Interactions between the plant hormones salicylic acid and jasmonic acid play an important role in the regulation of plant defense responses against pathogens and insects. This work provides mechanistic insight into this hormonal crosstalk by showing that salicylic acid antagonizes jasmonic acid–dependent defenses by targeting the transcriptional activator ORA59.
Abstract
Antagonism between the defense hormones salicylic acid (SA) and jasmonic acid (JA) plays a central role in the modulation of the plant immune signaling network, but the molecular mechanisms underlying this phenomenon are largely unknown. Here, we demonstrate that suppression of the JA pathway by SA functions downstream of the E3 ubiquitin-ligase Skip-Cullin-F-box complex SCFCOI1, which targets JASMONATE ZIM-domain transcriptional repressor proteins (JAZs) for proteasome-mediated degradation. In addition, neither the stability nor the JA-induced degradation of JAZs was affected by SA. In silico promoter analysis of the SA/JA crosstalk transcriptome revealed that the 1-kb promoter regions of JA-responsive genes that are suppressed by SA are significantly enriched in the JA-responsive GCC-box motifs. Using GCC:GUS lines carrying four copies of the GCC-box fused to the β-glucuronidase reporter gene, we showed that the GCC-box motif is sufficient for SA-mediated suppression of JA-responsive gene expression. Using plants overexpressing the GCC-box binding APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factors ERF1 or ORA59, we found that SA strongly reduces the accumulation of ORA59 but not that of ERF1. Collectively, these data indicate that the SA pathway inhibits JA signaling downstream of the SCFCOI1-JAZ complex by targeting GCC-box motifs in JA-responsive promoters via a negative effect on the transcriptional activator ORA59.
INTRODUCTION
Within their natural habitats, plants intimately interact simultaneously or sequentially with a broad range of microbial pathogens and insect herbivores with different lifestyles and invasion strategies. The evolutionary arms race between plants and their enemies provided plants with a highly sophisticated immune system that recognizes the invaders and responds by activating effective defenses (Jones and Dangl, 2006). The plant hormones salicylic acid (SA) and jasmonic acid (JA) play key roles in the regulation of the defense signaling network that is recruited upon perception of an invader (Pieterse et al., 2012). The hormonal blend that is produced upon pathogen or insect attack, the so-called signal signature, varies significantly in quantity, composition, and timing and depends greatly on the lifestyle and invasion strategy of the attacker (De Vos et al., 2005). Although there are exceptions, in general it can be stated that pathogens with a biotrophic lifestyle are more sensitive to SA-induced defenses, whereas necrotrophic pathogens and herbivorous insects are resisted through JA-mediated defenses (Glazebrook, 2005; Howe and Jander, 2008). Other growth regulators, such as ethylene (ET), abscisic acid, gibberellins, auxins, cytokinins, and brassinosteroids, are also implicated in the regulation of the plant immune signaling network (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012), indicating that the regulation of plant growth and defense is tightly linked.
Besides balancing the relative abundance of different hormones, intensive interplay between hormone signaling pathways emerged as an important regulatory mechanism by which the plant is able to tailor its immune response to the type of invader encountered (Reymond and Farmer, 1998; Verhage et al., 2010). For instance, in Arabidopsis thaliana, transcriptome analyses of wild-type and mutant plants challenged with different attackers revealed complex antagonistic and synergistic regulatory relationships between SA and JA signaling sectors of the plant immune signaling network (Glazebrook et al., 2003; De Vos et al., 2005; Sato et al., 2010). Such hormonal crosstalk is thought to optimize the immune response against single attackers that stimulate both the SA and the JA pathway or to prioritize one pathway over the other when plants are simultaneously or sequentially attacked by different enemies (Pieterse et al., 2012; Thaler et al., 2012). Several other hormones, such as ET, abscisic acid, gibberellic acid, and auxin, antagonistically or synergistically interact with the SA and JA pathways (Robert-Seilaniantz et al., 2011), adding yet another layer of complexity to the plant immune signaling network. Interestingly, successful pathogens and insect herbivores have been demonstrated to hijack hormone signal integration, either through the production of plant hormones, hormone mimics, or effectors that target hormone signaling components to manipulate the plant immune signaling network for their own benefit (Walling, 2008; Grant and Jones, 2009; Pieterse et al., 2012).
The SA and JA signaling sectors often act antagonistically. For instance, the JA-mimicking phytotoxin coronatine, which is produced by virulent Pseudomonas syringae bacteria, promotes virulence by suppressing effectual SA-dependent defenses in Arabidopsis and tomato (Solanum lycopersicum) (Brooks et al., 2005; Zheng et al., 2012). Conversely, many studies have demonstrated that endogenously accumulating SA antagonizes JA-dependent defenses, thereby prioritizing SA-dependent resistance over JA-dependent defense (Pieterse et al., 2012). Pharmacological experiments with Arabidopsis revealed that JA-responsive marker genes, such as PLANT DEFENSIN 1.2 (PDF1.2) and VEGETATIVE STORAGE PROTEIN2 (VSP2) are highly sensitive to suppression by SA (van Wees et al., 1999; Spoel et al., 2003). This antagonism between SA and JA signaling was observed in a large number of Arabidopsis accessions (Koornneef et al., 2008b), highlighting the potential significance of SA/JA crosstalk in nature. Many reports describe an antagonistic interaction between the SA and JA pathways, but synergistic interactions have been reported as well (Mur et al., 2006). Clearly, the kinetics of hormone production and signaling during the interaction of a plant with its enemies is highly decisive in the final outcome of the defense response (Koornneef et al., 2008b; Leon-Reyes et al., 2010a).
In Arabidopsis, the defense regulatory protein NONEXPRESSOR OF PR GENES1 (NPR1) was identified as a key signaling node in the regulation of SA/JA crosstalk because in mutant npr1-1 plants, the antagonistic effect of SA on PDF1.2 and VSP2 transcription was completely abolished (Spoel et al., 2003; Leon-Reyes et al., 2009). Several other molecular players in SA/JA crosstalk have been identified, including the mitogen-activated protein kinase MPK4 (Petersen et al., 2000), the lipase-like proteins ENHANCED DISEASE SUSCEPTIBILITY1 and PHYTOALEXIN-DEFICIENT4 (Brodersen et al., 2006), the fatty acid desaturase SUPPRESSOR OF SA INSENSITIVITY2 (Kachroo et al., 2003), glutaredoxin GRX480 (Ndamukong et al., 2007; Zander et al., 2010), and class II TGA and WRKY transcription factors (Li et al., 2004; Mao et al., 2007; Ndamukong et al., 2007; Leon-Reyes et al., 2010a; Zander et al., 2010; Gao et al., 2011). Mutation or ectopic expression of the corresponding genes often have contrasting effects on SA and JA signaling and on resistance against biotrophs and necrotrophs, indicating that these proteins are important regulators of SA/JA crosstalk. Although several regulatory proteins of SA/JA crosstalk have been identified, the molecular mechanism by which SA exerts its antagonistic effect on the JA pathway is still largely unknown.
JA and its structurally related oxylipin derivatives (collectively called jasmonates [JAs]) are lipid-derived compounds that upon pathogen or insect attack are rapidly synthesized via the oxylipin biosynthesis pathway (Gfeller et al., 2010). Previously, it was shown that SA-mediated suppression of the JA response is targeted at a position downstream of the JA biosynthesis pathway (Leon-Reyes et al., 2010b). Therefore, in this study, we set out to systematically scan the JA signaling pathway for potential targets of SA-mediated antagonism.
The JA signaling pathway is relatively well studied. Upon production, JA is rapidly conjugated to Ile via the activity of the JA conjugate synthase JAR1 (Staswick and Tiryaki, 2004), resulting in the biologically highly active form (+)-7-iso-jasmonoyl-l-Ile (JA-Ile) (Fonseca et al., 2009). The F-box protein CORONATINE INSENSITIVE1 (COI1) functions as a key regulator of JA signaling (Xie et al., 1998). coi1-1 mutant plants are unresponsive to JAs and show alterations in the level of resistance to different necrotrophic pathogens and insect herbivores (Van der Ent et al., 2009). COI1 is part of the E3 ubiquitin-ligase Skip-Cullin-F-box complex SCFCOI1 and functions together with JASMONATE ZIM domain (JAZ) transcriptional repressor proteins as a JA-Ile receptor (Browse, 2009; Yan et al., 2009; Sheard et al., 2010). Binding of JA-Ile to COI1 leads to ubiquitination and subsequent degradation of JAZ repressor proteins via the proteasome (Chini et al., 2007; Thines et al., 2007; Chung et al., 2009; Pauwels and Goossens, 2011). In resting cells, JAZ proteins act as transcriptional repressors of JA signaling by binding to positive transcriptional regulators, such as MYC2, MYC3, and MYC4 (Chini et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011), ETHYLENE INSENSITIVE3 (EIN3), and EIN3-LIKE1 (EIL1) (Zhu et al., 2011).
In Arabidopsis, the JAZ family of repressor proteins consists of 12 members with a similar structure containing a C-terminal Jas motif and an N-terminal ZIM domain (Chini et al., 2007; Thines et al., 2007). The Jas motif is important for interactions with COI1, the MYC transcription factors, and EIN3/EIL1 (Melotto et al., 2008; Chini et al., 2009; Zhu et al., 2011) and is required for JAZ protein breakdown upon perception of JA-Ile (Chung and Howe, 2009). In line with this, ectopically expressed JAZ proteins that lack the Jas domain are not targeted for proteasome-mediated degradation, resulting in a strong JA-insensitive phenotype (Chung and Howe, 2009). The N-terminal ZIM domain is important for mediating homo- and heterodimeric interactions between JAZ proteins (Chini et al., 2009; Chung and Howe, 2009). In addition, the ZIM domain interacts with NOVEL INTERACTOR OF JAZ that through its ETHYLENE RESPONSE FACTOR (ERF)-ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif, recruits the Groucho/Tup1-type corepressor TOPLESS, thereby preventing untimely activation of the JA pathway (Pauwels et al., 2010; Pauwels and Goossens, 2011). Also HISTONE DEACETYLASE6, which interacts with JAZ and EIN3/EIL1 transcription factor proteins, acts as a corepressor of the JA pathway (Zhu et al., 2011). In stimulated cells, the physical interaction between JAZ proteins, corepressors, and transcriptional activators is broken, which results in derepression of the JA pathway and activation of a large number of JA-responsive genes (Memelink, 2009; Pauwels and Goossens, 2011).
In Arabidopsis, two major branches of the JA signaling pathway are recognized: the MYC branch and the ERF branch. The MYC branch is controlled by MYC-type transcription factors that can bind to the G-box motif (CACGTG) and regulate expression of the marker gene VSP2 (Lorenzo et al., 2004; Dombrecht et al., 2007; Fernández-Calvo et al., 2011; Niu et al., 2011). The ERF branch, which requires both JA and ET signaling, is regulated by members of the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) family of transcription factors that bind to the GCC-box motif (AGCCGCC), such as ERF1 and ORA59 (for OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF domain protein 59), and regulate expression of the marker gene PDF1.2 (Lorenzo et al., 2003; Pré et al., 2008; Zhu et al., 2011). In general, the ERF branch is associated with enhanced resistance to necrotrophic pathogens (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003), whereas the MYC branch is associated with the wound response and defense against insect herbivores (Lorenzo et al., 2004; Kazan and Manners, 2008; Verhage et al., 2011).
