This study reveals a mechanism underlying chloroplast-actin filament dynamics during their disappearance processes, which are needed for the chloroplasts to produce a rapid avoidance response to strong blue light. These processes are specifically regulated by light intensity and position through phot2 and are different from those of cortical actin filaments.
Abstract
Phototropins (phot1 and phot2 in Arabidopsis thaliana) relay blue light intensity information to the chloroplasts, which move toward weak light (the accumulation response) and away from strong light (the avoidance response). Chloroplast-actin (cp-actin) filaments are vital for mediating these chloroplast photorelocation movements. In this report, we examine in detail the cp-actin filament dynamics by which the chloroplast avoidance response is regulated. Although stochastic dynamics of cortical actin fragments are observed on the chloroplasts, the basic mechanisms underlying the disappearance (including severing and turnover) of the cp-actin filaments are regulated differently from those of cortical actin filaments. phot2 plays a pivotal role in the strong blue light–induced severing and random motility of cp-actin filaments, processes that are therefore essential for asymmetric cp-actin formation for the avoidance response. In addition, phot2 functions in the bundling of cp-actin filaments that is induced by dark incubation. By contrast, the function of phot1 is dispensable for these responses. Our findings suggest that phot2 is the primary photoreceptor involved in the rapid reorganization of cp-actin filaments that allows chloroplasts to change direction rapidly and control the velocity of the avoidance movement according to the light’s intensity and position.
INTRODUCTION
Light is essential for photosynthesis; however, too much light can endanger plant survival (Kasahara et al., 2002; Sztatelman et al., 2010). To avoid the danger from excess light, plants have evolved photoprotective mechanisms, including light capture reduction by nastic leaf movements and/or chloroplast avoidance movements, adaptation to the light quality by a state transition, and dissipation of excess absorbed energy by the water-water cycle and/or violaxanthin cycle (Li et al., 2009; Takahashi and Badger, 2011). Among these mechanisms, the chloroplast avoidance response is crucial for plant survival (Wada et al., 2003; Suetsugu and Wada, 2007; Banaś et al., 2012) as evidenced by the fact that leaf necrosis and death have been observed in Arabidopsis thaliana phototropin2 (phot2) and chloroplast unusual positioning1 (chup1) mutant plants, which are deficient in this response (Kasahara et al., 2002). Chloroplasts change their positions in cells according to the quality, intensity, and position of the incident light; they move from the periclinal to the anticlinal wall in strong-intensity light (the avoidance response); conversely, they accumulate at the periclinal wall of palisade cells in low-intensity light (the accumulation response). This type of chloroplast movement is a common and dynamic phenomenon found in almost all of the land plants that have been tested to date (Wada et al., 2003; Suetsugu and Wada, 2009).
Phototropins (phot1 and phot2 in Arabidopsis) are UV-A/blue light receptors controlling a range of responses that serve to optimize the photosynthetic efficiency of plants (Kasahara et al., 2002; Takemiya et al., 2005). These include phototropic responses, chloroplast movement, stomatal opening, leaf flattening, and palisade cell development (Huala et al., 1997; Jarillo et al., 2001; Kagawa et al., 2001; Kinoshita et al., 2001; Sakai et al., 2001; Kozuka et al., 2011). With regard to chloroplast movement, phot1 and phot2 redundantly mediate the accumulation response, whereas phot2 alone mediates the avoidance response in Arabidopsis (Jarillo et al., 2001; Kagawa et al., 2001; Sakai et al., 2001). Phototropins possess highly conserved domains: two LOV (light, oxygen, or voltage) domains (LOV1 and LOV2) in the N terminus and a Ser/Thr kinase domain in the C terminus (Briggs et al., 2001). The light information perceived by the LOV domains is relayed to the kinase domain through conformational changes between these functional domains (Harper et al., 2003, 2004). Consequently, the activated kinase domain transfers the signals to the downstream targets (Christie et al., 2002; Kong et al., 2006, 2007; Inoue et al., 2008, 2011), although the signaling cascade that leads to chloroplast movement has not yet been identified.
Actin plays fundamental roles in a wide range of cellular functions throughout the plant life cycle, including cell division, cytoplasmic streaming, cell morphogenesis, and organelle motility (Shimmen and Yokota, 2004; Wada and Suetsugu, 2004; Hussey et al., 2006; Szymanski, 2009). As with most organelle movements in plants, the actin cytoskeleton also plays an important role in determining the intracellular positioning of chloroplasts and in directional chloroplast photorelocation movements (Oikawa et al., 2003; Kadota et al., 2009; Takagi et al., 2009; Suetsugu et al., 2010b). It has recently been reported that short chloroplast-actin (cp-actin) filaments are essential for chloroplast movements (Kadota et al., 2009; Kodama et al., 2010; Suetsugu et al., 2010b). Cp-actin filaments are localized along the chloroplast periphery on the plasma membrane face when chloroplasts are under weak light, and the filaments become relocalized to the leading edge of the chloroplasts before and during photorelocation in response to strong blue light (Kadota et al., 2009; Ichikawa et al., 2011; Kong and Wada, 2011; Yamashita et al., 2011; Tsuboi and Wada, 2012). Chloroplasts can change their direction of movement with a short lag time of several minutes (Tsuboi et al., 2009; Tsuboi and Wada, 2011). Therefore, the rapid reorganization of cp-actin filaments agrees well with the physiological relevance of chloroplast positioning and directional movement (Kong and Wada, 2011).
The velocity of the chloroplast avoidance movements is dependent on the fluence rate of blue light: A higher fluence rate induces a higher velocity. The velocity has also been shown to be dependent on the levels of phot2 in Arabidopsis (Kagawa and Wada, 2004; Kimura and Kagawa, 2009). Conversely, the velocity of the accumulation response is constant (∼0.3 µm/min in the fern Adiantum capillus-veneris and ∼1 µm/min in Arabidopsis) and is not dependent on the fluence rates of blue or red light in the prothallial cells of A. capillus-veneris (Kagawa and Wada, 1996; Tsuboi et al., 2009). Importantly, the velocity of the chloroplast movement is proportional to the difference in the amount of accumulated cp-actin filaments between the front and the rear regions of a chloroplast (Kadota et al., 2009). Therefore, the rapid disappearance of cp-actin filaments in the rear region and the accumulation of these filaments in the front region are critical to create an asymmetric arrangement of cp-actin filaments on a chloroplast, where the degree of asymmetry determines the velocity of the avoidance movement. The disappearance of cp-actin filaments is induced by strong blue light, a response specifically mediated by phot2 but not by phot1 (Kadota et al., 2009).
Actin filament rearrangements are generally mediated through the coordinated processes of filament nucleation, growth, severing, and turnover (Blanchoin et al., 2010). During these processes, rapid reorganization of cortical actin filaments is dominated by stochastic dynamics that feature random severing, filament buckling, and straightening events (Staiger et al., 2009). Although the mechanistic details are not yet understood, these processes are likely involved in the reversible appearance (nucleation and growth) and disappearance (severing and turnover) of cp-actin filaments to facilitate their rapid reorganization. Because asymmetric formation of cp-actin filaments is a prerequisite for directional and fast chloroplast movements, the rapid disappearance of cp-actin filaments is probably a primary step that promotes the avoidance of strong light in the chloroplasts.
Hangarter and colleagues recently identified an actin-bundling factor, THRUMIN1 that controls chloroplast movement by regulating actin reorganization at the plasma membrane in a phototropin-dependent manner (Whippo et al., 2011). However, they stated that the cp-actin filaments reported previously (Kadota et al., 2009; Suetsugu et al., 2010b; Kodama et al., 2010; Ichikawa et al., 2011; Kong and Wada, 2011) were not observed with their actin probes such as THRUMIN1-YFP (for yellow fluorescent protein) and LIFEACT-YFP, although blue light–dependent thick cytoplasmic actin cables were clearly present during chloroplast movement in response to microbeam irradiation (Whippo et al., 2011). Hence, these contradictory results from two independent groups need to be resolved in order to elucidate the underlying molecular mechanism of chloroplast movement.
To elucidate further the mechanism by which the chloroplast avoidance response is regulated, we first established a system for observation using confocal laser scanning microscopy (CLSM). This enabled us to easily and reliably observe cp-actin filaments visualized with various fluorescent protein–tagged actin binding proteins, such as mouse TALIN (Kadota et al., 2009), THRUMIN1 (Whippo et al., 2011), LIFEACT (Era et al., 2009), and the second actin binding domain of Arabidopsis FIMBRIN1 (fABD2) (Sheahan et al., 2004) under a controlled light environment. Next, we made higher resolution observations of cp-actin filaments using total internal reflection fluorescence microscopy (TIRFM). The direct observation of cp-actin filaments in mesophyll cells at high optical and temporal resolution revealed that cp-actin filament dynamics during their disappearance processes were different from those of cortical actin filaments. These processes are specifically regulated according to the light’s intensity and position through phot2 to allow chloroplasts to avoid strong light.
RESULTS
Actin Probes Have No Detrimental Effect on Chloroplast Photorelocation
There are numerous reports of detrimental effects of actin probes on actin dynamics and organization and/or biological responses in Arabidopsis (Ketelaar et al., 2004; Sheahan et al., 2004; Luesse et al., 2006; van der Honing et al., 2011). We therefore assessed chloroplast photorelocation movements in several actin probe lines expressing green fluorescent protein (GFP)-TALIN, GFP-fABD2 (Wang et al., 2008), LIFEACT-YFP (Era et al., 2009), THRUMIN1-GFP, and THRUMIN1-red fluorescent protein (RFP) compared with isogenic wild-type plants. Chloroplast photorelocation movements were evaluated using red light transmittance of rosette leaves under a series of light conditions (Wada and Kong, 2011). Red light transmittance in wild-type leaves decreased gradually (i.e., accumulation response) under low-intensity blue light (3.2 µmol m−2 s−1), while the transmittance increased (i.e., avoidance response) under higher intensity blue light (25 and 60 µmol m−2 s−1) (see Supplemental Figure 1A online). The red light transmittance changes in the actin probe lines were comparable to those of the wild type during the blue light–induced accumulation and avoidance responses (see Supplemental Figure 1A online). The speeds of the movements at the early stage after blue light onset were not statistically different between the wild type and the actin probe lines (P > 0.05) (see Supplemental Figure 1B online).
The THRUMIN1-GFP driven by the native THRUMIN1 promoter (TH1-G) complemented the thrumin1 mutant phenotypes (see Supplemental Figure 1C online). As previously reported by Whippo et al. (2011), the TH1-G transgenic line was fully functional in the accumulation response. However, the complementation of the avoidance response was slightly attenuated, at around 80% of that of the wild-type control under high-light conditions (see Supplemental Figure 1C online), compared with the 30% activity of the thrumin1 mutant. However, the movement speeds were not significantly different (P > 0.15) (see Supplemental Figure 1D online), suggesting that our transgenic lines complement the thrumin1 mutant phenotype effectively. Moreover, the THRUMIN1-GFP and -RFP lines in the wild-type background (TH1-G/WT and TH1-R/WT) exhibited typical chloroplast photorelocation movements as well (see Supplemental Figure 1E online). The TH1-G/WT and TH1-R/WT transgenic lines were fully functional in the accumulation response. However, the avoidance response was slightly attenuated in middle intensity blue light (25 µmol m−2 s−1) (P < 0.01) but normal under high-intensity blue light (60 µmol m−2 s−1) compared with the wild type (P > 0.01) (see Supplemental Figures 1E and 1F online). It should be noted that the cause of this inhibition is unclear, but the inhibition is overridden under higher intensity blue light (60 µmol m−2 s−1). Hence, these data collectively suggest that the actin probes in these lines have no quantifiable effect at least on high light–induced chloroplast photorelocation movements compared with isogenic wild-type Arabidopsis plants that do not express any fusion protein.
