The reorganization of the plant cortical microtubule cytoskeleton into a transverse coalignment was analyzed quantitatively in living cells using hormone induction. Transverse patterning initiates from the cell’s midzone, progressing bidirectionally toward the apex and base of the cell.
Abstract
The acentriolar cortical microtubule arrays in dark-grown hypocotyl cells organize into a transverse coaligned pattern that is critical for axial plant growth. In light-grown Arabidopsis thaliana seedlings, the cortical array on the outer (periclinal) cell face creates a variety of array patterns with a significant bias (>3:1) for microtubules polymerizing edge-ward and into the side (anticlinal) faces of the cell. To study the mechanisms required for creating the transverse coalignment, we developed a dual-hormone protocol that synchronously induces ∼80% of the light-grown hypocotyl cells to form transverse arrays over a 2-h period. Repatterning occurred in two phases, beginning with an initial 30 to 40% decrease in polymerizing plus ends prior to visible changes in the array pattern. Transverse organization initiated at the cell’s midzone by 45 min after induction and progressed bidirectionally toward the apical and basal ends of the cell. Reorganization corrected the edge-ward bias in polymerization and proceeded without transiting through an obligate intermediate pattern. Quantitative comparisons of uninduced and induced microtubule arrays showed a limited deconstruction of the initial periclinal array followed by a progressive array reorganization to transverse coordinated between the anticlinal and periclinal cell faces.
INTRODUCTION
Microtubules are cellular polymers composed of repeating α-β tubulin subunits assembled in a head-to-tail orientation to form hollow tubes (Ledbetter and Porter, 1964; Howard and Hyman, 2003). In most interphase animal cells, microtubules nucleate from γ-tubulin–containing complexes that are localized to a discrete organelle called the centrosome (Mazia, 1987; Doxsey, 2001; Bornens, 2002). By collecting the microtubule minus ends to the centrosome, animal cells create a radial array having the dynamic plus ends projecting outward toward the cell periphery. Plant microtubules also nucleate on γ-tubulin complexes (Liu et al., 1993, 1994; McDonald et al., 1993; Murata et al., 2005; Pastuglia et al., 2006), but unlike animals, flowering plants do not make centrioles or a centralized microtubule-organizing center (Newcomb, 1969). Interphase plant microtubules nucleate from distributed sites throughout the cell cortex, giving rise to a dynamic polymer network closely associated with the plasma membrane (Shaw et al., 2003; Murata et al., 2005; Nakamura and Hashimoto, 2009).
The organizational state of the plant cortical microtubule array influences the direction of plant cell expansion (Baskin, 2001; Lloyd, 2011). Hypocotyl and root cells extend axially to push the chlorophyll-bearing cotyledons into the sunlight and the primary root into the soil, respectively. The cortical microtubules in both cases organize into coaligned patterns that are transverse to the plant growth axis (Baskin, 2001). These microtubules, in turn, pattern cellulose deposition into the cell wall by guiding the plasma membrane–bound cellulose synthase complexes (Green, 1962; Giddings et al., 1980; Paredez et al., 2006; Chan et al., 2010). The transversely oriented cellulose fibers are proposed to retard turgor-driven radial cell swelling in favor of axial cell extension (Baskin, 2005). In the case of the dark-grown seedling hypocotyl, cells can extend to ∼500 times their original length by this mechanism in the absence of new cell divisions (Gendreau et al., 1997; Le et al., 2005). Exposing etiolated hypocotyl cells to light rapidly reorganizes the microtubule arrays, leading to a new pattern of cellulose deposition accompanying the switch from axial growth to radial thickening (Refrégier et al., 2004; Vandenbussche et al., 2005).
The mechanisms by which the cortical microtubules coalign and orient to the cell growth axis have been the subject of considerable speculation (Green, 1962; Ledbetter, 1982; Lloyd and Chan, 2002; Dixit and Cyr, 2004a; Ehrhardt and Shaw, 2006; Hashimoto and Kato, 2006; Baulin et al., 2007; Lucas and Shaw, 2008; Sedbrook and Kaloriti, 2008; Allard et al., 2010b; Eren et al., 2010; Ambrose et al., 2011). Early proposals, based on electron and immunofluorescence microscopy of fixed cells, focused on lateral sliding of the microtubules, possibly using motors to power the interactions between microtubules on the cell cortex (Hardham and Gunning, 1978; Lloyd and Wells, 1985; Cyr, 1994; Cyr and Palevitz, 1995; Wymer and Lloyd, 1996; Lloyd and Chan, 2002). Observations made in live cells found no evidence for lateral microtubule sliding, finding instead that polymers in hypocotyl cortical arrays remained attached to the cell cortex and exhibited a form of polymer treadmilling (Shaw et al., 2003; Ehrhardt and Shaw, 2006). The treadmilling motility facilitates cortical polymer interactions that often result in microtubule bundling (Shaw et al., 2003; Dixit and Cyr, 2004b) or changes to the polymerization state of the microtubule (Wightman and Turner, 2007). Plant microtubules were observed to nucleate at the cell cortex (Shaw et al., 2003; Murata et al., 2005) and either coalign to form a bundle or branch from an existing microtubule (Shaw et al., 2003; Murata et al., 2005; Nakamura et al., 2010). These live-cell observations of plant cortical microtubules in hypocotyl cells have provided a number of possible mechanisms that could contribute to transverse array organization (Ehrhardt, 2008; Lloyd and Chan, 2008; Lucas and Shaw, 2008).
Contemporary models for transverse microtubule array organization have focused on cell geometry and the potential self-organizing properties of the treadmilling microtubules. Self-organizing properties have generally been taken to be those that are a state property of all the microtubules in the system that can affect some change in the orientation of the microtubules within an array. Computational modeling was used to explore the proposal that angle-dependent association of treadmilling microtubules into bundles drives array coalignment (Wasteneys and Ambrose, 2009). These studies suggest that angle-dependent bundling plays a significant role but does not constitute a complete self-organizing system with reference to the cell axis (Baulin et al., 2007; Allard et al., 2010b; Eren et al., 2010). The addition of other phenomena, such as new microtubules entering from the cell’s longitudinal side walls or an increased probability of switching from growth to shortening (i.e., catastrophe) at microtubule intersections or at the apical or basal ends of the cell, have been introduced to improve the likelihood of convergence to a transverse coalignment (Baulin et al., 2007; Allard et al., 2010b; Eren et al., 2010; Sambade et al., 2012). Recent experimental evidence for localized catastrophes at apical and basal edges of the cell supports a model where longitudinally oriented microtubules are preferentially removed to leave a predominantly transverse array (Ambrose et al., 2011). In sum, the leading model to explain how plant microtubules become coaligned to a transverse orientation relies on a mechanism for actively removing the longitudinally oriented polymers going into, or emerging from, the apical and basal ends of the cell and self-organizing properties, such as bundling interactions between the remaining polymers (Ehrhardt and Shaw, 2006; Ambrose et al., 2011; Sambade et al., 2012).
A major impediment to testing these models and for determining the molecular and biophysical contributions to array organization has been the inability to routinely observe the array organizing into a transverse pattern. Light in general, and specifically from the microscope, actively signals the hypocotyl cells to form nontransverse arrays, making high-resolution imaging studies of cells organizing into a transverse coalignment somewhat inaccessible. Both auxin and gibberellic acid affect hypocotyl cell elongation, and both hormones alter microtubule array organization when exogenously applied (Shibaoka, 1974, 1993, 1994; Roberts et al., 1985; Ishida and Katsumi, 1991; Lloyd et al., 1995; Shibaoka and Takesue, 1999; Wenzel et al., 2000; Folta et al., 2003). Prior studies have clearly established that either hormone can promote microtubule array organization, though high-resolution temporal and spatial data have not been available from these studies for application to models for array organization (Takesue and Shibaoka, 1999; Wenzel et al., 2000). Using a combination of these hormones to synchronously induce light-grown Arabidopsis thaliana seedlings, we captured extensive time-lapse observations of the temporal and spatial changes in array organization leading to transverse array patterning in this important model system. Our quantitative analysis of these data provides a novel cell biological model for transverse patterning with array organization initiating as a band of transverse polymers at the cell midzone and progressively reorganizing toward the apical and basal cell faces.
