In contrast with animals, little is known about the regulation of HMGR, the rate-limiting enzyme of isoprenoid biosynthesis, in plants. Through the identification of second-site suppressors of the Arabidopsis dry2/sqe1-5 mutant, we found that the putative E3 ubiquitin ligase SUD1, likely involved in endoplasmic reticulum–associated degradation, is a regulator of HMGR activity.
Abstract
The 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) enzyme catalyzes the major rate-limiting step of the mevalonic acid (MVA) pathway from which sterols and other isoprenoids are synthesized. In contrast with our extensive knowledge of the regulation of HMGR in yeast and animals, little is known about this process in plants. To identify regulatory components of the MVA pathway in plants, we performed a genetic screen for second-site suppressor mutations of the Arabidopsis thaliana highly drought-sensitive drought hypersensitive2 (dry2) mutant that shows decreased squalene epoxidase activity. We show that mutations in SUPPRESSOR OF DRY2 DEFECTS1 (SUD1) gene recover most developmental defects in dry2 through changes in HMGR activity. SUD1 encodes a putative E3 ubiquitin ligase that shows sequence and structural similarity to yeast Degradation of α factor (Doα10) and human TEB4, components of the endoplasmic reticulum–associated degradation C (ERAD-C) pathway. While in yeast and animals, the alternative ERAD-L/ERAD-M pathway regulates HMGR activity by controlling protein stability, SUD1 regulates HMGR activity without apparent changes in protein content. These results highlight similarities, as well as important mechanistic differences, among the components involved in HMGR regulation in plants, yeast, and animals.
INTRODUCTION
For sessile organisms such as plants, metabolic plasticity is essential to survive in their changing environments (Nicotra et al., 2010). A good example of this plasticity is the thousands of isoprenoid compounds and derivatives that higher plants synthesize from the five-carbon building units isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (Bouvier et al., 2005). Plants synthesize IPP and dimethylallyl diphosphate by two independent pathways: the mevalonic acid (MVA) pathway, which produces cytosolic IPP (McGarvey and Croteau, 1995; Newman and Chappell, 1999); and the methylerythritol phosphate pathway, which is localized in the plastids (Eisenreich et al., 2001; Rodríguez-Concepción and Boronat, 2002). In higher plants, isoprenoids carry out numerous essential roles in developmental processes, including respiration, photosynthesis, growth, and reproduction, as well as adaptation to environmental challenges and involvement in plant defense mechanisms against different types of organisms (Tholl and Lee, 2011; Hemmerlin et al., 2012).
The main MVA-derived isoprenoid end products in plants are sterols, which are integral components of the membrane and are essential for plant growth and developmental processes. Other important MVA products are the steroid hormones brassinosteroids, dolichols, which are involved in protein glycosylation, and the prenyl groups used for protein prenylation and cytokinin biosynthesis (Benveniste, 2004; Phillips et al., 2006; Schaller, 2010). A number of studies over the years have shown the importance of correct sterol composition in plants because of their roles in embryonic pattern formation (Jang et al., 2000), cell division, elongation and polarity (Schrick et al., 2000; Willemsen et al., 2003; Men et al., 2008), vascular patterning (Carland et al., 2010), cellulose accumulation (Schrick et al., 2004), reactive oxygen species (ROS) production (Posé et al., 2009), and normal microRNA function (Brodersen et al., 2012). Still, little is known about the mechanisms and downstream targets by which isoprenoids in general, and sterols in particular, influence these processes (Boutté and Grebe, 2009; Clouse, 2002).
The enzyme 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) is considered the major rate-limiting enzyme controlling the metabolic flux in the early steps of the MVA pathway (Hemmerlin et al., 2012). The genome of Arabidopsis thaliana contains two differentially expressed HMGR genes, HMG1 and HMG2 (Enjuto et al., 1994), encoding three HMGR isoforms: HMGR1S (short isoform), HMGR1L (long isoform), and HMGR2. HMGR1S and HMGR1L are both encoded by the HMG1 gene and are identical in sequence, except for an N-terminal extension of 50 amino acid residues in HMGR1L (Lumbreras et al., 1995). HMGR1S has been proposed to have a housekeeping role, whereas HMGR1L and HMGR2 have a more specialized function, which might be required in particular cell types or at specific developmental stages (Suzuki et al., 2004, 2009). All plant HMGR variants are targeted to the endoplasmic reticulum (ER) and have the same topology in the membrane (Campos and Boronat, 1995). The diverged N-terminal region and the conserved catalytic domain are located in the cytosol, whereas only a short stretch of amino acids connecting the two transmembrane (TM) segments is in the ER lumen. Plant HMGR is modulated by a variety of developmental and environmental signals, and it has been proposed that major changes in HMGR activity are determined at the transcriptional level, whereas posttranscriptional regulation allows a finer and faster adjustment (Hemmerlin et al., 2012). In fact, evidence of posttranslational regulation of HMGR in Arabidopsis plants with enhanced or depleted flux through the sterol biosynthetic pathway has been obtained (Nieto et al., 2009). Similarly, inhibition of squalene epoxidase (SQE) activity in tobacco (Nicotiana tabacum) Bright Yellow-2 cells using terbinafine also triggers an increase in HMGR activity, even though it does not induce changes in the HMGR transcript levels (Wentzinger et al., 2002). Mechanistically, a protein phosphatase 2A (PP2A) has been recently identified as a negative regulator of Arabidopsis HMGR activity and protein levels (Leivar et al., 2011). Still, proteins involved in the posttranscriptional regulation of plant HMGR are mostly uncharacterized.
In plants, SQEs catalyze the conversion of squalene, the first committed precursor of essential MVA-derived isoprenoids, to 2,3-oxidosqualene (Rasbery et al., 2007; Posé et al., 2009; Schaller, 2010). The Arabidopsis drought hypersensitive2 (dry2/sqe1-5) mutant was identified by its extreme hypersensitivity to drought stress, altered stomatal responses, and root defects. Chemical analysis indicated that the dry2/sqe1-5 mutant has altered sterol composition in roots but wild-type sterol composition in shoots, indicating an essential role for SQE1 in root sterol biosynthesis. Importantly, the stomatal and root defects of the dry2/sqe1-5 mutant are associated with altered production of ROS, establishing a previously unknown link between the MVA pathway and ROS (Posé et al., 2009).
The dry2/sqe1-5 allele contains a point mutation in the 4th exon that produces a substitution of a conserved Gly by an Arg, resulting in reduced epoxidase activity (Posé et al., 2009). In contrast with the null alleles of SQE1 that are sterile, dry2/sqe1-5 plants are fertile, and this characteristic enables its use for genetic analyses. In this work, we used the hypomorphic dry2/sqe1-5 mutant allele to perform a genetic screen for second-site suppressor mutations to identify new genetic components regulating the MVA pathway. Several mutants (named sud for suppressors of dry2 defects) that reversed most of the dry2/sqe1-5 developmental phenotypes were isolated. As a result, we identified a regulatory element, SUD1, which encodes a protein with sequence and structural homology to the E3 ubiquitin ligases Degradation of α factor (Doa10) in yeast and TEB4 in humans (i.e., proteins that are involved in the endoplasmic reticulum–associated protein degradation [ERAD] pathway). Our results indicate that SUD1 functions as a positive posttranscriptional regulator of HMGR activity in Arabidopsis.
RESULTS
Phenotypic Characterization of the dry2 Suppressors
To identify undescribed elements that regulate the isoprenoid biosynthetic pathway, we performed a suppressor screening based on the recovery of the extreme drought hypersensitive phenotype of the previously characterized dry2 mutant affected in SQE1 (Posé et al., 2009; see Supplemental Figure 1 online). As a result, four independent mutants were selected and named sud, which maintained the reversion of the dry2 drought hypersensitive phenotype across multiple generations. Identification of the gene affected in the four suppressors indicated that the mutations were allelic (see below), and the mutants were subsequently designated dry2/sud1-1 to dry2/sud1-4.
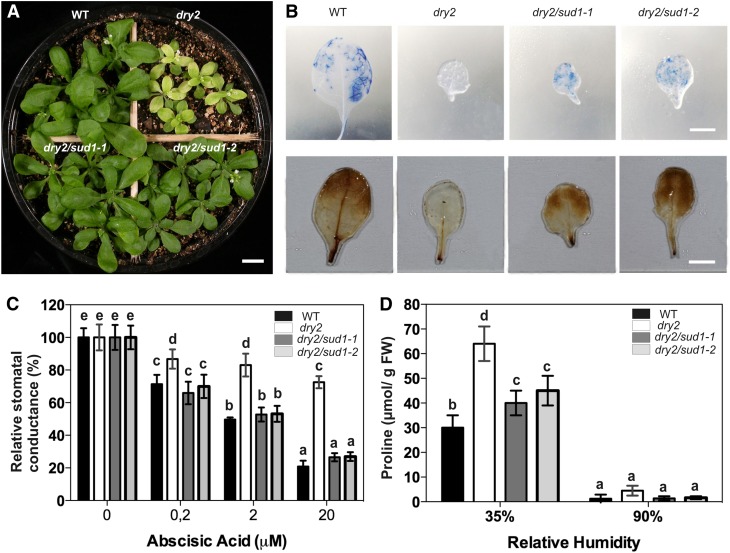
The recovery of the multiple dry2 phenotypic defects was further analyzed using the dry2/sud1-1 and dry2/sud1-2 alleles. The dry2/sud1-1 and dry2/sud1-2 mutants showed a restoration of leaf size and color observed in dry2 and rendered the mutant shoots undistinguishable from those of the wild type (Figure 1A). This phenotypic recovery of the shoots in the suppressors correlated with the reestablishment of hydrogen peroxide and O2− (superoxide) accumulation to wild-type levels (Figure 1B). Since the identification of the suppressors was based on the restoration of the dry2 extreme drought hypersensitivity, we expected that the defective abscisic acid (ABA) stomatal responses observed in dry2 would also be restored in the suppressors. As shown in Figure 1C, exogenous application of 20 μM ABA only caused an ∼20% reduction in the stomatal conductance of dry2 compared with the ∼80% reduction that occurred in wild-type, dry2/sud1-1, and dry2/sud1-2 plants. In addition, the Pro content in dry2/sud1-1 and dry2/sud1-2 was more similar to that of wild-type plants (Figure 1D). These results confirm that the recovery of the dry2 shoot phenotypes was associated with a restoration of the water relations in the suppressors.
