A nox mutant of Arabidopsis, devoid of xanthophylls, was reduced in photosystem I function due to enhanced protein turnover and impaired translation of the PsaA/PsaB messengers. These findings imply that xanthophylls, besides their role in light harvesting and photoprotection, are also needed for photosystem I core translation and stability and are thus indispensable for autotrophic growth.
Abstract
Carotenes, and their oxygenated derivatives xanthophylls, are essential components of the photosynthetic apparatus. They contribute to the assembly of photosynthetic complexes and participate in light absorption and chloroplast photoprotection. Here, we studied the role of xanthophylls, as distinct from that of carotenes, by characterizing a no xanthophylls (nox) mutant of Arabidopsis thaliana, which was obtained by combining mutations targeting the four carotenoid hydroxylase genes. nox plants retained α- and β-carotenes but were devoid in xanthophylls. The phenotype included depletion of light-harvesting complex (LHC) subunits and impairment of nonphotochemical quenching, two effects consistent with the location of xanthophylls in photosystem II antenna, but also a decreased efficiency of photosynthetic electron transfer, photosensitivity, and lethality in soil. Biochemical analysis revealed that the nox mutant was specifically depleted in photosystem I function due to a severe deficiency in PsaA/B subunits. While the stationary level of psaA/B transcripts showed no major differences between genotypes, the stability of newly synthesized PsaA/B proteins was decreased and translation of psaA/B mRNA was impaired in nox with respect to wild-type plants. We conclude that xanthophylls, besides their role in photoprotection and LHC assembly, are also needed for photosystem I core translation and stability, thus making these compounds indispensable for autotrophic growth.
INTRODUCTION
Carotenoids are a group of C40 pigments that contains extended conjugated double-bond systems conferring strong absorption of light and antioxidant properties. These compounds are widely distributed among taxa, including fungi, cyanobacteria, red and green algae, and land plants (Kull and Pfander, 1995; Britton et al., 2004). More than 700 different carotenoids have been described, and this structural diversity has likely evolved in relation to their many functions: They act as vitamins and hormones, as substrate for the synthesis of volatile products, and as colors in flowers and fruits (DellaPenna and Pogson, 2006; Cuttriss and Pogson, 2006). Furthermore, carotenoids play essential roles in higher plant photosynthesis, as components of the photosynthetic apparatus (Nelson and Ben Shem, 2004).
Carotenoids are composed of two major groups, namely, carotenes and their oxygenated derivatives, the xanthophylls. In higher plants, β-carotene binds to reaction center subunits of both photosystems, while xanthophylls are accessory pigments and structural elements of light-harvesting complexes (LHCs) (Bassi et al., 1993). Together with β-carotene, xanthophylls act as photoreceptors, absorbing light energy, which is used in photosynthetic electron transport. Also, they are photoprotectants of the photosynthetic apparatus from excess light (EL) and from the reactive oxygen species (ROS) generated during oxygenic photosynthesis (Niyogi, 2000; Dall’Osto et al., 2005; Havaux et al., 2007).
The xanthophyll composition of higher plants is remarkably conserved and includes four major xanthophylls: the β,ϵ-xanthophyll lutein and the three β,β-xanthophylls, violaxanthin (Viola), neoxanthin (Neo), and zeaxanthin (Zea; Demmig-Adams and Adams, 1992b). Recent studies on carotenoid biosynthetic mutants of Arabidopsis thaliana have improved our understanding of the biosynthetic pathway (Pogson et al., 1998; Fiore et al., 2006; Kim and DellaPenna, 2006; North et al., 2007; Dall’Osto et al., 2007b; Kim et al., 2009) (see Supplemental Figure 1 online). β-Carotene, the precursor of β,β-xanthophylls, is formed from lycopene by the symmetrical action of β-cyclase, while the β,ϵ-carotenoid α-carotene is formed by the combined action of both β-cyclase and ϵ-cyclase on lycopene (DellaPenna and Pogson, 2006), thus making a bifurcation in the pathway between the β,β- and β,ϵ-xanthophyll branches.
Hydroxylation of α-carotene produces lutein, the most abundant xanthophyll in leaves, while hydroxylation of β-carotene yields Zea, which is rapidly epoxidized to antheraxanthin and Viola and then converted to Neo (North et al., 2007). However, EL activates the deepoxidation of Viola, leading to the accumulation of Zea (Yamamoto et al., 1962; Demmig-Adams and Adams, 1992b). Biosynthesis of lutein requires one β- and one ϵ-ring hydroxylation of α-carotene, while synthesis of β-xanthophylls needs hydroxylation of both β-carotene β-rings. Arabidopsis mutants impaired in carotenoid biosynthesis showed that heme-containing cytochrome P450 hydroxylases CYP97A3 (LUT5) express a major β-carotene hydroxylase activity in vivo, while CYP97C1 (LUT1) is primarily involved in the hydroxylation of the ϵ ring of α-carotene (Tian et al., 2004; Kim and DellaPenna, 2006; Fiore et al., 2012). Non-heme di-iron monoxygenases CHY1 and CHY2 primarily catalyze the hydroxylation of β-carotene. Indeed, the chy1 chy2 double mutant showed an 80% reduction in β-carotene–derived xanthophylls (Tian et al., 2003; DellaPenna and Pogson, 2006). Recently, the expression of each carotenoid hydroxylase, as well as their biochemical activity, was assessed in vivo using knockout genotypes (Kim et al., 2009; Fiore et al., 2012).
A remarkable characteristic of higher plant xanthophylls is their similar spectral properties in the visible region (Kull and Pfander, 1995), implying that a similar light-harvesting activity operates across species. This evidence is apparently incoherent with the conservation of composition and relative abundance across plant taxa (Demmig-Adams and Adams, 1992a; Koniger et al., 1995), suggesting that each xanthophyll species serves a specific biological function. The phenotypes of Arabidopsis carotenoid biosynthetic mutants with altered xanthophyll composition suggest that the combination of constitutive contributions of lutein, Viola, and Neo, integrated by the action of Zea in EL, is the most functional solution for facing oxidative stress in highly variable natural conditions. Indeed, lack of lutein and/or Zea decreases the capacity for photoprotection in EL (Niyogi et al., 2001), while lack of Neo increases sensitivity to superoxide anions, formed in the Mehler’s reaction (Dall’Osto et al., 2007a). Depletion of Viola, as in the chy1 chy2 lut5 mutant, caused extreme photosensitivity (Dall’Osto et al., 2007b). Lutein bound to site L1 of all LHC proteins is essential for protein folding and for chlorophyll triplet quenching (Croce et al., 1999; Formaggio et al., 2001). Viola binds to L2 and/or V1 sites, while Neo has its own specific binding site called N1 (Liu et al., 2004; Caffarri et al., 2007; Pan et al., 2011). Zea is produced in EL and replaces Viola in LHC protein structure (Dall’Osto et al., 2005), leading to (1) enhancement of nonphotochemical quenching (NPQ) (Holt et al., 2005) and (2) the protection of thylakoid lipids from peroxidation (Baroli et al., 2003).
Although the above observations support the notion that each xanthophyll species has a specific role in photoprotection, their collected importance as a class of compounds distinct from carotenes has not been assessed so far. In this work, we isolated and characterized the Arabidopsis chy1 chy2 lut1 lut5 quadruple mutant (hereafter referred to as nox for the sake of brevity), which lacks all xanthophylls but retains carotenes. The nox mutant showed a complete depletion of LHC proteins, inactivation of NPQ, and high sensitivity to photooxidative stress with respect to the wild type. Besides, nox plants showed decreased photosynthetic electron transport and depletion of photosystem I (PSI), an unexpected feature, since β-carotene, the only carotenoid ligand in the core complex, is conserved in the mutant. PSI content declined due to enhanced protein turnover and impaired translation of the PsaA/PsaB messengers. Thus, carotenes alone were unable to sustain photosynthetic electron transport even in dim light. These findings imply that xanthophylls, besides their well-known role in light harvesting and photoprotection, are also essential for the biogenesis of the PSI core complex.
RESULTS
nox, a Quadruple Knockout Mutant of Arabidopsis, Accumulates α- and β-Carotene as the Only Carotenoids
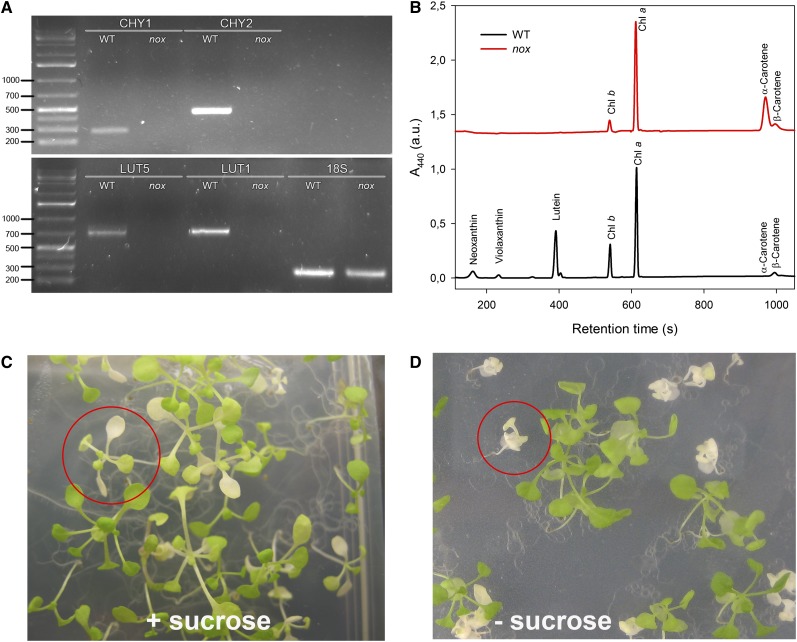
nox plants were obtained by crossing homozygous T-DNA mutants carrying insertions in genes encoding the two non-heme hydroxylases (chy1 and chy2), the ϵ-ring hydroxylase (lut1), and its paralog (lut5) (Fiore et al., 2006). Absence of the transcripts for the mutated genes was verified via RT-PCR (Figure 1A), while pigment composition of nox plants was verified by HPLC (Figure 1B): nox plants contained α- and β-carotene as the only carotenoids, but lacked xanthophylls. Homozygous knockout mutant seedlings developed white cotyledons and were lethal when grown on soil. They could only be maintained on medium supplemented with 3% Suc under low light (20 μmol photons m−2 s−1) (Figures 1C and 1D). Under sterile heterotrophic conditions, leaf morphology and size were unaltered in nox in comparison to wild-type plants (Figure 1C). Mutant seedlings were paler, with strongly reduced (∼17%) chlorophyll content per leaf area compared with wild-type plants; the chlorophyll a/b ratio was 7.5 in nox versus 2.9 in wild-type seedlings, while the chlorophyll/carotenoid ratio was less affected (Table 1).
Figure 1.
Genetic and Biochemical Characterization of Wild-Type and nox Arabidopsis Plants.
(A) RT-PCR amplification pattern of gene-specific transcripts. The 18S transcript was used as a control for the reverse transcriptase reaction. Details of primer sequences and expected sizes of the PCR products have been reported (Fiore et al., 2006). DNA ladder, 1 kb plus (75 to 20,000 bp). WT, the wild type.
(B) Analysis of pigment content of leaves from wild-type and nox plants. Separation of lipid-soluble pigments was based on HPLC analysis. Each chromatogram represents absorbance at 440 nm (A440) of pigments extracted from a dark-adapted leaf. Chromatograms were vertically shifted for better comparison. a.u., arbitrary units.
(C) and (D) Phenotype of mutant plants, grown either heterotrophically (C) or autotrophically (D) for 2 weeks. Representative mutant plants are circled.
[See online article for color version of this figure.]
Table 1. Pigment Content and PSII/PSI Functionality, Determined on the Leaves of Wild-Type and nox Arabidopsis Plants.
| Genotypes | Chlorophyll a/b | Chlorophyll/Carotenoid | Chlorophyll/m2 (mg) | Fv/Fm | t2/3−1 (·10−3, ms−1) | ΔAmax (705 nm, a.u.) |
|---|---|---|---|---|---|---|
| The wild type | 2.9 ± 0.1 | 3.5 ± 0.2 | 171.2 ± 4.2 | 0.77 ± 0.02 | 6.1 ± 1.1 | 68.1 ± 6.6 |
| nox | 7.5 ± 0.6* | 3.0 ± 0.3 | 31.3 ± 4.1* | 0.44 ± 0.09* | 1.8 ± 0.3* | 4.9 ± 1.2* |
Data are expressed as mean ± sd (n > 5), and significantly different values (Student’s t test, P < 0.05) with respect to the wild type are marked with an asterisk. a.u., arbitrary units.