Here, we provide evidence that SA suppresses the JA signaling pathway downstream of the SCFCOI1-JAZ machinery. Moreover, we show that the GCC-box motif, which is a binding site for AP2/ERF-type transcription factors such as ERF1 and ORA59, is overrepresented in JA-responsive promoters that are suppressed by SA and that this promoter motif is sufficient for SA-mediated suppression of JA-induced gene expression. Finally, we provide evidence that SA exerts a negative effect on the accumulation of the transcription factor ORA59, indicating that the antagonistic effect of SA on JA signaling is controlled at the level of transcriptional regulation.
RESULTS
SA-Mediated Antagonism of JA Signaling Is Not Targeted at JAZ Repressor Proteins
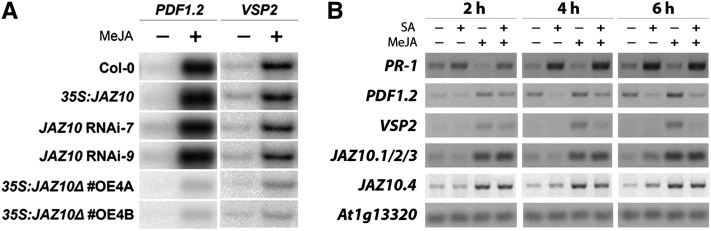
Since JAZ proteins are important negative regulators in the JA signaling pathway (Browse, 2009; Chung and Howe, 2009), they form a potential target for SA-mediated suppression of the JA response. Previously, a natural alternatively spliced form of JAZ10 (JAZ10.4) was identified that lacks the Jas domain and is therefore highly resistant to JA-induced degradation (Chung and Howe, 2009). We hypothesized that SA may antagonize the JA pathway by stimulating the production of this stable splice variant of JAZ10. First, we checked whether overexpression of JAZ10 without a functional Jas domain would lead to suppression of the JA pathway in our experimental setup. To this end, 5-week-old plants of wild-type Columbia-0 (Col-0) and 35S:JAZ10Δ #OE4A and 35S:JAZ10Δ #OE4B, which overexpress a truncated form of JAZ10 that lacks part of the Jas domain (Yan et al., 2007), were treated with methyl jasmonate (MeJA). As a control, the JAZ10 overexpressing line 35S:JAZ10 and the JAZ10 silenced lines JAZ10 RNAi-7 and JAZ10 RNAi-9 (Yan et al., 2007) were similarly treated. Twenty-four hours later, transcript levels of the marker genes PDF1.2 and VSP2 were assessed. Figure 1A shows that PDF1.2 and VSP2 transcripts accumulated to wild-type levels in 35S:JAZ10 and the JAZ10 RNA interference lines, confirming previous findings that increased or reduced abundance of one member of the JAZ protein family does not affect the JA response (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). However, exogenous application of MeJA to 35S:JAZ10Δ #OE4A and 35S:JAZ10Δ #OE4B resulted in only a weak activation of both PDF1.2 and VSP2 (Figure 1A), indicating that production of a single dominant-negative form of JAZ10 is in principle sufficient to suppress the MYC and the ERF branch of the JA pathway. To investigate whether SA differentially affects the accumulation of the four JAZ10 splice forms, we monitored JAZ10.1, JAZ10.2, JAZ10.3, and JAZ10.4 mRNA levels in 5-week-old Col-0 plants at different time points after treatment with SA, MeJA, or the combination of both. To this end, we used one set of primers specific for JAZ10.1, JAZ10.2, and JAZ10.3 and one set of primers specific for JAZ10.4. Figure 1B shows that, while SA-responsive PATHOGENESIS RELATED-1 (PR-1) gene expression was upregulated, MeJA-induced expression of PDF1.2 and VSP2 was suppressed after application of SA. In addition, MeJA induced the expression of JAZ10.1/2/3 at all time points tested. In contrast with PDF1.2 and VSP2, JAZ10.1/2/3 transcript levels were not affected by SA. The dominant-negative splice form JAZ10.4 followed a similar expression pattern as JAZ10.1/2/3 in all treatments. It can thus be concluded that SA does not suppress the JA pathway by overstimulating the production of the dominant-negative splice variant JAZ10.4.
Figure 1.
SA-Mediated Suppression of JA-Dependent Transcription Is Not Mediated via Production of a Dominant-Negative Splice Variant of JAZ10.
(A) MeJA-induced expression of PDF1.2 and VSP2 in JAZ10-modified transgenic lines. RNA gel blot analysis of PDF1.2 and VSP2 transcript levels in 5-week-old Col-0, 35S:JAZ10, JAZ10 RNAi-7, JAZ10 RNAi-9, 35S:JAZ10∆ #OE4A, and 35S:JAZ10∆ #OE4B plants. Leaves were harvested 24 h after treatment with 0.1 mM MeJA. Equal loading of RNA samples was confirmed using a probe for 18S rRNA but not included in the figure.
(B) RT-PCR to monitor expression of PR-1, PDF1.2, VSP2, and different JAZ10 splice variants in 5-week-old Col-0 plants after treatment with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Leaf tissue was harvested at indicated time points after treatment. Equal input was checked by monitoring the constitutively expressed gene At1g13320.
SA Does Not Inhibit Degradation of JAZ Proteins
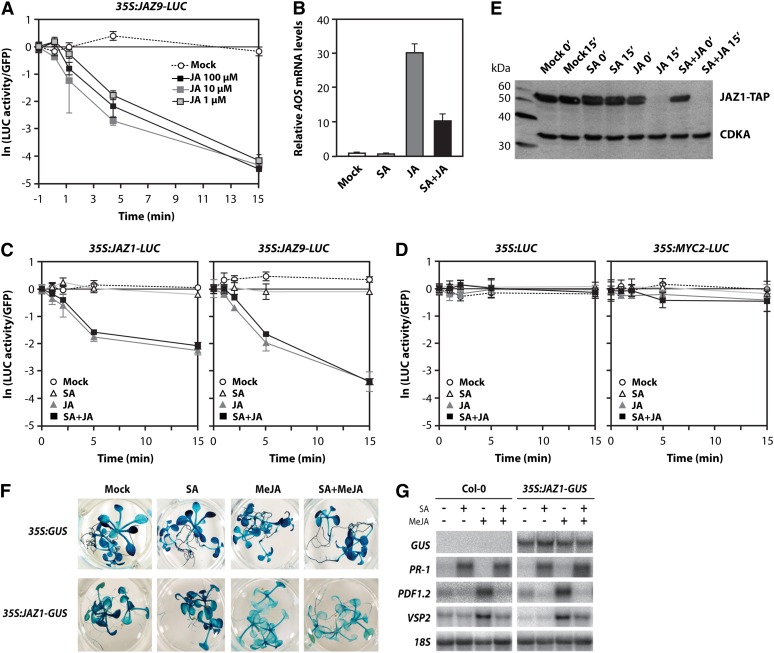
In the auxin signaling pathway, SA was shown to repress auxin signaling by stabilizing auxin repressor proteins (Wang et al., 2007). We hypothesized that a similar mechanism involving JAZ repressor proteins may play a role in SA/JA antagonism. To investigate this, we monitored the effect of SA on JA-induced degradation of JAZ proteins. We performed assays with stable Arabidopsis cell cultures overexpressing either JAZ1 or JAZ9 fused to the firefly luciferase-encoding reporter gene LUC. First, we checked the optimal JA concentration for the initiation of JAZ protein degradation in this assay. Therefore, an Arabidopsis cell suspension culture expressing 35S:JAZ9-LUC was treated with 0.001, 0.01, or 0.1 mM JA (Figure 2A). After 1, 2, 5, and 15 min, samples were taken and protein was isolated for LUC activity measurements. After application of JA, the amount of LUC activity decreased rapidly, indicating that JAZ proteins were quickly degraded (a half-life of ∼2.5 min), in contrast with the control where no degradation was observed (Figure 2A). To verify whether SA/JA crosstalk occurs in the Arabidopsis cell cultures within the time frame of 15 min in which the JAZ degradation assay was performed, we analyzed the expression of the early JA-responsive ALLENE OXIDE SYNTHASE (AOS) gene in cell cultures that were treated with SA and JA. Figure 2B shows that JA induced AOS transcription within 15 min after JA application. In the combination treatment, the AOS mRNA level was significantly suppressed, indicating that SA/JA crosstalk was functional in the cell culture assay. Next, we wanted to test the effect of SA on JAZ protein degradation. Therefore, we treated cell culture lines 35S:JAZ1-LUC and 35S:JAZ9-LUC with SA and JA and measured LUC activity at different time points after chemical treatment. Figure 2C shows that SA treatment had no effect on the stability of JAZ1-LUC and JAZ9-LUC. Moreover, degradation of JAZ1-LUC and JAZ9-LUC was similar upon application of JA or a combination of JA and SA, suggesting that SA has no effect on the JA-mediated degradation of these JAZ proteins. Similar results were obtained with 35S:JAZ2-LUC (see Supplemental Figure 1 online). To confirm that application of SA or JA did not interfere with the LUC reporter system, we tested two controls in our assays: 35S:LUC and 35S:MYC2-LUC. Neither JA nor SA affected LUC activity in these control lines, indicating that hormonal treatment did not affect the LUC reporter system per se (Figure 2D).
Figure 2.
JA-Mediated Degradation of JAZ Proteins Is Not Affected by SA.
(A) LUC activity in Arabidopsis cell suspension cultures expressing 35S:JAZ9-LUC and 35S:GFP, 0, 1, 2, 5, and 15 min after application of 0.1, 0.01, or 0.001 mM of JA. LUC activities were normalized to GFP internal controls. Three independent protein samples were measured per time point. The natural logarithm (ln) of the normalized LUC activities is depicted. Error bars represent standard errors (±se).
(B) Quantitative RT-PCR analysis of AOS mRNA levels in Arabidopsis cells 15 min after treatment with 0.001 mM JA, 0.01 mM SA, or a combination of these chemicals. The average of three replicas is depicted (±se).
(C) and (D) Luciferase activity in Arabidopsis cells expressing 35S:JAZ1-LUC, 35S:JAZ9-LUC, 35S:LUC, or 35S:MYC2-LUC. Cells were treated with 0.01 mM SA and 0.001 mM JA, which was applied 3 h after SA in the combination treatment. Samples were taken 0, 1, 2, 5, and 15 min after application of JA. LUC activities were normalized to GFP internal controls. Three independent protein samples were measured per time point. The natural logarithm of the normalized LUC activities is depicted (±se).
(E) Immunoblot analysis of the TAP-tagged JAZ1 protein levels in Arabidopsis cells expressing 35S:JAZ1-TAP, 0 and 15 min after treatment of the cells with 0.01 mM SA, 0.001 mM JA, or the combination of both chemicals. JAZ1-TAP was detected using an anti-PAP antibody. The constitutively expressed Cdc2 kinase (CDKA) served as an internal control and was detected with an anti-CDKA antibody.
(F) Histochemical staining of GUS activity in 2-week-old 35S:JAZ1-GUS and 35S:GUS seedlings. Twelve-day-old seedlings grown on MS agar plates were transferred to fresh medium containing 0.5 mM SA, 0.02 mM MeJA, or a combination of both chemicals and stained for GUS activity 24 h later.
