This work identifies a pair of evolutionarily conserved NOT2_3_5 domain–containing proteins that act as a scaffold to interact with polymerase II and some pri-miRNA processing proteins, thus promoting the transcription of protein-coding and MIR genes and facilitating efficient DCL1 recruitment in miRNA biogenesis.
Abstract
MicroRNAs (miRNAs) play key regulatory roles in numerous developmental and physiological processes in animals and plants. The elaborate mechanism of miRNA biogenesis involves transcription and multiple processing steps. Here, we report the identification of a pair of evolutionarily conserved NOT2_3_5 domain–containing-proteins, NOT2a and NOT2b (previously known as At-Negative on TATA less2 [NOT2] and VIRE2-INTERACTING PROTEIN2, respectively), as components involved in Arabidopsis thaliana miRNA biogenesis. NOT2 was identified by its interaction with the Piwi/Ago/Zwille domain of DICER-LIKE1 (DCL1), an interaction that is conserved between rice (Oryza sativa) and Arabidopsis thaliana. Inactivation of both NOT2 genes in Arabidopsis caused severe defects in male gametophytes, and weak lines show pleiotropic defects reminiscent of miRNA pathway mutants. Impairment of NOT2s decreases the accumulation of primary miRNAs and mature miRNAs and affects DCL1 but not HYPONASTIC LEAVES1 (HYL1) localization in vivo. In addition, NOT2b protein interacts with polymerase II and other miRNA processing factors, including two cap binding proteins, CBP80/ABH1, CBP20, and SERRATE (SE). Finally, we found that the mRNA levels of some protein coding genes were also affected. Therefore, these results suggest that NOT2 proteins act as general factors to promote the transcription of protein coding as well as miRNA genes and facilitate efficient DCL1 recruitment in miRNA biogenesis.
INTRODUCTION
MicroRNAs (miRNAs) are 20- to 25-nucleotide noncoding RNAs that negatively regulate gene expression at posttranscriptional levels either by mRNA cleavage or translational repression (Bartel, 2004; Kim, 2005; Filipowicz et al., 2008; Fabian et al., 2010). miRNAs act as universal regulatory factors that modulate gene expression in various biological processes in animals and plants, including developmental regulation and environmental adaptation (Lee et al., 1993; Kidner and Martienssen, 2005; Kim, 2005; Liu et al., 2005; Chen, 2009; Li et al., 2012; Sunkar et al., 2012). Elaborate biogenesis pathways have evolved to produce miRNAs (Chen, 2005; Jones-Rhoades et al., 2006; Xie et al., 2010).
In plants, the primary miRNAs (pri-miRNAs), which harbor an imperfect stem-loop structure, are predominately transcribed by DNA-dependent RNA polymerase II (Pol II) in coordination with the Mediator complex (Xie et al., 2005; Zheng et al., 2009; Kim and Chen, 2011; Kim et al., 2011). Prior to processing, the pri-miRNAs are captured by the RNA binding protein DAWDLE (DDL), which is presumed to stabilize pri-miRNAs and facilitate processing by DICER-LIKE1(DCL1). (Park et al., 2002; Yu et al., 2008). DCL1 sequentially processes pri-miRNAs to stem-loop precursors (pre-miRNAs) and eventually to miRNA/miRNA* duplexes in the nucleus (Kurihara and Watanabe, 2004); DCL1 processing involves the double-stranded RNA binding protein HYPONASTIC LEAVES1 (HYL1), the pri- and pre-mRNA binding protein TOUGH (Lu and Fedoroff, 2000; Han et al., 2004; Kurihara et al., 2006; Ren et al., 2012), and the zinc-finger protein SERRATE (SE) (Grigg et al., 2005; Yang et al., 2006; Laubinger et al., 2008). DCL1, HYL1, and SE colocalize in discrete nuclear bodies called D-bodies, where pri-miRNAs are processed (Fang and Spector, 2007; Liu et al., 2012).
In Saccharomyces cerevisiae, Cell Division Cycle 36 was identified as an essential negative transcriptional regulator that preferentially affected Tc-dependent transcription (in the absence of a conventional TATA box) from the HIS3 promoter and was thus referred to as NOT2 (for Negative on TATA less2) (Collart and Struhl, 1994). NOT2 is the core member of the CARBON CATABOLITE REPRESSION4 (CCR4)-NOT complex. In yeast, the CCR4-NOT complex profoundly and broadly affects mRNA metabolism at both transcriptional and posttranscriptional levels, including transcriptional repression and activation, mRNA decay and quality control, RNA export, translational repression, and protein ubiquitination (Denis and Chen, 2003; Collart and Timmers, 2004; Collart and Panasenko, 2012). Recently, NOT2 was found to bind RNA Pol II directly and promote transcriptional elongation, revealing the fundamental involvement of the CCR4-NOT complex in transcription (Kruk et al., 2011). The CCR4-NOT complex acts as the predominant mRNA deadenylase and is involved in mRNA decay as well as miRNA-directed mRNA degradation in yeast and animals (Daugeron et al., 2001; Tucker et al., 2001; Parker and Song, 2004; Fabian et al., 2010; Ito et al., 2011).
NOT2 is an evolutionarily conserved protein in eukaryotes (Anand et al., 2007). However, the function of NOT2 in plants remains largely unknown. The Arabidopsis thaliana genome has two highly similar NOT2 homologs with 60.0% identity and 70.8% consensus at the amino acid level, respectively, referred to as At-NOT2 and VIRE2-INTERACTING PROTEIN2 (VIP2) (Anand et al., 2007), which we refer to here as NOT2a and NOT2b for simplicity and clarity. It was reported that VIP2/NOT2b is required for Agrobacterium tumefaciens–mediated stable transformation in Nicotiana benthamiana and Arabidopsis (Anand et al., 2007).
Here, we report the identification of NOT2 proteins by their interaction with DCL1; in both rice (Oryza sativa) and Arabidopsis, NOT2 proteins are general transcriptional regulators essential for plant development. Multiple lines of evidence indicate that NOT2s act as a scaffold to interact with Pol II and some pri-miRNA processing proteins, thus coordinating MIR transcription and efficient DCL1 recruitment in Arabidopsis miRNA biogenesis.
RESULTS
NOT2 Is an Evolutionarily Conserved Protein That Directly Associates with Plant DCL1
Our previous work demonstrated that Os-DCL1 is a key enzyme in rice miRNA biogenesis (Liu et al., 2005). The Piwi/Ago/Zwille (PAZ) domain, a conserved functional domain of DCL1, recognizes the end of the double-stranded RNA, allowing DCL1 to measure and generate small RNAs of the correct size (MacRae et al., 2007). To further study the precise mechanism of rice miRNA biogenesis, we selected the PAZ domain as a bait to screen for potential interacting partners associated with DCL1. From this yeast two-hybrid screen, we identified the C-terminal region (305 to 622 amino acids) of Os-NOT2 (Os-NOT2p) as being represented by positive clones at the highest frequency. To further substantiate the interaction between Os-NOT2 and Os-DCL1, we used stable coexpression of AD-Os-DCL1 PAZ and BD-Os-NOT2p in yeast and found this combination activated the ADE2, HIS3, and lacZ reporter genes, further confirming the bona fide interaction between Os-DCL1 and Os-NOT2p (see Supplemental Figure 1A online).
To further characterize the biological roles of NOT2 in plants, we first used RNA interference (RNAi) to knock down Os-NOT2 in rice. The transgenic rice with reduced Os-NOT2 levels either could not regenerate from callus or became arrested at an early seedling stage (see Supplemental Figure 1B online), indicating that Os-NOT2 is vital for rice development. The lethality of Os-NOT2 RNAi in transgenic rice restricted further study of Os-NOT2; therefore, we next studied the function of NOT2 in Arabidopsis, based on the high sequence conservation of NOT2 proteins between rice and Arabidopsis (Anand et al., 2007).
