This study shows that two catalytic subunits of Arabidopsis protein phosphatase 6 (i.e., FyPP1 and FyPP3) negatively regulate abscisic acid responses through direct interaction with and dephosphorylation of ABI5, which subsequently regulates ABI5 stability and activity, seed germination, and postgermination seedling growth in Arabidopsis.
Abstract
The basic Leucine zipper transcription factor ABSCISIC ACID INSENSITIVE5 (ABI5) is a key regulator of abscisic acid (ABA)–mediated seed germination and postgermination seedling growth. While a family of SUCROSE NONFERMENTING1-related protein kinase2s (SnRK2s) is responsible for ABA-induced phosphorylation and stabilization of ABI5, the phosphatase(s) responsible for dephosphorylating ABI5 is still unknown. Here, we demonstrate that mutations in FyPP1 (for Phytochrome-associated serine/threonine protein phosphatase1) and FyPP3, two homologous genes encoding the catalytic subunits of Ser/Thr PROTEIN PHOSPHATASE6 (PP6), cause an ABA hypersensitive phenotype in Arabidopsis thaliana, including ABA-mediated inhibition of seed germination and seedling growth. Conversely, overexpression of FyPP causes reduced sensitivity to ABA. The ABA hypersensitive phenotype of FyPP loss-of-function mutants is ABI5 dependent, and the amount of phosphorylated and total ABI5 proteins inversely correlates with the levels of FyPP proteins. Moreover, FyPP proteins physically interact with ABI5 in vitro and in vivo, and the strength of the interaction depends on the ABI5 phosphorylation status. In vitro phosphorylation assays show that FyPP proteins directly dephosphorylate ABI5. Furthermore, genetic and biochemical assays show that FyPP proteins act antagonistically with SnRK2 kinases to regulate ABI5 phosphorylation and ABA responses. Thus, Arabidopsis PP6 phosphatase regulates ABA signaling through dephosphorylation and destabilization of ABI5.
INTRODUCTION
The phytohormone abscisic acid (ABA) is a major phytohormone that regulates plant growth and development, including seed dormancy, seed germination, seedling growth, and stomatal aperture, as well as plant responses to various abiotic and biotic stresses, such as drought, salt, and cold stresses and pathogen infection (Mauch-Mani and Mauch, 2005; Fujita et al., 2011; Hauser et al., 2011). Molecular genetic studies in Arabidopsis thaliana have led to the identification of a number of mutants affected in ABA signaling. Among them, the type-2C protein phosphatases ABSCISIC ACID INSENSITIVE1 (ABI1) and ABI2 were identified initially through the analysis of the dominant ABA-insensitive abi1-1 and abi2-1 mutants (Koornneef et al., 1982; Leung et al., 1994, 1997; Meyer et al., 1994). Subsequent characterization of loss-of-function alleles indicated that ABI1 and ABI2 negatively regulate many ABA responses, including inhibition of seed germination, seedling growth, and promotion of stomatal closure (Sheen, 1998; Gosti et al., 1999; Merlot et al., 2001). On the other hand, abi3, abi4, and abi5 were identified as recessive mutants that show ABA insensitivity during seed germination and early seedling development (Koornneef et al., 1982; Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000).
The ABI3, ABI4, and ABI5 genes encode B3-type, APETALA2 domain and basic Leucine zipper (bZIP)–type transcription factors (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). ABI5 is a protein that functions as a transcriptional activator by binding to an ABA-responsive element (ABRE; Giraudat, 1995; Busk and Pagès, 1998; Hattori et al., 2002), a conserved cis-acting element found in the promoters of many ABA-induced genes (Yamaguchi-Shinozaki and Shinozaki, 1994). Consistent with a dominant role in seed germination and seedling development, ABI3, ABI4, and ABI5 are expressed mainly in seeds, with only low levels of expression in vegetative tissues (Giraudat et al., 1992; Finkelstein et al., 1998; Finkelstein and Lynch, 2000).
In addition, a number of bZIP transcription factors involved in ABA signaling have been identified through characterization of the ABRE binding factors (ABFs; also referred to as AREBs), including ABF1, ABF2/AREB1, ABF3, ABF4/AREB2, and ABI5 (Guiltinan et al., 1990; Yamaguchi-Shinozaki and Shinozaki, 1994). ABF2/AREB1, ABF3, and ABF4/AREB2 have been reported to play key roles in the response to drought stress (Yoshida et al., 2010). Plants overexpressing ABF3 and ABF4 exhibit enhanced drought tolerance and altered expression of ABA/stress-regulated genes (Kang et al., 2002). ABF2/AREB1 regulates ABRE-dependent ABA signaling that enhances drought tolerance in vegetative tissues (Fujita et al., 2005). In addition, ABI5 is regulated by sugar and stress (Brocard et al., 2002; Arroyo et al., 2003), indicating that ABI5 also plays a role in stress response adjustment (Fujita et al., 2011).
Among these bZIP transcription factors, ABI5 plays a central role in regulating seed germination and postgermination seedling growth (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001, 2002). ABI5 protein levels are tightly regulated. They accumulate to high levels in dry seeds or young seedlings treated with ABA; however, after germination or removal of ABA from the seedlings, ABI5 proteins are rapidly degraded (Lopez-Molina et al., 2001; Piskurewicz et al., 2008), suggesting that the levels of ABI5 protein play an important role in regulating ABA signaling. In support of this notion, several mutants affected in ABI5 degradation show a hypersensitive ABA response (Stone et al., 2006; Lee et al., 2010). A REALLY INTERESTING NEW GENE-ANKYRIN E3 ligase, KEEP ON GOING (KEG), was identified as a negative regulator of ABA signaling that is required for ABI5 degradation (Stone et al., 2006; Liu and Stone, 2010). Lee et al. (2010) reported that DWD HYPERSENSITIVE TO ABA1 (DWA1) and DWA2 are negative regulators of ABA signaling and function as the substrate receptors for a CULLIN4 E3 ligase that targets ABI5 for degradation. The ABI5-interacting protein (AFP) was reported to facilitate ABI5 degradation through the 26S proteasome–dependent pathway (Lopez-Molina et al., 2003). However, a recent study proposed that NOVEL INTERACTOR OF JAZ and its related AFP proteins act as transcriptional corepressors of TOPLESS proteins to regulate ABA responses through direct interaction with ABI5 (Pauwels et al., 2010). ABI5 abundance is also regulated by SMALL UBIQUITIN-RELATED MODIFIER modification (Miura et al., 2009). The requirement of multiple E3 ligases (for ubiquitination and sumoylation) for regulation of ABI5 levels illustrates the complexity of ABI5 regulation and its importance in regulating ABA signaling in plants.
Given the importance of ABI5 protein levels in ABA signaling, it is striking to note that ABI5 overaccumulation is not sufficient to confer postgerminative growth arrest. Transgenic plants overexpressing ABI5 show increased ABI5 protein levels but grow normally in the absence of ABA (Lopez-Molina et al., 2001; Stone et al., 2006). Notably, additional higher molecular mass forms of ABI5 were observed in the keg-1 mutant but not observed in ABI5-overexpressing plants (Stone et al., 2006). Together with the previous observation that ABA is necessary to stimulate ABI5 activity (Lopez-Molina et al., 2001), it was speculated that the severe postgerminative growth arrest of keg-1 mutants might be caused by the accumulation of additional, presumably phosphorylated and active forms of ABI5 (Stone et al., 2006; Liu and Stone, 2010). Collectively, these data suggest that ABI5 is phosphorylated in response to ABA, and phosphorylation activates the transcription factor and enhances its stability and activity (Lopez-Molina et al., 2001; Piskurewicz et al., 2008).
Recent studies showed that three SUCROSE NONFERMENTING1-related protein kinases, SnRK2.2 (SRK2D), OPEN STOMATA1 (OST1/SnRK2.6/SRK2E), and SnRK2.3 (SRK2I), are responsible for phosphorylation of ABI5 and ABI5-like bZIP transcription factors in response to ABA (Kobayashi et al., 2005; Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009). The triple mutants of snrk2.2/2.3/2.6 are much more resistant to ABA and hypersensitive to drought stress than any single or double mutants (Furihata et al., 2006; Fujii and Zhu, 2009; Nakashima et al., 2009), indicating that SnRK2.2, SnRK2.6, and SnRK2.3 are positive regulators of ABA signaling and they have overlapping functions in ABA signaling. Given the importance of SnRK2-mediated phosphorylation and activation of ABI5 in the inhibition of seed germination, dephosphorylation and subsequent degradation of ABI5 should be critical for the initiation of seed germination. However, the phosphatase(s) responsible for dephosphorylation of ABI5 has not yet been identified. In addition, it is not yet clear whether other phosphatases in addition to those already characterized function in ABA signaling.
The Ser/Thr-specific phosphoprotein phosphatases (PPPs) execute the major phosphatase activities in eukaryotes (Olsen et al., 2006). The Arabidopsis genome encodes 26 catalytic (C) subunits of PPPs related to type 1 (PP1), type 2A (PP2A), PP2B, and the so-called novel phosphatases, including PROTEIN PHOSPHATASE4 (PP4), PP5, PP6, and PP7 (Farkas et al., 2007). The PP2A holoenzyme consists of an enzymatically active catalytic subunit (PP2Ac), a 65-kD regulatory A subunit (PP2A A), and a variable regulatory B subunit (PP2AB) (Farkas et al., 2007). Although there are high sequence similarities among the C subunits of PP6 and PP2A phosphatases, they were classified as different phosphatases because PP6 requires Zn2+ for its activity, whereas PP2A does not require any cation (Kim et al., 2002; Wang et al., 2007; Dai et al., 2012a). PPPs are ubiquitous enzymes in all eukaryotes, but their regulatory functions are largely unknown in plants (Farkas et al., 2007).
In a recent study, we showed that Arabidopsis FyPP1 (for Phytochrome-associated serine/threonine protein phosphatase1) and FyPP3, two homologous catalytic subunits of PP6, physically interact with SAL (for SAPS-domain like protein) and PP2AA proteins (RCN1/PP2AA1, PP2AA2, and PP2AA3) to form a PP6-type heterotrimeric holoenzyme that directly regulates the phosphorylation status of PIN-FORMED (PIN) proteins (auxin efflux carriers) and subsequently affects auxin efflux and plant development (Dai et al., 2012a, 2012b). In this study, we extend these findings by showing that FyPP also plays a critical role in regulating ABA signaling in Arabidopsis. We show that mutants without FyPP activity accumulate higher levels of ABI5 protein and display an ABA-hypersensitive phenotype in seed germination and postgermination seedling growth. We further show that FyPP proteins directly interact with and dephosphorylate ABI5. Genetic interaction studies demonstrate an antagonistic interaction between FyPP1/3 with SnRK2 in regulating ABA responses. Our findings suggest that FyPP1 and FyPP3 function as two negative regulators of ABA signaling through the dephosphorylation of ABI5.
RESULTS
FyPP Genes Negatively Regulate ABA Signaling
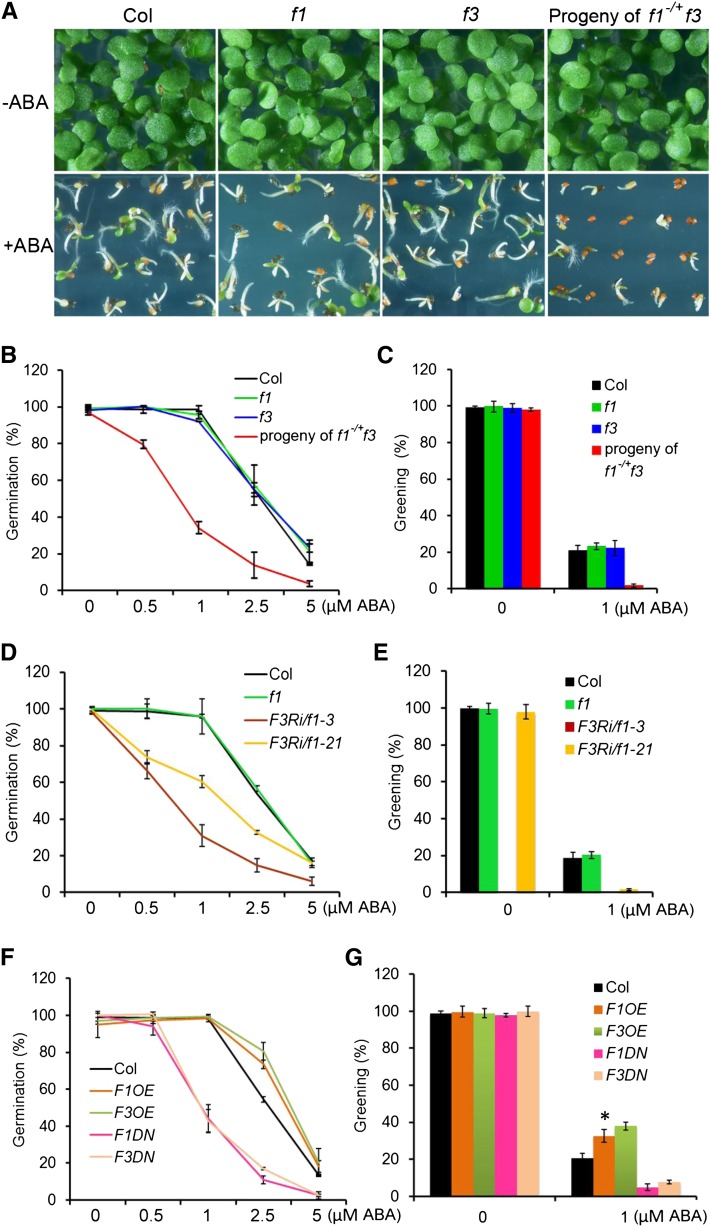
FyPP1 and FyPP3, the catalytic subunits of Arabidopsis PP6 phosphatases (PP6c) in Arabidopsis, share high sequence homology (99% amino acid identity) (Dai et al., 2012a). Publically available data (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007) show that FyPP1 and FyPP3 expression is upregulated by imbibition during seed germination, which is opposite to that of ABI5 (see Supplemental Figure 1 online), suggesting the involvement of FyPP1 and FyPP3 in regulating seed germination. To test this, we first investigated the germination of Columbia (Col; the wild type), fypp1 (f1), and fypp3 (f3) seeds and seeds from self-pollinated f1−/+ f3 plants (f1 is heterozygous and f3 is homozygous) on germination medium (GM) plates containing various concentrations of ABA. We used seeds from self-pollinated f1−/+ f3 plants in these experiments because the f1 f3 homozygous double mutant plants are completely infertile (Dai et al., 2012a). The results showed that both f1 and f3 single homozygous mutants had similar germination and greening percentages as Col at the tested ABA concentrations, while the seeds from self-pollinated f1−/+ f3 plants had much lower germination and greening percentages than Col at the tested ABA concentrations (Figures 1A to 1C; see Supplemental Figure 2 online). Genotyping analysis revealed that among the progeny of self-pollinated f1−/+ f3 plants incubated on 2.5 μM ABA plates, all geminated seeds were of the f3 genotype (n = 20 genotyped, 100%), while the nongermination seeds (n = 95 genotyped) consisted of three genotypes: f3 (12.6%), f1−/+ f3 (58%), and f1 f3 (29.4%). These observations indicate that seeds of the f1−/+ f3 and f1 f3 genotypes were hypersensitive to ABA.