RESULTS
Gibberellic Acid/Indole-3-Acetic Acid Induce Robust Transverse Cortical Microtubule Array Organization
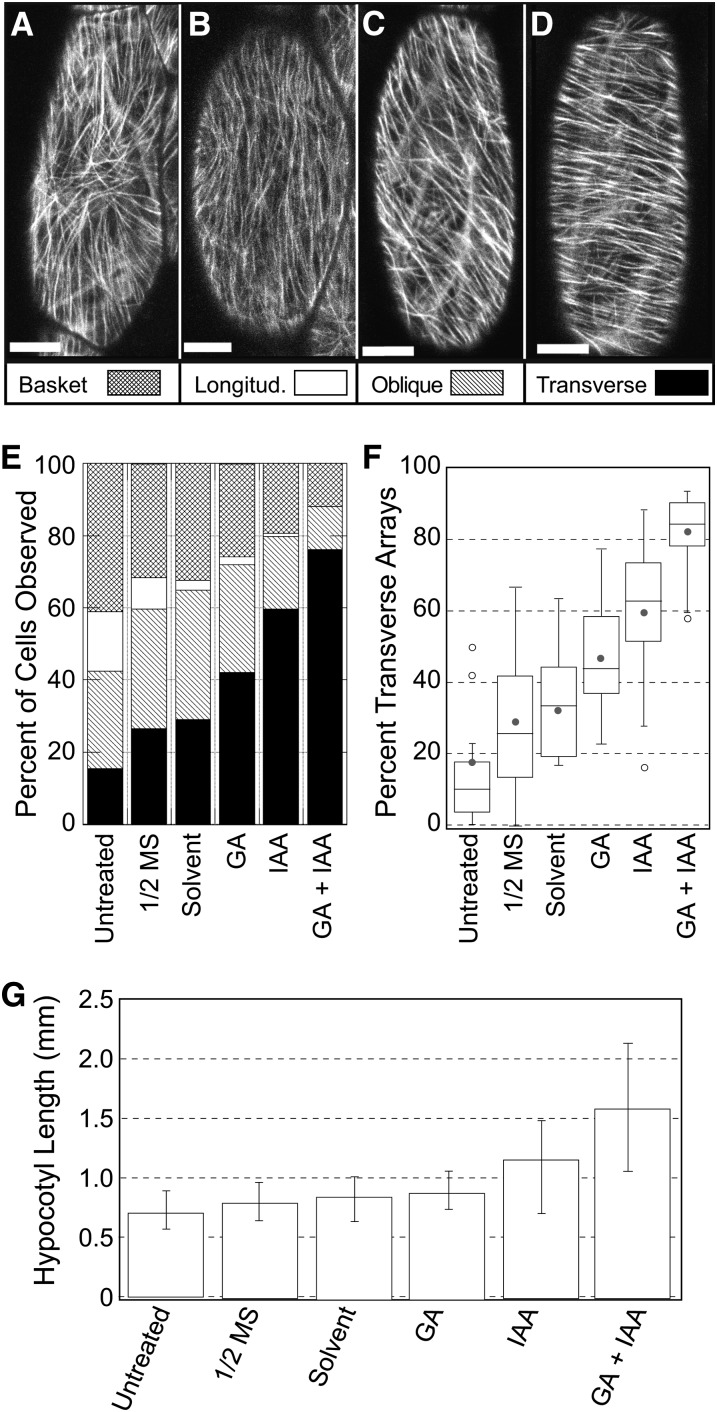
Our first goal was to establish a reproducible method for synchronously inducing light-grown Arabidopsis hypocotyl cells to form transverse coaligned microtubule arrays. We began by classifying the array patterns in untreated cells and determining the distribution of pattern classes in 6-d-old light-grown seedlings expressing a tubulin to green fluorescent protein (GFP):TuA6 transgene. Using reconstructed image composites of the entire hypocotyl length from confocal fluorescence microscopy, we visually classified the microtubule array organization for each cell from two adjacent hypocotyl cell files as one of four array patterns (Figures 1A to 1D). Arrays without a dominant coalignment typically had microtubules radiating from the interior of the outer (periclinal) cell face down the (anticlinal) sides of the cell, forming an inverted basket pattern (Figure 1A). Arrays with predominantly coaligned microtubules were classified as longitudinal (Figure 1B), oblique (Figure 1C), or transverse (Figure 1D) patterns, where oblique was 15° to 75° relative to the cell growth axis. Using the percentage of cells in each hypocotyl as a quantitative measure of array organization states (Figure 1E), we found that ∼60% of the cells in light-grown control plants showed a strong degree of microtubule coalignment at this developmental stage with ∼40% showing a less coaligned (i.e., basket) organization. We found fewer than 20% of the cells exhibited a transverse coalignment when taken directly from growth plates to the microscope (untreated in Figure 1F).
Figure 1.
GA4/IAA-Induced Cortical Microtubule Array Reorganization.
(A) to (D) Confocal microscopy images of 6-d-old light-grown epidermal hypocotyl cells showing basket (A), longitudinal (B), oblique (C), and transverse (D) microtubule array organization in GFP:TUA6-expressing plants. Bars = 10 µm.
(E) The percentage of epidermal hypocotyl cells exhibiting each array organization class for seedlings imaged directly from germination plates (untreated) or after 2 h in liquid culture with the associated treatment condition (bar pattern designations for array class displayed in [A] to [D]).
(F) Box and whisker quartile plots for the percentage of cells with transverse arrays at 2 h including the mean value (filled circle) and outlying data (open circles) for each treatment. Number of observations for (E) and (F), as plants/cells, were as follows: untreated (26/827), half-strength MS (15/376), solvent (13/397), GA4 (19/477), IAA (19/478), GA4 + IAA (15/476).
(G) The effect of all treatments on hypocotyl length was assayed by direct measurement of seedling hypocotyls after 72 h of continuous treatment. Bars represent mean values from >45 total plants pooled from three experiments for each treatment with variation displayed as the pooled sd.
Using this quantitative assay to determine the percentage of each array class, we examined the effects of auxin (indole-3-acetic acid [IAA]) and gibberellic acid (GA4) on array organization. Transfer of seedlings from agar growth plates to agar treatment plates was inconsistent and damaging to plants. We therefore transferred seedlings from agar plates into liquid culture containing hormones or solvent controls and incubated the seedlings for 2 h in the light at 22°C prior to imaging. For each treatment, we tabulated the total percentage of arrays falling into each pattern class (Figure 1E).
Transfer of seedlings into a liquid medium, with or without solvents, led to a small but significant (P < 0.01 comparing 22 untreated to 14 Murashige and Skoog [MS] plants) increase in the percentage of cells with transversely aligned microtubule arrays when compared with controls taken directly from growth plates (Figure 1F). Solvent treatment trended slightly higher for transverse arrays but was not significantly different (P > 0.05 comparing 13 solvent to 14 MS plants) from liquid medium alone (Figure 1F). These experiments establish that placing seedlings into a liquid medium will alter the distribution of microtubule array patterns but does not induce a single or predominant class of array pattern.
Treatment with 10 µM GA4 increased the relative percentage of cells with transverse arrays to near 50% at 2 h (Figure 1E). GA4 treatment appeared to reduce the percentage of basket-patterned arrays but had little effect on the total percentage of oblique arrays at 2 h. Treatment with 0.5 µM IAA resulted in hypocotyls having slightly more than 50% of cells exhibiting transversely coaligned microtubule arrays within 2 h (Figure 1E). All other array organizations were proportionately reduced. Both the GA4 (P = 0.01 comparing 18 GA4 to 14 solvent-treated plants) and IAA (P < 0.01 comparing 18 IAA to 14 solvent-treated plants) treatments were significant relative to solvent controls (Figure 1F).
The relatively modest increase in cells with transverse microtubule arrays and the variability between plants observed with GA4 or IAA used alone was not sufficient for robust quantitative assays or routine imaging of individual cells. We therefore tested induction using both hormones in combination and observed 75 to 85% of the epidermal hypocotyl cells in each plant to organize transverse array patterns by 2 h (Figures 1E and 1F). The effect of combining both hormones was significant (P < 0.01 comparing 15 GA4/IAA plants) relative to the addition of either hormone alone. Plants observed at 6 h had 75 to 85% transverse microtubule arrays, suggesting that this may be an upper limit for this treatment regime. These experiments establish a robust and repeatable method for synchronously inducing the microtubule arrays in wild-type light-grown Arabidopsis seedlings at 6 d after germination to organize from a variety of array patterns into a transverse coaligned array over a 2-h time course.
GA4/IAA Induce Significant Axial Extension of the Light-Grown Hypocotyl
Both GA4 and IAA have varying effects on hypocotyl growth, possibly related to the developmental stage and growth environment of the seedling (Shibaoka, 1974, 1994; Huang and Lloyd, 1999; Shibaoka and Takesue, 1999). The effect of single and combination treatments with GA4 and IAA on hypocotyl growth on 6-d-old light-grown Arabidopsis seedlings was determined by measuring hypocotyl length 72 h after hormone treatment. Light-grown seedlings were treated with 10 µM GA4, 0.5 µM IAA, or both hormones in combination starting at 6 d after germination. Hypocotyl length was longer for plants transferred to liquid culture with and without the solvent control (Figure 1G). Treatment of light-grown seedlings at this developmental stage with GA4 did not produce a substantive increase in hypocotyl length over solvent controls (P > 0.05 for 36 GA4 and 48 solvent-treated plants), but IAA treatment resulted in a significant change (P < 0.01 for 36 IAA-treated plants) with hypocotyls ∼130% as long as solvent control treated plants. Treatment with both hormones resulted in hypocotyls that were ∼200% as long as solvent controls, significantly longer (P < 0.01 for 48 GA4/IAA-treated plants) than control and IAA-treated seedlings (Figure 1G). These data indicate that the combined hormone treatment leads to a substantially higher percentage of hypocotyl cells with transverse coaligned microtubule arrays correlated with a substantial increase in axial hypocotyl extension.
Timing of Hormone-Induced Transverse Array Organization
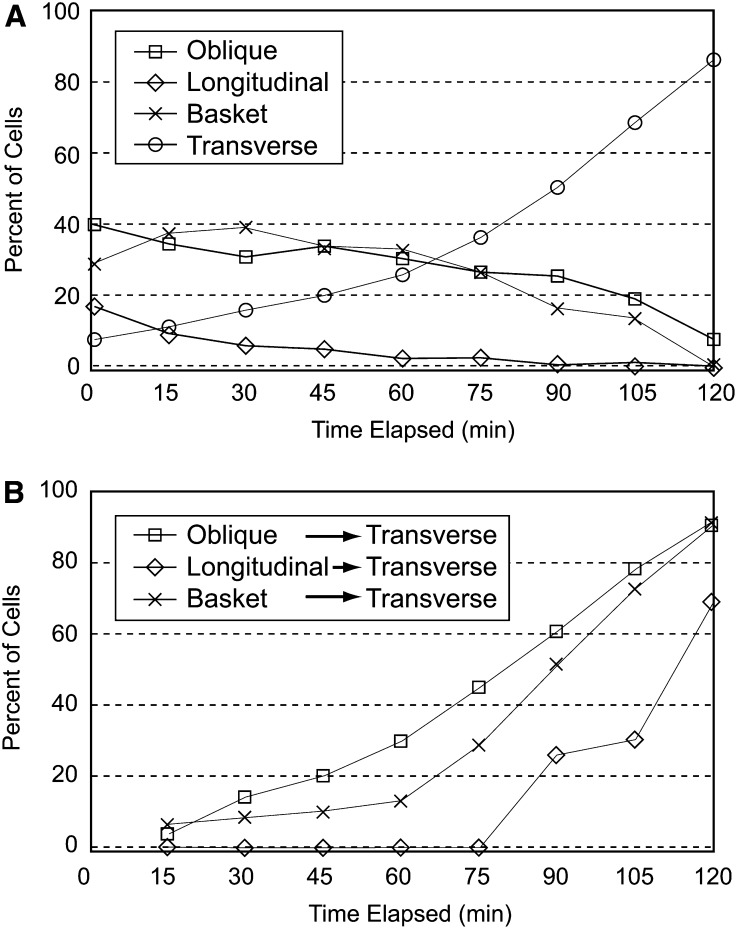
We examined the relative timing of microtubule array reorganization from each starting array pattern to a transverse coaligned pattern. We placed seedlings into liquid culture on slides containing a combination of 10 µM IAA and 0.5 µM GA4 and imaged hypocotyls every 15 min for 2 h. The percentage of hypocotyl cells observed in each array pattern at each time point was tabulated for ∼40 cells on the imaged side of each seedling from the time course (Figure 2A). The compiled data for cells with transverse arrays suggest that the induced reorganization occurs in two phases. There is an initial lag phase from 0 to 45 min where the number of cells classed as transverse increases relatively slowly. The lag was followed by a second, faster phase, established by 45 to 60 min after induction and continuing until 75 to 80% of the cells were transverse at 120 min (Figure 2A). The starting percentage of hypocotyl cells exhibiting a longitudinal array pattern (∼20%) showed an initial drop during the lag phase followed by a slow dissipation to zero by 2 h (Figure 2A). Although we observed a small initial rise in arrays classed as baskets, we found no obligate transition to an intermediate array pattern (e.g., oblique to basket) prior to achieving a transverse array organization.