Cp-Actin Filament Dynamics in Moving Chloroplasts
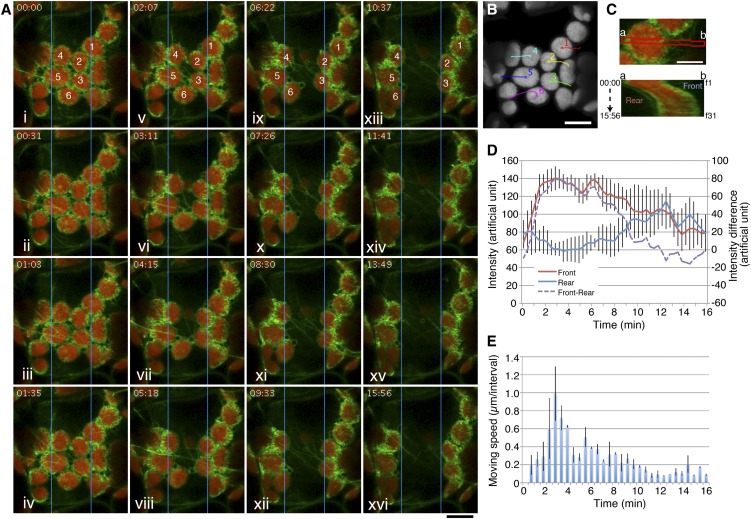
The cp-actin filament dynamics in intact palisade cells of the above actin probe lines were studied using CLSM. Time-lapse CLSM enabled us to observe the fluorescence of GFP-TALIN that decorated cp-actin filaments during the avoidance response at higher optical and temporal resolution than in our previous report (Kadota et al., 2009). GFP was excited by laser beam scans (wavelength peak at 488 nm), and GFP fluorescence was collected at ∼30-s time intervals (Figure 1A; see Supplemental Movie 1 online). In the first observations of the mesophyll cells, including the sample cells that were adapted to the low-intensity light under the identical culture conditions, fewer GFP signals for cp-actin filaments were observed on the edges of the chloroplasts. When a portion of a cell was irradiated with laser beam scans (wavelength peak at 458 nm), the chloroplasts moved out of the irradiated rectangular region (Figures 1A and 1B; see Supplemental Movie 1 online). During the avoidance response, drastic changes in the organization of cp-actin filaments were observed both inside and outside the irradiated region: Cp-actin filaments disappeared inside the irradiated region but appeared outside the irradiated region at the leading edge of the moving chloroplasts (Figure 1A).
Figure 1.
Dynamic Reorganization of Cp-Actin Filaments during the Chloroplast Avoidance Response.
(A) Reorganization of cp-actin filaments during the blue light–induced chloroplast avoidance response. Time-lapse images were collected at 31-s intervals for 15 min 56 s. The region in the central portion (5 × 50 µm) indicated by the blue rectangle was irradiated by 458-nm laser scans during the intervals between the image acquisitions. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The time points (minutes:seconds) at image acquisition are shown in the top left corner of the pictures. See also Supplemental Movie 1 online for the full time-lapse series. Bar = 10 μm.
(B) The paths of individual chloroplasts. The centers of representative chloroplasts, 1 to 6 in (A), were traced. The image for chlorophyll fluorescence shows the start position before the 458-nm laser scans. Bar = 10 μm.
(C) Kymograph analysis of the asymmetric cp-actin filaments on a moving chloroplast (chloroplast 1 in [A] and [B]). Thirty-one data points obtained from frames 1 (f1) to f31, which corresponded to 00:00 to 15:56 (minutes:seconds), respectively, at the red band (a and b in top panel) were subjected to kymograph analysis (bottom panel). Bar = 5 μm.
(D) Changes in the total cp-actin filament fluorescence at the front (brown line) and rear (blue line) regions of the moving chloroplasts. The intensities of the GFP fluorescence were calculated from kymographs like that shown in (C). The data were obtained from five chloroplasts from three different cells and are presented as mean ± sd. The intensity difference between the front and the rear regions is also shown by the dotted purple line.
(E) Velocity changes during the avoidance movement. The distances between two sequential time-lapse images of the same chloroplasts used for the kymograph analyses in (D) were calculated from the paths. The data are presented as mean ± sd.
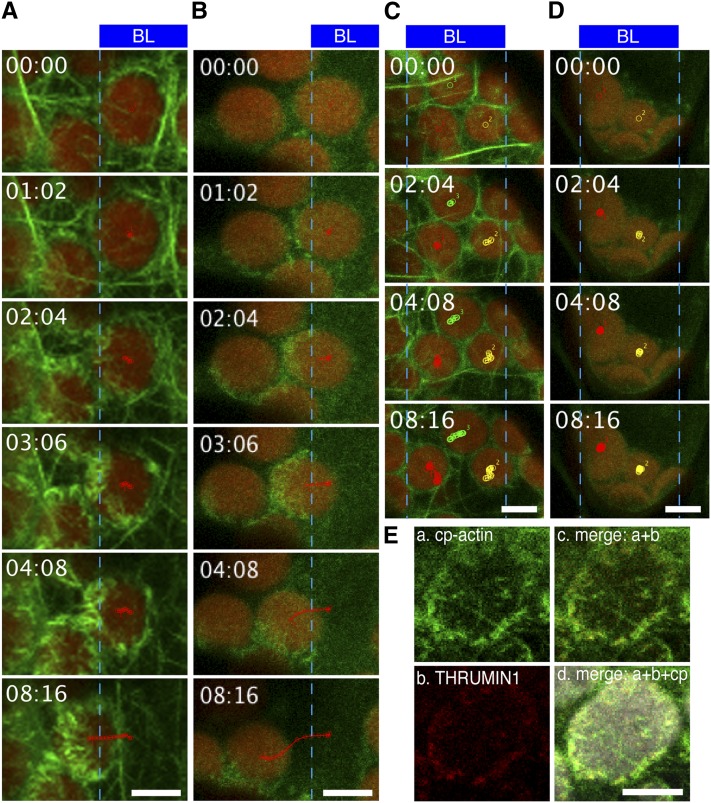
We further confirmed the reorganization of the cp-actin filaments under strong light in the LIFEACT-YFP and GFP-fABD2 lines. The fluorescent signals clearly indicated asymmetric cp-actin filaments on moving chloroplasts; however, the signals that were emitted from cp-actin filaments were faint in these lines, although cortical actin filaments were easily visualized (see Supplemental Figure 2 online). Compared with LIFEACT-YFP and GFP-fABD2 lines, the fluorescent signals of THRUMIN1-GFP obviously showed an asymmetric distribution of THRUMIN1 on chloroplasts during avoidance in response to strong blue light, as in the cells expressing GFP-TALIN (Figures 2A and 2B; see Supplemental Movie 2 online). The cp-actin filaments were not observed in thrumin1 mutant cells (Figure 2C; see Supplemental Movie 3 online). In addition, the THRUMIN1-GFP signal was not significant in chup1 mutant cells (Figure 2D; see Supplemental Movie 4 online), and the signals of THRUMIN1-RFP closely colocalized with the cp-actin filaments revealed by GFP-TALIN (Figure 2E), indicating that the fluorescent protein–tagged THRUMIN1 is also an effective probe for visualizing cp-actin filaments. The following data were obtained using the GFP-TALIN lines unless otherwise stated because it was the more adaptable probe for observing cp-actin filaments with our microscope configuration.
Figure 2.
Blue Light–Induced THRUMIN1 Association with Cp-Actin Filaments during the Chloroplast Avoidance Response.
(A) to (D) Serial images showing the chloroplast avoidance response induced by strong blue light (BL); the regions irradiated are indicated by the blue rectangles at the top part of the images. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The borders of irradiated and nonirradiated regions are shown with blue lines. Irradiation times (minutes:seconds) are shown at the top left corner. Selected tracks of moving chloroplasts are shown with red, green, or yellow lines. Reorganization of cp-actin filaments was visualized with GFP-TALIN in a wild-type cell (A) and a thrumin1 mutant cell (C), and with THRUMIN1-GFP in a wild-type cell (B) and a chup1 mutant cell (D). Note that cp-actin filaments are clearly visualized outside the irradiated region by both GFP-TALIN and THRUMIN1-GFP in wild-type cells but are not found or are very faint in both thrumin1 and chup1 mutant cells. See also Supplemental Movies 2 to 5 online for the full time-lapse series.
(E) Colocalization of cp-actin filaments visualized with GFP-TALIN and THRUMIN1-RFP. a, cp-actin filaments visualized with GFP-TALIN; b, THRUMIN1-RFP; c, merge of fluorescent images of GFP-TALIN and THRUMIN1-RFP; d, merge of fluorescent images of GFP-TALIN and THRUMIN1-RFP, and bright-field image of chloroplast (cp).
Bars = 5 µm.
The changes in cp-actin filaments during movement were analyzed using a kymograph that was obtained from the part of the chloroplast that is indicated with a red band in the top portion of Figure 1C. The total fluorescence of cp-actin filaments gradually increased in the front region of the moving chloroplast but decreased in the rear region (Figures 1C, bottom portion, and 1D; see Supplemental Figure 3 online). When the chloroplast moved out of the light-irradiated region, the chloroplast movement stopped; cp-actin filaments were polymerized even at the rear region of the chloroplast, and they surrounded the entire chloroplast at its periphery within a few minutes (Figures 1A and 1C). It is noteworthy that the intensity of chlorophyll autofluorescence did not change in either the front or the rear of the chloroplasts during the movement (see Supplemental Figure 3E online). Hence, the difference in GFP fluorescence is related to the reorganization of cp-actin filaments rather than simple bleaching of GFP fluorescence. Thus, strong light clearly causes reorganization of cp-actin filaments.
Cp-Actin Filaments Play a Vital Role in Determining the Direction and Velocity of Moving Chloroplasts
Next, we examined in detail the correlation between the reorganization of cp-actin filaments and the directional movement of chloroplasts. The velocity of moving chloroplasts gradually increased following the onset of strong light irradiation, and it reached its maximum after 2 min (Figure 1E). The speed of the avoidance in the irradiated region was correlated with the differences in fluorescence of GFP-tagged cp-actin filaments between the front and the rear regions of the chloroplasts (cf. Figures 1D, dotted line, and 1E). The greatest velocity of the chloroplasts was observed when the fluorescence difference between the two regions was the largest (Figures 1D and 1E). We also confirmed the same behaviors of THRUMIN1-GFP (see Supplemental Figure 4 online).
Although each chloroplast moves in a predictable direction depending on its spatial relation to the exciting microbeam (Tsuboi et al., 2009; Tsuboi and Wada, 2011), the timing and direction of chloroplast movement is not uniform, depending on the position in the irradiated region (light signal) and the distance from or relationship to neighboring chloroplasts. Hence, simply combining data from the chloroplasts under different circumstances can easily mask phenomena that are obvious from single-chloroplast analysis. In the following several studies, we undertook precise observations and analyses of representative chloroplasts under different conditions and show them as data from a single chloroplast. We confirmed the reproducibility of the phenomena by repeating the single chloroplast analyses at least three times.
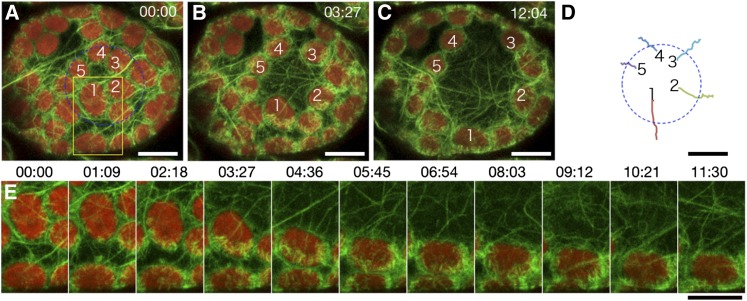
As shown in Figure 3 and Supplemental Movie 5 online, chloroplast number 1 in Figure 3 moved downward toward the irradiated circle edge obliquely instead of directly. Thus, this chloroplast did not travel the shortest route to reach the outside of the irradiated circle; however, the other chloroplasts (numbers 2 to 5) moved toward the outside of the irradiated circle by traveling approximately the shortest distance, each following a path perpendicular to a tangent of the irradiated circle (Figure 3D). During the avoidance response, the longitudinal axis of chloroplast number 1 altered from parallel to perpendicular to the direction of movement (counterclockwise by almost 90°) (Figure 3E). Interestingly, the cp-actin filaments were reformed so that they maintained their position on the leading edge of the chloroplast, even as the chloroplast changed its axis (Figure 3E). A similar behavior of cp-actin filaments during avoidance movement was also observed using the THRUMIN1-GFP (Figure 2B).
