This work identifies a role for the Arabidopsis Elongator complex subunit ELP2 in somatic DNA demethylation/methylation and suggests that ELP2-mediated epigenetic regulation plays a vital function in plant immune responses.
Abstract
The Arabidopsis thaliana Elongator complex subunit2 (ELP2) genetically interacts with NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), a key transcription coactivator of plant immunity, and regulates the induction kinetics of defense genes. However, the mechanistic relationship between ELP2 and NPR1 and how ELP2 regulates the kinetics of defense gene induction are unclear. Here, we demonstrate that ELP2 is an epigenetic regulator required for pathogen-induced rapid transcriptome reprogramming. We show that ELP2 functions in a transcriptional feed-forward loop regulating both NPR1 and its target genes. An elp2 mutation increases the total methylcytosine number, reduces the average methylation levels of methylcytosines, and alters (increases or decreases) methylation levels of specific methylcytosines. Interestingly, infection of plants with the avirulent bacterial pathogen Pseudomonas syringae pv tomato DC3000/avrRpt2 induces biphasic changes in DNA methylation levels of NPR1 and PHYTOALEXIN DEFICIENT4 (PAD4), which encodes another key regulator of plant immunity. These dynamic changes are blocked by the elp2 mutation, which is correlated with delayed induction of NPR1 and PAD4. The elp2 mutation also reduces basal histone acetylation levels in the coding regions of several defense genes. Together, our data demonstrate a new role for Elongator in somatic DNA demethylation/methylation and suggest a function for Elongator-mediated chromatin regulation in pathogen-induced transcriptome reprogramming.
INTRODUCTION
Immune responses are essential for both plants and animals to defend against microbial pathogens. Unlike animals, plants do not have any mobile cells specialized for defense but instead rely on individual cells to recognize pathogens and activate immune responses. In response to pathogen attack, plant cells reprogram their transcriptional profiles to mount a defense at the expense of normal cellular functions. The strength of the defense correlates with the kinetics and magnitude of the transcriptional changes. Suppressing or delaying pathogen-induced transcription by pathogenic effectors or mutations in the defense machinery compromises resistance (Tao et al., 2003; Jones and Dangl, 2006). Thus, it is crucial for plant cells to rapidly and efficiently reprogram transcription to fight infection.
In eukaryotic cells, RNA Polymerase II catalyzes the transcription of protein-encoding genes. A multitasking protein complex named Elongator was first identified as an interactor of hyperphosphorylated (elongating) RNA Polymerase II in yeast (Otero et al., 1999) and was later purified from human and Arabidopsis thaliana cells (Hawkes et al., 2002; Kim et al., 2002; Nelissen et al., 2010). Elongator consists of six subunits (Elongator complex subunit1 [ELP1]/ELONGATA2 [ELO2]/ABSCISIC ACID-OVERLY SENSITIVE1, ELP2, ELP3/ELO3, ELP4/ELO1, ELP5, and ELP6) that act together as a functional unit, with ELP1 and ELP2 serving as scaffolds for complex assembly, ELP3 being the catalytic subunit, and ELP4-6 forming an accessory complex. Loss of any Elongator subunit compromises its integrity, rendering the complex inactive (Versées et al., 2010). Elongator has been shown to function in several distinct cellular processes, including histone modification, tRNA modification, exocytosis, α-tubulin acetylation, and zygotic paternal genome demethylation (Hawkes et al., 2002; Huang et al., 2005; Rahl et al., 2005; Creppe et al., 2009; Okada et al., 2010). Mutations in yeast Elongator subunits lead to resistance to the zymocin γ-toxin subunit, defects in transcriptional silencing, and sensitivity to salt, caffeine, temperature, and DNA damaging agents (Otero et al., 1999; Jablonowski et al., 2001; Krogan and Greenblatt, 2001). In humans, Elongator deficiency causes familial dysautonomia, an autosomal recessive disease characterized by abnormally low numbers of neurons in the autonomic and sensory nervous systems (Anderson et al., 2001; Slaugenhaupt et al., 2001). In Arabidopsis, mutations of Elongator subunits result in pleiotropic effects, including hypersensitivity to abscisic acid, resistance to oxidative stress, severely aberrant auxin phenotypes, disease susceptibility, and altered cell cycle progression (Nelissen et al., 2005, 2010; Chen et al., 2006; Zhou et al., 2009; DeFraia et al., 2010; Xu et al., 2012).
The Elongator catalytic subunit ELP3 harbors a C-terminal histone acetyltranferase (HAT) domain and an N-terminal Cys-rich motif that resembles an iron-sulfur radical S-adenosylmethionine (SAM) domain (Chinenov, 2002; Winkler et al., 2002). ELP3 has intrinsic HAT activity and is capable of acetylating all four histones, whereas the six-subunit holo-Elongator predominantly acetylates Lys-14 of histone H3 and Lys-8 of histone H4 (Wittschieben et al., 1999; Winkler et al., 2002). Subsequently, the levels of acetylated histones H3 and H4 are reduced in yeast, human, and Arabidopsis elp mutants (Winkler et al., 2002; Close et al., 2006; Nelissen et al., 2010). Elongator may also have another catalytic function suggested by the fact that the archaea Methanocaldococcus jannaschii ELP3 binds and cleaves SAM (Paraskevopoulou et al., 2006). Indeed, a recent study indicated that the radical SAM domain of mouse ELP3, but not the HAT domain, is required for Elongator’s function in zygotic paternal genome demethylation (Okada et al., 2010), suggesting that mouse ELP3 may be a radical SAM protein catalyzing active DNA demethylation in zygotes. However, it is unknown whether Elongator functions in DNA demethylation in nondividing somatic cells and whether this activity is evolutionarily conserved in plants.