Figure 2.
Timing of Hormone-Induced Microtubule Array Reorganization.
Epidermal hypocotyl cells (n = 196 cells across six plants) were imaged after induction with 10 µM IAA and 0.5 µM GA4 at 15-min intervals for 120 min.
(A) The percentage of all cells exhibiting each array pattern class is plotted at each time interval. The percentage of cells with arrays classed as transverse shows an initially slow rise followed by a more rapid phase.
(B) The relative percentage of each array class that has reorganized to a transverse orientation was plotted for arrays starting as oblique, longitudinal, or basket patterns. Cells with initially oblique (∼40% of cells) arrays transited to transverse slightly ahead of cells with basket (∼32% of cells) array forms and longitudinal arrays (∼18% of cells) were slightly delayed when compared with both basket and oblique.
To determine if any starting array pattern was more rapidly reorganized to a transverse pattern, we examined the relative timing of cells transitioning from each starting array pattern to a final transverse array orientation (Figure 2B). Cells that started as oblique arrays reorganized to transverse at approximately the same rate as cells that started with basket-patterned arrays. These data indicate that the coalignment of microtubules found for the oblique arrays does not predispose them to a more rapid transition to transverse coalignment. Cells having initially longitudinal arrays were somewhat slower to be classed as transverse (15 min delay relative to other patterns) but ultimately became transverse in the same 2-h period as the other arrays (Figure 2B).
Transverse Array Organization Initiates in the Midzone of the Cell
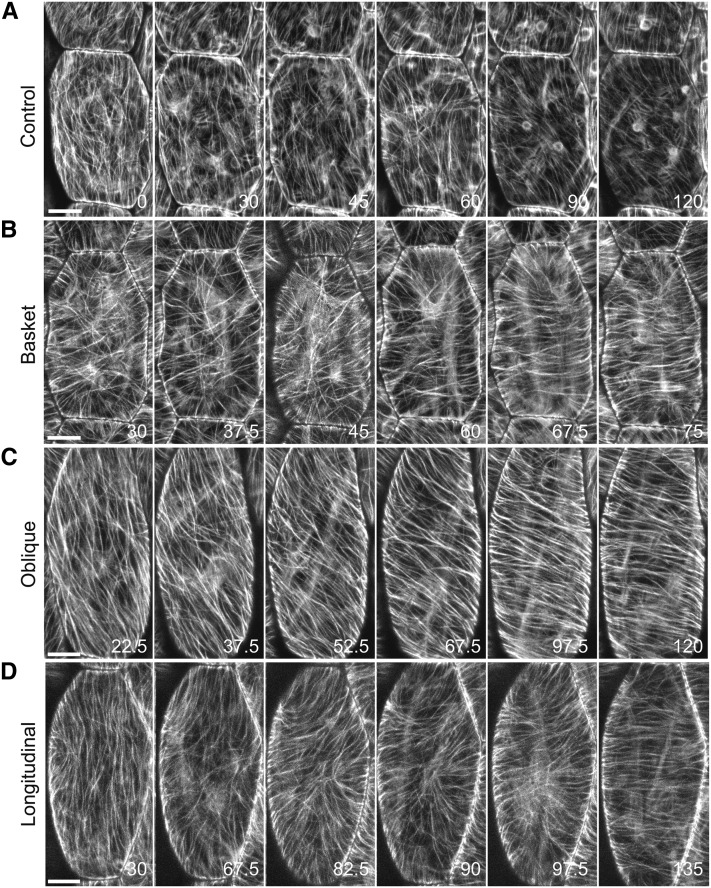
To determine how the pattern of microtubules changes over time to produce a transverse coalignment, we imaged individual hypocotyl cells expressing a GFP:TuA6 microtubule marker at 7.5- or 15-min intervals for 2.5 h using confocal laser scanning microscopy (Figure 3). Solvent-treated control cells showed occasional rearrangements of the cortical microtubules over the 120- to 150-min time period but showed no distinct organizational changes into a defined pattern on this time scale (Figure 3A). We examined the progression of changes to microtubule array organization in control cells (Figure 3A) and in hormone-treated cells that originated in basket (Figure 3B), oblique (Figure 3C), or longitudinal (Figure 3D) array patterns. Hormone-treated cells originating in a transverse coalignment simply maintained a transverse coalignment.
Figure 3.
Cortical Microtubule Reorganization from Basket, Oblique, or Longitudinal Patterns to Transverse over Time.
Confocal image projections of GFP:TUA6-labeled microtubules in epidermal hypocotyl cells grown for 6 d in continuous light. Time in minutes. Bars = 10 µm in (A) to (D).
(A) Selected images from a solvent control cell over 120 min of imaging. The array pattern changes over the course of observation but achieves no defined pattern of organization.
(B) to (D) Plants treated with GA4 and IAA.
(B) Cells with basket formed arrays are characterized by microtubules oriented perpendicular to the cell perimeter around the entire edge of the outer periclinal cell face. Transversely aligned microtubules appear across the middle of the cell between 45 and 60 min and progress toward the cell apex and base. Note the persistence of longitudinal microtubules at the cell apex and base.
(C) Oblique arrays appeared to rotate into a transverse position, progressively creating arrays of shallower angle relative to the cell growth axis. The middle zone of the cell becomes transverse ahead of the apex and base (67.5 min), creating a transient sigmoidal shape for the array polymers prior to becoming transverse.
(D) Reorganization of longitudinal arrays was characterized by the invasion of new microtubules from the lateral side faces of the cell (90 to 97.5 min) instead of the progressive shift to shallower angles.
Microtubule array organization to a transverse coalignment exhibited three consistent features that were independent of the starting array pattern. Single-cell imaging confirmed that no significant changes in array pattern appeared in the initial 30 to 45 min after hormone induction (Figures 3B to 3D). Transverse coalignment was initially observed around the midzone of the elongated hypocotyl cells and progressed in both an apical and basal direction. Finally, we observed that the apical and basal regions of the outer cell face were the last regions to reorganize into a transverse pattern. Persistent longitudinal polymers at the cell’s apical and basal ends often interacted with the advancing front of transverse microtubules, yielding curved polymers in the corners of the cell (e.g., Figure 3D, 82.5 to 97.5 min).
Cells with an initially oblique microtubule array progressed through a series of oblique organizational states, giving the appearance of a rotating array orientation (Figure 3C) as previously described (Roberts et al., 1985; Yuan et al., 1995; Wymer and Lloyd, 1996; Wymer et al., 1996). Using time-lapse image data taken at 3-min intervals, we observed new microtubule polymer appearing between the existing oblique microtubules and a progressive loss of the original oblique polymers (Figure 3C; see Supplemental Movie 1 online). We interpret the observations as showing new transversely oriented microtubules interacting with (i.e., bundling) the existing oblique microtubules at progressively shallower angles and therefore find that the microtubules are not sliding or rotating. Consistent with the other starting array patterns, we observed that the microtubules toward the midzone of the cell were initially more transversely aligned than the polymers toward the apical and basal ends of the cell. This gave the microtubules on the outer periclinal cell face a transiently sigmoidal pattern (Figure 3C, 52.5 to 67.5 min). We found that oblique arrays with a left-handed pitch reorganized to the right, and arrays with a right-handed pitch reorganized to the left, suggesting no structural bias in the organizing mechanism for these wild-type plants.
Cells with an initially longitudinal array organization developed transverse microtubules first at the lateral edges of the periclinal cell surface toward the center of the cell (Figure 3D). These microtubules polymerized transversely across the outer periclinal array and occasionally created a basket or focused to form a star-shaped array (Sambade et al., 2012) before establishing a transverse coalignment, consistent with the slight initial increase we observed in basket shaped arrays (Figure 2A). Longitudinally oriented microtubules toward the apical and basal cell faces persisted until the rest of the array had formed a transverse pattern (Figure 3D, 97.5 and 135 min). Longitudinal arrays differed from oblique arrays in that they did not typically show a progression of oblique arrays (i.e., apparent rotation) prior to be becoming transversely aligned. This was apparently due to the microtubules interacting with the longitudinal polymers at an angle that was too high to redirect microtubule polymerization into a bundle.
In sum, for arrays starting in a basket, oblique, or longitudinal pattern, no obvious changes in array pattern were observed during the first 30 to 45 min after induction (Figure 3), corresponding to the initial lag phase found when quantifying the percentage of cells in each array pattern over time (Figure 2). For each starting array pattern, we consistently found that transverse coalignment initiated at the midzone of the cell 30 to 45 min after induction and progressed to the apical and basal ends of the cell (Figures 3B to 3D). In the cases of the longitudinal and basket patterned arrays, the longitudinally oriented polymers at the apical and basal ends of the cell were clearly maintained throughout the first 90 to 120 min of induction and were the last elements of array pattern to change (Figures 3B to 3D).
