Winter annual and biennial plants use the prolonged period of winter cold as an environmental cue to ensure proper floral transition in early spring. This work demonstrates that a sophisticated regulatory network by two gene families coordinates the proper vernalization response in Arabidopsis.
Abstract
Vernalization is an environmentally induced epigenetic switch in which winter cold triggers epigenetic silencing of floral repressors and thus provides competence to flower in spring. Vernalization triggers the recruitment of chromatin-modifying complexes to a clade of flowering repressors that are epigenetically silenced via chromatin modifications. In Arabidopsis thaliana, VERNALIZATION INSENSITIVE3 (VIN3) and its related plant homeodomain finger proteins act together with Polycomb Repressive Complex 2 to increase repressive histone marks at floral repressor loci, including FLOWERING LOCUS C (FLC) and its related genes, by vernalization. Here, we show that VIN3 family of proteins nonredundantly functions to repress different subsets of the FLC gene family during the course of vernalization. Each VIN3 family protein binds to modified histone peptides in vitro and directly associates with specific sets of FLC gene family chromatins in vivo to mediate epigenetic silencing. In addition, members of the FLC gene family are also differentially regulated during the course of vernalization to mediate proper vernalization response. Our results show that these two gene families cooperated during the course of evolution to ensure proper vernalization response through epigenetic changes.
INTRODUCTION
The life cycle of plants is under the influence of changing environments. One important environmental cue is seasonal change. Periodic fluctuations of photoperiod and temperature serve as cues for plants to recognize changing seasons. Plants flower at a particular season during their life cycle as a result of adaptation. Proper timing of transition from vegetative to reproductive development ensures the survival of plant species. In some plant species, the prolonged cold of winter provides a crucial clue for plants to monitor seasonal changes. In biennial and winter-annual plants, exposure to prolonged period of winter cold renders plants responsive to inductive photoperiods, thus resulting in rapid flowering in the following spring. This process is known as vernalization (Sung and Amasino, 2005; Dennis and Peacock, 2007).
In the model plant Arabidopsis thaliana, the requirement for the vernalization response is mainly a result of FLOWERING LOCUS C (FLC) expression (Michaels and Amasino, 1999; Sheldon et al., 1999; Johanson et al., 2000). Vernalization results in the epigenetic silencing of FLC. FLC encodes a MADS box DNA binding protein that functions to repress genes encoding the floral integrators, including FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) (Michaels and Amasino, 1999; Sheldon et al., 1999; Lee et al., 2000; Samach et al., 2000; Hepworth et al., 2002; Helliwell et al., 2006; Searle et al., 2006). FLC antagonizes the effect of CONSTANS by directly binding to regulatory regions of FT and SOC1 (Searle et al., 2006). FLC exists as a small gene family that includes FLOWERING LOCUS M (FLM)/MADS AFFECTING FLOWERING1 (MAF1) and MAF2∼MAF5 (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001; Helliwell et al., 2006; Searle et al., 2006). Like FLC, other FLC clade members act as floral repressors when they are ectopically expressed (Ratcliffe et al., 2003), and some of them have been reported to be repressed by vernalization (Ratcliffe et al., 2003; Gu et al., 2009; Sheldon et al., 2009).
Mechanisms underlying vernalization-mediated gene silencing are well documented for the FLC repression (Schmitz and Amasino, 2007; Kim et al., 2009). Vernalization triggers a series of chromatin remodeling complexes that affect histone modifications at FLC chromatin (Bastow et al., 2004; Sung and Amasino, 2004). FLC mRNA expression is repressed during the cold exposure, and several repressive histone marks accumulate at FLC chromatin, including methylations at Histone H3 Lys 9 and Histone H3 Lys 27 (H3K27). The accumulation of these histone modifications at FLC chromatin depends on the activity of chromatin-remodeling complexes, including Polycomb Repression Complex 2 (PRC2). PRC2 is an evolutionarily conserved H3K27 methyltransferase. Increased enrichment of PRC2 at FLC chromatin by vernalization is critical for the stable repression of FLC (Wood et al., 2006; De Lucia et al., 2008).
PRC2 biochemically copurifies with members of the VERNALIZATION INSENSITIVE3 (VIN3) family of proteins, including VIN3, VIN3-LIKE1 (VIL1)/VERNALIZATION5 (VRN5) and VIL2/VERNALIZATION5/VIN3-LIKE1 (VEL1) (Wood et al., 2006; De Lucia et al., 2008). The VIN3 family of proteins contain a plant homeodomain (PHD) finger, and the association of PHD finger proteins with PRC2 has been proposed as a part of evolutionarily conserved mechanisms in Polycomb group (PcG)-mediated gene repression (De Lucia et al., 2008). Besides FLC, repressions of some other FLC clade members by vernalization, including FLM/MAF1, MAF2, and MAF3, also depend on the presence of VIN3 and other PRC2 components (Sung et al., 2006; Sheldon et al., 2009), raising the possibility of coordinated regulation of FLC clade through chromatin modifications by vernalization.
Targets for PcG-mediated gene repression are also targets for Trithorax group (TrxG)-mediated gene activation (Schuettengruber et al., 2007). PcG and TrxG antagonistically act on common targets, and their activities determine fates of transcriptional activities of their target genes during various developmental processes. TrxG activity mediates active histone marks, including methylations at Histone H3 Lys 4 (H3K4) and H3 Lys 36 (H3K36) (Kim et al., 2005; Jiang et al., 2009; Tamada et al., 2009; Ko et al., 2010). Multiple members of a Trithorax homolog family and a Trithorax-related family (i.e., ARABIDOPSIS HOMOLOG OF TRITHORAX1 [ATX1], ATX2, ARABIDOPSIS TRITHORAX-RELATED7 [ATXR7], and ATXR4) are redundantly required for the activation of FLC and other members of FLC family (Jiang et al., 2009; Tamada et al., 2009; Yun et al., 2012). In addition, EARLY FLOWERING IN SHORT DAYS (EFS) is required for methylations both at H3K4 and H3K36 (Kim et al., 2005; Ko et al., 2010). Some members of the FLC family are also regulated by these activating chromatin-remodeling complex components for their active transcription (Jiang et al., 2009; Tamada et al., 2009; Yun et al., 2012). Despite the clear involvement of TrxG-related components in the activation of FLC and its clade, their fates during vernalization-mediated repressions of FLC and its family member genes have not been addressed.
In this study, we show that all members of VIN3 family of proteins function to collectively repress different sets of members of FLC gene family during the course of vernalization. Contributions of VIN3 family of proteins to vernalization-mediated repression of FLC gene family members differ in their target specificities and in their timing to function during the course of vernalization. Members of the FLC gene family are also differentially expressed during the course of vernalization due to differential associations of PcG and TrxG on their chromatin and function to ensure a proper vernalization response. In addition, we show that VIN3 family proteins, PRC2, and Trithorax-like complexes are coordinated to mediate the vernalization response through coordinated repressions of FLC gene family members.
RESULTS
Expression Patterns of VIN3 Family of Proteins and FLC Gene Family during the Course of Vernalization
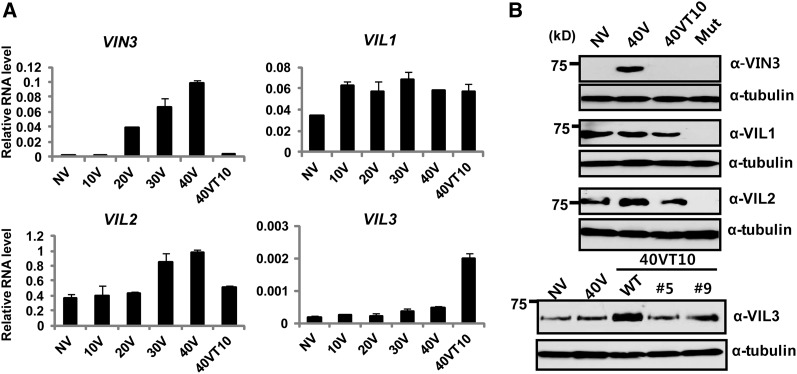
Previously, mRNA expression patterns of VIN3 gene family were determined using PCR during the course of vernalization (Sung et al., 2006; Greb et al., 2007). Using quantitative RT-PCR, we confirmed differential expression patterns among the VIN3 family (Figure 1A). VIN3 mRNA is expressed only during the vernalizing cold. VIL1/VRN5 mRNA is rather constitutively expressed, although there is a small increase by 10 d of cold. We also observed slight but significant upregulation of VIL2/VEL1 mRNA during vernalizing cold (30 and 40 d of cold). Interestingly, VIL3/VEL2 mRNA is distinctively increased after the cold exposure, suggesting its possible role in the vernalization after the cold exposure (Figure 1A). To explore roles of VIN3 family members in vernalization, we obtained antibodies against all members of VIN3 family of proteins (Figure 1B). Specificities of each antibody were tested using both recombinant proteins (see Supplemental Figure 1 online) and insertional mutants (for VIN3, VIL1/VRN5, and VIL2/VEL1) or knockdown lines (for VIL3/VEL2) (Figure 1B). Expression patterns of VIN3 family of proteins are largely correlated with their transcripts (Figure 1B), demonstrating that regulation of VIN3 family members mainly occurs at the transcription level.