Chloroplast ultrastructure was observed by transmission electron microscopy. Wild-type chloroplasts showed the characteristic organization with grana stacks interconnected by stroma membranes and small starch granules in most sections (see Supplemental Figures 2A and 2B online). Instead, the nox plastids were ∼2 times smaller and were depleted in thylakoids and starch granules. The plastids of the mutant had rudimentary thylakoids, lacking grana stacks, and consisted of stroma lamellae only, arranged into concentric monovesicles (see Supplemental Figures 2C and 2D online), resembling a sliced onion in section. It appears that thylakoid development was arrested at an early developmental stage of two disks grana.
nox Is Impaired in Photoprotection and Photosynthetic Electron Transport
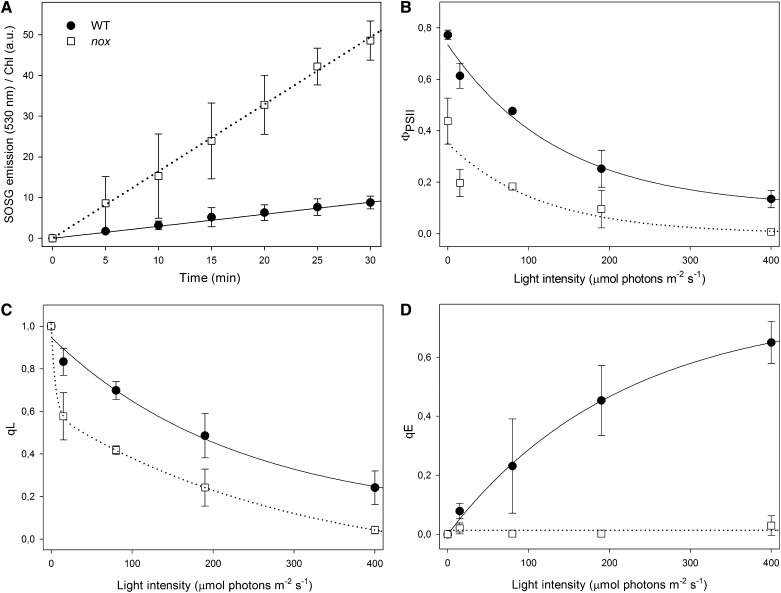
When photosynthetic organisms are exposed to EL, photooxidative stress occurs with the production of ROS in the chloroplast; in particular, singlet oxygen (1O2) derives from the reaction of chlorophyll triplets with molecular oxygen (Krieger-Liszkay, 2005). A feature of ROS is their high reactivity, leading to oxidative damage of biomolecules. To analyze the effect of missing xanthophylls on the yield of different ROS, we quantified the release of 1O2 directly in wild-type and nox leaves, infiltrated with singlet oxygen sensor green (SOSG), a 1O2-specific fluorescent probe. After illumination with moderate light intensity (200 μmol photons m−2 s−1, 22°C), nox leaves showed, at each time point, a far higher release of 1O2 per chlorophyll with respect to the wild type (Figure 2A). These results showed that xanthophyll-depleted nox leaves were impaired in the capacity of preventing 1O2 release compared with xanthophyll-containing wild-type leaves.
Figure 2.
Analysis of 1O2 Release and Chlorophyll Fluorescence during Photosynthesis in Wild-Type and nox Leaves.
(A) Wild-type and mutant leaves were infiltrated with SOSG, a fluorogenic probe that exhibits increased fluorescence yield upon reaction with 1O2. The increase in probe signal was monitored during illumination with red actinic light (200 μmol photon m−2 s−1, 600 < λ < 750 nm, 22°C); at each time point, the fluorescence signal was normalized to the leaf chlorophyll content. a.u., arbitrary units; WT, the wild type.
(B) to (D) Chlorophyll fluorescence was monitored in dark-adapted leaves during photosynthesis by following photosynthetic efficiency (ΦPSII) (B), PQ pool redox state (qL) (C), and the ΔpH-dependent component (qE) of thermal energy dissipation (NPQ) (D). Leaves were illuminated for 20 min, and parameters were determined during steady state photosynthesis. Symbols and error bars show mean ± sd (n > 3). Statistical analysis confirmed that parameters of nox leaves were always significantly different with respect to that of the wild type (Student’s t test, P < 0.05).
nox plants accumulate both α- and β-carotene, while wild-type leaves accumulate β-carotene only. Since both α- and β-carotene can bind to photosynthetic core complexes, it might be asked whether photosensitivity could be ascribed to the accumulation of α-carotene into PSI and photosystem II (PSII) core complexes. To address this question, 1O2 production (see Supplemental Figures 3A to 3C online) and photobleaching kinetics under EL (see Supplemental Figures 3D to 3F online) were measured on PSII and PSI complexes isolated from either wild-type and lut5 leaves, which accumulate β-carotene and α-carotene, respectively (Dall’Osto et al., 2007b). β-Carotene substitution in lut5 did not affect 1O2 yield or photobleaching kinetics when compared with complexes isolated from the wild type at all light intensities tested. On this basis, we can exclude that the enhanced sensitivity of the photosynthetic apparatus of nox is due to α-carotene accumulation, implying that it is rather attributed to the absence of xanthophylls.
Photosynthetic activity was investigated by measuring both PSII and PSI function in wild-type and nox leaves. The t2/3 of the chlorophyll fluorescence rise in DCMU-treated leaves was taken as a measure of the functional antenna size of PSII (Malkin et al., 1981). It was reduced by 70% in nox with respect to the wild type (Table 1), implying that the light-harvesting function is severely reduced in the absence of xanthophylls.
Further chlorophyll fluorescence analysis yielded insight into PSII function. Upon growth at 20 μmol photons m−2 s−1, nox mutants had Fv/Fm ratios (the maximal photochemical yield of PSII) of 0.44 versus 0.77 for the wild type (Table 1). Also, nox plants showed a decrease in the PSII photochemical efficiency (ФPSII) at all light intensities tested (Figure 2B); this parameter is equivalent to the quantum yield of linear electron flux through PSII and is linearly related to the yield of O2 evolution. The fraction of oxidized PSII acceptor QA (qL) was lower in nox compared with the wild type (Figure 2C). Finally, the capacity for heat dissipation (qE; measured as the rapidly reversible component of NPQ) was null at a light intensity up to 400 μmol photons m−2 s−1, while the wild type showed an increase in qE with increasing light intensity, implying that xanthophylls are needed for qE activity (Figure 2D).
It should be noted that, although reduced, a qL activity was detected in nox leaves, implying that photosynthetic electron flow through PSII was active, while a bottleneck is likely to be localized at later electron transfer steps. Further insight into the light-harvesting efficiency and electron transport activity downstream of QA was obtained by analyzing the fluorescence induction and relaxation in dark-adapted leaves. To test the hypothesis that electron transport is restricted in the QA to QB step in nox plants, QA reoxidation kinetics upon a single turnover flash were analyzed in order to assess the capacity for QA reoxidation by the plastoquinone (PQ) pool. In short, when PSII is excited by a very short flash of saturating light, QA is fully reduced to reach Fm and then the kinetic of Fv recovery in the dark is determined, which depends on PQ diffusing from the surrounding membrane domains to the PSII QB site and on the total amount of reducible PQ (Sane et al., 2003). Since similar fluorescence decay kinetics were measured in wild-type and nox leaves (see Supplemental Figure 4A online), no restrictions to the accessibility of the QB site to PQ are likely to be present in either genotype.
Comparison of the fluorescence induction curves showed that, upon switching from dark to light, the time needed for reaching the Fmax was longer in the mutant versus the wild type, consistent with the lower functional antenna size of nox plants (Table 1, t2/3−1). Nevertheless, fluorescence decline from Fmax to stationary fluorescence, evident after ∼10 s of continuous illumination in the wild type as a consequence of the activation of the Benson-Calvin cycle, was slower in the mutant (see Supplemental Figure 4B online). Taken together, these results suggest that the nonphotoautotrophic mutant phenotype is the consequence of a block in light-driven electron transport, which appears to occur at the level of cytochrome b6f, plastocyanin, or PSI. To further detail the origin of electron transport restriction, we proceeded to monitor PSI activity in vivo, by measuring the total amount of photooxidizable P700 in intact leaves. Thus, absorbance changes at 705 nm (ΔAmax) were measured upon far-red illumination, clearly showing that PSI efficiency was severely reduced in the nox mutant compared with the wild type (Table 1), suggesting that xanthophyll depletion might affect either PSI activity and/or accumulation of the complex.
nox Is Primarily Defective in PSI Accumulation
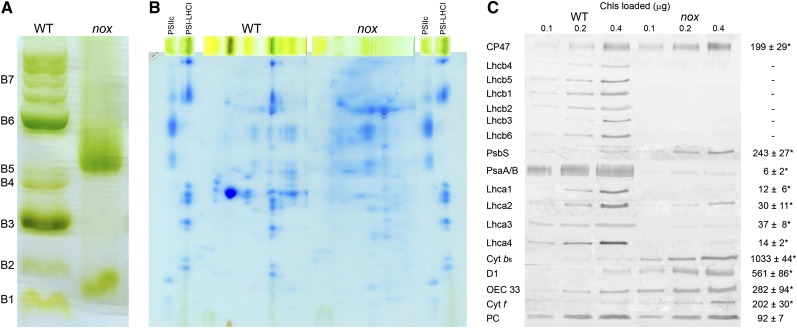
To investigate the state of PSI, pigment-protein complexes were separated in wild-type and mutant plants by nondenaturing Deriphat-PAGE, upon solubilization of thylakoid membranes with 0.6% dodecyl-α-d-maltoside (α-DM). Several major green bands were resolved in the wild type (Figure 3A): Four green bands with high apparent molecular mass were detected in the upper part of the gel (collectively marked as B7) containing undissociated PSII supercomplexes with different Lhcb complements; the PSI-LHCI complex was a major band (B6), while the components of the PSII-LHCII complexes migrated as multiple green bands with increasing mobility in the gel, namely, PSII core monomers (B5), the Lhcb4/Lhcb6/LHCII-M supercomplex (B4) (Dainese et al., 1992), the LHCII trimer (B3), and the Lhcb monomeric band (B2). Finally, a band (B1) migrated with the front and contained free pigments. The electrophoretic profile of nox thylakoid membranes was markedly different (Figure 3A): Only two green bands could be distinguished, corresponding to bands B5 and B1 of the wild type. To characterize their polypeptide composition, the green lanes from the first dimension were excised and further fractionated by denaturing SDS-PAGE in a second dimension (Figure 3B). LHC apoproteins, well resolved in the gel containing the wild-type sample, were not detectable in the case of nox. From the major band (B5) in nox, PSII subunits were separated, namely, D1/D2 and CP43/CP47. PSI core complex subunits (PsaA/PsaB) were not evident. Fluorometric measurements of wild-type and nox chloroplasts at 77K showed that the PSII emission band at 685 nm was present in the mutant, implying it originated from the PSII core rather than from Lhcb components. Moreover, the PSI-specific fluorescence band, the largest in amplitude in the wild-type spectrum, was strongly reduced in nox (see Supplemental Figure 5 online); the 733-nm peak emission was shifted to 728 nm in nox, while the band at 733 nm was reduced to a shoulder of the spectrum.
Figure 3.
Biochemical Characterization of Thylakoid Membranes from Wild-Type and nox Plants.
(A) Nondenaturing Deriphat-PAGE separation of pigmented thylakoid complexes from wild-type (WT) and nox plants. Numbering of green bands (B1 to B7) is reported.
(B) Two-dimensional resolution of thylakoid protein complexes. α-DM–solubilized thylakoid protein complexes purified from wild-type and nox leaves were fractionated by Deriphat-PAGE in the first dimension and SDS-PAGE (Tris-Tricine system) in the second. Gel slices containing purified PSII core complex (PSIIc) and PSI-LHCI complexes were loaded as reference.
