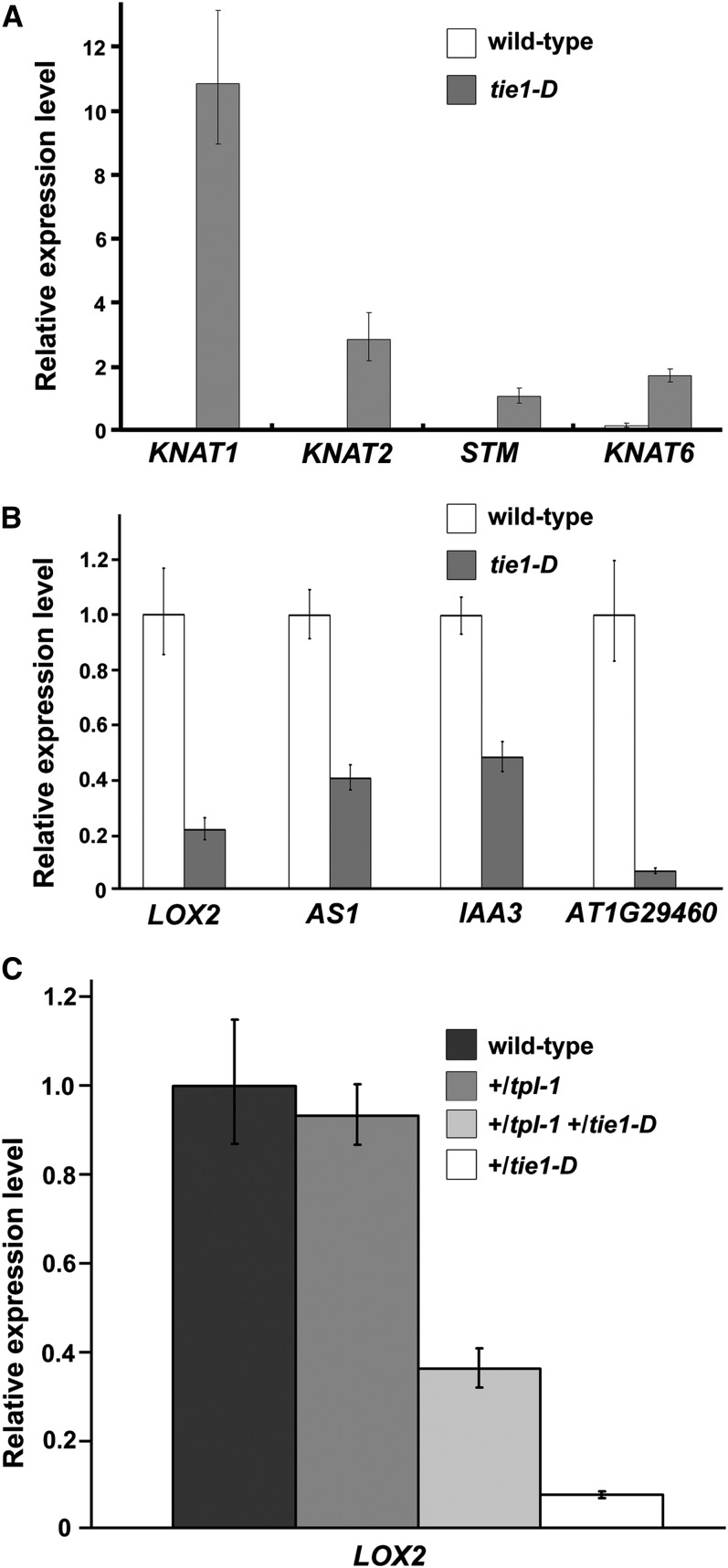
Expression of TCP Target Genes Was Altered in tie1-D
To further confirm the association of TIE1 with CIN-like TCP transcription factors, we examined the expression profiles of TCP-regulated genes when the TIE1 level was altered. Quantitative RT-PCR analysis showed that the expression of all the four class I KNOTTED-like homeobox (KNOX) genes comprising KNOTTED-LIKE FROM ARABIDOPSIS THALIANA (KNAT) 1, KNAT2, SHOOT MERISTEMLESS (STM) and KNAT2, the known target genes of TCP3 (Koyama et al., 2007), were upregulated in leaves of tie1-D (Figure 6A). Class I KNOX genes are well known to maintain the undifferentiated state of cells in the shoot apical meristem (Smith et al., 1992; Tsiantis and Hay, 2003; Hake et al., 2004; Byrne, 2005; Hay and Tsiantis, 2009). The upregulation of class I KNOX genes is consistent with the phenotype of tie1-D that the leaf cells are delayed in differentiation (Figures 1D to 1F). We also examined the expression of TCP direct target genes LIPOXYGENASE2 (LOX2), ASYMMETRIC LEAVES 1 (AS1), Indoleacetic acid-induced protein 3 (IAA3), and the SMALL AUXIN UP RNA gene At1g29460 in the leaves of tie1-D (Schommer et al., 2008; Koyama et al., 2010) and found that these four genes were significantly repressed (Figure 6B). To investigate whether the repression of these TCP direct target genes is released by the loss of function of TPL, we tested the expression of LOX2 in the leaves of the wild type, +/tpl-1, +/tpl-1 +/tie1-D double mutants, and +/tie1-D. As shown in Figure 6C, compared with that in leaves of +/tie1-D, the repression of LOX2 in +/tpl-1 +/tie1-D was largely released. These results again suggest that TIE1 plays essential roles in leaf development by regulating some important genes through interacting with TCP transcription factors and TPL/TPRs.