(G) RNA gel blot analysis of GUS, PR-1, PDF1.2, and VSP2 transcript levels in 2-week-old Col-0 and 35S:JAZ1-GUS plants that were treated with 0.5 mM SA, 0.02 mM MeJA, or a combination of both chemicals. Leaf tissue was harvested 24 h after chemical treatment for RNA analysis. Equal loading of RNA samples was checked using a probe for 18S rRNA.
[See online article for color version of this figure.]
In addition to the LUC reporter assay, we assessed the stability of JAZ1 by immunoblot analysis. A cell culture expressing 35S:JAZ1-TAP (for tandem affinity protein) was treated with SA, JA, or a combination of both chemicals. Samples were taken just prior to treatment (time point 0) and 15 min after treatment. Figure 2E shows that JAZ1-TAP was fully degraded 15 min after treatment of the cells with either JA or a combination of JA and SA. These results confirm that SA does not affect the JA-responsive degradation of JAZ proteins in Arabidopsis cell suspension cultures.
To check whether JA-mediated degradation of JAZ proteins is also not affected by SA in intact plants, we made use of the transgenic line 35S:JAZ1-GUS (for β-glucuronidase; Thines et al., 2007). Twelve-day-old 35S:GUS and 35S:JAZ1-GUS seedlings grown on Murashige and Skoog (MS) agar plates were transferred to fresh medium containing SA, MeJA, or a combination of both chemicals and stained for GUS activity 24 h later. Treatment of 35S:JAZ1-GUS seedlings with MeJA resulted in reduced GUS staining (Figure 2F), suggesting that the JAZ1-GUS protein was degraded upon induction of the JA pathway (confirming previous findings; Thines et al., 2007). Inclusion of SA in the medium had no effect on the MeJA-mediated degradation of JAZ1-GUS. Analysis of GUS, PR-1, PDF1.2, and VSP2 transcript levels in these seedlings showed that SA-mediated suppression of PDF1.2 and VSP2 gene expression was fully active (Figure 2G). Taken together, we conclude that the antagonistic effect of SA on JA signaling is not acting through the stabilization of JAZ proteins.
SA Antagonizes JA Signaling Downstream of SCFCOI1
The E3 ubiquitin-ligase SCFCOI1 complex plays a crucial role in the JA signaling pathway. Several Arabidopsis mutants with defects in one of the proteins in the SCFCOI1 complex show a reduced response to JA; moreover, the coi1-1 mutation of the JA receptor component COI1 completely abolishes JA-dependent responses (Feys et al., 1994; Xie et al., 1998; Devoto and Turner, 2005; Yang et al., 2009; Sheard et al., 2010). Though JAZ repressor proteins are not likely to be direct targets of SA in suppression of JA signaling, we cannot rule out that SA targets the SCFCOI1 complex, which functions upstream of JAZ. Therefore, to investigate whether this complex is a target for SA in suppression of the JA pathway, we made use of a transgenic line that overexpresses the AP2/ERF transcription factor ERF1 in the background of coi1-1 plants. In the JA-insensitive coi1-1 mutant, JA-dependent gene expression is completely blocked, but overexpression of ERF1 in this mutant background results in activation of PDF1.2 expression (Lorenzo et al., 2003), indicating that ERF1 is an activator of PDF1.2 transcription. We treated Col-0, coi1-1, 35S:ERF1, and 35S:ERF1/coi1-1 plants with SA, MeJA, or the combination of both and harvested the plants 24 h later. As expected, SA treatment resulted in the activation of PR-1 in all genotypes tested (Figure 3). PDF1.2 always shows a basal level of expression in our system (probably due to basal ET production), but MeJA treatment increased PDF1.2 expression in Col-0, and this induction could be suppressed by SA. By contrast, PDF1.2 expression was not visible in coi1-1 mutant plants under any of the conditions tested. Overexpression of ERF1 resulted in enhanced PDF1.2 expression upon mock treatment in both the Col-0 and coi1-1 genetic background, and MeJA treatment boosted the level of PDF1.2 transcription in plants expressing 35S:ERF1 in Col-0 background but did not further enhance PDF1.2 expression in 35S:ERF1/coi1-1 plants. Importantly, SA suppressed 35S:ERF1-mediated PDF1.2 transcription both in Col-0 and coi1-1 background, indicating that SA-mediated suppression of JA signaling acts downstream of the SCFCOI1 complex.
Figure 3.
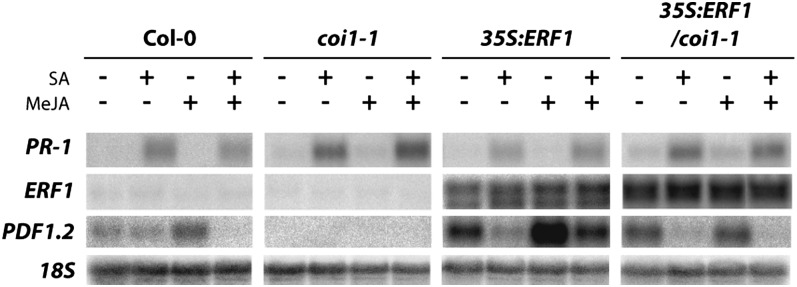
SA Can Suppress JA-Dependent Gene Expression Independent of SCFCOI1.
RNA gel blot analysis of PR-1, ERF1, and PDF1.2 expression in Col-0, coi1-1, 35S:ERF1, and 35S:ERF1/coi1-1 plants 24 h after treatment with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Equal loading of RNA samples was checked using a probe for 18S rRNA.
The SA/JA Crosstalk Transcriptome
The aforementioned results suggest that SA/JA crosstalk is predominantly regulated at the transcriptional level. To gain insight into the regulation and complexity of SA/JA crosstalk, we took a whole-genome transcript profiling approach to identify JA-responsive genes that are sensitive to SA-mediated suppression and to subsequently search for crosstalk-related cis-acting elements in the promoters of JA-responsive genes that are suppressed by SA. Three similar but fully independent SA/JA crosstalk experiments were performed with 5-week-old Col-0 plants that were treated with SA, MeJA, or a combination of both chemicals. The expression of the marker genes PR-1 and PDF1.2 was assessed in each biological replicate by RNA gel blot analysis. In all three experiments, SA induced PR-1 expression and suppressed MeJA-induced expression of PDF1.2 (see Supplemental Figure 2A online).
The transcript profile of each independent experiment was analyzed using Affymetrix ATH1 whole-genome GeneChips. A robust set of MeJA-responsive genes was identified by selecting genes that were statistically significant up- or downregulated in MeJA-treated plants compared with the mock-treated control (Student’s t test; P < 0.01), with an additional cutoff value of twofold. These selection criteria were met by 175 genes that were upregulated upon MeJA treatment (see Supplemental Data Set 1 online). Among these were genes involved in JA biosynthesis (LOX2, AOS, and OPR3), JA signal transduction (ERF6, ERF104, JAZ1, JAZ5, JAZ6, JAZ7, and JAZ9), and JA-dependent defenses (PDF1.2, Thi2.1, and VSP1). In addition, a group of 40 genes was downregulated by MeJA (see Supplemental Data Set 1 online). For SA-responsive genes, a similar selection procedure was followed, resulting in 50 SA-upregulated genes (including PR-1, WRKY18, WRKY38, and NIMIN1) and 15 SA-downregulated genes (see Supplemental Data Set 1 online).
To select for MeJA-induced genes that were suppressed by SA, we identified MeJA-upregulated genes that were at least 1.5-fold repressed by the combined treatment with SA and MeJA, compared with MeJA alone. In addition, we selected MeJA-downregulated genes that were significantly upregulated by SA and MeJA, compared with MeJA alone. These selection criteria resulted in the identification of 59 MeJA-inducible genes that were suppressed by SA and 15 MeJA-downregulated genes that were upregulated by SA (see Supplemental Data Set 2 online). Overall, 34% of all selected MeJA-responsive genes were affected by SA/JA crosstalk. Among the MeJA-inducible genes that were suppressed by SA were those encoding defense-related proteins PDF1.2a and PDF1.2b, confirming previous findings (van Wees et al., 1999; Spoel et al., 2003, 2007; Ndamukong et al., 2007; Leon-Reyes et al., 2009; Zander et al., 2010).
Promoter Analysis of MeJA-Responsive Genes That Are Suppressed by SA
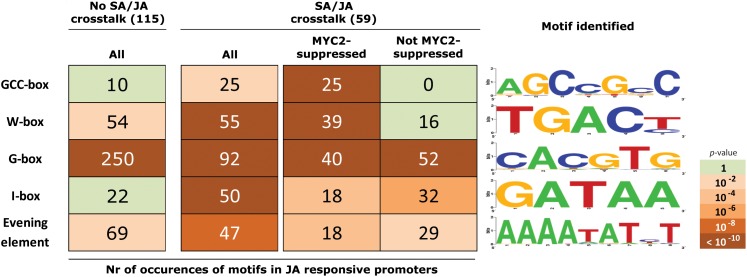
To search for cis-acting motifs with a putative role in SA/JA crosstalk, we performed an in silico analysis of the promoter sequences of the selected MeJA-inducible genes that, like the PDF1.2 marker gene, were suppressed by SA in the SA/MeJA combination treatment. We scanned the 1-kb sequences upstream of the 5′-untranslated regions of the 59 MeJA-inducible genes that were suppressed by SA (see Supplemental Data Set 2 online) using the method described by Breeze et al. (2011). Several promoter motifs were found significantly enriched in the promoters of the group of 59 SA/JA crosstalk genes: the GCC-box (AGCCGCC), the W-box (TGACY), the G-box (CACGTG), the I-box (GATAA), and the evening element (AAAATATCT) (Figure 4). The GCC-box and W-box motifs were found significantly enriched only in genes that are likely regulated by the ERF branch of the JA pathway (as their MeJA-activated expression level was increased in MYC2-impaired jin1-9 mutants compared with Col-0 plants; Dombrecht et al., 2007), whereas the G-box, the I-box, and the evening element were found significantly enriched in genes from both branches of the JA pathway (Figure 4). Together, these results point to specific roles for these motifs in SA-mediated suppression of different sets of JA-responsive genes. In the promoters of the 115 MeJA-inducible genes that were not suppressed by SA, the GCC-box and the I-box motifs were not overrepresented (Figure 4), suggesting that these elements are specifically targeted during SA/JA crosstalk.
Figure 4.
Promoter Elements Enriched in MeJA-Responsive Genes That Are Antagonized by SA.
Enriched promoter elements in the promoter sequences of the group of 59 SA/JA crosstalk genes that were upregulated by MeJA and suppressed by SA in the SA/MeJA combination treatment. The occurrence of enriched motifs was determined in the 1-kb sequences upstream of the 5′-untranslated regions. The set of 59 SA/JA crosstalk promoters was split into those that are suppressed by MYC2 and those that are not (Dombrecht et al., 2007). For comparison, the occurrence of the promoter motifs is also given for the 115 genes that were upregulated by MeJA but not suppressed by SA in the SA/MeJA treatment (No SA/JA crosstalk). Numbers represent the total number of occurrences of the given motif within the indicated set. The corresponding enrichment P values are color coded (green: not significant). The motif as found within all crosstalk promoters is depicted using Weblogo (Crooks et al., 2004).