Previous characterization of loss-of-function mutants of Arabidopsis ELP2 revealed that elp2 genetically interacts with a mutation in NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1)/NONINDUCIBLE IMMUNITY1/SALICYLIC ACID INSENSITIVE1 (SAI1), which encodes a key transcription coactivator regulating plant immunity (Cao et al., 1997; Ryals et al., 1997; Shah et al., 1997; DeFraia et al., 2010). Whereas NPR1 mostly affects the scale of defense gene expression, ELP2 regulates the kinetics of defense gene induction. Simultaneous removal of NPR1 and ELP2 completely compromises the resistance mediated by two different plant resistance (R) proteins, demonstrating the distinction between ELP2 and NPR1. At the transcriptional level, ELP2 regulates several NPR1 target genes, suggesting that ELP2 and NPR1 may have some overlapping functions. However, the mechanistic relationship between ELP2 and NPR1 in pathogen-induced transcriptional changes and how ELP2 regulates the kinetics of defense gene induction remain unclear.
Here, we performed in-depth characterization of the elp2 mutant using microarrays, chromatin immunoprecipitation, and genome-wide or locus-specific bisulfite sequencing. Our results show that ELP2 regulates the kinetics of pathogen-induced transcriptome reprogramming, maintains histone acetylation levels in several defense genes, modulates the genomic DNA methylation landscape, and influences pathogen-induced dynamic DNA methylation changes. Thus, Elongator plays an evolutionarily conserved role in DNA demethylation/methylation in plants and likely functions as an epigenetic regulator of plant immune responses.
RESULTS
The elp2 Mutation Exhibits a Broader and Stronger Impact Than npr1 on Pathogen-Induced Transcriptome Changes
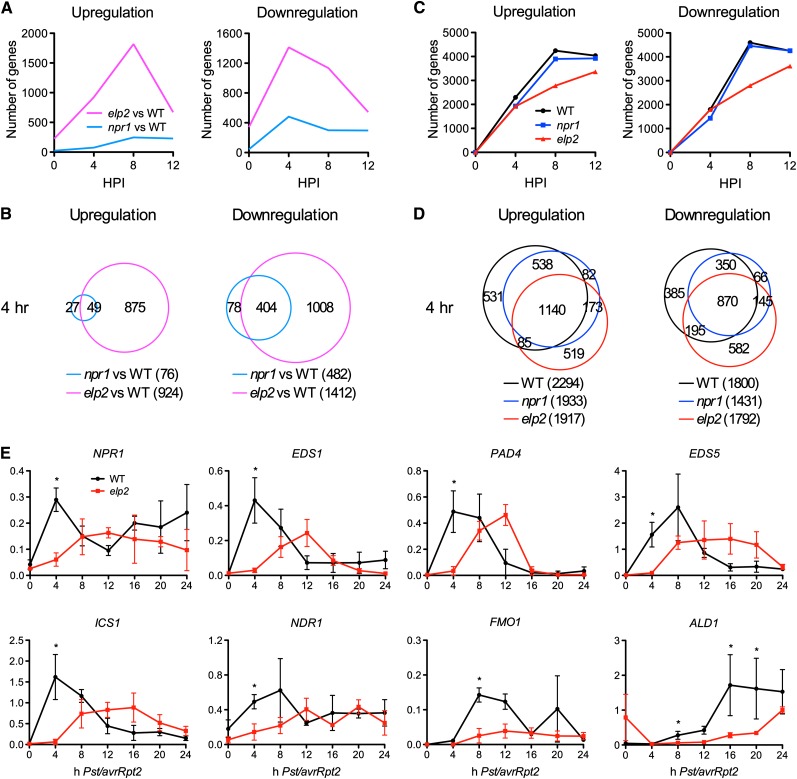
In order to identify and compare ELP2 target genes with those of NPR1 at the genome level, we performed a microarray experiment to monitor the avirulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) DC3000/avrRpt2-induced transcriptome changes in elp2, npr1, and the wild type (National Center for Biotechnology Information [NCBI] Gene Expression Omnibus series number GSE38986). Triplicate experiments were performed independently, and the data were analyzed to identify genes that were differentially expressed between elp2 or npr1 and the wild type. We used P values to identify differentially expressed candidate genes between elp2 and the wild type and then performed real-time quantitative PCR (qPCR) to verify the identified genes. Eight thus selected defense genes were all confirmed to be differentially expressed between elp2 and the wild type (Figure 1E); therefore, the P values computed for microarray analysis were not corrected for multiple testing. Genes that showed a twofold or larger difference in their expression levels with a low P value (≤0.05) were chosen for further analysis. Considerably more genes were differentially expressed between elp2 and the wild type than between npr1 and the wild type (Figure 1A). A total of 568, 2336, 2951, and 1218 genes were differentially expressed between elp2 and the wild type at 0, 4, 8, and 12 h after inoculation (hpi), respectively, whereas only 69, 558, 547, and 525 genes were differentially expressed between npr1 and the wild type. These results suggest that the elp2 mutation has a broader impact than npr1 on Pst DC3000/avrRpt2-induced transcriptome changes. Thus, ELP2 is a more general regulator of transcription than NPR1 in Pst DC3000/avrRpt2-induced immune response.
Figure 1.