Figure 6.
TIE1 Regulates TCP Target Genes.
(A) Relative expression levels of Class I KNOX genes in the leaves of tie1-D. The expression levels were normalized to the constitutive expression level of ACT8. The error bars represent the sd of three biological replicates.
(B) Relative expression levels of the known TCP direct target genes in the leaves of tie1-D. The expression levels of the genes in wild-type plants were set to 1.0. The error bars represent the sd of three biological replicates.
(C) Relative expression levels of LOX2 in +/tpl-1, +/tie1-D, and +/tpl-1 +/tie1-D double mutants. The expression levels of the genes in wild-type plants were set to 1.0. The error bars represent the sd of three biological replicates.
Genetic Interactions between TIE1 and TCPs
It is reported that miR319 is one of the main regulators of TCP transcription factors (Palatnik et al., 2003). Overexpression of miR319 in jaw-D mutants causes profound defects in leaf development. We recently identified a jaw-D mutant in which four copies of the CaMV 35S enhancer are located at 4125 bp upstream of the miR319b gene (see Supplemental Figure 6 online). The jaw-D mutant was designated as jaw-5D after previously reported jaw-D mutants (Palatnik et al., 2003). In the jaw-D mutant, the expression of TCP2, TCP3, TCP4, TCP10, and TCP24 was downregulated but not silenced completely, whereas that of TCP5, TCP13, and TCP17 was not affected (Palatnik et al., 2003). Because miR319 directly affects TCP mRNA levels and TIE1 represses TCP activities, we expected that tie1-D jaw-5D would genetically enhance each other. In contrast with the tie1-D mutant, which displayed serrated and curly leaves (Figures 7A to 7C and 7G to 7I) and jaw-5D, which also showed curvature and wavy margins on the leaves (Figures 7D and 7J) (Palatnik et al., 2003), the tie1-D jaw-5D double mutants exhibited much more severe phenotypes by producing highly serrated and deeply lobed leaves (Figures 7E, 7F, 7K, and 7L). Interestingly, the tie1-D jaw-5D double mutant frequently produced ectopic shoots in the sinus of serrations (Figures 7M to 7P), which was consistent with the observations that STM and other meristem-associated genes were upregulated in the leaves of tie1-D (Figures 6A). This phenotype is also similar to those observed in TCP3SRDX seedlings in which ectopic shoots are generated on cotyledons (Koyama et al., 2007). The observed synergistic interactions between tie1-D and jaw-5D suggest that TIE1 interacts with CIN-like TCPs and inhibits their activities.
Figure 7.
Genetic Interaction between TIE1 and TCPs.
(A) to (F) Twenty-four-day-old wild-type plant (A), heterozygous tie1-D (B), homozygous tie1-D (C), homozygous jaw-5D (D), +/tie1-D jaw-5D double mutant (E), and tie1-D jaw-5D double mutant (F). The double mutants displayed deeply lobed leaves. Bars = 1 mm.
(G) to (L) The close-up views of the 6th leaves from (A) to (F). Bars = 1 mm.
(M) to (P) Ectopic shoots were frequently produced in the sinus of leaf serrations in the double mutants. Bars = 1 mm in (M) to (O) and 100 µm in (P).
(M) and (O) Close-up views of ectopic shoots.
(N) Two leaves with ectopic shoots from 30-d-old +/tie1-D jaw-5D double mutants.
(P) Scanning electron micrograph of ectopic shoot from 30-d-old +/tie1-D jaw-5D double mutant.
(Q) and (R) Eighteen-day-old plants of tcp3 tcp4 tcp10 triple mutant ([Q], top) and TIE1mEARVP16 transgenic line in the tcp3 tcp4 tcp10 background ([R], top) and the scanning electron micrograph of their leaf adaxial epidermal cells (bottom). Bars = 1 mm (top) and 50 µm (bottom).
(S) The 15-d-old seedlings of 35S-mTCP4-10, GFP-TIE1-15 35S-mTCP4-10, and GFP-TIE1-15 transgenic plants (from left to right). Bar = 1 mm.
To further clarify the genetic interaction between TIE1 and TCPs, we transformed the TIE1mEARVP16 into tcp3 tcp4 tcp10 triple mutants. Five independent lines produced smaller leaves, which showed partial rescue of the larger leaves of tcp3 tcp4 tcp10 (Figures 7Q and 7R). Scanning electron microscopy analysis confirmed that the late differentiated cell phenotype of tcp3 tcp4 tcp10 leaves was also partially rescued (Figures 7Q and 7R), suggesting that overexpression of TIE1mEARVP16 probably activates other TCPs (e.g., TCP2, TCP5, TCP13, TCP17, and TCP24).
We then transformed 35S-mTCP4, in which a miR319-resistant mTCP4 was driven by a CaMV 35S promoter, into wild-type Arabidopsis to generate TCP4 overexpression lines that showed similar phenotypes as previously reported (Schommer et al., 2008). We crossed 35S-mTCP4 line (35S-mTCP4-10) to a TIE1 overexpression line (GFP-TIE1-15). The leaves of the double overexpression lines were similar to those of GFP-TIE1-15 (Figure 7R), indicating that the leaf phenotype of GFP-TIE1-15 is epistatic to that of 35S-mTCP4-10. These observations further support that TIE1 regulates leaf development by modifying the activities of TCPs.