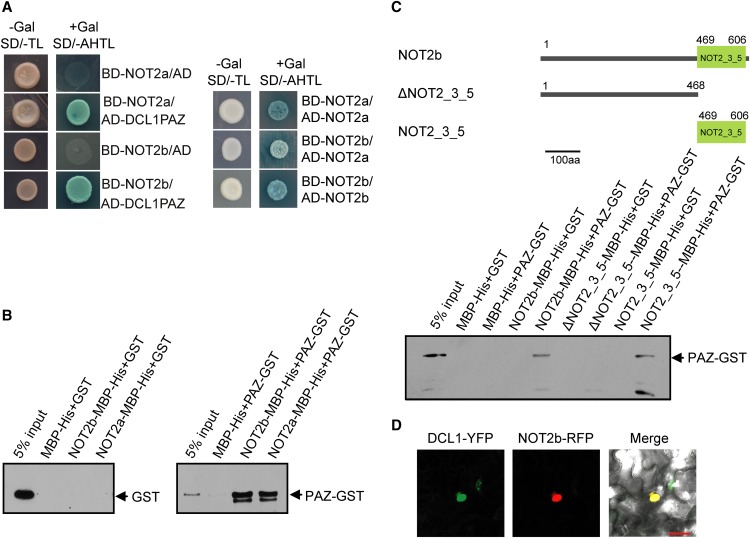
To determine whether the interactions between NOT2a or NOT2b and DCL1 exist in Arabidopsis, we used yeast two-hybrid assays. We generated the appropriate constructs with the Arabidopsis proteins and cotransformed them into yeast. We found that Arabidopsis NOT2a and NOT2b both interact with DCL1 PAZ in yeast (Figure 1A, left panels). To further verify this interaction, we performed pull-down assays using maltose binding protein (MBP)-His–tagged NOT2a or NOT2b as a bait and glutathione S-transferase (GST)–tagged DCL1 PAZ as a prey. Nickel pull-down and immunoblotting assays showed that both NOT2a and NOT2b capture DCL1 PAZ-GST efficiently in vitro compared with GST alone (Figure 1B).
Figure 1.
DCL1 and NOT2 Show Interaction in Vitro and Nuclear Localization.
(A) GAL4-based yeast two-hybrid assays for interactions between the Arabidopsis DCL1 PAZ domain and NOT2a or NOT2b (left panels) as well as between pairs of NOT2a/NOT2a, NOT2a/NOT2b, and NOT2b/NOT2b (right panels). AD and BD represent the plasmids encoding the fusions to the GAL4 activation domain and the DNA binding domain, respectively. Cotransformed yeast colonies were spotted on the selective SD medium minus Trp and Leu (-TL), then grown on SD medium minus adenine, His, Trp, and Leu (-AHTL) supplemented with 40 µg/mL X-α-Gal for β-galactosidase activity.
(B) In vitro pull-down assays between MBP-His–tagged NOT2a or NOT2b and GST-tagged DCL1 PAZ domain.
(C) In vitro pull-down assays between MBP-His–tagged full-length or truncated versions of NOT2b and GST-tagged DCL1 PAZ domain. The top panel shows schematic representations of NOT2b full-length (NOT2b), full-length NOT2 without the NOT2_3_5 domain (ΔNO2_3_5), and the NOT2_3_5 domain only (NOT2_3_5). Numbers indicate the positions of amino acid in the proteins. The bottom panel shows an immunoblot of proteins pulled down with the MBP-His tag recombinant protein (and input control) detected with anti-GST antibody. aa, amino acids.
(D) Subcellular localization of NOT2b and DCL1 in tobacco epidermal cells. RFP and YFP fluorescence are pseudocolored in red and green, respectively. Bar = 25 µm.
The similarities between NOT2a and NOT2b prompted us to test the relationship of these two proteins. Extensive interactions between NOT2a and NOT2a, NOT2b and NOT2b, as well as NOT2a and NOT2b were detected in yeast two-hybrid assays. The results indicated that both NOT2 proteins can form heterodimers or homodimers and may act as a functional unit in vivo (Figure 1A, right panels). In the following experiments, NOT2b is used as a representative to explore the function of NOT2 proteins.
We first used protein deletions to determine which domain of NOT2 interacts with DCL1. The NOT2_3_5 domain, a conserved motif of Os-NOT2 proteins, is responsible for interaction with the PAZ domain of Os-DCL1 according to our yeast two-hybrid assay (Figure 1A). The NOT2_3_5 domain of CNOT2 (human homolog of NOT2) has been found to act as a repressor of promoters in reporter gene activity assays using in vitro transient transfections of human cells (Zwartjes et al., 2004). Thus, we examined if the conserved domain of Arabidopsis NOT2 contributes to the interaction with DCL1. Three versions of MBP-His–tagged NOT2b, including full-length NOT2b (NOT2b), full-length NOT2 with the NOT2_3_5 domain deleted (△NOT2_3_5), and the NOT2_3_5 domain only (NOT2_3_5) (Figure 1C, top panels), were expressed for pull-down assays. The protein that lacks the NOT2_3_5 motif (△NOT2_3_5) did not show an interaction with DCL1 PAZ, but NOT2_3_5 itself completely retained this association (Figure 1C, bottom panel). The data thus indicate that the C-terminal NOT2_3_5 domain mediates the interaction with DCL1.
The interaction between DCL1 and NOT2b was also confirmed by examination of their subcellular locations, using protein fusions with fluorescent markers transiently expressed in epidermal cells of N. benthamiana. The yellow fluorescent protein (YFP)–tagged DCL1 localized in the nucleus, which is consistent with previous reports (Fang and Spector, 2007; Song et al., 2007). When DCL1-YFP and NOT2b–red fluorescent protein (RFP) were coexpressed, NOT2b signals clearly overlapped with those of DCL1-YFP (Figure 1D). Collectively, these results indicated that NOT2 proteins physically associate with DCL1 in rice and Arabidopsis and the NOT2_3_5 domain is critical for the direct interaction.
Mutation of NOT2s Causes Severe Male Gametogenesis Defects in Arabidopsis
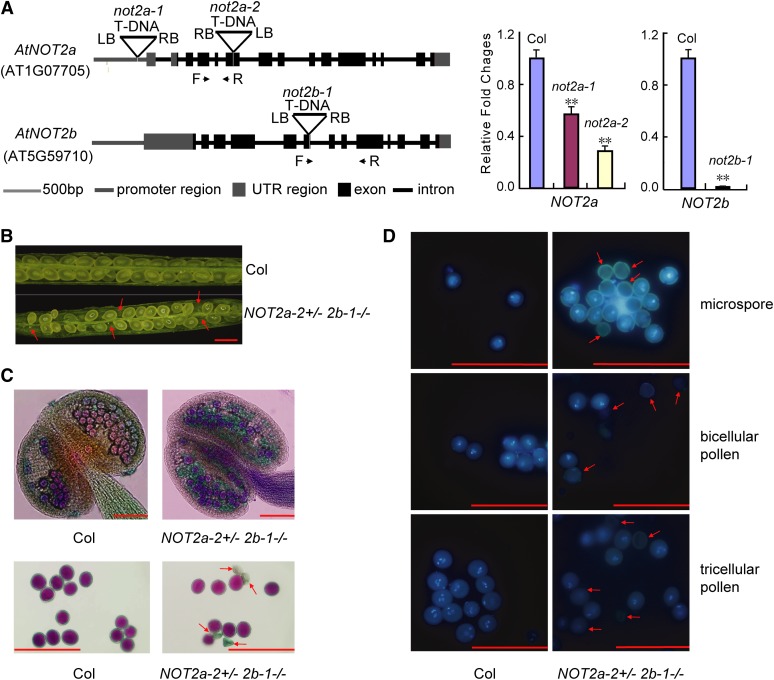
To determine the physiological roles of NOT2a and NOT2b, we identified the T-DNA insertion mutants not2a-1, not2a-2, and not2b-1 from the SALK or Nottingham Arabidopsis Stock Centre collections. not2b-1 is a null allele, and not2a-1 and not2a-2 are weak alleles with reduced NOT2a transcript levels (Figure 2A). Under normal growth conditions, the not2 single mutants did not display any obvious developmental defects. However, we failed to obtain the not2a-2 not2b-1 double mutant, implying that the homozygous double mutant is likely unable to survive.