(C) Immunotitration of photosynthetic subunits. Amounts of chlorophylls loaded on the gel were 0.1, 0.2, and 0.4 μg. The abundance of each subunit in nox (right column) was expressed as a percentage of the corresponding content in the wild type. The Arabidopsis genome encodes two plastocyanin isoforms (PetE1/2), which were resolved both in the wild type and nox and detected in the same relative abundance. Data are reported as mean ± sd (n = 3), and significantly different values (Student’s t test, P < 0.05) are marked (asterisk). Cyt, cytochrome; OEC 33, oxygen-evolving complex 33-kD subunit; PC, plastocyanin.
[See online article for color version of this figure.]
The level of selected thylakoid proteins, relative to the wild type on a chlorophyll basis, was determined by immunoblot analysis (Figure 3C): All Lhcb subunits were undetectable in nox, while the Lhca content was strongly reduced, amounting to ∼35% of wild-type values for Lhca2/3 and ∼15% for Lhca1/4. Plastocyanin content was the same in both genotypes, while subunits of the PSII core complex (D1, OEC33, and CP47), cytochrome b6f complex, and PsbS were present in higher amounts in nox with respect to the wild type. By contrast, the two major plastid-encoded PSI core subunits (PsaA and PsaB) were strongly decreased in nox, reaching ∼6% with respect to the wild-type level. One possible reason for the downregulation of protein level is destabilization due to missing cofactors, such as Fe-S centers, which was shown to be important for the accumulation of the PSI core complex (Amann et al., 2004; Yabe et al., 2004). To investigate whether the nox mutation affects iron-sulfur cluster assembly, we measured the amount of plastid Fe-S cluster-containing proteins. Ferredoxin (which contains a 2Fe-2S cluster), NDH-L and NDH-K (subunits of the NADPH-dependent dehydrogenase, a 4Fe-4S cluster protein), and the chloroplast HCF101 subunits (a scaffold protein for 4Fe-4S cluster assembly) were present in the same amount in wild-type and nox leaves (see Supplemental Figure 6 online).
To verify whether this dramatic effect on the steady state level of LHC and PSI pigment proteins was due to the presence of an unrelated mutation, we analyzed single knockout mutants for carotene hydroxylases by Deriphat-PAGE (see Supplemental Figure 7 online). Each of these mutants clearly exhibited a PSI-LHCI band.
Photoinhibition of PSI Is Enhanced during Short-Term EL Treatments of nox
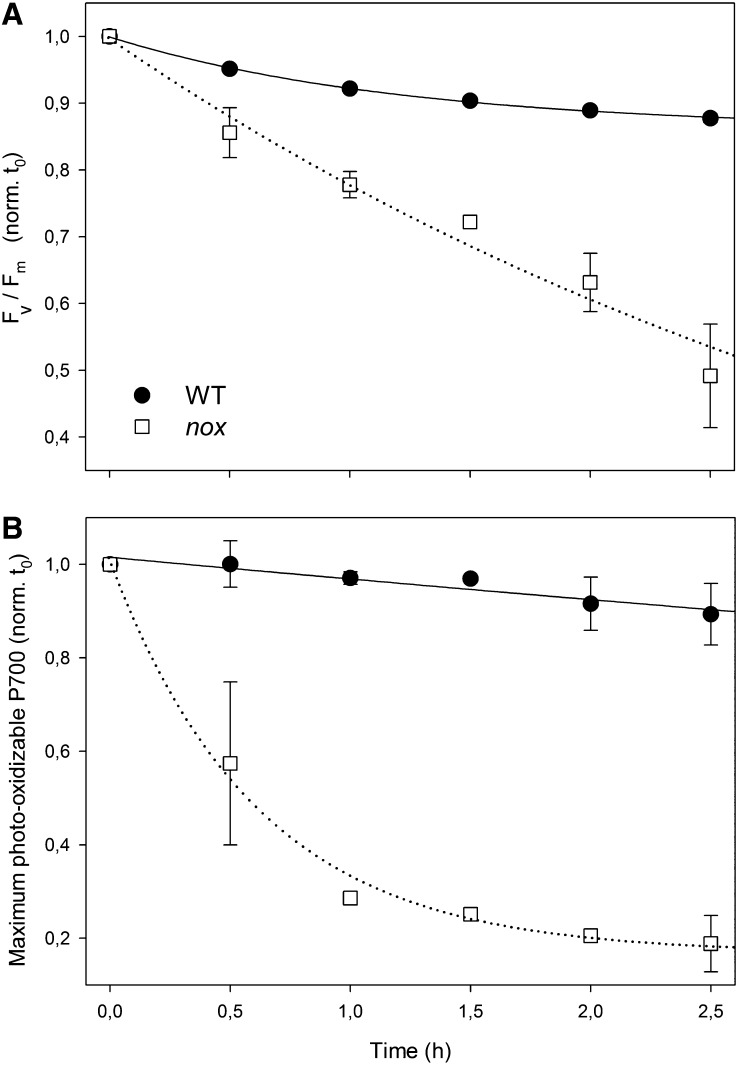
To investigate whether PSI depletion in nox plants was caused by an increased photosensitivity of PSI complexes under photooxidation, leaf discs from wild-type and mutant plants, grown in low light, were transferred to higher light (200 μmol photons m−2 s−1, room temperature) for 2.5 h; then, PSII and PSI activities were assessed by measuring the decrease in maximal quantum yield of PSII (Fv/Fm) or maximum photooxidizable P700 (ΔAmax) upon the treatment. Figure 4A shows that 2.5 h of treatment was effective in producing a faster PSII photoinhibition on nox leaves with respect to the wild type (photoinhibition half-time: 7.5 h for the wild type and 2.7 for nox). The results are consistent with the well-established role of xanthophylls in ensuring photoprotection of thylakoids. Surprisingly, PSI photoinhibition rate was even faster than that of PSII in the nox mutant (Figure 4B): At 200 μmol photons m−2 s−1, the half-time of PSI photoinhibition was 8.5 and 0.6 h for wild-type and nox leaves, respectively. Taken together, the above results show that nox plants are specifically affected in the PSI complex, leading to a strong downregulation of PSI core components and to a dramatic photosensitivity of PSI activity. It is particularly remarkable that PSI, normally the more stress resistant of the two photosystems, was indeed preferentially affected by light in the absence of xanthophylls.
Figure 4.
PSII and PSI Photoinhibition under Photooxidative Stress of the Wild-Type and nox Mutant Plants.
(A) Leaves excised from wild-type (WT) and nox and floating on water were treated at 200 μmol photons m−2 s−1 for 2.5 h, and the kinetics of the Fv/Fm decrease (PSII photoinhibition) was measured.
(B) PSI photoinhibition was quantified by measuring the decay kinetics of maximum photooxidizable P700 (ΔAmax at 705 nm) on detached leaves under high-light intensity (200 μmol photons m−2 s−1, 2.5 h). The maximum contents of P700 was determined on methyl viologen–treated leaves using a saturating flash (3000 μmol photons m−2 s−1) under a 520-μE far-red light background (Munekage et al., 2002). t0, time zero. Data are expressed as means ± sd (n > 3). Statistical analysis shows that values measured for nox leaves were always significantly different with respect to those for the wild type (Student’s t test, P < 0.05).
Lower Abundance of PSI in nox Leaves Resulted from a Combination of Slower Synthesis and Faster Degradation
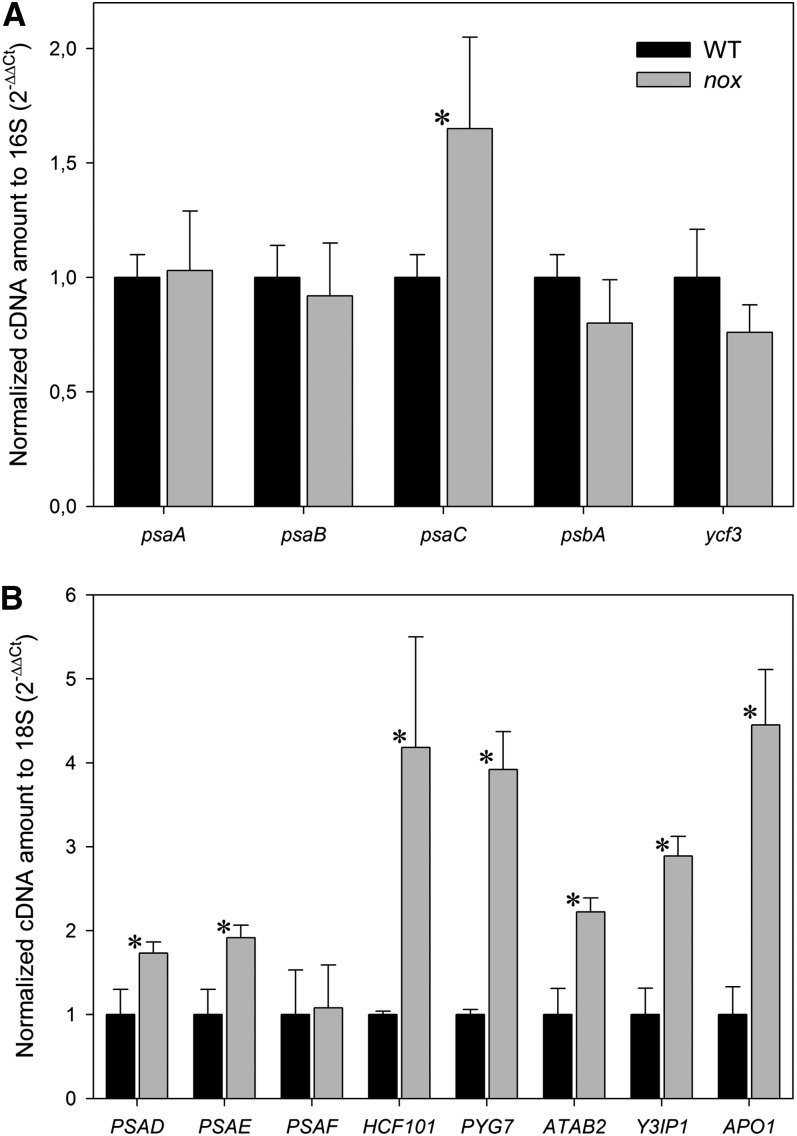
The low PSI content could be caused by increased degradation, as suggested by the above reported photodamage, or by impaired assembly, but also by a lower rate of synthesis, which in turn could be limited at different levels from transcription, to transcript maturation and/or translation. To determine the reasons for low PSI, the rate of these processes was determined. The level of transcripts encoding the main PSI subunits A, B, and C, as well as that encoding the PSII subunit D1 as a reference, was measured by quantitative real-time RT-PCR (qRT-PCR). Results are reported in Figure 5A: The transcript level of genes encoding PsaA, PsaB (PSI core subunits), and PsbA (PSII core subunit) were not significantly different in nox with respect to the wild type, while the PsaC transcript was upregulated in the mutant. Accumulation of transcripts of the nuclear-encoded subunits PsaD, E, and F, the depletion of which severely affected PSI accumulation (Haldrup et al., 2000; Varotto et al., 2000; Ihnatowicz et al., 2004), did not show any downregulation in nox with respect to the wild type (Figure 5B). Since KO of all other subunits of PSI (namely, PsaG-PsaL, PsaN, and PsaO) resulted in less severe PSI defects (Schöttler et al., 2011), these PSI subunits were not investigated in this study.
Figure 5.
qRT-PCR Transcript Profiles of Arabidopsis Genes Involved in PSI Biogenesis.
Transcript levels of genes encoding either PSI and PSII structural subunits or subunits involved in PSI assembly were determined using specific primers and random primers for cDNA production (see Methods for details). For individual genes, relative mRNA levels were normalized with respect to either the 16S (A) or the 18S (B) rRNA housekeeping transcript and then to wild-type (WT) levels. qRT-PCR was performed with three RNAs isolated independently from three different batches of mutant and wild-type plants. Data are expressed as mean ± sd (n = 3). Values significantly different (P < 0.05) from the wild type are marked with an asterisk.