Pathogen-Induced Transcriptome Changes in elp2.
(A) Dynamic changes in the numbers of genes that are differentially expressed between elp2 and the wild type (WT) and between npr1 and the wild type after Pst DC3000/avrRpt2 infection.
(B) Overlaps between the genes that are differentially expressed at 4 hpi between elp2 and the wild type and those between npr1 and the wild type.
(C) Dynamic changes in the numbers of genes that are up- or downregulated in the wild type, npr1, and elp2 after Pst DC3000/avrRpt2 infection.
(D) Overlaps among the genes that are up- or downregulated at 4 hpi in the wild type, npr1, and elp2.
(E) Expression of eight major defense genes in Pst DC3000/avrRpt2-infected wild-type and elp2 plants. The y axes indicate relative expression levels monitored by qPCR (results in [A] to [D] were from microarray analysis). Expression levels were normalized against UBQ5. The x axes indicate hours after Pst DC3000/avrRpt2 infection. Data represent the mean of three independent samples with sd. An asterisk indicates a significant difference between the wild type and elp2 (P < 0.05, t test).
To identity the degree of functional overlap between ELP2 and NPR1, we compared the differentially expressed genes in elp2 and npr1. Surprisingly, we found that a large number of genes influenced by NPR1 were also under the regulation of ELP2 (Figure 1B; see Supplemental Figure 1A online). ELP2 and NPR1, as positive regulators of defense genes, exhibited the most dramatic overlapping functions at 4 hpi when ELP2 was required for the full expression of ∼83.8% of the genes that were positively regulated by NPR1 (Figure 1B). These results indicate that ELP2 and NPR1 have significantly overlapping functions in plant immune responses.
To test whether ELP2 or/and NPR1 regulate the kinetics of pathogen-induced transcriptome changes, we queried the microarray data and identified genes that showed a twofold or higher induction or suppression with a low P value (≤0.05) in elp2, npr1, and the wild type. We found that all three genotypes exhibited dramatic transcriptional reprogramming upon Pst DC3000/avrRpt2 infection (Figure 1C). Interestingly, although both elp2 and npr1 significantly shifted their transcriptome profiles (Figure 1D; see Supplemental Figure 1B online), a dramatic effect on the kinetics of Pst DC3000/avrRpt2-induced transcriptome changes was seen only in elp2 (Figure 1C). While the numbers of genes up- or downregulated in npr1 and the wild type were highest at 8 hpi, in elp2, they were higher at 12 hpi. At 8 hpi, 3900 and 4461 genes in npr1, and 4242 and 4592 genes in the wild type were up- and downregulated, respectively, whereas only 2782 and 2796 genes in elp2 were up- and downregulated, respectively. Even at 12 hpi, the numbers of genes up- or downregulated in elp2 were still smaller than those in npr1 and the wild type (Figure 1C). Therefore, the elp2 genome responded more slowly to Pst DC3000/avrRpt2 infection than those of npr1 and the wild type, suggesting that the elp2 mutation has a larger impact than npr1 on pathogen-induced transcriptional reprogramming.
ELP2 Regulates NPR1 and Its Target Genes
As a more general regulator of transcription, ELP2 might regulate pathogen-induced transcriptome changes through major defense genes, such as NPR1 (Cao et al., 1997). To test this hypothesis, we analyzed the genes that were differentially expressed between elp2 and the wild type. Interestingly, the induction of NPR1 and many of the well-characterized major defense genes, such as ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), PHYTOALEXIN DEFICIENT4 (PAD4), EDS5/SA INDUCTION DEFICIENT1, ISOCHORISMATE SYNTHASE1 (ICS1)/SID2/EDS16, NON-RACE-SPECIFIC DISEASE RESISTANCE1 (NDR1), AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (ALD1), and FLAVIN-DEPENDENT MONOOXYGENASE1 (FMO1), was either delayed or decreased in elp2 compared with the wild type after Pst DC3000/avrRpt2 infection (Table 1) (Century et al., 1997; Falk et al., 1999; Jirage et al., 1999; Wildermuth et al., 2001; Nawrath et al., 2002; Song et al., 2004; Mishina and Zeier, 2006). To confirm the microarray results, we used real-time qPCR to monitor the induction of these genes in elp2 after DC3000/avrRpt2 infection. As shown in Figure 1E, induction of all eight genes was delayed and/or decreased in elp2, suggesting that ELP2 may function upstream of these major defense genes to regulate plant immune responses.
Table 1. Defense Genes That Are Differentially Expressed between elp2 and the Wild Type during Pst DC3000/avrRpt2 Infection.