Figure 1.
Expression Analysis of VIN3 Gene Family Members during the Course of Vernalization.
(A) mRNA expression analysis of VIN3 gene family members during the course of vernalization using real-time RT-PCR. NV, nonvernalized; 10V, 10 d of vernalization; 20V, 20 d of vernalization; 30V, 30 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. Relative fold change was determined by normalization with the levels of PP2A gene. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd. Primer sequence information used for real-time RT-PCR analysis is shown in Supplemental Table 1 online.
(B) Immunoblot analysis of VIN3 family of proteins during the course of vernalization. NV, nonvernalized; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. Corresponding insertional mutants were used as negative controls for VIN3, VIL1/VRN5, and VIL2/VEL1 immunoblots. Two independent RNAi lines (#5 and #9) were used as negative controls for VIL3/VEL2 immunoblots. WT, the wild type.
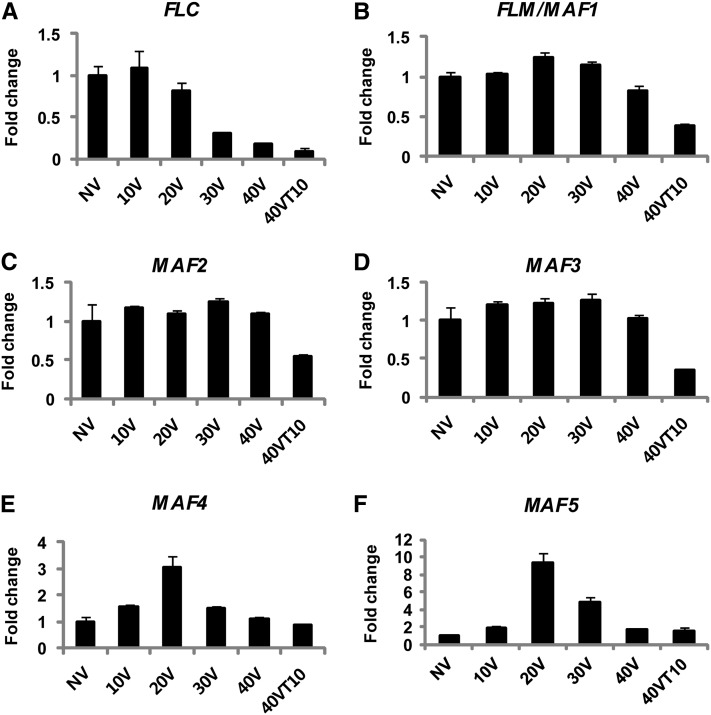
To date, FLC, FLM, MADS AFFECTING FLOWERING 2 (MAF2) and MADS AFFECTING FLOWERING 3 (MAF3) are considered to be repressed as a result of vernalization. However, there have been conflicting observations on their expression changes by vernalization (Ratcliffe et al., 2001, 2003; Sheldon et al., 2009). Furthermore, MADS AFFECTING FLOWERING 4 (MAF4) and MADS AFFECTING FLOWERING 5 (MAF5) are not considered to be repressed by vernalization because no significant change in their mRNA expressions by vernalization was observed in previous studies (Ratcliffe et al., 2001, 2003; Sheldon et al., 2009). The lack of significant change at their mRNA levels by vernalization also suggested that they are dispensable for vernalization-mediated accelerated flowering response. However, previous studies mainly determined the mRNA levels of the FLC gene family before and after vernalization but not with various durations of cold exposure.
In our analysis, we determined the mRNA expression levels of the FLC gene family following various durations of vernalization treatment (Figures 2A to 2F). As reported previously, FLC, FLM, MAF2, and MAF3 are repressed as a result of vernalization (Figure 2A to 2D). Interestingly, MAF4 and MAF5 show rather dynamic changes in their mRNA expression during the course of vernalization (Figures 2E and 2F). MAF4 and MAF5 transcripts are dramatically increased during early periods of the cold exposure and their expressions peak at 20 d of cold exposure (Figures 2E and 2F). Increased levels of MAF4 and MAF5 mRNA are eventually reduced to the levels comparable to those before cold exposure (Figures 2E and 2F), indicating that they may have a role during the early period of cold exposure.
Figure 2.
Expression Analysis of FLC Gene Family Members during the Course of Vernalization.
mRNA expression analysis of FLC gene family members during the course of vernalization using real-time RT-PCR. NV, nonvernalized; 10V, 10 d of vernalization; 20V, 20 d of vernalization; 30V, 30 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. Relative fold change was determined by normalization with the levels of the PP2A gene. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd. Primer sequence information used for real-time RT-PCR analysis is shown in Supplemental Table 1 online.
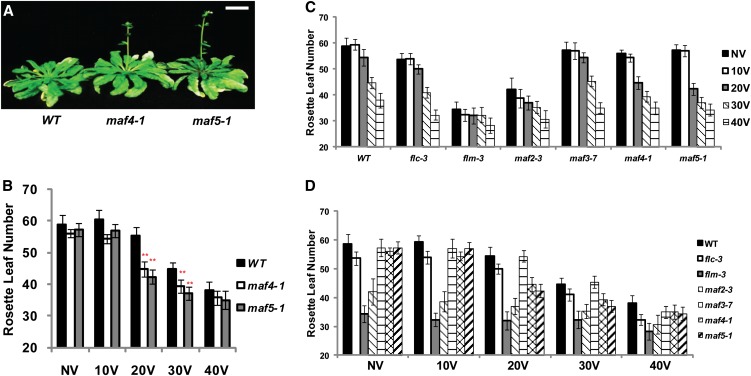
Precocious Vernalization Response in maf4 and maf5 Mutants
Our expression analysis indicates that MAF4 and MAF5 may contribute to vernalization response, especially during the early stage of vernalization (Figures 2E and 2F). To address the function of MAF4 and MAF5, we obtained insertional mutants for maf4 and maf5 (Figure 3A; see Supplemental Figure 2 online). We first focused on flowering times of plants with 20 d of cold treatment, when significant upregulation of MAF4 and MAF5 was observed (Figure 3A; see Supplemental Figure 2A online). With 20 d of cold treatment, maf4 and maf5 mutants flower earlier than the wild-type plants (Figures 3A and 3B; see Supplemental Figure 2A online). Earlier flowering of maf4 and maf5 mutants after 20 d of cold suggests that MAF4 and MAF5 may play a role in preventing precocious vernalization response.
Figure 3.
MAF4 and MAF5 Function to Prevent Precocious Vernalization Response.
(A) maf4 and maf5 mutants flower earlier than the wild-type (WT) plants in response to 20 d of cold exposure. Plants were grown under short-day conditions (8 h light and 16 h dark) after vernalization. Bar = 2 cm.
(B) Flowering times of maf4 and maf5 mutants compared with the wild type after various durations of cold exposure. For flowering time measurements, the number of rosette leaves was counted when bolts were ∼3 to 5 cm long. At least nine plants were used for each flowering time measurement.
(C) and (D) Flowering times of FLC gene family mutants compared with the wild type after various durations of cold exposure. NV, nonvernalized; 10V, 10 d of vernalization; 20V, 20 d of vernalization; 30V, 30 d of vernalization; 40V, 40 d of vernalization. All mutants are in the Columbia background, and flowering times were determined under short-day conditions (8 h light and 16 h dark) after the cold exposure. For flowering time measurements, the number of rosette leaves was counted when bolts were ∼3 to 5 cm long. At least nine plants were used for each flowering time measurement.
[See online article for color version of this figure.]
To test whether the earlier flowering observed in maf4 and maf5 mutants is unique among FLC gene family mutants, we determined flowering times with different durations of cold exposure in all FLC family mutants (Figures 3C and 3D). In the wild type, significant acceleration of flowering occurs only after 30 d of cold (Figures 3C and 3D). Prior to vernalization, maf4 and maf5 mutants do not show any significant difference in their flowering times compared with the wild type. Differences in flowering times of maf4 and maf5 mutants are apparent with 20 d of cold exposure, when the wild-type plants still do not show a significant vernalization response. After 40 d of vernalization, flowering times of maf4 and maf5 mutants are comparable to those of the wild-type plants, suggesting that the role of MAF4 and MAF5 is rather specific to a short period of cold exposure (i.e., 20 d of cold).
