CDKG1 directly regulates the splicing of CalS5 pre-mRNA for callose wall synthesis and pollen wall formation in Arabidopsis. CDKs are not only key regulators of the cell cycle, but also splicing regulators for other cellular processes, including pollen wall formation.
Abstract
Arabidopsis thaliana CYCLIN-DEPEDENT KINASE G1 (CDKG1) belongs to the family of cyclin-dependent protein kinases that were originally characterized as cell cycle regulators in eukaryotes. Here, we report that CDKG1 regulates pre-mRNA splicing of CALLOSE SYNTHASE5 (CalS5) and, therefore, pollen wall formation. The knockout mutant cdkg1 exhibits reduced male fertility with impaired callose synthesis and abnormal pollen wall formation. The sixth intron in CalS5 pre-mRNA, a rare type of intron with a GC 5′ splice site, is abnormally spliced in cdkg1. RNA immunoprecipitation analysis suggests that CDKG1 is associated with this intron. CDKG1 contains N-terminal Ser/Arg (RS) motifs and interacts with splicing factor Arginine/Serine-Rich Zinc Knuckle-Containing Protein33 (RSZ33) through its RS region to regulate proper splicing. CDKG1 and RS-containing Zinc Finger Protein22 (SRZ22), a splicing factor interacting with RSZ33 and U1 small nuclear ribonucleoprotein particle (snRNP) component U1-70k, colocalize in nuclear speckles and reside in the same complex. We propose that CDKG1 is recruited to U1 snRNP through RSZ33 to facilitate the splicing of the sixth intron of CalS5.
INTRODUCTION
Cyclin-dependent protein kinases (CDKs) belong to an important superfamily of Ser/Thr protein kinases that require the binding of a regulatory cyclin partner to induce their kinase activity (Morgan, 1997). Although CDKs were originally identified as key regulators of cell cycle transition (CDK1, 2, 4, and 6; Rane et al., 1999; Moore et al., 2003; Kozar et al., 2004), members in this family were also found to be involved in other cellular processes, such as activation of other CDKs (CDK7; Kaldis, 1999) and regulation of gene transcription by phosphorylating RNA polymerase II (CDK7, 8, and 9; Pinhero et al., 2004). Recent studies both in animal and plant cells suggest that a number of CDKs may function in RNA splicing regulation (Doonan and Kitsios, 2009). Human CDK11p110 interacts in vivo with two splicing factors, RNPS1 and 9G8, and promotes the splicing of β-globin pre-mRNA in vitro (Hu et al., 2003). CDK12 and CDK13 also alter the splicing pattern in cultured cells (Chen et al., 2006, 2007; Even et al., 2006). Recently, Arabidopsis thaliana CDKC2 was proposed to be involved in splicing regulation, as it was shown to colocalize with splicing factors and to modulate the distribution of spliceosome components (Kitsios et al., 2008).
RNA splicing is an important biological process, as most eukaryotic genes contain introns. For example, ∼79% of Arabidopsis genes have at least one intron (Schwartz et al., 2008). The splicing of pre-mRNA occurs in the spliceosome, which consists of pre-mRNA, five small nuclear ribonucleoprotein particles (snRNPs; U1, U2, U4/6, and U5), and other non-snRNP splicing factors, such as Ser/Arg (SR)–rich proteins (SR proteins; Krämer, 1996). An snRNP is composed of an snRNA and several related snRNP proteins, for example, U1 snRNA and U1-70K in U1 snRNP (Valadkhan and Jaladat, 2010). Spliceosome assembly is initiated by U1 snRNP recognition and binding of the 5′ splice site (5′ SS) of an intron, which is assisted by the SR-rich proteins (Cho et al., 2011). During early spliceosome assembly, a transient base-pairing forms between U1 snRNA and the 5′ SS, which could be stabilized by the binding of splicing factors to establish the U1 snRNP-pre-mRNA complex (Black, 2003). These splicing factors also include some SR protein family members, such as 9G8 and SC35. They are essential for the recognition of suboptimal splice sites (Kralovicova et al., 2011). Therefore, SR proteins are important early in spliceosome assembly (Shepard and Hertel, 2009). Several SR proteins interact with U1-70K, such as SRZ22 and SRZ21 (Golovkin and Reddy, 1998), SR33, and SR45 (Golovkin and Reddy, 1999).
In angiosperms, the pollen wall plays an important role in the pollen–stigma interaction and in the protection of pollen from environmental stresses (Heslop-Harrison, 1968; Meuter-Gerhards et al., 1999). The pollen wall consists of a simple inner intine layer, an intricate outer exine layer, and an outer pollen coat. Pollen wall development begins with primexine formation between the callose wall and the microspore membrane at the tetrad stage (Ariizumi and Toriyama, 2011). At this stage, four microspores, the products of a meiocyte, are enclosed inside a common callose envelope to form a tetrad. Callose synthase5 (CalS5; or glucan synthase-like2) is the main enzyme for the callose layer biosynthesis (Dong et al., 2005). Subsequently, primexine is deposited between the microspore plasma membrane and the callose wall. Defects in primexine formation result in male sterility or reduced male fertility (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Guan et al., 2008; Sun et al., 2013). After callose degradation, primexine develops into exine to form the pollen wall. Defects in callose synthesis affect primexine development, leading to aberrant exine pattern formation and male sterility (Dong et al., 2005; Nishikawa et al., 2005; Ariizumi and Toriyama, 2011; Chang et al., 2012).
Based on their cyclin binding motifs and phylogenetic analysis, Arabidopsis CDKs have been classified into seven categories and named as CDKA through CDKG (Menges et al., 2005; Umeda et al., 2005). CDKA and CDKB are direct regulators of cell cycle transitions (Boudolf et al., 2004; Iwakawa et al., 2006). CDKD and CDKF are CDK activation kinases, which regulate the activities of other CDKs (Shimotohno et al., 2004; Umeda et al., 2005; Hajheidari et al., 2012). CDKC and CDKE are related to the transcriptional regulation of RNA polymerase II (Barrôco et al., 2003; Wang and Chen, 2004; Cui et al., 2007). The CDKG category, comprising CDKG1 and CDKG2 with putative PLTSLRE cyclin binding motifs, is the least functionally defined (Menges et al., 2005). Here we show that CDKG1 plays an important role in regulating pre-mRNA splicing of the CalS5 gene. A loss-of-function mutation in CDKG1 resulted in aberrant callose deposition, defective pollen wall formation during microspore development, and severely reduced male fertility. Furthermore, our results suggest that CDKG1 interacts with splicing factor RSZ33; thus, it may play an important role in posttranscriptional regulation in Arabidopsis.
RESULTS
Plants with Loss of CDKG1 Function Show Dramatically Reduced Male Fertility
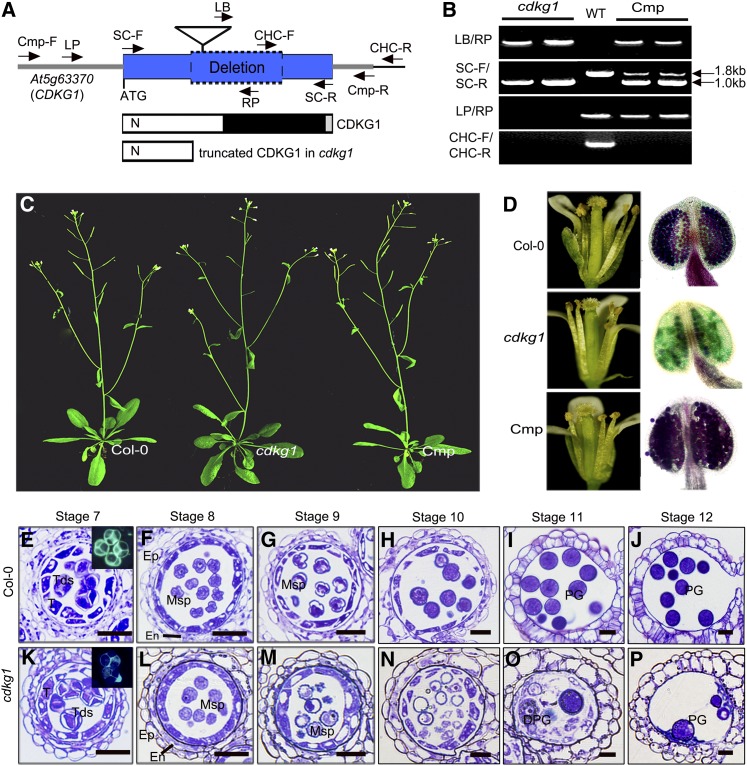
We performed functional analysis of CDKG1 in pollen wall formation and male fertility in Arabidopsis. We obtained a T-DNA insertion line of CDKG1 (SALK_075762, designated as cdkg1) from the ABRC (http://abrc.osu.edu/). The annotated T-DNA was stated to be inserted in the coding sequence (CDS) of CDKG1 (Figure 1A). However, PCR detection (Figure 1B) and sequencing analysis showed that the At5g63370 (CDKG1) CDS fragment from +605 to +1406 bp, together with the annotated T-DNA, was absent in this locus in cdkg1, although the T-DNA was still in the genome of cdkg1 (Figure 1A). The CDS for this truncated form of cdkg1 contained a premature stop codon (TGA, 607 to 609 bp) that would result in a truncated protein containing only a partial CDKG1 N terminus (Figure 1A). Therefore, this mutated protein is expected to lack CDK activity.