|
elp2/Wild Type |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 h |
4 h |
8 h |
12 h |
|||||||
| AGI Locus | Gene name | Log2 (FC) | P Value | Log2 (FC) | P Value | Log2 (FC) | P Value | Log2 (FC) | P Value | AGI Description |
| Major defense genes | ||||||||||
| At1g64280 | NPR1 | −0.494 | 0.001 | −1.783 | 0 | −0.389 | 0.001 | −0.074 | 0.287 | NONEXPRESSOR OF PR GENES1 |
| At5g45110 | NPR3 | −1.024 | 0.009 | −1.869 | 0.006 | – | n/a | – | n/a | NPR1-LIKE PROTEIN3 |
| At4g19660 | NPR4 | – | n/a | −1.435 | 0.006 | – | n/a | – | n/a | NPR1-LIKE PROTEIN4 |
| At4g16890 | SNC1 | – | n/a | −2.361 | 0 | – | n/a | – | n/a | SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
| At1g02450 | NIMIN1 | – | n/a | −3.318 | 0.017 | – | n/a | 1.142 | 0.002 | NIM-INTERACTING1 |
| At3g25882 | NIMIN2 | – | n/a | −2.493 | 0.034 | −1.099 | 0.001 | – | n/a | NIM-INTERACTING2 |
| At1g74710 | ICS1 | – | n/a | −4.154 | 0.001 | – | n/a | – | n/a | ISOCHORISMATE SYNTHASE1 |
| At4g39030 | EDS5 | – | n/a | −3.815 | 0.003 | – | n/a | – | n/a | ENHANCED DISEASE SUSCEPTIBILITY5 |
| At5g13320 | PBS3 | – | n/a | −3.979 | 0.008 | – | n/a | – | n/a | AVRPPHB SUSCEPTIBLE3 |
| At3g48090 | EDS1 | – | n/a | −2.208 | 0.007 | – | n/a | – | n/a | ENHANCED DISEASE SUSCEPTIBILITY1 |
| At3g52430 | PAD4 | – | n/a | −3.458 | 0.003 | – | n/a | 1.257 | 0.002 | PHYTOALEXIN DEFICIENT4 |
| At5g14930 | SAG101 | – | n/a | −1.055 | 0.013 | – | n/a | – | n/a | SENESCENCE-ASSOCIATED GENE101 |
| At4g23570 | SGT1A | – | n/a | −3.091 | 0 | – | n/a | 1.001 | 0.003 | Suppressor of G2 (Two) 1A |
| At4g14400 | ACD6 | −2.818 | 0.023 | −3.125 | 0.011 | −1.368 | 0 | – | n/a | ACCELERATED CELL DEATH6 |
| At2g13810 | ALD1 | – | n/a | −4.106 | 0.002 | −3.129 | 0.002 | −2.105 | 0 | AGD2-LIKE DEFENSE RESPONSE PROTEIN1 |
| At1g19250 | FMO1 | – | n/a | −4.486 | 0.004 | −2.561 | 0.005 | −1.447 | 0.001 | FLAVIN_DEPENDENT MOMOOXYGENASE1 |
| NPR1 target genes | ||||||||||
| At2g14610 | PR1 | −1.024 | 0.174 | 2.325 | 0.151 | −6.469 | 0 | −3.246 | 0.001 | PATHOGENESIS-RELATED GENE1 |
| At3g57260 | PR2 | −2.147 | 0.18 | – | n/a | −1.93 | 0 | −2.347 | 0.008 | PATHOGENESIS-RELATED GENE2 |
| At1g75040 | PR5 | −1.37 | 0 | −1.334 | 0.23 | −1.924 | 0 | – | n/a | PATHOGENESIS-RELATED GENE5 |
| At2g43570 | n.a. | −2.185 | 0.131 | – | n/a | −1.882 | 0.002 | – | n/a | Chitinase, putative |
| At4g12010 | n.a. | – | n/a | −1.31 | 0.006 | – | n/a | – | n/a | Disease resistance protein, putative |
| At4g34480 | n.a. | – | n/a | – | n/a | −1.01 | 0.003 | – | n/a | Glycosyl hydrolase family 17 protein |
| At5g43470 | RPP8 | – | n/a | −1.14 | 0.003 | – | n/a | – | n/a | RECOGNITION OF PERONOSPORA PARASITICA8 |
| At5g57550 | XTR3 | −1.77 | 0.001 | 1.332 | 0.018 | 1.362 | 0 | 1.256 | 0.015 | XYLOGLUCAN ENDOTRANSGLYCOSYLASE3 |
| At1g08450 | CRT3 | – | n/a | −1.653 | 0.003 | −1.293 | 0 | – | n/a | CALRETICULIN 3; calcium ion binding |
| At1g09210 | CRT2 | – | n/a | – | n/a | −1.755 | 0 | – | n/a | Calreticulin2 |
| At1g30900 | VSR6 | – | n/a | −2.128 | 0.023 | −1.8 | 0 | – | n/a | Vacuolar sorting receptor6 |
| At2g34250 | Sec61α | – | n/a | – | n/a | −1.161 | 0.106 | – | n/a | Protein transport protein, putative |
| At2g47470 | PDIL2-1 | – | n/a | – | n/a | −1.235 | 0 | – | n/a | PDI-LIKE2-1; thiol-disulfide exchange intermediate |
| At4g22670 | HIP1 | – | n/a | – | n/a | −1.084 | 0.029 | – | n/a | HSP70-INTERACTING PROTEIN1; binding |
| At4g24190 | SHD | – | n/a | – | n/a | −1.801 | 0 | – | n/a | SHEPHERD; ATP binding |
| At5g07340 | n.a. | – | n/a | – | n/a | −1.548 | 0.007 | – | n/a | Calnexin, putative |
| At5g42020 | BIP | – | n/a | – | n/a | −1.978 | 0.001 | – | n/a | LUMINAL BINDING PROTEIN; ATP binding |
| At5g61790 | CNX1 | – | n/a | – | n/a | −1.878 | 0 | – | n/a | Calnexin 1 |
| At4g31800 | WRKY18 | −1.377 | 0.012 | – | n/a | – | n/a | – | n/a | WRKY DNA binding protein 18 |
| At5g22570 | WRKY38 | −1.34 | 0.088 | – | n/a | −1.301 | 0.001 | – | n/a | WRKY DNA binding protein 38 |
| At4g23810 | WRKY53 | −1.144 | 0.006 | −1.777 | 0.004 | – | n/a | – | n/a | WRKY DNA binding protein 53 |
| At2g40750 | WRKY54 | −1.781 | 0.022 | −3.326 | 0.002 | – | n/a | – | n/a | WRKY DNA binding protein 54 |
| At3g01080 | WRKY58 | – | n/a | – | n/a | −1.374 | 0 | – | n/a | WRKY DNA binding protein 58 |
| At2g21900 | WRKY59 | – | n/a | −1.926 | 0 | −1.714 | 0 | 1.335 | 0.004 | WRKY DNA binding protein 59 |
| At1g80590 | WRKY66 | – | n/a | – | n/a | −1.365 | 0.005 | – | n/a | WRKY DNA binding protein 66 |
| At3g56400 | WRKY70 | −1.666 | 0.021 | −2.197 | 0.024 | – | n/a | – | n/a | WRKY DNA binding protein 70 |
AGI, Arabidopsis Genome Initiative; FC, fold change; n.a., not available; n/a, not applicable; –, less than twofold.