Microtubule Plus Ends Reveal Structural Properties of the Array
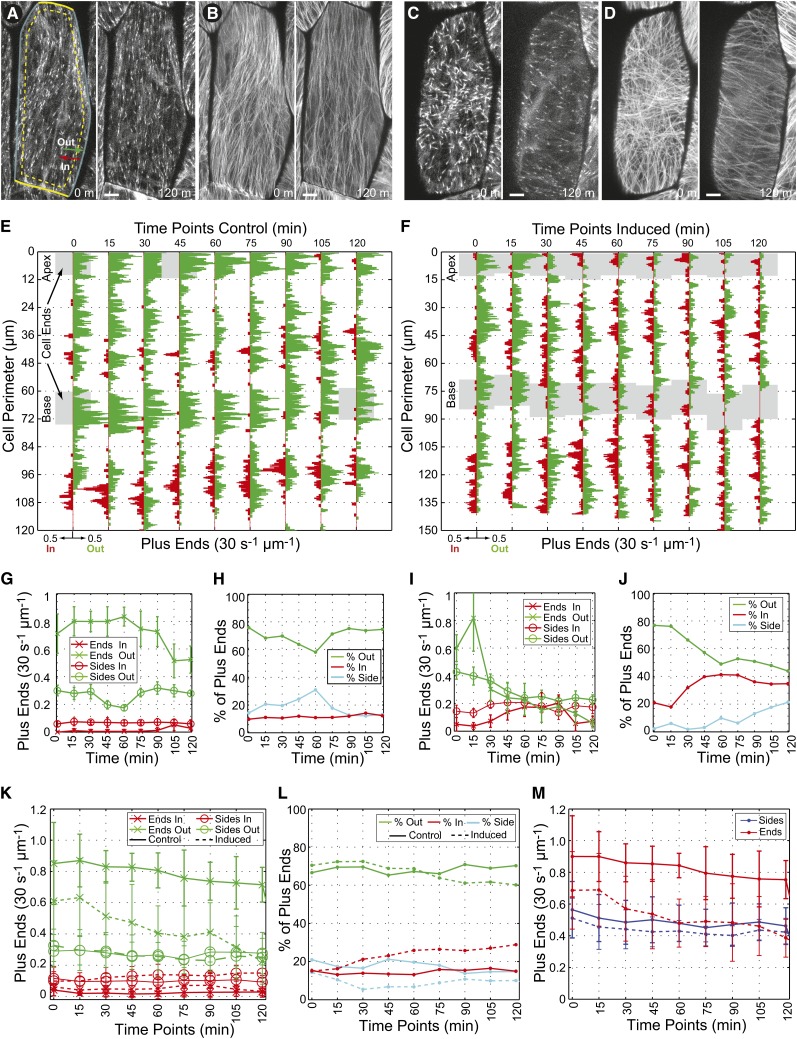
To investigate the underlying structural changes associated with the microtubule array reorganization into a transverse coalignment, we imaged cells expressing a GFP:AtEb1 probe (Figure 4) that labels polymerizing microtubule plus ends (Mathur et al., 2003). We imaged single hypocotyl cells every 15 min for 5-min durations over a time course of 2.5 h. To capture the entire curved surface of the outer periclinal cell face, we captured seven axial sections at 0.34-µm increments every 6 s and projected the information to a single image (Figures 4A and 4C). The pattern of microtubule array organization at each 15-min time point was evaluated by summing all of the projected frames together from each 5-min time-lapse subsegment (Figures 4B and 4D) to visualize the GFP:AtEb1 tracks formed by the microtubule polymerization trajectories (Figures 4A to 4D; see Supplemental Movies 2 to 4 online).
Figure 4.
Time-Lapse Imaging of GFP:AtEb1 in Hormone-Induced Cells Organizing into a Transverse Coalignment.
Confocal imaging of control ([A] and [B]) and hormone-induced ([C] and [D]) hypocotyl cells expressing a GFP:AtEb1 transgene marking the polymerizing microtubule plus ends. A single time point (projection of 5-µm depth) from 0 and 120 min ([A] and [C]) is paired with a summation image (sum of 50 projected frames taken at 6-s intervals) from 0 and 120 min ([B] and [D]) to show the trajectory of the microtubule plus ends and the representative pattern of microtubule array organization. Bars = 3 µm in (A) to (D). The density and orientation of plus-end polymerization was measured at the perimeter of the outer periclinal array (outline and dashed contour in [A]) at 15-min intervals ([E] to [M]). A 5-min time-lapse image series was acquired at 5-s intervals every 15 min for 120 min where the position and number of GFP:AtEb1 spots entering (In), exiting (out), or moving parallel to a cell edge (Side) was recorded within the ∼3-µm space delimited by the dashed contour line. The mean number of GFP:AtEb1 spots (i.e., microtubule plus ends) per micrometer of cell perimeter per 30-s time interval moving out of (green) or into (red) the outer periclinal array in a control (E) and an induced (F) cell are plotted for each 15-min time point in the 2-h time-lapse series (cells in [A] to [D] depicted in [E] and [F]). The cell perimeter was made linear in (E) and (F) with the cell’s apex and base shaded (gray). Note the spreading of incoming plus ends (red) on the lateral sides of the cell and the loss of plus ends at the apical and basal ends for the induced cell (F). Data from a single control (E) or treated (F) cell plotted numerically to show the change in the mean density ([G] and [I]) of plus ends polymerizing out of (green) or into (red) the outer periclinal array at the lateral sides (o) or ends (x) of the cell. The percentage of all plus ends in each orientation through time counted per time point in a single control (H) and treated (J) cell. The mean number of plus ends moving out of (green) or into (red) the periclinal array per 30 s ± sd (K) aggregated from six control (solid lines) and six treated (dashed lines) cells. The percentage of all plus ends in each orientation through time (L) aggregated from six control ([L], solid lines) and six hormone-induced ([L], dashed lines) cells. Combined density of polymerizing plus ends (In + Out) for six control ([M], solid lines) and six treated ([M], dashed lines) at the cell’s lateral sides (blue) and ends (red).
A visual evaluation of the time-lapse image data revealed that the cortical microtubule arrays in all uninduced cells are structured with the majority of plus ends polymerizing toward the cell perimeter (see Supplemental Movies 2 to 4 online). Hormone-induced cells with transversely coaligned arrays (Figure 4D) appeared to have lost this bias for edge-directed polymerization in the outer periclinal arrays (see Supplemental Movies 2 to 4 online), suggesting a significant change in the structure of the cortical array concurrent with the change in array organization.
To quantify the changes in underlying array structure over the course of array organization to transverse, we identified all GFP:AtEb1-labeled microtubule ends that were within 2 µm of the perimeter of the outer periclinal cell face (Figure 4A, cell perimeter and dashed contour lines) over sequential 30-s time intervals for each 5-min data set (seven intervals per 5-min time lapse; see Methods) at each 15-min interval. Using the time-lapse data, we marked each plus end as polymerizing outward from the periclinal cell surface to the anticlinal side faces, inward from the anticlinal cell faces to the outer periclinal array (Figure 4A, In or Out), or parallel to the adjacent cell face. The extracted data were plotted as the average number of plus ends (mean of seven 30-s intervals per 5-min time lapse) moving either in or out of the periclinal array per 1 (lateral) µm of cell perimeter at each 15-min time point (Figures 4E and 4F). Since the timing of reorganization differed slightly between different cells with different starting patterns (Figure 2), we present examples of the aggregate numerical averages of the plus-end densities and directions at the cell perimeter plotted for a single cell (Figures 4G to 4J) and as the average of multiple cells (Figures 4K to 4M).
Consistent with visual inspection of the time-lapse data from untreated control cells, our quantitative analysis revealed that 70 to 75% of the plus ends at the cell periphery were polymerizing out of the periclinal cell face with <20% of the plus ends coming into the periclinal face from the anticlinal side faces (Figures 4E, 4H, 4L, and 4M). The balance of microtubule ends (5 to 10%) were moving parallel to a perimeter side face and were counted for total density. The bias in polymerization trajectories was observed for all array pattern classes, including cells that were initially transverse. Microtubules polymerizing into the periclinal cell face were found in a few concentrated regions around the cell perimeter and rarely originated from the cell’s apical or basal ends (Figure 4E). We observed that the apical and basal ends of the cell maintained a relatively high density of polymerizing plus ends when compared with the side faces (Figures 4E, shaded regions, 4G, 4K, and 4M). These data show a substantial asymmetry (∼3:1) in the number of microtubule plus ends polymerizing out of the outer cell face relative to those entering the outer cell face, and, related to constructing models for array organization, we find relatively few microtubules originating from the apical and basal regions of the cell in untreated control plants, independent of the array pattern.
The addition of GA4/IAA to the GFP:Eb1-expressing plants resulted in reorganization of the array to a transverse coalignment (Figure 4D). We found that the density of microtubule plus ends measured at the cell perimeter declined by ∼30% within the first 5 min of hormone treatment and continued to decrease to <40% by 60 min after induction when compared with solvent-treated control cells (Figure 4F). The decrease in GFP:Eb1-labeled ends therefore occurred during the early lag phase of induction and prior to any substantial pattern changes observed with the GFP:TUA6 microtubule marker. The loss of polymerizing plus ends was most apparent at the apical and basal ends of the cell (Figure 4F, shaded regions), mostly due to the continued absence of microtubules entering the outer periclinal cell face from these positions. However, we observed that cells retained the longitudinally aligned polymers, likely to be microtubule bundles, at the apical and basal portions of the cell until the remainder of the cell became transverse.