Figure 1.
dry2/sud1-1 and dry2/sud1-2 Suppress the Shoot Defects of dry2.
(A) Shoot phenotype of 3-week-old wild-type (WT), dry2, dry2/sud1-1, and dry2/sud1-2 plants grown under standard long-day conditions. Bar = 0.5 cm.
(B) Accumulation of O2− (top panel) and hydrogen peroxide (bottom panel) in 3-week-old leaves of the wild type, dry2, dry2/sud1-1, and dry2/sud1-2. Plants were stained with nitroblue tetrazolium (top panel) and DAB (bottom panel). Compared with the wild type, dry2/sud1-1, and dry2/sud1-2, dry2 accumulates very low levels of O2− and hydrogen peroxide. Bars = 0.5 cm.
(C) Stomatal conductance of the wild type, dry2, dry2/sud1-1, and dry2/sud1-2. The measurements were made 4 h after spraying the indicated ABA concentrations. The suppressors show wild-type stomatal responses to exogenous ABA compared with dry2.
(D) Pro content in wild-type, dry2, dry2/sud1-1, and dry2/sud1-2 plants grown at 35 and 90% relative humidity. FW, fresh weight.
(C) and (D) Mean ± sd, n = 9; values with the same letter are not significantly different at P < 0.05. The experiments were repeated at least three times with similar results.
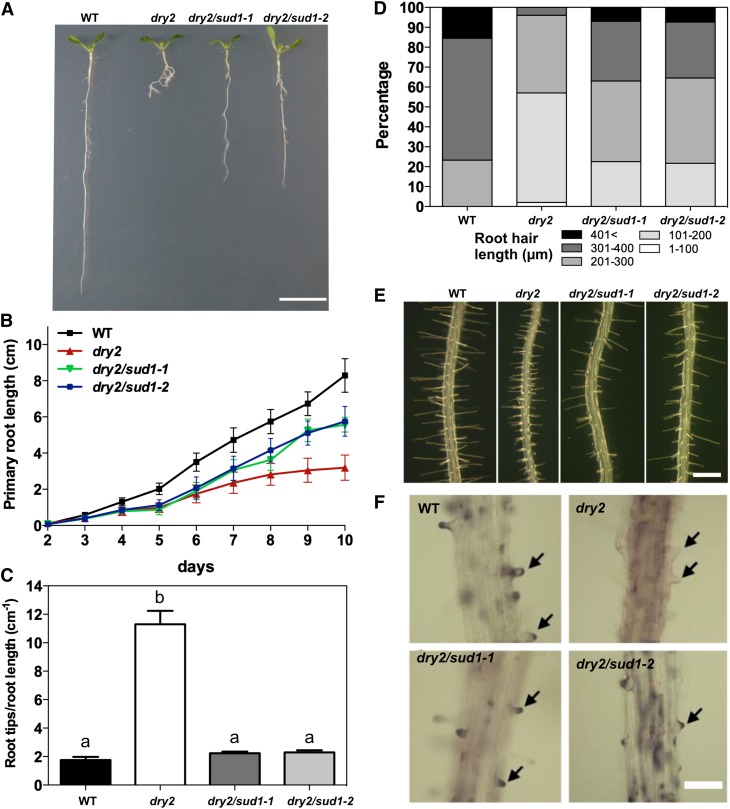
The primary root length of the dry2/sud1-1 and dry2/sud1-2 alleles was double than that of dry2, reaching ∼70% of the wild-type seedlings (Figures 2A and 2B). dry2/sud1-1 and dry2/sud1-2 also exhibited a decreased number of lateral roots compared with dry2 (Figures 2A and 2C). The striking defects in root hair length and morphology observed in dry2 (Posé et al., 2009) were also substantially restored in the suppressors (Figures 2D and 2E). Consistent with the rescue of the root hair growth defects, dry2/sud1-1 and dry2/sud1-2 showed wild-type ROS production at the bulge of the root hair tip (Figure 2F), in contrast with the aberrant dry2 ROS production caused by an ectopic localization of the NADPH oxidase C (AtrbohC) (Posé et al., 2009).
Figure 2.
The dry2/sud1 Mutants Partially Suppress the Root Elongation and Root Hair Growth Defects of the dry2 Mutant.
(A) Root developmental phenotypes of 10-d-old wild-type (WT), dry2, dry2/sud1-1, and dry2/sud1-2 mutant seedlings. Bar = 1 cm.
(B) Primary root growth during a 10-d period for the wild type, dry2, dry2/sud1-1, and dry2/sud1-2. Mean ± sd, n ≥ 30 roots per genotype.
(C) Root branching index for 10-d-old wild type, dry2, dry2/sud1-1, and dry2/sud1-2. The index was determined by counting the number of lateral root tips per length unit (cm) of primary root. Mean ± sd, n = 30 roots counted per genotype. Values with the same letter are not significantly different at P < 0.05.
(D) Root hair length distribution in 5-d-old wild-type, dry2, dry2/sud1-1, and dry2/sud1-2 (n ≥ 300 root hairs counted in a total of 30 roots per genotype).
(E) Morphologic root hair phenotype in 5-d-old wild type, dry2, dry2/sud1-1, and dry2/sud1-2. Bar = 500 μm.
(F) ROS staining in 5-d-old roots using DAB. Hydrogen peroxide (arrows) is localized at the tips of wild-type, dry2/sud1-1, and dry2/sud1-2 root hairs, but not in dry2. Bar = 200 μm.
[See online article for color version of this figure.]
All Four dry2 Suppressors Harbor Mutations in the SUD1 Gene
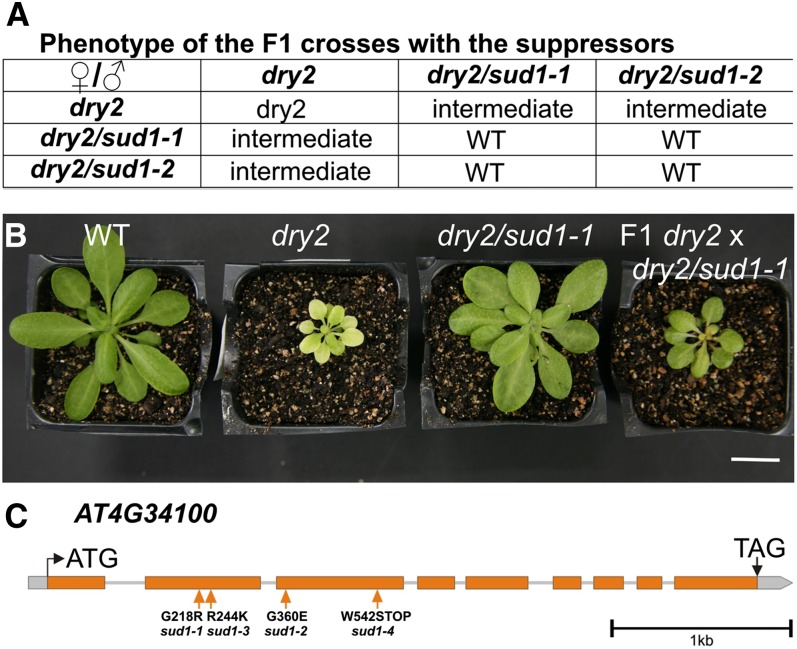
As a first step to determine the gene(s) affected by the sud mutations, we crossed dry2/sud1-1 and dry2/sud1-2 and performed an allelism test. As shown in Figure 3A, reciprocal crosses rendered progenies with wild-type phenotypes, suggesting that the sud1-1 and sud1-2 mutations were allelic. However, F1 plants from the backcross between the suppressors and dry2 showed an intermediate phenotype, indicating that the mutations were semidominant (Figures 3A and 3B). Based on these results, we could not directly infer whether the sud1-1 and sud1-2 mutations were allelic (Koornneef et al., 2006).
Figure 3.
Genetic Analyses of dry2/sud1-1 and dry2/sud1-2.
(A) Visual score of the phenotypes from the F1 of the corresponding crosses. WT, the wild type.
(B) Phenotypes of the wild type, dry2, dry2/sud1-1, and an F1 plant derived from a backcross between a dry2/sud1-1 and a dry2 plant. Bar = 2 cm.
(C) The four suppressor mutations are localized in the AT4G34100 gene. The intron/exon structure of the gene, the position of the mutations, and the resulting amino acids substitution of the four suppressors are depicted.
[See online article for color version of this figure.]
Next, we used a combination of map-based cloning and high-throughput sequencing to identify the sud1-1 mutation. For that purpose, the dry2 mutant allele (Landsberg erecta [Ler] ecotype) was crossed over seven generations into Columbia-0 (Col-0) ecotype to create an introgression line harboring the dry2 mutation in the (Col-0) background (dry2Col-0). Molecular markers demonstrated that dry2Col-0 was a near isogenic Col-0 line with the dry2 mutation, and this line displayed similar phenotypes as the original dry2 mutant (see Supplemental Figures 2A to 2C online). Since the dry2Col-0 line was a suitable parental line for map based cloning, an F2 population from the cross between dry2/sud1-1 and dry2Col-0 was generated. Fine-scale map-based cloning delimitated the region harboring the sud1-1 mutation in chromosome IV between the AT4G33970 and AT4G34250 loci (see Supplemental Figure 2D online). High-throughput sequencing and analysis of the region containing sud1-1 determined that the second-site mutation responsible for the dry2/sud1-1 suppression phenotype was a G-to-A substitution at nucleotide 652 relative to the ATG of the AT4G34100 gene (hereafter named SUD1). This nucleotide change caused a Gly218Arg substitution in the predicted SUD1 amino acid sequence (Figure 3C; see Supplemental Figure 2D online). Rough mapping of the other sud mutants using markers linked to sud1-1 indicated that all four sud mutations were located in the same region, and targeted sequencing of the SUD1 gene identified additional mutations on the three sud alleles. Thus, dry2/sud1-2 caused a Gly360Glu substitution, dry2/sud1-3 an Arg244Lys substitution, and dry2/sud1-4 a premature stop codon at position 542 in the predicted SUD1 protein (Figure 3C; see Supplemental Figure 2D online). The identification of four independent SUD1 mutant alleles in the suppressor screen demonstrates that mutations in the SUD1 gene are responsible for the phenotypic recovery of the dry2 defects.