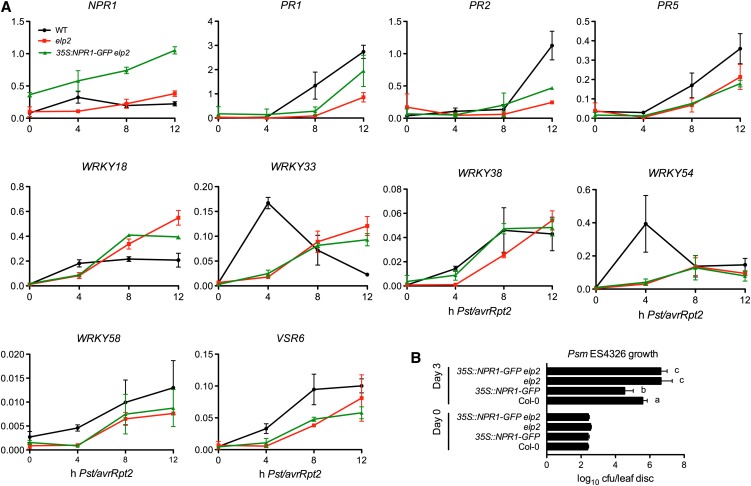
Since ELP2 regulates NPR1 induction, we expected that ELP2 would also regulate the expression of NPR1 target genes. Indeed, expression of the majority of the NPR1 target genes, which were identified in a previous study (Wang et al., 2005), were reduced in elp2 compared with the wild type at either one or more of the four time points after Pst DC3000/avrRpt2 infection (Table 1), suggesting that ELP2 is a key regulator of NPR1-mediated transcription. To find out how ELP2 is involved in the NPR1 transcriptional cascade, we crossed the well-characterized 35S:NPR1-GFP (for green fluorescent protein) transgene into elp2 and tested whether overexpression of NPR1-GFP could rescue the induction pattern of NPR1 target genes (Kinkema et al., 2000). As shown in Figure 2A, overexpression of NPR1-GFP did not restore the expression pattern of eight out of the nine tested NPR1 target genes. Furthermore, the heightened basal resistance conferred by overexpression of NPR1-GFP was completely suppressed by the elp2 mutation (Figure 2B) (Cao et al., 1998). These results suggest that ELP2 may also function independently of NPR1 to regulate the transcription of NPR1 target genes.
Figure 2.
Epistasis between elp2 and the 35:NPR1-GFP Transgene
(A) Expression of NPR1 and nine NPR1 target genes in Pst DC3000/avrRpt2-infected wild-type (WT), elp2, and 35S:NPR1-GFP elp2 plants. The y axes indicate relative expression levels. Expression levels were monitored using qPCR and normalized against UBQ5. The x axes indicate hours after Pst DC3000/avrRpt2 infection. Data represent the mean of three independent samples with sd.
(B) Growth of Psm ES4326 in wild-type, elp2, and 35S:NPR1-GFP elp2 plants. Data represent the mean of eight independent samples with sd. Different letters on the right of the bars indicate significant differences (P < 0.05, t test).
Experiments were repeated three times with similar results.