Figure 3.
Reconstruction of Cp-Actin Filaments on the Leading Edge of Chloroplasts during Avoidance.
(A) to (C) Serial images of chloroplasts during avoidance induced with 458-nm laser scans inside the blue circle (20 µm in diameter) during the intervals between the image acquisitions. Time-lapse images were collected at 34.5-s intervals for 12 min and 4 s. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The cp-actin filaments on the chloroplasts before (A), during (B), and after (C) the avoidance response are shown. The time points (minutes:seconds) at image acquisition are shown in the top right corner of the pictures. See also Supplemental Movie 5 online for the full time-lapse series. Bars = 10 µm.
(D) The paths of the individual chloroplasts that moved. The centers of the representative chloroplasts, 1 to 5 in (A) to (C), were traced. Bar = 10 µm.
(E) Enlarged and more precise time-lapse images of chloroplast 1 in the yellow inset of (A). Note that the reconstruction of cp-actin filaments always occurred in the front side of the moving chloroplast, even if the chloroplast turned during the movement. Bar = 10 µm.
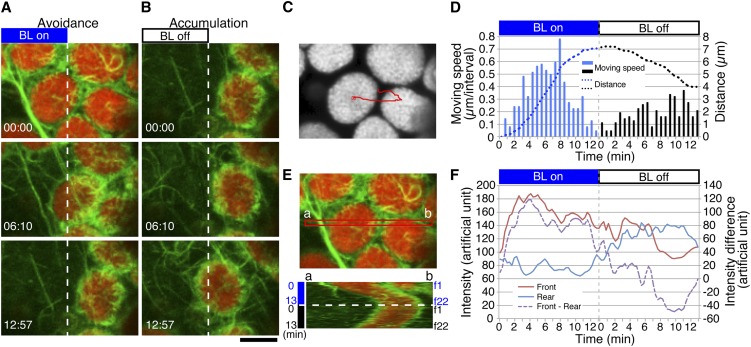
After the avoidance response induced by partial cell irradiation (Figure 4A), the chloroplasts moved back to the previously irradiated region (i.e., accumulation movement) within 1 to 2 min after the light was turned off (Figures 4B to 4D). During this movement, an asymmetric distribution of cp-actin filaments was generated by their reappearance on the leading edge without disappearing on the rear region (Figures 4B and 4E; see Supplemental Movie 6 online). In both the avoidance and accumulation movements, the reorganization of cp-actin filaments was correlated with the speed and direction of the chloroplast movement (Figures 4D and 4F).
Figure 4.
Reorganization of Cp-Actin Filaments during the Strong Blue Light–Induced Avoidance Response Followed by the Dark-Induced Accumulation Response.
(A) Reorganization of cp-actin filaments during the avoidance response. The avoidance response was induced with 458-nm laser scans in the left half of the picture (BL on) during the intervals between the image acquisitions. Time-lapse images were collected at 37-s intervals for 12 min and 57 s. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The time points (minutes:seconds) at image acquisition are shown in the bottom left corner of the pictures.
(B) Reorganization of cp-actin filaments during the accumulation response. After switching off the blue light (BL off), an accumulation response toward the previously irradiated region occurred following the avoidance response (A). The other details are the same as in (A). Note that the cp-actin filaments are observed at both the front and rear parts of the moving chloroplast. See also Supplemental Movie 6 online for the full time-lapse series of (A) and (B). Bar = 5 µm.
(C) The path of a chloroplast that showed avoidance followed by an accumulation response ([A] and [B]). The center of the chloroplast was traced. The image for chlorophyll fluorescence shows the starting position before blue light irradiation.
(D) Movement speed and distance of the chloroplast shown in (A) to (C). The other details are the same as in Figure 1E.
(E) Kymograph analysis of the asymmetric cp-actin filaments on a moving chloroplast. The data at the red band (a and b in the top panel) were subjected to kymograph analysis (bottom panel). Frames 1 (f1) to f22 correspond to 0 to 15 min, respectively. The other details are the same as in Figure 1C. Note that the cp-actin filaments are observed only at the front side during the avoidance response but at both the front and rear sides during the accumulation response.
(F) Changes in total fluorescence of cp-actin filaments in the front (brown line) and rear (blue line) regions of the moving chloroplasts. The intensities of the GFP fluorescence were calculated from the kymograph in (E). The intensity difference between the front and rear regions is shown by the dotted purple line.
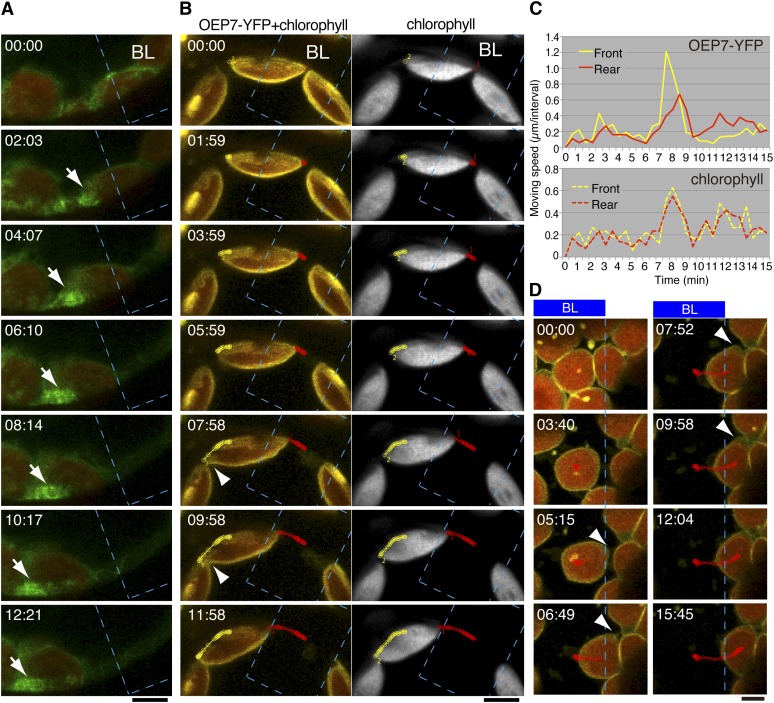
To examine the behavior of the chloroplast envelope during chloroplast movements, we produced transgenic plants expressing OEP7-YFP, which is specifically localized to the chloroplast outer envelope (Figure 5B; Lee et al., 2001; Oikawa et al., 2003). The chloroplast envelope, where cp-actin filaments (Figure 5A) were highly polymerized, extended toward the direction of movement (Figures 5B, left panel, and 5D; see Supplemental Movies 7 and 8 online). This extension occurred at the front edge but not at the rear edge (Figure 5C, top panel). One minute later, when the speed of extension at the front edge became maximal, the movement speed of the rear edge became maximal, indicating that the chloroplast reached its maximum speed when the envelope extension was largest. By contrast, no significant difference in extension and movement speed between the front and rear sides was detected from measurement of the chlorophyll fluorescence (Figures 5B, right panel, and 5C, bottom panel). These results show relationships between cp-actin filament formation at the leading edge of the moving chloroplast, the velocity of chloroplast movement, and the chloroplast envelope extension wherein the motive force for chloroplast movement is generated.
Figure 5.
Cp-Actin Filaments Pull the Chloroplast Envelope on the Leading Edge of Moving Chloroplasts.
(A) Reorganization of cp-actin filaments probed with GFP-TALIN at the interface between the moving chloroplast envelope and the plasma membrane. Time-lapse images during the avoidance response were collected at 31-s intervals for 12 min and 21 s. The region shown with a blue dotted line (BL) (10 × 15 µm) was irradiated using 458-nm laser scans during the intervals between the image acquisitions. The time points (minutes:seconds) at image acquisition are shown in the top left corner of the pictures. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. Note the side view of a chloroplast with a strong signal from cp-actin filaments on the leading edge. The GFP fluorescence for cp-actin filaments extends beyond that for chlorophyll (white arrow), suggesting that the pulling force was generated by the cp-actin filaments. See also Supplemental Movie 7 online for the full time-lapse series. Bar = 5 µm.
(B) Reorganization of the chloroplast envelope during the avoidance response. Time-lapse fluorescence images of OEP7-YFP, a chloroplast envelope–associated protein, and chlorophyll in a chloroplast viewed from the side were collected at 30-s intervals for 11 min and 58 s. The images are false-colored to indicate YFP (yellow) and chlorophyll (red in the left panels or gray in the right panels) fluorescence. The paths of the chloroplast envelope and chloroplast at the front (yellow color) and rear (red color) points were traced using the fluorescent signal of OEP7-YFP (left) and chlorophyll (right). The other details are the same as in (A). Note that the chloroplast envelope (arrowhead) facing the plasma membrane, but not chlorophyll, is extended at the leading edge of the moving chloroplast. See also Supplemental Movie 7 online for the full time-lapse series. Bar = 5 µm.
(C) The velocities for the front and rear edges of the chloroplast shown in (B) during an avoidance response. The chloroplast movement was traced based on the fluorescence of OEP7-YFP for the chloroplast envelope (top panel) and chlorophyll (bottom panel). The time points for the greatest velocity occurred earlier at the leading edge than at the trailing edge in the measurement of OEP7-YFP but are the same for those of chlorophyll fluorescence, indicating that the front side of chloroplast envelope was extended before the whole chloroplast moved.
(D) Serial images showing the extension of the chloroplast envelope on the leading edge during the avoidance response. Time-lapse images were collected at 31-s intervals for 15 min 45 s. Note that the envelope starts to extend when a part of the chloroplast comes out of the region irradiated with strong blue light (05:15), then the extension increases in the direction of movement (06:69 to 09:58) as shown with arrowheads. The other details are the same as in (B). See also Supplemental Movie 8 online for the full time-lapse series. Bar = 5 µm.
phot2 Is the Primary Photoreceptor for Cp-Actin Filament Dynamics
To accomplish a rapid avoidance movement, a large difference in the number of cp-actin filaments between the front and rear regions of chloroplast must be established; an increasing difference is correlated with an increasing speed of movement (Figures 1D and 1E; Kadota et al., 2009; Kodama et al., 2010). In spite of our efforts, however, tracking the disappearance of cp-actin filaments on moving chloroplasts was not amenable to detailed analysis with high temporal and optical resolution because the cp-actin filaments changed their location along with chloroplast movement and because the focal area frequently changed during movement and during the period of blue light irradiation. Therefore, we examined the disappearance of cp-actin filaments using stationary chloroplasts that were immobilized by irradiation with strong light (Figure 6).
Figure 6.
Reorganization of Cp-Actin Filaments Induced by Different Light Conditions.
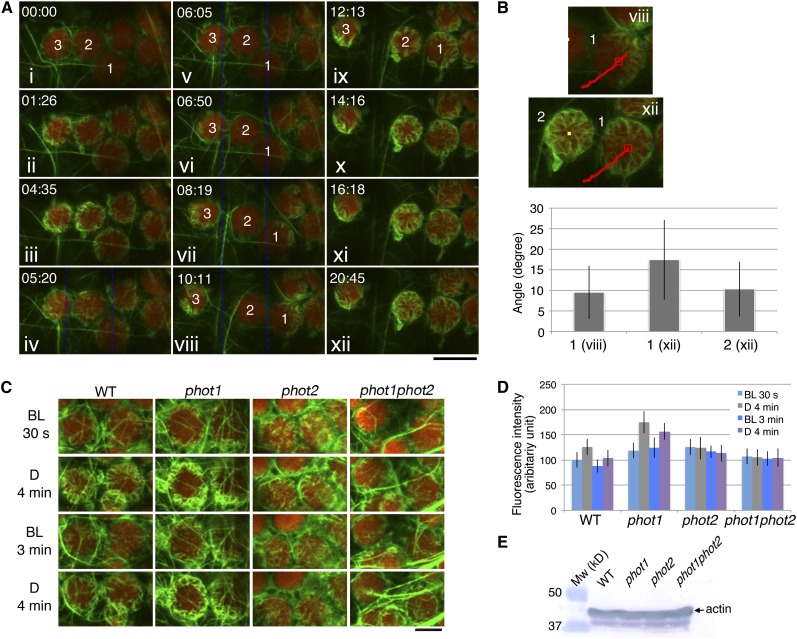
(A) Time-lapse images of the reorganization of cp-actin filaments under different light conditions. The 458-nm laser beam was used to scan the whole region shown here for 30 s before the first image was taken at 00:00 (i) for the depolymerization of all of the cp-actin filaments. The cells were then incubated in the dark until 04:35 (iii), except for a single image that was acquired at 01:26 (ii). The cp-actin filaments started to appear at the chloroplast periphery during the dark incubation. Subsequently, the region between the blue dotted lines was irradiated with a laser scan continuously until 10:11 (iv to viii). The chloroplasts on the left (3) and right (1) moved out of the beam with asymmetric cp-actin filaments, but the chloroplast (2) in the middle did not move, and its cp-actin filaments completely disappeared. After the laser beam was switched off at 10:11 (viii), the cp-actin filaments reappeared at the chloroplast periphery and extended toward the center of the chloroplast (ix to xii). The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The time points (minutes:seconds) at image acquisition are shown in the top left corner of the pictures. Note that the cp-actin filaments at the far right of the chloroplast are not clear because of the different focal plane of the CLSM. Bar = 10 µm.