Figure 2.
Male Gametophyte Development Is Affected in Severe not2 Mutants.
(A) Diagram of NOT2a and NOT2b genes and T-DNA insertion lines. Exons and introns are shown as black boxes and lines, respectively; gray boxes and lines represent 5′or 3′ untranslated regions (UTR) and promoter regions, respectively. The T-DNA insertion sites for each mutant are indicated by triangles. Transcript levels of NOT2a and NOT2b are detected by qRT-PCR in the indicated samples (right panels). Forward (F) and reverse (R) primer locations indicated by black arrows are shown. The relative fold changes were normalized to ACTIN. Error bars represent sd. Data are given as means sd of at least three independent biological replicates. Asterisks denote Student’s t test significance for difference between the indicated samples and the wild-type control: **P < 0.01.
(B) Seed development in the wild-type Col and NOT2a-2+/− 2b-1−/− mutant siliques. Red arrows indicate aborted ovules. Bar =1 mm.
(C) Alexander’s staining of anthers and mature pollen grains in the wild type (Col) and NOT2a-2+/− 2b-1−/− mutants. Viable pollen is stained purple and degenerated pollen is green. Bars = 100 µm.
(D) DAPI staining of microspores and bicellular and tricellular pollen in the wild type (Col) and NOT2a-2+/− 2b-1−/− mutants. Red arrows indicate degenerated pollen grains. Bars = 100 µm.
To confirm this hypothesis, we examined the fertility of NOT2a-2+/− 2b-1−/− plants, which should produce homozygous mutants as 25% of their progeny. Indeed, we found some aborted ovules in the siliques of NOT2a-2+/− 2b-1−/− (Figure 2B). We classified these mutants as weak, medium, and severe based on the length of siliques, which corresponded to the ratio of aborted ovules ranging from 32.4 to 67.1% (see Supplemental Table 1 online). We further examined whether the not2 mutant alleles were transmitted through the male and female gametophytes at the expected frequency. Statistical analysis of the progeny of reciprocal crosses to the wild type with NOT2a-2+/− 2b-1−/− or NOT2a-2−/− 2b-1+/− plants demonstrated that the transmission of the not2a-2 and not2b-1 alleles occurs at lower frequencies in both female and male gametophytes (Table 1). The female gametophyte transmission showed a moderate decrease from the expected 50 to 24.7% in NOT2a-2+/− 2b-1−/− plants (Table 1), indicating that both NOT2a and NOT2b are important for normal female gametophyte development in Arabidopsis. However, the transmission of not2a-2 and not2b-1 through the male gamete was 0.9 and 0%, respectively, instead of the expected 50% in the wild type, implying that absence of the homozygous double mutants is largely due to defects in male gametogenesis (Table 1). Also, staining the anthers of NOT2a-2+/− 2b-1−/− plants with Alexander’s stain (Alexander, 1969) revealed 43.7 to 10.6% nonviable pollen; in the wild type, there was <1% abnormal pollen (Figure 2C) (see Supplemental Table 2 online).
Table 1. Transmission Efficiency through Reciprocal Crosses.
| Progeny |
|||||||
|---|---|---|---|---|---|---|---|
| Parental Genotypes (Female × Male) Cross | NOT2a-2+/− | NOT2a-2+/+ | NOT2b-1+/− | NOT2b-1+/+ | Total | TEF | TEM |
| Wild type × NOT2a-2+/−2b-1−/− | 2 | 225 | 227 | 0 | 227 | NA | 0.9% |
| NOT2a-2+/− 2b-1−/− × wild type | 42 | 128 | 170 | 0 | 170 | 24.7% | NA |
| Wild type × NOT2a-2−/− 2b-1+/− | 126 | 0 | 0 | 126 | 126 | NA | 0% |
| NOT2a-2−/− 2b-1+/− × wild type | 172 | 0 | 45 | 127 | 217 | 25.9% | NA |
Reciprocal crosses among wild-type and NOT2a-2+/− 2b-1−/− or NOT2a-2−/− 2b-1+/− plants were used to determine the transmission efficiency of gametes from NOT2a-2 or NOT2b-1. We used PCR to determine the transmission efficiencies (TE). Transmission efficiency is calculated according to the following: TE = number of progenies with T-DNA insertion/number of progenies without T-DNA insertion × 100%; 50% of the reciprocal crosses is the expected value for the normal gamete transmission. TEF, female transmission efficiency; TEM, male transmission efficiency; NA, not applicable. P < 0.01 for TEM; P < 0.01 for TEF.
We next examined pollen formation in the mutants to determine the nature of the defect. During Arabidopsis microgametogenesis, meiosis of the microspore mother cell produces a tetrad of microspores. After release from the tetrad, each microspore undergoes an asymmetric cell division, pollen mitosis I, to generate a bicellular pollen grain containing a generative cell and a much larger vegetative cell. Then the generative cell performs a second cell division, pollen mitosis II, to give rise to a tricellular pollen (McCormick, 2004; Liu and Qu, 2008). To further determine at which stage(s) the aberrant pollen grains of NOT2a-2+/− 2b-1−/− were arrested, 4′,6-diamidino-2-phenylindole (DAPI) staining was applied to observe the pollen nuclei at different developmental stages. We observed degenerated and aberrant pollen grains that did not stain with DAPI at the tricellular, bicellular, and microspore stages in NOT2a-2+/− 2b-1−/− plants (Figure 2D), indicating that male gamete defects occur at or before pollen mitosis I. The functional roles of NOT2s in reproductive cells are also consistent with their high expression in carpels and stamens (see Supplemental Figure 2 online). These results suggest redundant roles of NOT2a and NOT2b in early male and female gametophyte development.
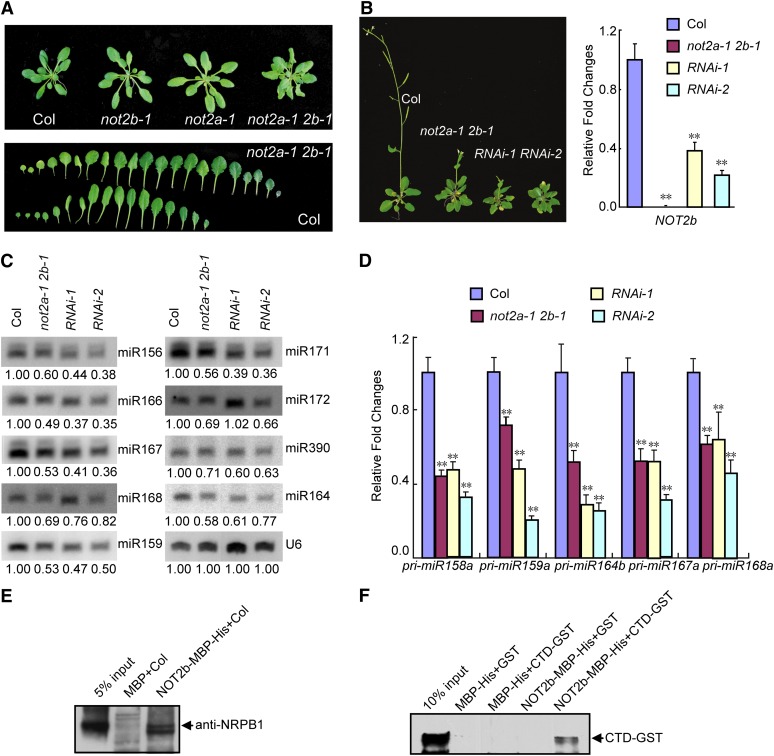
We also isolated a weak double mutant, not2a-1 not2b-1, which exhibits pleiotropic phenotypes, including crumpled and serrated rosette leaves and late flowering (Figures 3A and 3B). These phenotypes are similar to those of miRNA biogenesis pathway mutants, such as dcl1, hyl1, se, and cbp80/abh1 (Park et al., 2002; Vazquez et al., 2004; Laubinger et al., 2008). To further confirm the abnormal phenotypes of loss of function of both NOT2a and NOT2b, we generated transgenic plants with knockdown of NOT2b in the not2a-2 background. Two lines designated as RNAi-1 and RNAi-2 showed slightly enhanced phenotypes similar to not2a-1 not2b-1 (Figure 3B). These observations suggested that the defects in the not2a-1 not2b-1 mutants are due to loss of function of both NOT2a and NOT2b.