Figure 1.
Phenotypic Characterization of FyPP1 and FyPP3 Loss- and Gain-of-Function Mutants in ABA Responses.
(A) Germination and growth of Col, f1, and f3 seeds and seeds from self-pollinated f1−/+f3 plants incubated on GM plates with 0 (top panels) or 1 μM ABA (bottom panels) for 5 d. Seeds from self-pollinated f1−/+f3 plants were more sensitive to ABA than Col seeds.
(B) Germination of Col, f1, and f3 seeds and seeds from self-pollinated f1−/+f3 plants incubated on GM plates with varying concentrations of ABA (0, 0.5, 1, 2.5, and 5 μM) for 5 d. Seeds from self-pollinated f1−/+f3 plants were more sensitive to ABA than Col. Germination percentages were determined from three independent experiments, with more than 100 seeds per line for each experiment. Values are means ± sd.
(C) Greening of Col, f1, and f3 seeds and seeds from self-pollinated f1−/+f3 plants incubated on GM plates with 0 or 1 μM ABA for 5 d. Seeds from self-pollinated f1−/+f3 plants were more sensitive to ABA than Col seeds. Greening was determined with an average of >100 seeds from three independent experiments. Values are means ± sd.
(D) Germination of Col, f1, and F3Ri/f1 seeds incubated on GM plates with varying concentrations of ABA (0, 0.5, 1, 2.5, and 5 μM) for 5 d. All GM plates contained 0.1% ethanol. Ethanol-induced F3Ri/f1 seeds were more sensitive to ABA than Col or f1 seeds. Germination percentage was determined from three independent experiments, with more than 150 seeds per line for each experiment. Values are means ± sd.
(E) Greening of Col, f1, and F3Ri/f1 seeds incubated on GM plates (plus 0.1% ethanol) with 0 or 1 μM ABA for 5 d. Ethanol-induced F3Ri/f1 seeds were more sensitive to ABA than Col seeds or f1 seeds. Greening was determined with an average of >150 seeds from three independent experiments. Values are means ± sd.
(F) Germination of Col, F1DN, F3DN, F1OE, and F3OE seeds incubated on GM plates with varying concentrations of ABA (0, 0.5, 1, 2.5, and 5 μM) for 5 d. F1DN and F3DN seeds were more sensitive to ABA, while F1OE and F3OE seeds were less sensitive to ABA than Col. Germination percentages were determined from three independent experiments, with more than 100 seeds per line for each experiment. Values are means ± sd.
(G) Greening of Col, F1OE, F3OE, F1DN, and F3DN seeds incubated on GM plates with 0 or 1 μM ABA for 5 d. F1DN and F3DN seeds were more sensitive to ABA, while F1OE and F3OE were less sensitive to ABA than Col seeds. Greening was determined with an average of >100 seeds from three independent experiments. Values are means ± sd. Asterisks indicate the levels of statistical significance as determined by Student’s t test: *P < 0.01 versus Col.
We next investigated the root growth of f1 f3 single mutants and f1 f3 double mutants on GM plates containing 0 or 1 µM ABA. The f1 f3 double mutants have shorter roots and smaller/fused cotyledons than Col at the early seedling stage (Dai et al., 2012a; see Supplemental Figure 3 online), and we therefore chose f1 f3 seedlings for our stress experiment based on these phenotypes. We observed that the relative root growth of the f1 f3 mutants was much reduced compared with that of Col, whereas there were no obvious differences in the relative root growth of f1 or f3 single mutants and Col (see Supplemental Figure 4A online). Again, this observation indicates that the f1 f3 double mutants, but not the f1 or f3 single mutants, are hypersensitive to ABA.
To confirm our observations, we investigated the ABA responses of ethanol-inducible F3Ri/f1 lines, which were generated by introducing the AlcA-AlcR:FyPP3RNAi expression cassette into the fypp1 (f1) single mutant background (Dai et al., 2012a). We chose two F3Ri/f1 lines (F3Ri/f1-3 and F3Ri/f1-21) for our experiments. The results showed that after ethanol induction, the seed germination and greening percentages of both F3Ri/f1 lines were reduced compared with Col at the tested ABA concentrations, with F3Ri/f1-3 showing a more severe phenotype than F3Ri/f1-21 (Figures 1D and 1E). Interestingly, there was no greening of F3Ri/f1-3 seeds after 5 d on GM plates containing ethanol but not ABA (Figure 1E). RT-PCR analysis showed that the expression of FyPP3 was undetectable in F3Ri/f1-3 and dramatically reduced in F3Ri/f1-21, while ethanol itself had no effect on FyPP3 expression (see Supplemental Figure 5 online), indicating that the ABA hypersensitive phenotype of the F3Ri/f1 transgenic plants after ethanol induction is specifically due to the reduced expression of the FyPP3 gene. We also observed that the relative root growth of F3Ri/f1-21 on GM plates with ABA was much slower than that of Col after ethanol induction (see Supplemental Figure 4B online), further confirming that the F3Ri/f1 plants are hypersensitive to ABA.
To further investigate the function of FyPP1 and FyPP3 in ABA signaling, we investigated the seed germination and greening percentages of FyPP overexpressing (F1OE and F3OE) and FyPP dominant-negative (F1DN and F3DN) transgenic lines (Dai et al., 2012a). The results showed that the F1DN and F3DN seeds had much lower germination percentages after 1, 2.5, or 5 µM ABA treatment compared with Col seeds, while the F1OE and F3OE seeds had significantly higher germination percentages with 2.5 µM ABA treatment compared with Col seeds (Figure 1F). Additionally, the greening percentages of F1DN and F3DN seedlings were much lower, while the greening percentages of F1OE and F3OE seedlings were much higher than that of Col when treated with 1 µM ABA (Figure 1G). Thus, F1DN and F3DN transgenic lines showed a similar seed germination phenotype as the seeds from self-pollinated f1−/+ f3 plants and the F3Ri/f1 transgenic lines, while the F1OE and F3OE transgenic plants displayed the opposite seed germination phenotype as seeds from self-pollinated f1−/+ f3 plants and F3Ri/f1 transgenic lines. Taken together, these observations suggest that Arabidopsis FyPP1 and FyPP3 play critical roles in ABA signaling and negatively regulate ABA responses.
The ABA-Hypersensitive Phenotype of FyPP Loss-of-Function Mutants Is ABI5 Dependent
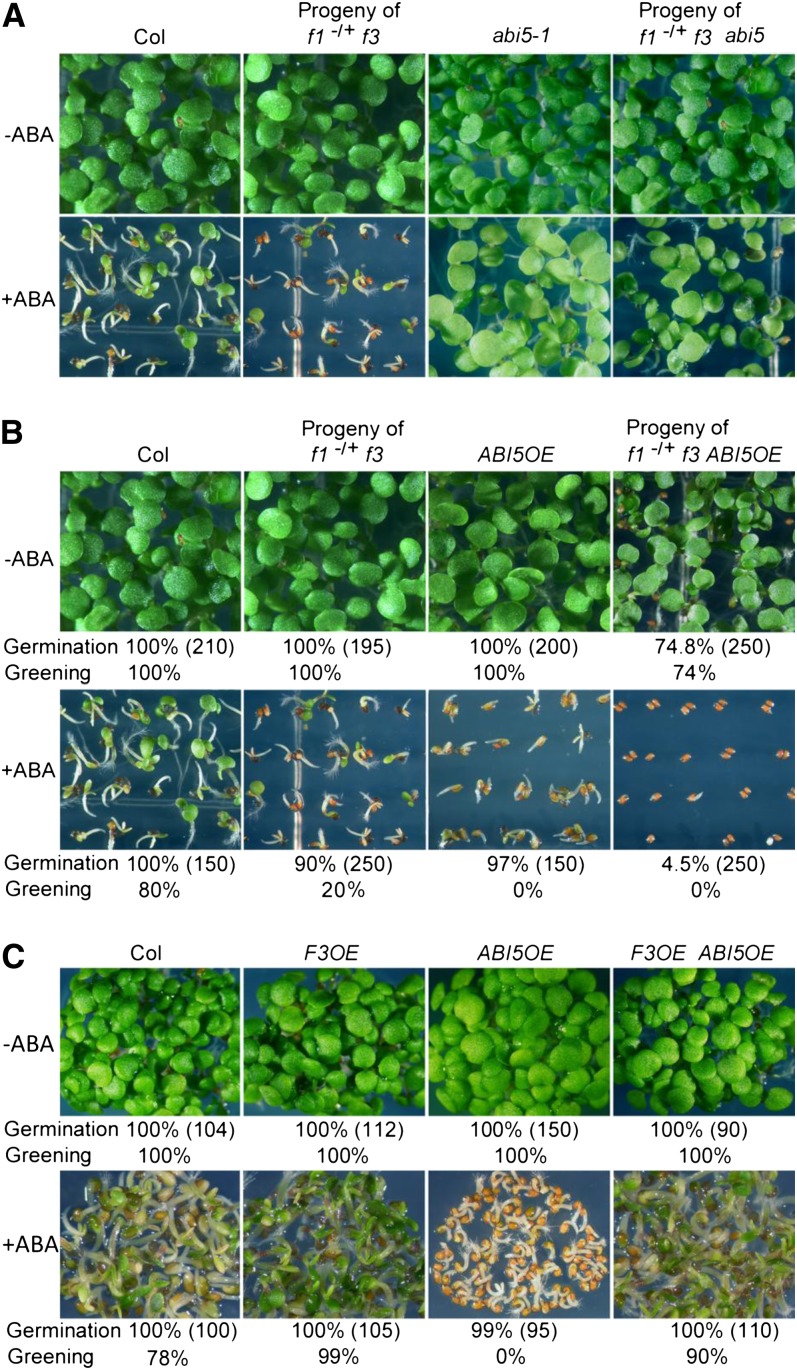
Since ABI5 is known as a key regulator of seed germination, we next tested the genetic interactions between FyPPs and ABI5. We introduced the f1 and f3 mutations into the abi5-1 mutant background (Finkelstein, 1994). We generated plants that were homozygous for f3 and abi5 but heterozygous for f1 (here referred to as f1−/+ f3 abi5-1) and obtained seeds from these self-pollinated plants in our germination assays. We observed that the seeds from self-pollinated f1−/+ f3 abi5-1 plants, self-pollinated f1−/+ f3 plants, and abi5-1 mutants, as well as the Col controls, all germinated and grew well on GM plates lacking ABA (Figure 2A; see Supplemental Figure 6A online). Seeds from self-pollinated f1−/+ f3 plants were hypersensitive to 0.5 µM ABA as expected, whereas seeds from selfed f1−/+ f3 abi5-1 plants were insensitive to 0.5 µM ABA, similar to abi5-1 (Figure 2A; see Supplemental Figures 6B and 6C online).
Figure 2.
Genetic Interaction between ABI5 and FyPPs.