(B) Orientation of the cp-actin filaments on the chloroplasts that showed an avoidance response (1, viii of [A]) and on the dark-adapted chloroplasts (1 and 2, xii of [A]). The angles of the cp-actin filaments with respect to the center of the chloroplast were measured at different time points. The red lines in the images indicate the route that the chloroplast traveled. The number of cp-actin filaments that were measured was 9 for chloroplast 1 of viii, 14 for chloroplast 1 of xii, and 15 for chloroplast 2 of xii. The data are presented as mean ± sd.
(C) Phototropin-dependent cp-actin filament dynamics. Cells of wild-type, phot1, phot2, and phot1 phot2 mutant leaves were irradiated with serial scans of a 458-nm laser for 30 s (BL, 30 s) and then incubated in the dark for 4 min (D, 4 min). Next, serial scans with 458- and 488-nm lasers for 3 min (BL, 3 min) were employed to induce the disappearance of the cp-actin filaments (see also the full time-lapse series in Supplemental Movie 9 online). Finally, the cells were incubated in the dark for 4 min (D, 4 min). The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. Note that the cp-actin filaments disappeared following the laser scan but appeared in the dark in both the wild type and the phot1 mutant but not in the phot2 and phot1 phot2 double mutant cells. Bar = 5 µm.
(D) Quantitative analysis of phototropin-dependent cp-actin filament dynamics. The fluorescence intensities of cp-actin filaments were measured in the wild-type, phot1, phot2, and phot1 phot2 mutant cells, representing the changes in the total fluorescence of cp-actin filaments shown in (C). The average fluorescence intensities of GFP-TALIN were obtained from different squares (1.6 × 1.6 µm) from five individual chloroplast edges. The data are presented as mean ± sd (n = 20 squares).
(E) Immunoblot analysis of the endogenous actin levels in the rosette leaves of the wild-type, phot1, phot2, and phot1 phot2 plants. Fifty micrograms of total protein was separated on a 10% SDS-PAGE gel and immunoblotted with an anti-actin monoclonal antibody (see Methods for details).
Before we performed the detailed analysis, we established an experimental system in which to observe the appearance and disappearance of cp-actin filaments under light irradiation and dark conditions. Serial scans over the entire viewing area with a 458-nm laser beam for 30 s induced the rapid disappearance of cp-actin filaments (Figure 6A, i). Following the laser beam treatment and during a subsequent dark incubation for 4 min and 35 s, a large number of cp-actin filaments appeared around the chloroplast periphery (Figure 6A, ii and iii). Subsequently, a partial cell irradiation that was provided by serial scans with the 458-nm laser beam in the irradiated region (rectangle in the solid line, Figure 6A, iv) induced avoidance movement by the chloroplasts that developed asymmetric cp-actin filament distribution as a consequence of the partial irradiation (Figure 6A, chloroplasts 1 and 3 in vi to viii). However, in the chloroplast that was located in the central portion of the irradiated region (Figure 6A, chloroplast 2), the cp-actin filaments disappeared completely, and the chloroplast remained under the strong light. When the microbeam irradiation laser was turned off, an increased number of thicker cp-actin filaments appeared at the chloroplast periphery (Figure 6A, ix to xii) and gradually grew toward the center of the chloroplast (Figures 6A, ix to xii, and 6B).
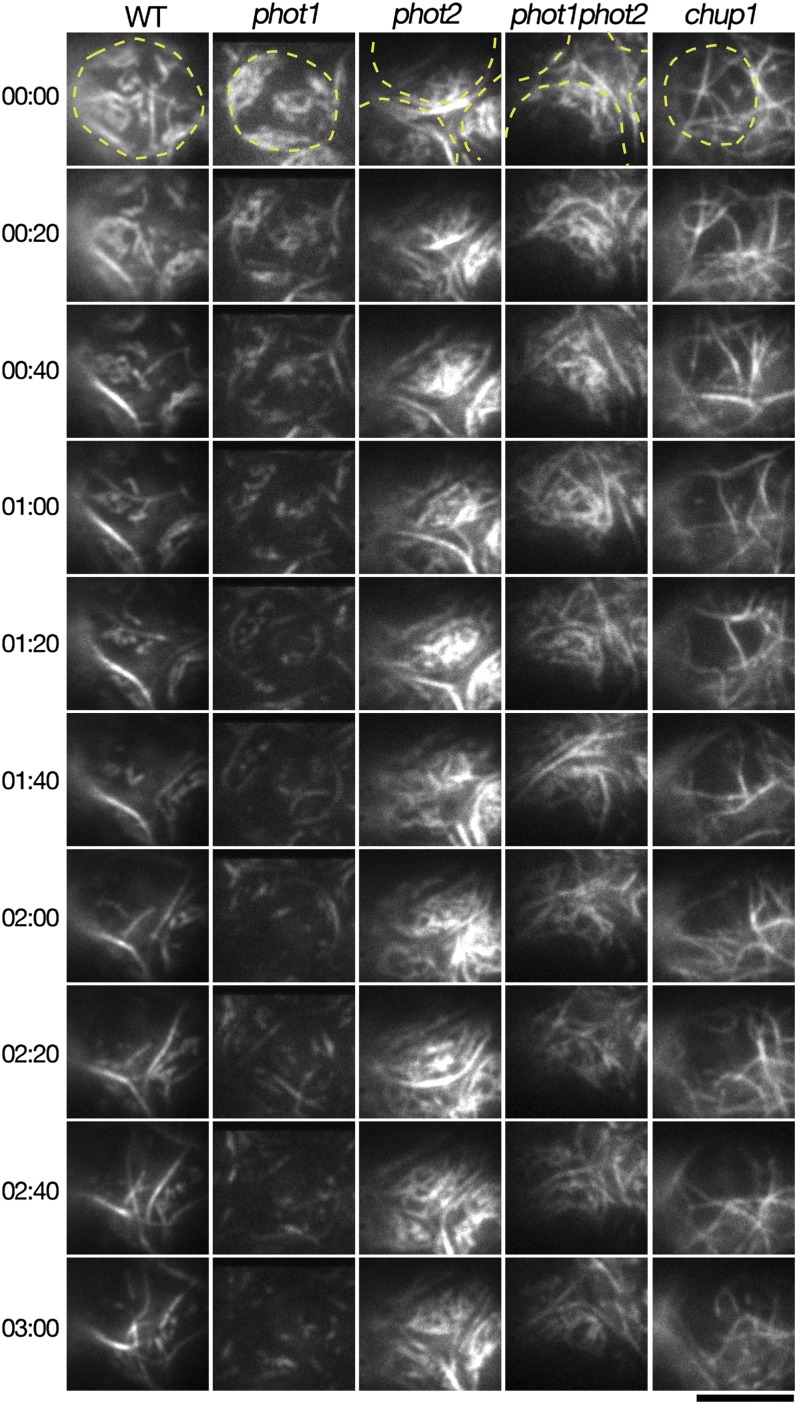
Through systematic observation, we reconfirmed the previous reports on the light-dependent cp-actin filament dynamics in phot1, phot2, and phot1 phot2 mutant cells (Kadota et al., 2009) with chup1 (Oikawa et al., 2003) mutant cells used as a control (see Supplemental Figure 5 online). The phot1 mutant cells exhibited normal cp-actin filament dynamics when compared with the wild type (see Supplemental Figure 5 online). However, in the phot2 cells, the chloroplasts that had accumulated in the irradiated region displayed slightly increased total fluorescence of cp-actin filaments compared with their states prior to strong blue light irradiation (see Supplemental Figure 5 online). By contrast, the phot1 phot2 double mutant cells exhibited strong GFP fluorescence from the cortical actin filaments but weak fluorescence from the cp-actin filaments (see Supplemental Figure 5 online). No cp-actin filaments were observed in the chup1 mutant cells, although the cortical actin filaments appeared normal (see Supplemental Figure 5 online).
We further assessed the functional roles of phot1 and phot2 in the reorganization of cp-actin filaments under strong light and in darkness. Disappearance of the cp-actin filaments from chloroplasts that were irradiated with serial 458-nm laser scans was observed in wild-type and phot1 cells, but it was not observed in phot2 and phot1 phot2 cells (Figures 6C and 6D, BL 3 min; see Supplemental Figure 6 online). The cp-actin filaments reappeared repeatedly after the 4-min dark incubation in wild-type and phot1 cells but not in phot2 and phot1 phot2 cells (Figures 6C and 6D, D 4 min). Immunoblot analysis with an anti-actin monoclonal antibody revealed that the amounts of total endogenous actin in the phot1, phot2, and phot1 phot2 mutant tissues were comparable to those in the wild type (Figure 6E), which indicates that the functional difference between phot1 and phot2 regarding cp-actin filament reorganization does not depend on endogenous actin levels in these mutant cells but depends on the regulatory functions of phot2 or phot1 and phot2 together.
Disappearance of Cp-Actin Filaments under Strong Blue Light
The disappearance of cp-actin filaments was analyzed with high temporal and optical resolution using the highly polymerized cp-actin filaments that were induced by strong light followed by dark incubation. The disappearance was analyzed by serial scans of CLSM with 488- and 458-nm laser beams, and data were acquired every 0.95 s. Under these conditions, the cp-actin filaments dispersed with rapid and random movements, and in the wild-type cells, the total fluorescence of cp-actin filaments gradually decreased to the initial levels (BL 30 s in Figure 6C) after 2 min (see Supplemental Figure 6 and Supplemental Movie 9 online). This disappearance of cp-actin filaments was also observed with THRUMIN1-GFP (see Supplemental Figure 7 online). However, the temporal and optical resolution of CLSM was not sufficient to analyze individual cp-actin filaments. Therefore, cp-actin filament dynamics were studied in mesophyll protoplasts using TIRFM, which provided almost real-time (five frames/second) acquisition, and the structural bases for each cp-actin filament were analyzed (Figure 7). In this analysis, the cp-actin filaments gradually disappeared in the wild-type and phot1 cells, but few significant changes were observed in the phot2 and phot1 phot2 cells (Figure 7; see Supplemental Figure 8 and Supplemental Movies 10 to 13 online). In addition, no cp-actin filaments were observed in the chup1 mutant cells, although the cortical actin filaments were dynamically reorganized (Figure 7; see Supplemental Movie 14 online).
Figure 7.
Phot2-Dependent Disappearance of Cp-Actin Filaments as Observed by TIRFM.
Time-lapse cp-actin filament images in the wild-type (WT), phot1, phot2, phot1 phot2, and chup1 mutant protoplasts were collected every 0.2 s using TIRFM and are shown at 20-s intervals. The disappearance of the cp-actin filaments was induced by continuous irradiation with 488-nm laser scans for GFP observation. The positions of each chloroplast are shown with dotted circles. Note that the actin filaments that were found on the chup1 chloroplast are not cp-actin but cortical actin filaments. See also the full time-lapse series in Supplemental Movie 10 online for wild-type, Supplemental Movie 11 online for phot1, Supplemental Movie 12 online for phot2, Supplemental Movie 13 online for phot1 phot2, and Supplemental Movie 14 online for chup1 protoplasts. Bar = 5 µm.