DISCUSSION
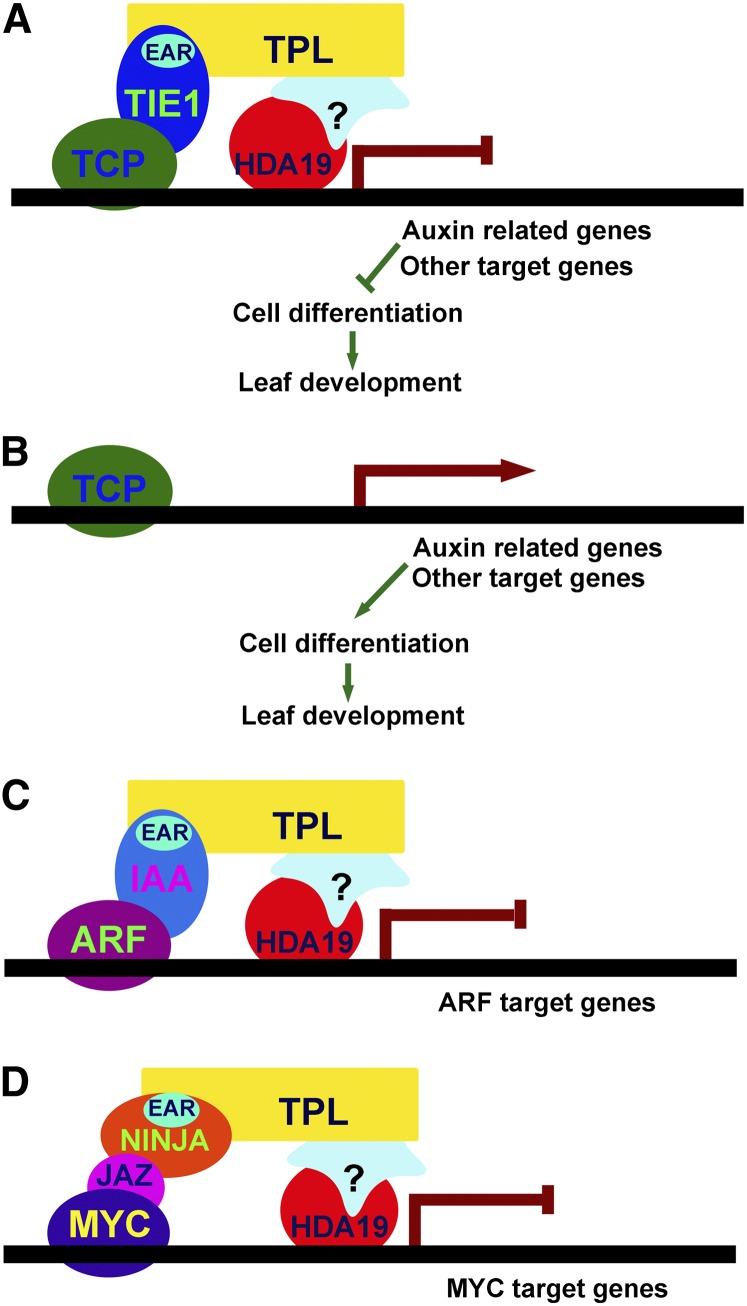
In this study, we identify TIE1, which encodes a novel EAR motif–containing protein and plays important roles in leaf development. Overexpression of TIEs caused hyponastic and serrated leaves, whereas disruption of TIEs led to epinastic leaves. The epidermal cells of mature leaves of the tie1-D mutant were less differentiated and smaller than those of the wild type, whereas the leaf epidermal cells in the TIE-deficient lines were differentiated earlier than those in wild-type controls. Therefore, we conclude that TIE1 controls leaf development by affecting cell differentiation. We further uncovered a molecular framework by which TIE1 regulates leaf development. TIE1, through its C-terminal EAR motif, recruits corepressors TPL/TPRs to repress the activities of TCP transcription factors that interact with the N-terminal domain of TIE1. We propose a model that TIE1 forms tertiary complexes with TCPs and TPL/TPRs to regulate leaf development (Figure 8A). The evidence is as follows. First, TIE1 is a transcription repressor with a monopartite nuclear localization signal and an EAR repressor motif (Dingwall et al., 1988; Ohta et al., 2001; Hiratsu et al., 2002). Second, yeast two-hybrid and BiFC analysis showed that TIE1 interacted with TPL/TPRs through the EAR motif, similar to other reported EAR-containing proteins (Szemenyei et al., 2008; Pauwels et al., 2010; Causier et al., 2012). Third, TIE1 physically and genetically interacted with CIN-like TCP transcription factors. Fourth, disruption of TIE1 could lead to single or fused cotyledons and epinastic true leaves similar to those observed in tpl-1 and TCP3 overexpression lines (Long et al., 2006; Koyama et al., 2007). Fifth, all of the eight CIN-like TCP genes have overlapping expression patterns with both TIE1 and TPL in the leaves (Koyama et al., 2007). Finally, quantitative real-time PCR analysis showed that TCP downstream genes LOX2, AS1, IAA3, and At1g29460 were regulated by TIE1. Taken together, this evidence demonstrates that TIE1 negatively regulate the activities of TCPs as a transcription repressor by recruiting the corepressor TPL/TPRs.
Figure 8.
A Working Model for TIE1 Function.
(A) In the shoot apical meristem and young leaves where TIE1 is expressed, TIE1 interacts with the TCP proteins through the N-terminal portion and also interacts with corepressor TPL/TPRs through the C-terminal EAR motif. The recruitment of TPL/TPRs to TCP proteins inhibits the expression of TCP target genes such as auxin-related genes and other genes that promote cell differentiation. Thus, the cells in young tissues remain undifferentiated.