Figure 1.
The cdkg1 Mutant Shows Reduced Male Fertility but Normal Vegetative Growth.
(A) A DNA fragment of 800 bp in CDKG1 (At5g63370) is deleted in cdkg1. The triangle shows the annotated T-DNA of SALK_075762. The left and the right arrows show the primers used for the genotyping and for the genetic complementation assay. The corresponding protein structure of CDKG1 and the truncated CDKG1 in cdkg1 are also shown. Blue box, the CDS; gray lines, the untranslated regions; blue box with dotted line, deleted portion; white box/N, the N-terminal extension; black box, the Ser/Thr protein kinase catalytic domain; gray box, the C-terminal extension.
(B) Genotype detection by PCR using four primer sets for cdkg1, wild type (WT; Col-0), and cdkg1 complementation transformants (Cmp).
(C) Thirty-five-day-old plants of the wild type (Col-0), cdkg1, and one complementation plant are shown.
(D) Open flowers (left panels) and stamens stained by Alexander’s stain (right panels) of the wild type, cdkg1, and a complementation line are shown.
(E) to (P) Semithin cross-section analysis for anther development of Col-0 ([E] to [J]) and cdkg1 ([K] to [P]), from stage 7 to stage 12. DPG, degraded pollen grain; En, endothecium; Ep, epidermis; Msp, microspore; PG, pollen grain; T, tapetum; Tds, tetrads. Bars = 50 μm.
cdkg1 plants displayed normal vegetative growth similar to the wild type (Columbia-0 [Col-0]; Figure 1C). Wild-type siliques contained 48.8 ± 11.4 seeds (n = 75) on average, whereas cdkg1 showed reduced fertility with only 5.35% seed set (2.61 ± 5.13 per silique, n = 400). The anther dehiscence and anther length of cdkg1 were similar to those of the wild type (Figure 1D). An Alexander’s staining (Alexander, 1969) assay for pollen viability showed the cdkg1 anthers contained very few purple-stained viable pollen grains (Figure 1D), which was consistent with its low fertility.
Backcrossing with the wild type (Col-0) resulted in F1 plants with normal fertility. In the F2 population, the male-sterile mutant phenotype was inherited in a Mendelian fashion, with 25% of the progeny displaying male sterility (98 of 409, χ2 = 0.235; P > 0.01), indicating that the reduced male fertility phenotype was caused by a single recessive nuclear mutation that was controlled sporophytically. For genetic complementation, a 3.6-kb genomic fragment consisting of 1300-bp upstream sequence, 1839-bp CDS, and 500-bp downstream sequence of CDKG1 (Figure 1A) was introduced into homozygous cdkg1 plants. In the T1 progeny, 17 of 23 transgenic lines showed restored fertility with normal pollen development and seed set (Figures 1C and 1D). This indicates that the deletion within CDKG1 was responsible for the mutant phenotype. There is another CDKG gene, CDKG2. However, the T-DNA insertion lines of this gene had no obvious phenotype, so we did not perform further analysis of the CDKG2 mutants.
To understand the details of the developmental defects in cdkg1 pollen development, we compared semithin cross sections of anthers of cdkg1 and the wild type. In Arabidopsis, anther development has been divided into 14 stages based on morphological characters (Sanders et al., 1999). Before stage 7 (tetrad stage), no detectable difference was observed between cdkg1 and the wild type. However, the microspores appeared to be shrunken in cdkg1 anthers at stage 7 (Figures 1E and 1K). At stage 8, most microspores of cdkg1 were relatively round rather than the typical angular shape of wild-type microspores (Figures 1F and 1L). At stage 9, wild-type microspores had a trivalvular shape with dense cytoplasm (Figure 1G), while most cdkg1 microspores were still round and cytoplasm leakage was observed from some microspores (Figure 1M). After stage 9, wild-type microspores gradually developed into mature pollen grains (Figures 1H to 1J). However, most cdkg1 microspores were gradually degraded after stage 9 (Figures 1N to 1P).
To obtain some clues about the role of CDKG1 in microspore development, we analyzed the expression pattern of CDKG1. Real-time RT-PCR analysis with total RNA from root, stem, rosette leaf, and inflorescence indicated that CDKG1 was preferentially expressed in leaf and inflorescence and weakly expressed in the other tissues examined (see Supplemental Figure 1A online). RNA in situ hybridization with wild-type anther cross sections showed that CDKG1 exhibited relatively higher expression at the meiosis and tetrad stages (see Supplemental Figure 1B online), suggesting that CDKG1 may play a role during these two stages.
cdkg1 Plants Are Defective in Pollen Wall Formation
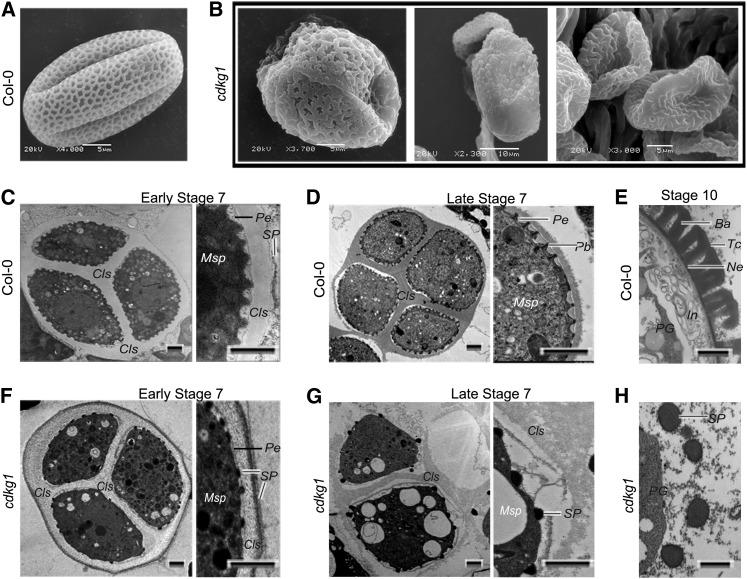
Several mutants defective in pollen wall formation also show round microspores at stage 8 (Ariizumi et al., 2004; Paxson-Sowders et al., 2001; Guan et al., 2008). Therefore, we investigated pollen wall pattern of cdkg1 via scanning electron microscopy analysis. The surviving pollen grains of cdkg1 exhibited abnormal exine pattern when compared with that of the wild type (Figures 2A and 2B). To further examine the reason for the pollen wall defect in cdkg1, we performed transmission electron microscopy (TEM). In the wild type, the primexine matrix was deposited between the callose wall and the microspore plasma membrane at the early tetrad stage, and the microspore membrane became regularly undulated (Figure 2C). However, in cdkg1, the primexine matrix was thinner and the regular plasma membrane undulation was not observed (Figure 2F). In the wild type, the callose wall was less electron dense at the early tetrad stage, a time when the electron-dense sporopollenin (or more probably its precursor) was deposited on the outside surface of the callose wall (Figure 2C). By contrast, in cdkg1, the callose wall was moderately electron dense, and flat sporopollenin particles were observed on the surface of the microspore plasma membrane at the early tetrad stage (Figure 2F). At the late tetrad stage of the wild type, the degenerating callose wall was electron dense, and the sporopollenin was deposited as rod-like probacula that were regularly distributed in the primexine matrix (Figure 2D). However, in cdkg1, dot-like sporopollenin particles were randomly distributed on the plasma membrane (Figure 2G). At stage 10, the exine composed of tecta and bacula was formed around wild-type microspores (Figure 2E), while this layer was absent in cdkg1, with only big sporopollenin granules around microspores (Figure 2H). These results suggest that the primexine formation is defective in cdkg1.
Figure 2.
Pollen Wall Development Is Defective in cdkg1.
(A) to (B) Scanning electron microscopy analysis.
(A) Typical pollen grain of the wild type with regular exine pattern. Bar = 5 μm.
(B) Typical surviving pollen grains of cdkg1 with irregular shape and abnormal exine pattern. Bars = 5 (left and right panels) or 10 μm (middle panel).
(C) to (H) TEM analysis. Pollen wall development of the wild type ([C] to [E]) and cdkg1 ([F] to [H]) at early stage 7 ([C] and [F]), late stage 7 ([D] and [G]), and stage 10 ([E] and [H]). Cls, callose; Msp, microspore; Pb, probacula; Pe, primexine; SP, sporopollenin particles or sporopollenin precursors. Bars = 1 μm.