[See online article for color version of this figure.]
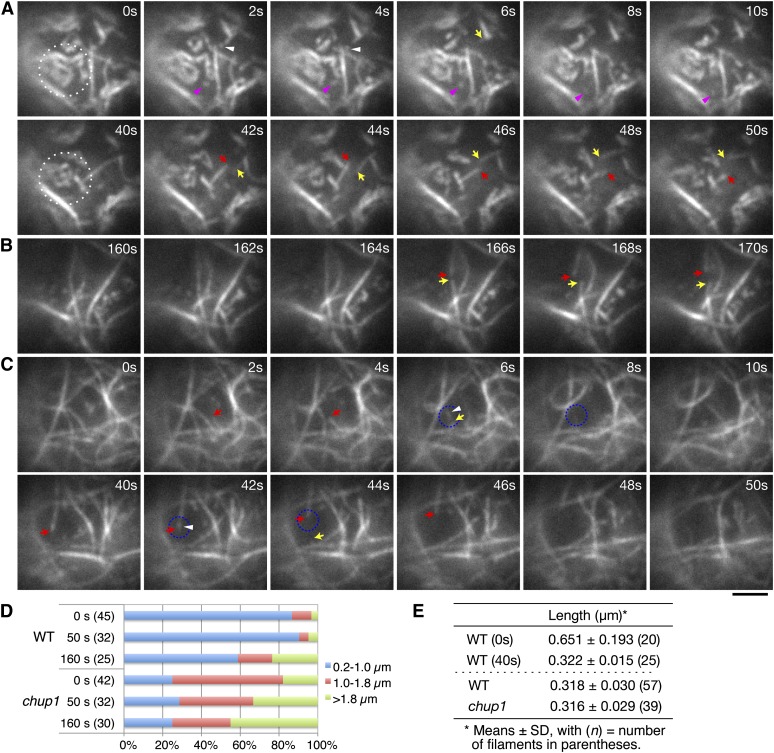
The cp-actin filaments that were present in the stationary chloroplasts were short and had a zigzag-shaped rough appearance under TIRFM. The thicker cp-actin filaments seemed to be composed of short filaments that were connected to each other to create longer actin filaments (Figures 7 and 8A), and the separation and reconnection of the short cp-actin filaments, which we designate as “remodeling,” was frequently observed at the corner of a zigzagged filament (see Supplemental Movies 10 to 13 online). Therefore, the length of the cp-actin filaments was measured from corner to corner as a unit length even if a cp-actin filament appeared to be a single, long filament. The cp-actin unit length was classified into one of three categories: from 0.2 to 1.0 µm, from longer than 1.0 to 1.8 µm, and longer than 1.8 µm (Figure 8D). At the start of the measurement (time 0), most of the cp-actin filaments (nearly 90%) were shorter than 1 µm in the wild-type cells (Figures 8A and 8D, 0 s). The population of 0.2- to 1.0-µm filaments, however, decreased gradually to 60%, while the population that was longer than 1.8 µm increased (Figures 8A and 8D). The short, independent fragments of the cp-actin filaments (that were continuously remodeled without permanent connection to other cp-actin filaments) had an average length of 0.651 ± 0.193 µm at 0 s, and their average length shortened to 0.322 ± 0.015 µm at 40 s (Figure 8E; see Supplemental Movie 10 online). Observation of cp-actin filament fragments revealed their rapid severing and random motility. However, no depolymerization (or shortening) at either end of the severed or independent cp-actin filament was observed (Figures 7 and 8A; see Supplemental Movies 10 and 11 online). This suggests that severing of the longer unit (0.651 µm) may produce two short units (0.322 µm). Although the severing and remodeling behavior of the cp-actin filaments were not completely abolished in the phot2 and phot1 phot2 mutant cells, the random motility of the fragmented cp-actin filaments was not observed in these mutant cells (Figure 7; see Supplemental Movies 12 and 13 online).
Figure 8.
Comparison of the Reorganization of the Cp-Actin and Cortical Actin Filaments as Observed by TIRFM.
(A) to (C) Actin filament dynamics in wild-type ([A] and [B]) and chup1 mutant (C) protoplasts. Time-lapse images were collected at 0.2-s intervals using TIRFM, and representative images are shown from the time points indicated. Disappearance of the cp-actin filaments was induced by continuous irradiation with 488-nm laser scans for GFP observation, and the cp-actin filaments in the region surrounded by the white dotted circle were carefully followed. Note that actin filaments observed in the chup1 mutant protoplast are not cp-actin filaments, but cortical actin filaments (C). The pink (A) and white ([A] and [C]) arrowheads indicate the small, fragmented cp-actin and cortical actin filaments, respectively. The arrows indicate the ends of the cortical actin filaments that depolymerized at both ends after severing. It is noted that remodeling of the actin filaments in the chup1 mutant protoplasts predominantly occurred via typical stochastic dynamics, including filament buckling and straightening events. See also the full time-lapse series in Supplemental Movie 10 online for the wild type and Supplemental Movie 14 online for the chup1 mutant. Bar = 2 µm.
(D) Quantitative analyses of the cp-actin filaments in the wild-type (WT) and the cortical actin filaments in the chup1 mutant protoplasts. The unit lengths of the actin filaments (see text for details) were measured at 0, 50, and 160 s of irradiation and sorted into three groups (0.2 to 1.0, >1.0 to 1.8, and >1.8 µm). The ratio among the three groups indicates that the short cp-actin filaments are abundant in wild-type cells. The number of cp-actin filaments measured is shown in parentheses.
(E) Average lengths of the cp-actin filaments and fragmented actin filaments. The average lengths of the cp-actin filaments in the wild-type cells were shortened by half between 0 and 40 s (top panel). The shortest lengths of the fragmented actin filaments that did not attach to the other actin filaments were similar for all of the cp-actin filaments (from the wild type) and the cortical actin filaments (from chup1) (bottom panel).
By contrast, in the chup1 mutant cells, long, smooth, and straight actin filaments were abundant, and the severed actin filaments became depolymerized at both ends (see the filament that is indicated with the red arrow in the bottom panel of Figure 8C; see Supplemental Movie 14 online). This depolymerization process was also observed for the long, straight actin filaments of the wild-type cells (see the filament that is indicated with the red arrows in Figures 8A and 8B). The length of cortical actin filaments was measured in the chup1 cells and classified into the three categories, as with the cp-actin filaments. The distance between the cross-points among the actin filaments was defined as the unit length of the cortical actin filaments. No significant change in the populations of the actin filament lengths was identified during the period of observation (Figure 8D).
Severing rates of cp-actin and cortical actin filaments were acquired from the actin filaments on the chloroplasts of the wild-type and chup1 cells, respectively. The severing rate of cp-actin filaments was significantly higher than that of cortical actin filaments, with 0.0529 ± 0.0101 breaks/µm/s (mean ± sd, n = 41) in the wild type and 0.0143 ± 0.0044 (n = 38) in chup1. Interestingly, the fragmented actin filaments exhibited very rapid random movements on the chloroplast envelope. The average length of the fragmented actin filaments that showed rapid and random movements was 0.318 ± 0.030 µm in the wild type (i.e., the cp-actin filaments) and 0.316 ± 0.029 µm in chup1 (i.e., the cortical actin filaments) (Figure 8E). Although the lengths of both types of fragmented actin filaments were quite similar, their lifetime was different. The filaments in the wild-type cells displayed a lifetime of ∼15.16 ± 7.88 s (mean ± sd, n = 36) on the chloroplast envelope before they disappeared from the chloroplast edges (Figure 8A, pink arrowheads). By contrast, the short fragmented actin fragments in the chup1 cells disappeared suddenly, within 2.33 ± 1.39 s (n = 33) of their production (Figure 8C, see the fragments that are surrounded by the blue, dotted circles).
Reduction of Severing and Random Motility of Cp-Actin Filaments under 2,3-Butanedione Monoxime Treatment
Treatment with 2,3-butanedione monoxime (BDM), a potent myosin ATPase inhibitor, was shown to inhibit chloroplast movement in the moss Physcomitrella patens and Arabidopsis; however, asymmetric rearrangements of cp-actin filaments could be clearly observed under strong light (Yamada et al., 2011; Yamashita et al., 2011). BDM also inhibited the random movements of severed cortical actin filaments in Arabidopsis seedlings (Staiger et al., 2009). Therefore, we examined the effect of BDM on cp-actin filament dynamics.
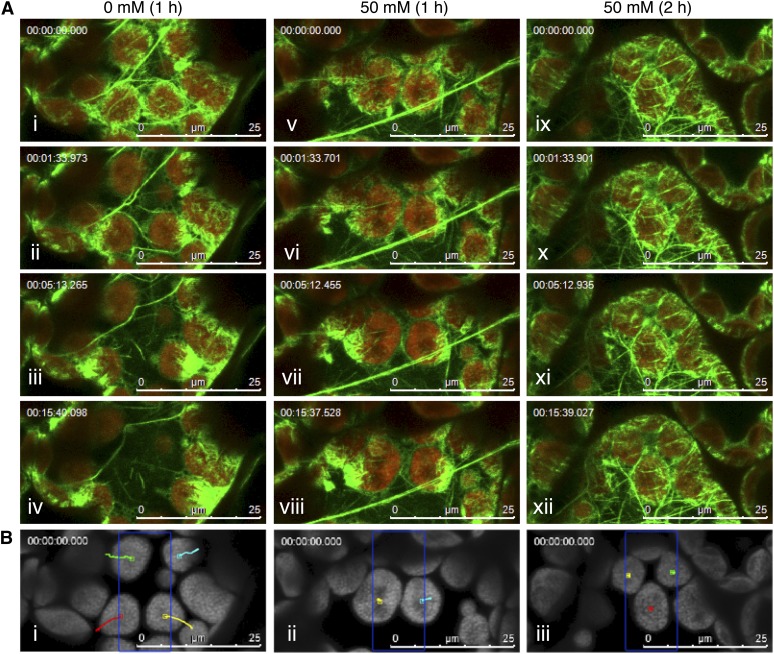
When Arabidopsis rosette leaves were treated with 50 mM BDM for 1 h, the chloroplast avoidance movement that was induced by strong microbeam irradiation was severely inhibited compared with that in the mock-treated cells (Figure 9B, i and ii). Although the asymmetric cp-actin filament formation was normal in mock-treated (Figure 9A, i to iv; see Supplemental Movie 15 online) and BDM-treated (Figure 9A, v to viii; see Supplemental Movie 16 online) cells, the cp-actin filament disappearance that followed serial confocal scans with 458- and 488-nm laser beams was very much delayed by BDM treatment (see Supplemental Movie 17 online). A 2-h treatment with 50 mM BDM prevented asymmetric cp-actin filament formation following strong microbeam irradiation as well as chloroplast avoidance movement, although the cp-actin filaments were clearly identifiable around the chloroplast periphery (Figures 9A, ix to xii, and 9B, iii; see Supplemental Movies 18 and 19 online).
Figure 9.
Inhibitory Effect of BDM on Blue Light–Induced Chloroplast Movement and Cp-Actin Filament Dynamics.
(A) Reorganization of the cp-actin filaments in leaves untreated (0 mM) (i to iv) or treated with 50 mM BDM for 1 h (v to viii) or 2 h (ix to xii). Time-lapse images of the avoidance response were collected at 31.2-s intervals for 15 min and 37 s under induction with 458-nm laser scans (in the 10 × 25-µm region indicated by the blue rectangle in [B]) during the intervals between the image acquisitions. The images are false-colored to indicate GFP (green) and chlorophyll (red) fluorescence. The time points at which the images were obtained are shown in the top left corner of the pictures. See also the full time-lapse series in Supplemental Movie 15 online for 0 mM (1 h), Supplemental Movie 16 online for 50 mM (1 h), and Supplemental Movie 17 online for 50 mM (2 h) BDM treatments. Bars = 25 µm.
(B) The paths of the individual chloroplast movements are shown as traces of the centers of the representative chloroplasts on the chlorophyll fluorescence images for 0 mM (i), 50 mM BDM for 1 h (ii), and 50 mM BDM for 2 h (iii). Bars = 25 µm.