ELP2 Regulates Histone Acetylation Levels in Several Defense Genes
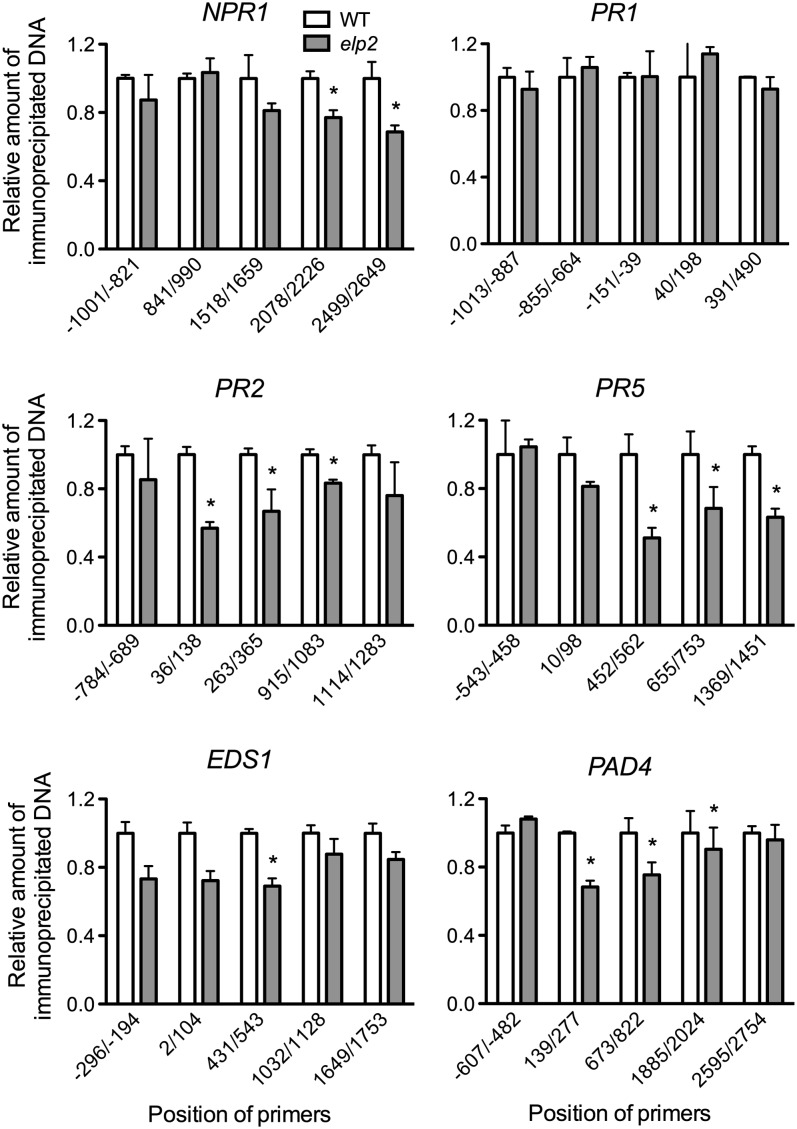
Elongator possesses HAT activity essential for maintaining normal histone acetylation levels in yeast, humans, and plants (Winkler et al., 2002; Close et al., 2006; Nelissen et al., 2010). To test whether ELP2 is required for maintaining normal (basal) histone acetylation levels in defense genes, we assessed the acetylation status of histone H3 in NPR1, PR1, PR2, PR5, EDS1, and PAD4 using chromatin immunoprecipitation. After formaldehyde cross-linking and cell lysis of elp2 and wild-type leaf tissues, histone-DNA complexes were immunoprecipitated using an antibody specific for histone H3 acetylated at Lys-9 and -14 (H3K9/14ac). Precipitated DNA was quantified using real-time qPCR to estimate the levels of histone H3K9/14ac. Interestingly, although the basal expression levels of NPR1, PR1, PR2, PR5, EDS1, and PAD4 were comparable in elp2 and the wild type (see Supplemental Figure 2 online), histone H3K9/14ac levels in the coding regions of these defense genes except PR1 were significantly lower in elp2 than in the wild type (Figure 3). Since histone acetylation is generally associated with transcriptional activation (Workman and Kingston, 1998), reduced basal histone acetylation levels may contribute to the delayed or/and decreased induction of defense genes in elp2.
Figure 3.
Histone H3 Acetylation Levels in Several Defense Genes.
The position of the primers is relative to the initiation ATG codon. The relative amount of immunoprecipitated chromatin fragments (as determined by real-time qPCR) from elp2 was compared with that from the wild type (WT; arbitrarily set to 1). Data represent the mean of three independent samples with sd. An asterisk indicates a significant difference between elp2 and the wild type (P < 0.05, t test). The experiment was repeated four times with similar results.
ELP2 Modulates DNA Methylation in Arabidopsis
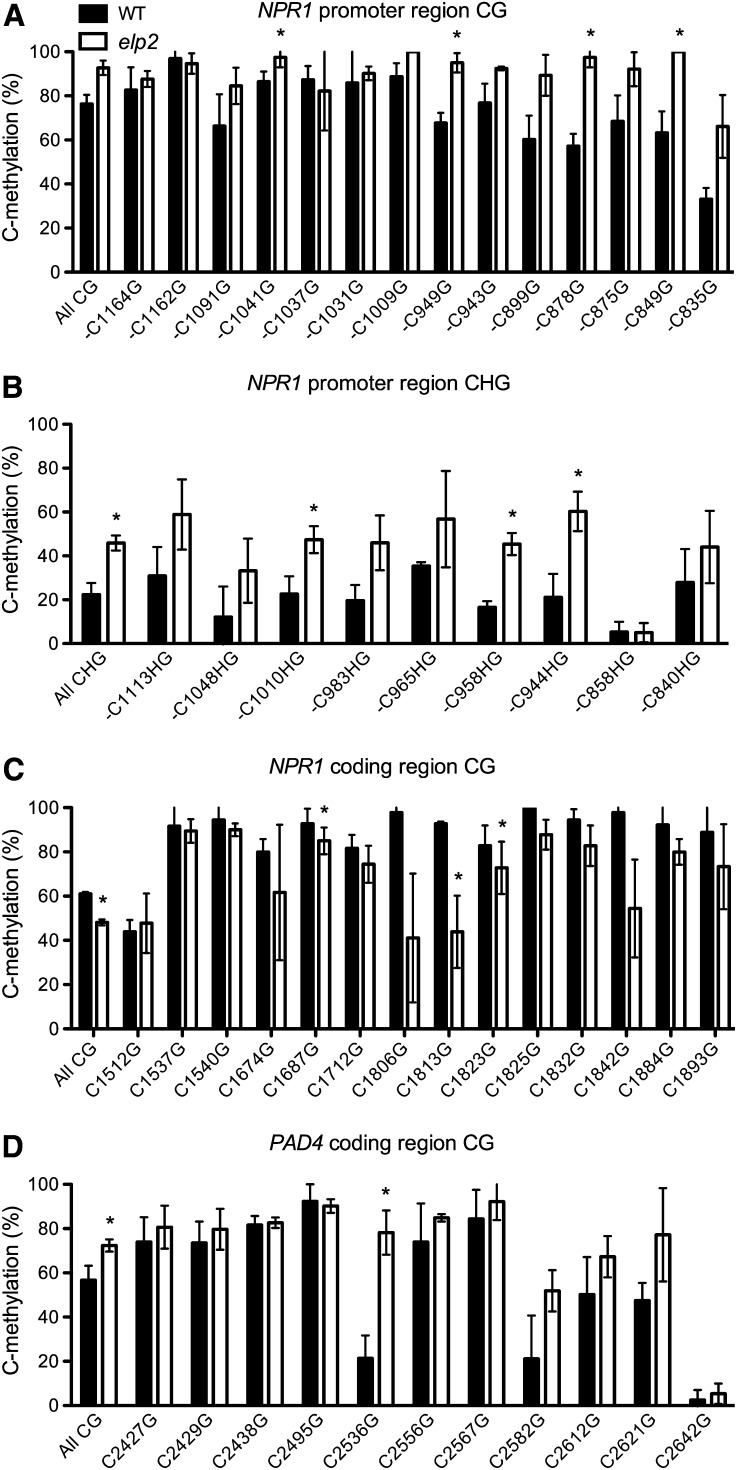
As Elongator plays a critical role in paternal genome demethylation in mouse zygotes (Okada et al., 2010), we asked whether Elongator modulates DNA methylation in Arabidopsis. To address this question, we first analyzed DNA methylation levels using bisulfite sequencing of several ELP2-regulated defense genes (see Supplemental Table 1 online). Consistent with the results reported previously (Lister et al., 2008), the cytosines in the sequenced regions of NPR1 and PAD4 were significantly methylated, while those in PR1, PR2, and PR5 (except three cytosines in PR2) were not methylated (Figure 4; see Supplemental Figure 3 online). Methylation occurred at CG, CHG, and CHH sites in the NPR1 promoter region (Figures 4A and 4B; see Supplemental Figure 3A online) but was largely restricted to CG