A previous study suggested that MAF2 acts to prevent earlier flowering by a suboptimal duration of the cold exposure, based on earlier flowering observed in maf2 mutants after a short period of cold exposure (∼2 weeks; Ratcliffe et al., 2003). Indeed, both maf1/flm and maf2 mutants also flower significantly earlier than the wild type after 20 d of cold exposure (Figures 3C and 3D; see Supplemental Figure 3 online). However, they flower significantly early even without vernalization and their flowering times are not significantly promoted by 20 d of cold exposure compared with nonvernalized controls (Figures 3C and 3D; see Supplemental Figure 3 online). Taken together, our results show that MAF4 and MAF5 specifically function to prevent a precocious vernalization response by suboptimal duration of cold exposure.
Divergent Contributions of VIN3 Gene Family to the Regulation of FLC Gene Family by Vernalization
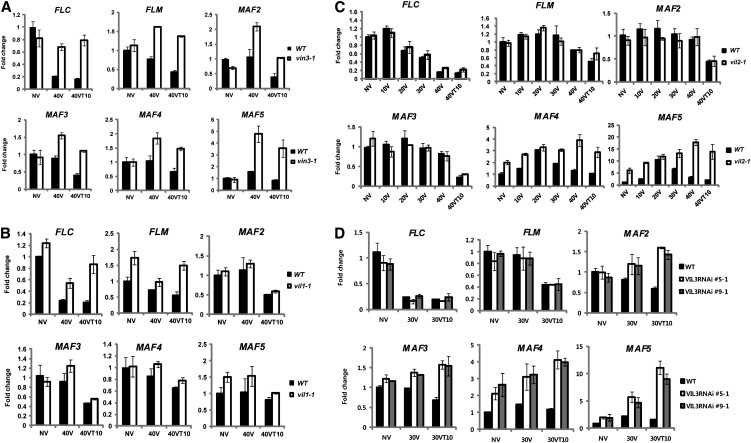
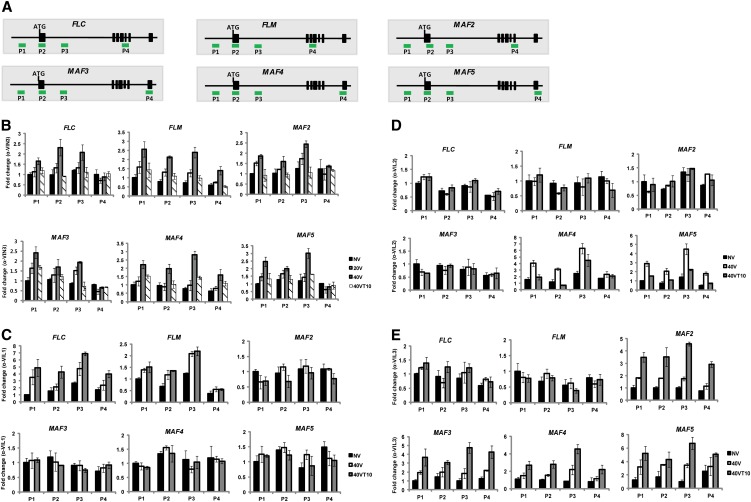
VIN3 and VIL1/VRN5 were previously identified as essential components for vernalization-mediated FLC and FLM/MAF1 repression (Sung et al., 2006; Greb et al., 2007). A mutation in VIN3 also compromises expression of some members of the FLC gene family (Sheldon et al., 2009). VIL2/VEL1 also biochemically copurifies with VIN3 and VIL1/VRN5 (De Lucia et al., 2008), but a mutation in VIL2/VEL1 does not have a significant effect on the vernalization response (Kim and Sung, 2010). Nonetheless, VIL2/VEL1 is required for the repression of MAF5 to regulate flowering time under short-day conditions (Kim and Sung, 2010). In addition, we observed dynamic expression patterns of both the VIN3 and FLC gene family during the course of vernalization (Figure 1). Their differential expressions during the course of vernalization indicate that the VIN3 family of proteins may act on various combinations of FLC gene family members. To address the involvement of the VIN3 gene family members in the vernalization-mediated regulation of the FLC gene family, we examined mRNA expression patterns of the FLC gene family members in all VIN3 family mutants during the course of vernalization (Figure 4).
Figure 4.
Misregulations of FLC Gene Family Members in VIN3 Gene Family Mutants during the Course of Vernalization.
(A) mRNA expression of FLC gene family members in vin3 mutants during the course of vernalization. WT, the wild type.
(B) mRNA expressions of FLC gene family members in vil1/vrn5 mutants during the course of vernalization.
(C) mRNA expressions of FLC gene family members in vil2/vel1 mutants during the course of vernalization.
(D) mRNA expressions of FLC gene family members in VIL3/VEL2 RNAi lines (#5 and #9) during the course of vernalization. NV, nonvernalized; 10V, 10 d of vernalization; 20V, 20 d of vernalization; 30V, 30 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. To show relative fold change, the value at nonvernalization of the wild type was converted to 1 and relative fold changes during the course of vernalization were presented. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
In vin3 mutants, all members of the FLC gene family are derepressed throughout the course of vernalization (Figure 4A). No significant repression of FLC gene family was observed both during the cold exposure and after vernalization (Figure 4A). Thus, VIN3 is required for the vernalization-mediated repression of all FLC gene family members.
Consistent with a previous report (Sung et al., 2006), both FLC and FLM are derepressed in vil1/vrn5 mutants compared with the wild type after vernalization (Figure 4B). Unlike in vin3 mutants, however, FLC and FLM are initially repressed during the cold exposure but are derepressed when plants return to warm growth temperature in vil1/vrn5 mutants (Figure 4B). Other members of the FLC gene family are not affected in vil1/vrn5 mutants, suggesting that VIL1/VRN5 specifically acts to maintain the repressed states of FLC and FLM after cold exposure.
In vil2/vel1 mutant plants, repression of FLC and FLM by vernalization still occurs (Figure 4C). During early periods of cold treatment, mRNA expression of MAF4 and MAF5 increase both in the wild type and in vil2/vel1 mutants. However, MAF4 and MAF5 are not repressed by longer periods of cold treatment in vil2/vel1 mutants (Figure 4C). Therefore, VIL2/VEL1 is required for the vernalization-mediated repressions of MAF4 and MAF5 during the cold.
As indicated earlier, expression of VIL3/VEL2 mRNA notably increase after vernalization (Figure 1), suggesting its role in the maintenance of vernalization response after the cold treatment. To address the role of VIL3/VEL2 in vernalization, we created RNA interference (RNAi) lines for VIL3/VEL2 (no T-DNA insertional mutant is available). We established multiple stable RNAi lines in which VIL3/VEL2 expression is reduced (see Supplemental Figure 4A online; Figure 1B). VIL3/VEL2 RNAi lines show reduced expression of VIL3/VEL2 mRNA without compromising expression of other members of the VIN3 gene family (see Supplemental Figure 5 online). VIL3/VEL2 RNAi plants do not exhibit a significant difference in flowering time under both long days and short days compared with the wild type prior to cold treatment (see Supplemental Figure 4B online). However, VIL3/VEL2 RNAi lines flower much later after the vernalization treatment (see Supplemental Figures 4C and 4D online). We selected two representative stable VIL3/VEL2 RNAi lines and determined mRNA expression levels of FLC gene family members (Figure 4D). VIL3/VEL2 RNAi lines showed significantly upregulated levels of MAF4 and MAF5 and to a lesser extent, MAF2 and MAF3, but not those of FLC and FLM/MAF1. Derepression of MAF2∼MAF5 is more evident after the cold (Figure 4D; see Supplemental Figure 4E online), indicating that VIL3/VEL2 is required for the maintenance of repressions of MAF2∼MAF5.
Direct Association of VIN3 Family of Proteins with Chromatin of FLC Gene Family Members during the Course of Vernalization
mRNA expression profiles of the FLC gene family members in VIN3 family mutants indicate that there are divergent contributions by VIN3 family proteins to the repression of the FLC gene family during the course of vernalization (Figure 4). To determine whether the effects of mutations in VIN3 family genes on the FLC gene family are due to their direct associations with their target chromatin, we performed chromatin immunoprecipitation (ChIP) using antibodies against the VIN3 family of proteins (Figure 5). VIN3 protein is transiently associated with all members of FLC gene family chromatin during the cold, when VIN3 is expressed (Figure 5B). Enrichments of VIN3 protein at FLC gene family chromatin increased only during the cold.