DISCUSSION
Functional Roles of Cp-Actin Filaments in Chloroplast Photorelocation Movements
Chloroplasts move using cp-actin filaments that are polymerized at the chloroplast periphery, although the mechanism by which the motive force is generated is not yet understood. However, it is clear that the ratio of the amounts of cp-actin filaments between the front and rear portions of the chloroplast is an important factor that determines the speed (Figures 1D and 1E; see Supplemental Figures 3 and 4 online; Kadota et al., 2009). Increased difference is correlated with increased speed of movement. A large difference can be established by intensive polymerization and depolymerization of cp-actin filaments in the front and rear regions of the chloroplast, respectively. Following strong light irradiation, the decrease in the overall cp-actin filament amount occurs earlier than the increase in the front region (Figures 1A and 5A; see Supplemental Figure 3 online; Kadota et al., 2009). Thus, disappearance of the cp-actin filaments at the trailing edge is critical for the avoidance movement.
Cp-actin filaments also play a role in determining the direction of the chloroplast movement. When a portion of a cell was irradiated with strong blue light, the chloroplasts moved away from the light beam by taking the shortest route rather than a detour route (Figure 3D; Tsuboi and Wada, 2011). During the process, cp-actin filaments always appeared at the leading edge of the chloroplast, regardless of their original positions. This indicates that the localization of the cp-actin polymerization site is important in determining the direction of movement. Moreover, when the chloroplasts returned to the previously irradiated region following the avoidance response (Figures 4B and 4C), the cp-actin filaments at the rear remained intact and did not disappear (Figures 4B and 4E). This is a phenomenon that could account for the low, constant speed of the accumulation response (Kagawa and Wada, 1996). Collectively, the results indicate that the difference in the amount of cp-actin between the front and rear edges of the moving chloroplasts determines the speed of the chloroplast movement and the position of this inequality determines the direction.
Motive Force for Chloroplast Photorelocation Movements
During the avoidance response, cp-actin filaments were highly abundant at the front region facing the plasma membrane (Figure 5A). Moreover, the chloroplast envelope was extended specifically in the front region of the moving chloroplast (Figures 5B and 5D). These data suggest that the motive force for the movement is generated at the front side where cp-actin filaments are intensively polymerized. To date, two different models of the mechanics of chloroplast motility, namely, myosin- and actin-based movements, have been discussed as the potential motive force generating system. Considering the roles of cp-actin filaments in chloroplast positioning and movement, rapid and dynamic reorganizations of cp-actin filaments are more likely to explain the actin-based chloroplast movement, although experimental substantiation is not yet available (see below).
It has been reported that BDM, as an inhibitor of myosin ATPase, does not affect the avoidance response but can block the accumulation response that occurs in the dark following an avoidance response induced by high-intensity blue light in Arabidopsis leaves (Paves and Truve, 2007). Under our conditions, however, BDM effectively inhibited not only the accumulation but also the avoidance responses (Figure 9; Yamada et al., 2011; Yamashita et al., 2011). Because the asymmetrically distributed cp-actin filaments were also reorganized during the accumulation response (Figure 4; Kadota et al., 2009), the fundamental mechanisms involving cp-actin filaments are the same both in accumulation and avoidance responses. Moreover, interference with the reorganization of the cp-actin filaments should be caused by the reduced random motility and severing activities (Figure 9; see Supplemental Movies 17 and 19 online) as previously shown for cortical actin filaments (Staiger et al., 2009).
Several myosins have been reported to be associated with the chloroplast envelope, and strong blue light–induced myosin displacement from the chloroplast envelope in a blue light–specific and phot2-dependent manner has been observed (Krzeszowiec and Gabryś, 2007). Therefore, it is plausible that myosin-dependent motility may be involved in the reorganization of cp-actin filaments via this myosin displacement. However, we cannot eliminate the possibility that a high concentration of BDM interferes with the chloroplast movements by acting on sites other than myosins, such as a Ca2+ channel (McCurdy, 1999). Genetic analyses of the Arabidopsis myosin VIII and XI families support this latter possibility. Chloroplast movements were normal even when multiple myosins were disrupted, although the motility and positioning of several organelles were severely impaired in these mutants (Avisar et al., 2008; Peremyslov et al., 2010; Suetsugu et al., 2010a; Ueda et al., 2010).
Distinct Structure, Dynamics, and Phototropin-Dependent Regulations of Cp-Actin Filaments
Our high-resolution analyses of the cp-actin filaments by TIRFM showed several structural and dynamic differences between the cp-actin filaments and the cortical actin filaments. The cp-actin filaments were short and zigzag shaped (Figure 8A), and they were remodeled, severed, and then fragmented into a smallest unit of an ∼0.3-µm length under strong light (Figure 8); however, depolymerization did not occur at the ends of the severed cp-actin filaments. The fragmented cp-actin filaments exhibited rapid random movements and disappeared frequently at the chloroplast edges (see Supplemental Movie 20 online). Conversely, the cortical actin filaments were long and smooth shaped; they displayed depolymerization at their severed ends, and the remodeling of the short actin array also featured filament buckling and straightening events throughout the cells (Figures 7 and 8), which were previously designated “stochastic dynamics” of cortical actin filaments (Staiger et al., 2009). The lifetime of the smallest units and the locations at which they disappeared were also different between the two types of actin filaments. The fragments of the cp-actin filaments were retained longer on the chloroplast envelope than were those of the cortical actin filaments at the plasma membrane (Figure 8). Collectively, these observations suggest that the cp-actin filaments are different from the cortical actin filaments in many aspects, as described above, and specific factors that are localized exclusively at the chloroplast envelope should be involved in their regulation (see next section).
The data support a model in which cp-actin filament dynamics are tightly regulated by the two phototropins phot1 and phot2. Of these, phot2 appears to function as the primary photoreceptor for the processes involved in cp-actin filament dynamics by regulating the severing activity and random movements (Figure 10). By contrast, phot1 functions in the severing activity and random motility only in a minor way (Figures 6C, 7, and 10). Therefore, strong blue light–induced severing activity and random motility of the cp-actin filaments are obviously involved in the asymmetric cp-actin distribution that is mediated by phot2 during the avoidance response. Logically, chloroplasts move more rapidly under stronger light in a fluence rate–dependent manner (Kagawa and Wada, 2004). In addition, the speed of this movement is dependent on the expression level of phot2 (Kagawa and Wada, 2004; Kimura and Kagawa, 2009). Indeed, the speed in a phot2 heterozygotic line was shown to be half that in wild-type and phot1 plants (Kagawa and Wada, 2004). Therefore, these physiological aspects of the strong light-induced avoidance response are consistent with phot2 functioning as the main photoreceptor for strong light-induced reorganizations of cp-actin filaments, specifically for disappearance of cp-actin filaments.
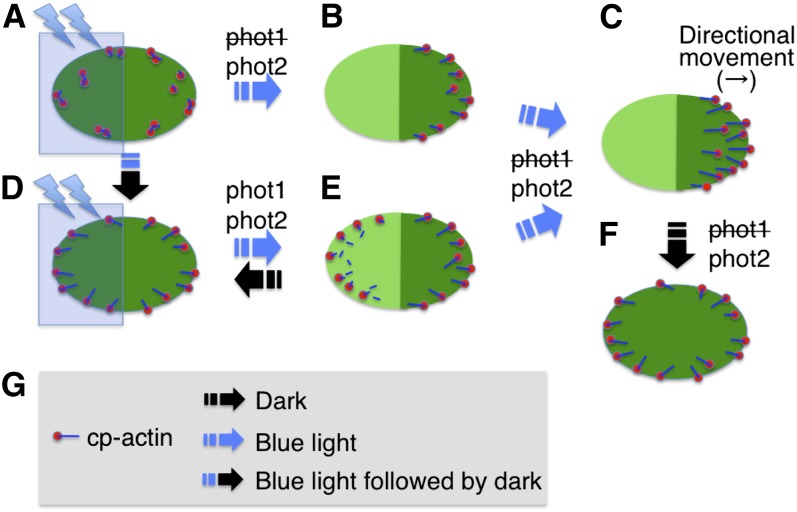
Figure 10.
Schematic Representation of Phototropin-Dependent Cp-Actin Filament Dynamics.
(A) Cp-actin filaments in low light–adapted cells.
(B) and (C) Asymmetric cp-actin filaments during the avoidance response induced by microbeam irradiation with strong blue light at the left half of the chloroplast as shown in (A). Cp-actin filaments disappeared when they were irradiated with strong blue light and reformed on the nonirradiated, front region of the chloroplast before and during movement.
(D) Cp-actin filaments appeared on the chloroplast envelope in the dark-adapted cells following irradiation with strong blue light on the whole region of the chloroplast.
(E) Asymmetrically located cp-actin filaments during the avoidance response. Rapid rearrangements of the cp-actin filaments were induced by microbeam irradiation with strong blue light at the left half of chloroplast as shown in (D). Cp-actin filaments disappeared via rapid severing and random motility in the strong microbeam-irradiated region but appeared via polymerization on the leading edge of the moving chloroplast.
(F) Rearrangement of the cp-actin filaments on the chloroplast envelope during dark adaption. The cp-actin filaments reappeared around the chloroplast periphery.
(G) Legends for a cp-actin filament and light conditions. The cp-actin filament (line in blue) is shown with a cp-actin nucleator (circle in red) located on the chloroplast envelope.
[See online article for color version of this figure.]
Downstream Factors Involved in Regulation of Cp-Actin Filaments
The cp-actin filaments entirely disappeared following whole-chloroplast irradiation with strong light, but they reappeared following dark incubation, regardless of chloroplast movement (Figures 6A and 6C). These processes must occur to mediate the rapid disappearance of the cp-actin filaments at the trailing edge and for the substantial polymerization of the cp-actin filaments at the leading edge of moving chloroplasts (Figure 10). During these processes, cp-actin nucleation would be activated during the dark incubation period or under weak light but would be inactivated under strong blue light. Furthermore, the elongation of the cp-actin filaments started at the chloroplast edge and progressed toward the center of the chloroplast (Figure 6B). Thus, the nucleation of cp-actin filaments is initiated at the chloroplast edge and the nucleation site is spatially and precisely regulated by the position and intensity of the incident light on the chloroplast envelope.
Recent molecular genetic analyses have contributed considerably to identification of the downstream signaling factors that are involved in the regulation of chloroplast photorelocation movements. Current data suggest that cp-actin filament dynamics are regulated by phototropins, CHUP1 (Kadota et al., 2009), KINESIN-LIKE PROTEIN FOR ACTIN-BASED CHLOROPLAST MOVEMENT1 (KAC1) and KAC2 (Suetsugu et al., 2010b), J-DOMAIN PROTEIN REQUIRED FOR CHLOROPLAST ACCUMULATION RESPONSE1 (Ichikawa et al., 2011), the WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT1/PLASTID MOVEMENT IMPAIRED2 complex (Kodama et al., 2010), and THRUMIN1 (this study). The signaling cascade is initiated by blue light–induced autophosphorylation activity of the phototropins, which consequently activate the downstream signaling components. These proteins are likely to be localized to or associated with the chloroplast periphery and/or the plasma membrane (Kong and Wada, 2011; Kong et al., 2013a, 2013b). Of the regulators mentioned above, CHUP1 plays a role in linking the chloroplast to the plasma membrane via its coiled-coil region (Oikawa et al., 2003, 2008). The ability of CHUP1 to bind to actin as well as profilin suggests that CHUP1 may play a pivotal role in the regulation of cp-actin filament dynamics (Oikawa et al., 2003; Schmidt von Braun and Schleiff, 2008; Kadota et al., 2009). The KAC proteins, which are also actin binding proteins, bind to F-actin and likely function much like CHUP1 (Suetsugu et al., 2010b). These data raise the intriguing possibility that both CHUP1 and the KAC proteins are involved in the regulation of cp-actin filaments by in some way regulating actin polymerization, nucleation, and/or maintenance.