(A) Germination and growth of seeds of Col, abi5, and self-pollinated f1−/+ f3 and f1−/+ f3 abi5 plants incubated on GM plates with 0 or 0.5 µM ABA for 5 d in white light. Seeds from self-pollinated f1−/+f3 plants showed increased sensitivity to ABA, while abi5-1 seeds were insensitive to ABA compared with Col controls. Seeds from self-pollinated f1−/+ f3 abi5-1 plants were insensitive to ABA, similar to abi5-1 seeds.
(B) Loss of FyPP1 and FyPP3 activity enhances the ABA-related phenotypes of ABI5OE seeds. Top panel shows seedlings grown on GM plates without ABA. The ungerminated seeds (25.2%) from self-pollinated f1−/+ f3 ABI5OE plants were further phenotyped and genotyped and confirmed to be f1 f3 ABI5OE homozygotes. Bottom panel shows the seedlings grown on GM plates containing 0.5 µM ABA. The germination and greening percentages of each line are shown at the bottom of each panel, and the number of seeds used for the calculation is shown in parentheses.
(C) Growth of Col, F3OE, ABI5OE, and F3OE ABI5OE seeds on GM plates with 0 or 0.5 µM ABA. Overexpression of FyPP3 rescued the ABA hypersensitivity phenotype of ABI5OE seeds. The germination and greening percentages of each line are shown at the bottom of each panel, and the number of seeds used for the calculation is shown in parentheses.
We next introduced the f1 and f3 mutations into transgenic plants overexpressing ABI5 (ABI5OE hereafter; Brocard et al., 2002). Similarly, we generated plants homozygous for f3 and ABI5OE but heterozygous for f1 (here referred to as f1−/+ f3 ABI5OE) and obtained seeds from these self-pollinated plants in our experiments. Without ABA treatment, seeds from both self-pollinated f1−/+ f3 plants and ABI5OE plants germinated and grew well (Figure 2B). Interestingly, we observed that 24.8% of seeds (n = 250) from self-pollinated f1−/+ f3 ABI5OE plants did not germinate after 5 d on GM plates (Figure 2B), although they eventually germinated after a longer incubation period (more than 7 d). Genotyping showed that these seeds were all f1 f3 ABI5OE homozygotes. After treatment with 0.5 µM ABA, the ABI5OE seedlings underwent growth arrest after 5 d, although their germination percentages were comparable to those of the seeds from self-pollinated f1−/+f3 plants and Col controls at this stage, while the germination percentage of the seeds from self-pollinated f1−/+ f3 ABI5OE plants was much lower that of the ABI5OE seeds, and the growth of seedlings segregated from f1−/+ f3 ABI5OE was more severely inhibited compared with the ABI5OE seedlings (Figure 2B). Genotyping analysis showed that after ABA treatment, all of the germinated seeds (n = 15 genotyped) from self-pollinated f1−/+ f3 ABI5OE plants were of the f3 ABI5 genotype, while the nongermination seeds (n = 85 genotyped) had three genotypes: f3 ABI5OE (20.5%), f1−/+ f3 ABI5OE (49%), and f1 f3 ABI5OE (30.5%). These observations indicate that the loss of FyPP1 and FyPP3 activity enhanced the sensitivity of ABI5OE seeds to ABA.
We also crossed F3OE transgenic lines with ABI5OE plants. As shown in Figure 2C, without ABA treatment, Col, F3OE, ABI5OE, and F3OE ABI5OE double mutants germinated and grew well, while in the presence of 0.5 µM ABA, the growth of the ABI5OE seedlings was more severely inhibited than that of the F3OE ABI5OE double mutants, F3OE seeds, and Col wild-type controls. These results suggest that enhanced FyPP activity can largely overcome the effects of ABI5 overexpression in response to ABA. Taken together, these data suggest that FyPP1 and FyPP3 act through the ABI5 pathway to regulate ABA responses.
FyPP1 and FyPP3 Directly Interact with ABI5
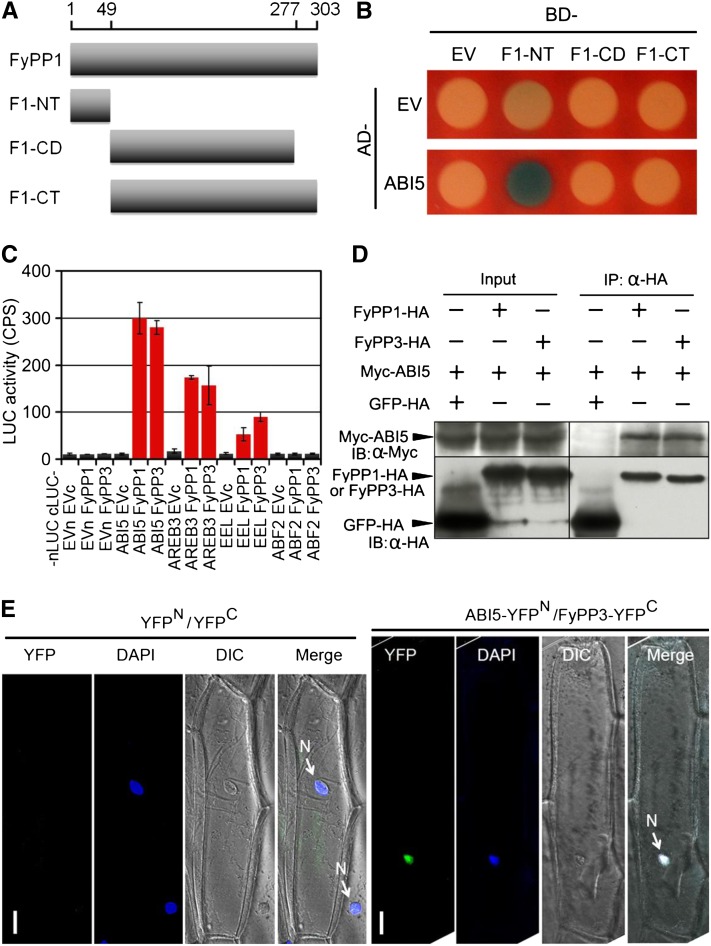
We next performed a series of experiments to test whether ABI5 could serve as a substrate of FyPP proteins. Yeast two-hybrid (Y2H) assays showed that both FyPP1 and FyPP3 specifically interacted with ABI5, but not with other key regulators of ABA signaling, including the ABA receptors (Pyr and Pyl1), the PP2C phosphatases (ABI1 and ABI2), the SnRK2 kinase (SnRK2.6/OST1), and the downstream transcription factors ABI3 and ABI4 (see Supplemental Figures 7 and 8 online). Domain deletion analysis showed that the N-terminal region of FyPP1 protein mediated the interaction between FyPP1 and ABI5 in yeast cells (Figures 3A and 3B; see Supplemental Figure 8 online). Moreover, luciferase complementation imaging (LCI) assays (Chen et al., 2008) showed that both cLUC-FyPP1 and cLUC-FyPP3 interacted with nLUC fusions of ABI5 and group I ABI5-like proteins (more similar to ABI5 protein, such as AREB3 and EEL), but not nLUC fusions of group II ABI5-like proteins (less similar to ABI5 protein, such as ABF2) (Figure 3C; see Supplemental Figure 9 online). Furthermore, coimmunoprecipitation (Co-IP) assays showed that Myc-ABI5 coimmunoprecipitated with FyPP1-HA and FyPP3-3HA, but not with green fluorescent protein (GFP)-HA in tobacco (Nicotiana tabacum) leaves (Figure 3D), further confirming the interaction between FyPPs and ABI5 in vivo.
Figure 3.
Protein–Protein Interactions between FyPPs and ABI5.
(A) Diagram of the FyPP1 protein constructs used in Y2H assays. F1-NT, FyPP1 N-terminal region (amino acids 1 to 49); F1-CD, FyPP1 catalytic domain (amino acids 50 to 277); F1-CT, FyPP1 C-terminal region (amino acids 50 to 303).
(B) ABI5 protein interacted with the F1-NT domain of FyPP1 protein in yeast cells. AD, B42 activation domain; BD, LexA DNA binding domain; EV, empty vector control.
(C) LCI assays showing that when fused with nLUC, ABI5 and group I ABI5-like proteins (AREB3 and EEL), but not group II protein (ABF2), interacted with both cLUC-FyPP1 and cLUC-FyPP3 in plant cells. Values are means ± sd, n = 3. EVc, cLUC empty vector; EVn, nLUC empty vector.
(D) Co-IP of ABI5 and FyPP1 or FyPP3 in plant cells. α-HA affinity matrix was used for immunoprecipitation (IP); α-HA and α-Myc antibodies were used for immunoblotting (IB). Input, total protein before immunoprecipitation.
(E) BiFC assays showing that AB15 and FyPP3 interacted in the nucleus. ABI5-YFPN and FyPP3-YFPC fusion proteins were expressed in onion epidermal cells through cobombardment. No YFP signal was observed in onion cells cobombarded with the YFPN (YFP protein N-terminal) and YFPC (YFP protein C-terminal) control plasmids. The nuclei were stained by DAPI (4',6-diamidino-2-phenylindole, blue). Bars = 50 μM.
To identify the subcellular localization of the interaction between FyPPs and ABI5, we performed bimolecular fluorescence complementation (BiFC) in onion (Allium cepa) epidermal cells. We observed strong yellow fluorescent protein (YFP) signals in the nuclei of onion cells bombarded with ABI5-YFPN (YFP N-terminal region) and FyPP3-YFPC (YFP C-terminal region) plasmids, but no YFP signal was detected in onion cells bombarded with the YFPN and YFPC control plasmids (Figure 3E). In Arabidopsis plants, the FyPP1 and FyPP3 proteins were detected in the nucleus, the plasma membranes, and the cytosol (see Supplemental Figure 10 online). These observations indicate that the interaction between ABI5 and FyPP3 occurs in the nucleus in planta. Taken together, these data indicate that there is a direct interaction between FyPPs and ABI5.
FyPP Directly Dephosphorylates ABI5 in Vitro
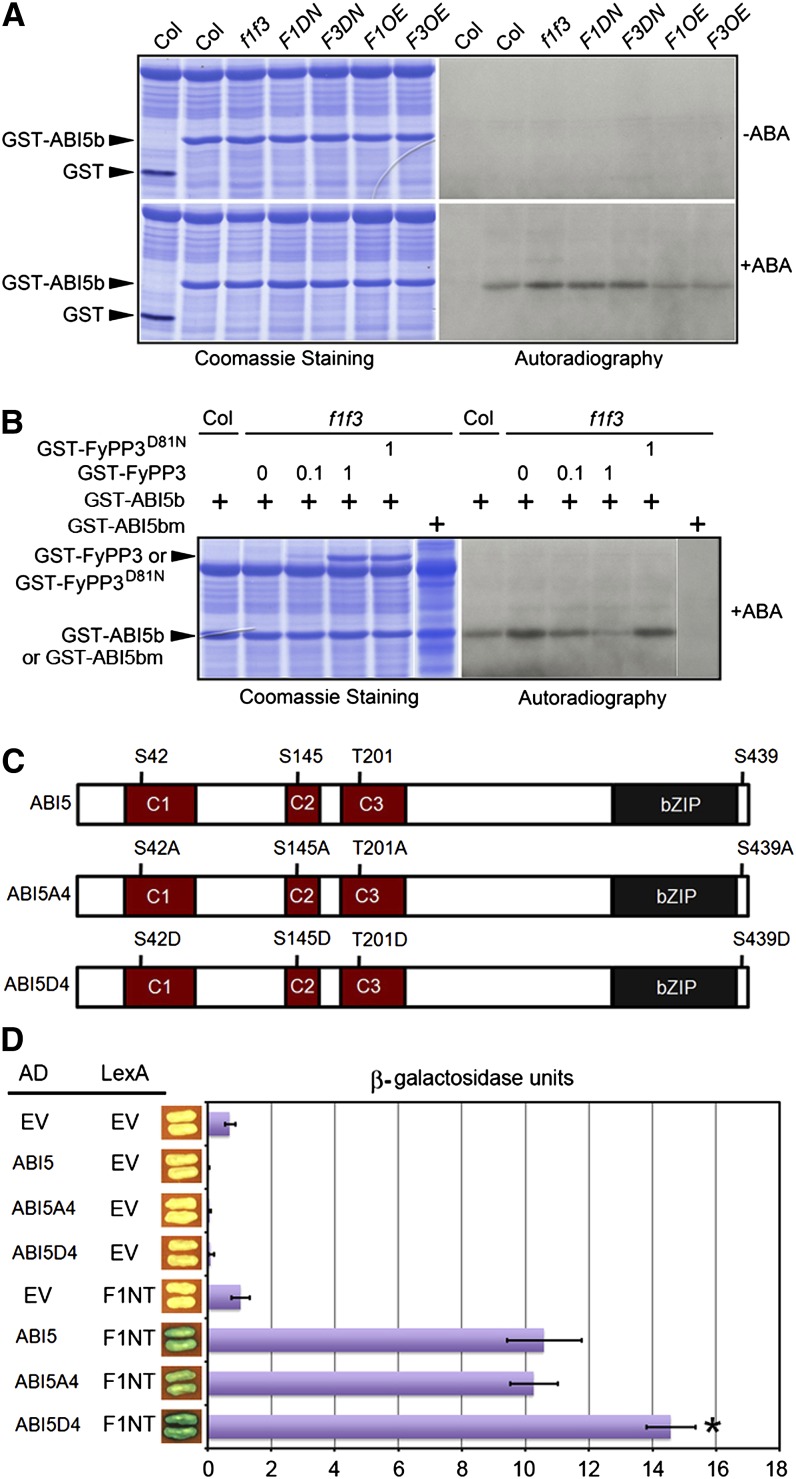
Since FyPP1 and FyPP3 directly interacted with ABI5, we next tested whether FyPP1 and FyPP3 could directly dephosphorylate ABI5. A previous study reported that the Ser119-Gln190 ABI5 fragment (ABI5b) is phosphorylated by the SnRK2.2/2.3/2.6 kinases (Nakashima et al., 2009). We therefore performed in vitro phosphorylation experiments using glutathione S-transferase (GST)–tagged ABI5b purified from Escherichia coli cells as the substrate and protein extracts prepared from Col, f1 f3, F1DN, F3DN, F1OE, and F3OE seedlings treated with ABA or with control solvent as the kinase sources. Equal amounts of protein extracts and substrates were coincubated in these assays. No phosphorylated GST-ABI5b was detected in any sample without ABA treatment (Figure 4A), indicating that phosphorylation of ABI5 is ABA dependent, which is consistent with previous reports (Fujii et al., 2007; Nakashima et al., 2009). However, after ABA treatment, the amounts of phosphorylated GST-ABI5b were higher in samples incubated with protein extracts from f1 f3, F1DN, and F3DN transgenic seedlings, while the amounts of phosphorylated GST-ABI5b were slightly lower in samples incubated with protein extracts from F1OE and F3OE seedlings, compared with Col (Figure 4A). These data suggested that there is a correlation between FyPP1/3 activity and phosphorylation of GST-ABI5b.