While the total density of GFP:Eb1-labeled plus ends measured at the cell periphery decreased, we observed that the density of plus ends polymerizing into the periclinal cell face from the anticlinal side faces increased slightly over time with the hormone induction (Figures 4I, 4K, and 4M). Coupled with the loss of outward polymerizing plus ends, we found that the relative percentage of all plus ends polymerizing into the periclinal cell face shows an average net increase from ∼15 to ∼35% of total density after hormone induction (Figures 4J and 4L). These quantitative data support the initial visual inspection, suggesting that the balance of microtubule plus ends moving into and out of the outer cortical array moves closer to a 1:1 ratio before the array becomes transversely coaligned. We observe that the majority of effect on the ratio of microtubules polymerizing into versus out of the array came from the reduction in polymerizing plus ends in the periclinal array and not through an increase in the anticlinal faces.
The microtubule plus ends polymerizing into the outer periclinal array prior to induction were typically concentrated into clumps and rarely originated from apical or basal cell faces (Figure 4). We found that the distribution of microtubule plus ends moving into the outer periclinal array changed dramatically after induction from concentrated clumps to a uniformly spaced distribution along the lateral sides of the cell (Figure 4E). The sites were further characterized by having microtubules polymerizing both into and out of the outer periclinal array. The total density of microtubule plus ends at the midzone did not appear to increase during array reorganization, but the spatial distribution of microtubules moving into the outer periclinal array had a tendency to spread bidirectionally from the cell’s midzone with an advancing front of transversely oriented polymers (Figure 4F). These data suggest that it is not a significant increase in the number or density of microtubules at the cell midzone that correlates with the appearance of the initial transverse microtubules, but rather the origin and orientation of these polymers relative to the cell axis. While hormones induced a redistribution of sites around the cell perimeter where microtubules entered the periclinal array, we found that the apical and basal ends of the cell were still excluded (Figure 4F).
Microtubules Polymerize across Cell Face Junctions
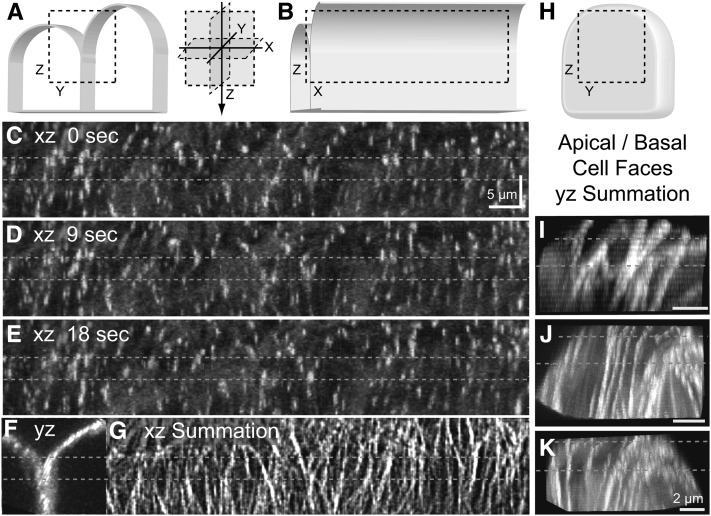
To determine if the redistribution of microtubule plus-end trajectories was occurring due to microtubules either preferentially initiating from, or terminating at, the edges or junctions between cell faces, we examined the GFP:Eb1 plus-end trajectories in the trans-facial regions (Figures 5A and 5B). Using three-dimensional (3D) confocal time-lapse microscopy, we created data sets that isolated the side faces of the cell (Figures 5C to 5E). When observed in cross section (Figure 5F), the hypocotyl cells did not show a sharp transition or edge between the outer periclinal and anticlinal cell faces. We therefore bracketed the region where adjacent cells form a junction and imaged to a position >3 µm below that position. We observed the GFP:Eb1 probe in cells 60 min after hormone induction traveling either from the outer periclinal cell face down the anticlinal side of the cell or from the anticlinal side face and into the outer periclinal array. It was not possible to unambiguously track individual GFP:Eb1-labeled microtubule ends due to the density of ends and bleed-through of GFP:Eb1 signal from the adjacent cell. Summing the 3D time-lapse data to visualize the GFP:Eb1 trajectory paths from the lateral face (Figure 5G) and the apical/basal face, we found that there were no apparent barriers to microtubule growth at the junction between adjacent cells and no obvious increase in the density of GFP:Eb1 origins (Figures 5C to 5E; see Supplemental Movie 5 online). We observed that many of the plus-end trajectories appear to follow the same tracks, implying the formation of microtubule bundles extending from the periclinal surface array to the anticlinal side face of the cell.
Figure 5.
Microtubule Polymerization Is Continuous between Periclinal and Anticlinal Hypocotyl Faces during Array Reorganization.
(A) Schematic of two adjacent hypocotyl cells viewed end on (yz projection) indicating the cross section of the imaged region of the lateral cell junction (dashed box).
(B) Rotation and cutaway of the schematic (xz projection) indicating the projected image volume used to assess the continuity of the microtubule polymerization trajectories in the lateral side faces of the cell.
(C) to (E) Confocal images of GFP:Eb1 taken at sequential axial (z axis) positions were combined and reprojected from the side (xz projections as in [B]) to create time-lapse images of microtubule plus-end trajectories.
(F) A rotation of the image data (yz projection as in [A]) indicating the position of the cell junction. Reference lines (dashed lines in [C] to [K]) bracket the cell junction.
(G) A summation of 100 consecutive side projected images (xz projections as in [B]) taken at 3-s intervals indicating the track or trajectory for the microtubule plus ends polymerizing across the cell junction.
(H) to (K) Summation images from time-lapse GFP:Eb1 data at the apical/basal cell face (schematic in [H]) for three cells at 60 min after hormone induction indicating continuous polymerization trajectories across trans-facial boundaries ([I] to [K]).
DISCUSSION
The formation of coaligned microtubule arrays organized into a transverse pattern relative to the plant growth axis has long been associated with the axial growth of plant cells (Green, 1962; Hepler and Newcomb, 1964; Cyr, 1994; Baskin, 2001; Wasteneys and Galway, 2003; Ehrhardt and Shaw, 2006; Hashimoto and Kato, 2006; Lloyd and Chan, 2008; Lloyd, 2011). Several models have been proposed to explain the molecular and biophysical factors required to achieve this special state of microtubule array organization. A significant limitation for these models has been the lack of time-resolved observations from cells that are transitioning into a transverse coaligned pattern. We developed a robust and reproducible protocol for synchronously inducing the cortical microtubules in light-grown hypocotyl cells to reorganize from their preexisting array patterns into a transverse coalignment. The induction method, combining two plant hormones previously shown to affect microtubule array organization (Shibaoka, 1974, 1993; Ishida and Katsumi, 1991; Lloyd et al., 1995; Shibaoka and Takesue, 1999), permits a direct quantitative comparison of treated and untreated plants under identical circumstances and at a temporal and spatial scale commensurate with microtubule turnover in the array. Using this approach, we evaluated the changes in microtubule array structure and pattern leading to transverse microtubule coalignment.
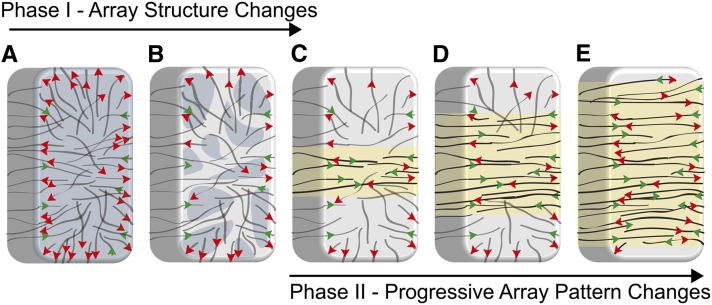
Our data reveal a progressive change in organization occurring in two principal phases (Figure 6). The initial response (Figures 6A and 6B) to hormones was a significant decrease in the number of GFP:Eb1-labeled microtubule plus ends occurring prior to any substantive change in the array pattern. A second phase (Figures 6C to 6E) was characterized by the appearance of transversely oriented microtubules around the midzone of the cell where transverse organization proceeded bidirectionally over a 30- to 60-min time course toward the apical and basal regions of the cell. The repatterning to transverse coalignment was marked by a notable transformation of the outer periclinal microtubule array structure. Specifically, we found that control cells had approximately three microtubules polymerizing toward the perimeter of that cell face for every one microtubule polymerizing toward the interior of the outer periclinal array. This remarkable bias in microtubule polymerization trajectory was progressively corrected after induction as the array reorganized from the cell’s midzone to become transverse (Figure 6B).
Figure 6.
Depiction of Hormone-Induced Cortical Array Reorganization to a Transverse Pattern.
Hypothetical hypocotyl cell with cortical microtubule array displayed (black lines) with polymerizing plus ends (arrowheads).
(A) Initial array organization with 3:1 ratio of outward (red arrowheads) to inward (green arrowheads) polymerizing plus ends.
(B) Hormone treatment induces a change in array structure observed as a decrease in the number of microtubule ends (mottled surface) on the outer periclinal cell face prior to significant changes to array pattern.
(C) to (E) Transverse microtubules initially appear at the cell midzone ([C]; yellow surface) and organize progressively ([D] and [E]) from the midzone to the apex and base of the cell.
We propose from our observations that the well-studied microtubule arrays on the outer periclinal cell face of these light-grown hypocotyl cells create patterns somewhat independently from the arrays on the other cell faces (Chan et al., 2011). We speculate that these array patterns contribute to cell wall construction that is required for radial hypocotyl thickening (Chan et al., 2007, 2010; Lloyd, 2011). Hormone induction altered the array structure from a system having 70 to 80% of the growing plus ends exiting the outer periclinal array, independent of array pattern, to a system with more balanced numbers of microtubules entering and exiting the outer periclinal array at the cell’s lateral side faces. We infer from our data that creation of a transverse coaligned microtubule array that is functional for axial cell growth requires coordination across multiple cell faces. We speculate that these globally coordinated transverse microtubule arrays are critical for producing a transverse cellulose microfibril alignment around the cell for axial growth (Baskin, 2001, 2005; Ehrhardt and Shaw, 2006; Lloyd, 2011).