Different Actin Probes for Cp-Actin Filaments
The imaging of the dynamic behavior of the actin cytoskeleton in living cells has contributed to the understanding of its diverse roles in chloroplast movements (Kong and Wada, 2011). Although actin filaments have not been visualized directly in living plant cells, several actin binding proteins, such as mouse TALIN, LIFEACT, and fABD2, have widely been used to visualize the actin cytoskeleton (Kost et al., 1998; Sheahan et al., 2004; Era et al., 2009; Vidali et al., 2009). In this study, we successfully visualized cp-actin filaments by visualizing fluorescent fusion proteins with TALIN, THRUMIN1, LIFEACT, and fABD2 (Figure 2; see Supplemental Figure 2 online), which had no detrimental effects on chloroplast movements (see Supplemental Figure 1 online). However, the GFP-TALIN efficiently decorated cp-actin filaments, whereas fABD2 decorated cortical actin filaments more efficiently than it decorated cp-actin filaments. By contrast, Whippo et al. (2011) did not detect any cp-actin filament-like structures with THRUMIN1-YFP, cyan fluorescent protein–TALIN, and LIFEACT-YFP fusion proteins. This discrepancy is probably related to technical factors during microscopy, such as laser beam intensity or the timing of observation, and/or expression levels of the fusion proteins. Because the structure and the characteristics of cp-actin filaments are different from those of cortical actin filaments, the binding affinity and/or the mechanism of binding of these proteins to each actin filament appear to be different. The visualization of a cp-actin–specific binding protein(s) would be preferable for the precise analysis of cp-actin filament dynamics.
The results presented in this study address many new details of the processes by which chloroplasts move, in particular during the avoidance response. Technical advances in microscopy have allowed us to probe these processes in far more depth than in the past. These results should form a basis for further critical studies of chloroplast movements, especially the avoidance response, which is essential for plant survival under high-light conditions.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana transgenic lines that express GFP-TALIN in different genetic backgrounds (the wild type, phot1, phot2, phot1 phot2, and chup1) were previously described (Kadota et al., 2009). The LIFEACT-YFP line (Era et al., 2009) was kindly provided by Takashi Ueda (The University of Tokyo). The GFP-fABD2, OEP7-YFP, THRUMIN1-GFP, and THRUMIN1-RFP transgenic lines were produced for this study (see below).
The genomic DNA fragments for OEP7pro:OEP7 and THRUMIN1pro:THRUMIN1 were amplified by PCR using specific primers and then cloned into the pPZP211/35S-nosT binary vector using the appropriate restriction enzyme sites (see details in Supplemental Figure 9 online). The binary vectors were used to transform wild-type (gl-1 background) Arabidopsis plants as previously described (Kong et al., 2006). The transgenic plants were selected on one-half-strength Murashige and Skoog medium that contained 35 mg/L kanamycin (Wako) and 100 mg/L claforan (Sanofi-Aventis). The T3 self-progeny of the homozygous lines were finally selected from the T2 plants, which showed a segregation ratio of 3:1 (kanamycin-resistant:-sensitive seeds). The kanamycin resistance cosegregated tightly with fluorescence of the appropriate fluorescent protein in every line. All of the analyses were performed using homozygous T3 plants. For the colocalization analysis of THRUMIN1, the TH1-R/WT line was crossed with the GFP-TALIN line, and the resulting F1 progeny was used. The transgenic lines for GFP-TALIN, TH1-G/WT, and TH1-R/WT were crossed with thrumin1-2 (Whippo et al., 2011). The resulting F3 progenies were used for the experiments. For microscopy, the seeds were sown directly on soil and grown in a plant growth room under 100 µmol m−2 s−1 white light (16 h)/dark (8 h) cycles at 23°C.
CLSM
The plant materials for microscopy observation of chloroplasts were prepared as previously described (Wada and Kong, 2011). Briefly, rosette leaves of 4-week-old plants were detached, and the leaves were deaerated by gentle evacuation using a syringe that was filled with an evacuation solution (0.01% Silwet L-77). The leaves were placed into a cuvette that was composed of two round cover slips that were supported by a ring-shaped silicon-rubber spacer.
The cp-actin filaments were visualized under a confocal microscope (SP5; Leica). The multi-Ar laser was set at 488 nm for GFP excitation and at 458 nm for the chloroplast avoidance response; the diode-pumped solid-state laser (561 nm) was used for RFP excitation. The fluorescent signals were captured through narrow bands: 500 to 550 nm for GFP, 580 to 630 nm for RFP, and 640 to 740 nm for chlorophyll. The images were captured with a lens (×63/1.20 W; Leica) at resolutions of 256 × 128, 256 × 256, or 512 × 512 using 4× digital zooming for 20 or 30 cycles. The appropriate rectangular or circular irradiated regions were scanned with the stimulating laser (458 nm, 2.8 µW) during intervals for the indicated times. The images were maximized using LAS imaging software (Leica Microsystems). The sequential images of cp-actin filament disappearance were captured at a resolution of 128 × 68 using 6× digital zooming for 3 min. The other microscopy conditions are described in the corresponding figure legends.
TIRFM
The cp-actin filament dynamics were observed in a total internal reflection fluorescence microscope using mesophyll protoplasts that were prepared from rosette leaves as previously described (Kadota et al., 2009). The protoplasts were loaded onto a round glass base dish (Iwaki) and covered with a 2% agar sheet to enlarge the face being observed against the cover slip (see Supplemental Figure 10 online). Variable-angle epifluorescence illumination was achieved using an objective-lens type total internal reflection fluorescence optical system (Tokunaga et al., 1997) that was mounted on an inverted microscope (Ti-E; Nikon) with an appropriate lens (×60/1.49 O; Nikon). After focusing on the first visible actin filaments at the protoplast periphery, the angle of laser illumination was adjusted for maximal contrast and signal-to-noise ratio. GFP and chlorophyll autofluorescence were excited with a 488-nm laser (Innova70C Spectrum; Coherent), and the emitted fluorescence was split in two by W-View Optics (A4313; Hamamatsu Photonics) with dichroic mirrors at a 550-nm cutoff wavelength (DML557; ASAHI Spectra) captured with a band-path filter (FF01-525/45; Semrock) for the GFP emission and a narrow band-path filter (647NB4; Omega Optical) for the chlorophyll autofluorescence. The fluorescent signals were recorded by an Electron Multiplying Charge-Coupled Device camera (iXon+, DU897BV; Andor Technology) at ∼200-ms exposure times.
Inhibitor Treatment
BDM (Sigma-Aldrich) was prepared as a 1 M stock solution in 50% DMSO and diluted to 50 mM in the evacuation solution. As a mock treatment, a 50% DMSO solution was added to the evacuation solution at the same dilution rate. Air spaces in the leaves were eliminated by gentle evacuation using a syringe that was filled with the solutions and then samples were incubated for the indicated times (1 or 2 h) under dim red light before observation.
Immunoblot Analysis
One hundred milligrams of rosette leaves were sampled from 3-week-old plants that were grown on one-half-strength Murashige and Skoog medium under 16-h/8-h (light/dark) conditions at 23°C; they were immediately frozen in liquid nitrogen and stored until use. The frozen tissue was homogenized with a glass homogenizer in 300 μL extraction buffer as previously described (Kong et al., 2006). The crude protein extract was clarified twice by centrifugation at 11,000g at 4°C for 15 min, and the resulting supernatant was used for immunoblot analysis. Fifty micrograms of protein was separated on a 10% SDS-PAGE gel, and actin was detected with MAbGPa, a monoclonal anti-actin antibody that was produced in mouse (Sigma-Aldrich), at a 1:1000 dilution. The MAbGPa antibody is uniformly reactive to all of the plant actin isoforms (Kandasamy et al., 2001).
Quantitative Analysis
The time-lapse images were obtained from more than three independent experiments. Of those, three representative time-lapse images were used for the quantitative analysis. The time-lapse images were cropped and analyzed using the ImageJ software program (http://rsbweb.nih.gov/ij/), which was populated with the appropriate plug-ins (see text for details).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL database under the following accession numbers: At3g25690 (CHUP1), At3G52420 (OEP7), At3g45780 (PHOT1), At5g58140 (PHOT2), and At1g64500 (THRUMIN1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Physiological Evaluation of Actin Probe Effects on Chloroplast Photorelocation Movements.
Supplemental Figure 2. Reorganization of Cp-Actin Filaments Visualized in Various Fluorescent Protein–Tagged Actin Binding Protein Lines.
Supplemental Figure 3. Relationship between Asymmetric Cp-Actin Filaments and the Velocity of Chloroplast Avoidance Movements.
Supplemental Figure 4. Changes in the Total Cp-Actin Filament Fluorescence at the Front (Brown Line) and Rear (Blue Line) Regions of the Moving Chloroplasts.
Supplemental Figure 5. Involvement of Phototropins in Asymmetric Cp-Actin Filament Formation during Chloroplast Avoidance Movements.
Supplemental Figure 6. Blue Light–Induced Disappearance of Cp-Actin Filaments in the Wild-Type and phototropin-Deficient Mutant Cells.
Supplemental Figure 7. Blue Light–Induced Disappearance of Cp-Actin Filaments probed with THRUMIN1-GFP in the Wild-Type Cells.
Supplemental Figure 8. Disappearance of the Cp-Actin Filaments under TIRFM.
Supplemental Figure 9. Schematic Diagram of the Binary Vectors Used in This Study.
Supplemental Figure 10. Mesophyll Protoplast Preparation for TIRFM.
Supplemental Movie 1. Reorganization of Cp-Actin Filaments during the Blue Light–Induced Chloroplast Avoidance Response.
Supplemental Movie 2. Reorganization of THRUMIN1-GFP during the Blue Light–Induced Chloroplast Avoidance Response.
Supplemental Movie 3. Reorganization of Cp-Actin Filaments in a thrumin1 Mutant Cell under a Condition Inducing the Blue Light–Induced Chloroplast Avoidance Response.
Supplemental Movie 4. Reorganization of THRUMIN1-GFP in a chup1 Mutant Cell under a Condition Inducing the Blue Light–Induced Chloroplast Avoidance Response.
Supplemental Movie 5. Reorganizations of Cp-Actin Filaments on the Leading Edge of Chloroplasts during an Avoidance Response.
Supplemental Movie 6. Reorganization of Cp-Actin Filaments during the Accumulation Response following the Avoidance Response.
Supplemental Movie 7. Reorganizations of Cp-Actin Filaments and the Chloroplast Envelope during the Avoidance Response in Side-Viewed Chloroplasts.
Supplemental Movie 8. Reorganization of Chloroplast Envelope proved by OEP7-YFP during the Avoidance Response.
Supplemental Movie 9. Disappearance of the Cp-Actin Filaments Captured by CLSM in a Wild-Type Cell.
Supplemental Movie 10. Cp-Actin Filament Dynamics Captured by TIRFM in a Wild-Type Cell.
Supplemental Movie 11. Cp-Actin Filament Dynamics Captured by TIRFM in a phot1 Mutant Cell.
Supplemental Movie 12. Cp-Actin Filament Dynamics Captured by TIRFM in a phot2 Mutant Cell.
Supplemental Movie 13. Cp-Actin Filament Dynamics Captured by TIRFM in a phot1 phot2 Mutant Cell.
Supplemental Movie 14. Cp-Actin Filament Dynamics Captured by TIRFM in a chup1 Mutant Cell.
Supplemental Movie 15. Reorganization of the Cp-Actin Filaments in Mock-Treated Cells for 1 h.
Supplemental Movie 16. Reorganizations of the Cp-Actin Filaments in 50 mM BDM-Treated Cells for 1 h.
Supplemental Movie 17. Cp-Actin Filament Dynamics Captured by CLSM in a Cell Treated with 50 mM BDM for 1 h.
Supplemental Movie 18. Reorganizations of the Cp-Actin Filaments in 50 mM BDM-Treated Cells for 2 h.
Supplemental Movie 19. Cp-Actin Filament Dynamics Captured by CLSM in a Cell Treated with 50 mM BDM for 2 h.
Supplemental Movie 20. Disappearance of the Fragmented Cp-Actin Filaments via the Chloroplast Edge.
Acknowledgments
We thank Takachika Tanaka for excellent technical assistance, Takashi Ueda for LIFEACT-YFP transgenic seeds, and Winslow R. Briggs for critical reading of the article. We also thank the ABRC for seed stocks. This work was supported in part by Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants 17084006 and 23120523 to M.W. and 21770050 to S.-G.K.) and the Japan Society for the Promotion of Science (Grant 20227001 to M.W.).