In addition, mRNA levels of genes encoding regulatory or assembly proteins for PSI biogenesis, such as hypothetical chloroplast RF3 (Ycf3, Boudreau et al., 1997; Albus et al., 2010), high chlorophyll fluorescence 101 (HCF101, Lezhneva et al., 2004), pale yellow green 7 (PYG7, Stöckel et al., 2006), Arabidopsis translation of PsaB mRNA (ATAB2, Barneche et al., 2006), Ycf3-interacting protein 1 (Y3IP1, Albus et al., 2010), and accumulation of photosystem one1 (APO1, Amann et al., 2004; Watkins et al., 2011), was the same in nox plants as in the wild type, or even higher (Figure 5B). Moreover, the amount of Ycf3 and Ycf4, assembly factors for PSI (Boudreau et al., 1997; Krech et al., 2012), was similar in both genotypes (see Supplemental Figure 6 online).
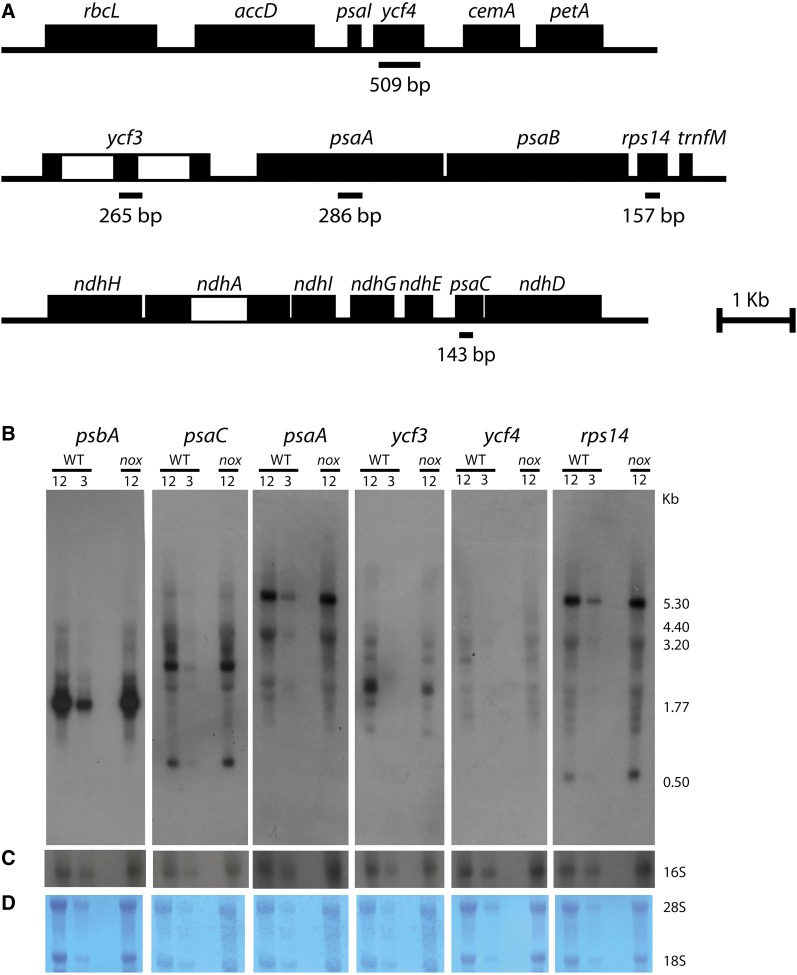
Levels of all transcripts analyzed remained unaffected or were upregulated by the nox mutation; thus, additional analysis through RNA gel blot hybridization was performed to investigate whether the PSI deficiency was related to altered mRNA maturation. Indeed, the chloroplast genome is mainly organized in polycistronic transcription units, and primary transcripts undergo a number of processing events that yield a population of RNA sequences. Mutations that affect the maturation pattern of the psaA/B operon (Meurer et al., 1998) or the pre-mRNA splicing of the PSI assembly factor ycf3 (Watkins et al., 2011) were shown to lead to a proportional loss of PSI.
A set of probes for plastid-encoded structural subunits of both PSI (psaA and psaB) and PSII (psbA) were used as well as probes for chloroplast genes involved in the PSI assembly process (ycf3 and ycf4) and for the ribosomal protein S14 (Figure 6). The transcripts that contain sequences of psaA/rps14, psaC, psbA, and ycf3/4 showed no major changes in abundance and/or length pattern in the wild type versus nox, thus ruling out the hypothesis of a lower maturation efficiency of these sequences upon xanthophyll depletion.
Figure 6.
RNA Gel Blot Analysis of Plastid-Encoded Transcripts in the Wild Type and nox.
(A) Genetic maps of the plastid operons and gene clusters analyzed. Introns of ycf3 and ndhA are shown as open boxes. The locations of probes used for RNA hybridization are marked below the genetic map together with their length in base pairs. Probes were cloned using primers described in Methods.
(B) RNA gel blot analysis of plastid-encoded genes of PSII (psbA), PSI (psaC and psaA), molecular chaperone-like factors involved in PSI assembly (ycf3 and ycf4), and of the ribosomal protein S14 (rps14) in the psaA/psaB/rps14 tricistronic transcript. Total RNA from the wild type (WT) and nox was electrophoresed under denaturing conditions, transferred to nylon membrane, and hybridized with the 32P-labeled probes described in (A), resulting in patterns similar to what was already reported in the literature (Lezhneva et al., 2004a; Meurer et al., 1998).
(C) Control hybridization with a probe for the plastid-encoded 16S rRNA.
(D) Staining with 0.1% toluidine blue of the cytoplasmic 28S and 18S rRNA on nylon membranes as a control of complete and homogenous transfer. For quantification, a dilution of wild-type RNA (corresponding to 12 and 3 µg of leaf RNA) were loaded and compared with transcripts in nox (12 µg RNA loaded per lane). The numbers indicate the size of the detected RNA species (in kilobases).
[See online article for color version of this figure.]
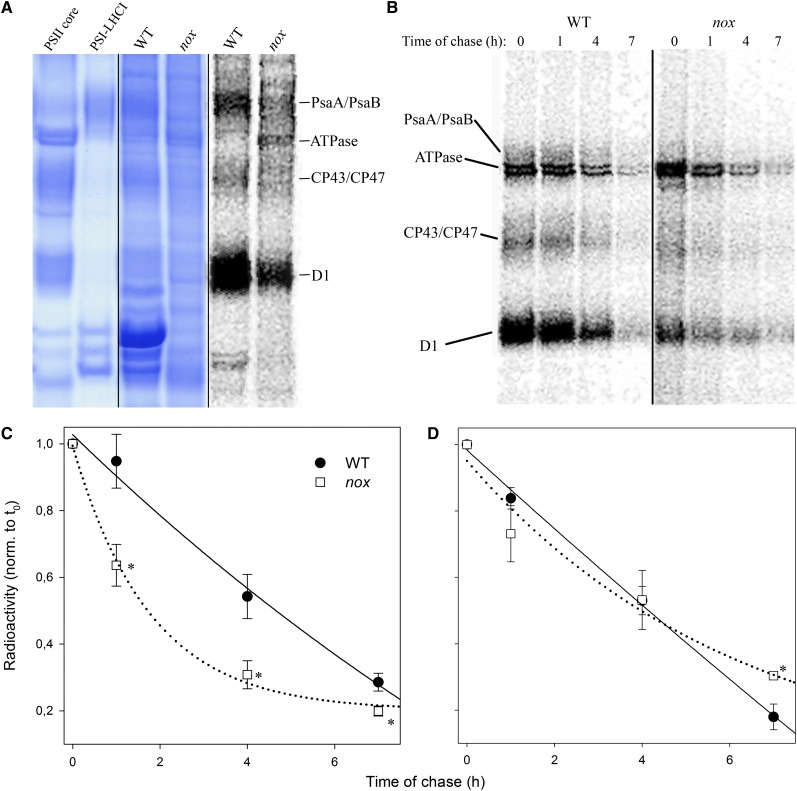
Furthermore, we monitored the synthesis and degradation rate of chloroplast-encoded thylakoid proteins by pulse labeling leaves with [35S]Met in the presence of cycloheximide, an inhibitor of cytoplasmic protein synthesis. As shown in Figure 7A, the incorporation of radioactivity into the α- and β-subunits of chloroplast ATP synthase was increased in the mutant with respect to the wild type; labeling of the PSII protein D1 was slightly reduced in the wild type, while incorporation of radioactivity in the proteins CP43 and CP47 was similar in nox and the wild type. As for PSI, accumulation of PsaA/PsaB proteins was strongly reduced. Pulse labeling for 30 min was followed by a chase with unlabeled Met to monitor the turnover rate of the chloroplast-encoded proteins (Figure 7B). Plotting the kinetics of decrease in 35S labeling during the chase showed that the turnover rates of PSII core protein D1 (Figure 7D) were similar in the wild type and nox mutant, whereas turnover of PsaA/PsaB was 3 times faster in the mutant with respect to the wild type (Figure 7C).
Figure 7.
In Vivo Protein Synthesis and Turnover of Chloroplast-Encoded Membrane Proteins from Wild-Type and nox Plants.
(A) Pulse labeling of thylakoid proteins. After pulse labeling of Arabidopsis leaves with [35S]Met in the presence of cycloheximide under low-light intensity (20 μmol photons m−2 s−1, room temperature), thylakoid membranes were isolated and proteins were separated by SDS-PAGE. The resolved proteins were visualized by autoradiography (left panel); image of Coomassie blue–stained gel is reported (central panel); unlabeled PSI core and PSI-LHCI complexes were separated on an SDS-PAGE gel as reference (right panel). WT, the wild type.
(B) Pulse and chase labeling of thylakoid proteins. A [35S]Met pulse labeling as in (A) (lane 0) was followed by chase in unlabeled medium for 1, 4, and 7 h under low light (20 μmol photons m−2 s−1, room temperature).
(C) and (D) Time course of PsaA/PsaB (C) and D1/D2 (D) turnover. Data are expressed as means ± sd (n > 3). Values significantly different (Student’s t test, P < 0.05) from the wild type are marked with an asterisk. norm., normalized.
[See online article for color version of this figure.]
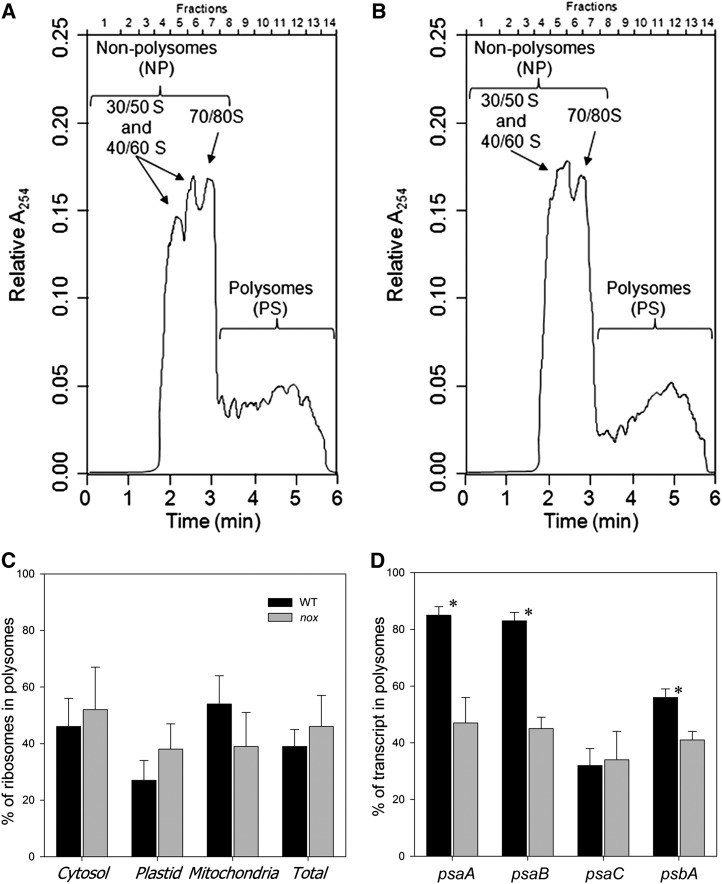
The above results suggest that impaired translation of PSI transcripts might contribute to limiting PSI accumulation. This was investigated by polysome analysis (Mathews et al., 2007) that consists of fractionating polysomes from wild-type and nox plants on Suc gradients by ultracentrifugation and assessing the partitioning of individual messages between fractions containing ribosome-free messages (plus monosomes) versus polysomal fractions. Figure 8 shows absorption profiles of ribonucleoprotein material that was extracted from wild-type (A) and nox (B) plants and harvested at the midpoint of the light period.