(B) In old leaves where TIE1 is expressed at low levels or not expressed, TCP activities would not be repressed by TIE1 and the target genes are activated to promote cell differentiation.
(C) The regulation of TCP transcriptional activities by TIE1 during leaf development is similar to the regulation of auxin response factor activities by AUX/IAAs in auxin signaling.
(D) The regulation of TCP transcriptional activities by TIE1 during leaf development resembles the modulation of MYC transcription factor activities by the combination of JAZ and NINJA in JA signaling.
Our findings reveal that TIE1 is a key transcriptional repressor that regulates leaf development by modulating the activities of TCP proteins. The CIN-like TCPs promote cell differentiation in leaves (Nath et al., 2003; Palatnik et al., 2003; Crawford et al., 2004; Koyama et al., 2007, 2010; Ori et al., 2007; Martín-Trillo and Cubas, 2010). Disruption of CIN-like TCP genes causes severe leaf abnormalities, including leaf curvature, increased leaf serration, and undifferentiated leaf epidermal cells. These phenotypes are similar to those observed in the tie1-D mutants (Nath et al., 2003; Palatnik et al., 2003; Efroni et al., 2008; Koyama et al., 2010). In young leaves, in which TIE1 is highly expressed, TIE1 may interact with both TCPs and TPL/TPRs to form transcriptional repressor complexes to repress the expression of TCP target genes (Figure 8A), thus preventing the cells in young tissues from undergoing differentiation. In mature leaves, TIE1 expression is decreased and the activities of TCP proteins may not be inhibited by TIE1. Therefore, the downstream genes of TCPs are activated to promote cell differentiation (Figure 8B). We found that the class I KNOX genes (e.g., STM, KNAT1, KNAT2, and KNAT6) are also regulated by TIE1. KNOX genes are critical to maintain the undifferentiated cell population in the shoot apical meristem, and repression of KNOX expression is crucial for formation of simple leaves (Tsiantis and Hay, 2003; Hake et al., 2004; Byrne, 2005; Hay and Tsiantis, 2009). Several essential regulators of KNOX genes have been identified. For instance, the polycomb group protein CLF represses the expression of KNOX genes in leaves (Katz et al., 2004; Schubert et al., 2006; Xu and Shen, 2008), and the MYB transcription factor AS1 acts together with LATERAL ORGAN BOUNDARY DOMAIN proteins AS2 and JAGGED LATERAL ORGANS (JLO) to repress KNOX expression (Phelps-Durr et al., 2005; Guo et al., 2008; Rast and Simon, 2012). AS1 also coordinates with auxin in the repression of KNAT1 (Hay et al., 2006). It was reported that class I KNOX gene expression was indirectly upregulated in the TCP-SRDX plants (Koyama et al., 2007). Therefore, it is reasonable to predict that the expression of class I KNOX genes will be changed in CIN-like TCP multiple knockout mutants. Our data in this study suggest that TIE1 regulates the expression of class I KNOX genes in leaf development, although it is an indirect regulator (i.e., by recruiting TPL/TPR corepressors to repress the activities of TCP transcription factors).
The molecular mechanism by which TIE1 regulates transcriptional activities of TCPs during leaf development resembles that used by AUXIN (AUX)/IAAs to control transcriptional activities of auxin response factors in auxin signaling and that used by Jasmonate ZIM-domain (JAZ) and Novel Interactor of JAZ (NINJA) to modulate activities of MYC transcription factors in JA signaling (Figures 8C and 8D) (Szemenyei et al., 2008; Pauwels et al., 2010). The common element in regulation of the activities of transcription factors in the above-described cases is the employment of an EAR motif–containing repressor. TIE1, AUX/IAA, and NINJA all contain an EAR motif that is used to recruit the transcriptional corepressors TPL/TPRs. The EAR-containing protein and TPL/TPR complex then repress the activities of a particular transcription factor (Figure 8). The EAR-TPL complex probably is widely used in various developmental processes and in hormone and stress signal transduction because there are 219 EAR-containing transcription regulators that belong to 21 families in Arabidopsis (Ohta et al., 2001; Hiratsu et al., 2002; Ciftci-Yilmaz et al., 2007; Kagale et al., 2010; Causier et al., 2012; Shyu et al., 2012). TIEs are novel EAR-containing factors that can serve as an adaptor similar to AUX/IAA proteins, bridging the interactions between TPL/TPRs and transcription factors.
The phenotypes of tie1-D were not completely identical to those observed in jaw-D, although in both mutants the TCP activities were downregulated. We speculate that, in addition to CIN-like TCP proteins, TIE1 may also bind to other transcription factors. Indeed, we identified other putative transcription factors that interact with TIE1 in a yeast two-hybrid screen using a GAL4-AD transcription factor library (Ou et al., 2011). These TIE1-interacting transcription factors include other TCPs, such as TCP18 and TCP20, basic helix-loop-helix proteins, and WRKY transcription factors. When and how TIE1 interacts with these transcription factors to regulate a specific developmental process need to be investigated in the future.