Callose Deposition Is Defective in cdkg1 Due to the Abnormal Splicing of CalS5 Pre-mRNA
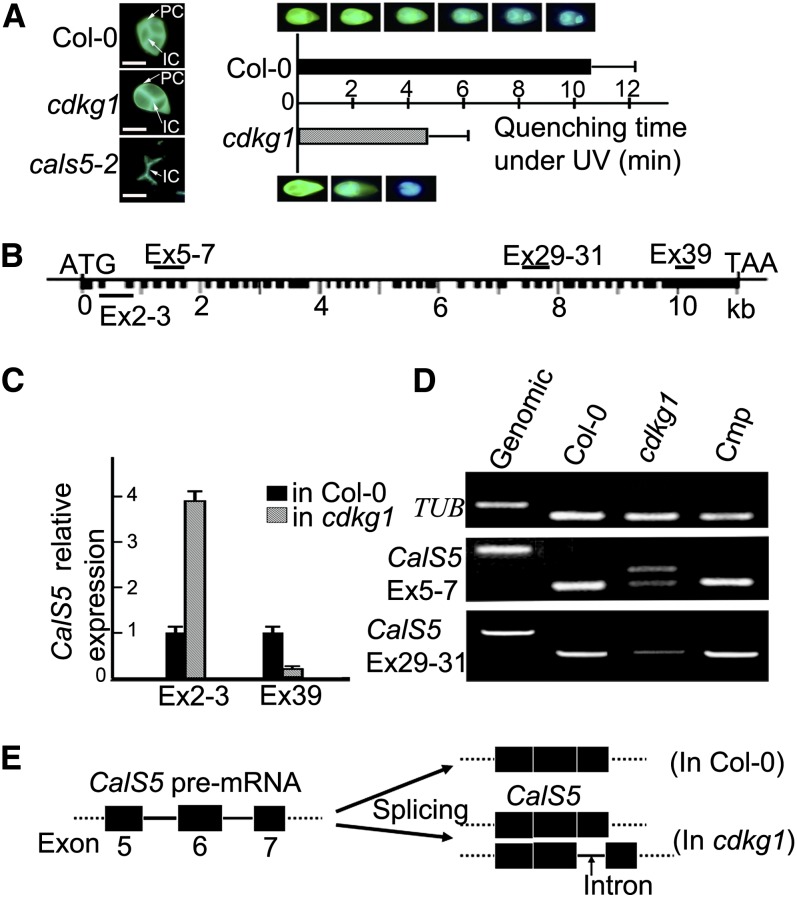
Since the callose wall is important in primexine formation (Nishikawa et al., 2005; Dong et al., 2005), we investigated callose deposition at the tetrad stage by aniline blue staining. The callose wall fluorescence of the cdkg1 tetrad was weaker than that of the wild type, especially that of the peripheral callose wall (Figure 3A). Callose staining of tetrad stage sections showed that the callose wall of cdkg1 was much thinner than that of the wild type (Figures 1E and 1K). This was further confirmed by a callose fluorescence bleaching test. For the 30 cdkg1 tetrads randomly selected from 30 tetrad-stage buds, the average fluorescence quenching time for the callose wall was much less than that for the wild type (Figure 3A). These results suggest that callose synthesis was significantly reduced in cdkg1. In Arabidopsis, CalS5 (Figure 3B) encodes the main synthase for the peripheral callose wall around meiocytes and tetrads (Dong et al., 2005), and this callose layer is almost lost in cals5 knockout mutants (Dong et al., 2005; Nishikawa et al., 2005; Figure 3A). We therefore investigated whether the expression of CalS5 was altered in cdkg1. Quantitative RT-PCR analysis using the primer set for exon2-3 of CalS5 showed 3.9-fold upregulation in cdkg1, whereas the primer set for exon 39 showed only 17% of the wild-type level (Figure 3C). This suggests that the full-length CalS5 transcript in cdkg1 is present at most at 17% of the level in the wild type.
Figure 3.
cdkg1 Shows Reduced Callose Synthesis and Abnormal pre-mRNA Splicing of CalS5.
(A) Examples of callose fluorescence in tetrads of Col-0, cdkg1, and cals5-2 (left panel). Callose fluorescence quenching (right panel). Error bars represent sd of the mean of 30 biological replicates. IC, interstitial callose; PC, peripheral callose. Bars = 20 μm.
(B) CalS5 gene structure. Black boxes, exons; black lines, introns; the name of the primer sets (e.g., Ex2-3) for gene expression analysis are shown.
(C) Real-time RT-PCR expression analysis of CalS5 in Col-0 and cdkg1, using the primer sets EX2-3 and Ex39. Error bars represent sd of the mean of three biological replicates.
(D) RT-PCR with different primer sets using the cDNAs of Col-0, cdkg1, and a complementation line (Cmp) TUB.
(E) Model of partial splicing of the sixth intron of CalS5 pre-mRNA in cdkg1, according to RT-PCR and the sequence analysis of the additional PCR product.
To further examine the abnormal transcription of CalS5 in cdkg1, additional primer pairs were designed for RT-PCR analysis. When the primer set for exons 5 to 7 was used, two PCR products were obtained in cdkg1. The smaller product was similar to the size of the PCR band from wild-type cDNA, while the larger one was in a size intermediate between the smaller one and the PCR product from the genomic DNA (Figure 3D). Sequencing showed that the larger product contained the 99-bp intron 6 of CalS5. Thus, the sixth intron was only partially spliced in cdkg1 (Figure 3E). A stop codon occurred inside intron 6, resulting in a truncated protein with only 254 amino acid residues for the abnormal CalS5 mRNA containing the intron. The other primer sets tested showed that other introns of CalS5 were spliced normally in cdkg1.
CalS5 Pre-mRNA Is Coimmunoprecipitated with CDKG1
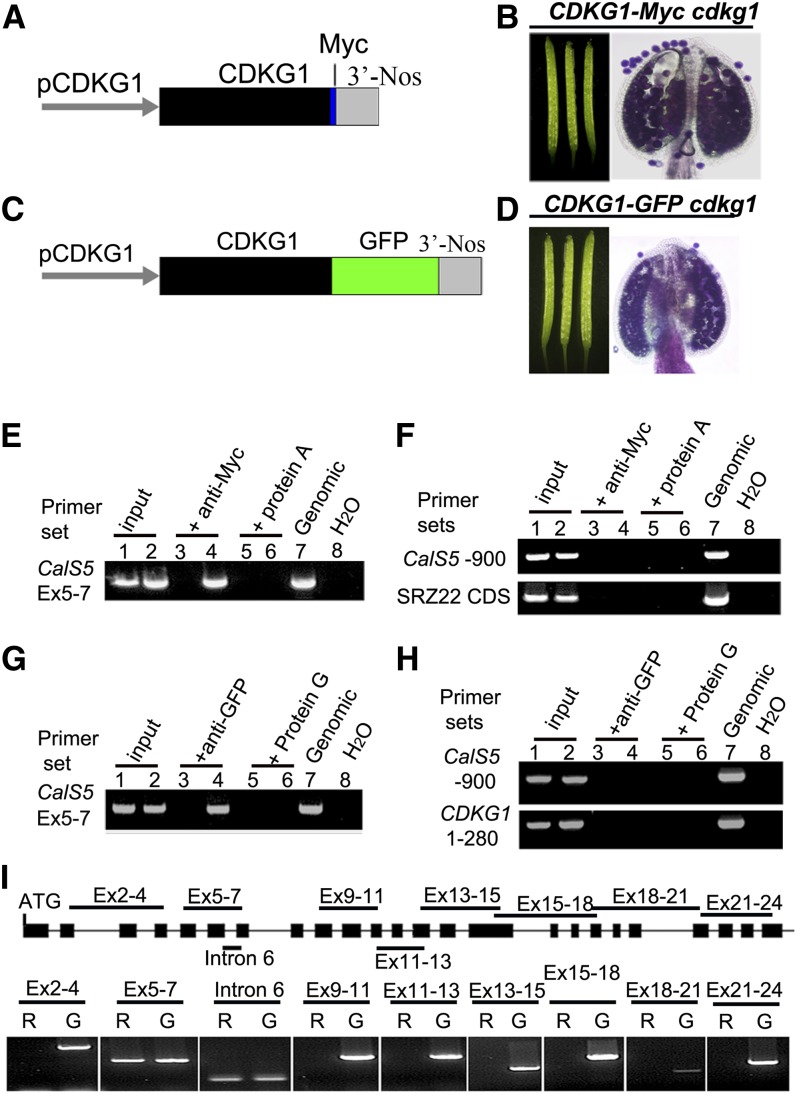
To understand whether CDKG1 is directly associated with intron 6 of CalS5 pre-mRNA, an RNA immunoprecipitation (RIP) assay was performed as described (Terzi and Simpson, 2009). We made two protein fusion constructs, one tagged with Myc, PCDKG1:CDKG1-Myc (Figure 4A), and one with green fluorescent protein (GFP), PCDKG1:CDKG1-GFP (Figure 4C). Each construct complemented the phenotypic defects when introduced into cdkg1 (Figures 4B and 4D), indicating that the two fusion proteins can fulfill the function of CDKG1. Inflorescences of these transgenic plants were used for the RIP assay. In the RIP assay with CDKG1-Myc, for the primer set spanning intron 6, an expected PCR product was detected in the anti-Myc RIP (Figure 4E, lane 4). This PCR product was absent in the anti-Myc RIP with the wild type (Figure 4E, lane 3) and in both mock RIPs (with magnetic beads only) of the transgenic plants and the wild type (Figure 4E, lane 5 and 6). We used the primer sets CalS5-900 and SRZ22-CDS as negative controls. No PCR product was obtained from any of these RIP cDNAs (Figure 4F). Similar results were observed when we used PCDKG1:CDKG1-GFP transgenic plants (Figures 4G and 4H). These results indicate that CDKG1 might bind to the CalS5 pre-mRNA. To investigate whether CDKG1 can bind to other parts of CalS5 pre-mRNA, a total of nine additional primer sets for CalS5 was designed for RIP RT-PCR analysis. PCR products were obtained only for primer sets covering intron 6 and exons 5 to 7 (Figure 4I). These results indicate that CDKG1 is associated with the CalS5 pre-mRNA around intron 6. Since CDKG1 is a protein without an obvious RNA recognition motif (RRM), we hypothesize that CDKG1 might bind to CalS5 pre-mRNA by interacting with some other proteins, such as splicing factors that have RRMs.