Figure 4.
FyPP Directly Dephosphorylates ABI5.
(A) In vitro kinase assay of GST-ABI5b (Ser119-Gln190). In the absence of ABA, there was no detectable phosphorylated GST-ABI5b when treated with the plant extracts derived from Col, f1 f3, F1DN, F3DN, F1OE, and F3OE seedlings. After treatment with ABA, there were increased amounts of phosphorylated GST-ABI5b when incubated with the plant extracts derived from f1 f3, F1DN, and F3DN seedlings, in contrast with the reduced abundance of phosphorylated GST-ABI5b when incubated with the plant extracts derived from F1OE and F3OE seedlings.
(B) In vitro dephosphorylation of GST-ABI5b by GST-FyPP3. GST-FyPP3 reversed the ABA-dependent dephosphorylation of GST-ABI5b treated with the plant extracts derived from f1 f3 seedlings. Increasing amounts of GST-FyPP3 decreased the amount of phosphorylated GST-ABI5b, while the inactive phosphatase (GST-FyPP3D81N) had no effect on the phosphorylation status of GST-ABI5b. A mutant form of ABI5 (GST-ABI5bS145A) was used as a negative control in the experiment. The amounts of GST-FyPP3 and GST-FyPP3D81N proteins used in the assay are indicated by the numbers (0, 0.1, and 1 μg).
(C) Schematic representation of the domain structure of the dephosphorylation mimic mutant ABI5A4 and the phosphorylation mimic mutant ABI5D4 used for the Y2H assays. The labeled Ser (S) and Thr (T) residues were mutated to Ala (A) or Asp (D), respectively.
(D) Y2H assays between FyPP1 N-terminal region (F1NT) and various ABI5 mutants shown in (A). The phosphorylation mimic mutant ABI5D4 showed enhanced interaction with F1NT. Values are means ± sd; n = 3. Asterisks indicate the levels of statistical significance as determined by Student’s t test: *P < 0.02 versus F1NT–ABI5 interaction. EV, empty vector control.
[See online article for color version of this figure.]
To confirm this observation, we performed in vitro phosphorylation/dephosphorylation assays. We incubated equal amounts of GST-ABI5b proteins and protein extracts from ABA-treated Col or f1 f3 seedlings. We added various amounts of purified wild-type PP6 phosphatase (GST-FyPP3) or PP6 null mutant (GST-FyPP3D81N; Dai et al., 2012a) to the samples mixed with protein extracts from f1 f3 seedlings and GST-ABI5b proteins. The results showed that the addition of exogenous GST-FyPP3, but not GST-FyPP3D81N, reduced the amounts of phosphorylated GST-ABI5b, and the more GST-FyPP3 protein added, the less phosphorylated GST-ABI5b detected (Figure 4B). Additionally, when we used mutant GST-ABI5b (ABI5bS145A or ABI5bm) as the substrate, we did not observe a phosphorylation band by the f1 f3 mutant extracts (Figure 4B). These data together suggest that FyPP1/3 proteins play a critical role in regulating the phosphorylation status of ABI5.
It was reported that the Ser/Thr sites in the conserved (Leu)xArgxxSer/Thr motif of ABI5 are the targets of SnRK2 kinases, including SnRK2.2, 2.3, and 2.6 (Furihata et al., 2006; Nakashima et al., 2009). To test whether these Ser/Thr sites mediate the interaction between ABI5 and FyPPs, we conducted site-directed mutagenesis and substituted these Ser/Thr sites with either Ala or Asp residues, thus generating the dephosphorylation mimic mutant (ABI5A4) or phosphorylation mimic mutant (ABI5D4) forms of ABI5 (Figure 4C). Y2H assays showed a stronger interaction between FyPP1 (F1NT) and ABI5D4 than the interaction between FyPP1 (F1NT) and wild-type ABI5 (Figure 4D; see Supplemental Figure 8 online). This observation suggests that phosphorylation of these Ser/Thr sites in the conserved (Leu)xArgxxSer/Thr motifs plays an important role in mediating the interaction between ABI5 and FyPP.
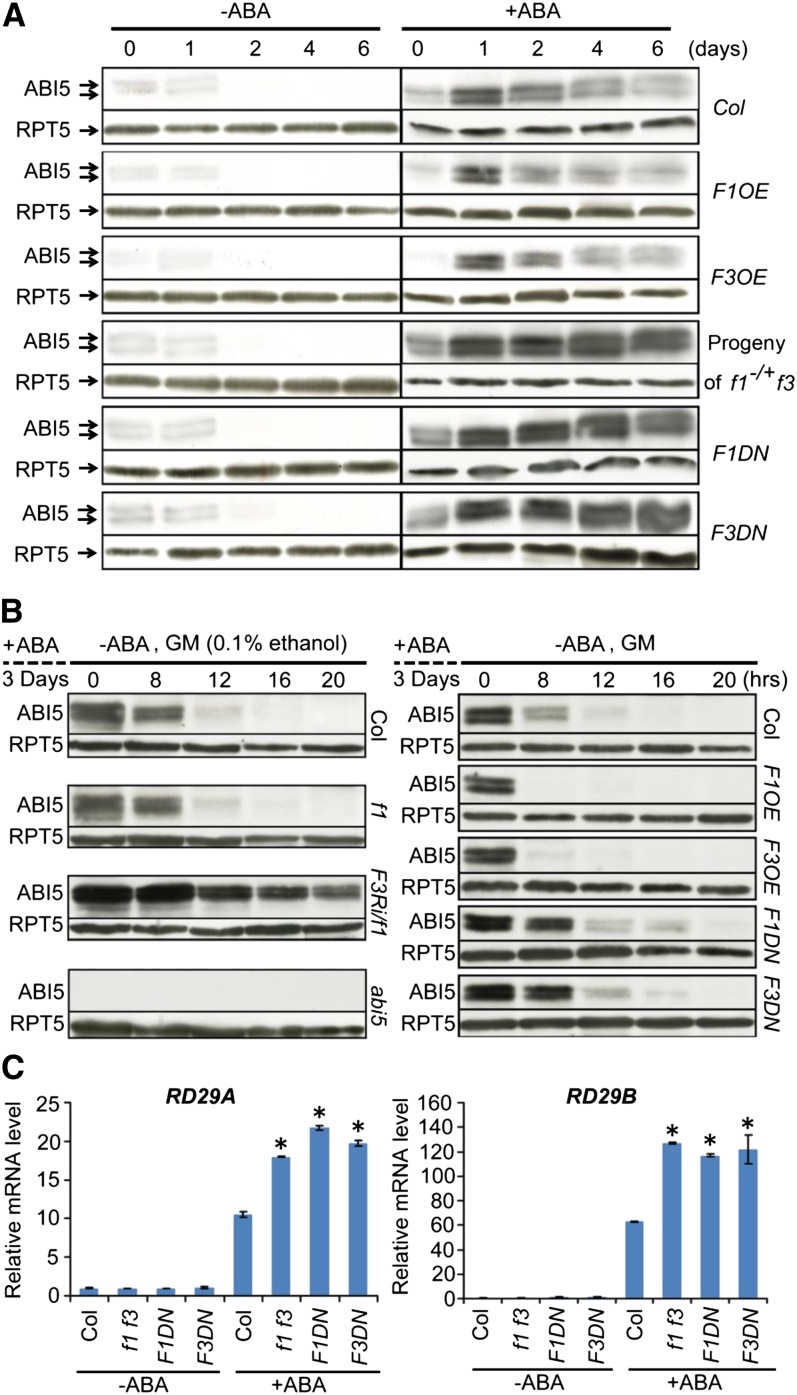
FyPP Regulates ABI5 Protein Stability
Phosphorylation plays a critical role in regulating protein stability (Hardtke et al., 2000; Karin and Ben-Neriah, 2000; Deshaies and Ferrell, 2001; Joo et al., 2008). We next tested whether FyPP1 and FyPP3 may also be involved in regulating ABI5 protein stability. We germinated Col, F1DN, F3DN, F1OE, and F3OE seeds and seeds from self-pollinated f1−/+f3 plants on GM plates containing 0 or 1 μM ABA. After varying incubation time (0, 1, 2, 4, and 6 d) under continuous white light, we extracted their total RNAs and proteins. We used the anti-ABI5–specific antibodies in the immunoblot analysis. Stone et al. (2006) detected 52.5-, 51-, and 50-kD ABI5 bands with the 52.5- and 50-kD bands predominating in immunoblot assays using this α-ABI5 antibody, whereas we detected two bands in this study and in the previous report (Figures 5A and 5B; Lee et al., 2010). The results showed that in the presence of ABA, ABI5 proteins were hyperaccumulated in the F1DN and F3DN seeds and seeds from self-pollinated f1−/+ f3 plants, but reduced in the F1OE and F3OE seeds, compared with Col (Figure 5A). Consistent with previous reports, ABI5 proteins were quickly degraded in germinated seeds due to diminished ABA levels (Figure 5A; Piskurewicz et al., 2008). Quantitative RT-PCR analysis showed that ABI5 expression was highly induced in all lines in response to ABA compared with the Col controls (see Supplemental Figure 11A online). These results suggest that the differential accumulation of ABI5 proteins in these plants is most likely due to posttranscriptional regulation. Consistent with this, seeds from self-pollinated f1−/+ f3 plants and F1DN and F3DN mutants had lower germination rates after ABA treatment, whereas F1OE and F3OE seeds had higher germination rates, compared with Col (see Supplemental Figures 2 and 12 online).
Figure 5.
Role of FyPP1 and FyPP3 in the Regulation of ABI5 Protein Stability.
(A) Immunoblot assays showing that more ABI5 protein accumulated in seeds of self-pollinated f1−/+ f3, F1DN, and F3DN lines but less accumulated in F1OE and F3OE seeds compared with Col seeds treated with ABA (1 μM) for various durations (0, 1, 2, 4, and 6 d). A total of 100 μg protein was loaded for each lane. All immunoblot assays were performed side-by-side under identical conditions. Arrows indicate the ABI5 protein bands. RPT5 was used as a loading control.
(B) Immunoblot assays showing that after ABA removal, ABI5 protein was more stable in F3Ri/f1, F1DN, and F3DN lines but more rapidly degraded in F1OE and F3OE seeds compared with Col seeds. The seeds were treated with ABA (5 μM) in white light for 3 d and then harvested at different time points after removal of the ABA. The medium used for each line is indicated on the top of each panel. abi5-1 mutant seeds were used as a negative control in the experiment. A total of 100 μg protein was loaded for each lane. All immunoblot assays were performed side-by-side under identical conditions. RPT5 was used as a loading control.
(C) Quantitative RT-PCR assay showing that the expression of RD29A and RD29B was hyperinduced in f1 f3, F1DN, and F3DN seedlings treated with ABA compared with ABA-treated Col. Five-day-old seedlings were incubated in 1× Murashige and Skoog liquid medium with ABA (100 μM) or control solvent (DMSO) for 1.5 h before harvest. Values are means ± sd; n = 3. Asterisks indicate the levels of statistical significance as determined by Student’s t test: *P < 0.01 versus Col.
[See online article for color version of this figure.]