The dynamic spatial and temporal regulation of leaf differentiation by CIN-TCPs plays important roles in leaf development (Ori et al., 2007; Koyama et al., 2010; Shleizer-Burko et al., 2011). Interestingly, alteration of TCP expression at different leaf developmental stages caused rather different leaf forms in Arabidopsis and tomato (Efroni et al., 2008; Shleizer-Burko et al., 2011), which indicates that the temporal and spatial TCP activities are important for leaf development. The molecular framework by which TIE1 regulates leaf development reveals a mechanism for modulation of TCP activities. This mechanism may enable plants to regulate TCP activities more flexibly and accurately.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 was used, except the tpl1-1 mutant is in the Landsberg erecta background. Gain-of-function mutants tie1-D and jaw-5D were obtained from our T-DNA insertion mutant collection (Qin et al., 2003). Arabidopsis seeds from the wild type, mutants, transgenic plants, and crossed plants were placed on half-strength Murashige and Skoog medium with or without 20 µg/mL dl-phosphinothricin or 50 µg/mL kanamycin. The plates were put at 4°C for 3 d to complete synchronization before being placed at 22 ± 2°C under long-day conditions (16-h light and 8-h dark) for 7 d. Green seedlings were transferred to soil and grown under the same conditions as described above. For the BiFC and TIE1 transcriptional activity assays, Nicotiana benthamiana was grown in soil at 22 ± 2°C under long day conditions.
PCR Analysis and Gene Expression Assays
All the primers used in this study are listed in Supplemental Table 1 online.
The flanking sequences of the T-DNA insertion site in tie1-D and jaw-5D were identified by thermal asymmetric interlaced PCR as previously described (Qin et al., 2003). The arbitrary primers (AD) and specific ones used in thermal asymmetric interlaced PCR were the same as those described previously (Qin et al., 2003). Primers DL1, P1, and P2 were used for cosegregation analysis of tie1-D; primers DL1, jaw-5D-P1, and jaw-5D-P2 were used for cosegregation analysis of jaw-5D. PCR was performed for 26 to 40 cycles (94°C for 30 s, 56 to 60°C for 30 s, and 72°C for 2 min).
For quantitative RT-PCR, total RNAs of the seedlings from wild-type, GFP-TIE1-15, tie1-D and tie1-455, or young or old leaves from 35-d-old or 40-d-old wild-type and tie1-D were extracted using TRIzol reagent (Invitrogen). Five micrograms of total RNA was reverse transcribed using the M-MLV kit (Invitrogen) in a reaction volume of 20 μL. The cDNA was diluted and used as a template for RT-PCR or quantitative RT-PCR. Quantitative RT-PCR was performed with three biological repeats using SYBR Green real-time PCR Master Mix (Toyobo) as described previously (Luo et al., 2011; Wang et al., 2012). PCR was performed at 95°C for 2 min, 94°C for 10 s, 55°C for 20 s, and 72°C for 20 s. The relative expression level of each gene was calculated using the 2−ΔΔCT (cycle threshold) method, and ACT8 was used as an internal control (Livak and Schmittgen, 2001).
Generation of Binary Constructs and Transformation
The coding region of TIE1 was isolated from the tie1-D cDNA by RT-PCR using primers TIE1-1 and TIE1-2. The DNA fragments were cloned into the EcoRV site of pBluescript SK+ to generate pBS-TIE1 (with ATG near the T7 primer) and pBS-TIE1r (with ATG near the T3 primer). Alternatively, the coding region of TIE1 was isolated from the tie1-D cDNA by RT-PCR using primers TIE1-3 and TIE1-4 and cloned into pDONRP2rP3 (Invitrogen) to generate pEN-R2-TIE1-L3. The CaMV 35S enhancer tetrad was amplified from pSKI015 with the primers Enh-1 and Enh-2 and cloned into pDONRP4P1r (Invitrogen) to generate pEN-L4-4Enh-R1 (Weigel et al., 2000). The 2790 bp of promoter between the ATG and 98 bp upstream of the T-DNA insertion site of tie1-D was amplified from Arabidopsis genomic DNA using the primers TIE1P-1 and TIE1P-2 and cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-TIE1P. Alternatively, primer pair TIE1P-3 and TIE1P-4 was used for amplification of the TIE1 promoter, which was cloned into pDONRP4P1r (Invitrogen) to generate pEN-L4-TIE1P-R1. For the generation of binary vectors, the HindIII-EcoRI fragment including the CaMV 35S promoter from pWM101 was introduced into pPZP111 (Hajdukiewicz et al., 1994) to generate pQG111. The overexpression construct of 35S-TIE1 was generated by ligation of the KpnI-PstI–digested fragments from pBS-TIE1 and KpnI-PstI–digested vector pQG111. The overexpression construct 4Enhancer-TIE1 was generated by LR reaction of four plasmids, including pK7m34GW, pEN-L4-4Enh-R1, pENTRY-TIE1P, and pEN-R2-TIE1-L3. To generate 35S-TIE3-RNAi construct, the full-length TIE3 coding region was amplified from the seedling cDNA using the primers TIE3-3 and TIE3-4. The fragment was cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-AsTIE3. 35S-TIE3-RNAi was generated by LR reaction with the plasmids pENTRY-AsTIE3 and pBIB-BASTA-35S-GWRNAi.