Figure 4.
CDKG1 Is Associated with the Intron 6 Region of CalS5 pre-mRNA.
(A) and (C) PCDKG1:CDKG1-Myc and PCDKG1:CDKG1-GFP constructs used for genetic complementation assays.
(B) and (D) siliques and stained anthers of cdkg1 plants bearing PCDKG1:CDKG1-Myc (B) or PCDKG1:CDKG1-GFP (D).
(E) and (F) RIP RT-PCR analysis of a PCDKG1:CDKG1-Myc line. Lanes 1, 3, and 5, the wild type; lanes 2, 4, and 6, PCDKG1:CDKG1-Myc line; lane 7, wild-type genomic DNA; lane 8, water.
(G) and (H) RIP RT-PCR analysis of a PCDKG1:CDKG1-GFP line. Lanes 1, 3, and 5, the wild type; lanes 2, 4, and 6, PCDKG1:CDKG1-GFP line; lane 7, wild-type genomic DNA; lane 8, water.
(I) RIP RT-PCR analysis of the RIP cDNA of the PCDKG1:CDKG1-Myc line using different primer pairs for CalS5. R, RIP cDNA; G, wild-type genomic DNA.
CDKG1 Interacts with Splicing Factor RSZ33
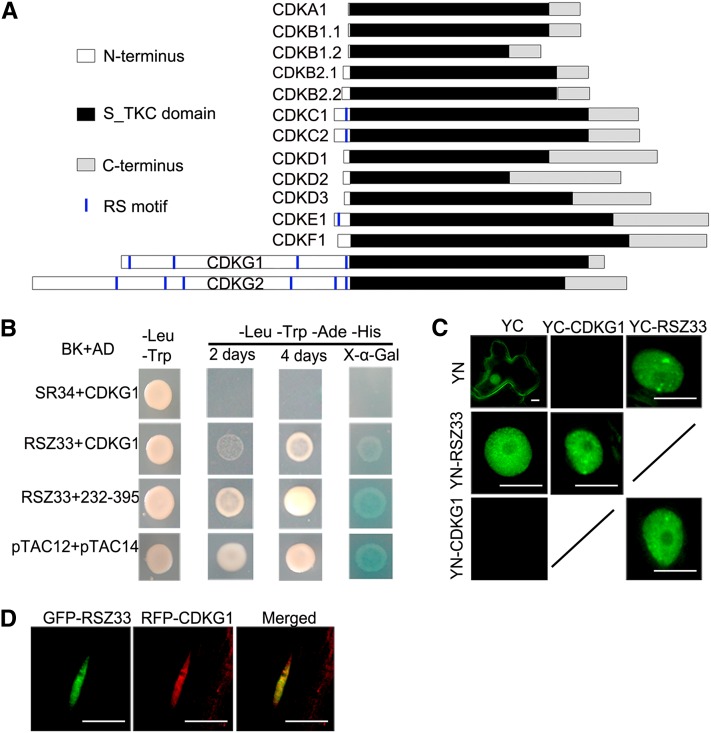
Several CDKs have been reported to be involved in pre-mRNA splicing, including human CDK11, CDK12, and CDK13 and Arabidopsis CDKC2 (Hu et al., 2003; Chen et al., 2006, 2007; Even et al., 2006; Kitsios et al., 2008). These CDKs share two common features: (1) they colocalize or interact with splicing factors such as SR proteins; and (2) they contain at least one RS motif in their N termini, for example, CDK11 (three RS motifs), CDK12 (20 RS motifs), CDC2L5/CDK13 (15 RS motifs), and CDKC2 (one RS motif). RS motifs are thought to mediate the interaction with splicing factor SR proteins (Wu and Maniatis, 1993; Zhu and Krainer, 2000). CDKG1 has an N terminus that is longer than those of CDKA to CDKF. CDKG1 contains four RS motifs in the N terminus (Figure 5A), including the last RS motif conserved in almost in all the CDKG1 orthologs (see Supplemental Figure 2A online). We also compared the core region (as defined by Ko et al., 2001) of CDKG1 with those of Arabidopsis CDKs and the mammalian CDKs related to splicing regulation. CDKG1 is more similar to the CDKs associated with splicing than the others, including CDKA, CDKB, and CDKF (see Supplemental Figure 2B online). In the phylogenetic tree of Arabidopsis CDKs, the CDKG clade is close to the CDKC clade (see Supplemental Figure 2C online). Therefore, as these CDKs are related to splicing regulation, CDKG1 might also interact with some SR proteins with one to two RRMs (Shepard and Hertel, 2009).
Figure 5.
CDKG1 Interacts with Splicing Factor RSZ33.
(A) Domains of 14 CDKs in Arabidopsis. S_TKC domain, Ser/Thr, protein kinase catalytic domain; RS motif, Ser/Arg motif.
(B) CDKG1 full length and an internal fragment both interact with RSZ33 in yeast cells; SR34+CDKG1 is a negative control, and pTAC12+pTAC14 is a positive control. BK, pGBK-T7; AD, pGADT7; 232-395, CDKG1 fragment amino acids 232 to 395.
(C) BiFC assay between CDKG1 and RSZ33 in tobacco leaf cells. YC, YFP C terminus; YN, YFP N terminus. Bars = 10 μm.
(D) mRFP-CDKG1 colocalizes with GFP-RSZ33. A root cell nucleus of a transgenic Arabidopsis plant expressing the two fusion proteins is shown. Bars = 10 μm.
We therefore performed yeast two-hybrid assays to find potential SR protein partners of CDKG1. The Arabidopsis genome contains 19 SR genes (Reddy, 2004). We cloned the CDSs of these SR genes into the pGBKT7 vector and the CDS of CDKG1 into the pGADT7 vector. The yeast clones bearing pGAD-CDKG1 and pGBK-RSZ33 were able to grow on the selective medium (Trp-, Leu-, adenine-, and His-), although their growth was slower than that of the positive control, containing pTAC12 and pTAC14 (Gao et al., 2011). These clones showed positive X-α-Gal reaction (Figure 5B). These results show that CDKG1 interacts with RSZ33 in yeast.
To confirm and visualize the interaction between RSZ33 and CDKG1 in plant cells, a bimolecular fluorescence complementation (BiFC) assay was performed based on split yellow fluorescent protein (YFP; Hu et al., 2002). CDKG1 and RSZ33 were fused with the N and C terminus of YFP, respectively, and transiently coexpressed in tobacco (Nicotiana tabacum) leaves. For two combinations (YN-CDKG1+YC-RSZ33 and YC-CDKG1+YN-RSZ33), YFP signals were observed in the nucleus, with relatively stronger signals in nuclear speckles. No YFP signal was observed for the other two combinations, YN+YC-CDKG1 and YC+YN-CDKG1 (Figure 5C). The BiFC assay confirmed the interaction between CDKG1 and RSZ33. When the monomeric red fluorescent protein (mRFP)–tagged CDKG1 (mRFP-CDKG1) and GFP-RSZ33 were coexpressed in transgenic plants, we also observed their colocalization in nuclear speckles (Figure 5D), where most SR proteins localize (Fang et al., 2004). This colocalization assay therefore supports the interaction between the two proteins.