It was reported that ABI5 is highly induced and stabilized by ABA but is degraded rapidly after the removal of ABA (Lopez-Molina et al., 2001). To investigate how FyPPs function in this process, we treated Col, f1, F3Ri/f1-21, F1DN, F3DN, F1OE, and F3OE seeds with 5 µM ABA for 3 d and then removed the ABA and grew the seeds on GM plates. We harvested samples after various incubation time points (0, 8, 12, 16, and 20 h) for protein and RNA extraction. Immunoblot analysis showed that after the removal of ABA, ABI5 was more abundant in the FyPP loss-of-function mutants, such as F1DN, F3DN, and F3Ri/f1-21 seeds, after ethanol induction, but this protein was more rapidly degraded in the F1OE and F3OE seeds compared with Col (Figure 5B). Gene expression assays showed that ABI5 mRNA was inhibited to comparable levels in all seeds after removal of ABA (see Supplemental Figures 11B to 11E online). Together, these observations support the notion that FyPPs are essential for posttranscriptional regulation of ABI5 in response to ABA removal.
We next investigated the effect of ABA on the expression of ABI5-regulated ABA-responsive genes in the FyPP loss-of-function mutants. Two RD genes, RD29A and RD29B, have ABREs in their promoters and are transactivated by ABI5 (Nakashima et al., 2006). Finkelstein et al. (2005) also showed dramatically reduced ABA inducibility of RD29B in abi5 seedlings. Our quantitative RT-PCR assays showed that the expression of both RD29A and RD29B was upregulated in Col, f1 f3, F1DN, and F3DN mutants after ABA treatment, with a hyperinduction of RD29A and RD29B in the FyPP loss-of-function mutants (Figure 5C). These observations suggest that the ABA hypersensitivity of FyPP loss-of-function mutants was due to the hyperaccumulation of ABI5 proteins, resulting in upregulation of downstream ABA-responsive gene expression.
FyPP1 and FyPP3 Function Antagonistically with SnRK2 in Mediating ABA Signaling
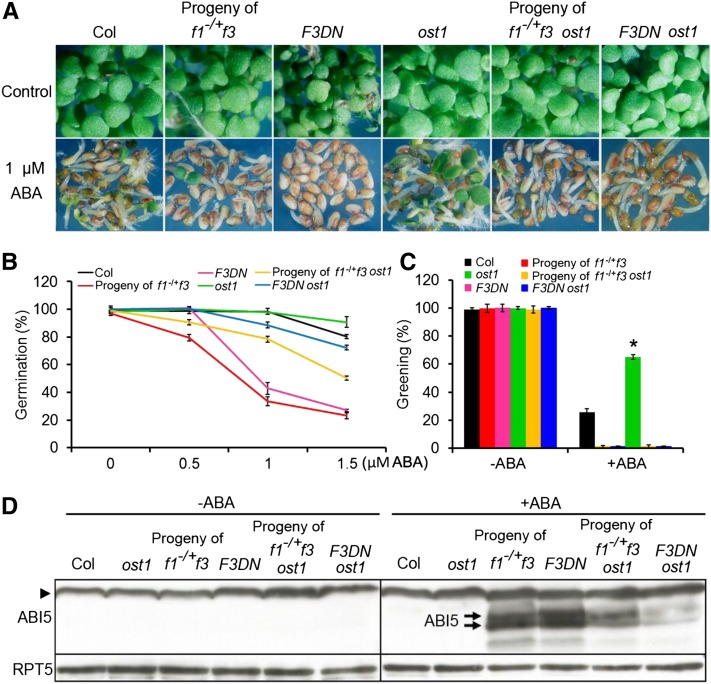
'Because FyPP/PP6 phosphatases and SnRK2 kinases show antagonistic effects on ABA signaling, we were interested in understanding the genetic interactions between SnRK2 and FyPPs. OST1 (SnRK2.6) plays a critical role in mediating various ABA responses, and its loss-of-function mutant (ost1 or snrk2.6) displays a hypersensitive phenotype in response to drought stress and resistance to ABA during seed germination (Li et al., 2000; Mustilli et al., 2002; Yoshida et al., 2006; Zheng et al., 2010). Additionally, the single mutants of snrk2.2 and snrk2.3 do not display obvious ABA-related phenotypes, whereas ost1 is more resistant to ABA during seed germination than the wild type (Fujii et al., 2007; Zheng et al., 2010). We therefore introduced the ost1 mutation into f1−/+ f3 and F3DN mutant backgrounds. We generated plants homozygous for f3 and ost1 but heterozygous for f1 (here referred to as f1−/+ f3 ost1) and obtained seeds from these self-pollinated plants for our germination/growth assays. Our results showed that seeds from self-pollinated f1−/+ f3 plants and F3DN mutants were sensitive to ABA as expected, while seeds from self-pollinated f1−/+ f3 ost1 plants or the F3DN ost1 double mutants were more tolerant to ABA than their f1−/+ f3 or F3DN parental lines, respectively (Figures 6A and 6B). On the other hand, compared with the ost1 mutants, seeds from self-pollinated f1−/+ f3 ost1 plants and the F3DN ost1 double mutants were still sensitive to ABA (Figures 6A to 6C). Consistent with this observation, the ABI5 protein levels in ABA-treated seeds of self-pollinated f1−/+ f3 ost1 plants and F3DN ost1 double mutants were reduced compared with ABA-treated f1−/+ f3 or F3DN parental lines, respectively (Figure 6D). These results suggest that ost1 attenuates the ABA hypersensitivity of seeds from self-pollinated f1−/+f3 and F3DN and that FyPPs function antagonistically with OST1/SnRK2 kinases in mediating ABA signaling.
Figure 6.
Antagonistic Relationship between OST1 (SnRK2.6) and FyPPs.
(A) Germination and growth of Col, ost1, F3DN, and ost1 F3DN seeds and seeds from self-pollinated f1−/+ f3 and f1−/+ f3 ost1 plants incubated on GM plates with or without ABA. Seeds were incubated under white light for 5 d.
(B) Germination of Col, ost1, F3DN, and ost1 F3DN seeds and seeds from self-pollinated f1−/+ f3 and f1−/+ f3 ost1 plants. Seeds were grown under white light for 5 d on GM plates with the indicated concentrations of ABA. Germination was determined with an average of >100 seeds from three independent experiments. Values are means ± sd.
(C) Greening of Col, ost1, F3DN, and ost1 F3DN seeds and seeds from self-pollinated f1−/+ f3 and f1−/+ f3 ost1 plants incubated on GM plates with (1 μM) or without ABA treatment for 5 d. Greening was determined with an average of >100 seeds from three independent experiments. Values are means ± sd. Asterisks indicate the levels of statistical significance as determined by Student’s t test: *P < 0.01 versus Col.
(D) Immunoblot analysis of ABI5 accumulation in kinase- and phosphatase-deficient mutants. In the absence of ABA, there was no detectable ABI5 protein in Col, ost1, f1−/+ f3, F3DN, f1−/+ f3 ost1, and F3DN ost1 seedlings. After ABA treatment (1 μM, 8 d), ABI5 (indicated by the arrows) hyperaccumulated in seeds of self-pollinated f1−/+ f3 or F3DN lines, in contrast with the reduced accumulation of ABI5 in seeds of self-pollinated f1−/+ f3 ost1 plants and F3DN ost1 lines. There was no detectable ABI5 protein in Col and ost1 seedlings at this stage. The arrowhead indicates the nonspecific band recognized by the ABI5 antibody. Arrows indicate the ABI5 bands. RPT5 was used as a loading control. A total of 100 μg protein was loaded for each lane.
ABA Promotes the Degradation of FyPP Proteins in Seedlings
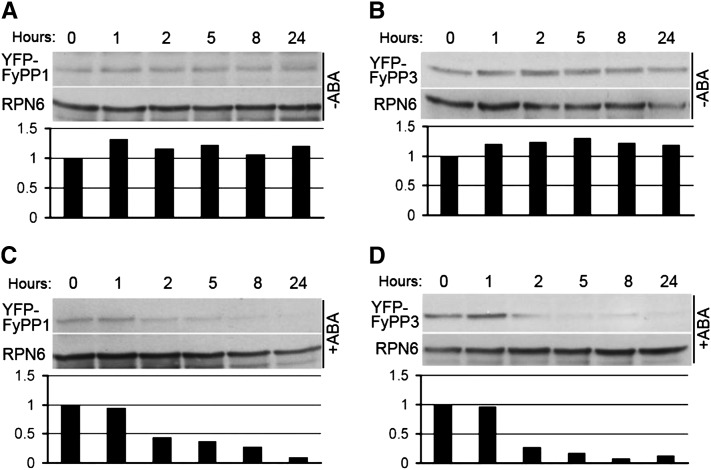
Since FyPP loss-of-function mutants are hypersensitive to ABA, we were interested in understanding how ABA regulates FyPP1 and FyPP3 protein stability. We treated 4-d-old F1OE and F3OE seedlings with the protein synthesis inhibitor cycloheximide for 15 h and then added ABA or the control solvent DMSO. We harvested the samples after different incubation time (0, 1, 2, 5, 8, and 24 h) and performed immunoblot assays. The results showed that the YFP-FyPP1 protein levels in F1OE seedlings and the YFP-FyPP3 protein levels in F3OE seedlings decreased after treatment with ABA, but not after the addition of DMSO (Figures 7A to 7D). These observations suggest that ABA promotes degradation of FyPP proteins.
Figure 7.
Degradation of FyPP1 and FyPP3 Protein upon ABA Treatment.
(A) YFP-FyPP1 protein levels in seedlings treated with control solvent (DMSO) for different durations (0, 1, 2, 5, 8, and 24 h).
(B) YFP-FyPP3 protein levels in seedlings treated with control solvent (DMSO) for different durations (0, 1, 2, 5, 8, and 24 h).
(C) YFP-FyPP1 protein levels in seedlings treated with ABA for different durations (0, 1, 2, 5, 8, and 24 h).
(D) YFP-FyPP3 protein levels in seedlings treated with ABA for different durations (0, 1, 2, 5, 8, and 24 h).
The relative amounts of YFP-FyPP proteins are shown below the immunoblot assays; 100 μg total protein was loaded for each lane.
DISCUSSION
In this study, we characterized the function of FyPP1 and FyPP3, two homologous genes encoding the catalytic subunits of PP6 in ABA signaling. We showed that the FyPP dominant-negative and f1 f3 double mutants are ABA hypersensitive. FyPP1 and FyPP3 protein levels are downregulated by ABA. We also showed that FyPP1 and FyPP3 interact with ABI5 in yeast and in planta and directly dephosphorylate ABI5 in vitro. Furthermore, we demonstrated that the ABA-hypersensitive phenotype of the f1 f3 mutants is ABI5 dependent, and FyPPs function antagonistically with SnRK2 in regulating ABI5 phosphorylation and ABA signaling.
FyPP1 and FyPP3 Function as Negative Regulators of ABA Signaling
In this study, we collected several lines of evidence supporting the hypothesis that FyPP1 and FyPP3 play a negative role in regulating ABA signaling. First, the f1 f3 double mutants, FyPP dominant-negative mutants, and ethanol-induced F3Ri/f1 lines all showed a hypersensitive response to ABA during seed germination and root growth. The ABA-hypersensitive phenotype of these FyPP loss-of-function mutants closely resembles other loss-of-function mutants of negative regulators of ABA signaling, such as ABI1, ABI2, PP2Ac2, KEG, DWA1, and DWA2 (Sheen, 1998; Gosti et al., 1999; Merlot et al., 2001; Stone et al., 2006; Pernas et al., 2007; Lee et al., 2010; Liu and Stone, 2010). Second, two representative ABA-responsive genes, RD29A and RD29B, are hyperinduced in the f1 f3 double mutants and FyPP dominant-negative mutants after ABA treatment. Third, the F1OE and F3OE lines showed increased tolerance to ABA during germination, which is opposite to the ABA response of FyPP loss-of-function mutants. Notably, the levels of FyPP1 and FyPP3 proteins are downregulated by ABA, suggesting a possible feedback loop between ABA signaling and FyPP stability/activity.