SUD1 Is Homologous to E3 Ubiquitin Ligases from Yeast and Mammals Involved in ERAD
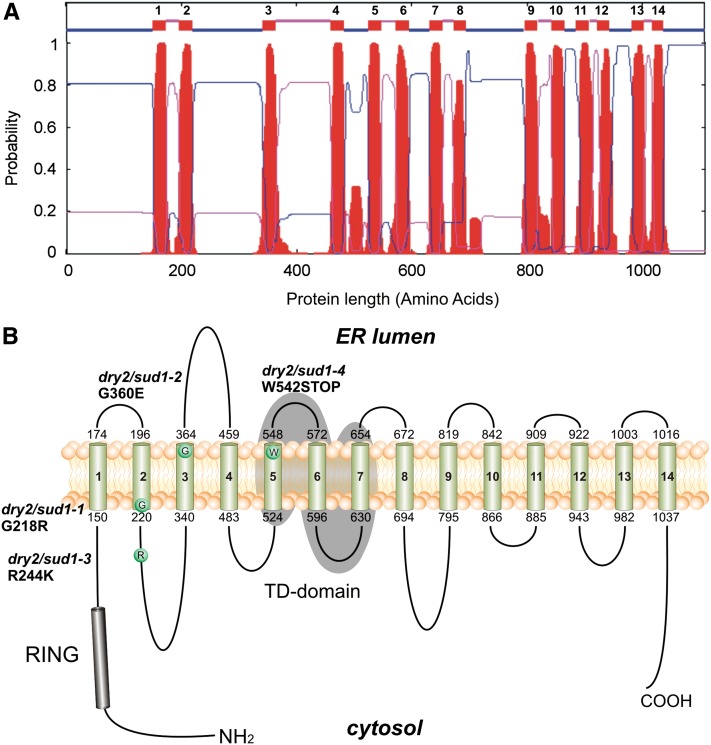
The Arabidopsis SUD1 locus AT4G34100 has been recently reported as ECERIFERUM9, a gene involved in cuticular wax biosynthesis (Lü et al., 2012), but little is known about the molecular mechanisms underlying SUD1 activity. SUD1 is predicted to encode a large protein of 1108 amino acids with a molecular mass of ∼123 kD. SUD1 contains a Really Interesting New Gene-variant (RING-v) domain (C4HC3 RING-finger domain) near the N terminus (Stone et al., 2005; Lü et al., 2012) and 14 putative TM domains (Lü et al., 2012) (Figure 4A). The SUD1 RING-v domain shares high similarity to that of the E3 ubiquitin ligases TEB4 (57% amino acid identity) and Doa10 (49% amino acid identity). TEB4 and Doa10 are components of the ERAD complex involved in the quality control of ER proteins in human and yeast, respectively (Hassink et al., 2005; Kreft et al., 2006; Kreft and Hochstrasser, 2011). SUD1 also displays a high degree of similarity in an internal conserved segment of ∼130 residues called TD (TEB4-Doa10) present in all Doa10 orthologs (Swanson et al., 2001). Thus, the TD domain (TMs 5, 6, and 7) of SUD1 has 45 and 31% amino acid identity to those of TEB4 and Doa10, respectively (Lü et al., 2012).
Figure 4.
Topology Model for SUD1 Protein.
(A) Hydrophobicity plot of Arabidopsis SUD1 predicted by TMHMM2.0. The plot shows the posterior probabilities of inside/outside/TM segments. The 14 TM segments are depicted as numbered small boxes near the top of the figure; lines connected to the bottom edge of these boxes represent cytosolic loops, and those connected to the top edges depict luminal loops.
(B) The 14 TM domains of SUD1 are numbered and represented by cylinders and are the result of the TMHMM2.0 prediction shown in (A). Both protein termini are represented facing the cytosol based on the experimentally determined Doa10 topology (Kreft et al., 2006). The amino acid substitution of a given sud1 allele is depicted as a circle. Positions of the amino acid substitutions and the protein variants are indicated. The RING domain (residues 68 to 115) at the N terminus is shown as a black cylinder, and the conserved TD domain (residues 524 to 654) is highlighted by gray shading.
[See online article for color version of this figure.]
Next, we generated a topological model for SUD1 using sequence alignments and data from the experimental validation available for the homologous Doa10 (Kreft and Hochstrasser, 2011). For that purpose, we selected multiple SUD1 homologous proteins that complied with the following criteria: (1) the conserved N terminus RING-v domain, (2) the internal conserved TD domain, and (3) at least 10 predicted TM domains (Swanson et al., 2001). As a result of the topological analysis, the N terminus RING-v domain (and hence the putative ligase activity of SUD1) and the C terminus were predicted to face the cytosol (Figure 4), a similar disposition to that of Doa10 (Kreft and Hochstrasser, 2011). This model was also used to locate the putative position of the amino acid residues affected in the different sud1 mutant alleles. Thus, the mutations in the sud1-1 and sud1-2 alleles affected residues located at the transition between a TM domain and a hydrophilic loop. The mutation in sud1-3 was located in the second cytosolic loop, and the mutation in sud1-4 produced a premature stop codon at the end of the TM5 domain (Figure 4B).
Additionally, an alignment between SUD1 and SUD1 homologous proteins of several plant species was performed using the plant comparative genomics resource PLAZA database (http://bioinformatics.psb.ugent.be/plaza/; Proost et al., 2010). As shown in Supplemental Figure 3 online, the alignment of Arabidopsis SUD1 protein with the most homologous SUD1 proteins from several dicots (Vitis vinifera, Populus trichocarpa, Medicago truncatula, Lotus japonicus, and Glycine max) and monocots (Brachypodium distachyon, Oryza sativa, and Zea mays) showed striking sequence conservation. From the alignment, we inferred that the amino acid substitutions in all suppressors occurred in conserved residues among monocots and dicots (see Supplemental Figure 3 online).
Suppression of the dry2 Defects Occurs without Recovery in the Composition of Major Sterols
dry2 and the wild type have similar sterol compositions in shoots but significantly different sterol composition in roots (Posé et al., 2009) (Table 1). Since it has been proposed that the developmental defects in sterol biosynthetic mutants are the result of structural defects due to sterol depletions (Babiychuk et al., 2008; Men et al., 2008), we determined whether the suppression of dry2 phenotypes was associated with a recovery in sterol content in roots. Sterol profiling using gas chromatography–mass spectrometry analysis was performed separately in the shoots and roots of wild-type, dry2, and dry2/sud1-1 seedlings. As shown in Table 1, wild-type, dry2, and dry2/sud1-1 shoots presented similar bulk sterol compositions. By contrast, the bulk sterol composition in dry2/sud1-1 roots was similar to that of dry2 and showed significant differences relative to the wild type. We therefore concluded that the reversion of the dry2/sud1-1 root defects was not due to a recovery of major sterols to wild-type levels.
Table 1. Mass Spectral Analysis of Sterols and Squalene from the Wild Type, dry2, and dry2/sud1-1.
| Root | Shoot | |||||
|---|---|---|---|---|---|---|
| Sterol | Wild Type | dry2 | dry2/sud1-1 | Wild Type | dry2 | dry2/sud1-1 |
| Cycloartenol | 37 ± 6a | 104 ± 22b | 82 ± 23b | 28 ± 2a | 28 ± 6a | 26 ± 5a |
| 24-Methylenecycloartenol | 46 ± 1a | 86 ± 12b | 102 ± 8b | 48 ± 2a | 44 ± 3a | 47 ± 9a |
| Isofucosterol | 63 ± 4a | 41 ± 9a | 51 ± 25a | 63 ± 3a | 69 ± 4a | 66 ± 13a |
| Sitosterol | 2579 ± 144b | 1518 ± 58a | 1617 ± 91a | 2234 ± 101ab | 2052 ± 34a | 2302 ± 148b |
| Stigmasterol | 904 ± 10b | 304 ± 53a | 317 ± 19a | 60 ± 3a | 65 ± 13a | 69 ± 6a |
| Campesterol | 407 ± 3ab | 499 ± 75b | 263 ± 107a | 422 ± 27a | 439 ± 38a | 423 ± 50a |
| Cholesterol | 32 ± 12a | 18 ± 6a | 24 ± 7a | 46 ± 4a | 45 ± 1a | 48 ± 4a |
| Squalene | 10 ± 4a | 1206 ± 207b | 75 ± 13a | 10 ± 2a | 15 ± 4a | 13 ± 5a |
Values are given in μg g−1 dry weight. Mean ± sd, n = 3; values with the same letter are not significantly different at P < 0.05.
The recovery of the dry2 defects by the sud1-1 mutation without a recovery of sterol composition was further investigated by analyzing whether sud1-1 was able to suppress the developmental defects of more severe sterol-deficient mutants. The selected mutants were cpi1-1, which causes the loss of function of the cyclopropylsterol isomerase gene (Lovato et al., 2000; Men et al., 2008), and fackel, which is mutated in a sterol C-14 reductase (Jang et al., 2000; Schrick et al., 2000). As shown in Supplemental Figure 4 online, no phenotypic recovery was observed when sud1-1 was introduced in the cpi1-1 and fk-x224 mutants. These combined results support the notion that the reversion of the dry2 phenotype by sud1 is not concomitant with changes in the sterol content.
A Root-Derived Long-Distance Signal Causes the dry2 Shoot Phenotypic Defects
Consistent with the identification of SQE1 as the main SQE enzyme in roots and with the sterol profiling results for dry2 (Rasbery et al., 2007; Posé et al., 2009), we observed a dramatic accumulation of the substrate squalene in roots but not shoots of the dry2 mutant (Table 1). Interestingly, dry2/sud1-1 roots showed a significant reduction of squalene relative to dry2 (Table 1), suggesting that dry2 root defects could be caused by an accumulation of squalene and/or isoprenoid intermediates upstream of SQE1.