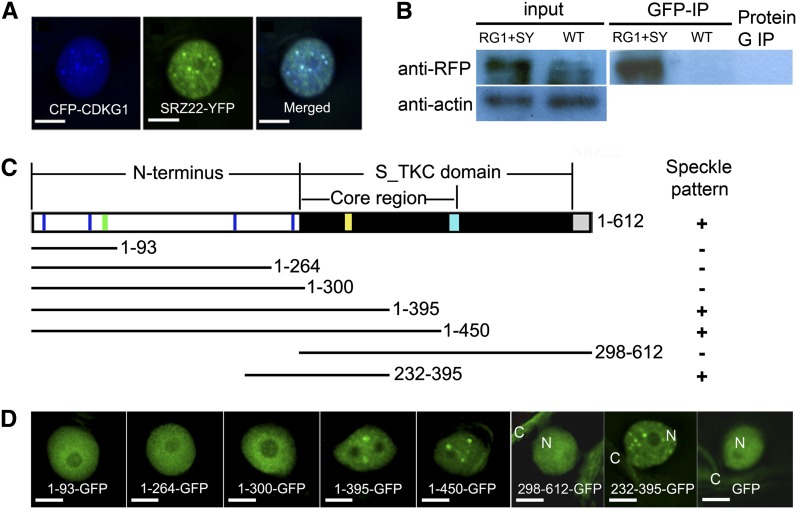
CDKG1 Is Targeted to Splicing Machinery and Is Dependent on the Region Including the Conserved RS Motif
RSZ33 is known to interact with SRZ22 and SRZ21 (Lopato et al., 2002), which further interact with U1-70k, a component of the U1 snRNP (Golovkin and Reddy, 1998). This suggested that CDKG1 might be targeted to the splicing machinery through physical and functional interaction with RSZ33. To further verify the involvement of CDKG1 in the splicing machinery, we performed colocalization and coimmunoprecipitation assays with CDKG1 and SRZ22. When coexpressed in tobacco leaf epidermal cells, both the cyan fluorescent protein (CFP)–tagged CDKG1 (CDKG1-CFP) and SRZ22-YFP were observed in the nucleoplasm, primarily in nuclear speckles (Figure 6A). We obtained transgenic Arabidopsis lines expressing both P35S:mRFP-CDKG1 and P35S:SRZ22-YFP. Nuclear extracts from the transgenic plants were immunoprecipitated using an anti-GFP antibody. Immunoblot analysis of the immunoprecipitated products using an anti-RFP antibody detected a protein band consistent with the size of the mRFP-CDKG1 fusion protein (Figure 6B). This coimmunoprecipitation assay suggests that CDKG1 and SRZ22 reside in the same complex in nuclear speckles.
Figure 6.
CDKG1 Is Targeted to Splicing Machinery in a Manner Dependent on the Region Including the Conserved RS Motif.
(A) CDKG1-CFP colocalizes with SRZ22-YFP in tobacco leaf cells. Bars = 5 μm.
(B) mRFP-CDKG1 is coimmunoprecipitated with SRZ22-YFP. Left panels, inputs. The inputs detected by anti-RFP antibody are nuclear extracts and the inputs detected by anti-β-actin antibody (down) are total soluble proteins. Right panel, nuclear extracts detected by anti-RFP antibody that were immunoprecipitated by anti-GFP magnetic beads or by magnetic beads alone. RG1, mRFP-CDKG1; SY, SRZ22-YFP; WT, the wild type.
(C) Diagrammatic representation of CDKG1 and the fragments fused with GFP. Blue line, RS motifs; green line, nuclear localization signal; yellow line, PLTSLRE motif; cyan box, T-loop; white box, N terminus; black box, S_TKC domain; gray box, C terminus.
(D) The cellular localization of CDKG1-GFP fragments in tobacco leaf cells. C, cytoplasm; N, nucleus. Bars = 5 μm.
To define the region in CDKG1 responsible for nuclear speckle localization, we made several constructs using GFP fused with the CDKG1 fragments (Figure 6C). CDKG1 contains a nuclear localization signal in its N terminus, according to the PredictNLS server (Cokol et al., 2000; https://rostlab.org/owiki/index.php/PredictNLS). When fused with N-terminal fragments containing various RS motifs, CDKG1 1 to 93 (RS motifs 1 and 2), 1 to 264 (RS motifs 1 to 3), and 1 to 300 (all four RS motifs), diffuse GFP signals were observed in the nuclei of the epidermal cells (Figure 6D). Similarly, for CDKG1 298-612-GFP lacking the CDKG1 N terminus, the diffuse GFP signals were observed in the cytoplasm and nucleus, similar to the localization pattern of free GFP (Figure 6D). These results suggest that both the N terminus and the S_TKC domain are required for its nuclear speckle localization. The other three fusion proteins, including 1-395-GFP, 1-450-GFP, and 232-395-GFP, showed nuclear speckle localization (Figure 6D), similar to full-length CDKG1-CFP. The CDKG1 fragment includes the peptide from 232 to 395, which showed a stronger interaction with RSZ33 than did the full-length CDKG1 (Figure 5B). This 232 to 395 fragment contains a partial core region and an N-terminal RS motif (the fourth motif in the N-terminal region), which is conserved in almost all orthologs of CDKG1 in other eukaryotes (see Supplemental Figure 2A online). The core region also includes some critical residues for the function of a CDK, including the putative substrate binding pocket. These results suggest that the fragment 232 to 395 including the conserved RS motif is sufficient to define the CDKG1 nuclear speckle localization and to interact with RSZ33.
DISCUSSION
CDKG1 is one of the two CDKG genes in the Arabidopsis CDK gene family, with RS motifs in its N terminus. The cdkg1 is a knockout mutant, presumably due to a T-DNA–induced deletion (Figure 1A). Similar T-DNA–induced deletions have been reported in dozens of T-DNA lines (Wang, 2008). Here, we present that CDKG1 regulates pollen wall formation (Figure 2) and male fertility (Figure 1C). Molecular and biochemical evidence shows that CDKG1 interacts with a splicing factor (Figures 5B to 5D) to regulate the pre-mRNA splicing of CalS5 (Figures 3D and 3E), the major enzyme for callose synthesis and pollen wall development. Double mutant analysis of cdkg1 cals5-2 (see Supplemental Figure 3 online) suggests that CDKG1 may also regulate some other genes involved in pollen wall formation and plant fertility.
CDKG1 Interacts with RSZ33 to Regulate the Splicing of Cals5 Pre-mRNA
We present several lines of evidence, including yeast two-hybrid, BiFC, and colocalization assays, to support the interaction between CDKG1 and splicing factor RSZ33 (Figures 5B to 5D). RSZ33 is known to interact with SRZ22 and SRZ21 (Lopato et al., 2002), which further interact with U1-70K, a component of U1 snRNP (Golovkin and Reddy, 1998). Additionally, CDKG1 is also colocalized with SRZ22 in tobacco leaves and coimmunoprecipitated with this SR protein in transgenic Arabidopsis (Figures 6A and 6B). The RIP assay showed that CDKG1 is associated with the pre-mRNA of CalS5 (Figures 4E to 4I). Therefore, it is likely that CDKG1 binds to the CalS5 pre-mRNA indirectly through SRZ33, a splicing factor with one RRM (Shepard and Hertel, 2009).
The sixth intron of CalS5 is a rare type of intron with a GC 5′ SS. A GC 5′ SS is intrinsically weaker than the major GT 5′ SS because the T>C substitution introduces a mismatch in the base pairing between the splice site and U1 snRNA (Kralovicova et al., 2011). For this rare type of intron, various SR proteins bind to the U1 snRNP-pre-mRNA complex to stabilize the base pairing (Black, 2003). Human SR proteins 9G8 and SC35 can promote the splicing of introns with the GC 5′ SS (Kralovicova et al., 2011). In cdkg1, the low splicing efficiency of the CalS5 intron 6 (Figure 3D) verifies the weakness of this GC 5′ SS. This suggests that U1 snRNP binding to the sixth intron of CalS5 is not stable. The normal splicing of this intron in the wild type and in cdkg1 complementation lines suggests that CDKG1 is recruited to stabilize the binding of U1 snRNP to the region of CalS5 intron 6. This facilitates spliceosome removal of this rare type of intron. Kralovicova et al. (2011) reported that the flanking sequences could enhance the splicing efficiency of an intron with the GC 5′ SS. Therefore, both CDKG1 and the flanking sequences are important for efficient splicing of the introns with a GC 5′ SS.
CDKG1 Affects Pollen Wall Formation during Microsporogenesis
CDKG1 is preferentially expressed in leaves and inflorescences with relatively low expression in root and stem (see Supplemental Figure 1A online). However, only a defect in male reproduction was observed in cdkg1. This is similar to some other genes with extensive expression patterns that are involved in microsporogenesis, such as CDKA (Iwakawa et al., 2006) and NPU (Chang et al., 2012). It is probable that a lack of gene redundancy in anthers allows observation of the cdkg1 phenotype during anther development. CDKG1 may be involved in other biological processes that cannot be observed morphologically in other tissues. CalS5 shows highest expression in meiocytes, tetrads, microspores, and mature pollen (Dong et al., 2005; Nishikawa et al., 2005). The relatively higher expression of CDKG1 at the meiosis and the tetrad stages (see Supplemental Figure 1B online) is consistent with the regulation of CalS5 pre-mRNA splicing during these stages.