The PP2A and PP2A-like PPPs, such as PP4 and PP6, are major Ser/Thr phosphatases in most eukaryotic cells. The in vivo activities of these PPPs are regulated by a set of regulatory subunits. In a previous study, we showed that FyPP1 or FyPP3 physically interacts with SAL (SAP domain proteins) and PP2AA proteins (RCN1 or PP2AA1, PP2AA2, and PP2AA3) to form a PP6-type heterotrimeric holoenzyme complex that directly interacts with and dephosphorylates a subset of PIN proteins to regulate polar auxin transport and plant development. We demonstrated that FyPPs, SAL, and PP2A As have comparable roles in regulating auxin transport and plant development. In addition, there is a synergistic interaction between FyPP1 (or 3) (catalytic subunit), RCN1 (PP2AA1), and SAL1 in the regulation of plant development (Dai et al., 2012a). Intriguingly, RCN1 was previously reported as a positive regulator of ABA signaling, as rcn1 mutants show reduced ABA sensitivity in seed germination and ABA-induced gene expression (Kwak et al., 2002). The ABA-insensitive phenotype of rcn1 is similar to that of FyPP overexpression lines but opposite to that of the FyPP loss-of-function mutants. Notably, another recent study reported that overexpression of PP2Ac2 causes an ABA-insensitive phenotype, while the pp2ac2 loss-of-function mutant shows an ABA-hypersensitive phenotype (Pernas et al., 2007), suggesting that PP2Ac also plays a negative regulatory role in ABA signaling. As we previously demonstrated, RCN1 and FyPPs can assemble into functional PP6 heterotrimeric holoenzyme complexes (Dai et al., 2012a). It is not completely clear now why the loss-of-function mutants of PP2AA (such as RCN1) and FyPP1/FyPP3 display opposite ABA phenotypes. At least two possible scenarios can be envisaged. (1) PP2AA proteins (PP2AA1 or RCN1, PP2AA2, and PP2AA3) are promiscuous and can function as the regulatory subunits for both PP2A and PP6 phosphatase holoenzymes (Dai et al., 2012a), and the loss-of-function phenotype of RCN1 might be due to the combined effects on PP2A and PP6 activities, rather than simply reflecting the effect on PP6 alone. (2) It is possible that the A regulatory subunits (such as RCN1) may exert different regulatory effects (activation or inhibition) on the phosphatase activity toward different substrates or different developmental tissues or signaling pathways. For example, it has been shown that α4 is a common regulatory subunit of PP2A, PP4, and PP6 in mammals. Binding of α4 to PP2Ac inhibits its activity on phosphorylated 4E-BP1 (Nanahoshi et al., 1998), while addition of α4 to PP2Ac promotes PP2A activity when using myelin basic protein as the substrate (Murata et al., 1997). This variation in α4 effects on PP2Ac activity also occurs in different cell lines. For example, overexpression of α4 in COS-1 cells results in increased activities of cellular PP2A, whereas in COS7 cells, α4 overexpression has differential effects on the phosphorylation of endogenous phosphoproteins (Prickett and Brautigan, 2006; Nien et a., 2007). In addition, when using myelin basic protein as the substrate, the activity of PP6 bound to α4 is severely reduced in contrast with the increased activity of PP2A bound to α4, suggesting that a single regulatory subunit can recognize two kinetically identical catalytic subunits to induce different allosteric effects that alter enzyme activity (Prickett and Brautigan, 2006). Characterization of other regulatory subunits of PP2A and PP6 (such as the SAL proteins) and their genetic interactions with the catalytic subunits will help clarify these issues.
FyPP1 and FyPP3 Regulate ABI5 Phosphorylation and Stability
Phosphorylation and degradation are two important posttranslational modifications of proteins. The posttranslational nature of both processes ensures a rapid response to exogenous and endogenous cues without going through the more time-consuming transcriptional regulation. Such a mechanism is widely used in plants in response to many stresses and environmental signals. In previously characterized examples, phosphorylation of a substrate in response to a specific signal mostly serves as a recognition tag for an E3-ubiquitin ligase, which facilitates degradation of the marked protein (Hardtke et al., 2000; Karin and Ben-Neriah, 2000; Deshaies and Ferrell, 2001). For example, a group of basic helix-loop-helix transcription factors called phytochrome-interacting factors (PIFs) promote etiolation and repress photomorphogenesis in darkness. Photoactivation of phytochrome induces intranuclear phosphorylation of the PIFs and their subsequent proteasome-mediated degradation. These data indicate that phytochrome-induced phosphorylation of target proteins may represent the primary intermolecular signaling action of the activated photoreceptor. The resultant proteolysis of the PIFs triggers the transition from skotomorphogenic to photomorphogenic development (deetiolation), upon initial exposure of seedlings to light, by directly altering the expression of PIF target genes (J. Li et al., 2011). There are also cases in which phosphorylation protects the protein from proteasome-mediated degradation while dephosphorylation promotes protein degradation. For example, phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6 leads to the accumulation of 1-aminocyclopropane-1-carboxylic acid synthase proteins and the induction of ethylene synthesis (Joo et al., 2008). Similarly, it was reported that PP2A dephosphorylates BRI1, leading to decreased BRI1 abundance and decreased brassinosteroid signaling (Wu et al., 2011).
ABI5 is known to play a key role in regulating seed germination and seedling growth. In dry seeds, high levels of ABA promote SnRK2-dependent phosphorylation and stabilization of ABI5 proteins (Fujii et al., 2007; Nakashima et al., 2009). However, during seed germination, ABI5 proteins are rapidly degraded due to the diminished ABA content (Piskurewicz et al., 2008). Furthermore, the ABA-induced inhibition of seed germination and the severity of ABA-triggered postgermination growth arrest are correlated with increased ABI5 protein levels (Lopez-Molina et al., 2001). These studies indicate that phosphorylation stabilizes ABI5 and that the abundance of ABI5 protein plays an important role in regulating seed germination and postgermination growth. As phosphorylated ABI5 proteins are stable and functional, dephosphorylation of ABI5 should be an essential prerequisite for the degradation of ABI5 and the initiation of seed germination. Our results provide substantial evidence that FyPP1 and FyPP3 plays a key role in promoting dephosphorylation and subsequent degradation of ABI5, allowing seed germination and postgermination growth to occur. First, our genetic interaction studies showed that the ABA hypersensitivity of f1 f3 double mutants during seed germination was dependent on ABI5 function, and overexpression of FyPP3 in the ABI5OE background largely rescued the ABA-related phenotypes of ABI5OE seeds (Figure 2). Second, we showed that after ABA treatment, the f1 f3, F1DN, and F3DN mutants accumulated higher levels of ABI5 protein (Figure 5A). This observation suggests that the activity of FyPPs is necessary for maintaining proper ABI5 levels in response to ABA. Third, we showed that after removal of ABA, the ABI5 protein is more stable in FyPP loss-of-function mutants, but it is more quickly degraded in the FyPP overexpression lines (Figure 5B), further suggesting that FyPP activities play a critical role in regulating ABI5 stability and ABA signaling. Fourth, both FyPP1 and FyPP3 interacted with ABI5 in yeast and plant cells, and these interactions happened in the nucleus (Figure 3; see Supplemental Figure 10 online). Furthermore, phosphorylated ABI5 interacted better than nonphosphorylated ABI5 with FyPP1 protein in yeast cells, indicating the substrate specificity of FyPP1 and FyPP3 toward ABI5 (Figures 4C and 4D). Finally, the plant extracts from f1 f3, F1DN, and F3DN exhibited enhanced phosphorylation activity on ABI5 proteins in an ABA-dependent manner, and the exogenous wild-type FyPP3 proteins, but not the phosphatase null mutant FyPP3D81N, reversed the phosphorylation activity of f1 f3 plant extracts on ABI5 proteins (Figure 4). Together, these data support the notion that FyPP1 and FyPP3 act to dephosphorylate ABI5, thus promoting degradation of ABI5 and abrogating the inhibitory effect of ABA on seed germination and postgermination growth.
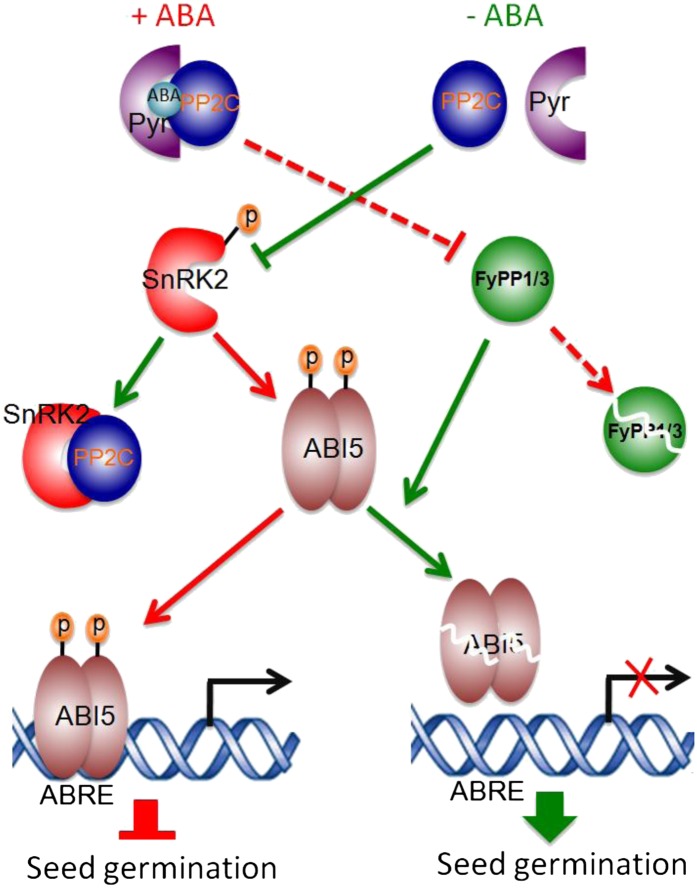
Reversible phosphorylation of proteins mediated by kinases and phosphatases plays essential roles in regulating plant growth and development, including signaling by various phytohormones, such as auxin, ethylene, and brassinosteroids (Michniewicz et al., 2007; Kamiyoshihara et al., 2010; Tang et al., 2011), by modulating the stability and activity of key intermediates in various signaling pathways. The virtual elimination of ABA responses in the snrk2.2/2.3/2.6 triple mutant (Fujii et al., 2007; Nakashima et al., 2009) and the hypersensitivity of the f1 f3, F3Ri/f1, F1DN, and F3DN mutants to ABA (Figure 1) suggest that the SnRK2 kinases and FyPP/PP6 phosphatases might function antagonistically in ABA signaling. It is known that phosphorylation of ABI5 is necessary for ABI5 function because overexpression of ABI5 alone is not sufficient to suppress seed germination (Lopez-Molina et al., 2001). Our data show that loss of FyPP function is sufficient to trigger ABI5-dependent inhibition of seed germination (Figure 2B), which is very similar to the previous report that overexpression of a SnRK2 kinase (PKABA1) in the ABI5-OE background is sufficient to activate ABI5 protein and suppress seed germination (Piskurewicz et al., 2008). Phosphorylation of ABI5 by SnRK2 kinase is ABA dependent (Fujii et al., 2007; Nakashima et al., 2009), while the FyPP loss-of-function mutants, such as f1 f3, F1DN, and F3DN, showed enhanced ABA-dependent phosphorylation of ABI5 proteins (Figure 4). These observations suggest that the loss of FyPP activity and the overproduction of SnRK2 activity produced similar effects on ABI5 phosphorylation and function. Additionally, the genetic interactions between FyPPs and SnRK2.6 kinase indicate that snrk2.6 mutants attenuated the ABA hypersensitivity phenotypes of the FyPP loss-of-function mutants (Figure 6), again suggesting an antagonistic role of FyPP and SnRK2.6 kinase in mediating ABA signaling. Taken together, these data provide strong evidence for the following model: SnRK2 kinase-mediated phosphorylation acts to promote the stability and function of ABI5 upon the perception of ABA, while FyPP/PP6-mediated dephosphorylation promotes the degradation and inactivation of ABI5 after the removal of ABA (Figure 8). Thus, SnRK2- and FyPP/PP6-mediated phosphorylation and dephosphorylation of ABI5 function as a molecular switch that regulates ABA signaling.
Figure 8.
A Model Showing the Antagonistic Interaction between SnRK2 Kinases and FyPP/PP6 Phosphatases in ABA Signaling.
In the presence of ABA (+ABA), ABA mediates the formation of an ABA receptor-ABA-PP2C phosphatase complex (such as Pyr-ABA-PP2C complex). This causes the autophosphorylation and activation of the downstream SnRK2 kinases, including SnRK2.2, SnRK2.3, and SnRK2.6. Meanwhile, ABA promotes the degradation of the PP6 catalytic subunits, including FyPP1 and FyPP3. The active SnRK2 kinases then phosphorylate and activate the downstream transcription factors, such as ABI5, which further activate the expression of ABRE-containing genes and repress seed germination. By contrast, in the absence of ABA (-ABA), PP2C phosphatases persist, allowing them to inhibit SnRK2 kinase activity by forming PP2C-SnRK2 protein complexes and dephosphorylating SnRK2 kinases. Simultaneously, FyPP/PP6 perceives this signal and dephosphorylates ABI5, triggering its degradation and altering the expression of ABRE-containing genes, therefore promoting seed germination. The red lines indicate events happening in the presence of ABA. The green lines indicate events happening in the absence of ABA. The dashed lines indicate events happening through unknown mechanisms. p, phosphate.
It is intriguing to note that previous studies showed that a subfamily of Mg2+-dependent Ser/Thr protein phosphatases (PP2Cs), including ABI1, ABI2, and HAB1, physically interact with SnRK2 kinases, including SnRK2.2, SnRK2.3, and SnRK6/OST1, and dephosphorylate a Ser residue in the kinase activation loop in the absence of ABA (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Soon et al., 2012). When ABA is present, ABA associates with the PYR/PYL/RCAR family of ABA receptors to promote the binding of these receptors to the catalytic site of PP2Cs. This inhibits their phosphatase activity and subsequently activates SnRK2 kinases through autophosphorylation (Ma et al., 2009; Park et al., 2009; Soon et al., 2012). The active SnRK2s relay the ABA signal to downstream effectors, such as ABI5/bZIP transcriptional factors (Cutler et al., 2010; Hubbard et al., 2010). Protein–protein interaction assays between ABI1 and the key transcriptional regulators involved in ABA signaling, such as ABI3, ABI4, and ABI5, failed to detect binding to PP2C (Nakamura, et al., 2001). Thus, the ABA signaling pathway appears to involve at least two distinct families of phosphatases. The main role of PP2Cs is most likely to act as a hub in mediating early ABA signaling events, modulating the phosphorylation and activity of SnRK2 kinases, whereas FyPP/PP6 specifically dephosphorylates ABI5 (and possibly other ABI5-like bZIP transcription factors) and promotes its degradation, causing subsequent changes in gene expression and ABA responses (Figure 8). Interestingly, our LCI assays failed to detect protein–protein interaction between FyPP proteins and group II ABI5-like bZIP transcription factors involved in the drought response, such as ABF2, ABF3, and ABF4 (Figure 3C). Despite this observation, we cannot exclude the possibility that FyPP1/3 may also regulate these bZIP transcription factors through the help of additional regulatory factors, as the interaction between phosphatase and substrate in vivo is usually dynamic and sometimes needs the help of other regulators (Shi, 2009). It will be interesting to investigate how the phosphorylation and activity of these ABI5-like bZIP transcription factors are regulated in future studies.