Figure 5.
VIN3 Family of Proteins Are Enriched at Their Specific Target Chromatins of FLC Gene Family Members during the Course of Vernalization.
(A) Schematic representation of FLC gene family genomic regions (not scaled) and relative position of primers used in ChIP assays shown (P1 to P4 for each gene).
(B) VIN3 is enriched at all FLC gene family chromatins during the cold exposure.
(C) VIL1/VRN5 enriches at FLC and FLM chromatins, predominantly after the cold exposure.
(D) VIL2/VEL1 is enriched at MAF4 and MAF5 chromatins, predominantly during the cold exposure.
(E) VIL3/VEL2 is enriched at MAF2, MAF3, MAF4, and MAF5 chromatins, predominantly after the cold exposure. NV, nonvernalized; 20V, 20 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
[See online article for color version of this figure.]
Unlike VIN3, VIL1/VRN5 enrichment is observed during the cold, and the increased enrichment persists even after the cold at FLC and FLM/MAF1 chromatin (Figure 5C). We could not observe significant enrichment of VIL1/VRN5 at MAF2∼MAF5 chromatin (Figure 5C), consistent with the lack of derepression of MAF2∼MAF5 in vil1/vrn5 mutants (Figure 4B). Consistent with the observation that MAF4 and MAF5 are derepressed in vil2/vel1 mutants, enrichment of VIL2/VEL1 protein at MAF4 and MAF5 chromatin is increased especially during the cold (Figure 5D), suggesting its role in establishing repression of MAF4 and MAF5 during the exposure to vernalizing cold.
mRNA expression analysis of FLC gene family in VIL3/VEL2 RNAi lines indicated that VIL3/VEL2 functions to maintain repressed states of MAF4∼MAF5 and to a lesser degree MAF2∼MAF3 (Figure 4D). To determine whether VIL3/VEL2 directly binds to these FLC family chromatins, we performed ChIP using antibody against VIL3/VEL2 (Figure 5E). Consistent with expression data, VIL3/VEL2 accumulates at MAF2, MAF3, MAF4, and MAF5 chromatin but not at FLC and FLM/MAF1 chromatin. Enrichment of VIL3/VEL2 at its target chromatin is persistent even after vernalizing cold, suggesting its role in maintaining the repression of MAF2∼MAF5 after the cold (Figure 5E). Taken together, all members of VIN3 family protein directly associate with specific subsets of FLC gene family chromatin at certain times during the course of vernalization to coordinate repression of FLC gene family members.
VIN3 Family Proteins Preferentially Bind to H3K9me2
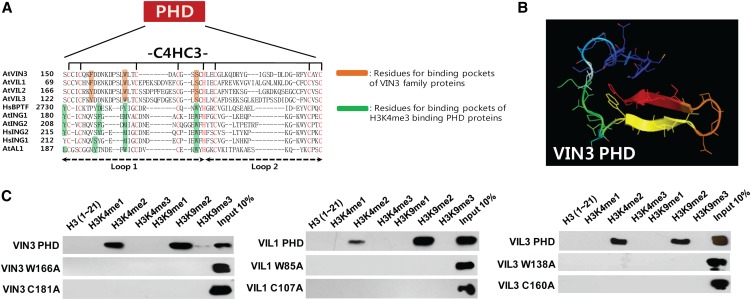
VIN3 family of proteins commonly contains a PHD finger motif (Figures 6A and 6B). The PHD motif can bind to various histone tails, ranging from nonmethylated histone H3 (Lan et al., 2007) to methylated histone H3 Lys 9 (Karagianni et al., 2008) and methylated histone H3 Lys 36 (Shi et al., 2007). In addition, we reported that VIL2/VEL1 preferentially binds to the H3K9me2 histone peptide (Kim and Sung, 2010). To identify histone modifications to which the VIN3 family of proteins bind, we performed in vitro histone peptide binding assays (Shi et al., 2006; Wysocka et al., 2006) using a series of synthesized histone peptides with various modifications (Figure 6C). All of the VIN3 family of proteins preferentially bind to Histone H3 Lys 9 dimethylated (H3K9me2) peptides, although they also exhibit significant binding to dimethylated Histone H3 Lys 4 (H3K4me2). H3K9me2 is a modified histone mark that is enriched in repressed chromatins, including FLC after vernalization (Bastow et al., 2004; Sung and Amasino, 2004).
Figure 6.
VIN3 Family of Proteins Preferentially Binds to H3K9me2 Peptides in Vitro.
(A) Multiple alignments of PHD fingers with known three-dimensional structure compared with the PHD fingers of VIN3 family of proteins.
(B) Predicted three-dimensional structure of the PHD finger of VIN3.
(C) In vitro histone peptides pull-down assay using recombinant VIN3 family of proteins. Full list of synthesized histone peptides used in this study are described in Supplemental Table 2 online.
To address the specificity of the binding in detail, the putative tertiary structure of VIN3 PHD finger domain was simulated using the SWISS-MODEL program (Figure 6B; see Supplemental Figure 5 online). The predicted structure of VIN3 PHD domain contained two antiparallel β-sheets, commonly found in the PHD domain structure (Sanchez and Zhou, 2011). A hydrophobic aromatic ring residue is required to produce specific π-interaction with the polar methyl charge of the Lys (K) histone residue in PHD finger domain (Sanchez and Zhou, 2011). Based on a predicted structure of VIN3 PHD motif, Phe-157, Trp-166, and Ser-175 residues of VIN3 appear to form an aromatic cage that could serve as a docking site for a modified histone (Figure 6B; see Supplemental Figure 6 online). These residues are conserved in all members of the VIN3 family of proteins (Figure 6A). To test if these cage-forming residues are important for modified histone binding, we introduce mutations to one of essential residues of this cage (Trp to Ala) in the PHD domain of VIN3, VIL1/VRN5, and VIL3/VEL2. These mutations completely abolished the histone peptide binding of the PHD fingers of the VIN3 family of proteins (Figure 6C). Not surprisingly, disruption of the zinc fingers itself of VIN3 family proteins by the substitution of one of critical Cys residue with Ala residues completely abolished binding of VIN3 family proteins (Figure 6C). Thus, we concluded that the VIN3 family of proteins have a common binding preference for H3K9me2 histone mark and require a binding pocket structure consisting of two hydrophobic aromatic residues (Phe/Tyr and Trp) and one polar residue (Ser) in the PHD finger for H3K9me2 histone mark.
PRC2-Mediated Repression of FLC Clade Chromatin
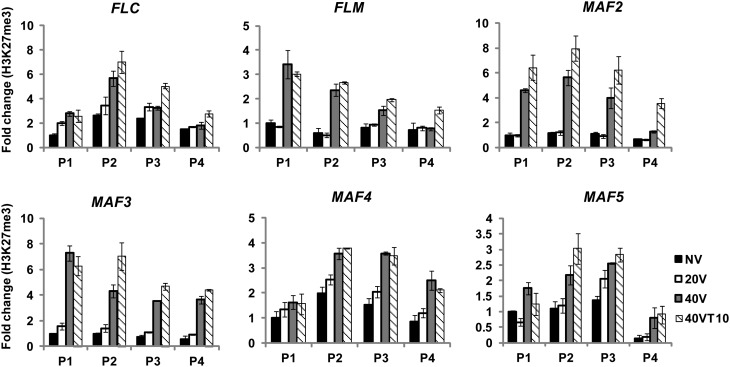
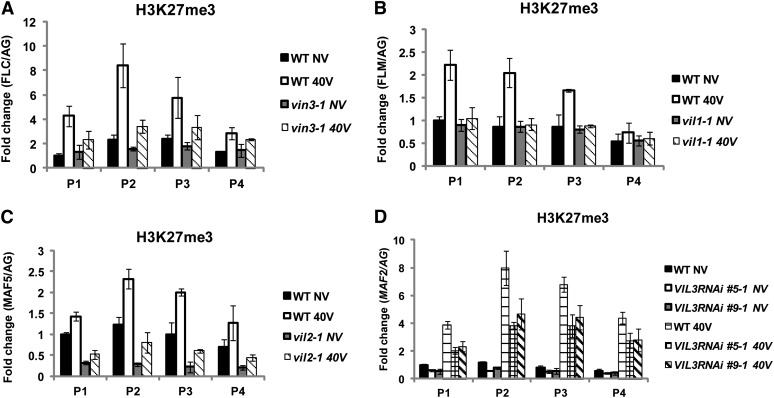
Members of the VIN3 protein family copurify with components of PRC2 (Wood et al., 2006; De Lucia et al., 2008). PRC2 enriches at FLC chromatin by vernalization and mediates the increase in levels of H3K27me3 at FLC chromatin, which signifies the stable silencing of FLC (Schubert et al., 2006; Wood et al., 2006; De Lucia et al., 2008; Jiang et al., 2008). As discussed earlier, all members of the FLC gene family undergo vernalization-mediated repression. Thus, we first determined levels of Histone H3 Lys27 trimethylation (H3K27me3) at chromatin of FLC gene family members during the course of vernalization (Figure 7). The H3K27me3 enrichment increases not only at FLC chromatin but also at all other members of FLC gene family chromatins as a result of vernalization (Figure 7). Furthermore, as is the case for the previously known roles of VIN3 and VIL1/VRN5 (Figures 8A and 8B), other VIN3 family proteins are also required for the H3K27me3 enrichment at their respective target FLC gene family member chromatin (Figures 8C and 8D), suggesting that all VIN3 family of proteins are required for the enrichment of H3K27me3.