Pollen development includes several cell division processes: male meiosis, tapetal cell endoreproduction, and pollen mitosis. We analyzed these cell division processes with 4′,6-diamidino-2-phenylindole staining (Ross et al., 1996). No obvious defects were found in cdkg1 in these cell cycle events. During meiosis I and II, the nuclear division and chromosome distribution in cdkg1 were indistinguishable from those in the wild type. In cdkg1, the tapetal cells contained two nuclei at the meiosis stage, and the surviving pollen grains contained three nuclei (see Supplemental Figure 4 online). These data indicate that CDKG1 is not or not solely involved in cell cycle regulation of anther cells during anther development.
Pollen wall development initiates at the tetrad stage with primexine formation beneath the callose wall. At this stage, the callose wall may also act as a molecular filter to protect the haploid microspores in tetrads from the influence of the diploid tissues in anther (Heslop-Harrison and Mackenzie, 1967). CalS5 is responsible for callose layer synthesis (Dong et al., 2005). Both knockout (Dong et al., 2005) and reduced expression of CalS5 (Nishikawa et al., 2005) cause an exine developmental defect. cdkg1 showed downregulated CalS5 expression (Figure 3C) and improper callose deposition around tetrads (Figure 3A), which are quite similar to those in CalS5 mutants. Based on the molecular filter function of callose wall and the TEM results (Figures 2C and 2D), we speculated that at the early tetrad stage in the wild type, the sporopollenin or its precursors can hardly pass through the dense callose wall until the callose wall begins to be degenerated at the late tetrad stage, as indicated by the electron density of callose wall and the probacula formation (Figures 2C and 2D). By contrast, in cdkg1 tetrads, the sporopollenin seems to be deposited around microspores in advance (Figure 2F). It is possible that the lower callose content (Figure 3A) results in a bigger mesh size of the molecular filter of the cdkg1 callose wall. Therefore, it cannot protect the sporopollenin or its precursors from invading at the early tetrad stage, when the primexine, the mold for exine formation, is not fit to accept them. Simultaneously, the primexine is also thinner in cdkg1 tetrads than in the wild type, even at the late tetrad stage (Figures 2F and 2D). CDKG1 may regulate the synthesis and/or the deposition of primexine. It was proposed that callose could conceivably trap primexine subunits, increasing their local concentration and preventing them from diffusing into the anther locule (Ariizumi and Toriyama, 2011). Probably the bigger mesh size of the molecular filter of the callose wall in cdkg1 leads to leakage of the primexine materials. In cdkg1, the probacula do not have enough space to develop because of the thinner primexine. As a result, sporopollenin is deposited randomly into flat particles (Figure 2F) and then into incomplete dot-like structure around the microspores of cdkg1 (Figures 2G and 2H). Finally, most developing microspores of cdkg1 undergo degradation after stage 9 (Figures 1M to 1P) as they lose protection from the complete and regular exine layer (Figure 2H).
In cdkg1, the CalS5 5′ region was upregulated to 3.9-fold and its 3′ region was downregulated to ∼17% of the wild-type level (Figure 3C). One possibility for the abnormal expression is a pause in gene transcription owing to the disturbed splicing as reported (Bentley, 1999; Kitsios et al., 2008). Alternatively, the abnormal CalS5 transcript might undergo noncoding mRNA decay due to the premature termination codon in the nonspliced intron 6. Previous studies revealed that abnormal mRNA containing a premature termination codon could be degraded by the noncoding mRNA decay mechanism (Culbertson and Leeds, 2003; Isken and Maquat, 2007). Both of the above possibilities could result in reduced expression of mature CalS5 and the accumulation of the 5′ region of CalS5 mRNA in cdkg1 (Figure 3C).
The phenotype rescue by CalS5 CDS is important in revealing the relationship between the cdkg1 phenotype and the abnormal CalS5 splicing. When we obtained the full-length CDS of CalS5, all of the fragments were mutated. This is in agreement with a previous report that T-DNA constructs containing a full-length genomic copy of CalS5 are lethal in Agrobacterium tumefaciens (Nishikawa et al., 2005). Therefore, we constructed double mutants to analyze the relationship between CDKG1 and CalS5 in this work. Both cdkg1 and CalS5 knockout mutants were able to yield a limited number of seeds. If CalS5 is the only target of CDKG1 in microsporogenesis, the phenotype of cdkg1 cals5-2 should be the same as that of cals5-2. However, the double mutant had a more severe phenotype (i.e., almost complete male sterility and pollen loss; see Supplemental Figure 3 online). This suggests that CDKG1 may regulate some other genes important for primexine development beyond CalS5. In Arabidopsis, DEX1, NEF1, RPG1, and NPU are involved in primexine formation (Paxson-Sowders et al., 2001; Ariizumi et al., 2004; Guan et al., 2008; Chang et al., 2012; Sun et al., 2013), but the splicing patterns of these genes were not altered in cdkg1, and their transcription levels were similar to those in the wild type (see Supplemental Figure 5 online). CDKG1 may therefore regulate some novel gene(s) important for pollen wall formation and plant fertility.
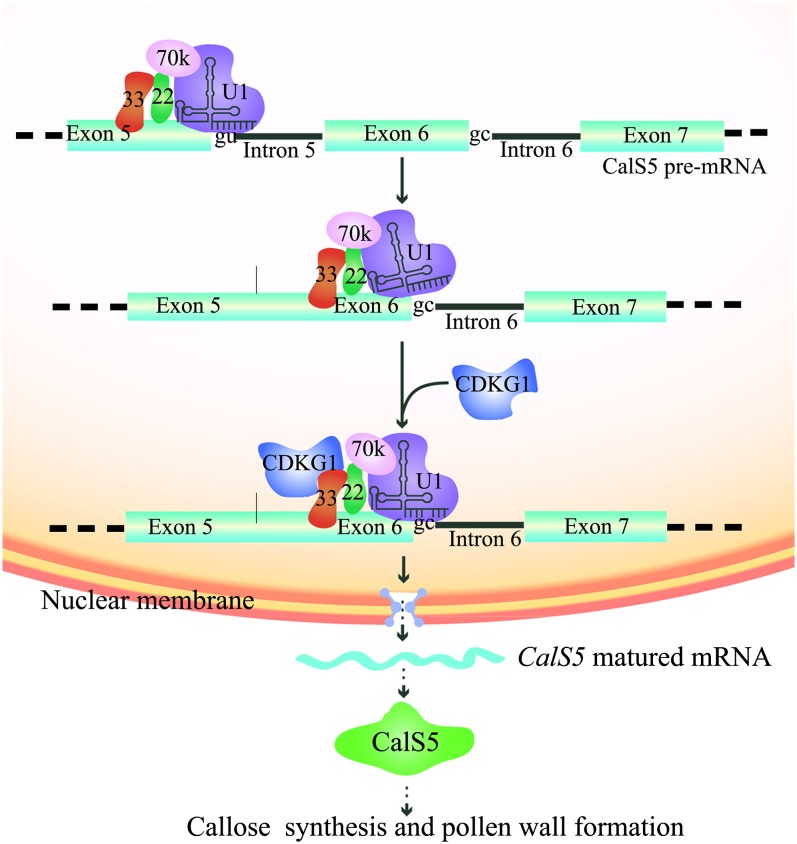
Putative Working Model of CDKG1 Function in Pre-mRNA Splicing
We propose a working model for CDKG1 function in pre-mRNA splicing of CalS5 (Figure 7). Normally, introns with the GT 5′ SS are spliced without the help of CDKG1. For rare introns with the GC 5′ SS, such as intron 6 of CalS5, CDKG1 is recruited to U1 snRNP through RSZ33 and SRZ22 to stabilize the base paring between the GC 5′ SS and U1 snRNA. This recruitment facilitates the efficient splicing of intron 6. The mature CalS5 mRNA is then transferred to the cytoplasm for translation of CalS5.
Figure 7.
Model for CDKG1 in Regulation of pre-mRNA Splicing of CalS5 and Pollen Wall Development.
For an intron with the GT 5′ SS, such as intron 5 of CalS5, U1 snRNP can recognize the splice site and initiate the removal of this intron efficiently without the help of CDKG1. For CalS5 intron 6 with the GC 5′ SS, U1 snRNP can’t recognize the intron efficiently because of the mismatch between the GC 5′ SS and U1 snRNA. The recruitment of CDKG1 through RSZ33 to the U1 snRNP stabilizes the binding of U1 snRNP to the region around intron 6 and facilitates the efficient splicing of intron 6. The matured CalS5 mRNA is transferred to the cytoplasm for translation of CalS5 protein, which is responsible for callose synthesis and pollen wall formation. 33, RSZ33; 22, SRZ22; 70K, U1-70k; U1, U1 snRNP.
[See online article for color version of this figure.]
METHODS
Plant Growth Conditions
The Arabidopsis thaliana plants used in this study are in a Col-0 background. Seeds were sown on vermiculite and allowed to imbibe for 3 d at 4°C. Plants were grown under long-day conditions (16 h light/8 h dark) in an ∼22°C growth room. Transgenic plants were generated via Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998) and selected on PNS (Plant Nutritional Solution) media containing 20 mg/L hygromycin B and/or 50 mg/L kanamycin. Transient expression for subcellular localization and BiFC assays in tobacco (Nicotiana tabacum) leaves were performed as previously described (Hu et al., 2002).