Previously, PDF1.2 promoter deletion constructs fused to the GUS reporter gene were tested for their ability to show SA/JA crosstalk (Spoel et al., 2003). A truncated PDF1.2 promoter that consisted of only 311 bp upstream of the ATG start codon was sufficient for SA/JA crosstalk. Interestingly, this part of the PDF1.2 promoter includes the I-box and the GCC-box (Spoel et al., 2003). Site-directed mutagenesis of the I-box in the PDF1.2 promoter did not alter its response to SA, JA, or both chemicals, indicating that the I-box motif is not an essential regulatory element in the SA/JA antagonism (see Supplemental Figure 3 online). Therefore, we further focused our investigations on the involvement of the GCC-box motif in the regulation of SA/JA crosstalk.
The GCC-Box Motif Is Sufficient for SA-Mediated Suppression of JA-Responsive Gene Transcription
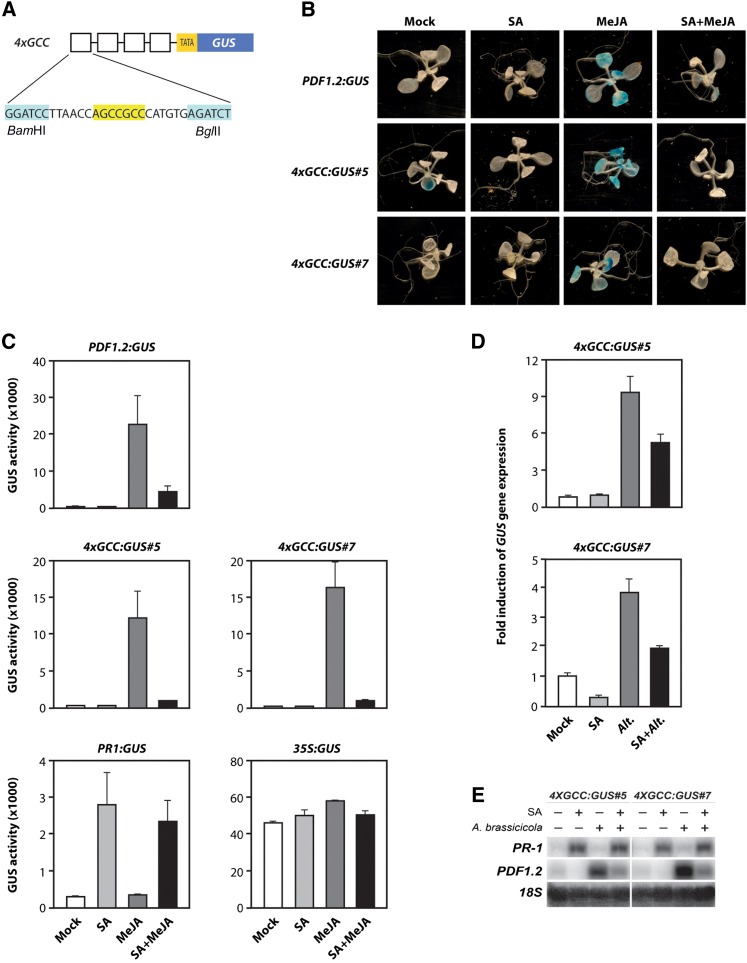
The GCC-box in the PDF1.2 promoter was shown to be required and sufficient for induction of gene expression by MeJA and ET (Brown et al., 2003; Zarei et al., 2011). Therefore, it provides an attractive target for SA-mediated suppression of JA-responsive gene expression. To test this, we used transgenic 4xGCC:GUS lines containing four copies of the GCC-box fused to a minimal 35S promoter and the GUS reporter gene (Figure 5A). Two-week-old seedlings grown on MS agar plates were transferred to MES buffer containing SA, MeJA, or a combination of both chemicals. In addition, a low dose of the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (0.002 mM) was added to all treatments to enhance the expression of PDF1.2:GUS and GCC:GUS. Twenty-four hours later, samples were taken for the analysis of GUS activity. PDF1.2:GUS, 4xGCC:GUS line #5, and 4xGCC:GUS line #7 showed induced GUS activity after application of MeJA (Figure 5B), confirming previous findings (Zarei et al., 2011). Interestingly, SA was able to suppress the induction of the 4xGCC promoter by MeJA. Quantitative analysis of GUS activity in PDF1.2:GUS and the 4XGCC:GUS lines yielded similar results: The PDF1.2 and the 4XGCC promoters were activated by MeJA and suppressed by SA (Figure 5C). The SA-responsive PR-1:GUS line and the 35S:GUS line were used as controls (Figure 5C). Together, these results indicate that the GCC-box is sufficient for both transcriptional activation by MeJA and suppression by SA.
Figure 5.
The GCC-Box Is Sufficient for SA-Mediated Suppression of JA-Induced Gene Expression.
(A) Schematic representation of the 4XGCC:GUS construct.
(B) Histochemical staining of GUS activity in 2-week-old seedlings of PDF1.2:GUS, 4XGCC:GUS line #5, and 4XGCC:GUS line #7. Two-week-old seedlings grown on MS agar plates were transferred to MES buffer solution containing 0.5 mM SA, 0.1 mM MeJA, or a combination of both chemicals. All treatments contained 0.002 mM ACC. Plants were stained for GUS activity 24 h later.
(C) Quantitative analysis of GUS activity in 2-week-old seedlings of PDF1.2:GUS, 4XGCC:GUS#5, 4XGCC:GUS#7, PR-1:GUS, and 35S:GUS. Error bars represent ± se of three independent biological replicates.
(D) Analysis of GUS gene expression in 5-week-old 4XGCC:GUS#5 and 4XGCC:GUS#7 plants that were mock treated or inoculated with the fungus A. brassicicola (Alt.) and dipped in 1 mM SA 24 h later. Leaf tissue was harvested 24 h after treatment with SA for the analysis of GUS expression. Error bars represent ± se of three replicates.
(E) RNA gel blot analysis of PR-1 and PDF1.2 transcription in the same plant material as used in (D). Equal loading of RNA samples was checked using a probe for 18S rRNA.
To further substantiate this finding, we investigated whether SA is able to suppress the activation of the 4XGCC promoter when induced by a pathogen. Therefore, we inoculated 4XGCC:GUS lines #5 and #7 with the necrotrophic pathogen Alternaria brassicicola and treated half of the plants with SA 24 h later. Plants were harvested for gene expression analysis 24 h after SA treatment. A. brassicicola induced the expression of both 4xGCC:GUS (Figure 5D) and PDF1.2 (Figure 5E). In addition, SA suppressed pathogen-induced 4xGCC:GUS and PDF1.2 expression, corroborating our finding that the GCC-box is sufficient for SA-mediated suppression of JA-induced gene expression.
SA Targets ORA59 Accumulation
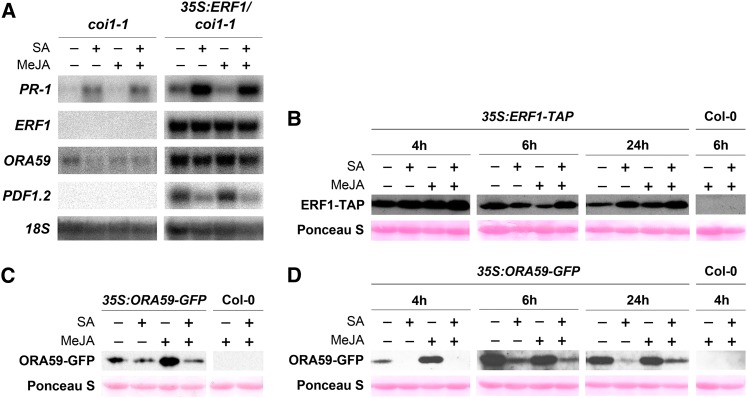
The GCC-box is a binding site for members of the family of AP2/ERF transcription factors (Hao et al., 1998), such as ERF1 and ORA59, which both have an important role in activation of PDF1.2 (Lorenzo et al., 2003; Pré et al., 2008; Zarei et al., 2011). Since suppression of PDF1.2 expression by SA can take place downstream of COI1, and independent of ERF1 mRNA accumulation (Figure 3), we hypothesized that SA/JA crosstalk could be mediated through reduction of ORA59 mRNA levels. To investigate this, ORA59 transcription was monitored in plants ectopically expressing 35S:ERF1 in the coi1-1 mutant background. Like PDF1.2, ORA59 transcription was activated by 35S:ERF1 in the coi1-1 background (Figure 6A). However, in contrast with 35S:ERF1-mediated PDF1.2 transcription, 35S:ERF1-induced ORA59 expression was not suppressed by SA. Hence, SA-mediated suppression of PDF1.2 expression can function independently of both ERF1 and ORA59 mRNA accumulation.
Figure 6.
SA Negatively Affects ORA59 Protein Accumulation.
(A) RNA gel blot analysis of PR-1, ERF1, ORA59, and PDF1.2 expression in coi1-1 and 35S:ERF1/coi1-1 plants 5 h after treatment with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Equal loading of RNA samples was checked using a probe for 18S rRNA.
(B) Immunoblot analysis of the TAP-tagged ERF1 protein levels in 5-week-old 35S:ERF1-TAP plants treated with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals and harvested at indicated times after treatment. Wild-type Col-0 was included as a negative control. ERF1-TAP was detected using an anti-PAP antibody. Ponceau S–stained ribulose-1,5-bisphosphate carboxylase/oxygenase was included as a loading control.
(C) and (D) immunoblot analysis of the GFP-tagged ORA59 protein levels in 11-d-old 35S:ORA59-GFP plants treated with 0.5 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Plants were harvested at 6 h after treatment (C) or 4, 6, and 24 h after treatment (D). Wild type Col-0 was included as a negative control. ORA59-GFP was detected using an anti-GFP antibody. Ponceau S–stained ribulose-1,5-bisphosphate carboxylase/oxygenase was included as a loading control.
[See online article for color version of this figure.]
Next, it was postulated that SA could interfere with the production or stability of ERF1 or ORA59. To investigate this, 5-week-old 35S:ERF1-TAP plants were treated with SA, MeJA, or a combination of both hormones, and accumulation of ERF1-TAP was assessed. Figure 6B shows that none of the hormone treatments affected the accumulation of ERF1-TAP in comparison to the control treatment. Ectopic expression of 35S:ERF1-TAP constitutively activated PDF1.2, which was hyperinduced by MeJA and suppressed by SA (see Supplemental Figure 4A online), confirming the findings presented in Figure 3. These results indicate that ERF1 accumulation is not a target for SA-mediated suppression of the JA pathway.
Next, we tested the effect of SA and/or MeJA on the accumulation of ORA59. To this end, 11-d-old 35S:ORA59-GFP (for green fluorescent protein) plants were treated with SA, MeJA, or a combination of both hormones, after which ORA59-GFP protein levels were assessed. Markedly, SA had a negative effect on accumulation of the ORA59-GFP protein (Figure 6C). Similar results were obtained in a time-course experiment, in which SA had a negative effect on accumulation of ORA59-GFP at all time points tested (Figure 6D). Contrary to 35S:ERF1-TAP plants in which PDF1.2 was strongly suppressed by SA, such a negative effect of SA on 35S:ORA59-GFP–driven PDF1.2 expression was not observed in 35S:ORA59-GFP plants (see Supplemental Figure 4B online). This is in line with findings that showed that overexpression of ORA59 can overrule the ability of SA to suppress JA-induced gene expression (Leon-Reyes et al., 2010a). In two out of five experiments, we did not detect the negative effect of SA on ORA59-GFP accumulation, suggesting that this effect can be modulated by so far unidentified environmental conditions. Overall, these results indicate that SA can negatively affect ORA59 protein accumulation, which can provide an explanation for the antagonistic effect of SA- on JA-responsive gene expression in wild-type plants.