Since dry2 shoots showed no differences in terms of bulk sterols or squalene accumulations with the wild type, we questioned whether squalene or other isoprenoid intermediates generated in the dry2 roots could move toward the shoots, causing the observed phenotypes. In order to investigate this possibility, we performed micrografting experiments using wild-type, dry2, and dry2/sud1-1 seedlings. The root and shoot of the grafted plants were genotyped by sequencing the corresponding DRY2 and SUD1 alleles. As expected, control grafted plants (i.e., Ler scion/Ler rootstock and dry2 scion/dry2 rootstock) showed wild-type and dry2 phenotypes, respectively (Figure 5A). Importantly, dry2 scion onto both Ler rootstock (Figure 5A) and dry2/sud1-1 rootstocks (Figure 5B) showed a wild-type phenotype, suggesting that a root-derived signal was causing the dry2 shoot defects. Despite multiple attempts, we were unable to obtain a viable graft using wild-type or sud1-1 scions onto dry2 rootstocks. We therefore could not evaluate the effect of the dry2 root-derived signal in healthy scions.
Figure 5.
Suppression of dry2 Shoot (Scion) Defects by Wild-Type and dry2/sud1-1 Rootstocks.
A total of 20 viable plants per graft combination were analyzed, and a representative plant per combination is depicted after 18 d of growth in soil. Bottom panels show the plants after soil removal to visualize the root phenotype. Bar = 2 cm.
(A) dry2 shoot (scion) recovers the wild -type phenotype when grafted onto wild-type Ler rootstock. Self-grafted wild-type (Ler) and dry2 were used as controls.
(B) dry2 shoot (scion) recovers the wild-type phenotype when grafted onto dry2/sud1-1 rootstock. Self-grafted dry2/sud1-1 and dry2 were used as controls.
[See online article for color version of this figure.]
sud1 Mutations Suppress dry2 Root Defects by Downregulating HMGR Activity
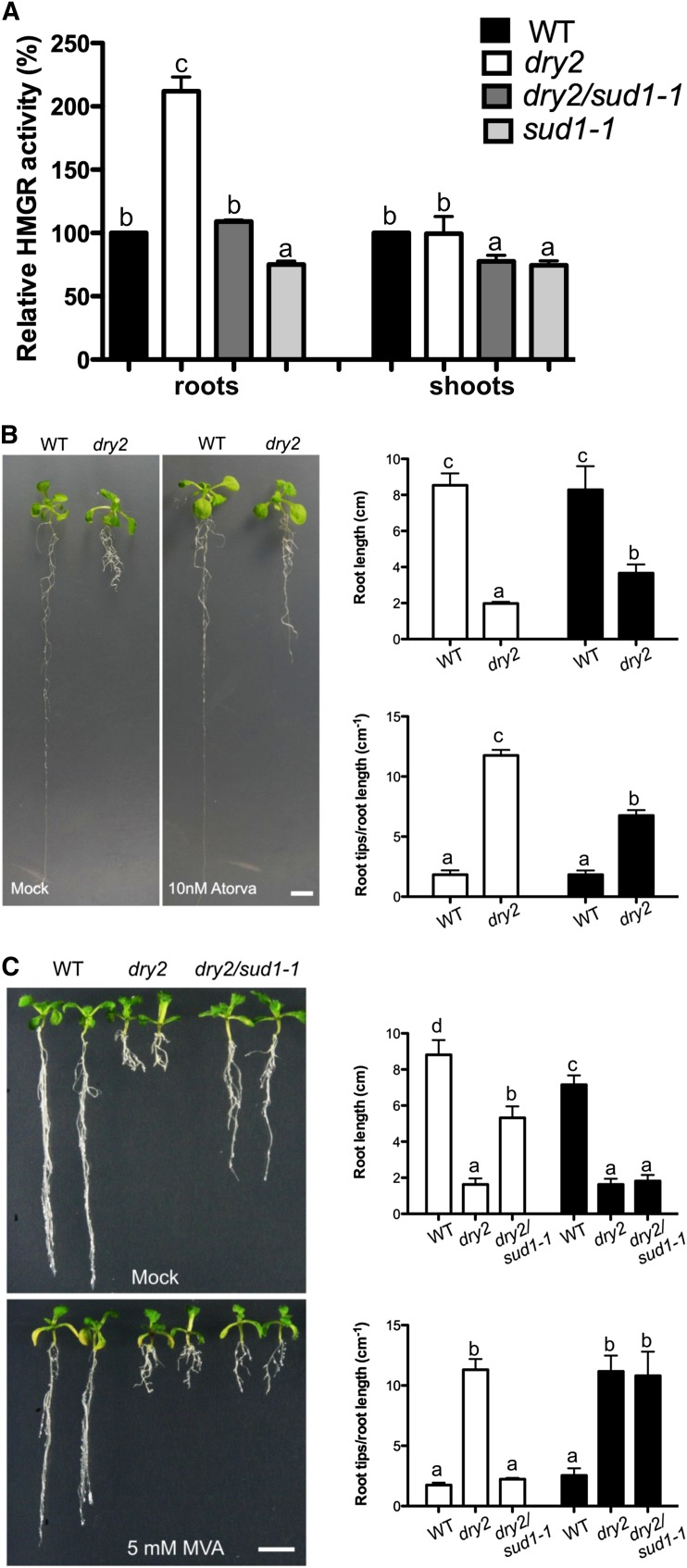
It is known that in addition to the accumulation of squalene, the reduction of SQE activity causes a compensatory increase of HMGR activity (Wentzinger et al., 2002; Posé et al., 2009). As shown in Figure 6A, dry2 roots increased the HMGR activity approximately twofold, while no differences were found in shoots. Importantly, in the roots of dry2/sud1-1, the HMGR activity returned to near wild-type levels, whereas in shoots, HMGR activity decreased to ∼0.7-fold that in shoots of both the wild type and dry2 (Figure 6A).
Figure 6.
HMGR Activity Changes Correlate with the Phenotypes Observed in dry2 and dry2/sud1-1.
(A) HMGR activity measurement in roots and shoots of 15-d-old wild-type (WT), dry2, dry2/sud1-1, and sud1-1 seedlings (mean ± sd, n = 3; each measurement corresponds to a pool of ≥100 seedlings). Values with the same letter are not significantly different at P < 0.05. The experiment was repeated at least three times with similar results.
(B) Inhibition of HMGR activity partially recovers dry2 roots defects. Wild-type and dry2 seeds were germinated and grown on MS plates for 4 d. Seedlings were then transferred to MS supplemented with 10 nM of the HMGR inhibitor atorvastatin and grown for additional 2 weeks (mean ± sd, n = 30; values with the same letter are not significantly different at P < 0.05). The experiment was repeated three times with similar results. Bar = 1 cm.
(C) dry2/sud1-1 roots phenocopy dry2 in the presence of MVA. The wild type, dry2, and dry2/sud1-1 were germinated and grown on MS plates for 4 d. Seedlings were then transferred to MS medium supplemented with MVA and grown for two additional weeks (mean ± sd, n = 30; values with the same letter are not significantly different at P < 0.05). The experiment was repeated three times with similar results. Bar = 1 cm.
[See online article for color version of this figure.]
To further investigate whether the reduction of HMGR activity could cause the recovery of the dry2 roots, we used atorvastatin, a specific inhibitor of HMGR activity. After testing several concentrations, we selected 10 nM atorvastatin. As shown in Figure 6B, this concentration did not have apparent effects on wild-type root growth or branching but substantially improved both phenotypes in dry2. This result supports the notion that a reduction in HMGR activity in the dry2/sud1-1 suppressor was responsible for the recovery of the dry2 root phenotypic defects. We next treated wild-type, dry2, and dry2/sud1-1 seedlings with MVA, the product of the reaction catalyzed by HMGR. As shown in Figure 6C, a 5 mM MVA treatment caused a mild reduction in wild-type root elongation, while no visible changes were observed in dry2 roots. However, the same treatment abolished the sud1-1 suppressive effect of the dry2 root defects, phenocopying dry2 roots (Figure 6C). This result indicates that bypassing HMGR activity by adding the product of the HMGR reaction prevents the recovery of the dry2 phenotype in a dry2/sud1-1 background.
Genetic Analysis of the Regulation of HMGR Activity by SUD1
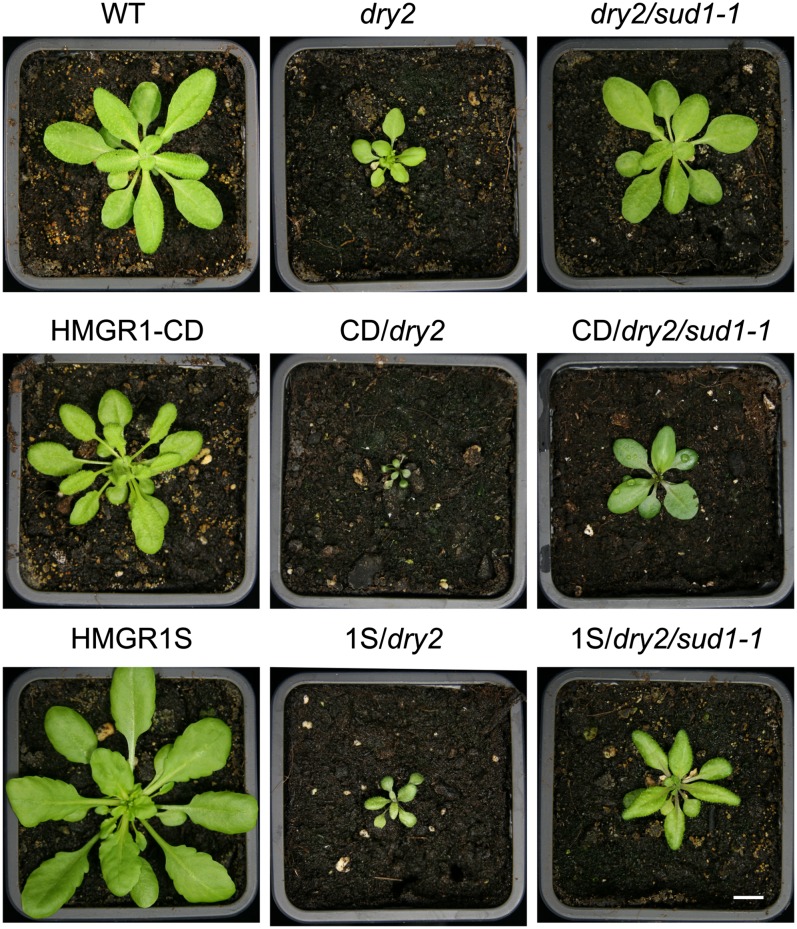
To further investigate the link between HMGR activity and SUD1, the dry2/sud1-1 mutant was crossed with transgenic lines overexpressing the catalytic domain of the HMGR1 (HMGR1-CD) and the short isoform of HMGR1 containing the TM domains (HMGR1S) (see Supplemental Figure 5 online). HMGR1-CD and HMGR1S lines show an ∼10-fold and an approximately threefold increase in HMGR activity compared with the wild type, respectively (Manzano et al., 2004). As shown in Figure 7, the enhanced HMGR activity in the wild-type background only caused slight growth inhibition in the HMGR1-CD line and no visible effect in the HMGR1S line (Manzano et al., 2004). By contrast, the HMGR1-CD/dry2 combination greatly enhanced the growth inhibition defects of dry2 generating dwarf plants (Figure 7). The HMGR1S/dry2 combination also showed enhanced growth inhibition compared with dry2, but this effect was less drastic than that observed in the HMGR1-CD/dry2. In fact, viable seeds were obtained in this genotype, while HMGR1-CD/dry2 plants died before reaching maturity (Figure 7). These results indicate that the increase in HMGR activity in the presence of a dry2 mutation accounts for the severity of the observed developmental phenotypes. Importantly, when the sud1-1 mutation was introduced into HMGR-CD/dry2 and HMGR1S/dry2, there was an important recovery of the defective phenotypes to such an extent that HMGR-CD/dry2/sud1-1 plants were fertile (Figure 7).