Figure 3.
MiRNA Biogenesis Is Compromised in Weak not2 Mutants.
(A) Phenotypes of 5-week-old plants (top panel) and rosette leaves arranged (from left to right) on the developmental axis (bottom panel) in the wild type (Col) and the weak not2a-1 not2b-1 double mutant (not2a-1 2b-1).
(B) Phenotypes of transgenic lines with RNAi knockdown of NOT2b in the not2a-2 background (left panel). The expression levels of NOT2b, measured by qRT-PCR, in the indicated samples are displayed in the right panel. The transcript levels are presented as the ratio of the value relative to that of the wild type (Col). Error bars represent sd. Data are given as means and sd of at least three independent biological replicates. **P < 0.01 by Student’s t test.
(C) The accumulation of various miRNAs as detected by RNA hybridization in Col, the not2a-1 not2b-1 mutant (not2a-1 2b-1), and two RNAi lines of NOT2b in the not2a-2 background. U6 serves as a loading control. The numbers below each lane represent the intensity ratio of each signal relative to the wild type (Col).
(D) qRT-PCR analysis of pri-miRNA levels at five MIR gene loci in indicated samples. The relative fold changes were normalized to ACTIN. Data are given as means sd of at least three independent biological replicates. **P < 0.01 by Student’s t test.
(E) NOT2b coprecipitated with Pol II by pull-down assays in vitro. Twelve-day-old seedlings of wild-type crude protein extract were used as prey. Pol II was analyzed by immunoblotting using antibodies against the RNA Pol II largest subunit, NRPB1.
(F) In vitro pull-down assays between MBP-His–tagged full-length NOT2b and GST-tagged C-terminal domain (CTD) of NRPB1. The GST-tagged CTD was detected by anti-GST antibody.
[See online article for color version of this figure.]
NOT2 Is Required for the Biogenesis of MiRNAs
The combined evidence that NOT2 binds DCL1 and that the phenotype of not2a-1 not2b-1 is reminiscent of miRNA biogenesis pathway mutants prompted us to test whether miRNAs were affected by inactivation of NOT2s. Indeed, we found that in not2a-1 not2b-1 or RNAi lines, all nine tested miRNAs were reduced in abundance by 1.4- to 2.7-fold relative to the wild type (Figure 3C).
miRNA biogenesis is a sequential process, proceeding mainly through three steps: transcription of pri-miRNA, pri-miRNA processing to pre-miRNA, and pre-miRNA processing to mature miRNA. To determine how miRNA biogenesis was affected by NOT2, we first examined the levels of pri-miRNAs. Quantitative RT-PCR revealed that the levels of pri-miRNAs at five MIR loci (MIR158a, MIR159a, MIR164b, MIR167a, and MIR168a) were reduced by nearly twofold in not2a-1 not2b-1 and RNAi lines (Figure 3D). This observation is in contrast with dcl1, hyl1, se, and cbp80/cbp20 mutants, in which pri-miRNA transcripts accumulate to much higher levels than the wild type (Grigg et al., 2005; Kurihara et al., 2006; Gregory et al., 2008). This suggested that NOT2a and NOT2b are likely involved in transcriptional regulation of MIR genes.
NOT2s Promote MIR Transcription
To confirm that NOT2s regulate miRNA transcription, we used a β-glucuronidase (GUS) reporter to test the effect of mutation of NOT2s on GUS expression driven by the MIR172a promoter (Yu et al., 2008). The transgenic plants containing pMIR172a:GUS were crossed to not2a-1 not2b-1. The individuals containing pMIR172a:GUS, along with either NOT2a-1+/− 2b-1+/− or not2a-1 not2b-1, were isolated from the F2 population for GUS staining. GUS expression was markedly decreased in not2a-1 not2b-1 compared with NOT2a+/− 2b+/− (see Supplemental Figure 3 online, top panels). By contrast, GUS expression driven by the promoter of a protein coding gene, PISTILLATA (PI) (Chen et al., 2000), did not exhibit reduction in the two genotypes (see Supplemental Figure 3 online, bottom panels). These results indicated that NOT2s promote MIR transcription.
Next, we tested how NOT2s regulate MIR transcription. MIR genes are predominately transcribed by Pol II (Xie et al., 2005; Borchert et al., 2006); we therefore examined the interaction between NOT2b and Pol II by pull-down assays. The purified MBP-His–tagged NOT2b protein on beads was incubated with a crude protein extract of wild-type Arabidopsis seedlings, and immunoblotting was conducted to determine whether Pol II was retained by the beads. Pol II (detected by anti-NRPB1 [RNA Pol II largest subunit] antibody) was coprecipitated with MBP-His–tagged NOT2b but not with MBP-His only (Figure 3E). Furthermore, a pull-down experiment was performed using MBP-His–tagged NOT2b (NOT2b-MBP-His) as the bait and GST-tagged C-terminal domain of NRPB1 (designated as CTD-GST) as the prey. Nickel pull-down and immunoblotting showed retention of NOT2b-MBP-His along with CTD-GST but not with the GST control, indicating that NOT2b interacts directly with the CTD of Pol II (Figure 3F). This result provided further support for the interaction between NOT2 and Pol II. Thus, we conclude that NOT2s promote MIR expression via interacting with Pol II.