TIE1mEAR was amplified from pBS-TIE1 using primers TIE1-1 and TIE1mEAR-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-TIE1mEAR or using primers TIE1-1 and TIE1mEARN-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-TIE1mEARN in which ATG near T3 primer direction and without the stop codon. Alternatively, primer pair of TIE1topo-1 and TIE1mEAR-2 was used to amplify TIE1mEAR and the fragment was cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-TIE1mEAR. TIE1ΔEARN was amplified from pBS-TIE1 using primers TIE1-1 and TIE1ΔEARN-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-TIE1ΔEARN in which ATG was near T3 primer direction and without the stop codon. VP16 was amplified from pTA7002 (Aoyama and Chua, 1997) using primers VP16-1 and VP16-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-VP16. The dominant-negative constructs 35S-TIE1mEARVP16 and 35S-TIE1ΔEARVP16 were generated by ligation of the XmaI-PstI–digested vector pQG111, XmaI-SalI–digested fragment from pBS-TIEmEARN or pBS-TIEΔEARN, and SalI-PstI–digested fragment from pBS-VP16. TIE1P-TIE1mEAR was generated by LR reaction with the three plasmids, including pK7m24GW, pEN-L4-TIE1P-R1, and pENTRY-TIE1mEAR.
To test subcellular localization, TIE1 coding regions with or without stop codon were amplified from pBS-TIE1 using primers TIE1topo-1 and TIE1-2 or TIE1topoN-2 and cloned into pENTR/D-TOPO to generate pENTRY-TIE1 and pENTRY-TIE1N. pENTRY-TIE1 was cloned into pB7WGF2 using LR reaction to generate GFP-TIE1, and pENTRY-TIE1N was cloned into pB7FWG2 using LR reaction to generate TIE1-GFP.
For examination of TIE1 expression, TIE1P-GUS was generated by LR reaction between the plasmids pENTRY-TIE1P and pBGWFS7. To examine TPL expression in leaves, the TPL 2.8-kb promoter was amplified from Arabidopsis genomic DNA with the primers TPLP-1 and TPLP-2 and cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-TPLP. The pTPLP-GUS was generated by LR reaction between pENTRY-TPLP and pHGWFS7.
To generate TCP-TIE1C constructs, the coding region of TCP10, TCP17 was amplified from cDNA of wild-type plants by RT-PCR using primer pairs of TCP10-1 and TCP10N-2, and TCP17-1 and TCP17N-2. The fragments were cloned into pENTRY/D-TOPO to generate pENTRY-TCP10N and pENTRY-TCP17N, respectively. The CaMV 35S promoter was amplified from pQG111 using primers 35S-4 and 35S-5 and cloned into pDONRP4P1r (Invitrogen) to generate pEN-L4-35S-R1. The sequence coding C terminus of TIE1 was amplified from pBS-TIE1 using primers TIE1-5 and TIE1-4 and cloned into pDONRP2rP3 (Invitrogen) to generate pEN-R2-TIE1C-L3. TCP10-TIE1C and TCP17-TIE1C constructs were generated by LR reactions of plasmids, including pK7m34GW, pEN-L4-35S-R1, pENTRY-TCP10N, or pENTRY-TCP17N and pEN-R2-TIE1C-L3.
To generate the TCP overexpression line, the CaMV 35S promoter was amplified from pWM101 with the primers 35SB4-1 and 35SB1R-2 and cloned into pDONRP4P1r (Invitrogen) to generate pEN-L4-35S-R1. The full-length coding region of TCP4 was amplified from Arabidopsis cDNA by RT-PCR and cloned into pENTRY/D-TOPO to generate pENTRY-TCP4. Then, the point mutation was introduced using primers TCP4m-1 and TCP4m-2 to generate pENTRY-mTCP4. 35S-mTCP4 was generated by LR reaction with pK7m24GW, pEN-L4-35S-R1, and pENTRY-mTCP4.
Constructs were transformed into Agrobacterium tumefaciens GV3101/pMP90 and then into Arabidopsis as described previously (Qin et al., 2005).
Staining and Microscopy
The histochemical GUS staining was performed as described previously (Qin et al., 2005). Briefly, tissues from TIE1P-GUS or TPLP-GUS transgenic lines were immerged in a staining buffer containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide. Samples were vacuumed for 10 min and then put in a 37°C incubator overnight. The staining buffer was removed, and the samples were cleared using 70% ethanol before microscopy.
For 4′,6-diamidino-2-phenylindole (DAPI) staining, leaves were dissected and soaked in 1 µg/mL DAPI solution. Nuclear localization observation was performed using a confocal laser scanning microscope (Leica TCS SPE confocal microscope).
For scanning electron microscopy, the 5th mature leaf from 60-d-old wild-type and tie1-D plants and 5th leaves from 27-d-old wild-type and TIE1mEAR-7 transgenic lines were isolated and fixed in the FAA buffer containing 50% ethanol, 6% glacial acetic acid, and 5% formaldehyde for 4 h at 25°C. Serial ethanol dehydration and isoamyl acetate substitution were then performed. Leaves were dried at critical point in liquid CO2. Samples were analyzed using a scanning electron microscope (Jeol JSM-6610LV) as described in the user manual. The areas of leaf epidermal cells were analyzed using SPOT software (SPOT Imaging Solutions). About 100 cells in the given region of leaves were measured by SPOT software, and the frequency of cells with different cell size was calculated.
Yeast Two-Hybrid Assays
To test the interaction between TIE1 and TPL family proteins, the N terminus of TPL or TPRs including the C terminus to lissencephaly homology domain were amplified from Arabidopsis cDNA by RT-PCR using primer pairs TPL-1 and TPL-2b, TPR1-1 and TPR1-2b, TPR2-1 and TPR2-2b, TPR3-1 and TPR3-2b, and TPR4-1 and TPR4-2b, respectively. The products were cloned into pENTR/D-TOPO to generate pENTRY-NTPL and pENTRY-NTPRs. The bait constructs pDEST32-NTPL and pDEST32-NTPRs were generated by LR reaction between these plasmids and pDEST32 (Invitrogen), respectively. The prey constructs pDEST22-TIE1, pDEST22-TIE2, pDEST22-TIE3, and pDEST22-TIE4 were generated by LR reaction between pENTRY-TIE1, pENTRY-TIE2, pENTRY-TIE3, and pENTRY-TIE4 and pDEST22 (Invitrogen). Bait plasmids and prey plasmids or the blank pDEST22 were cotransformed into yeast strain AH109 (Clontech), respectively.