Figure 7.
Phenotypic Analysis of Plants Overexpressing HMGR1S and HMGR1-CD in dry2 and dry2/sud1-1 Backgrounds.
Wild-type (WT), dry2, and dry2/sud1-1 plants are in the Ler background ecotype. The transgenic HMGR1-CD or HMGR1S plants are in Col glabrous background. The resulting F2 from the crosses are Col-Ler hybrids. Plants were grown for 3 weeks in soil under long-day conditions. All plants were fertile with the exception of CD/dry2 plants. Bar = 1 cm.
[See online article for color version of this figure.]
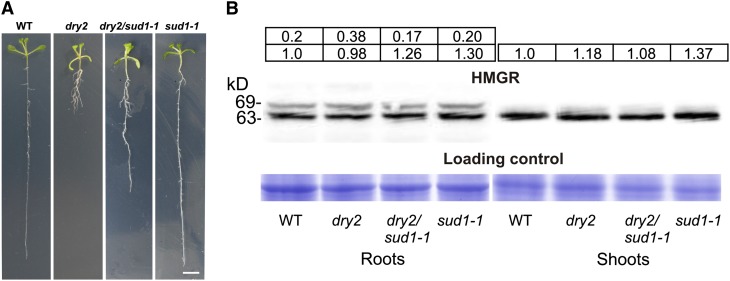
Based on these results, we propose a model in which the increase of HMGR activity concomitant to the reduction of SQE1 activity is mainly responsible for the observed developmental phenotypes in the dry2 background. In this scenario, mutations in the positive regulator of HMGR SUD1 cause the reversion of the dry2 developmental defects by decreasing HMGR activity. An obvious question was whether the regulation of HMGR activity by SUD1 was dependent on the presence of the dry2 mutation. Therefore, the sud1-1 mutation was segregated from dry2 by backcrossing dry2/sud1-1 to the wild type. As shown in Figure 8A, the single sud1-1 mutant did not show any obvious phenotypic difference compared with wild-type plants, except by a glossy-like phenotype in shoots reminiscent of the phenotypes described for the loss-of-function cer9-1 and cer9-2 mutants allelic to sud1 (Lü et al., 2012; see Supplemental Figure 6 online). As shown in Figure 6A, HMGR activity in the sud1-1 single mutant was ∼0.75-fold lower than that of the wild type in both shoots and roots, indicating that SUD1 is a positive regulator of HMGR activity acting independently of the dry2 mutation.
Figure 8.
sud1-1 Mutation Produces No Visible Phenotype and No Changes in HMGR Protein Content.
(A) Phenotype of wild-type (WT), dry2, dry2/sud1-1, and sud1-1 seedlings grown for 15 d on MS medium under long-day conditions. Bar = 1 cm.
(B) Protein gel blot analysis of HMGR protein in 15-d-old roots and shoots of the wild type, dry2, dry2/sud1-1, and sud1-1. Intensities of the HMGR protein bands (top panel) and the Coomassie blue–stained gel (bottom panel) were quantified using ImageJ software (http://rsb.info.nih.gov/ij). The normalized HMGR protein levels expressed as relative abundance to the amount of the HMGR1S isoform in wild-type plants (arbitrarily set at 1) is shown at the top of each lane. Image shows the results from one representative experiment. Four independent experiments were performed with similar results.
[See online article for color version of this figure.]
Regulation of HMGR Activity by SUD1 Does Not Involve Changes in Protein Content
Because SUD1 contains a RING-v domain putatively involved in ubiquitination (Stone et al., 2005), we used protein gel blot analysis to investigate whether the regulation of HMGR activity mediated by SUD1 involves changes in HMGR protein content. The immunospecific antibodies used in the analysis were raised against the catalytic domain of HMGR1 (Manzano et al., 2004; Leivar et al., 2005). As shown in Figure 8B, the analysis of HMGR protein content in root samples revealed two bands of ∼63 and 69 kD, corresponding to the HMGR1S and HMGR1L isoforms, respectively, whereas in shoot samples, only the 63-kD protein band was detected. In both cases, no significant differences in HMGR protein content among the wild type, dry2, dry2/sud1-1, and sud1-1 were observed (Figure 8B). This result, together with results from previous pharmacological studies (Wentzinger et al., 2002; Nieto et al., 2009), indicates that the variations in HMGR activity in the different genetic backgrounds occur without changes in the total HMGR protein content.
DISCUSSION
Mutations in the sterol biosynthetic SQE1 gene produce multiple developmental defects, but in contrast with null alleles of SQE1 (Rasbery et al., 2007), the hypomorphic sqe1-5 allele is fully fertile (Posé et al., 2009). This characteristic lends itself to the use of dry2/sqe1-5 as a genetic tool to identify processes that otherwise would be concealed. To find components regulating isoprenoid biosynthesis and/or signaling in Arabidopsis, we performed genetic screening for suppressors of dry2. Here, we report the analysis of four suppressors and show that all mutations affect the At4g34100 gene encoding a protein with a RING-v domain, found in ubiquitin E3 ligases, subsequently named SUD1. Based on phylogenetic and structural similarities, it is proposed that SUD1 is an Arabidopsis orthologous protein of yeast Doa10 and mammalian TEB4, which are involved in the ERAD-C pathway. Our physiological, molecular, biochemical, and genetic analyses strongly support that sud1 recovers the dry2 defects through the reversion of the enhanced HMGR activity of dry2 to wild-type levels. Thus, our study uncovers SUD1 as a regulator of HMGR activity in the plant isoprenoid biosynthetic pathway.
The Accumulation of an MVA-Derived Signal in Roots Causes the dry2 Phenotypes
Despite its dramatic phenotypic defects, sterol analysis of dry2 showed only moderate changes in major bulk sterols in roots and no significant changes in shoots, compared with the wild type. Interestingly, sterol analysis of dry2/sud1-1 roots revealed a similar composition to that of dry2, indicating that the dry2 phenotypes cannot simply be explained by structural defects caused by the reduction in bulk sterols. Moreover, dry2 shoots display wild-type characteristics when grafted onto wild-type rootstocks, suggesting that a toxic mobile signal that originated in the dry2 roots is responsible for the observed dry2 shoot phenotypes.
Although it is tempting to speculate that the squalene accumulated in dry2 roots is the mobile signal responsible for the phenotypes, we argue against this notion because plants deal with excess endogenously produced or exogenously added squalene by storing it as remobilizable cytosolic lipid droplets without obvious phenotypic defects (Wentzinger et al., 2002; Bouvier-Navé et al., 2010). Thus, we propose an alternative model whereby the accumulation of toxic intermediates, or derivatives acting upstream of squalene, is responsible for the observed dry2 developmental phenotypes. In fact, three independent experiments, including (1) the inhibition of HMGR with atorvastatin that partially improved the dry2 root defects, (2) the HMGR bypass with MVA that caused dry2/sud1-1 (but not the wild type) to phenocopy dry2, and (3) the overexpression of HMGR that enhanced the dry2 phenotypes, suggest that the dry2 mobile signal(s) is not only triggered by the reduction of SQE1 activity but also by the concomitant upregulation of HMGR activity.
Supporting our model, the presence of toxic MVA-derived intermediates associated with HMGR activity changes has been reported in Insig double knockout mice that show developmental defects linked to enhanced HMGR activity (Engelking et al., 2006). As was true for dry2, the developmental defects of the mice were ameliorated with the use of HMGR inhibitors (Engelking et al., 2006). Interestingly, the Insig knockout mice are not an isolated example. Nonsterol MVA-derived compounds upstream of squalene have been linked to the regulation of HMGR protein content in mammals, yeast, and plants. Thus, the degradation of mammalian HMGR is accelerated by the addition of farnesol, geranylgeraniol, and its precursor geranylgeranyl diphosphate (Correll et al., 1994; Meigs et al., 1996; Räikkönen et al., 2010). geranylgeranyl diphosphate is also known to regulate the degradation of HMGR2p in yeast (Garza et al., 2009). Surprisingly, the effect of farnesol on plant HMGR activity seems to be different from that in mammals because the addition of subtoxic concentrations of farnesol to tobacco Bright Yellow-2 cells had a drastically stimulatory effect on HMGR activity (Hemmerlin and Bach, 2000).