AUTHOR CONTRIBUTIONS
S.-G.K. and M.W. designed the experiments and analyzed the data. S.-G.K. and Y.A. performed the experiments. S.-G.K. and N.S. prepared the plant materials. Y.A. and T.Y. contributed new analytic/computational tools. S.-G.K. and M.W. wrote the article.
Glossary
- cp-actin
chloroplast-actin
- YFP
yellow fluorescent protein
- CLSM
confocal laser scanning microscopy
- TIRFM
total internal reflection fluorescence microscopy
- GFP
green fluorescent protein
- RFP
red fluorescent protein
- BDM
2,3-butanedione monoxime
References
- Avisar D., Prokhnevsky A.I., Makarova K.S., Koonin E.V., Dolja V.V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146: 1098–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaś A.K., Aggarwal C., Łabuz J., Sztatelman O., Gabryś H. (2012). Blue light signalling in chloroplast movements. J. Exp. Bot. 63: 1559–1574 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: A team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723 [DOI] [PubMed] [Google Scholar]
- Briggs W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13: 993–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J.M., Swartz T.E., Bogomolni R.A., Briggs W.R. (2002). Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32: 205–219 [DOI] [PubMed] [Google Scholar]
- Era A., Tominaga M., Ebine K., Awai C., Saito C., Ishizaki K., Yamato K.T., Kohchi T., Nakano A., Ueda T. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper S.M., Christie J.M., Gardner K.H. (2004). Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry 43: 16184–16192 [DOI] [PubMed] [Google Scholar]
- Harper S.M., Neil L.C., Gardner K.H. (2003). Structural basis of a phototropin light switch. Science 301: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Huala E., Oeller P.W., Liscum E., Han I.S., Larsen E., Briggs W.R. (1997). Arabidopsis NPH1: A protein kinase with a putative redox-sensing domain. Science 278: 2120–2123 [DOI] [PubMed] [Google Scholar]
- Hussey P.J., Ketelaar T., Deeks M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57: 109–125 [DOI] [PubMed] [Google Scholar]
- Ichikawa S., Yamada N., Suetsugu N., Wada M., Kadota A. (2011). Red light, Phot1 and JAC1 modulate Phot2-dependent reorganization of chloroplast actin filaments and chloroplast avoidance movement. Plant Cell Physiol. 52: 1422–1432 [DOI] [PubMed] [Google Scholar]
- Inoue S.-I., Kinoshita T., Matsumoto M., Nakayama K.I., Doi M., Shimazaki K.-I. (2008). Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. USA 105: 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S.-I., Matsushita T., Tomokiyo Y., Matsumoto M., Nakayama K.I., Kinoshita T., Shimazaki K.-I. (2011). Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiol. 156: 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Gabryś H., Capel J., Alonso J.M., Ecker J.R., Cashmore A.R. (2001). Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410: 952–954 [DOI] [PubMed] [Google Scholar]
- Kadota A., Yamada N., Suetsugu N., Hirose M., Saito C., Shoda K., Ichikawa S., Kagawa T., Nakano A., Wada M. (2009). Short actin-based mechanism for light-directed chloroplast movement in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 13106–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S., Kato T., Tabata S., Okada K., Wada M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291: 2138–2141 [DOI] [PubMed] [Google Scholar]
- Kagawa T., Wada M. (1996). Phytochrome- and blue-light-absorbing pigment-mediated directional movement of chloroplasts in dark-adapted prothallial cells of fern Adiantum as analyzed by microbeam irradiation. Planta 198: 488–493 [Google Scholar]
- Kagawa T., Wada M. (2004). Velocity of chloroplast avoidance movement is fluence rate dependent. Photochem. Photobiol. Sci. 3: 592–595 [DOI] [PubMed] [Google Scholar]
- Kandasamy M.K., Gilliland L.U., McKinney E.C., Meagher R.B. (2001). One plant actin isovariant, ACT7, is induced by auxin and required for normal callus formation. Plant Cell 13: 1541–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Kagawa T., Oikawa K., Suetsugu N., Miyao M., Wada M. (2002). Chloroplast avoidance movement reduces photodamage in plants. Nature 420: 829–832 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Anthony R.G., Hussey P.J. (2004). Green fluorescent protein-mTalin causes defects in actin organization and cell expansion in Arabidopsis and inhibits actin depolymerizing factor’s actin depolymerizing activity in vitro. Plant Physiol. 136: 3990–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Kagawa T. (2009). Blue light-induced chloroplast avoidance and phototropic responses exhibit distinct dose dependency of PHOTOTROPIN2 in Arabidopsis thaliana. Photochem. Photobiol. 85: 1260–1264 [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M., Shimazaki K.-I. (2001). Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414: 656–660 [DOI] [PubMed] [Google Scholar]
- Kodama Y., Suetsugu N., Kong S.-G., Wada M. (2010). Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 19591–19596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong S.-G., Kinoshita T., Shimazaki K.-I., Mochizuki N., Suzuki T., Nagatani A. (2007). The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses. Plant J. 51: 862–873 [DOI] [PubMed] [Google Scholar]
- Kong S.-G., Suzuki T., Tamura K., Mochizuki N., Hara-Nishimura I., Nagatani A. (2006). Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 45: 994–1005 [DOI] [PubMed] [Google Scholar]
- Kong S.-G., Wada M. (2011). New insights into dynamic actin-based chloroplast photorelocation movement. Mol. Plant 4: 771–781 [DOI] [PubMed] [Google Scholar]
- Kong S.-G., Kagawa T., Wada M., Nagatani A. (2013a). A carboxy-terminal membrane association domain of phototropin 2 is necessary for chloroplast movement. Plant Cell Physiol. 54: 57–68 [DOI] [PubMed] [Google Scholar]
- Kong S.-G., Suetsugu N., Kikuchi S., Nakai M., Nagatani A., Wada M. (2013b). Both phototropin 1 and 2 localize on the chloroplast outer membrane with distinct localization activity. Plant Cell Physiol. 54: 80–92 [DOI] [PubMed] [Google Scholar]
- Kost B., Spielhofer P., Chua N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kozuka T., Kong S.-G., Doi M., Shimazaki K.-I., Nagatani A. (2011). Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 23: 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeszowiec W., Gabryś H. (2007). Phototropin mediated relocation of myosins in Arabidopsis thaliana. Plant Signal. Behav. 2: 333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Wakao S., Fischer B.B., Niyogi K.K. (2009). Sensing and responding to excess light. Annu. Rev. Plant Biol. 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Luesse D.R., DeBlasio S.L., Hangarter R.P. (2006). Plastid movement impaired 2, a new gene involved in normal blue-light-induced chloroplast movements in Arabidopsis. Plant Physiol. 141: 1328–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCurdy D.W. (1999). Is 2,3-butanedione monoxime an effective inhibitor of myosin-based activities in plant cells? Protoplasma 209: 120–125 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., Kanegae T., Niwa Y., Kadota A., Wada M. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15: 2805–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa K., Yamasato A., Kong S.-G., Kasahara M., Nakai M., Takahashi F., Ogura Y., Kagawa T., Wada M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 148: 829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paves H., Truve E. (2007). Myosin inhibitors block accumulation movement of chloroplasts in Arabidopsis thaliana leaf cells. Protoplasma 230: 165–169 [DOI] [PubMed] [Google Scholar]
- Peremyslov V.V., Prokhnevsky A.I., Dolja V.V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22: 1883–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T., Kagawa T., Kasahara M., Swartz T.E., Christie J.M., Briggs W.R., Wada M., Okada K. (2001). Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. USA 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt von Braun S., Schleiff E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227: 1151–1159 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T., Yokota E. (2004). Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16: 68–72 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Wada M. (2007). Chloroplast photorelocation movement mediated by phototropin family proteins in green plants. Biol. Chem. 388: 927–935 [DOI] [PubMed] [Google Scholar]
- Suetsugu N., Dolja V.V., Wada M. (2010a). Why have chloroplasts developed a unique motility system? Plant Signal. Behav. 5: 1190–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu, N., and Wada, M.(2009). Chloroplast photorelocation movement. In The Chloroplast: Interactions with the Environment, A.S. Sandelius and H. Aronsson, eds (Berlin, Heidelberg: Springer), pp. 235–266. [Google Scholar]
- Suetsugu N., Yamada N., Kagawa T., Yonekura H., Uyeda T.Q.P., Kadota A., Wada M. (2010b). Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 8860–8865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztatelman O., Waloszek A., Banaś A.K., Gabryś H. (2010). Photoprotective function of chloroplast avoidance movement: In vivo chlorophyll fluorescence study. J. Plant Physiol. 167: 709–716 [DOI] [PubMed] [Google Scholar]
- Szymanski D.B. (2009). Plant cells taking shape: New insights into cytoplasmic control. Curr. Opin. Plant Biol. 12: 735–744 [DOI] [PubMed] [Google Scholar]
- Takagi S., Takamatsu H., Sakurai-Ozato N. (2009). Chloroplast anchoring: its implications for the regulation of intracellular chloroplast distribution. J. Exp. Bot. 60: 3301–3310 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Badger M.R. (2011). Photoprotection in plants: A new light on photosystem II damage. Trends Plant Sci. 16: 53–60 [DOI] [PubMed] [Google Scholar]
- Takemiya A., Inoue S., Doi M., Kinoshita T., Shimazaki K. (2005). Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Kitamura K., Saito K., Iwane A.H., Yanagida T. (1997). Single molecule imaging of fluorophores and enzymatic reactions achieved by objective-type total internal reflection fluorescence microscopy. Biochem. Biophys. Res. Commun. 235: 47–53 [DOI] [PubMed] [Google Scholar]
- Tsuboi H., Wada M. (2011). Chloroplasts can move in any direction to avoid strong light. J. Plant Res. 124: 201–210 [DOI] [PubMed] [Google Scholar]
- Tsuboi H., Wada M. (2012). Distribution pattern changes of actin filaments during chloroplast movement in Adiantum capillus-veneris. J. Plant Res. 125: 417–428 [DOI] [PubMed] [Google Scholar]
- Tsuboi H., Yamashita H., Wada M. (2009). Chloroplasts do not have a polarity for light-induced accumulation movement. J. Plant Res. 122: 131–140 [DOI] [PubMed] [Google Scholar]
- Ueda H., Yokota E., Kutsuna N., Shimada T., Tamura K., Shimmen T., Hasezawa S., Dolja V.V., Hara-Nishimura I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107: 6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Honing H.S., van Bezouwen L.S., Emons A.M., Ketelaar T. (2011). High expression of Lifeact in Arabidopsis thaliana reduces dynamic reorganization of actin filaments but does not affect plant development. Cytoskeleton (Hoboken) 68: 578–587 [DOI] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kagawa T., Sato Y. (2003). Chloroplast movement. Annu. Rev. Plant Biol. 54: 455–468 [DOI] [PubMed] [Google Scholar]
- Wada, M., and Kong, S.-G. (2011). Analysis of chloroplast movement and relocation in Arabidopsis In Chloroplast Research in Arabidopsis: Methods and Protocols, R.P. Jarvis, ed (Totowa, NJ: Humana), pp. 87–102. [DOI] [PubMed] [Google Scholar]
- Wada M., Suetsugu N. (2004). Plant organelle positioning. Curr. Opin. Plant Biol. 7: 626–631 [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Yoo C.M., Blancaflor E.B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536 [DOI] [PubMed] [Google Scholar]
- Whippo C.W., Khurana P., Davis P.A., DeBlasio S.L., DeSloover D., Staiger C.J., Hangarter R.P. (2011). THRUMIN1 is a light-regulated actin-bundling protein involved in chloroplast motility. Curr. Biol. 21: 59–64 [DOI] [PubMed] [Google Scholar]
- Yamada N., Suetsugu N., Wada M., Kadota A. (2011). Phototropin-dependent biased relocalization of cp-actin filaments can be induced even when chloroplast movement is inhibited. Plant Signal. Behav. 6: 1651–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H., Sato Y., Kanegae T., Kagawa T., Wada M., Kadota A. (2011). Chloroplast actin filaments organize meshwork on the photorelocated chloroplasts in the moss Physcomitrella patens. Planta 233: 357–368 [DOI] [PubMed] [Google Scholar]