DISCUSSION
Plant immunity is regulated by a complex network of cross-communicating signaling pathways. The plant hormones SA and JA play a crucial role in controlling plant defenses that are triggered after pathogen or insect attack. The SA and JA signaling pathways are often mutually antagonistic, but the outcome of the signal interaction greatly depends on the context in which they are activated (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012; Thaler et al., 2012). In this study, we investigated the molecular mechanism by which SA exerts its antagonistic effect on the JA signaling pathway. Therefore, we systematically tested different components of the JA signaling pathway to identify the site of action of SA-mediated antagonism.
JAZ Proteins
Chung and Howe (2009) identified a naturally occurring alternative splice variant of JAZ10 that completely lacks the Jas domain (JAZ10.4) and is therefore insensitive to JA-induced degradation. We hypothesized that SA targets the JA pathway via increased production of this dominant-negative splice variant of JAZ10. However, no SA-induced expression of JAZ10.4 could be detected, while PDF1.2 and VSP2 gene expression was still suppressed by SA (Figure 1B), making it unlikely that SA/JA crosstalk is regulated via enhanced production of JAZ10.4.
Previously, it was shown that SA inhibits the auxin signaling pathway through the stabilization of members of the auxin/indole-3-acetic acid family of transcriptional repressors (Wang et al., 2007). In analogy to JAs, auxins induce gene expression through direct physical interaction with TIR1-like F-box proteins in the SCFTIR1-complex, which in turn target the auxin/indole-3-acetic acid family of transcriptional repressors for degradation via the proteasome (Gray et al., 2001). Hence, we postulated that the antagonistic effect of SA on JA signaling might similarly function via the stabilization of JAZ transcriptional repressor proteins. The fact that overexpression of one JAZ protein that is partially resistant to JA-induced degradation is sufficient to suppress JA-dependent gene expression (Figure 1A) supported the hypothesis that stabilization of JAZ proteins by SA may be a plausible mechanism for the antagonistic effect of SA on JA signaling. However, we found that SA had no effect on the stability of JAZ proteins JAZ1, JAZ2, and JAZ9, neither in cell suspension cells nor in whole plants that ectopically expressed JAZ-LUC or JAZ-GUS reporter fusion proteins (Figure 2; see Supplemental Figure 1 online). Since most of the 12 JAZ proteins of Arabidopsis are likely to exert similar and overlapping functions in the JA signaling pathway, we conclude that it is very unlikely that SA-mediated suppression of the JA response functions through the stabilization of JAZ repressor proteins.
SCFCOI1
The E3 ubiquitin-ligase SCFCOI1 complex plays a crucial role in the regulation of the JA response as it targets JAZ transcriptional repressor proteins for degradation upon perception of biologically active JAs (Browse, 2009; Chung et al., 2009). To investigate its requirement for SA/JA crosstalk, we also monitored the effect of SA on PDF1.2 expression that was activated in the JA-insensitive coi1-1 mutant background through ectopic expression of 35S:ERF1. The AP2 domain/ERF transcription factor ERF1 is a positive regulator of PDF1.2 (Lorenzo et al., 2003). Ectopic expression of ERF1 strongly activated PDF1.2, even in the coi1-1 background that is fully blocked in JA signaling (Lorenzo et al., 2003) (Figure 3). Exogenous application of SA readily suppressed ERF1-mediated PDF1.2 transcription in the mutant coi1-1 background (Figure 3). These results clearly demonstrate that SA exerts its antagonistic effect independent of the E3 ubiquitin-ligase SCFCOI1-JAZ machinery in the JA signaling pathway.
The SA/JA Crosstalk Transcriptome
Because we found that SA is likely to target the JA pathway at the level of gene transcription, we first established the SA/JA crosstalk transcriptome using Affymetrix ATH1 GeneChips (see Supplemental Figure 2 and Supplemental Data Sets 1 and 2 online). The effect of SA and MeJA on gene expression has been analyzed in several small- and large-scale microarray studies in Arabidopsis and Sorghum bicolor (Schenk et al., 2000; Salzman et al., 2005). In addition, global expression phenotyping of signaling-defective mutants of the SA and JA pathways has been exploited to investigate the network of regulatory interactions among different defense signaling pathways (Glazebrook et al., 2003). These expression profiling studies revealed one-way and mutual antagonism as well as synergistic effects between SA- and JA-dependent signaling pathways. Similar SA/JA signal interactions were also apparent in the SA/JA crosstalk transcriptome of this study, but because an in-depth functional analysis of these microarray data was not the focus of this study, we will not discuss it here. For this study, we used the SA/JA crosstalk transcriptome to search for regulatory motifs that are overrepresented in the JA-responsive promoters that are sensitive to suppression by SA. We identified 175 genes that were significantly induced by MeJA, 59 of which were significantly downregulated by SA (see Supplemental Data Sets 1 and 2 online). In silico analysis of the 1-kb promoter region of the MeJA-inducible genes that were suppressed by SA revealed that the G-box element CACGTG (Myc/ABRE element), the W-box TGACY, the evening element AAAATATCT, the I-box GATAA, and the GCC-box AGCCGCC were significantly overrepresented (Figure 4), suggesting that these elements may be involved in the regulation of the SA/JA antagonism.
The GCC-Box
Spoel et al. (2003) tested PDF1.2 promoter deletion constructs fused to the GUS reporter gene for their ability to show SA/JA crosstalk. Deletion of the PDF1.2 promoter up to 311 bp upstream of the ATG start codon did not interfere with the ability of the promoter to be induced by MeJA and suppressed by SA. Since this part of the PDF1.2 promoter includes the I-box and the GCC-box, we focused on the involvement of these promoter elements in the regulation of SA/JA crosstalk (Spoel et al., 2003). Site-directed mutagenesis of the I-box motif in the PDF1.2 promoter did not alter the response to SA, MeJA, or both chemicals, demonstrating that the I-box motif is not essential for crosstalk (see Supplemental Figure 3 online). The GCC-box remained an interesting candidate for crosstalk regulation, as this element is essential and sufficient for MeJA responsiveness of the PDF1.2 promoter (Brown et al., 2003; Zarei et al., 2011). To investigate whether the antagonistic effect of SA on the JA response is targeted at the GCC-box in JA-responsive genes, we tested the effect of MeJA and SA on the responsiveness of 4XGCC:GUS reporter lines. We confirmed the findings of Zarei et al. (2011) that the GCC-box tetramer is sufficient for transcriptional activation by JA. Moreover, we showed that the GCC-box is sufficient for the downregulation of JA-responsive gene expression by SA (Figure 5).
ERF1 and ORA59
The GCC-box is a binding site for members of the AP2/ERF family of transcription factors (Hao et al., 1998), such as ERF1 and ORA59, which are both important activators of PDF1.2 (Lorenzo et al., 2003; Pré et al., 2008; Zarei et al., 2011). Therefore, we were interested in the effect of SA on the regulation of these transcriptional activators. Since the suppressive effect of SA on PDF1.2 was independent of both ORA59 and ERF1 mRNA levels in this study (Figures 3 and 6A), we hypothesized that antagonistic action of SA in the JA pathway involves an effect of SA on ERF1 or ORA59 protein levels. To this end, we monitored ERF1-TAP and ORA59-GFP protein accumulation in 35S:ERF1-TAP and 35S:ORA59-GFP plants, respectively. In the 35S:ERF1-TAP line, ERF1-TAP accumulation was not affected by SA, while PDF1.2 gene expression was induced by MeJA and suppressed by SA (Figure 6B; see Supplemental Figure 4A online). In the 35S:ORA59-GFP line, by contrast, ORA59-GFP protein accumulation was strongly reduced by SA. This result indicates that SA negatively affects ORA59 protein accumulation, which may explain the antagonistic effect of SA on JA-responsive PDF1.2 gene expression. Despite the fact that SA reduced ORA59 accumulation in 35S:ORA59-GFP plants, PDF1.2 gene expression could not be suppressed by SA in these plants (Figures 6C and 6D; see Supplemental Figure 4B online). Leon-Reyes et al. (2010a) reported previously that overexpression of ORA59 negatively affects the ability of SA to suppress PDF1.2 gene expression. This finding highlights the importance of ORA59 in the outcome of the SA/JA antagonism. However, in the 35S:ORA59-GFP line, reduction of ORA59-GFP protein levels upon SA treatment apparently does not directly result in a decrease of PDF1.2 gene expression levels (see Supplemental Figure 4B online). A plausible explanation for this could be that in 35S:ORA59-GFP plants, the level of ORA59-GFP protein is still too high after SA treatment to allow suppression of PDF1.2, but in wild-type plants, SA-induced suppression of ORA59 accumulation is sufficient to suppress PDF1.2.
Mode of Action of SA/JA Signal Interaction
Our results indicate that the antagonistic effect of SA on the JA response functions downstream of the SCFCOI1-JAZ complex at the level of gene transcription. We found that the GCC-box is sufficient for transcriptional activation by JA and suppression by SA and suggest that SA/JA crosstalk via the GCC-box is mediated through SA-mediated suppression of ORA59 protein accumulation. Like PDF1.2, the promoter of ORA59 contains a GCC-box and can be suppressed by SA (Zander et al., 2012). However, we showed that this is not required for SA-mediated suppression of JA-dependent PDF1.2 gene expression, as in the ERF1 overexpression line the level of ORA59 transcript is not affected by SA while the at same time the PDF1.2 expression level is reduced by SA (Figure 6A). Nevertheless, suppression of ORA59 expression may contribute to the overall antagonistic effect of SA on JA signaling.
In future research, it will be interesting to find out how SA can target ORA59 protein levels. One possibility is that SA stimulates proteasome-mediated degradation of ORA59 through posttranslational modification of the protein. Proteasome-mediated turnover of transcriptional (co)activators has been reported as a common mechanism for regulation of transcriptional activity (Spoel et al., 2010; Moore et al., 2011). The ET-responsive transcription factor EIN3 was shown to be continuously targeted to the proteasome via the SCFEBF1/2 ubiquitin ligase in the absence of ET (Guo and Ecker, 2003; Potuschak et al., 2003; Gagne et al., 2004). In addition, phosphorylation and subsequent proteasome-mediated turnover plays an important role in the activity of the transcriptional coactivator NPR1 (Spoel et al., 2009). Moreover, the AP2/ERF transcription factor ERF#104 is phosphorylated by MPK6, resulting in stabilization of the protein, which likely leads to enhanced transcription of ERF#104 target genes (Bethke et al., 2009). In parallel to an effect of SA on the ORA59 protein, SA might interfere with JA-dependent transcription via production of transcriptional repressors that can bind to the GCC-box. Transcription factors belonging to the AP2/ERF family share a common DNA binding motif (Nakano et al., 2006); therefore, family members that can function as transcriptional repressors, such as the EAR domain–containing AP2/ERFs, are putative candidates with a role in SA/JA crosstalk.