To verify the interactions between TIE1 and CIN-like TCPs, all the CIN-like TCP genes were cloned into pDEST22 as preys. Bait DBD-TIE1 plasmids and prey plasmids of pDEST22-TCPs or the blank pDEST22 were cotransformed into yeast strain AH109, respectively.
Medium supplemented with SD-Leu-Trp-His and 5 mM 3-amino-1,2,4 triazole was used for selection.
Transient Expression Analysis in Leaves of N. benthamiana
For generation of 35S-UAS-GUS reporter, Gal4UAS was amplified from pTA7002 (Aoyama and Chua, 1997) using primers Gal4UAS-1 and Gal4UAS-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-Gal4UAS; 35S promoter without TATA box was amplified from pBI121 using primers 35S-1 and 35S-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-35S; 35S minimum promoter, GUS, and NOS terminator were amplified from pBI121 using primers 35S-3 and NosT-2 and cloned into the EcoRV site of pBluescript SK+ to generate pBS-GUST. The reporter construct 35S-UAS-GUS was generated by ligation of four fragments: the HindIII-EcoRI fragment from pBI121, the HindIII-SacII fragment from pBS-35S, the SacII-NcoI fragment from pBS-Gal4UAS, the NcoI-EcoRI fragment from pBS-GUST. G4BD, Gal4BD-TIE1, Gal4BDTIE1mEAR, and G4DBD-TIE1ΔEAR were amplified from pYF503 series used in the transactivation activity assays in yeasts using primers of G4BD-1, G4BD-2, TIE1-2, TIE1mEAR-2, and TIE1ΔEAR-2. The products were cloned into pENTR/D-TOPO (Invitrogen) to generate pENTRY-G4BD, pENTRY-G4BD-TIE1, pENTRY-G4BDTIE1mEAR, and pENTRY-G4BD- TIE1ΔEAR, respectively. Then, the four entry plasmids were used in LR reaction with pK2GW7 to generate the effector constructs.
The plasmids of reporter and effector constructs were transformed into Agrobacterium GV3101/pMP90, and then different effectors were coinfiltrated with the reporter 35S-UAS-GUS and pCam-P19 into leaves of N. benthamiana as described previously (Voinnet et al., 2003). After incubation in dark for 24 h and then in light for 72 h, the leaves were used for histochemical GUS staining.
BiFC Assays
Full-length coding sequences of TCP10 and TPL were amplified from Arabidopsis cDNA by RT-PCR using primers of TPL-1, TPL-2, TCP10-1, and TCP10-2. The products were cloned into pENTRY/D-TOPO to generate pENTRY-TCP10 and pENTRY-TPL. cCFP-TIE1 or nYFP-TIE1 was generated by LR reactions between pcCFPxGW or pnYFPxGW (Ou et al., 2011) and pENTRY-TIE1. cCFP-TPL and nYFP-TCP10 were generated from LR reactions between pcCFPxGW or pnYFPxGW and pENTRY-TPL or pENTRY-TCP10. BiFC analysis was performed as described previously (Ou et al., 2011) with some modifications.
These plasmids were transformed into Agrobacterium GV3101/pMP90. The Agrobacterium harboring cCFP-TIE1 or nYFP-TIE1 was coinfiltrated with pCam-P19 and nYFP-TCP10 or cCFP-TPL into the leaves of N. benthamiana. The empty pcCFPxGW and pnYFPxGW vectors were used as negative controls. After incubation in dark for 24 h and then in light for 72 h, the leaves were dissected for observation under the microscope (Leica SPE).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative, Rice Genome Annotation Project, or GenBank/EMBL databases under the following accession numbers: TIE1, At4g28840; TIE2, At2g20080; TIE3, At1g29010; TIE4, At2g34010; TPL, At1g15750; TPR1, At1g80490; TPR2, At3g16830; TPR3, At5g27030; TPR4, At3g15880; TCP2, At4g18390; TCP3, At1g53230; TCP4, At3g15030; TCP5, At5g60970; TCP10, At2g31070; TCP13, At3g02150; TCP17, At5g08070; TCP24, At1g30210; LOX2, At3g45140; AS1, At2g37630; and IAA3, At1g04240.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phenotypes of Dissected Leaves from tie1-D.
Supplemental Figure 2. TIE1 Interacts with Other TPL Family Proteins through the EAR Motif, but Not with SAP18.
Supplemental Figure 3. TIE3 and TIE4 May Have Redundant Functions with TIE1 and Disruption of TIE Genes Led to Epinastic Leaves.
Supplemental Figure 4. Dominant-Negative Disruption of TIE1 Caused Epinastic Leaves.
Supplemental Figure 5. Examination of Activation Activity of TIE1 in Yeasts.
Supplemental Figure 6. Identification of jaw-5D.
Supplemental Table 1. The Primer List Used in This Study.