Phenotype Characterization and Microscopy
Plant photos were taken with a Cybershot T-20 digital camera (Sony). Flower pictures were taken using an Olympus dissection microscope with an Olympus digital camera. Alexander’s staining and 4′,6-diamidino-2-phenylindole staining were performed as described (Alexander, 1969; Ross et al., 1996). Callose staining and semithin sections were performed as described in a previous report (Zhang et al., 2007). For the callose fluorescence bleaching assay, the inflorescences were fixed in Carnoy’s fixative for 3 h. After rinsing with phosphate buffer, pH 7.2, three times, the inflorescences were stained with 0.1% aniline blue at 4°C for 3 d. The tetrads were isolated from the anthers of the inflorescences and kept under UV. The quenching time was recorded when the callose fluorescence of each tetrad disappeared. Pictures were taken with an Olympus BX-51 microscope (Olympus).
For scanning electron microscopy examination, the opened flowers of the wild type and cdkg1 were dissected and the dehiscent anthers were coated with 8-nm gold. Observation and photography were performed on a JSM-840 microscope (Jeol). TEM samples were prepared as previously described (Zhang et al., 2007). Sections were observed on an H-600 transmission electron microscope (Hitachi).
For cdkg1 complementation, the 3.6-kb CDKG1 genomic fragment was amplified using LA-Taq polymerase (Takara) with primer set CMP-F/CMP-R. The oligonucleotides used for complementation and other assays in this study are listed in Supplemental Table 1 online. Primers were synthesized by Life Technologies. After verification by sequencing (Genomics), the fragment was cloned into the pCAMBIA1300 binary vector (Cambia) and subsequently introduced into homozygous cdkg1 plants. For cdkg1 background verification, SC-F/SC-R primers were used to validate DNA deletion of CDKG1 for the transformants, LP/RP primers were used to detect either CDKG1 genomic sequence or transgenic complementation fragment, and the genomic-specific primers CHC-F/CHC-R were used to validate the homozygous cdkg1 background. As CHC-R primer was designed 180 bp downstream of CMP-R primer, PCR with the CHC-F/CHC-R primer set was not able to amplify a 1.7-kb fragment in homozygous plants even if the complementation fragment was integrated into the genome.
Expression Analysis
RNA extraction was performed using Trizol (Life Technologies) following the protocol in the user’s manual. A total of 30 cycles of PCR was performed for RT-PCR if not mentioned. Col-0 and cdkg1 inflorescence cDNAs were used for the expression analysis of CDKG1, CalS5, and the pre-mRNA splicing of CalS5. Real-time quantitative PCR were performed using gene-specific primers and iQ SYBR Green Master Mix (Bio-Rad) on the ABI 7300 platform (Applied Biosystems). The experiment was repeated thrice and data were averaged. β-Tubulin gene was used as an internal normalization control. Fold change in gene expression were calculated using the ΔΔCt (cycle threshold) values.
Subcellular Localization, Colocalization Analysis, and BiFC Assay
GFP, CFP, YFP, and mRFP fluorescence was detected under an Olympus IX70 inverted microscope system. For GFP fusion, CDKG1 and its fragments were amplified from Arabidopsis wild-type genomic DNA with KOD polymerase (Toyobo). The fragments were cloned into the binary vector pMON530-GFP or pCAMBIA1300-35S-GFP, driven by the 35S promoter. These constructs were introduced into wild-type plants or transiently expressed in tobacco leaf via Agrobacterium strain ASE or GV3101 as described (Clough and Bent, 1998; Hu et al., 2002).
For CFP and mRFP fusions, the CDKG1 CDS was amplified with KOD polymerase (Takara) using the primer set G1-Bam-F/ G1-Sal-R. The CDKG1 CDS was cloned into the binary vectors pC131-35S-CFP and pCAMBIA1300-35S-mRFP. SRZ22 CDS was amplified from Arabidopsis wild-type inflorescence cDNA with the primer set SRZ22-Kpn-Eco-F/SRZ22-Bam-Sal-R and then cloned into pMON530-YFP. The two constructs were introduced into wild-type plants or transiently expressed in tobacco leaves as described above.
For BiFC assays, the CDSs of CDKG1 and RSZ33 were cloned into pCAMBIA1300-35S-YN and pCAMBIA1300-35S-YC vectors and transiently expressed in tobacco leaf cells as described by Hu et al. (2002).
In Vivo Coimmunoprecipitation
In vivo coimmunoprecipitation assay was performed as previously described (Fill et al., 2008). The leaves of 4-week-old transgenic plants of the wild type and 35S:SRZ22-YFP+35S:mRFP-CDKG1 were used as protein sources. After fixation, grinding, and sonication treatment, nuclear proteins were extracted from 4 g of leaves. Protein G–coupled magnetic beads (Life Technologies) and an anti-GFP antibody (Cwbiotech) were used for immunoprecipitation. Eluted proteins were run on SDS-PAGE, immunoblotted using the Pierce Fast Western Blot Kit (Thermo Scientific), and probed with an anti-RFP (MBL).
RIP Assay
The 3.1-kb CDKG1 genomic sequence before the terminal code was amplified with the primer set PG1-Eco-F/G1-Sal-R and cloned into pCAMBIA1300-Myc and pCAMBIA1300-GFP, following a 3′-Nos sequence. The RIP assay was performed as described (Terzi and Simpson 2009). Briefly, 3 to 5 g of inflorescence from transgenic and wild-type (as control) plants was fixed, and the total soluble nuclear extracts were isolated after sonication. A portion of each nuclear extract was immunoprecipitated with the corresponding antibody-coupled magnetic protein G beads (Life Technologies) or protein G beads as control. Sixty microliters of each nuclear extract was stored at −70°C for input preparation. RNAs were isolated from the immunoprecipitated products and each input. The resulting cDNAs were used for RT-PCR analysis. The anti-Myc antibody was obtained from Genomics.
Yeast Two-Hybrid Analysis
Yeast two-hybrid assay was performed using the Clontech two-hybrid system according to the manufacturer’s instructions. The CDS of CDKG1 and its fragment 232 to 395 were amplified and cloned into the pGADT7 vector. The CDSs of 19 SR proteins were cloned into pGBKT7 vector individually. The pGADT7-CDKG1 was cotransformed into the yeast strain AH109 with the pGBKT7-SR individually. The transformants were screened on supplemented synthetic dextrose medium lacking Leu and Trp or on supplemented synthetic dextrose medium lacking Leu, Trp, His, and adenine hemisulfate salt with X-α-Gal. The positive clones were verified both by CDKG1-specific primers and by SR protein–specific primers.
Phylogenetic Analysis
The multiple sequence alignment of full-length protein sequences was performed using the ClustalW tool online (http://www.genome.jp/tools/clustalw/) and displayed using Boxshade (http://www.ch.embnet.org/software/BOX_form.html). The alignment is available as Supplemental Data Set 1 online. Phylogenetic trees were constructed and tested by MEGA4.0 based on the neighbor-joining method (bootstrap method with 1000 trials was used to calculate the statistical support for the nodes).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: CDKG1 (At5g63370), CalS5 (At2g13680), RSZ33 (At2g37340), SRZ22 (At4g31580), CDKA1 (At3g48750), CDKB1;1 (At3g54180), CDKB1;2 (At2g38620), CDKB2;1 (At1g76540), CDKB2;2 (At1g20930), CDKC1 (At5g10270), CDKC2 (At5g64960), CDKD1 (At1g73690), CDKD2 (At1g66750), CDKD3 (At1g18040), CDKE1 (At5g63610), CDKF1 (At4g28980), and CDKG2 (At1g67580).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Pattern of CDKG1.
Supplemental Figure 2. CDKG1 Shows Similarity to Those CDKs Related to Splicing Regulation.
Supplemental Figure 3. The Phenotype of Double Mutant cdkg1 cals5-2.
Supplemental Figure 4. Cell Divisions Related to Pollen Development Are Normal in cdkg1.
Supplemental Figure 5. The Splicing and Expression of Primexine-Related Genes in cdkg1.
Supplemental Table 1. List of PCR Primers and Their Usage in This Study.
Supplemental Data Set 1. The Alignment of CDK family members in Arabidopsis.
Acknowledgments
We thank the ABRC (Ohio State University) for Arabidopsis seeds from T-DNA insertion lines, Yu-Da Fang from the Institute of Plant Physiology and Ecology (Shanghai, China) for his help in the colocalization and BiFC analysis, and Sheila McCormick (USDA/Agricultural Research Service, University of California, Berkeley) and Peter Breslin (Loyola University) for their critical reading and editing of this article. This work was supported by a grant from the National Science Foundation of China (30925007), by the Program of Shanghai Subject Chief Scientist (11XD1403900), and by the Leading Academic Discipline Project of Shanghai Municipal Education Commission (J50401).