NOT2 Is Associated with Key Factors in MiRNA Biogenesis
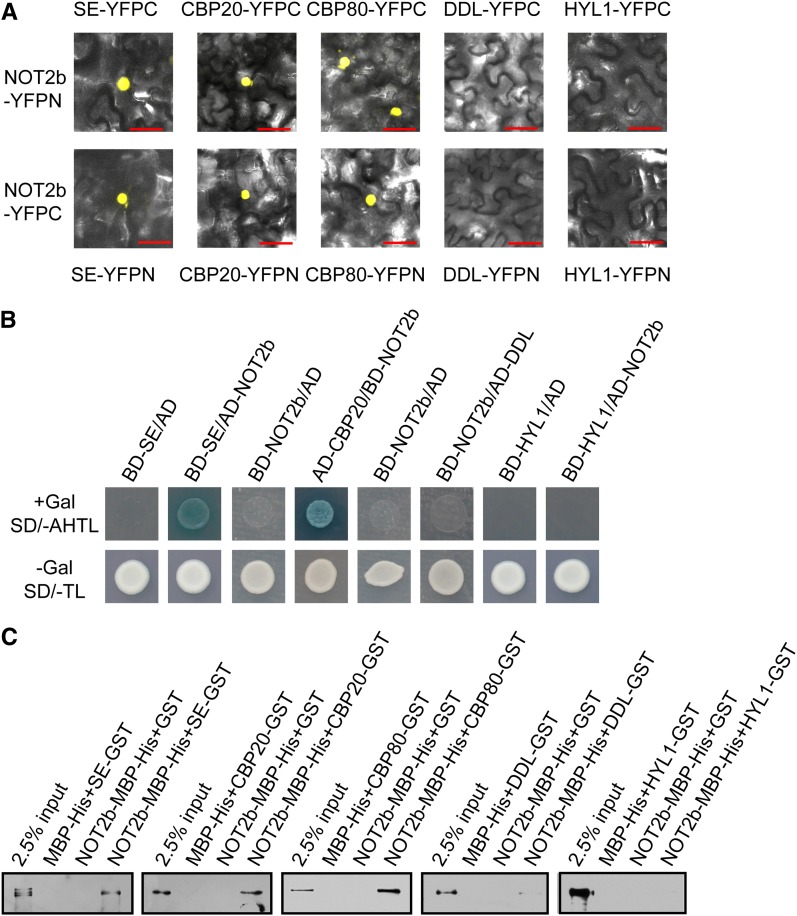
In Arabidopsis, pri-miRNAs are transcribed by Pol II and subsequently form fold-back structures to recruit proteins necessary for their posttranscriptional processing. DCL1, HYL1, and SE interact and colocalize with each other, forming a pri-miRNA processing and/or storage complex, or center (Fang and Spector, 2007). The observation that NOT2 physically associated with DCL1 and Pol II suggested that NOT2 might act as a scaffold to bridge miRNA transcription and DCL1 recruitment. We used bimolecular fluorescence complementation (BiFC) to determine whether NOT2 can also interact with other proteins involved in pri-miRNA processing (Walter et al., 2004). As a positive control, we found that HYL1 interacts with SE in discrete foci in the nucleoplasm (see Supplemental Figure 4A online), which is consistent with previously published observations (Fang and Spector, 2007). Intriguingly, we observed that intense BiFC fluorescence appeared when NOT2b was coexpressed with SE, CBP20, and CBP80 (Figure 4A), but BiFC fluorescence was not observed for the NOT2b/DDL and NOT2b/HYL1 pairs (Figure 4A). Independent assays by yeast two-hybrid and pull down also confirmed that NOT2 associated with SE, CBP20, and CBP80, but not with DDL or HYL1 (Figures 4B and 4C). Consistent with previous studies, we also observed that CBP20 interacts with CBP80 in vivo and thus is part of the CBP complex (CBC) (see Supplemental Figure 4B online, top panels) (Izaurralde et al., 1994). Additionally, we showed that CBP20, but not CBP80, binds SE in vivo and in vitro, which further supports the hypothesis that CBC and SE share common molecular functions (Laubinger et al., 2008) (see Supplemental Figures 4B to 4D online). These data suggested that CBP20 may affect miRNA biogenesis by binding SE directly. Therefore, the CBC might be required for forming a pri-miRNA processing center by interacting with SE directly or indirectly.
Figure 4.
NOT2b Interacts with Some Pri-miRNA Processing Factors.
The interactions between NOT2b and SE, CBP20, CBP80, DDL, and HYL1 were examined by BiFC (A), yeast two-hybrid (B), and pull-down (C) assays. For BiFC, protein partners fused to an N-terminal fragment (YFPN) or C-terminal fragment of YFP (YFPC) are indicated. Bars = 50 µm.
Impairment of NOT2s Affects DCL1 Localization
DCL1, HYL1, CBP20/80, and SE proteins are involved in pri-miRNA processing and they interact with each other directly or indirectly. NOT2 protein interacts with DCL1, SE, CBP20, and CBP80; therefore, we proposed that NOT2 may assist in the recruitment of these components to form an efficient processing center.
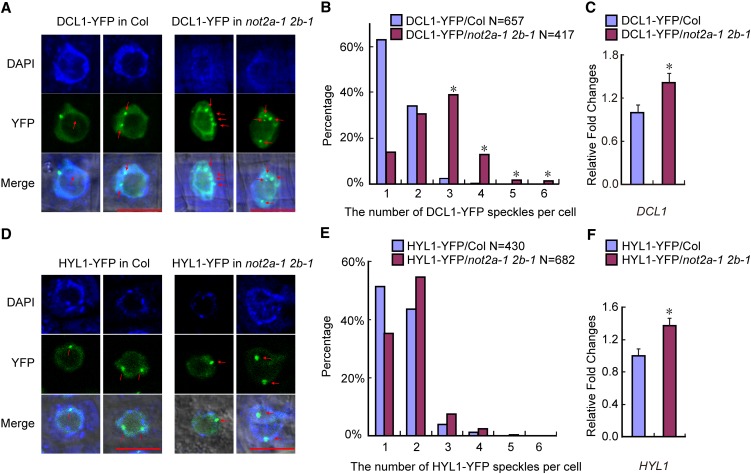
To test this hypothesis, we examined the effects of NOT2 impairment on the localization of two major miRNA processing components, DCL1 and HYL1. A previous study revealed that DCL1-YFP–containing bodies distribute throughout the nucleoplasm and ∼79% of plant cells harbor two or fewer DCL1-YFP speckles in wild-type Arabidopsis (Fang and Spector, 2007). We used the F2 population generated from transgenic plants expressing DCL1-YFP or HYL1-YFP crossed to the not2a-1 not2b-1 double mutant and counted the number of DCL1-YFP or HYL1-YFP bodies in each root cell in the wild type and not2a-1 not2b-1 backgrounds. In the wild type, most cells contained one or two DCL1-YFP speckles (Figure 5A, left panels), consistent with a previous report (Fang and Spector, 2007). By contrast, we observed three to four DCL1-YFP speckles in not2a-1 not2b-1 root cells (Figure 5A, right panels). Statistical analysis based on observations of 657 and 417 nuclei of the wild type and not2a-1 not2b-1, respectively, showed a significant increase in DCL1-YFP speckle number in the not2 double mutant (Figure 5B). By contrast, the numbers of HYL1-YFP speckles were not significantly increased in not2a-1 not2b-1 compared with that in the wild type root cells (Figures 5D and 5E). We also analyzed DCL1-YFP and HYL1-YFP expression and both showed equivalent induction (∼1.5-fold, P < 0.05) in not2a-1 not2b-1 (Figures 5C and 5F), which rules out the possibility that the increased number of DCL1-YFP speckles was due to upregulation of DCL1-YFP expression. Based on its positive regulatory role on MIR transcription and its interaction with multiple miRNA processing factors, we propose that NOT2 coordinates the recruitment of DCL1 bodies onto pri-miRNAs subsequent to MIR transcription; loss of NOT2 probably leads to the mislocalization of its direct interaction partner DCL1 but not HYL1, which does not interact with NOT2 directly.
Figure 5.
Localization of DCL1-YFP, but Not HYL1-YFP, Was Affected in not2a-1 not2b-1.
(A) Representative confocal images of transgenic DCL1-YFP fluorescence in the wild type (Col) and not2a-1 not2b-1 (not2a-1 2b-1). Root cells from the meristematic zone were analyzed. Red arrows indicate the DCL1 bodies. Bars = 10 µm.
(B) The percentage distribution of DCL1-YFP speckle numbers per cell in the wild type (Col) and not2a-1 not2b-1 (not2a-1 2b-1). The x axis represents the number of DCL1-YFP speckles per cell, and the y axis represents the percentage of cells with the corresponding DCL1-YFP speckle numbers. “N” represents the numbers of analyzed root cells. *P < 0.05, which was calculated by Wilcoxon-Matt-Whitney test.
(C) qRT-PCR analysis of DCL1 expression level in DCL1-YFP transgenic plants. Data are given as means and sd of at least three independent biological replicates. *P < 0.05 by Student’s t test.
(D) Representative confocal images of transgenic HYL1-YFP fluorescence in the wild type (Col) and not2a-1 not2b-1 (not2a-1 2b-1). Root cells from the meristematic zone were analyzed. Red arrows indicate the HYL1 bodies. Bars = 10 µm.
(E) The percentage distribution of HYL1-YFP speckle numbers per cell in the wild type (Col) and not2a-1 not2b-1 (not2a-1 2b-1). The x axis represents the number of HYL1-YFP speckles per cell, and the y axis represents the percentage of cells with the corresponding HYL1-YFP speckle numbers. “N” represents the numbers of analyzed root cells.
(F) qRT-PCR analysis of HYL1 expression in HYL1-YFP transgenic plants. Data are given as means sd of at least three independent biological replicates. *P < 0.05 by Student’s t test.