Acknowledgments
We thank Yunde Zhao (University of California at San Diego), Yuxian Zhu (Peking University), and Hongwei Guo (Peking University) for discussions and valuable suggestions. We also thank Jeff Long (Salk Institute) for kindly providing the seeds of tpl-1, Tomotsugu Koyama (Kyoto University) for the seeds of tcp3 tcp4 tcp10 triple mutants, Yunde Zhao (University of California at San Diego) for jaw-5D seeds, and Jia Li (Lanzhou University) for the vector pBIB-BASTA-35S-GWRNAi. This work was supported by the National Key Basic Research Program of the People’s Republic of China (Grant 973 2009CB941503), by the National Natural Science Foundation of China (Grant 30970248), and by Program for New Century Excellent Talents in University from the Ministry of Education of China (Grant NCET-09-0188). This work was also partially supported by the 111 Project (B06001).
AUTHOR CONTRIBUTIONS
G.Q. conceived the project. G.Q. and Q.T. designed the experiments. G.Q., Q.T., D.G., B.W, F.Z., C.P., H.J., and J.Z. performed the experiments. Q.T., G.Q., D.G., T.W., H.G., and L.-J.Q. analyzed the data. G.Q. and L.-J.Q. wrote the article.
Glossary
- miR319
microRNA319
- IAA
indole-3-acetic acid
- CaMV
cauliflower mosaic virus
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- JA
jasmonic acid
- BiFC
bimolecular fluorescence complementation
- DAPI
4′,6-diamidino-2-phenylindole
References
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Byrne M.E. (2005). Networks in leaf development. Curr. Opin. Plant Biol. 8: 59–66 [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S., Morsy M.R., Song L., Coutu A., Krizek B.A., Lewis M.W., Warren D., Cushman J., Connolly E.L., Mittler R. (2007). The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Crawford B.C., Nath U., Carpenter R., Coen E.S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Dingwall C., Robbins J., Dilworth S.M., Roberts B., Richardson W.D. (1988). The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107: 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Blum E., Goldshmidt A., Eshed Y. (2008). A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M.C. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S., Smith H.M., Holtan H., Magnani E., Mele G., Ramirez J. (2004). The role of knox genes in plant development. Annu. Rev. Cell Dev. Biol. 20: 125–151 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2009). A KNOX family TALE. Curr. Opin. Plant Biol. 12: 593–598 [DOI] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. (2008). A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Ohta M., Matsui K., Ohme-Takagi M. (2002). The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22: 3574–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen E.E., Fox S., de Reuille P.B., Kennaway R., Bensmihen S., Avondo J., Calder G.M., Southam P., Robinson S., Bangham A., Coen E. (2012). Generation of leaf shape through early patterns of growth and tissue polarity. Science 335: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Li H., Tiwari S.B., Hagen G., Guilfoyle T.J. (2011). Identical amino acid substitutions in the repression domain of auxin/indole-3-acetic acid proteins have contrasting effects on auxin signaling. Plant Physiol. 155: 1252–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Luo Y., Qin G., Zhang J., Liang Y., Song Y., Zhao M., Tsuge T., Aoyama T., Liu J., Gu H., Qu L.J. (2011). D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 23: 1352–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Masuda H.P., Cabral L.M., De Veylder L., Tanurdzic M., de Almeida Engler J., Geelen D., Inzé D., Martienssen R.A., Ferreira P.C., Hemerly A.S. (2008). ABAP1 is a novel plant Armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath U., Crawford B.C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Nishal B., Tantikanjana T., Sundaresan V. (2005). An inducible targeted tagging system for localized saturation mutagenesis in Arabidopsis. Plant Physiol. 137: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Ou B., Yin K.Q., Liu S.N., Yang Y., Gu T., Wing Hui J.M., Zhang L., Miao J., Kondou Y., Matsui M., Gu H.Y., Qu L.J. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4: 546–555 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen E., Wu X., Schommer C., Schwab R., Carrington J.C., Weigel D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., Carrington J.C., Weigel D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps-Durr T.L., Thomas J., Vahab P., Timmermans M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17: 2886–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G., Gu H., Zhao Y., Ma Z., Shi G., Yang Y., Pichersky E., Chen H., Liu M., Chen Z., Qu L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G.J., et al. (2003). Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci. 165: 941–949 [Google Scholar]
- Rast M.I., Simon R. (2012). Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24: 2917–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J.F., Aggarwal P., Chételat A., Cubas P., Farmer E.E., Nath U., Weigel D. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shleizer-Burko S., Burko Y., Ben-Herzel O., Ori N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138: 695–704 [DOI] [PubMed] [Google Scholar]
- Shyu C., Figueroa P., Depew C.L., Cooke T.F., Sheard L.B., Moreno J.E., Katsir L., Zheng N., Browse J., Howe G.A. (2012). JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24: 536–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.G., Greene B., Veit B., Hake S. (1992). A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116: 21–30 [DOI] [PubMed] [Google Scholar]
- Steiner E., Efroni I., Gopalraj M., Saathoff K., Tseng T.S., Kieffer M., Eshed Y., Olszewski N., Weiss D. (2012). The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.P., Galbraith D.W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60: 241–257 [DOI] [PubMed] [Google Scholar]
- Sun S.J., Guo S.Q., Yang X., Bao Y.M., Tang H.J., Sun H., Huang J., Zhang H.S. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61: 2807–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tsiantis M., Hay A. (2003). Comparative plant development: The time of the leaf? Nat. Rev. Genet. 4: 169–180 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang W.Y., Zhang L., Xing S., Ma Z., Liu J., Gu H., Qin G., Qu L.J. (2012). Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J. Genet. Genomics 39: 81–92 [DOI] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Shen W.H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18: 1966–1971 [DOI] [PubMed] [Google Scholar]