Mechanistic Models for Transverse Microtubule Coalignment
Earlier observations of microtubules in hormone-treated cells, using immunofluorescence or microinjected fluorescent tubulins, proposed that array organization occurred through movements of existing polymers through intermediate patterns rather than through direct polymerization into a new pattern (Roberts et al., 1985; Yuan et al., 1995; Wymer and Lloyd, 1996; Wymer et al., 1996). More recent models for transverse array patterning, drawing from observations in live cells, propose that organization is driven by preferential loss of longitudinally oriented microtubules at the cell’s apical and basal ends (Ehrhardt and Shaw, 2006; Baulin et al., 2007; Allard et al., 2010a, 2010b; Eren et al., 2010; Ambrose et al., 2011; Sambade et al., 2012). These models propose that if microtubules at the cell’s apical and basal ends are disallowed from entering the outer periclinal array or are selectively destroyed through preferential catastrophes at cell edges (Ambrose et al., 2011), then the remaining microtubules will passively form a transverse coalignment. The degree of microtubule coalignment for the remaining microtubules is hypothesized to improve through the action of angle-dependent bundling (Allard et al., 2010b; Eren et al., 2010), intrabundle versus branched microtubule nucleation (Nakamura et al., 2010; Kirik et al., 2012), or through an increase in the number of microtubules entering from the cell’s lateral side faces (Sambade et al., 2012). Computer simulations have been developed to show the plausibility of these mechanisms for organizing an existing microtubule population into a transverse coalignment (Baulin et al., 2007; Allard et al., 2010b; Eren et al., 2010; Ambrose et al., 2011; Sambade et al., 2012).
Our repeated observations of hormone-induced microtubule array transitions from a variety of initial patterns to a transverse coalignment shed significant light on what mechanisms create a global transverse pattern. We observed a progressive change in the array pattern, somewhat consistent with previous findings from GA4- or IAA-treated cells (Roberts et al., 1985; Ishida and Katsumi, 1991; Lloyd et al., 1995; Yuan et al., 1995; Wymer and Lloyd, 1996; Wymer et al., 1996; Shibaoka and Takesue, 1999). However, we demonstrate that the reorganization does not occur through reshuffling or sliding of existing microtubules. For example, we clearly show that the previously posited rotation of cortical array microtubules (Roberts et al., 1985; Yuan et al., 1995; Wymer and Lloyd, 1996; Wymer et al., 1996) corresponds to a situation where newly appearing transverse microtubules are interacting with an initially oblique array to form microtubule bundles of increasingly shallow transverse orientation. The relatively long time period (∼60 min) required for transverse patterning, relative to polymer turnover (Shaw et al., 2003; Shaw and Lucas, 2011), indicates that the transitional patterns observed after induction likely arise through creation of new microtubules and not through reorganization of the existing polymers.
Our observations in hypocotyl cells do not support current models for transverse patterning that rely substantively on the targeted destruction of longitudinal microtubules at the cell’s apical and basal ends. Our naive expectation was that if the microtubules were preferentially removed at the apical and basal ends of the cell, hormone treatment would induce a loss of longitudinal microtubules at the cell’s ends, leading to a transverse reorganization initiating from the ends of the cell. Contrary to this expectation, our data show that the longitudinal microtubules at the cell’s apex and base were consistently the last elements of the original array pattern to disappear (Figures 6D and 6E). Furthermore, we consistently observed that transverse array organization initiated at the midzone of the cell and progressed to the apex and base, independent of the original array pattern (Figures 6C to 6E). These observations were most clearly observed for the ∼20% of cells that started with a predominantly longitudinal microtubule coalignment. New microtubules emerging from the lateral anticlinal sides of the cell invaded the central midzone portion of the outer periclinal array before any changes were observed in the pattern of longitudinal microtubules running the length of the cell (Figures 3 and 6C to 6E). Based on these observations, we conclude that transverse array organization is driven primarily by an active process that creates new transverse microtubules rather than by passive retention of transverse microtubules in cells where the longitudinal microtubules are directly targeted for destruction (Figure 6).
Our findings suggest that the apical and basal ends of these axially extended hypocotyl cells have distinct properties for creating or maintaining microtubules. We found relatively few GFP:Eb1-labeled ends entering the outer periclinal array from the apical and basal cell faces. Critically, this phenomenon was observed in both the control cells and in hormone-induced cells. This suggests that the apical and basal cell faces, at this developmental stage, are not contributing significantly to the population of longitudinal microtubules in the outer periclinal array. Nearly all of the longitudinally oriented microtubules toward the apex and base of the outer periclinal cell face were polymerizing toward the cell edges. We infer from our quantitative observations that the majority of these longitudinal microtubules must then be originating from positions on the outer periclinal cell face. Given our observation that hormone induction rapidly depleted the total number of plus ends on the outer periclinal cell face, we speculate that the cell is ultimately cutting off the supply of these longitudinal microtubules rather than targeting the existing polymers for destruction. We found that the reorganization to transverse was marked by the progressive elimination of the edge-ward polymerization bias along the lateral sides of the cell (Figures 6C to 6E). We infer that the apical and basal cell faces have important properties because this progressive correction stops and does not extend into those cell faces to produce microtubules that polymerize into the outer periclinal array.
Our discovery that the cell progressively organizes from the midzone to the ends of the cell, regardless of initial array pattern, suggests a mechanism that is spatially regulated by the cell. We therefore propose that a global transverse coalignment does not arise strictly due to self-organizing properties imposed globally on the microtubules. We suspect that changing some of the known microtubule behaviors, such as branched nucleation or severing at microtubule crossover positions, should have important consequences for creating or maintaining an array pattern. But we propose that some of these phenomena are likely to be deployed in a spatially distinct manner to affect transverse array patterning and not simply changed on a global scale. How different microtubule-related activities are spatially controlled, and precisely which activities are required for transverse patterning, remain to be determined.
Working Model for Transverse Array Organization
Keeping in mind that exogenously applied hormone is not a naturally occurring event for the plant, we propose that the temporal sequence of events leading to transverse coalignment shows us one pathway among several possible pathways to the transverse coaligned state. Comparing the induced transverse arrays to the four general pattern classes in the untreated cells, we point to the distribution of positions along the cell’s lateral sides where microtubules are polymerizing both into and out of the outer periclinal array as the key difference in array structure (Figure 6). Our quantitative evaluation of GFP:Eb1-labeled polymer ends suggests that this change in array structure arises not through a significant rise in the total number of plus ends emerging from the side faces, but more directly from their redistribution along the lateral cell faces. Therefore, our data are in partial agreement with conclusions drawn from prior work suggesting that new microtubules coming from the side faces of the cells would help drive transverse coalignment (Sambade et al., 2012). That work looked almost exclusively at arrays that began in a longitudinal coalignment and concluded that all arrays form an obligate star pattern prior to cell growth. We interpret our data, examining a full spectrum of starting array patterns, as indicating that all cells initiate transverse patterning toward the midzone of the cell before that patterning progresses to the cell’s ends. In the specific case of cells with initially longitudinal arrays (∼20% of population), the early part of the reorganization at the midzone likely corresponds to the previously characterized star pattern. However, in the more general case of cells that do not start with a longitudinal array (∼80% of population), those cells do not reorganize into a star or basket pattern as an obligate intermediate step toward becoming transverse. The new microtubules creating the transverse pattern interact with the existing array pattern and eventually dominate the organization.
We speculate that the hormone-induced reorganization of arrays into a transverse coalignment is primarily driven by a redistribution of microtubule nucleation sites. The 3:1 bias in microtubules polymerizing edge-ward in the outer periclinal arrays strongly suggests that microtubule nucleations are more prominent on this cell face than on the anticlinal cell faces in untreated cells. This proposal is consistent with, and may possibly explain, prior microtubule dynamics measurements indicating more total polymerization (∼2:1) than depolymerization (Shaw et al., 2003; Shaw and Lucas, 2011). A strong bias for microtubule nucleation in the outer periclinal cell face, with concomitant microtubule extinction of the treadmilling polymers more prominent on other cell faces, could explain both the observed asymmetry in plus-end trajectories and the apparent non-steady state microtubule dynamics measurements. We note that this positive drift coefficient (Maly, 2002), and nucleation rates in general, are dealt with somewhat arbitrarily in current computer simulations for array organization (Baulin et al., 2007; Allard et al., 2010b; Eren et al., 2010; Hawkins et al., 2010; Tindemans et al., 2010; Deinum et al., 2011; Sambade et al., 2012).
Alternatively, the bias for edge-ward polymerization could be due to cell edge effects (Ambrose et al., 2011; Zhang et al., 2011), such as an upregulation of catastrophe or microtubule severing at the perimeter of these untreated cells (Burk et al., 2001; Wasteneys and Ambrose, 2009; Nakamura et al., 2010). If microtubules underwent a cycle of preferential catastrophe or severing at the edge of the periclinal/anticlinal cell face, followed by a high probability of rescue to polymerization at points within the periclinal array, we would expect to count more apparent plus ends polymerizing toward the cell edge than coming from the cell edge. While our 3D time-lapse observations of the GFP:EB1 probe at the cell’s side and end faces revealed no visible interruption of plus-end polymerization trajectory at cell edges, we cannot discount the possibility of an increased catastrophe frequency within the anticlinal side faces of the cell (Oda and Fukuda, 2012).
In summary, we propose that transverse coalignment of the microtubules occurs in these cells primarily owing to an influx of transversely oriented microtubules from the lateral side faces of the cell initiating at the cell midzone and progressing bidirectionally to the apex and base (Figure 6). How the transverse polymerization trajectory is set for these microtubules is not known, but we speculate that they are following the relatively transverse orientation of the anticlinal side facing arrays (Crowell et al., 2011) and eventually bridge across the outer periclinal array to form transverse antiparallel bundles. We propose that an increase or redistribution of the nucleation sites for these polymers, at the expense of microtubules in all other orientations (i.e., not just transverse) originating on the outer periclinal cell face, promotes the antiparallel microtubule bundles transverse to the cell’s axis (Figures 6C to 6E). We speculate that the progression of transverse array organization from the middle of the cell to the ends may be due to a gradient in signaling for microtubule nucleation in the lateral anticlinal cell faces and influenced by a lag in the disassembly of the longitudinal polymer bundles in the apical and basal regions of the cell.