Figure 8.
Polysome Loading Analysis of Wild-Type and nox Mutant Plants.
(A) and (B) Representative examples of the distribution of ribosomes in a Suc gradient for the wild type (A) and nox (B). RNA was measured as absorbance at 254 nm (A254). Ribosome subunits, free ribosomes, and monosomes are on the left and increasingly large polysomes toward the right of the display.
(C) Estimated proportion of ribosomes on polysomes for the wild type (WT; black bars) and nox (gray bars). The ribosome number in each fraction was calculated by determining the amount of the small subunit rRNA transcript for cytosolic, plastid, and mitochondrial ribosomes.
(D) Ribosome occupancy for the psaA, psaB, psaC, and psbA transcripts for the wild type (black bars) and nox (gray bars). The percentage of each transcript in the polysomes fractions was calculated using the absolute concentration of the transcript in the NP and PS fractions as (PS/[NP+PS]) and are given as numbers in the figures. The concentration of the transcripts was measured by qRT-PCR using random primers for cDNA production and specific PCR primers for the transcripts (see Methods for details). The results are given as mean ± sd (n = 4). Values significantly different (Student’s t test, P < 0.05) from the wild type are marked with an asterisk.
Based on the absorption readings (Figures 8A and 8B), no major differences were apparent in the polysome profiles, implying that the level of translation was comparable in the wild-type and nox plants (∼34% of ribosomes were on polysomes for both). When using the ribosome concentration determined by qRT-PCR (Figure 8C), the wild-type and nox plants showed no significant difference in the total ribosome levels and on polysomes (39 and 46%, respectively). These results indicate that the global translational activity is similar in wild-type and nox plants. Also, the significant amount of free ribosome subunits and monosomes indicates that the availability of ribosomes does not seem to be a limiting factor for the global translation efficiency in nox and wild-type plants. A key parameter of the translational status of a gene is the fraction of its transcripts engaged in translation, defined as the transcript’s ribosomal occupancy (Mathews et al., 2007): The greater the percentage of an mRNA that is associated with polysomes, the greater its translational efficiency. When the polysome occupancy of psaA, psaB, psbA, and psaC was analyzed, a strong reduction of the psaA/psaB transcripts (38%) and a weak but significant reduction of the psbA transcript (15%) was evidenced in the case of nox samples with respect to the wild type (Figure 8D), while the psaC transcript level was similar in the two genotypes. These results suggest that the translation initiation of PSI core subunits in nox plants is downregulated in response to xanthophyll depletion.
DISCUSSION
In this work, we analyzed the modifications of the photosynthetic apparatus in the Arabidopsis carotenoid biosynthetic mutant chy1 chy2 lut1 lut5 (nox) that accumulates α- and β-carotene only. Knockout of the four hydroxylase genes completely abolished xanthophyll biosynthesis, thus confirming that CHY1, CHY2, LUT1, and LUT5 constitute the full complement of carotenoid hydroxylases in Arabidopsis (Kim et al., 2009). The biogenesis of the photosynthetic apparatus was strongly affected in nox plants, and this resulted in reduced photosynthetic electron transport and increased photosensitivity. This was mainly due to depletion in Lhcb proteins and of the PSI complex in mutant plants, to such an extent that nox is unable to sustain photoautotrophic growth in low light and rapidly undergoes photoinhibition in moderate light. Thus, xanthophylls appear to be essential not only for photoprotection but also for biogenesis of the photosynthetic machinery.
Reduced Xanthophyll Content Negatively Affects Photoprotection
The photoprotective role of carotenoids in photosynthesis has been widely investigated (Havaux and Niyogi, 1999; Davison et al., 2002; Baroli et al., 2003; Dall’Osto et al., 2007b). Photoprotection is performed both by carotenes (Wang et al., 2004; Telfer, 2005) and xanthophylls (Formaggio et al., 2001). A complete lack of xanthophylls in nox plants results in a strongly enhanced photosensitivity with respect to wild-type plants, as evidenced by the higher release of 1O2 in high light (Figure 2A) and faster photoinhibition (Figure 4) in mutant leaves. A major feature of this mutant is the depletion in LHC proteins (Figure 3), consistent with the evidence that xanthophylls are needed for folding of LHC in vitro (Paulsen 1997). However, immunotitration of pigment-protein complexes in nox shows that a reduced level of Lhca proteins is maintained in the mutant, as also supported by the red-shifted fluorescence emission spectra at 77K, typical of LHCI complexes (see Supplemental Figure 5 online), while Lhcb proteins are virtually absent. This effect is likely due to the inability of carotenes to replace xanthophylls in stabilizing Lhcb (Plumley and Schmidt, 1987). Instead, Lhca can bind carotenes and form functional complexes (Morosinotto et al., 2002; Mozzo et al., 2006).
The photosensitive phenotype of nox plants implies that functional LHC complexes are essential for photoprotection, consistent with previous reports (Havaux et al., 2007; Dall’Osto et al., 2010). Indeed, while 1O2 production from purified PSII core complexes is independent from the xanthophyll content, LHC from wild-type thylakoids produce far less 1O2 with respect to that from xanthophyll-depleted mutants (Dall’Osto et al., 2007b), implying that LHC proteins folded in the presence of sufficient xanthophyll levels act in preventing production and/or in scavenging 1O2 (Dall’Osto et al., 2010). One might wonder if the lack of antenna proteins alone could explain the impaired photoautotrophic growth in nox plants. An extreme reduction in LHC proteins is also obtained with the ch1 mutation (Espineda et al., 1999), in which assembly of LHC is prevented. The ch1 mutant can still grow on soil and is not photoinhibited in moderate light (Havaux et al., 2007); thus, despite a reduction in antenna size comparable to that of nox, ch1 can perform photoautotrophic growth. We conclude that reduced LHC content cannot be the only reason for the extreme light sensitivity of nox.
The capacity for excess energy dissipation into heat (qE mechanism) is strongly reduced in the mutant (Figure 2D). Since the level of PsbS (Li et al., 2002) is not reduced in nox plants relative to the wild type (Figure 3), qE depletion can thus be attributed to a lack of PsbS interacting partners, namely, the Lhcb proteins (Ahn et al., 2008; Avenson et al., 2008; Betterle et al., 2009). All together, these results support the correlation between xanthophyll content and qE amplitude, previously suggested on the basis of antisense inhibition of β-hydroxylation (Pogson and Rissler, 2000). As for the role in photoprotection, the npq4 mutation abolished qE in Arabidopsis (Li et al., 2000) but has a modest impact on the photosensitivity in vivo (Li et al., 2002), just a fraction of the effect observed upon decreasing the level of xanthophylls (Niyogi et al., 2001; Dall’Osto et al., 2007b). We thus conclude that the differential resistance of wild-type versus nox plants cannot be attributed to qE depletion. Rather, the decreased efficiency of photosynthetic electron transport, higher photoinhibition, and lethality in soil of the nox plants are to be ascribed to a severe deficiency in PSI.
nox Plants Fail to Accumulate Normal Levels of PSI Core Complexes
The immunotitration of pigment-protein complexes in mutant thylakoids showed that PSII core subunits and cytochrome b6f accumulated in nox, while PsaA/B subunits were almost absent (Figure 3). The decrease in the xanthophyll/carotenoid ratio was shown to cause a proportional decrease in the abundance of PSI core units with respect to PSII (Fiore et al., 2012). Here, we show that nox leaves fail to accumulate PSI complexes, thus confirming that xanthophylls are needed for PSI biogenesis. This result is surprising, since (1) there is no evident reason for the preferential effect of xanthophyll depletion on PSI versus PSII core complexes, and (2) PSI core complexes bind chlorophyll a and carotene as the only pigments (Nelson and Ben Shem, 2004), which are not limited in nox thylakoids (Figure 1B).
Therefore, how does xanthophyll depletion contribute to the lower accumulation of PSI in nox plants with respect to the wild type?
One possibility is that the assembly/stability of the PSI core might be limited by the amount of LHCI available, which has a photoprotective role on PSI (Alboresi et al., 2009) and modulates plant fitness (Ganeteg et al., 2004). Since xanthophyll depletion in nox plants limits the total amount of LHC, this might enhance photooxidative stress, consistent with the increased PSI core degradation in nox mutant (Figure 7). Nevertheless, several Arabidopsis mutants strongly depleted in LHCI have been described (Klimmek et al., 2005; Havaux et al., 2007), and all of them maintain a functional PSI core complex.
Alternatively, the steady state level of PSI and PSII core complexes could be limited by chlorophyll availability; indeed, a coregulation of chlorophyll and carotenoid biosynthesis has been reported (Härtel et al., 1997). Nevertheless, the lower content of chlorophylls in carotenoid biosynthesis mutants is mainly due to destabilization of LHC (Paulsen, 1997), while chlorophyll biosynthesis mutants showed that the PSI level is less affected than in nox (Tottey et al., 2003). Therefore, we conclude that the nox phenotype is unlikely to be due to limitation in chlorophyll supply, and the contribution of xanthophylls to the modulation of PSI level might be substantially different from the indirect control of the chlorophyll biosynthetic rate.
Comparison between nox and Other Mutants Affected in the Biogenesis of PSI
The phenotype of nox leads to the conclusion that xanthophyll depletion might have further targets that interfere with thylakoid development, since additional features than that expected from xanthophyll depletion became evident, namely, (1) the albino phenotype of nox and the inability to grow photoautotrophically (Figure 1); (2) the poor development of the thylakoid membrane system; (3) the small size of the chloroplasts (Figure 2); and (4) the depletion in PSI core complex (Figure 3). The above phenotypic traits are consistent with a primary lesion leading to PSI depletion. In fact, similar symptoms are found in PSI-deficient mutants (Lezhneva and Meurer, 2004; Stöckel et al., 2006; Schöttler et al., 2011).
The PSI supercomplex has a chimeric nature: Some of its subunits are encoded by the nuclear genome and others by the chloroplast genome. The initial event in PSI assembly involves subunits encoded by the plastome: The formation of the heterodimer PsaA/PsaB is followed by association of PsaC, which is needed for further binding of PsaD and PsaE (Antonkine et al., 2003) and then a number of smaller subunits are added. While PSI in plants can accumulate in the absence of most small subunits, it is well known that mutants devoid of the PsaA/B dimer are unable to assemble a functional PSI core complex, although some peripheral subunits or detached Lhca can accumulate in the thylakoids (Redding et al., 1999; Lezhneva and Meurer, 2004).
Some of the available PsaA/B mutants are either defective in the transcription of the psaA-psaB-rps14 operon (Hein et al., 2009) or in its stability (Lezhneva and Meurer, 2004); others are impaired in the translation of the dimeric complex or cannot accumulate a stable heterodimer (Stöckel et al., 2006; Albus et al., 2010) or are affected in intron splicing of chloroplast mRNAs, including those encoding molecular chaperone-like complexes, such as Ycf3 (Landau et al., 2009; Ye et al., 2012); others are defective in the stable assembly of several [4Fe-4S] cluster–containing complexes of chloroplasts, including the PsaA/B dimer (Schwenkert et al., 2010).
Several lines of evidence suggest that the accumulation of the chloroplast-encoded PSI subunits is mainly controlled at the level of translation (Lezhneva and Meurer, 2004; Schöttler et al., 2011). Indeed, only severe reductions in the amount of the psaA/B transcript lead to reduced PSI accumulation. Even in nox plants, the lower accumulation of PsaA/B subunits appears to be related to posttranscriptional regulatory events, since the psaA and psaB messenger levels are not affected with respect to the wild type (Figure 5).
Moreover, maturation of plastid mRNAs was shown to have a regulatory role in plastid gene expression (Stern et al., 2010), as suggested by mutations such as pac-2, that affect the maturation pattern of psaA/B messenger and lead to strong depletion of PSI (Meurer et al., 1998). However, the polycistronic transcripts analyzed (Figure 6) showed no major changes in abundance and distribution pattern in the wild type versus nox, thus ruling out the hypothesis of an effect of xanthophyll depletion on the maturation efficiency of these sequences.
Rather, a reduction in PsaA/B synthesis (Figure 8) and its accelerated degradation (Figure 7) both appear to be the major reasons for the absence of the PSI core complex in nox.