dinucleotides in the NPR1 and PAD4 coding regions (Figures 4C and 4D). Interestingly, DNA methylation levels were generally higher in elp2 than in the wild type except at the NPR1 coding region, where DNA methylation levels were lower in elp2. In the NPR1 promoter and the PAD4 coding regions, methylation levels at several specific methylcytosines were significantly higher in elp2 than in the wild type (Figures 4A, 4B, and 4D). These results indicate that ELP2 modulates basal DNA methylation levels in NPR1 and PAD4, which could suggest that ELP2 regulates defense gene expression through DNA methylation.
Figure 4.
DNA Methylation Levels in Several Defense Genes.
(A) and (B) DNA methylation levels at CG sites (A) and CHG sites (B) of the NPR1 promoter region in elp2 and the wild type (WT).
(C) and (D) DNA methylation levels at CG sites in the coding region of NPR1 (C) and PAD4 (D) in elp2 and the wild type.
DNA samples were extracted from three biological replicates of each genotype. After bisulfite conversion and PCR amplification, the PCR products were cloned into pGEM-T easy vector. A total of 45 independent clones were sequenced for each genotype (15 for each DNA sample). The 15 clones from the same DNA sample were used to calculate methylation levels, which were then used for statistical analysis. Data represent the mean of three independent samples with sd. An asterisk indicates a significant difference between elp2 and the wild type (P < 0.05, t test).
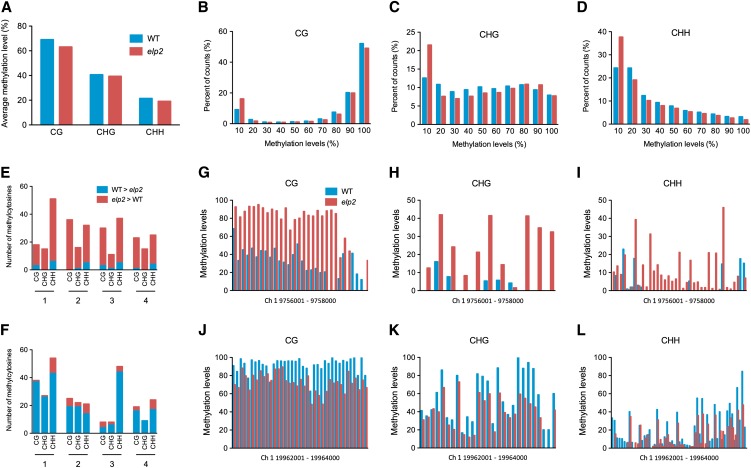
To reveal broad effects of ELP2 on DNA methylation profiles, we generated genome-scale DNA methylation maps of elp2 and the wild type using bisulfite deep sequencing (BS-Seq) (NCBI Short Read Archive accession number SRA055073). A total of 58,976,071 and 48,090,948 sequence reads were mapped in elp2 and the wild type, respectively, of which 24,234,987 and 14,930,215 were unique (nonclonal); and a total of 3,850,501 cytosines were covered by at least 10 reads in both elp2 and the wild type. Based on the reads aligned to the unmethylated chloroplast genome that was isolated and sequenced in conjunction with the nuclear genome, the average nonconversion rate of the bisulfite conversion was 0.28% and the average thymidine-to-cytosine sequencing error rate was 0.30% (see Supplemental Table 2 online). Using 0.58% (0.28% plus 0.30%) as a measure of the false methylcytosine discovery rate, a binomial probability distribution was used to calculate the minimum sequence depth at a cytosine position at which a methylcytosine could be called while maintaining a false positive rate below 5%. From the 3,850,501 cytosines, 2,204,921 were identified as methylcytosines in at least one genotype, which accounts for 5.12% of all genomic cytosine. These methylcytosines were used for further analysis.