Several candidate proteins for the SA-mediated suppression of JA-responsive gene expression have been described. Several WRKY transcription factors are thought play a role in SA/JA crosstalk (Pieterse et al., 2012). Expression of a large part of the WKRY transcription factors is SA responsive (Dong et al., 2003), and certain WRKYs have been described as important components in transcriptional regulation of SA-responsive gene expression (Wang et al., 2006; van Verk et al., 2011). A recent report showed that SA-mediated suppression of JA-induced PDF1.2 gene expression was abolished in wrky50 single and wrky50 wrky51 double knockout mutants, indicating that these WRKYs play an important role in SA/JA crosstalk (Gao et al., 2011). Although we did find an overrepresentation of the WRKY binding site (W-box) in promoters of SA/JA crosstalk genes (Figure 4), the PDF1.2 promoter does not contain a W-box element. This suggests that WRKYs act indirectly in the regulation of transcription of PDF1.2. The mechanism by which the W-box in promoters of JA-dependent genes plays a role in the suppression of these genes by SA remains to be elucidated.
In addition to WRKYs, TGA transcription factors are implicated in SA-mediated suppression of JA-dependent gene expression (Ndamukong et al., 2007; Leon-Reyes et al., 2010a; Zander et al., 2010). Like WRKYs, TGA transcription factors are important in transcriptional regulation of SA-responsive gene expression (Zhang et al., 2003; Kesarwani et al., 2007). In addition, Zander et al. (2010) demonstrated that the tga2 tga5 tga6 triple mutant shows reduced JA/ET-responsive transcription of PDF1.2, indicating that in the absence of SA, class II TGA factors function as positive regulators of JA/ET-responsive gene transcription. Interestingly, both the tga2 tga5 tga6 triple mutant and the tga2 tga3 tga5 tga6 quadruple mutant were shown to be insensitive to SA-mediated suppression of JA/ET-responsive expression of PDF1.2 (Ndamukong et al., 2007; Leon-Reyes et al., 2010a; Zander et al., 2010), suggesting that TGAs may have an important function in SA-mediated suppression of JA-responsive gene expression. TGA2 binds to the TGACG motif in the promoter of PDF1.2 (Spoel et al., 2003). However, deletion of the TGACG motif does not affect PDF1.2 promoter activity (Spoel et al., 2003; Zander et al., 2010), suggesting that, like WRKY transcription factors, also TGA factors act indirectly in the regulation of JA/ET-responsive transcription of PDF1.2 (e.g., via a yet unknown protein that controls PDF1.2 promoter activity). Future research will be focused on the identity of novel components of the SA pathway that antagonize JA-dependent activation of GCC-box-containing promoters as they will be key in unraveling the molecular basis of the SA/JA signal interaction. A working model for the mode of action of SA/JA signal interaction on GCC-box-containing promoters is given in Figure 7.
Figure 7.
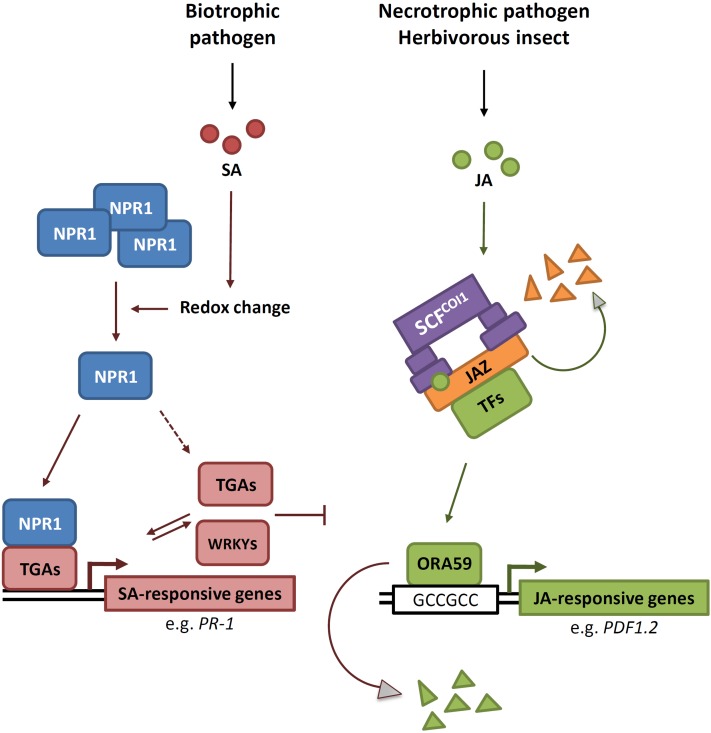
Model for SA/JA Signal Interaction on GCC-Box-Containing Promoters of JA-Responsive Genes.
Infection by a biotrophic pathogen results in the accumulation of SA and monomerization of NPR1 through SA-mediated redox changes in the cell. Monomeric NPR1 is then translocated into the nucleus where it interacts with TGA transcription factors, ultimately leading to the activation of SA-responsive genes. Expression of a large set of WRKY genes is induced by SA, some of which can regulate SA-responsive gene expression. Wounding, such as that caused by insect feeding or infection by a necrotrophic pathogen, results in the accumulation of JA. Binding of JA to the SCFCOI1-E3 ubiquitin-ligase complex leads to degradation of JAZ transcriptional repressor proteins via the proteasome, which results in the release of transcriptional activators, such as MYC2, 3, and 4, and EIN3 and EIL1. Subsequently, AP2/ERF transcription factors, such as ORA59 and ERF1, are induced that activate the ERF branch of the JA pathway. Binding of ERFs to the GCC-box induces JA-responsive gene expression, which can be suppressed by SA in an SCFCOI1-JAZ–independent manner. The GCC-box is sufficient for SA-mediated suppression of JA-induced gene expression. SA can negatively affect ORA59 protein accumulation, which provides an explanation for the antagonistic effect of SA- on JA-responsive gene expression in wild-type plants. Since mutations in certain TGAs and WRKYs impair SA/JA crosstalk, TGAs and WRKYs may play a direct or indirect regulatory role in this process. Solid arrows and inhibition lines indicate established activities or accumulation of compounds; dashed arrows represent hypothesized connections. Red lines indicate activities mediated by the SA pathway; green lines indicate activities mediated by the JA pathway.
Although the GCC-box is sufficient for SA-mediated suppression of JA-responsive gene expression, many JA responsive genes do not contain such a motif (e.g., VSP2) but nevertheless are suppressed by SA. Interestingly, we have also found an overrepresentation of the G-box in promoters of SA/JA crosstalk genes. The G-box or G-box-like sequences are commonly found in promoters that are activated by JA, such as VSP2, and are binding sites for the JA-dependent transcription factor MYC2 (Memelink, 2009). It is tempting to speculate that a similar scenario as described above for the suppression of GCC-box-containing promoters is functional for MYC2-regulated G-box containing promoters, but this remains to be investigated.
METHODS
Plant Material
Seeds of Arabidopsis thaliana wild-type accession Col-0 and the mutants and transgenic lines (all in Col-0 background) coi1-1 (Feys et al., 1994), 35S:JAZ10 (originally called At5g13220.1; Yan et al., 2007), JAZ10 RNAi-7 (originally called At5g13220 RNAi-7; Yan et al., 2007), JAZ10 RNAi-9 (originally called At5g13220 RNAi-9; Yan et al., 2007), 35S:JAZ10Δ #OE4A (originally called At5g13220 OE4A; Yan et al., 2007), 35S:JAZ10Δ #OE4B (originally called At5g13220 OE4B; Yan et al., 2007), 35S:JAZ1-GUS (Thines et al., 2007), 35S:ERF1 (Lorenzo et al., 2003), 35S:ERF1-1/coi1-1 (Lorenzo et al., 2003), 35S:ERF1-TAP, 35S:ORA59-GFP, PDF1.2:GUS (Koornneef et al., 2008a), PDF1.2ΔIbox:GUS, PR-1:GUS (Koornneef et al., 2008a), 35S:GUS (PG15) (Koornneef et al., 2008a), 4XGCC:GUS #5 (Zarei et al., 2011), and 4XGCC:GUS #7 (Zarei et al., 2011) were sown in quartz sand. After 2 weeks, seedlings were transferred to 60-mL pots containing a sand/potting soil mixture that was autoclaved twice for 20 min and further cultivated in a growth chamber with an 8-h day (24°C) and 16-h night (20°C) cycle at 70% relative humidity for another 3 weeks as described (van Wees et al., 1999). For experiments with in vitro–grown plants, seedlings were grown on plates containing Murashige and Skoog (MS) medium, pH 5.7, supplemented with 10 g·L−1 Suc and 0.8% (w/v) plant agar.
Construction of Transgenic Plants
For the construction of the I-box knockout line, the 1.2-kb PDF1.2 (At5g44420) promoter fragment was amplified by PCR from genomic DNA of Col-0 plants using the PDF1.2 FW2 and PDF1.2 RV2 primers (see Supplemental Figure 3 online). The PDF1.2 promoter fragment was first cloned into the pCR-Blunt II-TOPO vector (Invitrogen). The I-box motif (5′-GATAAG-3′) was mutagenized to an EcoRI recognition sequence (5′-GAATTC-3′) to facilitate identification of mutagenized transformants using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer’s protocol. Primers PDF1.2∆Ibox FW and PDF1.2∆Ibox RV were designed for the introduction of the desired mutation. The mutated PDF1.2ΔIbox promoter fragment was ligated into the pGREENII 0229-GUS binary vector (Hellens et al., 2000), using the SpeI and PstI recognition sites.
For construction of the 35S:ERF1-TAP line, the TAP insert was excised from pBS1479 (Puig et al., 2001) with BamHI and cloned into pC1300intB-35SnocBK (accession number AY560326) digested with BgIII. pC1300intB-35SnocBK is a derivative of the binary vector pCAMBIA1300 carrying a cauliflower mosaic virus (CaMV) 35S expression cassette. The ERF1 (At3g23240) open reading frame lacking the stop codon (ERF1-ΔSTOP) was amplified by PCR using the primers ERF1 FW1 and ERF1 RV1 and cloned in pGEM-T Easy (Promega). The ERF1 open reading frame was excised from pGEM-T Easy with SalI-XhoI and cloned into pC1300intB-35SnocBK-TAP.
For construction of the 35S:ORA59-GFP line, ORA59 (At1g06160) was amplified by PCR with the primer set ORA59 FW1 and ORA59 RV1 and cloned in pGEM-T Easy such that the XhoI site flanked the SpeI site. The ORA59-ΔSTOP insert was excised with SalI-SpeI and cloned into pTH2SN (a derivative of pTH2; Kuijt et al., 2004). ORA59-ΔSTOP was excised from pTH2SN with SalI-NcoI and cloned into pTH2 (Chiu et al., 1996; Niwa et al., 1999). The GFP expression cassette was transferred from pTH2 as a HindIII-EcoRI fragment to pCAMBIA1300 (accession number AF234296).
Arabidopsis plants were transformed using the floral dip method as described (Clough and Bent, 1998; Koornneef et al., 2008a). Transformed seedlings were selected as described (Harrison et al., 2006).