At present, the functional basis for the relation between impaired translation of PsaA/B and the lack of xanthophylls is unclear. Among mutants affected in PsaA/B synthesis, nox has common features with the maize (Zea mays) mutant cps2, which shows an impaired association of the psaA/B transcripts into functional polysomes (Barkan, 1993; Amann et al., 2004) or to Arabidopsis atab2, in which psaA/B transcripts are poorly associated with polysomes (Barneche et al., 2006). Expression of the translation initiation factor ATAB2 is upregulated in the mutant (Figure 5), thus ruling out its involvement in the nox phenotype. Thus, it seems reasonable to speculate that xanthophylls might be needed for the initiation/elongation phases of psaA and psaB translation or targeting of the nascent chains to the thylakoid membranes.
However, even assuming that PSI accumulation is mainly regulated by the translation of the two subunits PsaA/B, a 50% reduction in the psaA/B ribosome occupancy in nox chloroplasts relative to the wild type is unlikely to account for a 95% reduction in PSI accumulation. Rather, the faster degradation rate of radiolabeled PsaA/PsaB (Figure 7) suggests a further block at the level of PSI assembly: In fact, if newly synthesize polypeptides are not assembled into a functional complexes, they are rapidly degraded, even though their synthesis proceeds at normal levels (Barkan et al., 1995).
Moreover, PSI is one of the most elaborate macromolecular complexes of the chloroplast, having 17 protein subunits and nearly 200 cofactors (Amunts et al., 2010) that need to be assembled in a concerted manner. Although a number of auxiliary factors involved in these tasks have been described, a complete understanding on how the molecular machinery works is still lacking. The transcription of genes encoding auxiliary proteins for PSI assembly described to date (Schöttler et al., 2011) was found to occur at similar levels in wild-type and nox plants or to be upregulated in the mutant (Figure 5). Otherwise, xanthophyll depletion in nox could impair the capacity for the biogenesis or assembly of [4Fe-4S] clusters (Amann et al., 2004; Lezhneva et al., 2004), possibly as a side effect of the enhanced release of ROS in nox chloroplasts. However, the mutant accumulates normal amounts of other [2Fe-2S] and [4Fe-4S] cluster–containing proteins, such as ferredoxin and the NDH complex (see Supplemental Figure 6 online). Finally, ribosomal loading of psaC is similar in wild-type and nox chloroplasts (Figure 7), despite the fact that the PsaC protein contains two [4Fe-4S] clusters (Amunts et al., 2010).
At the moment, we can only speculate as to which specific steps of PSI biogenesis are supported by xanthophylls. One possibility is that xanthophylls have a role in mediating (transitory) molecular interactions between photosystem subunits or with their putative chaperones (e.g., Ycf3, Ycf4, and Y3IP1). Alternatively, early events of cotranslational insertion in the thylakoid membrane of the PsaA subunit might require that a complex forms with xanthophylls, which is missing in nox.
Evidence for carotenoid-derived, mobile signaling molecules that regulate several aspects of plant physiology has emerged in recent years: Abscisic acid, strigolactone, and β-cyclocitral (Gomez-Roldan et al., 2008; Ramel et al., 2012) are examples of water-soluble carotenoids catabolites that have been previously found to regulate nuclear gene expression, apical dominance, and branching. Whether or not these signals affect the expression of the molecular mechanism underlying PSI assembly is currently unknown. Application of analytical approaches for the identification of protein interaction partners present in wild-type psaA-psaB-rps14 polysomes and absent in nox will be crucial to identify xanthophyll-dependent assembly subunits involved in PSI biogenesis.
METHODS
Plant Materials and Growth Conditions
Wild-type plants of Arabidopsis thaliana ecotype Columbia and mutants lut1, lut5, chy1, and chy2 were obtained from NASC (The European Arabidopsis Stock Centre). The T-DNA knockout lines used are lut1 (Salk 129724), lut5 (Salk 116660), chy1 (SAIL 49A07), and chy2 (SAIL 1242B12). The chy1 chy2 lut1 lut5 quadruple mutant (nox) was isolated by crossing single mutant plants. Surface-sterilized seeds were sown on Petri dishes containing 0.3% (w/v) solidified plant-agar supplemented with 0.5% Murashige and Skoog nutrients and 2% (w/v) Suc. Seedlings were grown for 3 weeks with a 16-h photoperiod at a photon flux density of ∼20 μmol photons m−2 s−1, at 22°C. Since nox seedlings were unable to produce a mature inflorescence, the mutation was maintained in the heterozygous state.
Thylakoid Isolation and Sample Preparation
Unstacked thylakoid membranes were isolated as previously described (Bassi et al., 1988). Membranes from wild-type and lut5 plants, corresponding to 400 µg of chlorophyll, were solubilized with 0.7% β-dodecylmaltoside and then fractionated by ultracentrifugation in a 0.1 to 1 M Suc gradient (Caffarri et al., 2009).
Pigment Analyses
Pigments were extracted from leaves with 80% acetone and then separated and quantified by HPLC (Gilmore and Yamamoto, 1991).
Gel Electrophoresis and Immunoblotting
SDS-PAGE analysis was performed with the Tris-Tricine buffer system (Schägger and von Jagow, 1987). For immunotitration, thylakoid samples corresponding to 0.1, 0.2, and 0.4 μg of chlorophylls were loaded for each sample and electroblotted on nitrocellulose membranes. Filters were incubated with specific antibodies and were detected with alkaline phosphatase–conjugated antibody. To avoid any deviation between different immunoblots, samples were compared only when loaded in the same gel. Nondenaturing Deriphat-PAGE was performed following the method described previously (Peter et al., 1991) with modifications reported by Havaux et al. (2004). Thylakoids concentrated at 1 mg/mL chlorophylls were solubilized with 0.8% α-DM, and 10 or 25 μg of chlorophylls was loaded in each lane.
Analysis of Chlorophyll Fluorescence and P700 Redox State
NPQ of chlorophyll fluorescence, PQ redox state (qL), and PSII yield (ΦPSII) were measured at room temperature (22°C) with a PAM 101 fluorometer (Walz) according to the equation defined by Van Kooten and Snel (1990). Calculation of the ΔpH-dependent component of chlorophyll fluorescence quenching (qE) was performed as described previously (Walters and Horton, 1995). Fluorescence kinetics were measured with a home-built setup, in which leaves were vacuum infiltrated with 10−5 M DCMU 3.0 and excited with green light at 520 nm (Luxeon, Lumileds), and the emission was measured in the near far red (Rappaport et al., 2007). The t2/3 of the fluorescence rise was taken as a measure of the functional antenna size of PSII (Malkin et al., 1981). The reoxidation kinetics of QA were measured as the decay of chlorophyll a fluorescence with the PAM fluorometer. Saturating single-turnover flashes obtained from a single-turnover flash unit (Heinz Walz XE-ST) were used to convert all QA to QA−; the variable fluorescence decay, reflecting the reoxidation of QA−, was detected at a 20-μs resolution. Data from 12 recordings were averaged. Changes in the redox state of P700 were measured on leaves vacuum infiltrated with methyl viologen (2 mM), by monitoring absorbance at 705 nm with a JTS-100 (BioLogic Instruments); the maximum content of photooxidizable P700 (ΔAmax) was determined on methyl viologen–treated leaves using a saturating flash (3000 μmol photons m−2 s−1) under a 520 μmol m−2 s−1 far-red light background (Munekage et al., 2002).
Electron Microscopy
Intact leaf fragments from wild-type and nox mutant leaves were fixed, embedded, and observed in thin section as previously described (Sbarbati et al., 2004).
Spectroscopy
Steady state spectra were obtained using samples in 10 mM HEPES, pH 7.5, 0.06% α(β)-DM, and 0.2 M Suc. Absorption measurements were performed using a SLM-Aminco DW-2000 spectrophotometer. Fluorescence emission spectra were measured using a Jobin-Yvon Fluoromax-3 spectrofluorometer.
Measurements of ROS Production
ROS production in leaves was measured with SOSG, a fluorescent probe highly selective for 1O2 (Flors et al., 2006). Analysis of 1O2 release in leaves or purified complexes was performed as previously described (Dall’Osto et al., 2010).
Photobleaching Assay
The photobleaching kinetics of pigment-protein complexes was measured as described (Croce et al., 1999). Samples were cooled at 10°C. Initial and maximal absorbance was set in order to have the same absorbance area (corresponding to ∼5 μg chlorophylls/mL) in the wavelength range of the actinic light (600 < λ < 750 nm).
Determination of the Sensitivity to Photooxidative Stress
High light treatment was performed for 2.5 h at 200 μmol photons m−2 s−1, room temperature, on detached leaves floating on water. Light was provided by 150-W halogen lamps (Focus3; Prisma). Decay kinetics of Fv/Fm (Havaux et al., 2004) and maximum content of photooxidizable P700 (ΔAmax at 705 nm) (Sonoike, 2011) were recorded during illumination to assess the photoinhibition of PSII and PSI, respectively.
qRT-PCR
qRT-PCR was performed using 0.5 µg total RNA from each sample. The RNA was reverse transcribed using random primers with MoMLV reverse transcription reagents (Promega protocol). The cDNA was quantified using a Qbit fluorometer (Invitrogen), diluted, and used for quantitative PCR amplifications with specific primers. Each qRT-PCR was performed with SYBR Green fluorescence detection in a quantitative PCR thermal cycler (ABI Prism 7300; Applied Biosystems). Each reaction was prepared using 5 μL from a 0.2 ng/μL dilution of cDNA derived from the reverse transcription, 10 μL of SYBR Green PCR Master Mix (Applied Biosystems), and 0.5 µM forward and reverse primers in a total volume of 25 µL. The cycling conditions were as follows: 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Melting curve analysis was performed to identify nonspecific PCR products and primer dimers. Transcript levels of genes coding for PsaD, E, and F were determined with the following primers: PSAD-f, 5′-AACAGGAGGAGCTGCGATAA-3′; PSAD-r, 5′-CTCCATCTTTCGGGTGAAGA-3′; PSAE-f, 5′-CCGCTAAGGCTAAACCTCCT-3′, PSAE-r, 5′-ATTCGCGTAATTCACCTTGG-3′; PSAF-f, 5′-GACGGTTTACCGCACTTGAT-3′; and PSAF-r, 5′-CGGAAGATGATCCGACTAGC-3′. Transcript levels of genes coding for either PSI and PSII structural subunits, or subunits involved in PSI assembly, were determined using specific primers reported previously (Amann et al., 2004; Lezhneva and Meurer, 2004; Barneche et al., 2006; Stöckel et al., 2006; Albus et al., 2010).
In Vivo Protein Synthesis of Chloroplast-Encoded Membrane Proteins
In vivo labeling of leaf proteins was performed as previously described (Sun et al., 2010). Leaves from 3-week-old plants were preincubated in 20 μg/mL cycloheximide for 30 min and radiolabeled with 100 μCi/mL [35S]Met in 20 μg/mL cycloheximide, at a light intensity of 20 μmol photons m−2 s−1 for 30 min at room temperature, followed by a chase with cold Met. Thylakoid membranes were isolated and the proteins were separated by SDS-PAGE, and then the gels were stained and dried and the radioactivity was quantified using ImageMaster (Pharmacia Biotech).
RNA Gel Blot Analysis
Total RNAs were isolated using the RNeasy mini kit (Qiagen). RNAs were quantified and their quality was assessed using a 2100 Bioanalyzer (Agilent). Approximately 12 μg of RNAs from the wild type and nox (a dilution of 3 μg for the wild type was also loaded) were fractionated on 1% agarose-formaldehyde denaturing gels. The gels were blotted on positively charged Hybond N+ membranes (GE Healthcare) in 10× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). Total RNAs were immobilized on membranes by UV cross-linking and then stained with 0.1% toluidine blue. The DNA probes were labeled with [32P]CTP by random priming using Ready to Go DNA labeling beads (-dCTP) (GE Healthcare). Unincorporated nucleotides were removed with an Illustra AutoSeq G-50 Dye Terminator Removal Kit (GE Healthcare). The membranes were hybridized overnight at 42°C in ULTRAhyb buffer (Ambion) in the presence of 106 cpm/mL of labeled probe. After hybridizations, unhybridized probes were removed by washing the membranes in 2× SSC containing 0.1% SDS at 42°C. Autoradiography was then performed using Kodak XAR-5 films. DNA probes were stripped by incubating the membranes in boiling 0.1% aqueous SDS solution. All the filters were reprobed with the 16S rRNA probe. The DNA sequences corresponding to the probes adopted for the hybridizations were cloned in pGEM-T Easy (Promega) and sequenced. The probes were obtained by PCR amplification with the following primers: ycf3-F, 5′-ATGTCGGCTCAATCTGAAGG-3′, and ycf3-R, 5′-CTCCTTGTTGAATGGCCTGT-3′ for the ycf3 probe; and ycf4-F, 5′-TAACGGGGTCTCGAAAAACA-3′, and ycf4-R, 5′-TTCAATTGGTACACGCAAGAA-3′, for ycf4. The probes for psaA, psaC, psbA, and rps14 were all cloned as previously described (Lezhneva and Meurer, 2004).