Despite the similarities in the regulation of HMGR by a nonsterol MVA-derived molecule in different species, the mechanisms that regulate HMGR activity in Arabidopsis seem to operate at a different level from those in yeast and animals. Thus, our study and a previous report (Nieto et al., 2009) have shown that both the genetic and pharmacological block of Arabidopsis SQE activity leads to upregulation of HMGR activity without changing HMGR protein amounts, while in yeast and animals, HMGR activity depends on protein stability.
Structural Characteristics of SUD1
All sud1 alleles show a similar phenotypic recovery of dry2 phenotypes, including sud1-4, which is caused by a premature stop codon at the fifth TM domain. The sud1 alleles also show a glossy-like phenotype in leaves reminiscent of the cer9 mutants in the same locus. Because cer9 has been reported to be a loss-of-function mutant (Rashotte et al., 2004; Lü et al., 2012), and the premature stop codon is much more downstream in cer9-2 than in sud1-4, we presume that sud1 are also loss-of-function alleles. Interestingly, the cer9 alleles are recessive while all sud1 alleles are semidominant with respect to the dry2 mutation, which suggests that SUD1 regulation of HMGR activity is dose dependent. Thus, the heterozygous, SUD1/sud1 genotype is unable to produce enough SUD1 protein to fully reproduce the dry2 phenotypes.
The phylogenetic analysis and the structural features of SUD1 suggest that this protein might function as one ortholog of yeast Doa10 and human TEB4 in Arabidopsis (Carvalho et al., 2006; Kreft et al., 2006; Kreft and Hochstrasser, 2011); these proteins are involved in the quality control that degrades misfolded ER proteins (Swanson et al., 2001). However, despite multiple attempts, we failed to complement the yeast doa10 mutant with SUD1 because this protein is highly unstable in yeast. This result is not entirely surprising because efforts to perform complementation of yeast doa10 with TEB4 have also been unsuccessful despite the fact that yeast Doa10 and human TEB4 are orthologous proteins (Kreft et al., 2006).
When we analyze SUD1 plant homologs, we find a striking conservation of SUD1 sequence with homologous proteins from dicots and monocots. Indeed, amino acid substitutions in all suppressors occur in plant conserved residues. Thus, sud1-1 and sud1-2 result in Gly218Arg and Gly360Glu substitutions that change small nonpolar residues for basic and acidic residues, respectively. Interestingly, both residues are located at the transition between a TM segment and a hydrophylic loop. Interruption of TM helices by a short nonhelical segment containing Pro, Gly, and/or Ser residues has also been observed in many classes of transporters, including amino acid antiporters (Gao et al., 2009), neurotransmitter-sodium symporters (Yamashita et al., 2005), and sodium-independent transporters (Schulze et al., 2010). Interruption of helical structures exposes main-chain carbonyl oxygen and nitrogen atoms for hydrogen bonding and ion coordination, aspects that are essential for proper function (Yamashita et al., 2005). The mutation in sud1-3 results in the Arg244Lys substitution. These two amino acids are chemically related, and it would be expected that its substitution did not cause important changes. However, phylogenetically distant plant species, such as monocots and dicots, maintain a conserved Arg around position 244, suggesting an important role for this specific residue in SUD1 function.
Regulation of HMGR Activity by SUD1
A wealth of information about ERAD comes from yeast and mammals (Vembar and Brodsky, 2008; Smith et al., 2011). The HRD pathway (ERAD-L and ERAD-M) is involved in the degradation of misfolded ER-luminal and intramembrane domains; HRD genes were identified in a genetic screening for regulators of HMGR degradation (hence the name HRD, for HMGR reductase degradation) (Hampton et al., 1996). The finding that feedback regulation of sterol synthesis in mammalian and yeast cells uses the ERAD machinery (Hampton, 2002) illustrates co-option of the basic quality control mechanism for regulatory processes and reveals potential functions in cell-to-cell signaling. ERAD-regulated HMGR proteins, such as those from yeast and mammals, contain the known sterol sensing domain motif consisting of five consecutive TM spans (Goldstein et al., 2006; Theesfeld et al., 2011). However, HMGR from plants contain two predicted TM domains (see Supplemental Figure 5 online) (Campos and Boronat, 1995), therefore lacking any potential sterol sensing domain motif. Surprisingly, following a nontargeted screening for plant HMGR regulators, we identified SUD1, a likely ERAD component. Our first explanation for this was that HMGR stability was regulated by ERAD, either directly by SUD1 or through a compensatory increase of the HRD pathway in sud1 mutants. However, protein gel blot analyses indicated that SUD1 did not exert its function by regulating HMGR proteins levels. Because SUD1 likely encodes an E3 ubiquitin ligase, another plausible explanation is that a negative regulator of HMGR is being degraded in a SUD1-dependent manner in dry2, so the loss of SUD1 function would impair this degradation, leading to the recovery of HMGR activity to wild-type levels.
Transcriptional versus Translational Regulation of HMGR
It has been proposed that major changes in HMGR activity in plants would be determined at the transcriptional level, whereas posttranslational control would allow a finer and faster adjustment (Chappell, 1995). Whereas transcriptional modulation of HMGR has been demonstrated in many plant systems, evidence for mechanisms regulating HMGR activity at the posttranslational level is scarce. Thus, Nieto et al. (2009) have shown that metabolic perturbations by enhancing or depleting the flux through the sterol pathway in Arabidopsis causes a compensatory response in HMGR activity, without changes in transcript or protein levels, and Flores-Pérez et al. (2010) reported that the inactivation of the Arabidopsis WD protein PRL1 leads to reduced HMGR activity with no changes in transcript and protein levels. This effect could be related to the ability of PRL1 to interact and inhibit the activity of the Arabidopsis SNF1-related protein kinases (SnRK1) AKIN10 and AKIN11 (Bhalerao et al., 1999), presumably targeting them for ubiquitination and proteasomal degradation (Lee et al., 2008). Since plant SnRK1 phosphorylates and inactivates HMGR (Dale et al., 1995; Sugden et al., 1999), the loss of PRL1 function would result in increased SnRK1 activity followed by HMGR phosphorylation and the subsequent reduction of HMGR activity. It has also been demonstrated that HMGR activity is negatively regulated by PP2A-mediated dephosphorylation (Leivar et al., 2011). Therefore, SnRK1 and/or PP2A, regulators of Arabidopsis HMGR activity, are candidates to act as mediators of SUD1 regulation of HMGR activity. An alternative possibility is that SUD1 might produce the direct monoubiquitination of HMGR, thereby increasing its activity, as has been reported for other proteins (Schnell and Hicke, 2003).
Overall, using genetic, physiological, biochemical, and molecular approaches, we show that SUD1, a likely component of the Arabidopsis ERAD-C pathway, is a positive regulator of HMGR activity. Future research should help clarify the mechanistic basis for the ERAD regulation of HMGR activity in plants and what signals are implicated in this regulation.
METHODS
Plant Material and Growth Conditions
Unless stated otherwise, the Arabidopsis thaliana plants used in this study were either grown on soil or in Petri dishes using an environmental chamber set for long-day lighting conditions (16 h light/8 h dark) and a temperature of 22°C. For in vitro assays, surface-sterilized and cold-stratified Arabidopsis seeds were sown onto Murashige and Skoog (MS) phytagel-solidified medium (MS salts, 30 g L−1 Suc, and 7 g L−1 phytagel [Sigma-Aldrich], pH 5.7). For chemical treatments, the appropriate amounts of filter sterilized chemical stock solutions were added to cooled autoclaved growth medium. The dry2 (Posé et al., 2009), cpi1-1 (Schrick et al., 2000), and fk-x224 (Men et al., 2008) mutants and the HMGR1-CD and HMGR1S overexpressing lines (Manzano et al., 2004) have been previously described.
Genetic Screen for Second-Site Suppressor Mutations of dry2
The dry2 seeds were mutagenized by imbibition in 75 mM ethyl methanesulfonate (Sigma-Aldrich) for 4 h at room temperature. After washing thoroughly with water for complete ethyl methanesulfonate removal, the mutagenized seeds (M1) were sown on soil and grown under high humidity conditions. The M2 seeds were harvested as 131 independent pools (each pool corresponding to 50 M1 plants). For the identification of dry2 suppressors, the M2 seeds were grown on soil under low watering conditions. Suppressors with enhanced drought tolerance compared with dry2 plants were visually identified and selected for further analysis. The SQE1-DRY2 gene of each candidate suppressor was amplified by PCR using DRY2-specific primers and sequenced in order to confirm the presence of the dry2 mutation as described by Posé et al. (2009). The primers sequences for genotyping were DRY2 SEQ F, 5′-ATTGTTCTCGGTTGGGTGAG-3′, and DRY2 SEQ R, 5′-GATTGCAGTTCTCTAGGACCAA-3′, and internal primer to sequence DRY2 SEQ2, 5′-TCAAAGAATGCGGGAGAAAG-3′.
Detection of ROS
Hydrogen peroxide was visually detected in leaves using the 3,3′-diaminobenzidine (DAB; Sigma-Aldrich) substrate as described previously (Orozco-Cardenas and Ryan, 1999). DAB was also used for in situ detection of hydrogen peroxide in roots from seedlings grown on phytagel-solidified medium (Carol et al., 2005). For in situ detection of superoxide in leaves, the nitroblue tetrazolium (NBT Color Development Substrate; Promega) staining method (Jabs et al., 1996) was used. In all cases, stained leaves were imaged under dark-field illumination using a Leica MZ FLII stereomicroscope.
Whole-Plant Stomatal Conductance and Determination of Pro Content
Leaf stomatal conductance to water vapor was measured in 25-d-old leaves grown under short-day lighting conditions (8 h light/16 h dark) using a Leaf Porometer Model SC-1 (Decagon Services). Measurements were performed after spraying the leaves with 0, 0.2, 2, or 20 μM of ABA (Sigma-Aldrich) dissolved in a 0.1% Tween 20 solution. Pro was extracted and quantified as described previously (Borsani et al., 2002).
Root Measurements
Root measurements were performed according to the procedure described by Posé et al. (2009). Briefly, seeds were grown vertically on phytagel-solidified MS medium for 5 and 10 d for roots hair and root elongation measurements, respectively. For root elongation assays, primary root length pictures were taken daily using a Nikon Coolpix 4500 camera attached to a MZ FLII stereomicroscope (Leica). Quantitative measurements were made using Image J software (http://rsb.info.nih.gov/ij/). Root branching was determined by counting the number of root tips per length unit (cm) of primary root. Root hair length was measured in the differentiation zone as described by Posé et al. (2009).