NOT2 Proteins Act as General Factors to Affect Transcription
The association between NOT2 and Pol II raised the question of whether NOT2s affect pri-miRNAs specifically or act as general factors in transcription. To address this question, we performed high-throughput RNA sequencing (RNA-seq) using RNA samples from not2a-1 not2b-1 and wild-type seedlings. The data quality is summarized in Supplemental Table 3 online. Compared with wild-type Columbia (Col), several MIR genes were significantly reduced, which is consistent with our quantitative RT-PCR (qRT-PCR) analysis (see Supplemental Figure 5A online; Figure 3D). We also found 634 downregulated and 366 upregulated genes in not2a-1 not2b-1, respectively (P < 0.01, fold change > 2) (see Supplemental Data Set 1 and Supplemental Figure 5B online). The expression levels of some downregulated genes were validated in not2a-1 not2b-1 mutants and RNAi lines (see Supplemental Figure 5C online). We found that the expression of 14 selected genes (with low P value and high expression) was significantly reduced in the mutant or RNAi lines, indicating that NOT2 might be a general factor associated with Pol II, promoting transcription at both MIR and protein coding genes.
DISCUSSION
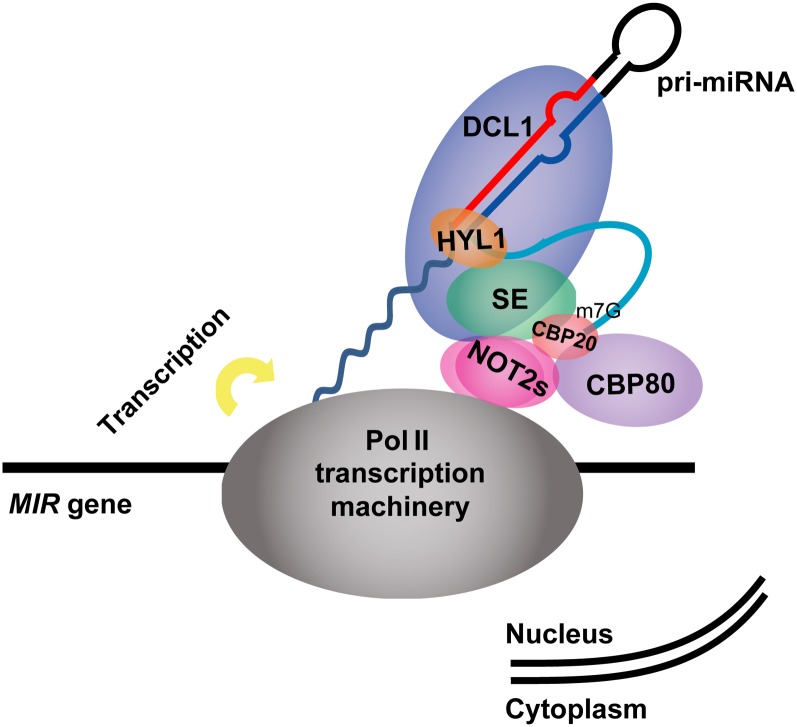
In this study, the evolutionarily conserved NOT2 proteins were shown to bind DCL1 in both the monocot rice and the dicot Arabidopsis. Inactivation of NOT2 in rice caused seedling death. In Arabidopsis, the two NOT2 paralogs NOT2a and NOT2b were found to form homo- or heterodimers. Double mutants of strong alleles of not2a and not2b displayed defects in male gametogenesis, indicating an essential role of NOT2 proteins in plant development; double mutants of weak alleles of not2a not2b phenocopied mutants in genes in the miRNA biogenesis pathway, with decreased levels of mature miRNAs. Unlike most miRNA biogenesis mutants, not2a-1 not2b-1 double mutants exhibited a decrease in pri-miRNA levels, which may be mainly attributed to a general role of NOT2 in transcription. Such a role is supported by the interaction between NOT2 and Pol II and the reduction of mRNA levels in protein coding genes in the not2a-1 not2b-1 double mutant. Besides DCL1 and Pol II, our results also revealed that NOT2 associated with the pri-miRNA processing factors CBP20, CBP80, and SE. In addition, we observed that the DCL1-YFP but not the HYL1-YFP speckle number in each cell significantly increased in not2a-1 not2b-1, which may represent mislocalization of DCL1-YFP, the NOT2-interacting protein. Thus, we propose that NOT2, in addition to serving as a general factor to promote transcription, plays a role in pri-miRNA processing by coupling it to MIR transcription (Figure 6).
Figure 6.
Summary of NOT2 Functions in MiRNA Biogenesis.
NOT2 promotes MIR expression at the transcriptional level. After transcription, pri-miRNAs form a stem-loop structure, which probably acts as a platform for pri-miRNA posttranscription processing. NOT2 proteins interact with Pol II and multiple miRNA biogenesis factors, including DCL1, CBP20, CBP80, and SE. Therefore, NOT2s may act as a scaffold to link MIR gene transcription and posttranscriptional processing.
NOT2 Proteins Affect Pol II–Dependent Transcription in Arabidopsis
In yeast, recent evidence shows that CCR4-NOT acts in mRNA metabolism (Collart and Panasenko, 2012), in which NOT2 binds Pol II directly and promotes transcriptional elongation of protein coding genes (Kruk et al., 2011). We show that pri-miRNA levels were reduced and transcript levels of hundreds of protein-coding genes were also affected in the not2a not2b double mutant, suggesting that NOT2 proteins are required for proper levels of Pol II–dependent protein-coding and noncoding gene transcription. The changes in the expression of these genes could be attributed to the interaction between NOT2 and Pol II, although they could be indirect effects caused by the pleiotropic developmental defects of the not2a not2b double mutant.
NOT2 Links Pri-miRNA Transcription and Processing Factor Recruitment in Arabidopsis
At miRNA loci, besides interacting with Pol II and thus regulating MIR gene transcription, NOT2 proteins also interact with the processing factors DCL1, SE, and CBPs. This suggests that NOT2s may act as a scaffold to link MIR gene transcription and DCL1 recruitment, thus making pri-miRNA production and subsequent posttranscription processing more coordinated and efficient. This hypothesis is supported by the effect on DCL1 localization of NOT2 impairment. D-bodies are highly dynamic structures visualized as foci within the nuclear compartment, where pri-miRNA processing and/or storage/assembly occur (Fang and Spector, 2007). In wild-type plants, DCL1 is enriched in the D-body and 79% of cells harbor two or fewer D-bodies per cell (Fang and Spector, 2007). The number of D-bodies significantly increased to nearly three to four per cell in not2a-1 not2b-1, which may reflect mislocalization of DCL1 and impairment of DCL1 recognition of pri-miRNAs. Consistent with this notion, HYL1, another major component in D-bodies, was not significantly affected by loss of NOT2 function. The different effects between these two could be due to the fact that DCL1 interacts with NOT2 directly, whereas HYL1 does not. It is still unclear whether the DCL1-containing foci in not2a-1 not2b-1 are fully functional for miRNA processing or whether the increased number of DCL1-containing speckles compensates for impaired processing function.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used in this study were in the Col ecotype. Plants were grown under long photoperiod conditions (16 h light/8 h dark) at 23°C. Seedlings were grown on Murashige and Skoog plates containing 3% Suc and 0.8% agar. The mutants of not2a-1 (SALK_062057), not2b-1 (SALK_058645), and not2a-2 (GABI_104B08) were obtained from SALK (ABRC) and the European Arabidopsis Stock Centre collections, respectively. The mutant cbp80 (SALK_016753) was described previously (Deng et al., 2010). Rice (Oryza sativa) transformation and regeneration were performed as previously published (Liu et al., 2007). The DCL1-YFP or HYL1-YFP/not2a-1 not2b-1 lines were obtained by crossing DCL1-YFP or HYL1-YFP/Col (Fang and Spector, 2007) with not2a-1 not2b-1 plants. In the F2 population, plants with the genotype NOT2a+/+ NOT2+/+ and NOT2a−/− NOT2−/− were selected for further observation. pMIR172a:GUS and pPI:GUS (Yu et al., 2008) were introduced into not2a-1 nto2b-1 by the same approach.