METHODS
Plant Materials
Seed was sterilized in 100% ethanol for 10 s before planting on 0.3% agar (Sigma-Aldrich) plates containing half-strength MS medium and no sugar (Sigma-Aldrich). We exposed the plated seed to 2 h of light, wrapped the plates in foil, and stored them for 2 d at 4°C to synchronize germination. The foil was removed and the plates were incubated vertically at 22°C under continuous light for 6 d prior to imaging. Wild-type Arabidopsis thaliana (Columbia-0) plants expressing a 35S-GFP:TUA6 transgene were used for visualizing microtubules and a 35S-Eb1:GFP expressing line for visualizing polymerizing microtubule plus ends (Mathur et al., 2003).
Hormone Treatments
Seedlings were taken from agar germination plates after 6 d at 22°C and placed into liquid half-strength MS medium for treatment. A 10 mM GA4 (Sigma-Aldrich) stock dissolved in 1:1 (v/v) of ethanol and distilled water was made and plants were treated with a 10 μM final concentration. A 0.1 mM IAA (Sigma-Aldrich) stock dissolved in 100% DMSO (Sigma-Aldrich) was made and plants were treated with a 0.5 μM final concentration. For observations made at 2 h, plants were placed in 5 mL of liquid and moved to a cover glass with the same medium immediately prior to imaging. For time-lapse experiments, plants were positioned onto a glass slide with 3∼0.5 mL of medium and sealed between slide and cover glass using silicon vacuum grease (Dow Corning) to form a chamber.
Confocal Microscopy
Hypocotyl cells were imaged using a Leica SP5 confocal laser scanning microscope with either a ×40 1.2–numerical aperture (NA) or a ×63 1.2-NA water emersion objective lens. Confocal volume imaging of the apical and basal side faces of the cell was accomplished with a Nikon A1 confocal laser scanning microscope using a ×40 1.2-NA water immersion lens. The GFP-based probes were excited using a 488-nm laser line, and excitation was collected using the spectrophotometric detector.
To capture array organization in whole hypocotyls, the hypocotyls were imaged in sections (typically four) beginning with the root/hypocotyl junction and progressing apically to the nascent meristem. Optical sections for reconstructed image projections were taken at 0.25- to 0.34-μm steps. Two-dimensional projections of GFP:TUA6 data sets were created by converting Leica files into tiff format (Imaris; Bitplane) and subsequently creating a summation or standard deviation projection using ImageJ (NIH, Wayne Rasband). Photoshop (Adobe Software) was then used to linearly scale the contrast for presentation. Images showing the entire hypocotyl were composited by hand in Illustrator (Adobe Software).
To determine the number and direction of polymerizing microtubule plus ends at time intervals during array organization, Eb1:GFP in control and GA4 and IAA cotreated seedlings were imaged. Data were collected at 15-min intervals starting at the time when plants were placed into the liquid inducing media on the microscope slide and terminating 2.5 h later. Plants saw exposure to hormone for no more than 3 min before the first time point. The seedlings were continuously imaged for the first 5 min of every 15-min interval. A series of seven frames, taken at 0.5-µm steps along the microscope’s optical axis (upper 3 to 4 µm of the cell), was taken every 6 s to produce the 3D time-lapse data sets.
For evaluation of microtubule ends polymerizing in the lateral side and end faces of the cell, we used the Leica confocal laser scanning microscope to collect 2- to 5-min duration time-lapse movies of 6-d-old GFP:Eb1-expressing plants at 3.1- to 5.9-s intervals. A narrow XY field of view was used to capture only the side or end face of interest from the outer to the inner periclinal cell faces. Images were taken every 0.2 µm to a minimum depth of 10 µm from the top surface of the cell.
Data Analysis
For assaying the percentage of cells with transverse cortical arrays, the two-dimensional projections were given to two individuals for single blind evaluation using the same classing key (Figure 1) for scoring array types. Array organization was scored in two adjacent cell files along the entire hypocotyl length (∼28.0 ± 3.1 cells) to avoid any bias in organization for cell file. Counts for each array class from different counters were averaged for final reporting.
A series of MATLAB (The Mathworks) scripts was developed and implemented as a graphical user interface for assessing the GFP:Eb1 trajectories and for computing summary statistics. Prior to analysis, each of the 3D data sets was projected to a single image creating 50 or more time-lapse image frames at 6-s intervals for each 5-min image series. Each 50-frame data set of the nine data sets comprising the 2-h time lapse was used separately for analysis. Our first goal was to measure the average number of microtubule plus ends polymerizing into or out of the outer periclinal array during each 15-min interval. Our second goal was to relate any change in number or ratio of trajectories to the spatial position on the cell perimeter. To accomplish these goals, a line was traced around the perimeter of the cell using the computer pointing device and a contour was automatically generated inside the cell creating a zone where GFP:Eb1 spots were counted one time. Each 5-min time-lapse sequence was broken into seven sequential time periods with five images per period. Using the computer pointing device, each GFP:Eb1 spot in the perimeter delimited zone was marked only once by hand, and the trajectory of the spot was recorded as either moving “in” to the periclinal array, “out” of the periclinal array, or “sideways” relative to the cell edge. The trajectory assignment was made by looping the five images in the time period to visualize polymerization direction. The width of the counting zone contour was set to ∼3 µm such that GFP:Eb1 spots were not double counted across the seven time periods. Summary statistics for each 5-min time period were therefore calculated as the average number of microtubule plus ends per 1-µm linear distance of cell perimeter that were traversing the cell perimeter per 30 s with a number of seven time periods per 5 min. To validate the data transcription, one control and one experimental cell were counted for all nine time points by two different individuals, where the resulting differences in the number and direction of GFP:Eb1 spots were found to be negligible.
For determining the spatial distributions of each spot, the marked GFP:Eb1 position was projected onto the hand-drawn perimeter line for each cell and the measured pixels/micrometer value was used to convert and scale the data. The apical and basal regions of each cell were marked by hand in the MATLAB graphical user interface from known orientations of the cells in the images.
For side and end face dynamics, the data were projected side-on (xz or yz projections) using 2.1- to 2.5-µm thick slices of the 3D time-lapse data set to evaluate the GFP:Eb1 trajectory. For examining tracks, data sets from sequential 3D projections were summed to a single image (Imaris).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Movie 1. Short Interval Imaging of an Oblique Array Transitioning to Transverse.
Supplemental Movie 2. Microtubule Plus-End Trajectories Indicate Dramatic Changes to the Array Architecture after Hormone Induction.
Supplemental Movie 3. Microtubule Plus-End Trajectories Indicate Dramatic Changes to the Array Architecture after Hormone Induction.
Supplemental Movie 4. Microtubule Plus-End Trajectories Indicate Dramatic Changes to the Array Architecture after Hormone Induction.
Supplemental Movie 5. Microtubules Polymerize Continuously between the Periclinal and Anticlinal Cell Faces.
Acknowledgments
We thank Roger Hangarter for advice regarding the hormone treatments and growth conditions and Jim Powers at the Indiana University Light Microscopy Imaging Center for help with the microscopy. We acknowledge support for this work from the National Science Foundation (MCB-0920555 and MCB-1157982 to S.L.S.).
AUTHOR CONTRIBUTIONS
L.V. designed and executed the majority of experiments. J.R.L. performed the original hormone induction experiments. A.E. measured and correlated the array orientation assays. S.D. performed the hypocotyl length assays. S.L.S. developed the project and wrote the article.