Ribosome Loading Analysis
Polysomes were fractionated from crude leaf extracts as described (Piques et al., 2009) but using 100 to 150 mg fresh weight. The gradients were fractionated using a programmable density gradient fractionation system (Teledyne Isco) to continuously record absorbance at 254 nm. Polysome levels were determined from the area under the polysome profile after subtracting gradient baseline absorbance. The area of each polysome profile was normalized to an equal value to account for differences in sample loading. Levels of nonpolysomes (NPs; gradient region containing mRNP complexes, 40/60S and 30/50S ribosome subunits, and 70/80S monosomes) and polysomes (PSs; gradient region containing more than two ribosomes per mRNA) were determined by calculating the corresponding peak areas of gradient regions. Areas corresponding to the NP and PS fractions were reported as a percentage of the total area under the profile.
Purification of Polysomal RNA and Quantification of Transcript Levels by qRT-PCR
Fractions corresponding to NS (1 to 8) and PS (9 to 14) were combined, and the total RNA was purified from these fractions separately by ethanolic precipitation using the RNeasy plant mini kit (Qiagen) as described (Piques et al., 2009). For absolute quantification of the transcript levels in the NP and PS fractions, a mix of six artificial poly(A)+ RNAs (ArrayControl RNA spikes; Applied Biosystems/Ambion) were spiked into the NP and PS fractions before RNA purification. The concentrations of the spike-in controls were adjusted to make a calibration curve with a dynamic range from 9.0E+11 to 6.0E+07 copy number per gram fresh weight. RNA concentration and integrity were measured with a NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies) and an Agilent-2100 bioanalyzer using RNA 6000 NanoChip (Agilent Technologies). The cDNA synthesis, using 200 ng of total RNA, quality control of the synthesized cDNA, the qRT-PCR reactions, and data analysis were performed as described (Piques et al., 2009). All standard curves, derived from the six spike-in controls, had R2 values higher than 0.99 and were used to calculate the concentration of the transcripts as copy g−1 fresh weight. The primers for the PsaA, PsaB, PsaC, and PsbA were designed according to Lezhneva and Meurer (2004), and the list of the primers for the other genes used is in Supplemental Table 1 online.
Estimation of the Ribosome Content
The ribosome number within NP and PS fractions was calculated by determining the amounts of cytosolic, plastid, and mitochondrial rRNA small subunits by qRT-PCR on the basis that each of these rRNAs corresponds to one ribosome (Piques et al., 2009).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At4G25700 (chy1), At5G52570 (chy2), At3G53130 (lut1), and At1G31800 (lut5).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Biosynthetic Pathway of Carotenoids in Arabidopsis thaliana.
Supplemental Figure 2. Transmission Electron Micrographs of Plastids from Leaf Mesophyll Cells of the Arabidopsis Wild-Type and nox Mutant Plants.
Supplemental Figure 3. Photoprotection Capacity of α- versus β-Carotene Binding Complexes.
Supplemental Figure 4. Decay and Induction Kinetics of PSII Fluorescence.
Supplemental Figure 5. Low-Temperature Fluorescence Emission Spectra of Chloroplasts.
Supplemental Figure 6. Analysis of Polypeptide Composition of Wild-Type and nox Thylakoid Membranes.
Supplemental Figure 7. Biochemical Characterization of Single Knockout Mutants of Arabidopsis.
Supplemental Table 1. Quantification of Transcript Levels on Polysomal RNA by qRT-PCR.
NOTE ADDED IN PROOF
The reader is cautioned that the nox mutant name used in this paper, short for “no xanthophylls”, is being used for convenience to describe the chy1 chy2 lut1 lut5 quadruple mutant, which has mutations in all four genes encoding carotenoid hydroxylases in Arabidopsis and is devoid of xanthophylls. It does not describe a new gene or protein name, and should not be confused with Arabidopsis NOX proteins, which are NADPH oxidases.
This work was supported by the Marie Curie Actions–Networks for Initial Training Harvest (Grant PITN-GA-2009-238017) and by Ministero delle Politiche Agricole, Alimentari e Forestali BioMassVal (Grant 2/01/140). We thank Paolo Bernardi and Andrea Sbarbati (University of Verona) for help in sample preparation for electron microscopy. We thank Anita Zamboni (University of Verona) for help on PCR analysis and Nicola Tamassia (University of Verona) for valuable advice regarding ImageMaster. Antibodies against HCF101, NdhH, and NdhK were kind gifts of Jörg Meurer (Ludwig-Maximilians-Universität, Munich, Germany) and Toshiharu Shiakani (Kyoto University, Kyoto, Japan). We thank Alejandra M. Landau (Instituto Nacional de Tecnologia Agropecuaria, Castelar, Argentina) and Ralph Bock (Max-Planck-Institut für Molekulare Pflanzenphysiologie, Potsdam-Golm, Germany) for providing antibodies against Ycf3 and Ycf4, respectively. We thank Mark Stitt for his support while conducting the polysome experimental work.
AUTHOR CONTRIBUTIONS
L.D. isolated single knockout Arabidopsis mutants, carried out the crossings, and performed a biochemical and physiological characterization of their photosynthetic apparatus. M.P. carried out ribosome loading analysis. S.C. was involved in the photooxidative treatments and stress measurements and in data analysis. M.R. and A.A. carried out qRT-PCR and the assay for protein synthesis of chloroplast-encoded membrane proteins. B.M. and A.A. performed RNA blot analysis. R.B. and L.D. conceived the study, participated in its design and coordination, and drafted the article.
Glossary
- LHC
light-harvesting complex
- EL
excess light
- ROS
reactive oxygen species
- Zea
zeaxanthin
- Viola
violaxanthin
- Neo
neoxanthin
- NPQ
nonphotochemical quenching
- PSI
photosystem I
- 1O2
singlet oxygen
- SOSG
singlet oxygen sensor green
- PSII
photosystem II
- Fv/Fm
maximal photochemical yield of PSII
- PQ
plastoquinone
- α-DM
dodecyl-α-d-maltoside
- qRT-PCR
quantitative real-time RT-PCR
- NP
nonpolysome
- PS
polysome
References
- Ahn T.K., Avenson T.J., Ballottari M., Cheng Y.C., Niyogi K.K., Bassi R., Fleming G.R. (2008). Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science 320: 794–797 [DOI] [PubMed] [Google Scholar]
- Alboresi A., Ballottari M., Hienerwadel R., Giacometti G.M., Morosinotto T. (2009). Antenna complexes protect photosystem I from photoinhibition. BMC Plant Biol. 9: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albus C.A., Ruf S., Schöttler M.A., Lein W., Kehr J., Bock R. (2010). Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22: 2838–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Toporik H., Borovikova A., Nelson N. (2010). Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonkine M.L., Jordan P., Fromme P., Krauss N., Golbeck J.H., Stehlik D. (2003). Assembly of protein subunits within the stromal ridge of photosystem I. Structural changes between unbound and sequentially PS I-bound polypeptides and correlated changes of the magnetic properties of the terminal iron sulfur clusters. J. Mol. Biol. 327: 671–697 [DOI] [PubMed] [Google Scholar]
- Avenson T.J., Ahn T.K., Zigmantas D., Niyogi K.K., Li Z., Ballottari M., Bassi R., Fleming G.R. (2008). Zeaxanthin radical cation formation in minor light-harvesting complexes of higher plant antenna. J. Biol. Chem. 283: 3550–3558 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Voelker R., Mendel-Hartvig J., Johnson D., Walker M. (1995). Genetic analysis of chloroplast biogenesis in higher plants. Physiol. Plant. 93: 163–170 [Google Scholar]
- Barneche F., Winter V., Crèvecoeur M., Rochaix J.D. (2006). ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 25: 5907–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroli I., Do A.D., Yamane T., Niyogi K.K. (2003). Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell 15: 992–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi R., Pineau B., Dainese P., Marquardt J. (1993). Carotenoid-binding proteins of photosystem II. Eur. J. Biochem. 212: 297–303 [DOI] [PubMed] [Google Scholar]
- Bassi R., Rigoni F., Barbato R., Giacometti G.M. (1988). Light-harvesting chlorophyll a/b proteins (LHCII) populations in phosphorylated membranes. Biochim. Biophys. Acta 936: 29–38 [Google Scholar]
- Betterle N., Ballottari M., Zorzan S., de Bianchi S., Cazzaniga S., Dall’osto L., Morosinotto T., Bassi R. (2009). Light-induced dissociation of an antenna hetero-oligomer is needed for non-photochemical quenching induction. J. Biol. Chem. 284: 15255–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton, G., Liaaen-Jensen, S., and Pfander, H. (2004). Carotenoids Hand Book. (Basel, Switzerland: Birkhauser Publishing). [Google Scholar]
- Caffarri S., Kouril R., Kereïche S., Boekema E.J., Croce R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28: 3052–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffarri S., Passarini F., Bassi R., Croce R. (2007). A specific binding site for neoxanthin in the monomeric antenna proteins CP26 and CP29 of photosystem II. FEBS Lett. 581: 4704–4710 [DOI] [PubMed] [Google Scholar]
- Croce R., Weiss S., Bassi R. (1999). Carotenoid-binding sites of the major light-harvesting complex II of higher plants. J. Biol. Chem. 274: 29613–29623 [DOI] [PubMed] [Google Scholar]
- Cuttriss, A.J., and Pogson, B.J. (2006). Carotenoids. In The Structure and Function of Plastids, R.R.Wise and J.K. Hoober, eds (Dordrecht, The Netherlands: Springer), pp. 315–334. [Google Scholar]
- Dainese, P., Santini, C., Ghiretti-Magaldi, A., Marquardt, J., Tidu, V., Mauro, S., Bergantino, E., and Bassi, R. (1992). The organization of pigment-proteins within photosystem II. In Research in Photosynthesis, Vol. II, N. Murata, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 13–20. [Google Scholar]
- Dall’Osto L., Caffarri S., Bassi R. (2005). A mechanism of nonphotochemical energy dissipation, independent from PsbS, revealed by a conformational change in the antenna protein CP26. Plant Cell 17: 1217–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., Havaux M., Bassi R. (2010). Enhanced photoprotection by protein-bound vs. free xanthophyll pools: A comparative analysis of chlorophyll b and xanthophyll biosynthesis mutants. Mol. Plant 3: 576–593 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L., Cazzaniga S., North H., Marion-Poll A., Bassi R. (2007a). The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell 19: 1048–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Fiore A., Cazzaniga S., Giuliano G., Bassi R. (2007b). Different roles of α- and β-branch xanthophylls in photosystem assembly and photoprotection. J. Biol. Chem. 282: 35056–35068 [DOI] [PubMed] [Google Scholar]
- Davison P.A., Hunter C.N., Horton P. (2002). Overexpression of beta-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418: 203–206 [DOI] [PubMed] [Google Scholar]
- DellaPenna D., Pogson B.J. (2006). Vitamin synthesis in plants: Tocopherols and carotenoids. Annu. Rev. Plant Biol. 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W.W. (1992a). Carotenoid composition in sun and shade leaves of plants with different life forms. Plant Cell Environ. 15: 411–419 [Google Scholar]
- Demmig-Adams B., Adams W.W. (1992b). Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 599–626 [Google Scholar]
- Espineda C.E., Linford A.S., Devine D., Brusslan J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 10507–10511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A., Dallosto L., Cazzaniga S., Diretto G., Giuliano G., Bassi R. (2012). A quadruple mutant of Arabidopsis reveals a β-carotene hydroxylation activity for LUT1/CYP97C1 and a regulatory role of xanthophylls on determination of the PSI/PSII ratio. BMC Plant Biol. 12: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore A., Dall’osto L., Fraser P.D., Bassi R., Giuliano G. (2006). Elucidation of the beta-carotene hydroxylation pathway in Arabidopsis thaliana. FEBS Lett. 580: 4718–4722 [DOI] [PubMed] [Google Scholar]
- Flors C., Fryer M.J., Waring J., Reeder B., Bechtold U., Mullineaux P.M., Nonell S., Wilson M.T., Baker N.R. (2006). Imaging the production of singlet oxygen in vivo using a new fluorescent sensor, Singlet Oxygen Sensor Green. J. Exp. Bot. 57: 1725–1734 [DOI] [PubMed] [Google Scholar]