Identification of the sud1-1 Suppressor Mutation
The dry2 mutant in the Ler background was crossed into the Col-0 ecotype for seven generations to generate a nearly isogenic dry2Col-0 line for map-based cloning. An F2 mapping population was created from a cross between the dry2/sud1-1 (Ler) and the introgressed dry2Col-0 lines. A total of 120 F2 plants displaying the suppression phenotypes conferred by the dry2/sud1-1 mutations were used for rough mapping. For fine mapping, a total of 2400 chromosomes were analyzed to locate the SUD1 locus in a 117-kb region at the bottom of chromosome IV with 36 candidate genes. All information regarding the genetic markers used in the map-based cloning was obtained from The Arabidopsis Information Resource (http://www.Arabidopsis.org/). The entire genome of dry2/sud1-1 was sequenced using high-throughput sequencing with the Illumina platform. Reads were filtered with Fastx-Toolkit software (http://hannonlab.cshl.edu/fastx_toolkit/index.html) and mapped with the Arabidopsis genome sequence version TAIR10 using the Burrows-Wheeler Alignment Tool (Li and Durbin, 2009). Polymorphisms for the 117-kb candidate region were analyzed with Samtools and Bcftools using regions with at least 5× depth coverage (Li et al., 2009) and filtered using the sequence information for the Ler ecotype available at 1001 Genomes (http://1001genomes.org/). After filtering, two nonsynonymous mutations in the AT4G34100 and AT4G34135 loci were identified. The identification of four independent suppressor alleles with mutations in the AT4G34100 locus confirmed the identity of the AT4G34100 locus as SUD1.
Informatic Tools Used for Functional Characterization of SUD1
The InterPro database (http://www.ebi.ac.uk/Tools/pfa/iprscan/) was used to search for conserved domains of SUD1 protein. The National Center for Biotechnology Information BLASTp tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) was used to identify putative SUD1 orthologs using the predicted proteome from Saccharomyces cerevisiae (taxid4932), Homo sapiens (taxid9606), Mus musculus (taxid10090), Drosophila melanogaster (taxid7227), and Caenorhabditis elegans (taxid6239).
The TMHMM2.0 program (www.cbs.dtu.dk/services/TMHMM/) was used to predict the putative TM domain topology of SUD1 based on the hydrophobicity plot (Krogh et al., 2001). The plant comparative genomics resource PLAZA (http://bioinformatics.psb.ugent.be/plaza/) was used to search for SUD1 homologous protein sequences in different plant species (Proost et al., 2010). Protein sequence alignment of Arabidopsis SUD1 with homologous proteins from other plant species was performed with the software ClustalW2 available online from the European Bioinformatics Institute (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Default values were used for all parameters, including Gonnet protein weight matrix, gap open of 10, gap extension of 0.20, gap distance of 5, no end gaps, should read: no iteration, numiter of 1, and clustering neighbor-joining no iteration, numiter of 1, and clustering neighbor-joining (Gonnet et al., 1992).
Sterol and Squalene Analysis and Determination of HMGR Activity
Fifteen-day-old seedlings grown in phytagel-solidified MS medium were used for sterol, squalene, and HMGR activity measurements. A pool of ≥100 seedlings per genotype was used per each measurement. Since HMGR activity and isoprenoid biosynthesis are regulated by light conditions (Learned, 1996; Rodríguez-Concepción et al., 2004), shoot and roots were collected and measured separately at 3 h from the start of the light period. Quantification of total sterol content and determination of sterol profiles were performed as previously reported (Masferrer et al., 2002). HMGR activity was assayed as described (Nieto et al., 2009). In our assays, one unit of HMGR activity is defined as the amount of enzyme that converts 1 pM of 3-hydroxy-3-methylglutaryl CoA into MVA per min and mg of protein at 37°C.
Arabidopsis Grafting
Four-day-old seedlings grown vertically were transferred to a 0.22-μm sterile filter (Millipore) in contact with half-strength MS medium containing 0.6% (w/v) phytagel (Sigma-Aldrich). After 3 d, seedlings were grafted in a wedge graft (Y shape) under sterile conditions, as described by Turnbull et al. (2002). Afterwards, the grafted plants were grown for seven additional days under humid conditions. Successful grafts were transferred to soil, and the grafting unions were confirmed by sequencing analysis of the dry2 mutant allele of shoots and roots as described above.
Determination of HMGR Protein Levels
HMGR protein levels were determined by immunoblot analysis using a rabbit polyclonal antibody raised against the catalytic domain of Arabidopsis HMGR1 (Manzano et al., 2004; Leivar et al., 2005). The antibody was used at 1:1000 dilution, and the secondary antibody (horseradish peroxidase anti-rabbit IgG; Sigma-Aldrich) was diluted at 1:14,000. For immunoblot analysis, total root and shoot protein were loaded onto 10% acrylamide SDS gels. Immunoblot images were developed with Advanced ECL (GE Healthcare) and exposed to an x-ray film for 30 s to 1 min. Coomassie Brilliant Blue staining was used to confirm equal loading.
Statistical Analysis
Statistical analysis was performed using the Statgraphic Centurion program (Statpoint Technologies). The significance of differences was determined by analysis of variance (for three or more samples) or a t test (for two samples).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: SQE1 (At1g58440), SUD1 (At4g34100), CPI1 (At5g50375), FK (At3g52940), and HMGR1 (At1g76490).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Identification of Suppressor Lines Recovering the dry2 Drought Hypersensitivity.
Supplemental Figure 2. Map-Based Cloning of sud1-1.
Supplemental Figure 3. Protein Sequence Alignment of SUD1 with Homologs from Other Plant Species.
Supplemental Figure 4. The sud1-1 Mutation Does Not Improve the Phenotypic Defects of the Sterol Biosynthesis Mutants cpi1-1 and fk-x224.
Supplemental Figure 5. Schematic Representation of the HMGR Protein Versions Overexpressed in the HMGR1S and HMGR1-CD Transgenic Lines.
Supplemental Figure 6. sud1-1 Leaves but Not the Wild Type Show Glossy-Like Appearance.
Acknowledgments
We thank Pedro Carvalho for helpful suggestions and Colin Turnbull for advice on grafting experiments. This work was supported by grants from Ministerio de Ciencia e Innovación (cofinanced by the European Regional Development Fund) to M.A.B. (BIO2011-23859 and CSD2007-00057), A.F. (BIO2009-06984 and CSD2007-00036), and O.B. from Universidad de la República-Comisión Sectorial de Investigación Científica (Grupo 418). V.G.D. was supported by a Formación del Personal Investigador fellowship from Ministerio de Educación y Ciencia (BIO2005-04733), and V.A.-S. was supported by Fundação para a Ciência e a Tecnologia (FCT) (Grant SFRH/BD/38583/2007).
AUTHOR CONTRIBUTIONS
V.G.D. and V.A.-S. performed the physiological, biochemical, and genetic experiments. D.P. identified the suppressors and performed an initial characterization. M.A. performed sterols and squalene analysis and HMGR activity measurements. A.B. conducted the genomic analysis for SUD1 identification. H.A. performed the ROS analyses. A.E. performed genetic crosses for SUD1 identification. O.B. designed and performed the atorvastatin experiments. V.G.D., A.R., V.V., R.M.T., and M.A.B. designed the research. V.G.D.,V.A.-S., A.R., A.F., and M.A.B. wrote the article.