Glossary
- IAA
indole-3-acetic acid
- GA4
gibberellic acid
- MS
Murashige and Skoog
- NA
numerical aperture
- 3D
three-dimensional
References
- Allard J.F., Ambrose J.C., Wasteneys G.O., Cytrynbaum E.N. (2010a). A mechanochemical model explains interactions between cortical microtubules in plants. Biophys. J. 99: 1082–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard J.F., Wasteneys G.O., Cytrynbaum E.N. (2010b). Mechanisms of self-organization of cortical microtubules in plants revealed by computational simulations. Mol. Biol. Cell 21: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Allard J.F., Cytrynbaum E.N., Wasteneys G.O. (2011). A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215: 150–171 [DOI] [PubMed] [Google Scholar]
- Baskin T.I. (2005). Anisotropic expansion of the plant cell wall. Annu. Rev. Cell Dev. Biol. 21: 203–222 [DOI] [PubMed] [Google Scholar]
- Baulin V.A., Marques C.M., Thalmann F. (2007). Collision induced spatial organization of microtubules. Biophys. Chem. 128: 231–244 [DOI] [PubMed] [Google Scholar]
- Bornens M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14: 25–34 [DOI] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., Ye Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13: 807–827 [PMC free article] [PubMed] [Google Scholar]
- Chan J., Calder G., Fox S., Lloyd C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9: 171–175 [DOI] [PubMed] [Google Scholar]
- Chan J., Crowell E., Eder M., Calder G., Bunnewell S., Findlay K., Vernhettes S., Höfte H., Lloyd C. (2010). The rotation of cellulose synthase trajectories is microtubule dependent and influences the texture of epidermal cell walls in Arabidopsis hypocotyls. J. Cell Sci. 123: 3490–3495 [DOI] [PubMed] [Google Scholar]
- Chan J., Eder M., Crowell E.F., Hampson J., Calder G., Lloyd C. (2011). Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light-grown Arabidopsis hypocotyls. J. Cell Sci. 124: 1088–1094 [DOI] [PubMed] [Google Scholar]
- Crowell E.F., Timpano H., Desprez T., Franssen-Verheijen T., Emons A.-M., Höfte H., Vernhettes S. (2011). Differential Regulation of Cellulose Orientation at the Inner and Outer Face of Epidermal Cells in the Arabidopsis Hypocotyl. Plant Cell 23: 2592–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr R.J. (1994). Microtubules in plant morphogenesis: Role of the cortical array. Annu. Rev. Cell Biol. 10: 153–180 [DOI] [PubMed] [Google Scholar]
- Cyr R.J., Palevitz B.A. (1995). Organization of cortical microtubules in plant cells. Curr. Opin. Cell Biol. 7: 65–71 [DOI] [PubMed] [Google Scholar]
- Deinum E.E., Tindemans S.H., Mulder B.M. (2011). Taking directions: The role of microtubule-bound nucleation in the self-organization of the plant cortical array. Phys. Biol. 8: 056002. [DOI] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004a). The cortical microtubule array: From dynamics to organization. Plant Cell 16: 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Cyr R. (2004b). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16: 3274–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S. (2001). Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2: 688–698 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W. (2008). Straighten up and fly right: Microtubule dynamics and organization of non-centrosomal arrays in higher plants. Curr. Opin. Cell Biol. 20: 107–116 [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Shaw S.L. (2006). Microtubule dynamics and organization in the plant cortical array. Annu. Rev. Plant Biol. 57: 859–875 [DOI] [PubMed] [Google Scholar]
- Eren E.C., Dixit R., Gautam N. (2010). A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol. Biol. Cell 21: 2674–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folta K.M., Pontin M.A., Karlin-Neumann G., Bottini R., Spalding E.P. (2003). Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 36: 203–214 [DOI] [PubMed] [Google Scholar]
- Gendreau E., Traas J., Desnos T., Grandjean O., Caboche M., Höfte H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T.H., Jr, Brower D.L., Staehelin L.A. (1980). Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 84: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P.B. (1962). Mechanism for plant cell morphogensis. Science 138: 1404–1405 [DOI] [PubMed] [Google Scholar]
- Hardham A.R., Gunning B.E. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77: 14–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Kato T. (2006). Cortical control of plant microtubules. Curr. Opin. Plant Biol. 9: 5–11 [DOI] [PubMed] [Google Scholar]
- Hawkins R.J., Tindemans S.H., Mulder B.M. (2010). Model for the orientational ordering of the plant microtubule cortical array. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 82: 011911. [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Newcomb E.H. (1964). Microtubules and fibrils in cytoplasm of coleus cells undergoing secondary wall deposition. J. Cell Biol. 20: 529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. (2003). Dynamics and mechanics of the microtubule plus end. Nature 422: 753–758 [DOI] [PubMed] [Google Scholar]
- Huang R.F., Lloyd C.W. (1999). Gibberellic acid stabilises microtubules in maize suspension cells to cold and stimulates acetylation of alpha-tubulin. FEBS Lett. 443: 317–320 [DOI] [PubMed] [Google Scholar]
- Ishida K., Katsumi M. (1991). Immunofluorescence microscopical observation of cortical microtubule arrangement by gibberellin in d5 mutant of Zea mays. Plant Cell Physiol. 32: 409–417 [Google Scholar]
- Kirik A., Ehrhardt D.W., Kirik V. (2012). TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24: 1158–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le J., Vandenbussche F., De Cnodder T., Van Der Straeten D., Verbelen J.P. (2005). Cell elongation and microtubule behavior in the Arabidopsis hypocotyl: Responses to ethylene and auxin. J. Plant Growth Regul. 24: 166–178 [Google Scholar]
- Ledbetter M.C. (1982). The role of microtubules in plant cell wall growth. Recent Adv. Phytochem. 16: 125–150 [Google Scholar]
- Ledbetter M.C., Porter K.R. (1964). Morphology of microtubules of plant cell. Science 144: 872–874 [DOI] [PubMed] [Google Scholar]
- Liu B., Joshi H.C., Wilson T.J., Silflow C.D., Palevitz B.A., Snustad D.P. (1994). γ-Tubulin in Arabidopsis: Gene sequence, immunoblot, and immunofluorescence studies. Plant Cell 6: 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Marc J., Joshi H.C., Palevitz B.A. (1993). A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104: 1217–1228 [DOI] [PubMed] [Google Scholar]
- Lloyd, C. (2011). Dynamic microtubules and the texture of plant cell walls. Int. Rev. Cell Mol. Biol. 287: 287–329. [DOI] [PubMed]
- Lloyd C., Chan J. (2002). Helical microtubule arrays and spiral growth. Plant Cell 14: 2319–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C., Chan J. (2008). The parallel lives of microtubules and cellulose microfibrils. Curr. Opin. Plant Biol. 11: 641–646 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Shaw P.J., Warn R.M., Yuan M. (1995). Gibberellic-acid-induced reorientation of cortical microtubules in living plant cells. J. Microsc. 181: 140–144 [Google Scholar]
- Lloyd C.W., Wells B. (1985). Microtubules are at the tips of root hairs and form helical patterns corresponding to inner wall fibrils. J. Cell Sci. 75: 225–238 [DOI] [PubMed] [Google Scholar]
- Lucas J., Shaw S.L. (2008). Cortical microtubule arrays in the Arabidopsis seedling. Curr. Opin. Plant Biol. 11: 94–98 [DOI] [PubMed] [Google Scholar]
- Maly I.V. (2002). Diffusion approximation of the stochastic process of microtubule assembly. Bull. Math. Biol. 64: 213–238 [DOI] [PubMed] [Google Scholar]
- Mathur J., Mathur N., Kernebeck B., Srinivas B.P., Hülskamp M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13: 1991–1997 [DOI] [PubMed] [Google Scholar]
- Mazia D. (1987). The chromosome cycle and the centrosome cycle in the mitotic cycle. Int. Rev. Cytol. 100: 49–92 [DOI] [PubMed] [Google Scholar]
- McDonald A.R., Liu B., Joshi H.C., Palevitz B.A. (1993). Gamma-tubulin is associated with a cortical-microtubule-organizing zone in the developing guard cells of Allium cepa L. Planta 191: 357–361 [DOI] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., Hasebe M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7: 961–968 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12: 1064–1070 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Hashimoto T. (2009). A mutation in the Arabidopsis gamma-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci. 122: 2208–2217 [DOI] [PubMed] [Google Scholar]
- Newcomb E.H. (1969). Plant microtubules. Annu. Rev. Plant Physiol. 20: 253–288 [Google Scholar]
- Oda Y., Fukuda H. (2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495 [DOI] [PubMed] [Google Scholar]
- Pastuglia M., Azimzadeh J., Goussot M., Camilleri C., Belcram K., Evrard J.L., Schmit A.C., Guerche P., Bouchez D. (2006). Gamma-tubulin is essential for microtubule organization and development in Arabidopsis. Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G., Pelletier S., Jaillard D., Höfte H. (2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135: 959–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts I.N., Lloyd C.W., Roberts K. (1985). Ethylene-induced microtubule reorientations - Mediation by helical arrays. Planta 164: 439–447 [DOI] [PubMed] [Google Scholar]
- Sambade A., Pratap A., Buschmann H., Morris R.J., Lloyd C. (2012). The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell 24: 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook J.C., Kaloriti D. (2008). Microtubules, MAPs and plant directional cell expansion. Trends Plant Sci. 13: 303–310 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., Ehrhardt D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Shaw S.L., Lucas J. (2011). Intrabundle microtubule dynamics in the Arabidopsis cortical array. Cytoskeleton (Hoboken) 68: 56–67 [DOI] [PubMed] [Google Scholar]
- Shibaoka H. (1974). Involvement of wall microtubules in gibberellin promotion and kinetin inhibition of stem elongation. Plant Cell Physiol. 15: 255–263 [Google Scholar]
- Shibaoka H. (1993). Regulation of gibberellins of the orientation of cortical microtubules in plant cells. Aust. J. Plant Physiol. 20: 461–470 [Google Scholar]
- Shibaoka H. (1994). Plant hormone-induced changes in the orientation of cortical microtubules: Alterations in the cross-linking between microtubules and the plasma membrane. Annu. Rev. Plant Physiol. 45: 527–544 [Google Scholar]
- Shibaoka H., Takesue K. (1999). Auxin-induced longitudinal-to-transverse reorientation of cortical microtubules in nonelongating epidermal cells of azuki bean epicotyls. Protoplasma 206: 27–30 [Google Scholar]
- Takesue K., Shibaoka H. (1999). Auxin-induced longitudinal-to-transverse reorientation of cortical microtubules in nonelongating epidermal cells of azuki bean epicotyls. Protoplasma 206: 27–30 [Google Scholar]
- Tindemans S.H., Hawkins R.J., Mulder B.M. (2010). Survival of the aligned: Ordering of the plant cortical microtubule array. Phys. Rev. Lett. 104: 058103. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Verbelen J.P., Van Der Straeten D. (2005). Of light and length: Regulation of hypocotyl growth in Arabidopsis. Bioessays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Ambrose J.C. (2009). Spatial organization of plant cortical microtubules: Close encounters of the 2D kind. Trends Cell Biol. 19: 62–71 [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Galway M.E. (2003). Remodeling the cytoskeleton for growth and form: An overview with some new views. Annu. Rev. Plant Biol. 54: 691–722 [DOI] [PubMed] [Google Scholar]
- Wenzel C.L., Williamson R.E., Wasteneys G.O. (2000). Gibberellin-induced changes in growth anisotropy precede gibberellin-dependent changes in cortical microtubule orientation in developing epidermal cells of barley leaves. Kinematic and cytological studies on a gibberellin-responsive dwarf mutant, M489. Plant Physiol. 124: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R., Turner S.R. (2007). Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 52: 742–751 [DOI] [PubMed] [Google Scholar]
- Wymer C., Lloyd C. (1996). Dynamic microtubules: Implications for cell wall patterns. Trends Plant Sci. 1: 222–228 [Google Scholar]
- Wymer C.L., Wymer S.A., Cosgrove D.J., Cyr R.J. (1996). Plant cell growth responds to external forces and the response requires intact microtubules. Plant Physiol. 110: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Warn R.M., Shaw P.J., Lloyd C.W. (1995). Dynamic microtubules under the radial and outer tangential walls of microinjected pea epidermal cells observed by computer reconstruction. Plant J. 7: 17–23 [DOI] [PubMed] [Google Scholar]
- Zhang D., et al. (2011). Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 13: 361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]