Figure 7.
The Enrichment of H3K27me3 at Chromatin of FLC Gene Family Members as a Result of Vernalization.
The level of H3K27me3 enrichment at chromatin of FLC gene family members during the course of vernalization was determined by ChIP assays. Relative positions of primers used for each chromatin are depicted in Figure 5A. NV, nonvernalized; 20V, 20 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. To show relative fold change, the value at nonvernalization of the wild type was converted to 1 and relative fold changes during the course of vernalization were presented. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
[See online article for color version of this figure.]
Figure 8.
Increased H3K27me3 Enrichments at Chromatins of FLC Gene Family Members by Vernalization Depends on VIN3 Gene Family Members.
(A) The level of H3K27me3 enrichment at FLC chromatin between the wild type (WT) and vin3 mutants as a result of vernalization.
(B) The level of H3K27me3 enrichment at FLM chromatin between the wild type and vil1/vrn5 mutants as a result of vernalization.
(C) The level of H3K27me3 enrichment at MAF5 chromatin between the wild type and vil2/vel1 mutants as a result of vernalization.
(D) The level of H3K27me3 enrichment at MAF2 chromatin between the wild type and VIL3/VEL2 RNAi lines (#5 and #9) as a result of vernalization. Relative positions of primers used for each chromatin are depicted in Figure 5A. NV, nonvernalized; 40V, 40 d of vernalization. AGAMOUS (AG) was used as a control for normalization. To show relative fold change, the value at nonvernalization of the wild type was converted to 1 and relative fold changes during the course of vernalization were presented. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
[See online article for color version of this figure.]
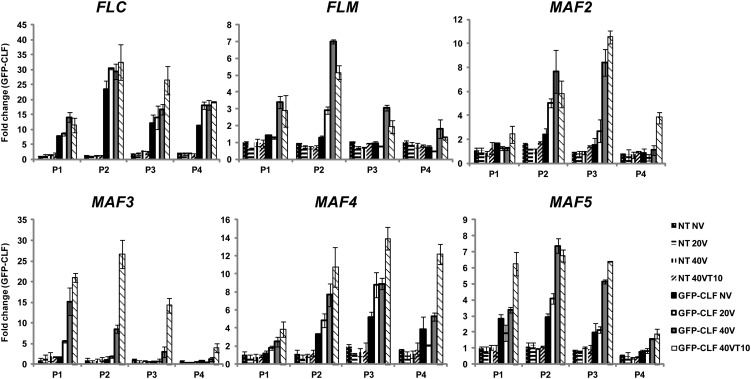
To test whether PRC2 directly associates with FLC gene family chromatin, we used green fluorescent protein (GFP)-CLF transgenic plants to trace PRC2 enrichment at FLC gene family member chromatin (Figure 9). Consistent with the increase in H3K27me3, all FLC gene family chromatin showed significant accumulation of GFP-CLF during the course of vernalization. It is interesting to note that PRC2 is enriched above the background level at FLC, MAF4, and MAF5 chromatins prior to vernalization (compared with nontransgenic lines), and the PRC2 enrichment is further increased by vernalization. On the other hand, FLM/MAF1, MAF2, and MAF3 chromatins are enriched with PRC2 only by vernalization (Figure 9). This observation is also consistent with a genome-wide H3K27me3 enrichment study (Zhang et al., 2007). Taken together, our results show that all FLC gene family chromatins undergo PRC2-mediated H3K27me3 accumulation in a VIN3 family protein-dependent manner by vernalization.
Figure 9.
Vernalization-Mediated Enrichments of PRC2 at Chromatins of FLC Gene Family Members.
ChIP assay using GFP-CLF transgenic plants to address the enrichment of CLF at chromatins of FLC gene family members during the course of vernalization. Nontransgenic plants (NT) were compared for the specificity of precipitation. Relative positions of primers used for each chromatin are depicted in Figure 5A. NV, nonvernalized; 20V, 20 d of vernalization; 40V, 40 d of vernalization; 40VT10, 40 d of vernalization followed by 10 d of normal growth temperature. Real-time PCR experiments were performed with three biological replicates and error bars indicate sd. AG was used as a control. First, relative enrichment of each fragment was calculated based on comparison to quantitative PCR using INPUT material. Second, relative enrichment was calculated by comparison to the AG proximal promoter control (Schubert et al., 2006). To show relative fold change, the value of nonvernalization time point in nontransgenic plants was converted to 1 and relative fold changes during the course of vernalization were presented.
[See online article for color version of this figure.]
Changes in Histone Modification at FLC Gene Family Chromatin
To address the in vivo implications of the in vitro histone binding preference by VIN3 family proteins (Figure 6), we examined histone modifications at FLC gene family chromatin during the course of vernalization in the wild type and VIN3 family mutants. Although VIN3 family proteins are required for the enrichment of H3K27me3 at their target chromatin (Figure 7), VIN3 family proteins do not exhibit binding to methylated H3K27 peptides (see Supplemental Figure 7 online). We examined the level of enrichment of H3K4me2 (a modification that VIN3 family of protein binds in vitro) at chromatin of FLC gene family members during the course of vernalization. We did not observe a significant difference at the level of enrichment of H3K4me2 by vernalization at chromatin of FLC gene family members (see Supplemental Figure 8 online). Furthermore, no significant difference was observed in the level of enrichment of H3K4me2 between the wild type and vin3 family mutants (see Supplemental Figure 8 online), suggesting that the level of H3K4me2 does not play a significant role in vernalization-mediated FLC gene family repression.
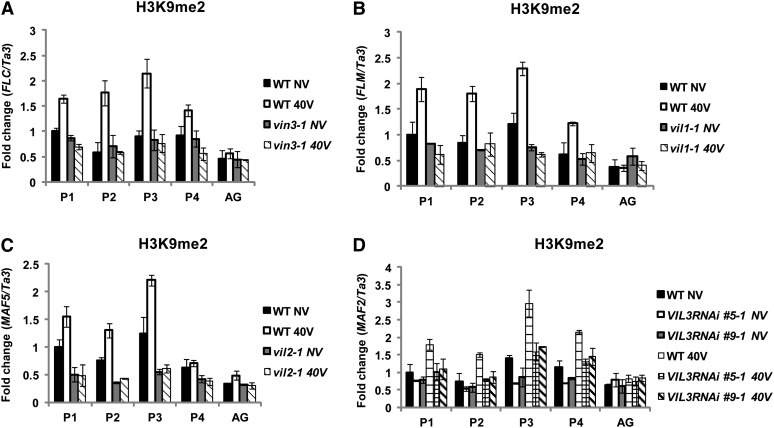
On the other hand, the level of enrichment of H3K9me2 (another modification that VIN3 family of protein binds in vitro) increases as a result of vernalization at FLC gene family chromatin (Figures 10A to 10D). Furthermore, levels of H3K9me2 showed significant differences between the wild type and vin3 family mutants at their representative target chromatin by vernalization (Figures 10A to 10D). Enrichment of H3K9me2 at FLC gene family chromatin is rather unique among PRC2 target loci. For example, another well-known H3K27me3-enriched AGAMOUS chromatin (Schubert et al., 2006) does not show significant enrichment of H3K9me2 at its chromatin (Figure 10). Thus, our results indicate that VIN3 family proteins are required for the vernalization-mediated enrichment of H3K9me2 at FLC gene family chromatin through their affinity to H3K9me2.
Figure 10.
Increased H3K9me2 Enrichments at Chromatins of FLC Gene Family Members by Vernalization Depends on VIN3 Gene Family Members.
AG was used as a negative control. Ta3 was also used as a control for normalization
(A) The level of H3K9me2 enrichment at FLC chromatin between the wild type (WT) and vin3 mutants as a result of vernalization.