- Formaggio E., Cinque G., Bassi R. (2001). Functional architecture of the major light-harvesting complex from higher plants. J. Mol. Biol. 314: 1157–1166 [DOI] [PubMed] [Google Scholar]
- Ganeteg U., Külheim C., Andersson J., Jansson S. (2004). Is each light-harvesting complex protein important for plant fitness? Plant Physiol. 134: 502–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore A.M., Yamamoto H.Y. (1991). Zeaxanthin formation and energy-dependent fluorescence quenching in pea chloroplasts under artificially mediated linear and cyclic electron transport. Plant Physiol. 96: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Haldrup A., Simpson D.J., Scheller H.V. (2000). Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J. Biol. Chem. 275: 31211–31218 [DOI] [PubMed] [Google Scholar]
- Härtel H., Lokstein H., Dörmann P., Grimm B., Benning C. (1997). Changes in the composition of the photosynthetic apparatus in the galactolipid-deficient dgd1 mutant of Arabidopsis thaliana. Plant Physiol. 115: 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Dall’osto L., Bassi R. (2007). Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 145: 1506–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M., Dall’Osto L., Cuiné S., Giuliano G., Bassi R. (2004). The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. J. Biol. Chem. 279: 13878–13888 [DOI] [PubMed] [Google Scholar]
- Havaux M., Niyogi K.K. (1999). The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 96: 8762–8767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein P., Stöckel J., Bennewitz S., Oelmüller R. (2009). A protein related to prokaryotic UMP kinases is involved in psaA/B transcript accumulation in Arabidopsis. Plant Mol. Biol. 69: 517–528 [DOI] [PubMed] [Google Scholar]
- Holt N.E., Zigmantas D., Valkunas L., Li X.P., Niyogi K.K., Fleming G.R. (2005). Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Ihnatowicz A., Pesaresi P., Varotto C., Richly E., Schneider A., Jahns P., Salamini F., Leister D. (2004). Mutants for photosystem I subunit D of Arabidopsis thaliana: Effects on photosynthesis, photosystem I stability and expression of nuclear genes for chloroplast functions. Plant J. 37: 839–852 [DOI] [PubMed] [Google Scholar]
- Kim J., DellaPenna D. (2006). Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 103: 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Smith J.J., Tian L., Dellapenna D. (2009). The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 50: 463–479 [DOI] [PubMed] [Google Scholar]
- Klimmek F., Ganeteg U., Ihalainen J.A., van Roon H., Jensen P.E., Scheller H.V., Dekker J.P., Jansson S. (2005). The structure of higher plant LHCI: In vivo characterisation and structural interdependence of the Lhca proteins. Biochemisty 44: 3065–3073 [DOI] [PubMed] [Google Scholar]
- Koniger M., Harris G.C., Virgo A., Winter K. (1995). Xanthophyll-cycle pigments and photosynthetic capacity in tropical forest species - A comparative field study on canopy, gap and understory plants. Oecologia 104: 280–290 [DOI] [PubMed] [Google Scholar]
- Krech K., Ruf S., Masduki F.F., Thiele W., Bednarczyk D., Albus C.A., Tiller N., Hasse C., Schöttler M.A., Bock R. (2012). The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 159: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A. (2005). Singlet oxygen production in photosynthesis. J. Exp. Bot. 56: 337–346 [DOI] [PubMed] [Google Scholar]
- Kull, O., and Pfander, H. (1995). List of new carotenoids. In Carotenoids: Isolation and Analysis, S.L.-J.a.H.P.e.G. Britton, ed (Basel, Switzerland: Birkhauser Publishing), pp. 295–317. [Google Scholar]
- Landau A.M., Lokstein H., Scheller H.V., Lainez V., Maldonado S., Prina A.R. (2009). A cytoplasmically inherited barley mutant is defective in photosystem I assembly due to a temperature-sensitive defect in ycf3 splicing. Plant Physiol. 151: 1802–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Amann K., Meurer J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Meurer J. (2004). The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 38: 740–753 [DOI] [PubMed] [Google Scholar]
- Li X.P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Li X.P., Muller-Moule P., Gilmore A.M., Niyogi K.K. (2002). PsbS-dependent enhancement of feedback de-excitation protects photosystem II from photoinhibition. Proc. Natl. Acad. Sci. USA 99: 15222–15227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Yan H., Wang K., Kuang T., Zhang J., Gui L., An X., Chang W. (2004). Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428: 287–292 [DOI] [PubMed] [Google Scholar]
- Malkin S., Armond P.A., Mooney H.A., Fork D.C. (1981). Photosystem II photosynthetic unit sizes from fluorescence induction in leaves. Correlation to photosynthetic capacity. Plant Physiol. 67: 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, M.B., Sonenberg, N., and Hershey, J.W.B. (2007). Origins and principles of translational control. In Translational Control in Biology and Medicine, M.B. Mathews, N. Sonenberg, and J.W.B. Hershey, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 1–40. [Google Scholar]
- Meurer J., Grevelding C., Westhoff P., Reiss B. (1998). The PAC protein affects the maturation of specific chloroplast mRNAs in Arabidopsis thaliana. Mol. Gen. Genet. 258: 342–351 [DOI] [PubMed] [Google Scholar]
- Morosinotto T., Castelletti S., Breton J., Bassi R., Croce R. (2002). Mutation analysis of Lhca1 antenna complex. Low energy absorption forms originate from pigment-pigment interactions. J. Biol. Chem. 277: 36253–36261 [DOI] [PubMed] [Google Scholar]
- Mozzo M., Morosinotto T., Bassi R., Croce R. (2006). Probing the structure of Lhca3 by mutation analysis. Biochim. Biophys. Acta 1757: 1607–1613 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. (2002). PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Nelson N., Ben Shem A. (2004). The complex architecture of oxygenic photosynthesis. Nature 5: 1–12 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3: 455–460 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K., Shih C., Soon Chow W., Pogson B.J., Dellapenna D., Björkman O. (2001). Photoprotection in a zeaxanthin- and lutein-deficient double mutant of Arabidopsis. Photosynth. Res. 67: 139–145 [DOI] [PubMed] [Google Scholar]
- North H.M., De Almeida A., Boutin J.P., Frey A., To A., Botran L., Sotta B., Marion-Poll A. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50: 810–824 [DOI] [PubMed] [Google Scholar]
- Pan X., Li M., Wan T., Wang L., Jia C., Hou Z., Zhao X., Zhang J., Chang W. (2011). Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat. Struct. Mol. Biol. 18: 309–315 [DOI] [PubMed] [Google Scholar]
- Paulsen H. (1997). Pigment ligation to proteins of the photosynthetic apparatus in higher plants. Physiol. Plant. 100: 760–768 [Google Scholar]
- Peter, G.F., Takeuchi, T., and Thornber, J.P. (1991). Solubilization and two-dimensional electrophoretic procedures for studying the organization and composition of photosynthetic membrane polypeptides. Methods: A Companion to Methods in Enzymology 3: 115–124.
- Piques M., Schulze W.X., Höhne M., Usadel B., Gibon Y., Rohwer J., Stitt M. (2009). Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis. Mol. Syst. Biol. 5: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumley F.G., Schmidt G.W. (1987). Reconstitution of chlorophyll a/b light-harvesting complexes: Xanthophyll-dependent assembly and energy transfer. Proc. Natl. Acad. Sci. USA 84: 146–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Niyogi K.K., Björkman O., DellaPenna D. (1998). Altered xanthophyll compositions adversely affect chlorophyll accumulation and nonphotochemical quenching in Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 95: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B.J., Rissler H.M. (2000). Genetic manipulation of carotenoid biosynthesis and photoprotection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Ginies C., Soubigou-Taconnat L., Triantaphylidès C., Havaux M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport F., Béal D., Joliot A., Joliot P. (2007). On the advantages of using green light to study fluorescence yield changes in leaves. Biochim. Biophys. Acta 1767: 56–65 [DOI] [PubMed] [Google Scholar]
- Redding K., Cournac L., Vassiliev I.R., Golbeck J.H., Peltier G., Rochaix J.-D. (1999). Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J. Biol. Chem. 274: 10466–10473 [DOI] [PubMed] [Google Scholar]
- Sane P.V., Ivanov A.G., Hurry V., Huner N.P., Oquist G. (2003). Changes in the redox potential of primary and secondary electron-accepting quinones in photosystem II confer increased resistance to photoinhibition in low-temperature-acclimated Arabidopsis. Plant Physiol. 132: 2144–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbarbati A., Merigo F., Benati D., Tizzano M., Bernardi P., Osculati F. (2004). Laryngeal chemosensory clusters. Chem. Senses 29: 683–692 [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Albus C.A., Bock R. (2011). Photosystem I: Its biogenesis and function in higher plants. J. Plant Physiol. 168: 1452–1461 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Netz D.J.A., Frazzon J., Pierik A.J., Bill E., Gross J., Lill R., Meurer J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoike K. (2011). Photoinhibition of photosystem I. Physiol. Plant. 142: 56–64 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stöckel J., Bennewitz S., Hein P., Oelmüller R. (2006). The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141: 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.W., Ouyang M., Guo J.K., Ma J.F., Lu C.M., Adam Z., Zhang L.X. (2010). The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. Plant J. 62: 240–249 [DOI] [PubMed] [Google Scholar]
- Telfer A. (2005). Too much light? How beta-carotene protects the photosystem II reaction centre. Photochem. Photobiol. Sci. 4: 950–956 [DOI] [PubMed] [Google Scholar]
- Tian L., Magallanes-Lundback M., Musetti V., DellaPenna D. (2003). Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15: 1320–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Musetti V., Kim J., Magallanes-Lundback M., DellaPenna D. (2004). The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc. Natl. Acad. Sci. USA 101: 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S., Block M.A., Allen M., Westergren T., Albrieux C., Scheller H.V., Merchant S., Jensen P.E. (2003). Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. USA 100: 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kooten O., Snel J.F.H. (1990). The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 25: 147–150 [DOI] [PubMed] [Google Scholar]
- Varotto C., Pesaresi P., Meurer J., Oelmüller R., Steiner-Lange S., Salamini F., Leister D. (2000). Disruption of the Arabidopsis photosystem I gene psaE1 affects photosynthesis and impairs growth. Plant J. 22: 115–124 [DOI] [PubMed] [Google Scholar]
- Walters R.G., Horton P. (1995). Acclimation of Arabidopsis thaliana to the light environment: Changes in photosynthetic function. Planta 197: 306–312 [DOI] [PubMed] [Google Scholar]
- Wang Y.L., Mao L.S., Hu X.C. (2004). Insight into the structural role of carotenoids in the photosystem I: A quantum chemical analysis. Biophys. J. 86: 3097–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Rojas M., Friso G., van Wijk K.J., Meurer J., Barkan A. (2011). APO1 promotes the splicing of chloroplast group II introns and harbors a plant-specific zinc-dependent RNA binding domain. Plant Cell 23: 1082–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe O.T., Morimoto K., Kikuchi S., Nishio K., Terashima I., Nakai M. (2004). The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16: 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H.Y., Nakayama T.O., Chichester C.O. (1962). Studies on the light and dark interconversions of leaf xanthophylls. Arch. Biochem. Biophys. 97: 168–173 [DOI] [PubMed] [Google Scholar]
- Ye J.-W., Gong Z.-Y., Chen C.-G., Mi H.-L., Chen G.-Y. (2012). A mutation of OSOTP 51 leads to impairment of photosystem I complex assembly and serious photo-damage in rice. J. Integr. Plant Biol. 54: 87–98 [DOI] [PubMed] [Google Scholar]