Construction of Transgenic Cell Suspension Cultures
For the JAZ degradation assays in cell suspension cultures, the plasmid pEN-L4-2-R1 holding the CaMV 35S promoter, pEN-R2-LUC-L3, and entry clones holding JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ9 (At1g70700), or MYC2 (At1g32640) open reading frame without stop codon were recombined by MultiSite Gateway LR reaction using pKCTAP as destination vector essentially as described (Karimi et al., 2007). The T-DNA in the latter vector additionally expresses GFP under control of the rolD promoter (Van Leene et al., 2007). Plasmids were transfected into Agrobacterium tumefaciens strain C58 (pMP90) by electroporation. The Arabidopsis PSB-D cell suspension culture used in this study was maintained and transformed with the plasmids as described previously (Van Leene et al., 2007). Transformed cells were directly selected in liquid medium. For the experiment shown in Figure 2E, entry clones pEN-L4-2-R1 holding the CaMV 35S promoter, pEN-R2-GStag-L3 holding a GS-TAP tag, and pDONR221-JAZ1 (without stop codon) were recombined by MultiSite Gateway LR reaction using pKCTAP as destination vector. Subsequently, the construct was introduced in the Arabidopsis PSB-D cell culture as described above.
Chemical Treatments of Intact Plants
Plants were treated with SA and/or MeJA by dipping the leaves into a solution containing 0.015% (v/v) Silwet L77 (Van Meeuwen Chemicals) and either 1 mM SA (Mallinckrodt Baker), 0.1 mM MeJA (Serva, Brunschwig Chemie), or a combination of these chemicals as described previously (Spoel et al., 2003). Control treatments (mock) were dipped into a solution containing 0.015% (v/v) Silwet L77.
For the experiments shown in Figures 2F and 2G, chemical induction of plants grown on MS medium was performed by transferring 12-d-old plate-grown seedlings to fresh MS medium supplemented with 0.5 mM SA, 0.02 mM MeJA, or a combination of these chemicals (Spoel et al., 2003). For the experiments shown in Figure 5, 2-week-old plate-grown plants were transferred to 24-well plates containing 1.5 mL MES buffer (5 mM MES and 1 mM KCl, pH 5.7) per well. Five seedlings were used per sample. Twenty-four hours after transfer to MES buffer, 0.5 mL MES buffer supplemented with SA and/or MeJA was added to the seedlings, resulting in final concentrations of 0.5 mM SA and 0.1 mM MeJA, respectively. In addition, ACC was added to all wells resulting in a final concentration of 0.002 mM. Seedlings were harvested 24 h after induction treatment and immersed in GUS staining solution or frozen in liquid nitrogen and used for quantitative GUS activity measurement. For the experiments shown in Figure 6, 11-d-old plate-grown plants were transferred to 24-well plates containing 1.5 mL MES buffer per well and were treated as described above, with exception of the addition of ACC. The seedlings were harvested at different time points after induction treatment and immediately frozen in liquid nitrogen. In all cases, MeJA was added to the solutions from a 1000-fold concentrated stock in 96% ethanol. To the solutions without MeJA, a similar volume of 96% ethanol was added (end concentration of 0.1%).
Alternaria brassicicola Inoculation
A. brassicicola strain MUCL 20297 was grown on potato dextrose agar (Difco Laboratories) plates for 2 weeks at 22°C. Subsequently, conidia were collected as described (De Vos et al., 2005). Plants were inoculated when 5 weeks old by applying 5-μL droplets of half-strength potato dextrose broth containing 5 × 105 spores per mL, as described previously (Leon-Reyes et al., 2009). After inoculation plants were kept at 100% relative humidity for optimal fungal germination.
RNA Extraction, RNA Gel Blot, and Quantitative Real-Time PCR Analysis
For RNA extraction, at least five plants per treatment were harvested at the time points indicated. RNA isolation and RNA gel blot analysis was performed as described (van Wees et al., 1999). RNA gel blots were hybridized with gene-specific probes for PR-1 (At2g14610), PDF1.2 (At5g44420), and VSP2 (At5g24770) as described (van Wees et al., 1999; Pozo et al., 2008). Probes for the GUS reporter gene, the genes ERF1 (At3g23240) and ORA59 (At1g06160) and 18S rRNA were made by PCR amplification on cDNA using the following primers: GUS FW1 and GUS RV1, ERF1 FW2 and ERF1 RV2, ORA59 FW2 and ORA59 RV2, and 18S FW and 18S RV. After hybridization with [α-32P]dCTP-labeled probes, blots were exposed for autoradiography. Signal intensities of probes were quantified using a Bio-Rad Molecular Imager FX with Quantity One software (Bio-Rad).
For the experiment described in Figure 1B, expression of PR-1 (At2g14610), PDF1.2 (At5g44420), VSP2 (At5g24770), JAZ10.1/2/3 (At5g13220.1/2/3), JAZ10.4 (At5g13220.4), and the constitutively expressed gene At1g13320 was determined by RT-PCR. Fermentas RevertAid H minus reverse transcriptase was used to convert DNA-free total RNA into cDNA. The following gene-specific primers were used for amplification (see Supplemental Table 1 online): PR-1 FW and PR-1 RV, PDF1.2 FW1 and PDF1.2 RV1, VSP2 FW1 and VSP2 RV1, JAZ10.1/2/3 FW1 and JAZ10.1/2/3 RV1, and JAZ10.4 FW1 and RV1 as described by Chung and Howe (2009), and At1g13320 FW1 and RV1 as described by Czechowski et al. (2005).
AOS gene expression in Arabidopsis cell suspension cells (Figure 2) was analyzed by quantitative RT-PCR (qRT-PCR) as described by Pauwels et al. (2008) using the AOS-specific primers AOS FW1 and AOS RV1 (see Supplemental Table 1 online). Δ-CT (for cycle threshold) relative quantification with multiple reference gene normalization was performed with the qBase program (medgen.ugent.be/qbase). The reference genes used for normalization were At1g69280, At4g17300, At3g25800, and At1g04300, as described (Pauwels et al., 2008). For the experiments shown in Figure 5 and Supplemental Figure 4 online, gene expression was analyzed by qRT-PCR as described by Verhage et al. (2011), with some modifications. Fermentas RevertAid H minus reverse transcriptase was used to convert DNA-free total RNA into cDNA. The following primers were used to analyze expression of PR-1, PDF1.2, ORA59-GFP, and the GUS reporter gene (see Supplemental Table 1 online): PR-1 primers as described in the paragraph above, PDF1.2 FW3 and PDF1.2 RV3, GFP FW and GFP RV, and GUS FW2 and GUS RV2. The reference gene used for normalization of the genes of interest was At1g13320 as described (Czechowski et al., 2005).
JAZ Degradation Assay in Cell Suspension Cultures
Transformed cell cultures were grown for several weeks in the absence of kanamycin before protein degradation assays were performed. Fresh cell cultures were grown for 1 week after subculturing before use in the JAZ degradation assay. For crosstalk experiments, 0.001 mM JA (Duchefa) and/or 0.01 mM SA was added to the cells, which were subsequently harvested at multiple time points by vacuum filtration. Samples were immediately frozen in liquid nitrogen and ground using a Retsch MM300 shaker. Subsequently, proteins were extracted using LUC extraction buffer (100 mM KPO4, pH 7.8, 1 mM EDTA, 7 mM β-mercaptoethanol, 1 mM PMSF, and 1 complete protease inhibitor tablet [Roche] per 10 mL) as described (Salmon et al., 2008). The supernatant was used for subsequent measurements of GFP fluorescence and LUC activity. GFP fluorescence was used to normalize for variations in protein extraction. Half-life calculations were performed as described (Dreher et al., 2006) with modifications. For each sample individually, a LUC activity (l)/GFP fluorescence (g) ratio was calculated and divided by an average l/g value for the control samples (i.e., the first time point without JA) to generate a normalized l/g value. For graphic presentation, the natural log of the normalized l/g value was determined and plotted in function of time.
Protein Extraction and Immunoblot Analysis
For the experiment shown in Figure 2E, protein extraction and immunoblot analysis were performed as described by Hemerly et al. (1995). For the detection of JAZ1-CTAP, a 1:2500 dilution of the peroxidase antiperoxidase (PAP) soluble complex antibody (Sigma-Aldrich) was used. As an internal control for loading of the SDS-PAGE gel and transfer of proteins to the membrane, the constitutively accumulating protein CDKA was detected using a primary anti-CDKA antibody (1:2500 dilution) and a secondary peroxidase-conjugated anti-rabbit antibody (GE Healthcare) (1:10,000 dilution).
For experiments shown in Figure 6, protein was extracted by resuspension of frozen and ground seedlings in protein extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.2% Triton X-100, 6 mM β-mercaptoethanol, 5 mM EDTA, protease inhibitor cocktail for plant cell and tissue extracts 1:100 [Sigma-Aldrich], and 50 µM MG132 [Z-Leu-Leu-Leu-Al; Sigma-Aldrich]). Samples were centrifuged for 10 min at 4°C, and supernatant containing soluble protein was harvested. Protein concentration was determined using Bio-Rad protein assay. Immediately after isolation, the soluble protein fraction was transferred to SDS sample buffer (Laemmli, 1970). Fifteen micrograms of protein was separated by SDS-PAGE (10% acrylamide) as described (Laemmli, 1970). Next, proteins were electroblotted onto nitrocellulose membrane (Amersham Hybond ECL; GE Healthcare). Nitrocellulose membranes were blocked overnight at 4°C in 5% skim milk (Elk) in TBST (500 mM Tris-HCl, pH 7.5, 1.5 mM NaCl, and 0.05% Tween 20). Next, membranes were incubated for 2.5 h at room temperature with PAP antibody (rabbit; Sigma-Aldrich) diluted 1:1000 in 5% skim milk in TBST or anti-GFP antibody (mouse, monoclonal; Roche) diluted 1:800 in 5% skim milk in TBST. Membranes incubated with PAP antibody were washed three times for 10 min with TBST and one time for 10 min with TBS (500 mM Tris-HCl, pH 7.5, and 1.5 mM NaCl), after which protein was detected as described below. Membranes incubated with anti-GFP antibody were washed four times for 10 min in TBST and incubated for 1 h at room temperature with goat anti-mouse IgG, horseradish peroxidase conjugate (Novagen/Merck) diluted 1:5000 in 5% skim milk in TBST. Next, membranes were washed three times 10 min with TBST and one time for 10 min with TBS. Proteins were detected on Kodak Biomax XAR films (Sigma-Aldrich) using Super Signal Pico Chemiluminescent Substrate and Super Signal Femto Chemiluminescent Substrate (Thermo Scientific) mixed in 3:1 ratio. As an internal control for loading of the SDS-PAGE gel and transfer of proteins to the membrane, membranes were stained with Ponceau S (0.1% Ponceau S and 5% acetic acid).
GUS Assays
In the histochemical GUS assay, GUS activity was assessed by transferring seedlings to a GUS staining solution (1 mM X-Gluc, 100 mM NaPi buffer, pH 7.0, 10 mM EDTA, and 0.1% [v/v] Triton X-100). After vacuum infiltration and overnight incubation at 37°C, the seedlings were destained by repeated washes in 70% ethanol (Spoel et al., 2003). For the quantitative GUS assays, protein was isolated from frozen plant material and quantitative GUS activity measurement was performed as described (Pré et al., 2008).