(B) The level of H3K9me2 enrichment at FLM chromatin between the wild type and vil1/vrn5 mutants as a result of vernalization.
(C) The level of H3K9me2 enrichment at MAF5 chromatin between the wild type and vil2/vel1 mutants as a result of vernalization.
(D) The level of H3K9me2 enrichment at MAF2 chromatin between the wild type and VIL3/VEL2 RNAi lines (#5 and #9) as a result of vernalization. Relative positions of primers used for each chromatin are depicted in Figure 5A. NV, nonvernalized; 40V, 40 d of vernalization. To show relative fold change, the value at nonvernalization of the wild type was converted to 1 and relative fold changes during the course of vernalization were presented. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
[See online article for color version of this figure.]
Behavior of Activating Chromatin-Modifying Complexes during the Course of Vernalization
One of the characteristic changes at FLC chromatin upon vernalization is the loss of Histone Lys 4 trimethylation (H3K4me3) enrichment (Bastow et al., 2004; Sung and Amasino, 2004; Sung et al., 2006; Greb et al., 2007; De Lucia et al., 2008). Multiple activating chromatin modifying complexes have been identified to be responsible for H3K4me3 enrichment at FLC chromatin and, thus, the active state of FLC prior to vernalization (Kim et al., 2005; Jiang et al., 2009; Tamada et al., 2009; Ko et al., 2010). Although the resulting decrease in levels of H3K4me3 enrichment at FLC chromatin by vernalization is well known (Bastow et al., 2004; Sung and Amasino, 2004; Sung et al., 2006; Greb et al., 2007; De Lucia et al., 2008), the fate of activating complexes during the course of vernalization has not been addressed.
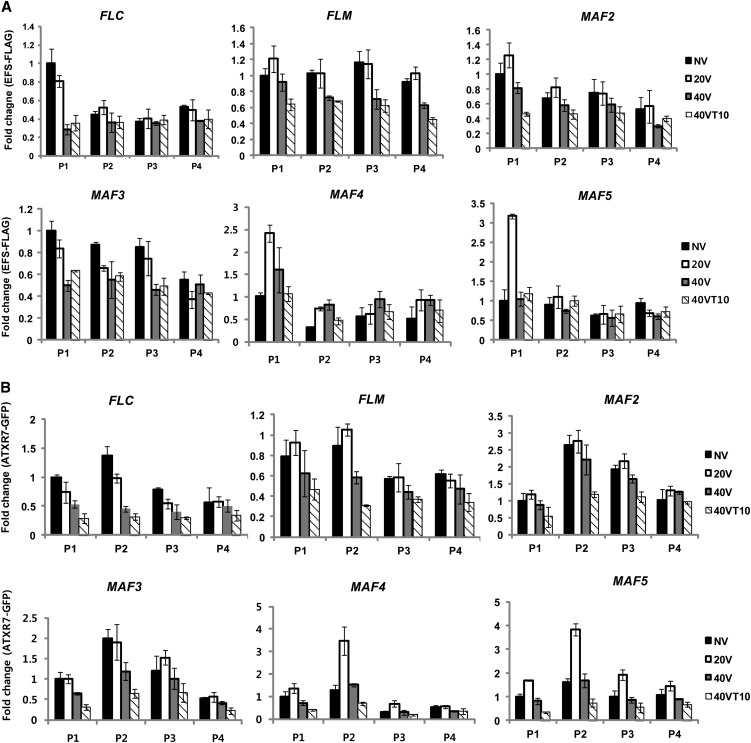
We tested subsets of known activating chromatin modifying complex components affecting FLC activation for their roles in the regulation of FLC gene family during the course of vernalization. First, we measured expression levels of FLC gene family between the wild type and mutants for known components of activating chromatin modifying complexes, including efs, atx1-1, atxr7-1, and atx1-1 atxr7-1, during the course of vernalization (see Supplemental Figure 9 online). EFS encodes a SET domain protein that mediates both H3K4me3 and H3K36me2 at FLC chromatin prior to the cold (Ko et al., 2010) and is also required for activation of some other members of FLC gene family (Kim et al., 2005; see Supplemental Figure 9 online). ATX1 and ATXR7 are Trithorax-type SET domain proteins, which mediate H3K4 methylation in Arabidopsis. They are redundantly required for the activation of FLC and some other members of the FLC gene family (see Supplemental Figure 9 online). It is also likely that additional members of Trithorax-type SET domain proteins are redundantly involved in the activation of FLC and other members of FLC gene family (Yun et al., 2012). Most members of FLC gene family are downregulated in efs mutants or in atx1 atxr7 double mutants during the course of vernalization, although there are variations in degrees of severity (see Supplemental Figure 9 online). Interestingly, upregulations of MAF4 and MAF5 with 20 d of cold treatment are significantly reduced in efs and atx1 atxr7 mutants, suggesting that EFS, ATX1, and ATXR7 play roles in the transient activation of MAF4 and MAF5 during the early periods of cold exposure.
Although it has been documented that EFS and some Trithorax-type methyltransferases directly bind to FLC chromatin to mediate H3K4me3 and to activate FLC (Tamada et al., 2009; Ko et al., 2010), it has not been determined whether these components are removed from FLC chromatin by vernalization (when FLC is silenced). Thus, we used epitope-tagged transgenic lines for EFS and ATXR7 to monitor these activating chromatin-modifying complexes at FLC gene family chromatin during the course of vernalization (Figure 11; see Supplemental Figures 10 and 11 online). Before vernalization, both EFS and ATXR7 showed enrichment at chromatin of all FLC gene family members, especially around their respective transcriptional start site regions, which is consistent with previous reports (Tamada et al., 2009; Ko et al., 2010). By vernalization, enrichments of EFS and ATXR7 at FLC gene family chromatins are reduced (Figures 11A and 11B), suggesting that activating chromatin-modifying complexes are replaced by repressive chromatin complexes (i.e., PRC2) at chromatin of FLC gene family members as a result of vernalization (Figure 9). It is interesting to note that enrichments of EFS and ATXR7 at MAF4 and MAF5 chromatin are transiently increased by 20 d of cold when their expressions peak (Figure 4). Therefore, MAF4 and MAF5 chromatin becomes enriched with activating chromatin-modifying complexes by short-term cold, but the increased enrichment of EFS and ATXR7 during early period of cold exposure is eventually replaced by increased enrichments of PRC2 (Figure 9).
Figure 11.
Dynamic Changes in Enrichments of EFS and ATXR7 at Chromatins of FLC Gene Family Members during the Course of Vernalization.
ChIP assay using EFS-FLAG and ATXR7-GFP transgenic plants to address the enrichments of EFS and ATXR7 at chromatins of FLC gene family members during the course of vernalization. Real-time PCR experiments were performed with three biological replicates, and error bars indicate sd.
[See online article for color version of this figure.]
DISCUSSION
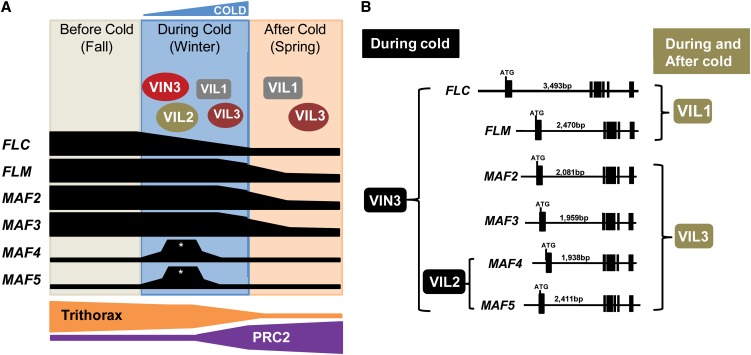
Our study shows that all members of VIN3 and FLC gene families function to mediate the vernalization response in Arabidopsis. Among members of VIN3 gene family, VIN3 appears to be a master regulator in vernalization, since it is required for repression of all members of the FLC gene family (Figure 4A). Consistent with its expression profile, VIN3 only functions during the cold exposure (i.e., VIN3 protein associates with chromatin of the FLC gene family only during cold exposure) and appears to be required for initial repression of FLC gene family during the cold (Figure 5B). VIL2/VEL1 acts redundantly in part with VIN3, as it is required for initial repression of MAF4 and MAF5 (Figure 4C), and VIL2/VEL1 protein is transiently enriched at MAF4 and MAF5 chromatin during the cold exposure (Figure 5D). Unlike VIN3 and VIL2/VEL1, VIL1/VRN5 and VIL3/VEL2 function even after the cold exposure. VIL1/VRN5 and VIL3/VEL2 appear to contribute to H3K27me3 and H3K9me2 enrichments both during and after cold (Figures 5C, 5E, and 12). Thus, it is likely that there is functional divergence among VIN3 family of proteins in their temporal involvement in vernalization. Furthermore, there are clear differences in their target specificities (Figure 12).