Glossary
- IPP
isopentenyl diphosphate
- MVA
mevalonic acid
- ROS
reactive oxygen species
- HMGR
3-hydroxy-3-methylglutaryl-CoA reductase
- ER
endoplasmic reticulum
- SQE
squalene epoxidase
- ERAD
endoplasmic reticulum–associated protein degradation
- ABA
abscisic acid
- Ler
Landsberg erecta
- Col-0
Columbia-0
- TM
transmembrane
- MS
Murashige and Skoog
- DAB
3,3′-diaminobenzidine
References
- Babiychuk E., Bouvier-Navé P., Compagnon V., Suzuki M., Muranaka T., Van Montagu M., Kushnir S., Schaller H. (2008). Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. USA 105: 3163–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55: 429–457 [DOI] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert K., Bakó L., Okrész L., Szabados L., Muranaka T., Machida Y., Schell J., Koncz C. (1999). Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl. Acad. Sci. USA 96: 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Cuartero J., Valpuesta V., Botella M.A. (2002). Tomato tos1 mutation identifies a gene essential for osmotic tolerance and abscisic acid sensitivity. Plant J. 32: 905–914 [DOI] [PubMed] [Google Scholar]
- Boutté Y., Grebe M. (2009). Cellular processes relying on sterol function in plants. Curr. Opin. Plant Biol. 12: 705–713 [DOI] [PubMed] [Google Scholar]
- Bouvier F., Rahier A., Camara B. (2005). Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44: 357–429 [DOI] [PubMed] [Google Scholar]
- Bouvier-Navé P., Berna A., Noiriel A., Compagnon V., Carlsson A.S., Banas A., Stymne S., Schaller H. (2010). Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 152: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Sakvarelidze-Achard L., Schaller H., Khafif M., Schott G., Bendahmane A., Voinnet O. (2012). Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 1778–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos N., Boronat A. (1995). Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell 7: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland F., Fujioka S., Nelson T. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153: 741–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol R.J., Takeda S., Linstead P., Durrant M.C., Kakesova H., Derbyshire P., Drea S., Zarsky V., Dolan L. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T.A. (2006). Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Chappell J. (1995). The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiol. 107: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S. (2002). Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14: 1995–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll C.C., Ng L., Edwards P.A. (1994). Identification of farnesol as the non-sterol derivative of mevalonic acid required for the accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 269: 17390–17393 [PubMed] [Google Scholar]
- Dale S., Arró M., Becerra B., Morrice N.G., Boronat A., Hardie D.G., Ferrer A. (1995). Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur. J. Biochem. 233: 506–513 [DOI] [PubMed] [Google Scholar]
- Eisenreich W., Rohdich F., Bacher A. (2001). Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci. 6: 78–84 [DOI] [PubMed] [Google Scholar]
- Engelking L.J., Evers B.M., Richardson J.A., Goldstein J.L., Brown M.S., Liang G. (2006). Severe facial clefting in Insig-deficient mouse embryos caused by sterol accumulation and reversed by lovastatin. J. Clin. Invest. 116: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuto M., Balcells L., Campos N., Caelles C., Arró M., Boronat A. (1994). Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc. Natl. Acad. Sci. USA 91: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez U., Pérez-Gil J., Closa M., Wright L.P., Botella-Pavía P., Phillips M.A., Ferrer A., Gershenzon J., Rodríguez-Concepción M. (2010). Pleiotropic regulatory locus 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Mol. Plant 3: 101–112 [DOI] [PubMed] [Google Scholar]
- Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009). Structure and mechanism of an amino acid antiporter. Science 324: 1565–1568 [DOI] [PubMed] [Google Scholar]
- Garza R.M., Tran P.N., Hampton R.Y. (2009). Geranylgeranyl pyrophosphate is a potent regulator of HRD-dependent 3-hydroxy-3-methylglutaryl-CoA reductase degradation in yeast. J. Biol. Chem. 284: 35368–35380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein, J.L., DeBose-Boyd, R.A., and Brown, M.S. (2006). Protein sensors for membrane sterols. Cell 124: 35–46. [DOI] [PubMed]
- Gonnet G.H., Cohen M.A., Benner S.A. (1992). Exhaustive matching of the entire protein sequence database. Science 256: 1443–1445 [DOI] [PubMed] [Google Scholar]
- Hampton R.Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14: 476–482 [DOI] [PubMed] [Google Scholar]
- Hampton R.Y., Gardner R.G., Rine J. (1996). Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassink G., Kikkert M., van Voorden S., Lee S.-J., Spaapen R., van Laar T., Coleman C.S., Bartee E., Früh K., Chau V., Wiertz E. (2005). TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 388: 647–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A., Bach T.J. (2000). Farnesol-induced cell death and stimulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase activity in tobacco cv bright yellow-2 cells. Plant Physiol. 123: 1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerlin A., Harwood J.L., Bach T.J. (2012). A raison d’être for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 51: 95–148 [DOI] [PubMed] [Google Scholar]
- Jabs T., Dietrich R.A., Dangl J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Jang J.-C., Fujioka S., Tasaka M., Seto H., Takatsuto S., Ishii A., Aida M., Yoshida S., Sheen J. (2000). A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 14: 1485–1497 [PMC free article] [PubMed] [Google Scholar]
- Koornneef M., Alonso-Blanco C., Stam P. (2006). Genetic analysis. Methods Mol. Biol. 323: 65–77 [DOI] [PubMed] [Google Scholar]
- Kreft S.G., Hochstrasser M. (2011). An unusual transmembrane helix in the endoplasmic reticulum ubiquitin ligase Doa10 modulates degradation of its cognate E2 enzyme. J. Biol. Chem. 286: 20163–20174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft S.G., Wang L., Hochstrasser M. (2006). Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI). J. Biol. Chem. 281: 4646–4653 [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Learned R.M. (1996). Light suppresses 3-Hydroxy-3-methylglutaryl coenzyme A reductase gene expression in Arabidopsis thaliana. Plant Physiol. 110: 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Gusmaroli G., Charron J.B.F., Yoon H.J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Antolín-Llovera M., Ferrero S., Closa M., Arró M., Ferrer A., Boronat A., Campos N. (2011). Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell 23: 1494–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., González V.M., Castel S., Trelease R.N., López-Iglesias C., Arró M., Boronat A., Campos N., Ferrer A., Fernàndez-Busquets X. (2005). Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 137: 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovato M.A., Hart E.A., Segura M.J.R., Giner J.-L., Matsuda S.P.T. (2000). Functional cloning of an Arabidopsis thaliana cDNA encoding cycloeucalenol cycloisomerase. J. Biol. Chem. 275: 13394–13397 [DOI] [PubMed] [Google Scholar]
- Lü S., Zhao H., Des Marais D.L., Parsons E.P., Wen X., Xu X., Bangarusamy D.K., Wang G., Rowland O., Juenger T., Bressan R.A., Jenks M.A. (2012). Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras V., Campos N., Boronat A. (1995). The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 8: 541–549 [DOI] [PubMed] [Google Scholar]
- Manzano D., Fernández-Busquets X., Schaller H., González V.C., Boronat A., Arró M., Ferrer A. (2004). The metabolic imbalance underlying lesion formation in Arabidopsis thaliana overexpressing farnesyl diphosphate synthase (isoform 1S) leads to oxidative stress and is triggered by the developmental decline of endogenous HMGR activity. Planta 219: 982–992 [DOI] [PubMed] [Google Scholar]
- Masferrer A., Arró M., Manzano D., Schaller H., Fernández-Busquets X., Moncaleán P., Fernández B., Cunillera N., Boronat A., Ferrer A. (2002). Overexpression of Arabidopsis thaliana farnesyl diphosphate synthase (FPS1S) in transgenic Arabidopsis induces a cell death/senescence-like response and reduced cytokinin levels. Plant J. 30: 123–132 [DOI] [PubMed] [Google Scholar]
- McGarvey D.J., Croteau R. (1995). Terpenoid metabolism. Plant Cell 7: 1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meigs T.E., Roseman D.S., Simoni R.D. (1996). Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J. Biol. Chem. 271: 7916–7922 [DOI] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.-D., Hartmann M.-A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Newman J.D., Chappell J. (1999). Isoprenoid biosynthesis in plants: Carbon partitioning within the cytoplasmic pathway. Crit. Rev. Biochem. Mol. Biol. 34: 95–106 [DOI] [PubMed] [Google Scholar]
- Nicotra A.B., Atkin O.K., Bonser S.P., Davidson A.M., Finnegan E.J., Mathesius U., Poot P., Purugganan M.D., Richards C.L., Valladares F., van Kleunen M. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Nieto B., Forés O., Arró M., Ferrer A. (2009). Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry 70: 53–59 [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M., Ryan C.A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 96: 6553–6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D.R., Rasbery J.M., Bartel B., Matsuda S.P. (2006). Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9: 305–314 [DOI] [PubMed] [Google Scholar]
- Posé D., Castanedo I., Borsani O., Nieto B., Rosado A., Taconnat L., Ferrer A., Dolan L., Valpuesta V., Botella M.A. (2009). Identification of the Arabidopsis dry2/sqe1-5 mutant reveals a central role for sterols in drought tolerance and regulation of reactive oxygen species. Plant J. 59: 63–76 [DOI] [PubMed] [Google Scholar]
- Proost S., Van Bel M., Sterck L., Billiau K., Van Parys T., Van de Peer Y., Vandepoele K. (2010). PLAZA: A comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbery J.M., Shan H., LeClair R.J., Norman M., Matsuda S.P.T., Bartel B. (2007). Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J. Biol. Chem. 282: 17002–17013 [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., Jenks M.A., Ross A.S., Feldmann K.A. (2004). Novel eceriferum mutants in Arabidopsis thaliana. Planta 219: 5–13 [DOI] [PubMed] [Google Scholar]
- Räikkönen J., Mönkkönen H., Auriola S., Mönkkönen J. (2010). Mevalonate pathway intermediates downregulate zoledronic acid-induced isopentenyl pyrophosphate and ATP analog formation in human breast cancer cells. Biochem. Pharmacol. 79: 777–783 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Boronat A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M., Forés O., Martinez-García J.F., González V., Phillips M.A., Ferrer A., Boronat A. (2004). Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H. (2010). Sterol and steroid biosynthesis and metabolism in plants and microorganisms. Comprehensive Natural Products II Chemistry and Biology 1: 755–787 [Google Scholar]
- Schnell J.D., Hicke L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278: 35857–35860 [DOI] [PubMed] [Google Scholar]
- Schrick K., Fujioka S., Takatsuto S., Stierhof Y.-D., Stransky H., Yoshida S., Jürgens G. (2004). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38: 227–243 [DOI] [PubMed] [Google Scholar]
- Schrick K., Mayer U., Horrichs A., Kuhnt C., Bellini C., Dangl J., Schmidt J., Jürgens G. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14: 1471–1484 [PMC free article] [PubMed] [Google Scholar]
- Schulze S., Köster S., Geldmacher U., Terwisscha van Scheltinga A. C., Kühlbrandt W. (2010). Structural basis of Na(+)-independent and cooperative substrate/product antiport in CaiT. Nature 467: 233–236 [DOI] [PubMed] [Google Scholar]
- Smith M.H., Ploegh H.L., Weissman J.S. (2011). Road to ruin: Targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C., Donaghy P.G., Halford N.G., Hardie D.G. (1999). Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 120: 257–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kamide Y., Nagata N., Seki H., Ohyama K., Kato H., Masuda K., Sato S., Kato T., Tabata S., Yoshida S., Muranaka T. (2004). Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 37: 750–761 [DOI] [PubMed] [Google Scholar]
- Suzuki M., Nakagawa S., Kamide Y., Kobayashi K., Ohyama K., Hashinokuchi H., Kiuchi R., Saito K., Muranaka T., Nagata N. (2009). Complete blockage of the mevalonate pathway results in male gametophyte lethality. J. Exp. Bot. 60: 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R., Locher M., Hochstrasser M. (2001). A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 15: 2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld, C.L., Pourmand, D., Davis, T., Garza, R.M., and Hampton R.Y. (2011). The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem. 286: 26298–26307 [DOI] [PMC free article] [PubMed]
- Tholl, D., and Lee, S. (2011). Terpene specialized metabolism in Arabidopsis thaliana The Arabidopsis Book 9:e0143. doi:10.1043/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C.G.N., Booker J.P., Leyser H.M. (2002). Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. (2008). One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9: 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzinger L.F., Bach T.J., Hartmann M.A. (2002). Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme a reductase. Plant Physiol. 130: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Friml J., Grebe M., van den Toorn A., Palme K., Scheres B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Singh S.K., Kawate T., Jin Y., Gouaux E. (2005). Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437: 215–223 [DOI] [PubMed] [Google Scholar]