Yeast Two-Hybrid, Nickel Pulldown, and Immunoblotting
For yeast two-hybrid assays, the full-length or partial coding regions of Os-DCL1 PAZ, DCL1 PAZ, NOT2a, NOT2b, CBP20, and DDL were PCR amplified using the primers listed in Supplemental Table 4 online. The fragments were fused in frame with the GAL4 DNA binding domain (pGBKT7/BD/XF117) or activation domain (pGADT7/AD/XF118). All the recombinant plasmid pairs were cotransformed into the yeast strain AH109. Yeast transformation was performed following the manufacturer’s handbook (Clontech Yeast Protocols Handbook). The cotransformed yeast clones were first grown on SD/-Leu/-Trp medium and subsequently plated on SD/-Ade/-His/-Leu/-Trp medium containing 40 µg/mL X-α-Gal.
For pull-down assays, the coding sequences of NOT2a and NOT2b were inserted in frame into the expression vector pMAL-C2-MBP (XF510) as the bait, and the DCL1 PAZ, CBP20, CBP80, DDL, and Pol II NRPB1 CTD coding sequences were cloned into the expression vector pGEX4T-1(XF156) or pMAL-C2-GST (XF760) as prey proteins. pMAL-C2-MBP and pMAL-C2-GST are a family of compatible ligation-independent cloning vectors, which are suitable for construction of recombinant protein expression vectors independent of ligation (Eschenfeldt et al., 2009). The primer sequences are listed in Supplemental Table 4 online. All the MBP-His–tagged or GST-tagged recombinant proteins were affinity purified from Escherichia coli BL21 codon-plus RIL strain (BL21 CP, Stratagene) with extra copies of E. coli argU, ileY, and LeuW tRNA genes. Nickel pull-down assays were performed as previously described (Yan et al., 2007), except that His-tag Dynabeads (101.04D; Invitrogen) were used. Briefly, the pellet of BL21 with expressed proteins was resuspended in 1× His binding buffer (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 0.01% Tween 20). Then, the culture lysate was incubated with 50 μL (2 mg) Dynabeads on a rotator for 5 to 10 min at room temperature. The beads with immobilized bait proteins were washed thoroughly four times with 300 μL binding buffer and resuspended with 1× pull-down buffer (3.25 mM sodium phosphate, pH 7.4, 70 mM NaCl, and 0.01% Tween 20). GST-tagged prey proteins were purified by glutathione sepharose beads and eluted with buffer containing 10 mM reduced glutathione. Prior to incubating with prey proteins, equal amounts of baits were blocked with 10% BSA in 1× His binding buffer. Equal amounts (2 µg) of the bait and prey recombinant proteins were incubated in 1× pull-down buffer for 1 h at 4°C, and after extensive washing (1× binding buffer with 400 mM NaCl and 0.5% ICA-630), the bound GST fusion proteins were separated by 10% SDS-PAGE and subjected to immunoblotting assay using α-GST primary antibody and subsequently the horseradish peroxidase–conjugated mouse secondary antibody (31430; Pierce).
For detection of the interaction between NOT2 and Pol II, wild-type Arabidopsis total crude extract was used as prey. Proteins were separated with NuPAGE Novex 3 to 8% Tris-Acetate Gel (EA0375BOX; Invitrogen), then immunoblotting analysis was performed using primary antibodies against NRPB1 (ab5408; Abcam) and the horseradish peroxidase–conjugated anti-mouse secondary antibody (31430; Pierce).
Histochemical GUS Staining
NOT2a/b genomic DNA regions were PCR amplified and cloned into the Gateway pENTR/D-TOPO vector (11791-020 and 45-0218; Life Technologies) according to the manufacturer’s instructions. The resulting plasmids were recombined with pGWB3 (XF1379) (for GUS fusion of NOT2a and NOT2b). Histochemical GUS staining was performed according to the standard procedure (Jefferson et al., 1987) with a modified buffer, which contains 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid cyclohexylammonium salt, 50 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 2 mM potassium ferrocyanide, 2 mM potassium ferricyanide, and 10 mM EDTA. Plant tissues were incubated in the buffer at 37°C in the dark. Prior to observation, tissues were cleared with ethanol.
RNA Sequencing and Data Analysis
Total RNAs were extracted from 12-d-old seedlings of the wild type and not2a-1 not2b-1 using TRIzol reagent (DP405-02; Tiangen). Polyadenylated RNAs were isolated using a Dynabeads mRNA purification kit (610-06; Invitrogen). RNA-seq libraries were prepared using the Illumina Directional mRNA-Seq library prep v1.5 protocol and sequenced on an Illumina GAII to generate high-quality single-end reads of 50 nucleotides in length. The raw reads with strand direction were aligned to the TAIR10 genome using TopHat (Trapnell et al., 2009), allowing up to six mismatches. Only uniquely mapped reads were used for subsequent analysis. The gene expression levels were measured in reads per kilobase of exon model per million mapped reads (Mortazavi et al., 2008). The differentially expressed genes were identified using DEGseq with parameter as twofold change and P < 0.01 (Wang et al., 2010).
Quantitative PCR and Small RNA Gel Blotting
Total RNAs were extracted from 12-d-old seedling plants with TRIzol reagent (Tiangen), and the first-strand cDNA was reversed transcribed with anchored oligo(dT) using GoldScript (C81401190; Invitrogen) according to the manufacturer’s instructions. The qRT-PCR was performed in the CFX96 real-time system (Bio-Rad). Primers for detecting pri-miRNAs were used as previously described (Gregory et al., 2008). Primer information is listed in Supplemental Table 4 online. Student’s t test significance was calculated as previously published (Niu et al., 2012), except for the statistical analysis in Figures 5B and 5E, in which the P value was calculated by Wilcoxon-Matt-Whitney test.
Small RNAs were enriched by 8 M LiCl from 20 to 30 μg total RNAs for at least 2 h at 4°C. RNA hybridization was performed as previously described (Liu et al., 2005) with minor modifications. The enriched small RNAs were size separated by 15% SDS-PAGE containing 7 M Urea under denaturing conditions and transferred electrophoretically to Hybond-NX nylon membrane (RPN303T; GE Healthcare), then subjected to chemical cross-linking as previously described (Pall and Hamilton, 2008). Antisense oligonucleotides for specific miRNAs were used as probes for detecting miRNAs, which were 5′-end labeled ([γ-32P]ATP) by T4 polynucleotide kinase (M0201; New England Biolabs). The probe sequences are listed in Supplemental Table 5 online. Radioactive signals were quantified with ImageQuant 5.2, and the relative quantitative values were normalized to U6.
Pollen Observation Using Alexander’s and DAPI Staining
For pollen analysis, Alexander’s stain was used as previously described (Alexander, 1969). Pollen DAPI staining was performed using DAPI solution (0.1 M sodium phosphate, pH 7.0, 1 mM EDTA, 0.1% Triton X-100, and 0.5 mg/mL DAPI). Alexander’s staining and DAPI fluorescence were observed on a microscope equipped with a charge-couple device camera (Olympus DP70).
BiFC Assays and Subcellular Localization
BiFC assays were performed as described (Fang and Spector, 2007). The coding regions of NOT2b, DDL, CBP20, and CBP80 were inserted into pCAMBIA1300-YFPN (XF1698) or pCAMBIA1300-YFPC (XF1699), respectively. The recombinant constructs were transformed into the Agrobacterium tumefaciens strain EHA105 and the Agrobacterium cultures were injected into Nicotiana benthamiana leaves. The transfected tobacco leaves were grown at 25°C for 2 to 3 d before observation. The tobacco epidermal cells were then observed with a Leica TCS SP5 confocal laser scanning microscope.