FyPP1 and FyPP3 Act in the Nexus of Multiple Signaling Pathways
It was previously reported that Arabidopsis FyPP1 and FyPP3 play an essential role in regulating flowering time. Recombinant FyPP efficiently dephosphorylates oat (Avena sativa) phytochrome A in a spectral form–dependent manner, and overexpression of FyPP caused delayed flowering while reducing the expression of FyPP-accelerated flowering in Arabidopsis (Kim et al., 2002). In a recent study, we showed that FyPP1 or FyPP3 interacts with the SAL proteins and the A subunits of PP2A (PP2AAs) to form PP6 heterotrimeric holoenzymes that act antagonistically with the PID/AGC3 kinases in the regulation of PIN-FORMED (PIN) protein phosphorylation, subsequently regulating directional auxin transport and root development (Dai et al., 2012a). H. Li et al. (2011) also reported that FyPP1 and PID mediated the switch of PIN1 polar localization in the regulation of pavement cell development in Arabidopsis leaves. In this work, we have shown that FyPP1 and FyPP3 act as negative regulators in ABA signaling. They function antagonistically with SnRK2 kinases to modulate the reversible phosphorylation of ABI5 and subsequently regulate seed germination and postgermination development. Thus, it appears that coupled with various kinases, the FyPP/PP6 phosphatases play a multifaceted role and may function in the nexus of multiple signaling pathways to regulate diverse plant developmental processes by mediating the reversible phosphorylation of key developmental regulators. Further studies of these phosphatases will undoubtedly provide more molecular and biochemical insights into the roles and functional modes of PPP phosphatases in the regulation of plant development and responses to environmental stimuli.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana lines fypp1 (f1), fypp3 (f3), f1−/+ f3, f1 f3, F3Ri/f1, F1OE, F3OE, F1DN, and F3DN lines were described previously (Dai et al., 2012a). The ost1 (or snrk2.6, SALK_008068; Zheng et al., 2010) mutant is in the Col background and was obtained from the SALK Institute. abi5-1 mutants and ABI5OE transgenic lines were reported previously (Finkelstein, 1994; Brocard et al., 2002). The F3Ri/f1 lines were generated by introducing an ethanol-inducible expression cassette AlcA-AlcR:FyPP3RNAi into fypp1 backgrounds as described previously (Dai et al., 2012a). More F3Ri/f1 lines were screened in this study based on the expression of FyPP3 after ethanol induction. Two types of F3Ri/f1 lines were identified by RT-PCR: FyPP3 undetectable (e.g., F3Ri/f1-3) and FyPP3 weakly expressed (e.g., F3Ri/f1-21). F3Ri/f1-3 and F3Ri/f1-21 were then used in this study. Because the f1 f3 double mutants have shorter roots and smaller/fused cotyledons (see Supplemental Figure 3 online; Dai et al., 2012a), we were able to choose f1 f3 plants for ABA treatment. Seeds were sterilized as described (Dai et al., 2012a). Arabidopsis seedlings were grown as previously described (Lee et al., 2010). Specifically, for the assays shown in Supplemental Figure 4 online, seeds were germinated and grown vertically on GM (Lee et al., 2010) plates (with 0.1% ethanol for Supplemental Figure 4B online) for 3 d under continuous white light at 22°C in a growth chamber after stratification for 4 d in darkness. The seedlings were transferred onto fresh GM plates (with 0.1% ethanol for Supplemental Figure 4B online) with 0 or 1 μM ABA. The positions of the primary root tips were marked. The seedlings were then grown vertically for four more days in the same growth chamber, and the additional root growth was measured with a ruler. The percentage of relative root elongation was calculated with three replicates based on control plants grown on unsupplemented media. To minimize the effect of harvesting time on seed germination, all seed batches compared in this study were harvested on the same day from plants grown in the same growth chamber with identical environmental conditions.
Because f1 f3 homozygous plants are completely sterile (Dai et al., 2012a), seeds from self-pollinated f1−/+ f3 plants (f1 is heterozygous, and f3 is homozygous), f1−/+ f3 abi5-1 (homozygous for f3 and abi5 but heterozygous for f1), f1−/+ f3 ABI5OE (homozygous for f3 and ABI5OE but heterozygous for f1), and f1−/+ f3 ost1 (homozygous for f3 and ost1 but heterozygous for f1) were used for the seed germination assays (for Figures 1, 2, and 6; see Supplemental Figures 2 and 6 online), immunoblot assays (for Figures 5 and 6), and quantitative RT-PCR assay (for Supplemental Figure 11 online). The progeny produced from these selfed plants were of mixed genotypes, including the following: a mixture of f3, f1−/+ f3, and f1 f3, with a theoretical segregation ratio of 1:2:1 for self-pollinated f1−/+ f3 plants; a mixture of f3 abi5-1, f1−/+ f3 abi5-1, and f1 f3 abi5-1 genotypes, with a theoretical segregation ratio of 1:2:1 for self-pollinated f1−/+ f3 abi5-1 plants; a mixture of f3 ABI5OE, f1−/+ f3 ABI5OE, and f1 f3 ABI5OE genotypes, with a theoretical segregation ratio of 1:2:1 for self-pollinated f1−/+ f3 ABI5OE plants; and a mixture of f3 ost1, f1−/+ f3 ost1, and f1 f3 ost1 genotypes, with a theoretical segregation ratio of 1:2:1 for self-pollinated f1−/+ f3 ost1 plants.
Generation of Constructs
For Y2H assays, full-length coding sequences (CDSs) of Pyr, Pyl1, ABI1, ABI2, ABI3, ABI4, ABI5, and OST1 were amplified by RT-PCR with primers Pyr-F/R, Pyl1-F/R, ABI1-F/R, ABI2-F/R, ABI3-F/R, ABI4-F/R, ABI5-F1/R1, and OST1-F/R, respectively; with EcoRI or XhoI sites at the end of F or R primers, respectively, for cloning Pyr, ABI2, ABI5, and OST1 fragments; EcoRI or SalI sites at the end of F or R primers, respectively, for cloning Pyl1; MfeI or XhoI sites at the end of F or R primers, respectively, for cloning ABI1; MfeI or SalI sites at the end of F or R primers, respectively, for cloning ABI3 and ABI4. EcoRI- and XhoI-digested Pyr, ABI2, ABI5, and OST1 fragments were then inserted into pEG and pJG vectors digested with the same enzymes to generate pEG-Pyr, pEG-ABI2, pEG-ABI5, pEG-OST1, pJG-Pyr, pJG-ABI2, pJG-ABI5, and pJG-OST1 plasmids. The Pyl1 fragment digested with EcoRI and SalI was inserted into pEG and pJG vectors digested with the same enzymes to generate pEG-Pyl1 and pJG-Pyl1 plasmids, respectively. The ABI1 fragment digested with MfeI and XhoI was inserted into the pEG and pJG vectors digested with the same enzymes to generate pEG-ABI1 and pJG-ABI1 plasmids, respectively. ABI3 and ABI4 fragments digested with MfeI and SalI were inserted into pEG and pJG vectors digested with the same enzymes to generate pEG-ABI3 and pEG-ABI4, and pJG-ABI3 and pJG-ABI4 plasmids, respectively. The ABI5A4 and ABI5D4 mutant genes were generated using the same strategy used to clone FyPP1D81N (Dai et al., 2012a). ABI5A4 and ABI5D4 fragments digested with EcoRI and XhoI were inserted into pEG and pJG vectors digested with the same enzymes to generate pEG-ABI5A4 and pEG-ABI5D4, and pJG-ABI5A4 and pJG-ABI5D4 plasmids, respectively. pEG-FyPP1, pEG-F1NT, pEG-F1CD, pEG-F1CT, pEG-FyPP3, pJG-FyPP1, pJG-F1NT, pJG-F1CD, pJG-F1CT, and pJG-FyPP3 plasmids were generated as described (Dai et al., 2012a).
For LCI assays, the full-length CDSs of ABI5, AREB3, EEL, and ABF2 were amplified by PCR with the primer pairs ABI5-F1/R1, AREB3-F/R, EEL-F/R, and ABF2-F/R with KpnI or XhoI sites at the end of F or R primers for ABI5 and KpnI or SalI sites at the end of F or R primers for AREB3, EEL, and ABF2, respectively. ABI5 was digested with KpnI and XhoI and AREB3, EEL, and ABF2 were digested with KpnI and SalI and then inserted into the pCAMBIA1300-cLUC and -nLUC vectors (Chen et al., 2008) to generate pCAMBIA-ABI5-nLUC, pCAMBIA-AREB3-nLUC, pCAMBIA-EEL-nLUC, pCAMBIA-ABF2-nLUC, pCAMBIA-cLUC-ABI5, pCAMBIA-cLUC-AREB3, pCAMBIA-cLUC-EEL, and pCAMBIA-cLUC-ABF2. pCAMBIA-FyPP1-nLUC, pCAMBIA-FyPP3-nLUC, pCAMBIA-cLUC-FyPP1, and pCAMBIA-cLUC-FyPP3 plasmids were generated as described previously (Dai et al., 2012a).
For BiFC assays, the CDS of ABI5 was amplified by PCR with the primer pair ABI5-F3/R3 with SalI and BamHI sites at the end of F and R primers, respectively. ABI5 digested with SalI and BamHI was then cloned into the pY2N vector containing the C-terminal (156 to 239 amino acids) domain of the YFP fluorescent protein (YFPC) to generate the pY2N-ABI5 plasmid. The Y1C-FyPP3 plasmid was generated as described (Dai et al., 2012a).
For Co-IP assays, ABI5 was amplified with ABI5-F2/R2 with BamHI and SpeI at the end of F and R primers, respectively. ABI5 digested with BamHI and SpeI was then inserted into the pCAMBIA-Myc vector (from Fang Chen, Yale University) digested with the same enzymes to generate pCAMBIA-Myc-ABI5. pCAMBIA-FyPP1-3HA and pCAMBIA-FyPP3-3HA were generated as described (Dai et al., 2012a).
For in vitro protein expression, ABI5b fragment (ABI5 Ser119-Gln190; Nakashima et al., 2009) was amplified by PCR with the primer pair ABI5b-F/R, with EcoRI and XhoI sites at the end of F and R primers, respectively. The ABI5bm (ABI5bS145A) mutant fragment was generated using the same strategy used to clone FyPP1D81N (Dai et al., 2012a). ABI5b and ABI5bm digested with EcoRI and XhoI was inserted into the pGEX-4T-1 vector digested with the same enzymes to generate pGEX-ABI5b and PGEX-ABI5bm. pGEX-FyPP3 and pGEX-FyPP3D81N plasmids were generated as described (Dai et al., 2012a).
All genes/fragments were confirmed by sequencing. The primers used in PCR cloning are shown in Supplemental Table 1 online.
Germination Assays
The germination of Arabidopsis seeds was described previously (Piskurewicz et al., 2008). Seed germination was determined based on the appearance of an embryonic axis (i.e., radicle) protrusion, as observed under a microscope. Seedling greening was determined based on the appearance of green cotyledons in a seedling.
Y2H Assays
Y2H assays were conducted as described previously (Yang et al., 2005; Dai et al., 2012a).
LCI Assays
The LCI assays were performed as described (Chen et al., 2008; Dai et al., 2012a).
BiFC Assays
The BiFC assays were described previously (Bracha-Drori et al., 2004; Shen et al., 2009; Dai et al., 2012a).
Co-IP Assays
Various plasmids were transformed into Agrobacterium tumefaciens strain GV2260 as previously described (Dai et al., 2012a). Various bacterial strains were coinfiltrated into young tobacco (Nicotiana tabacum) leaves as previously described (Chen et al., 2008) and grown for 3 d. Protein extraction and co-IP were performed as described (Dai et al., 2012a). Immunoprecipitation products were separated by electrophoresis using 10% acrylamide gels, and the target proteins were detected by protein gel blots using α-HA or α-Myc antibodies (Roche) at a dilution of 1:1000 in 5% milk.