Figure 12.
Schematic Representations of Regulatory Networks between VIN3 and FLC Gene Families.
(A) FLC, FLM, MAF2, and MAF3 are repressed by vernalization through coordinated activities of VIN3 family of proteins and PRC2. MAF4 and MAF5 are transiently activated during an early period of cold exposure (asterisks) but eventually repressed by VIN3 family of proteins and PRC2.
(B) Target specificities of VIN3 family of proteins.
[See online article for color version of this figure.]
Although VIN3 is enriched at all FLC gene family chromatin, other members of the VIN3 gene family are involved in the repressions of only specific members of the FLC gene family. Despite differences in their temporal and target specificities, they are all required for repression of their respective target FLC gene family. PRC2 biochemically copurifies with members of VIN3 family of proteins, suggesting they act together to mediate epigenetic silencing of the FLC gene family. Consistent with this notion, all FLC gene family chromatin is eventually enriched with PRC2 (Figure 9), and PRC2 is shown to mediate H3K27me3 enrichment. It should be noted that we could not observe significant binding of VIN3 family of proteins to H3K27me3 peptides in vitro (see Supplemental Figure 7 online), although all members of the VIN3 gene family are required for enrichment of H3K27me3 at their respective target chromatin (Figure 7). Instead, all of the VIN3 family of proteins binds to H3K9me2, another histone modification that is enriched at chromatin of the FLC gene family by vernalization. Furthermore, the H3K9me2 enrichment by vernalization depends on the presence of VIN3 gene family (Figure 10). Thus, the VIN3 family of proteins likely provides positive feedback between two repressive histone modifications at their target chromatins in part through the PRC2 activity. In addition, VIN3 family proteins bind to an H3K4me2 in in vitro binding assay. It has been reported that unlike H3K4me3, H3K4me2 is not coupled with gene activation in Arabidopsis (Zhang et al., 2009). Consistent with this, we did not observe any significant change at the level of H3K4me2 at chromatin of FLC gene family. Although levels of H3K4me2 enrichment are not changed by the extended cold exposure, it is possible that H3K4me2 binding preference by VIN3 family of proteins may play a role in facilitating FLC repression upon vernalization.
As the VIN3 gene family shows functional divergences among its members, the FLC gene family also exhibits functional divergence among its members. In particular, MAF4 and MAF5 function to prevent a precocious vernalization response. maf4 and maf5 mutant plants respond to a suboptimal, short period of cold exposure (Figure 3A; see Supplemental Figure 2A online). One characteristics of vernalization response is that it is a response to a prolonged period of cold exposure. It is not known how plants measure the duration of cold to trigger vernalization response. Our results indicate that MAF4 and MAF5 function, at least in part, to ensure plants to respond to a long period of cold (i.e., winter) to promote flowering. A previous report showed that MAF2 may function to prevent a precocious vernalization response (Ratcliffe et al., 2003). In our analysis, however, maf2 mutants flower much earlier even prior to vernalization and they do not significantly respond to short-term cold (Figure 3B). Furthermore, transient increases in levels of mRNA expression of MAF4 and MAF5 by a short-term cold (Figures 2E and 2F) suggest that MAF4 and MAF5 are specifically responsive to short-term cold to prevent a precocious vernalization response.
Although there is no net reduction in MAF4 and MAF5 transcripts after vernalization, MAF4 and MAF5 chromatins undergo dramatic changes during the course of vernalization. Initial increases in their transcription by short-term cold are correlated with increased enrichments of activating chromatin remodeling complexes. Further cold exposure eventually triggers enrichments of VIL2/VEL1, VIL3/VEL2, and PRC2 at MAF4 and MAF5 chromatins and, thus, enrichments of repressive histone marks. Furthermore, these changes at chromatin depend on two VIN3-related proteins, VIL2/VEL1 and VIL3/VEL2. Mutations in VIL2/VEL1 and VIL3/VEL2 result in derepressions of MAF4 and MAF5 after vernalization (Figures 5D and 5E). Therefore, eventual repressions of MAF4 and MAF5 by vernalization should be considered as part of vernalization-mediated repression.
Dynamic expression profiles of the FLC gene family during the course of vernalization coincide with differential enrichments of chromatin-modifying complexes, including VIN3 family proteins, PRC2, and Trithorax-type activating complexes. In a recent study, the PRC2 enrichment at FLC chromatin by vernalization is mediated by a long noncoding RNA, COLDAIR (Heo and Sung, 2011). Here, we describe diverse recruitment of the VIN3 family of proteins, which are associated with PRC2. How VIN3 family proteins achieve their target specificity remains to be addressed. Our study presents a detailed analysis of the dynamics of repressive and activating chromatin remodeling complexes at multiple environmentally regulated loci. Related but yet distinct functions among members of gene families show that plants have evolved sophisticated regulatory networks among members of gene families to interpret an environmental input (i.e., winter cold) to developmental transitions (i.e., flowering). Our thorough analysis will also be helpful to use this regulatory network between VIN3 and FLC gene families as a model to address mechanistic details of epigenetic regulation of developmental genes.
METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis thaliana were germinated on agar plates for 10 d in short-day conditions and vernalized at 4°C under short days. Seedling plants were transplanted to soil or harvested for total RNA isolation. Vernalization treatments were as previously described (Kim et al., 2010). In short, nonvernalized seedlings were grown for 11 d after germination under short-day conditions. For 40VT0 and 40VT10 plants, seedlings were grown for 7 and 4 d, respectively, after seed germination and then seedlings were subjected to cold treatment. Samples were harvested for experiments right after the cold treatment (40VT0) or grown for 10 d more (40VT10) under short days. Plants were grown at 22°C in different photoperiods for measurement of flowering time as described in each experiment. All of the Arabidopsis plants used in his study were in the Columbia background except the GFP-CLF transgenic line (CaMV35S:GFP:CLF in clf;FRI in Wassilewskija background). Information on mutants used in this study is shown in Supplemental Table 3 online.
Development of Transgenic Lines
For GFP:MAF4 and MAF5:GFP transgenic lines, cDNA of each MAF4 and MAF5 was amplified and cloned into pENTR-D TOPO cloning vector (Invitrogen) followed by LR reaction into the plant expression Gateway vector pB7FWG2,0 using LR enzyme (Invitrogen). Each construct was transformed into maf4-1 (SALK_028506) (Gu et al., 2009) and maf5-1 (SALK_095092) mutants. For VIL3/VEL2 RNA knockdown lines, 317 bp (+1798 to 2114) of 3′ region of VIL3/VEL2 was amplified and digested with AscI-SwaI and BamHI-SpeI restriction enzymes, respectively, and cloned into pFGC1008 RNAi vector to produce double-stranded RNA structure. Construct was transformed into Columbia-0 wild-type plants. All transgenic lines were selected on selective antibiotic medium and were confirmed by PCR. Stable homozygous T3 transgenic plants were used for experiments. The cauliflower mosaic virus 35 promoter-driven GFP-CLF construct (Schubert et al., 2006) was previously described and used for this study. ATXR7-GFP (Tamada et al., 2009) and EFS-FLAG (Ko et al., 2010) constructs including the native promoter regions were previously described and used for ChIP assay.
Production of Polyclonal Antibodies
Amino acids sequences of VIN3 family proteins were analyzed with the OptimumAntigen design tool (GenScript) to identify good antigen sequences. Selected synthetic peptide antigens of VIN3 family proteins were injected into rabbit for production of polyclonal antibodies (GenScript). The synthetic peptide sequences used as antigens are as follows: VIN3 (493-KRDIYKGKQGGNK), VIL1/VRN5 (464-DDAVSNGRRKNNN), VIL2/VEL1 (473-PKKPSSKNEDNNSP), and VIL3/VEL2 (75- EIIPSPKRQKRDLV). Synthesized peptides were injected into rabbit, and resulting antiserum were further purified by affinity purification (GenScript).
Real-Time Quantitative RT-PCR Analyses
Total RNA was isolated using a Plant RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. Before reverse transcription reaction, DNase I (Invitrogen) treatment was performed for 30 min at 37°C to eliminate contaminated genomic DNA from total RNAs. First-strand cDNA synthesis was performed on 2 µg of RNA using the M-MLV System (Promega). Real-time quantitative RT-PCR was performed using SYBR green dye reaction mixture (Thermo Scientific) on an ABI prism 7900HT sequence detection system (Life Technologies). Each sample was quantified in triplicate, and the relative transcript level of each gene was determined by normalization of the resulting expression levels versus that of PP2A, as described (Czechowski et al., 2005). Error bars indicate standard deviation from triplicate samples. For primer sequences used in real time RT-PCR analyses, see Supplemental Table 1 online.