AUTHOR CONTRIBUTIONS
X.-Y.H. and Z.-N.Y. designed the research. X.-Y.H., J.N., M.-X.S. J.Z., J.-F.G., J.Y., and Q.Z. performed the research. X.-Y.H. and Z.-N.Y. analyzed the data. X.-Y.H. and Z.-N.Y. prepared the article.
Glossary
- CDK
cyclin-dependent protein kinase
- snRNP
small nuclear ribonucleoprotein particle
- SR
Ser/Arg
- 5′ SS
5′ splice site
- CDS
coding sequence
- Col-0
Columbia-0
- TEM
transmission electron microscopy
- RIP
RNA immunoprecipitation
- GFP
green fluorescent protein
- RRM
RNA recognition motif
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- mRFP
monomeric red fluorescent protein
- CFP
cyan fluorescent protein
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Barrôco R.M., De Veylder L., Magyar Z., Engler G., Inzé D., Mironov V. (2003). Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell. Mol. Life Sci. 60: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. (1999). Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11: 347–351 [DOI] [PubMed] [Google Scholar]
- Black D.L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Boudolf V., Vlieghe K., Beemster G.T., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.S., Zhang C., Chang Y.H., Zhu J., Xu X.F., Shi Z.H., Zhang X.L., Xu L., Huang H., Zhang S., Yang Z.N. (2012). No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 158: 264–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.H., Wang Y.C., Fann M.J. (2006). Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cell. Biol. 26: 2736–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.H., Wong Y.H., Geneviere A.M., Fann M.J. (2007). CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem. Biophys. Res. Commun. 354: 735–740 [DOI] [PubMed] [Google Scholar]
- Cho S., Hoang A., Sinha R., Zhong X.Y., Fu X.D., Krainer A.R., Ghosh G. (2011). Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 108: 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair R., Rost B. (2000). Finding nuclear localization signals. EMBO Rep. 1: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Fan B., Scholz J., Chen Z. (2007). Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell 19: 1388–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M.R., Leeds P.F. (2003). Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13: 207–214 [DOI] [PubMed] [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328 [DOI] [PubMed] [Google Scholar]
- Doonan J.H., Kitsios G. (2009). Functional evolution of cyclin-dependent kinases. Mol. Biotechnol. 42: 14–29 [DOI] [PubMed] [Google Scholar]
- Even Y., Durieux S., Escande M.L., Lozano J.C., Peaucellier G., Weil D., Genevière A.M. (2006). CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J. Cell. Biochem. 99: 890–904 [DOI] [PubMed] [Google Scholar]
- Fang Y., Hearn S., Spector D.L. (2004). Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell 15: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fill B.K., Qiu J.L., Petersen K., Petersen M., Mundy J. (2008). Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc. 2008: .pdb.prot5049 [DOI] [PubMed] [Google Scholar]
- Gao Z.P., Yu Q.B., Zhao T.T., Ma Q., Chen G.X., Yang Z.N. (2011). A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (1998). The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell 10: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem. 274: 36428–36438 [DOI] [PubMed] [Google Scholar]
- Guan Y.F., Huang X.Y., Zhu J., Gao J.F., Zhang H.X., Yang Z.N. (2008). RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 147: 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajheidari M., Farrona S., Huettel B., Koncz Z., Koncz C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24: 1626–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1968). Pollen wall development. The succession of events in the growth of intricately patterned pollen walls is described and discussed. Science 161: 230–237 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison J., Mackenzie A. (1967). Autoradiography of soluble [2-14-C]thymidine derivatives during meiosis and microsporogenesis in Lilium anthers. J. Cell Sci. 2: 387–400 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu D., Mayeda A., Trembley J.H., Lahti J.M., Kidd V.J. (2003). CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278: 8623–8629 [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L.E. (2007). Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 21: 1833–1856 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831 [DOI] [PubMed] [Google Scholar]
- Kaldis P. (1999). The cdk-activating kinase (CAK): From yeast to mammals. Cell. Mol. Life Sci. 55: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitsios G., Alexiou K.G., Bush M., Shaw P., Doonan J.H. (2008). A cyclin-dependent protein kinase, CDKC2, colocalizes with and modulates the distribution of spliceosomal components in Arabidopsis. Plant J. 54: 220–235 [DOI] [PubMed] [Google Scholar]
- Ko T.K., Kelly E., Pines J. (2001). CrkRS: A novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 114: 2591–2603 [DOI] [PubMed] [Google Scholar]
- Kozar K., Ciemerych M.A., Rebel V.I., Shigematsu H., Zagozdzon A., Sicinska E., Geng Y., Yu Q., Bhattacharya S., Bronson R.T., Akashi K., Sicinski P. (2004). Mouse development and cell proliferation in the absence of D-cyclins. Cell 118: 477–491 [DOI] [PubMed] [Google Scholar]
- Kralovicova J., Hwang G., Asplund A.C., Churbanov A., Smith C.I., Vorechovsky I. (2011). Compensatory signals associated with the activation of human GC 5′ splice sites. Nucleic Acids Res. 39: 7077–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. (1996). The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409 [DOI] [PubMed] [Google Scholar]
- Lopato S., Forstner C., Kalyna M., Hilscher J., Langhammer U., Indrapichate K., Lorković Z.J., Barta A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277: 39989–39998 [DOI] [PubMed] [Google Scholar]
- Menges M., de Jager S.M., Gruissem W., Murray J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Meuter-Gerhards A., Riegart S., Wiermann R. (1999). Studies on sporopollenin biosynthesis in Curcurbita maxima (DUCH)-II: the involvement of aliphatic metabolism. J. Plant Physiol. 154: 431–436 [Google Scholar]
- Moore J.D., Kirk J.A., Hunt T. (2003). Unmasking the S-phase-promoting potential of cyclin B1. Science 300: 987–990 [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1997). Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Nishikawa S., Zinkl G.M., Swanson R.J., Maruyama D., Preuss D. (2005). Callose (β-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxson-Sowders D.M., Dodrill C.H., Owen H.A., Makaroff C.A. (2001). DEX1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 127: 1739–1749 [PMC free article] [PubMed] [Google Scholar]
- Pinhero R., Liaw P., Bertens K., Yankulov K. (2004). Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur. J. Biochem. 271: 1004–1014 [DOI] [PubMed] [Google Scholar]
- Rane S.G., Dubus P., Mettus R.V., Galbreath E.J., Boden G., Reddy E.P., Barbacid M. (1999). Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in beta-islet cell hyperplasia. Nat. Genet. 22: 44–52 [DOI] [PubMed] [Google Scholar]
- Reddy A.S. (2004). Plant serine/arginine-rich proteins and their role in pre-mRNA splicing. Trends Plant Sci. 9: 541–547 [DOI] [PubMed] [Google Scholar]
- Ross K.J., Fransz P., Jones G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4: 507–516 [DOI] [PubMed] [Google Scholar]
- Sanders P.M., Anhthu Q.B., Weterings K., McIntire K.N., Hsu Y., Lee P.Y., Troung M.T., Beals T.P., Goldberg R.B. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11: 297–322 [Google Scholar]
- Schwartz S.H., Silva J., Burstein D., Pupko T., Eyras E., Ast G. (2008). Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 18: 88–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard P.J., Hertel K.J. (2009). The SR protein family. Genome Biol. 10: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno A., Umeda-Hara C., Bisova K., Uchimiya H., Umeda M. (2004). The plant-specific kinase CDKF;1 is involved in activating phosphorylation of cyclin-dependent kinase-activating kinases in Arabidopsis. Plant Cell 16: 2954–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, M.X., Huang, X.Y., Yang, J., Guan, Y.F., and Yang, Z.N. (2013). Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. doi: 10.1007/s00497-012-0208-1 [DOI] [PubMed] [Google Scholar]
- Terzi L.C., Simpson G.G. (2009). Arabidopsis RNA immunoprecipitation. Plant J. 59: 163–168 [DOI] [PubMed] [Google Scholar]
- Umeda M., Shimotohno A., Yamaguchi M. (2005). Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol. 46: 1437–1442 [DOI] [PubMed] [Google Scholar]
- Valadkhan S., Jaladat Y. (2010). The spliceosomal proteome: At the heart of the largest cellular ribonucleoprotein machine. Proteomics 10: 4128–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chen X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.H. (2008). How effective is T-DNA insertional mutagenesis in Arabidopsis? J. Biochem. Tech. 1: 11–20 [Google Scholar]
- Wu J.Y., Maniatis T. (1993). Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75: 1061–1070 [DOI] [PubMed] [Google Scholar]
- Zhang Z.B., Zhu J., Gao J.F., Wang C., Li H., Zhang H.Q., Zhang S., Wang D.M., Wang Q.X., Huang H., Xia H.J., Yang Z.N. (2007). Transcription factor AtMYB103 is required for anther development by regulating microspore release from tetrads and exine formation in Arabidopsis. Plant J. 52: 528–538 [DOI] [PubMed] [Google Scholar]
- Zhu J., Krainer A.R. (2000). Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 14: 3166–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]