Sample Preparation and Microarray Data Collection
For isolation of RNA, whole rosettes from Col-0 plants were mock treated or treated with 1 mM SA, 0.1 mM MeJA, or a combination of both as described above. Leaf tissue was harvested 28 h after treatment and immediately frozen in liquid nitrogen. RNA was prepared from three independent biological experiments and purified using RNeasy Plant Mini Kit columns (Qiagen Benelux). RNA samples were analyzed for quality using a lab-on-a-chip RNA Nano Chip assay (Agilent Technologies). Probe preparation and hybridization to Arabidopsis ATH1 full-genome GeneChips (Affymetrix) were performed by ServiceXS and the Affymetrix Service Station of Leiden University Medical Center.
Expression Profiling and Promoter Analysis
The obtained Arabidopsis ATH1 microarray CEL files were normalized using an Empirical Bayes GC Robust Multi-array Average (GCRMA) background adjustment, quantile normalization, and Median Polish summarization (Wu et al., 2004). For analysis of differentially expressed genes, the log2-transformed expression values of the three independent biological experiments were compared between treatments using a two-sample, two-tailed Student’s t test. To identify overrepresented promoter elements of SA/JA crosstalk genes, the approach as described by Breeze et al. (2011) was applied with the following minor modifications. In total, 881 promoter elements were obtained from the JASPAR (Sandelin et al., 2004), PLACE (Higo et al., 1999), and TRANSFAC (Matys et al., 2006) databases. The 1-kb upstream regions of 33,602 genes were obtained from the TAIR10 release of the Arabidopsis genome (www.Arabidopsis.org). The 100-million-bp random sequence generated by a third-order Markov model was learned from the whole Arabidopsis genome (Chromosome 1-5; TAIR10 release). The top k nonoverlapping hits within the 1-kb upstream region were optimized within the range 1 to 10 for minimum binomial P value.
Accession Numbers
Arabidopsis Genome Initiative numbers for genes described in this article are listed in Supplemental Table 1 online. All microarray data are deposited in NASCArrays under experiment reference number NASCARRAYS-684 (http://www.affymetrix.Arabidopsis.info).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. JA-Mediated Degradation of JAZ2 Is Not Affected by SA.
Supplemental Figure 2. The SA/JA Crosstalk Transcriptome of Arabidopsis.
Supplemental Figure 3. The I-Box Motif Is Not Required for SA/JA Crosstalk.
Supplemental Figure 4. SA- and JA-Responsive Gene Expression in 35S:ERF1-TAP and 35S:ORA59-GFP Plants.
Supplemental Table 1. Arabidopsis Genome Initiative Numbers and Primers Used in This Study for Cloning and Studying Expression of Arabidopsis Genes.
Supplemental Data Set 1. MS Excel File with Normalized Expression Levels, Fold-Change Information, AGI Numbers, and TIGR Annotation of the Selected MeJA- and SA-Responsive Genes.
Supplemental Data Set 2. MS Excel File with Normalized Expression Levels, Fold-Change Information, AGI Numbers, and TIGR Annotation of the Selected SA/JA Crosstalk Genes.
Acknowledgments
We thank Ruth Joosten, Hans van Pelt, Jan Geerinck, Robin van den Bossche, Hana Návarová, Wouter Jansen, Jordi Boshoven, and Anja van Dijken for technical assistance. This work was supported by VICI Grant 865.04.002 and VIDI Grant 11281 of the Netherlands Organization of Scientific Research and ERC Advanced Grant 269072 of the European Research Council.
AUTHOR CONTRIBUTIONS
D.V.d.D., A.L.-R., A.K., A.G., J.M., T.R., S.C.M.V.W., and C.M.J.P. designed the research. D.V.d.D., A.L.-R., A.K., N.R., L.P., and A.P.K. performed research. M.V.V., A.G., T.R., and J.M. contributed new analytic/computational/etc. tools. D.V.d.D., A.L.-R., A.K., M.V.V., L.P., A.G., J.M., T.R., S.C.M.V.W., and C.M.J.P. analyzed data. D.V.d.D., A.L.-R., A.K., S.C.M.V.W., and C.M.J.P. wrote the article.
Glossary
- SA
salicylic acid
- JA
jasmonic acid
- ET
ethylene
- JA-Ile
(+)-7-iso-jasmonoyl-l-Ile
- Col-0
Columbia-0
- MeJA
methyl jasmonate
- GUS
β-glucuronidase
- MS
Murashige and Skoog
- ACC
1-aminocyclopropane-1-carboxylic acid
- CaMV
cauliflower mosaic virus
- qRT-PCR
quantitative RT-PCR
- GFP
green fluorescent protein
- PAP
peroxidase antiperoxidase
References
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Bethke G., Unthan T., Uhrig J.F., Pöschl Y., Gust A.A., Scheel D., Lee J. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. USA 106: 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M.-A., Shokat K.M., Rietz S., Parker J., Mundy J. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47: 532–546 [DOI] [PubMed] [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Brown R.L., Kazan K., McGrath K.C., Maclean D.J., Manners J.M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132: 1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R. (2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Chung H.S., Howe G.A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21: 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H.S., Niu Y., Browse J., Howe G.A. (2009). Top hits in contemporary JAZ: An update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. (2004). WebLogo: A sequence logo generator. Genome Res. 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M., Van Oosten V.R., Van Poecke R.M.P., Van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Métraux J.P., Van Loon L.C., Dicke M., Pieterse C.M.J. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Devoto A., Turner J.G. (2005). Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant. 123: 161–172 [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.X., Chen C.H., Chen Z.X. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Dreher K.A., Brown J., Saw R.E., Callis J. (2006). The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J.F., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. (2004). Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. USA 101: 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q.-M., Venugopal S., Navarre D., Kachroo A. (2011). Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A., Dubugnon L., Liechti R., Farmer E.E. (2010). Jasmonate biochemical pathway. Sci. Signal. 3: cm3. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Chen W., Estes B., Chang H.-S., Nawrath C., Métraux J.-P., Zhu T., Katagiri F. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Grant M.R., Jones J.D.G. (2009). Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guo H.W., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hao D.Y., Ohme-Takagi M., Sarai A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273: 26857–26861 [DOI] [PubMed] [Google Scholar]
- Harrison S.J., Mott E.K., Parsley K., Aspinall S., Gray J.C., Cottage A. (2006). A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hemerly A., Engler Jde.A., Bergounioux C., Van Montagu M., Engler G., Inzé D., Ferreira P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14: 3925–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15: 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: Toward an integrated view. Plant Physiol. 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F.C., Van Loon L.C., Pieterse C.M.J. (2008b). Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147: 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Verhage A., Leon-Reyes A., Snetselaar R., Van Loon L.C., Pieterse C.M.J. (2008a). Towards a reporter system to identify regulators of cross-talk between salicylate and jasmonate signaling pathways in Arabidopsis. Plant Signal. Behav. 3: 543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt S.J.H., Lamers G.E.M., Rueb S., Scarpella E., Ouwerkerk P.B.F., Spaink H.P., Meijer A.H. (2004). Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol. Biol. 55: 781–796 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A., Du Y., Koornneef A., Proietti S., Körbes A.P., Memelink J., Pieterse C.M.J., Ritsema T. (2010a). Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 23: 187–197 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A., Spoel S.H., De Lange E.S., Abe H., Kobayashi M., Tsuda S., Millenaar F.F., Welschen R.A.M., Ritsema T., Pieterse C.M.J. (2009). Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A., Van der Does D., De Lange E.S., Delker C., Wasternack C., Van Wees S.C.M., Ritsema T., Pieterse C.M.J. (2010b). Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232: 1423–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Brader G., Palva E.T. (2004). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sánchez-Serrano J.J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P., Duan M., Wei C., Li Y. (2007). WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 48: 833–842 [DOI] [PubMed] [Google Scholar]
- Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34 (Database issue): D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J. (2009). Regulation of gene expression by jasmonate hormones. Phytochemistry 70: 1560–1570 [DOI] [PubMed] [Google Scholar]
- Moore J.W., Loake G.J., Spoel S.H. (2011). Transcription dynamics in plant immunity. Plant Cell 23: 2809–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A.J., Kenton P., Atzorn R., Miersch O., Wasternack C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I., Abdallat A.A., Thurow C., Fode B., Zander M., Weigel R., Gatz C. (2007). SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P., Browse J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Hirano T., Yoshimoto K., Shimizu M., Kobayashi H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18: 455–463 [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C.M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Pozo M.J., Van Der Ent S., Van Loon L.C., Pieterse C.M.J. (2008). Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 180: 511–523 [DOI] [PubMed] [Google Scholar]
- Pré M., Atallah M., Champion A., De Vos M., Pieterse C.M.J., Memelink J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001). The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229 [DOI] [PubMed] [Google Scholar]
- Reymond P., Farmer E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Salmon J., Ramos J., Callis J. (2008). Degradation of the auxin response factor ARF1. Plant J. 54: 118–128 [DOI] [PubMed] [Google Scholar]
- Salzman R.A., Brady J.A., Finlayson S.A., Buchanan C.D., Summer E.J., Sun F., Klein P.E., Klein R.R., Pratt L.H., Cordonnier-Pratt M.M., Mullet J.E. (2005). Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 138: 352–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engström P., Wasserman W.W., Lenhard B. (2004). JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 32 (Database issue): D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Tsuda K., Wang L., Coller J., Watanabe Y., Glazebrook J., Katagiri F. (2010). Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P.M., Kazan K., Wilson I., Anderson J.P., Richmond T., Somerville S.C., Manners J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Johnson J.S., Dong X. (2007). Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. USA 104: 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z.L., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Tada Y., Loake G.J. (2010). Post-translational protein modification as a tool for transcription reprogramming. New Phytol. 186: 333–339 [DOI] [PubMed] [Google Scholar]
- Staswick P.E., Tiryaki I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T., Whiteman N.K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G.H., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Van der Ent S., Van Wees S.C.M., Pieterse C.M.J. (2009). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588 [DOI] [PubMed] [Google Scholar]
- Van Leene J., et al. (2007). A tandem affinity purification-based technology platform to study the cell cycle interactome in Arabidopsis thaliana. Mol. Cell. Proteomics 6: 1226–1238 [DOI] [PubMed] [Google Scholar]
- Van Verk M.C., Bol J.F., Linthorst H.J.M. (2011). Prospecting for genes involved in transcriptional regulation of plant defenses, a bioinformatics approach. BMC Plant Biol. 11: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees S.C.M., Luijendijk M., Smoorenburg I., van Loon L.C., Pieterse C.M.J. (1999). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41: 537–549 [DOI] [PubMed] [Google Scholar]
- Verhage A., Van Wees S.C.M., Pieterse C.M.J. (2010). Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 154: 536–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A., Vlaardingerbroek I., Raaymakers C., Van Dam N.M., Dicke M., Van Wees S.C.M., Pieterse C.M.J. (2011). Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling L.L. (2008). Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A.H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17: 1784–1790 [DOI] [PubMed] [Google Scholar]
- Wu Z.J., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Kloepper J.W., Ryu C.M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14: 1–4 [DOI] [PubMed] [Google Scholar]
- Zander M., Chen S., Imkampe J., Thurow C., Gatz C. (2012). Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 5: 831–840 [DOI] [PubMed] [Google Scholar]
- Zander M., La Camera S., Lamotte O., Métraux J.-P., Gatz C. (2010). Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 61: 200–210 [DOI] [PubMed] [Google Scholar]
- Zarei A., Körbes A.P., Younessi P., Montiel G., Champion A., Memelink J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.L., Tessaro M.J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15: 2647–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.-Y., Spivey N.W., Zeng W., Liu P.-P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]