ChIP–Quantitative PCR Analyses
Whole seedling plants were harvested for grinding. ChIP assays were performed as previously described (Kim and Sung, 2010). For primer sequences, see Supplemental Table 1. Quantitative PCR reaction was done using SYBR green dye reaction mixture (Thermo Scientific) according to the manufacturer’s instructions. Relative enrichment of each fragment was calculated based on comparison to quantitative PCR using INPUT material. Second, relative enrichment was calculated by comparison to each listed control region. All ChIP assays were performed at least three times from at least two biological replicates and produced similar results. Error bars indicate sd from triplicate samples. Quantitative PCR reaction was performed on 7900HT Fast Real-Time PCR system (Life Technologies).
In Vitro Histone Peptide Pull-Down Assay
cDNAs of full-length and PHD-only VIN3 family genes were amplified by RT-PCR and cloned into the pVP13 (Thao et al., 2004) expression vector. Recombinant full-length and PHD-only VIN3 family proteins were purified using a nickel-nitrilotriacetic acid (Ni-NTA) column (Qiagen). Biotinylated histone peptides were purchased from Millipore (see Supplemental Table 2 online). After pull down, proteins were transferred using a mini trans-blot electrophoresis transfer cell apparatus (Bio-Rad). His-tag antibody (Santa Cruz Biotech) was used for immunoblot analysis. Commercial antibodies used in this study are listed in Supplemental Table 4 online.
Prediction of Putative Tertiary Structure of the VIN3 PHD Domain
Tertiary structure prediction for the PHD finger domain of VIN3 was performed using the Swiss-Model website (http://swissmodel.expasy.org/) that simulates tertiary structure based on the reported protein structure of a sequence with similarity to it (Arnold et al., 2006). The acquired PDB file was graphically visualized using Pymol version 1.4.0 program (www.pymol.com). Briefly, a template sequence of VIN3 PHD domain (loop 1 region, CICQKFDDNKDPSLWLTCDACGSSCHGRCVGILQSEAELIDEYV) was submitted into the template identification step, and then predicted structure was produced based on parent template PDB file (2ri7A) in automatic modeling mode of the Swiss-Model website.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VIN3 (At5g57380), VIL1/VRN5 (At3g24440), VIL2/VEL1 (At4g30200), VIL3/VEL2 (At2g18880), FLC (At5g10140), FLM/MAF1 (At1g77080), MAF2 (At5g65050), MAF3 (At5g65060), MAF4 (At5g65070), MAF5 (At5g65080), ATX1 (At2g31650), ATXR7 (At5g42400), and EFS (At1g77300). Germplasm used was maf4-1 (SALK_028506) and maf5-1 (SALK_095092).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Immunoblots Using Recombinant VIN3 Family of Proteins.
Supplemental Figure 2. Flowering Times of maf4 and maf5 Mutants.
Supplemental Figure 3. Flowering Times of maf2 Mutants by Various Durations of Cold Exposure.
Supplemental Figure 4. VIL3 Knockdown Lines Compromise Vernalization Response.
Supplemental Figure 5. Specificity of VIN3 Family Mutants.
Supplemental Figure 6. Predicted Histone Binding Pocket of the VIN3 PHD Finger Motif.
Supplemental Figure 7. In Vitro Histone Peptides Pull-Down Assay Using Recombinant VIN3 Family Proteins.
Supplemental Figure 8. H3K4me2 Enrichment at Chromatin FLC Gene Family Chromatin.
Supplemental Figure 9. Expression Analysis of the FLC Gene Family during the Course of Vernalization in Activating Chromatin-Modifying Complex Components.
Supplemental Figure 10. Dynamic Changes in Enrichment of EFS at Chromatins of FLC Gene Family during the Course of Vernalization.
Supplemental Figure 11. Dynamic Changes in Enrichment of ATXR7 at Chromatins of FLC Gene Family during the Course of Vernalization.
Supplemental Table 1. Primer Sequences Used in This Study.
Supplemental Table 2. Histone Peptides Used for in Vitro Pull-Down Assay.
Supplemental Table 3. Information on Mutants Used in This Study.
Supplemental Table 4. Commercial Antibodies Used for This Study.
Acknowledgments
We thank Richard Amasino for providing ATXR7-GFP transgenic line seeds and Yoo-Sun Noh for EFSFLAG transgenic line seeds. This work was supported by the National Science Foundation (IOS 0950785) and by the National Institutes of Health (R01 GM100108A1).
AUTHOR CONTRIBUTIONS
D.-H.K. and S.S. conceived and designed the experiments, analyzed the data, and wrote the article. D.-H.K. performed the experiments.
Glossary
- PHD
plant homeodomain
- PcG
Polycomb group
- TrxG
Trithorax group
- RNAi
RNA interference
- ChIP
chromatin immunoprecipitation
References
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.S., Peacock W.J. (2007). Epigenetic regulation of flowering. Curr. Opin. Plant Biol. 10: 520–527 [DOI] [PubMed] [Google Scholar]
- Greb T., Mylne J.S., Crevillen P., Geraldo N., An H., Gendall A.R., Dean C. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Gu X., Jiang D., Wang Y., Bachmair A., He Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57: 522–533 [DOI] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192 [DOI] [PubMed] [Google Scholar]
- Heo J.B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hepworth S.R., Valverde F., Ravenscroft D., Mouradov A., Coupland G. (2002). Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Gu X., He Y. (2009). Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Karagianni P., Amazit L., Qin J., Wong J. (2008). ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol. Cell. Biol. 28: 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim, D.H., and Sung, S. (2010). The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc. Natl. Acad. Sci. USA 107: 17029–17034. [DOI] [PMC free article] [PubMed]
- Kim D.H., Zografos B.R., Sung S. (2010). Vernalization-mediated VIN3 induction overcomes the LIKE-HETEROCHROMATIN PROTEIN1/POLYCOMB REPRESSION COMPLEX2-mediated epigenetic repression. Plant Physiol. 154: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.Y., He Y., Jacob Y., Noh Y.S., Michaels S., Amasino R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Mitina I., Tamada Y., Hyun Y., Choi Y., Amasino R.M., Noh B., Noh Y.S. (2010). Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J. 29: 3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Collins R.E., De Cegli R., Alpatov R., Horton J.R., Shi X., Gozani O., Cheng X., Shi Y. (2007). Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 448: 718–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Suh S.S., Park E., Cho E., Ahn J.H., Kim S.G., Lee J.S., Kwon Y.M., Lee I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Kumimoto R.W., Wong B.J., Riechmann J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Nadzan G.C., Reuber T.L., Riechmann J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sanchez R., Zhou M.M. (2011). The PHD finger: A versatile epigenome reader. Trends Biochem. Sci. 36: 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Amasino R.M. (2007). Vernalization: A model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim. Biophys. Acta 1769: 269–275 [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Scortecci K.C., Michaels S.D., Amasino R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26: 229–236 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C.C., Finnegan E.J., Peacock W.J., Dennis E.S. (2009). Mechanisms of gene repression by vernalization in Arabidopsis. Plant J. 59: 488–498 [DOI] [PubMed] [Google Scholar]
- Shi X., et al. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442: 96–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., et al. (2007). Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 282: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2005). Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56: 491–508 [DOI] [PubMed] [Google Scholar]
- Sung S., Schmitz R.J., Amasino R.M. (2006). A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev. 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada Y., Yun J.Y., Woo S.C., Amasino R.M. (2009). ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21: 3257–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao S., Zhao Q., Kimball T., Steffen E., Blommel P.G., Riters M., Newman C.S., Fox B.G., Wrobel R.L. (2004). Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J. Struct. Funct. Genomics 5: 267–276 [DOI] [PubMed] [Google Scholar]
- Wood C.C., Robertson M., Tanner G., Peacock W.J., Dennis E.S., Helliwell C.A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., Wu C., Allis C.D. (2006). A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Yun J.Y., Tamada Y., Kang Y.E., Amasino R.M. (2012). Arabidopsis trithorax-related3/SET domain GROUP2 is required for the winter-annual habit of Arabidopsis thaliana. Plant Cell Physiol. 53: 834–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernatavichute Y.V., Cokus S., Pellegrini M., Jacobsen S.E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. (2007). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14: 869–871 [DOI] [PubMed] [Google Scholar]