For detection of NOT2b subcellular localization, the coding sequence of NOT2b was engineered into pCAMBIA1300-RFP (XF0954). The detailed primer sequences are listed in Supplemental Table 4 online. Agrobacterium strains containing NOT2b-RFP and DCL1-YFP were used to cotransfect N. benthamiana. The visualization of DCL1-YFP in Arabidopsis was observed using the Zeiss LSM 710 NLO confocal laser scanning microscope.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Os02g54210 (Os-NOT2), At1g07705 (NOT2a), At5g59710 (NOT2b), At3g20550 (DDL), At1g09700 (HYL1), At2g27100 (SE), At2g13540 (ABH1/CBP80), At5g44200 (CBP20), At1g01040 (DCL1), and At 4g35800 (Pol II NRPB1). The RNA-seq data were deposited in the Gene Expression Omnibus database under accession number GSE42901.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification and Functional Analysis of Os-NOT2 in Rice.
Supplemental Figure 2. Tissue-Specific Expression of NOT2a and NOT2b.
Supplemental Figure 3. pMIR172a:GUS and pPI:GUS Expression.
Supplemental Figure 4. CBP20 Interacts with SE in Vivo and in Vitro.
Supplemental Figure 5. Analysis of Differentially Expressed Genes and qRT-PCR Validation.
Supplemental Table 1. Statistical Analysis of Aborted Ovules per Silique in NOT2a-2+/− 2b-1−/−.
Supplemental Table 2. Statistical Analysis of Abnormal Pollen in NOT2a-2+/− 2b-1−/−.
Supplemental Table 3. Summary of High-Throughput RNA Sequencing Analysis for Col and not2a-1 not2b-1.
Supplemental Table 4. Primer Sequences.
Supplemental Table 5. Probe Sequences for RNA Gel Blots.
Supplemental Data Set 1. List of Downregulated and Upregulated Genes.
Acknowledgments
We thank Dr. Bairu Zhang (Institute of Genetics and Developmental Biology) for taking images on the Zeiss LSM 710 NLO confocal laser scanning microscope. We thank Miltos Tsiantis (University of Oxford) for providing se-1 seeds; Dr. Yuke He (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing GST-HYL1 and GST-SE plasmids; Dr. Yuda Fang (Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for providing DCL1-YFP, HYL1-YFP, SE-YFPN/C, and HYL1-YFPN/C plasmids; and the Arabidopsis Biological Resource Center (ABRC) and the European Arabidopsis Stock Centre for providing SALK and GABI T-DNA insertion lines. This work was supported by the Transgenic Project (Grant 2011ZX08009-001 to X.S.), the National Basic Research Program of China (Grant 2009CB941500 to X.F.C.), and by the National Natural Science Foundation of China (Grant 90919033 to X.C. and 31123007 and 31210103901 to X.F.C.).
AUTHOR CONTRIBUTIONS
L.W., X.M.C., and X.F.C. designed the research. L.W. performed the research. X.L., X.S., S.C., X.C., and C.C. provided technical assistance. L.G. performed large-scale data analysis. X.M.C. and X.F.C. analyzed data. LW., X.S., and X.F.C. wrote the article.
Glossary
- miRNA
microRNA
- pri-miRNA
primary miRNA
- Pol II
polymerase II
- PAZ
Piwi/Ago/Zwille
- RNAi
RNA interference
- GST
glutathione S-transferase
- YFP
yellow fluorescent protein
- RFP
red fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- GUS
β-glucuronidase
- CBC
CBP complex
- RNA-seq
RNA sequencing
- Col
Columbia
- qRT-PCR
quantitative RT-PCR
- BiFC
bimolecular fluorescence complementation
References
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Anand A., Krichevsky A., Schornack S., Lahaye T., Tzfira T., Tang Y., Citovsky V., Mysore K.S. (2007). Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Borchert G.M., Lanier W., Davidson B.L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13: 1097–1101 [DOI] [PubMed] [Google Scholar]
- Chen X. (2005). MicroRNA biogenesis and function in plants. FEBS Lett. 579: 5923–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.M., Riechmann J.L., Jia D.X., Meyerowitz E. (2000). Minimal regions in the Arabidopsis PISTILLATA promoter responsive to the APETALA3/PISTILLATA feed back control do not contain a CArG box. Sex. Plant Reprod. 13: 85–94 [Google Scholar]
- Collart M.A., Panasenko O.O. (2012). The Ccr4—Not complex. Gene 492: 42–53 [DOI] [PubMed] [Google Scholar]
- Collart M.A., Struhl K. (1994). NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8: 525–537 [DOI] [PubMed] [Google Scholar]
- Collart M.A., Timmers H.T. (2004). The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Daugeron M.C., Mauxion F., Séraphin B. (2001). The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu L., Liu C., Lu T., Lu F., Lu Z., Cui P., Pei Y., Wang B., Hu S., Cao X. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C.L., Chen J. (2003). The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Eschenfeldt W.H., Lucy S., Millard C.S., Joachimiak A., Mark I.D. (2009). A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Sonenberg N., Filipowicz W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Fang Y., Spector D.L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866 [DOI] [PubMed] [Google Scholar]
- Grigg S.P., Canales C., Hay A., Tsiantis M. (2005). SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437: 1022–1026 [DOI] [PubMed] [Google Scholar]
- Han M.H., Goud S., Song L., Fedoroff N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Takahashi A., Morita M., Suzuki T., Yamamoto T. (2011). The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell 2: 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. (1994). A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Bartel D.P., Bartel B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Kidner C.A., Martienssen R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8: 38–44 [DOI] [PubMed] [Google Scholar]
- Kim V.N. (2005). MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Chen X. (2011). The plant Mediator and its role in noncoding RNA production. Front. Biol. 6: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk J.A., Dutta A., Fu J., Gilmour D.S., Reese J.C. (2011). The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Takashi Y., Watanabe Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12: 206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Watanabe Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Sachsenberg T., Zeller G., Busch W., Lohmann J.U., Rätsch G., Weigel D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 8795–8800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854 [DOI] [PubMed] [Google Scholar]
- Li J., Terzaghi W., Deng X.W. (2012). Genomic basis for light control of plant development. Protein Cell 3: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li P., Li X., Liu C., Cao S., Chu C., Cao X. (2005). Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol. 139: 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Qu L.J. (2008). Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Mol. Plant 1: 564–574 [DOI] [PubMed] [Google Scholar]
- Liu Q., Shi L., Fang Y. (2012). Dicing bodies. Plant Physiol. 158: 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bai X., Wang X., Chu C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Lu C., Fedoroff N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12: 2351–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae I.J., Zhou K., Doudna J.A. (2007). Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 14: 934–940 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004). Control of male gametophyte development. Plant Cell 16 (suppl.): S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Niu L., Lu F., Zhao T., Liu C., Cao X. (2012). The enzymatic activity of Arabidopsis protein arginine methyltransferase 10 is essential for flowering time regulation. Protein Cell 3: 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G.S., Hamilton A.J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Song H. (2004). The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Ren G., Xie M., Dou Y., Zhang S., Zhang C., Yu B. (2012). Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 12817–12821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Han M.H., Lesicka J., Fedoroff N. (2007). Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc. Natl. Acad. Sci. USA 104: 5437–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Li Y.F., Jagadeeswaran G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9): 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. (2001). The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Vazquez F., Gasciolli V., Crété P., Vaucheret H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Zhang X. (2010). DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26: 136–138 [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Khanna K., Ruan S. (2010). Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 21: 790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Zhang Y., Niu L., Yuan Y., Cao X. (2007). Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem. J. 408: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Liu Z., Lu F., Dong A., Huang H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47: 841–850 [DOI] [PubMed] [Google Scholar]
- Yu B., Bi L., Zheng B., Ji L., Chevalier D., Agarwal M., Ramachandran V., Li W., Lagrange T., Walker J.C., Chen X. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. USA 105: 10073–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwartjes C.G., Jayne S., van den Berg D.L., Timmers H.T. (2004). Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J. Biol. Chem. 279: 10848–10854 [DOI] [PubMed] [Google Scholar]