Protein Isolation and Immunoblot Analysis
Plant proteins were isolated with extraction buffer containing 50 mM Tris-Cl, pH 7.5, 6 mM NaCl, 1 mM MgCl2, 1 mM PMSF, 1× protease inhibitor cocktail (Sigma-Aldrich), and 1% Nonidet P-40. For Figure 7, 4-d-old F1OE and F3OE seedlings were treated with the protein synthesis inhibitor cycloheximide for 15 h, and then ABA (0.2 mM) or control solvent (DMSO) was added. Samples were then harvested after different incubation times. Yeast proteins were extracted using the Y-PER Yeast Protein Extraction Reagent (Thermo Scientific) according to the manufacturer’s instructions. To determine the protein concentration, various amounts of BSA protein were added to 1× Bradford protein assay buffer (Bio-Rad) and incubated at room temperature for 5 min before reading the A600 value. A standard curve of A600 versus concentration of BSA was then generated. The total protein concentrations were determined by extrapolating the A600 values of the sample proteins in 1× Bradford solution against the BSA standard curve. The extracts were mixed with 2× SDS sample buffer, boiled for 5 min, and then separated on 10% SDS protein gels. The membrane transfer and immunoblot assays were performed as described previously (Lee et al., 2010). For Figure 7, α-GFP antibodies (Invitrogen) were used at a dilution of 1:1000, and α-RPN6 (Chen et al., 2006) antibodies were used at a 1:2000 dilution. For Figure 5, α-ABI5 antibodies were used at a 1:1000 dilution. α-RPT5 antibodies (Kwok et al., 1999) were used at a 1:1500 dilution. For Supplemental Figure 8 online, α-LexA (Abcam) and α-B42 (Sigma-Aldrich) antibodies were used at a 1:2000 dilution.
In Vitro Phosphorylation Assays
GST and recombinant GST-FyPP3, GST-FyPP3D81N, and GST-ABI5b proteins were expressed in Escherichia coli strain BL21 and purified as described previously (Park et al., 2008). Six-day-old seedlings were treated with 100 µM ABA or control solvent DMSO for 30 min. Samples were harvested into liquid nitrogen. Total proteins were extracted with 1× kinase/phosphatase buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 5 mM MgCl2, and 1 mM Zn2+), plus 1× protease inhibitor and 1 mM PMSF. In vitro kinase assays with plant extracts were performed essentially as described previously (Michniewicz et al., 2007; Dai et al., 2012a) with a few modifications. For Figures 4A and 4B, 2 µg of GST, GST-ABI5b, or GST-ABI5bm (ABI5b145A) proteins and 25 µg of plant seedling extracts were mixed in 1× kinase/phosphatase buffer, 1× protease inhibitor, 1 mM PMSF, and 1× ATP solution (100 μM ATP and 1 μCi [γ-32P]ATP) in a total volume of 50 μL. Various amounts of exogenous GST-FyPP3 or GST-FyPP3D81N fusion proteins were added to the reactions shown in Figure 4B. The samples were incubated at 30°C for 30 min, and the reactions were stopped by adding 5× loading buffer and boiling for 5 min. The products were separated by electrophoresis using 12% acrylamide gels. The gels were stained, dried, and then visualized by exposure to x-ray films.
RNA Isolation and RT-PCR/Real-Time PCR (Quantitative PCR) Analysis
Samples were harvested in liquid nitrogen. Total RNAs were isolated using RNeasy plant mini kits (Qiagen). One microgram of total RNA of each sample was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer’s introductions. RT-PCR was performed as described (Lee et al., 2010). For real-time PCR, 50 ng of cDNAs was used for each reaction using the SYBR Green kit according to the manufacturer’s introductions in an Applied Biosystems real-time PCR machine. Expression levels were normalized to that of an actin gene. All quantitative PCR experiments were independently performed in triplicate, and representative results were shown. The primers for RT-PCR and quantitative PCR are shown in Supplemental Table 1 online.
Confocal Observations
For Supplemental Figure 10 online, seedlings were grown to 4 d old and the roots were then harvested for confocal observation. GFP fluorescence was observed with a Carl Zeiss LSM510 confocal microscope.
Accession Numbers
Sequence data from this study can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g50370 (FyPP1), At3g19980 (FyPP3), At4g17870 (Pyr), At5g46790 (Pyl1), At4g26080 (ABI1), At5g57050 (ABI2), At3g24650 (ABI3), At2g40220 (ABI4), At2g36270 (ABI5), At4g33950 (OST1), At2g41070 (EEL), At3g56850 (AREB3), At3g44460 (DPBF2), At1g49720 (ABF1) At1g45249 (ABF2), At4g34000 (ABF3), At3g19290 (ABF4), At5g52310 (RD29A), and At5g52300 (RD29B).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of FyPP1, FyPP3, and ABI5 during Seed Germination.
Supplemental Figure 2. Germination Rates of Col, f1, and f3 Seeds and Seeds from Self-Pollinated f1−/+ f3 Plants Treated with or without ABA at Various Concentrations.
Supplemental Figure 3. Phenotypes of f1 f3 Homozygotes at the Seedling Stage.
Supplemental Figure 4. Relative Root Growth of f1 f3 and Ethanol-Induced F3Ri/f1 Mutants after ABA Treatment.
Supplemental Figure 5. Expression of FyPP3 in F3Ri/f1 Lines.
Supplemental Figure 6. Germination Phenotypes of Col and abi5-1 Seeds and Seeds from Self-Pollinated f1−/+ f3 and f1−/+ f3 abi5-1 Plants.
Supplemental Figure 7. Yeast Two-Hybrid Assays between FyPP1/3 and Several Key Regulators in ABA Signaling.
Supplemental Figure 8. Expression of Fusion Proteins in the Yeast Cells for Yeast Two-Hybrid Assays in This Work.
Supplemental Figure 9. Sequence Alignment of the bZIP Transcription Factors.
Supplemental Figure 10. Subcellular Localization of YFP-FyPP1 and YFP-FyPP3 in Arabidopsis Roots.
Supplemental Figure 11. Expression Analysis of ABI5 mRNA.
Supplemental Figure 12. Germination rates of Col, F1OE, F3OE, F1DN, and F3DN Seeds Treated with or without ABA.
Supplemental Table 1. List of the Primer Sequences Used in This Study.
Acknowledgments
We thank Tian Xu (Medical School, Yale University) for providing Xenogen IVIS spectrum equipment for the LCI assay, Hong-Gu Kong (Boyce Thompson Institute for Plant Research) for the pBIN61-GFP-3HA vector, Eric Lam (Rutgers, The State University of New Jersey) for the pZM104-inducible binary vector, Jian-Min Zhou (National Institute of Biological Science, Beijing) for sharing the pCAMBIA-nLUC and -cLUC vectors, Ning Wei (Yale University) for critical reading and comments on the article, and Richard Viestra (University of Wisconsin) for providing the ABI5 antibody. This work was supported by funds from the National Science Foundation (MCB-1004808, IOS-0954313, and IOS-1026630 to H.W.) and the National Institutes of Health (GM47850 to X.W.D.), by grants from the Next-Generation BioGreen 21 Program (No. PJ00901001), Rural Development Administration, Republic of Korea, and from Pusan National University Research Grant 2011 (to J.-H.L.).
AUTHOR CONTRIBUTIONS
M.D., X.W.D., and H.W. designed the research. M.D. cloned the constructs, generated the transgenic plants, established the mutants and crosses, and performed in vitro phosphorylation assay, protein–protein interaction assays, and expression analysis. F.C. performed BiFC assays. Q.X., T.M., K.M., J.-H.L., C.D.N., L.G., W.T., and J.W. carried out genotyping and some expression analysis. M.D. and H.W. wrote the article.
Glossary
- ABA
abscisic acid
- bZIP
basic Leu zipper
- ABRE
ABA-responsive element
- PPP
phosphoprotein phosphatase
- Col
Columbia
- GM
germination medium
- Y2H
yeast two-hybrid
- LCI
luciferase complementation imaging
- Co-IP
coimmunoprecipitation
- GFP
green fluorescent protein
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- GST
glutathione S-transferase
- PIF
phytochrome-interacting factor
- CDS
coding sequence
References
- Arroyo A., Bossi F., Finkelstein R.R., León P. (2003). Three genes that affect sugar sensing (abscisic acid insensitive 4, abscisic acid insensitive 5, and constitutive triple response 1) are differentially regulated by glucose in Arabidopsis. Plant Physiol. 133: 231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427 [DOI] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Pagès M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., Zheng N., Deng X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dai M., Terzaghi W., Wang H. (2012b). Multifaceted roles of Arabidopsis PP6 phosphatase in regulating cellular signaling and plant development. Plant Signal. Behav. 8: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., et al. (2012a). A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell 24: 2497–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J., Ferrell J.E., Jr (2001). Multisite phosphorylation and the countdown to S phase. Cell 107: 819–822 [DOI] [PubMed] [Google Scholar]
- Farkas I., Dombrádi V., Miskei M., Szabados L., Koncz C. (2007). Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12: 169–176 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S.S., Lynch T.J., Thomas T.L., Rock C.D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59: 253–267 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5: 756–771 [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Wang M.L., Lynch T.J., Rao S., Goodman H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J. (1995). Abscisic acid signaling. Curr. Opin. Cell Biol. 7: 232–238 [DOI] [PubMed] [Google Scholar]
- Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F., Goodman H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F., Beaudoin N., Serizet C., Webb A.A., Vartanian N., Giraudat J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan M.J., Marcotte W.R., Jr., andQuatrano R.S. (1990). A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda K., Osterlund M.T., Oyama T., Okada K., Deng X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T., Totsuka M., Hobo T., Kagaya Y., Yamamoto-Toyoda A. (2002). Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hauser F., Waadt R., Schroeder J.I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21: R346–R355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kamiyoshihara Y., Iwata M., Fukaya T., Tatsuki M., Mori H. (2010). Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J. 64: 140–150 [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. (2000). Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu. Rev. Immunol. 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Kang J.G., Yang S.S., Chung K.S., Song P.S., Park C.M. (2002). A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14: 3043–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Reuling G., Karssen C.M. (1982). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61: 377–383 [Google Scholar]
- Kwak J.M., Moon J.H., Murata Y., Kuchitsu K., Leonhardt N., DeLong A., Schroeder J.I. (2002). Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok S.F., Staub J.M., Deng X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoon H.J., Terzaghi W., Martinez C., Dai M., Li J., Byun M.O., Deng X.W. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22: 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J., Bouvier-Durand M., Morris P.C., Guerrier D., Chefdor F., Giraudat J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lin D., Dhonukshe P., Nagawa S., Chen D., Friml J., Scheres B., Guo H., Yang Z. (2011). Phosphorylation switch modulates the interdigitated pattern of PIN1 localization and cell expansion in Arabidopsis leaf epidermis. Cell Res. 21: 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X.Q., Watson M.B., Assmann S.M. (2000). Regulation of abscisic acid-induced stomatal closure and anion channels by guard cell AAPK kinase. Science 287: 300–303 [DOI] [PubMed] [Google Scholar]
- Liu H., Stone S.L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17: 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B., Mauch F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8: 409–414 [DOI] [PubMed] [Google Scholar]
- Merlot S., Gosti F., Guerrier D., Vavasseur A., Giraudat J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25: 295–303 [DOI] [PubMed] [Google Scholar]
- Meyer K., Leube M.P., Grill E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Wu J., Brautigan D.L. (1997). B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. USA 94: 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A.C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26: 627–635 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Nanahoshi M., Nishiuma T., Tsujishita Y., Hara K., Inui S., Sakaguchi N., Yonezawa K. (1998). Regulation of protein phosphatase 2A catalytic activity by alpha4 protein and its yeast homolog Tap42. Biochem. Biophys. Res. Commun. 251: 520–526 [DOI] [PubMed] [Google Scholar]
- Nien W.L., Dauphinee S.M., Moffat L.D., Too C.K. (2007). Overexpression of the mTOR alpha4 phosphoprotein activates protein phosphatase 2A and increases Stat1alpha binding to PIAS1. Mol. Cell. Endocrinol. 263: 10–17 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127: 635–648 [DOI] [PubMed] [Google Scholar]
- Park H.J., Ding L., Dai M., Lin R., Wang H. (2008). Multisite phosphorylation of Arabidopsis HFR1 by casein kinase II and a plausible role in regulating its degradation rate. J. Biol. Chem. 283: 23264–23273 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M., García-Casado G., Rojo E., Solano R., Sánchez-Serrano J.J. (2007). A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J. 51: 763–778 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett T.D., Brautigan D.L. (2006). The alpha4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J. Biol. Chem. 281: 30503–30511 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1998). Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc. Natl. Acad. Sci. USA 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Zhou Z., Feng S., Li J., Tan-Wilson A., Qu L.J., Wang H., Deng X.W. (2009). Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell 21: 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2009). Serine/threonine phosphatases: Mechanism through structure. Cell 139: 468–484 [DOI] [PubMed] [Google Scholar]
- Soon F.F., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335: 85–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Chevalier, D., Larue, C., Cho, S.K., and Walker, J.C. (2007). The protein phosphatases and protein kinases of Arabidopsis thaliana The Arabidopsis Book 5: e0106, doi/10.1199/tab.0106. 10.1199/tab.0106 [DOI] [PMC free article] [PubMed]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Wang X., Li X., Kamiya Y., Otegui M.S., Chory J. (2011). Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 4: ra29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Umezawa T., Mizoguchi T., Takahashi S., Takahashi F., Shinozaki K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zheng Z., et al. (2010). The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 153: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]