Although some methylcytosines were identified in only one genotype, the majority (72.8%) of the methylcytosines in all sequence contexts (CG, CHG, and CHH) were identified in both elp2 and the wild type (see Supplemental Figure 4 online). The average methylation levels of methylcytosines in the CG, CHG, and CHH contexts in elp2 were 5.94, 1.27, and 2.34% lower than those in the wild type, respectively (Figure 5A). In all sequence contexts, significantly more methylcytosines in elp2 displayed a low percentage (<10%) of methylation than in the wild type (Figures 5B to 5D). However, although the distribution patterns of methylcytosine were similar in elp2 and the wild type (see Supplemental Figure 5 online), more methylcytosines were identified in elp2 than in the wild type (see Supplemental Figures 4 and 5 online). Methylcytosine differences between elp2 and the wild type were evident in all sequence contexts with more prominent differences being identified in the CHG and CHH contexts (see Supplemental Figure 5 online). The hypermethylated regions in elp2 were evenly distributed along the chromosomes except for the centromeric regions (see Supplemental Figure 5 online). We scanned the methylcytosines on chromosome 1 and identified several regions where DNA methylation levels in elp2 and the wild type differed significantly (Figures 5E and 5F). In each of these regions, the patterns of DNA methylation in all sequence contexts varied dramatically between elp2 and the wild type (Figures 5G to 5L). We observed that the DNA methylation pattern of PAD4 revealed by the genome-wide BS-Seq was similar to that detected by traditional bisulfite sequencing (Figure 4D; see Supplemental Figure 6 online), which validated the BS-Seq method employed in this study. Taken together, our results showed that the elp2 mutation increased the total number of methylcytosines, decreased average methylation levels of methylcytosines, and modulated (either increased or decreased) methylation levels of specific cytosines, suggesting that ELP2 is an epigenetic regulator modulating both DNA demethylation and methylation.
Figure 5.
Genomic DNA Methylation Profiles of elp2.
(A) Average genome-wide DNA methylation levels of elp2 and the wild type (WT).
(B) to (D) Distribution of methylation percentage in the sequence context of CG (B), CHG (C), and CHH (D) in elp2 and the wild type The x axes are divided into 10 individual bins that correspond to methylation levels. The y axes are the percentage of total counts for each respective bin.
(E) and (F) Regions on chromosome 1 where more methylcytosines are hypermethylated in either elp2 (E) or the wild type (F). Regions 1 to 4 correspond to nucleotides 8,390,001 to 8,392,000, 568,001 to 570,000, 9,756,001 to 9,758,000, and 3,876,001 to 3,878,000, respectively (E), or nucleotides 19,962,001 to 1,996,400, 11,318,001 to 11,320,000, 16,434,001 to 16,436,000, and 27,932,001 to 27,934,000, respectively (F).
(G) to (L) Methylation levels of the methylcytosines in all sequence contexts in two regions on chromosome 1.
ELP2 Is Required for Pathogen-Induced Dynamic DNA Methylation Changes in NPR1 and PAD4
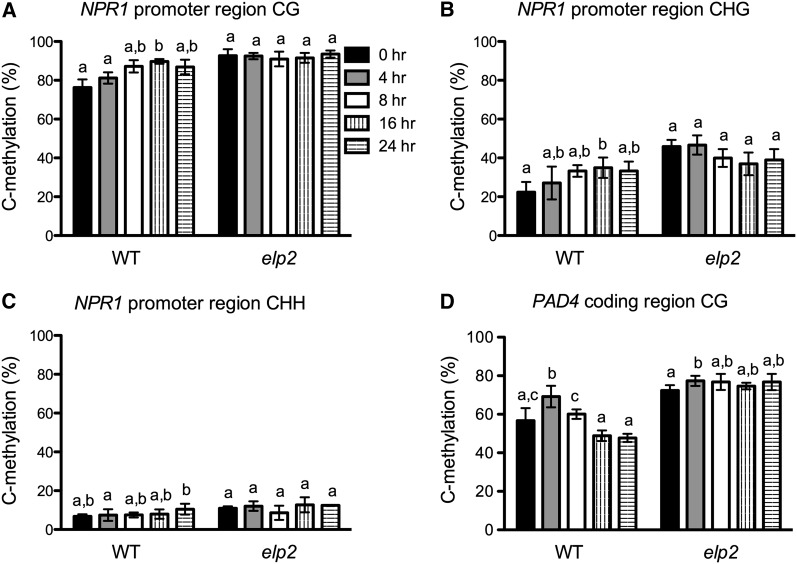
Pathogen infection has been shown to cause DNA hypomethylation in Arabidopsis (Pavet et al., 2006; Agorio and Vera, 2007; Dowen et al., 2012). Since ELP2 modulates basal DNA methylation levels, we asked whether ELP2 is involved in pathogen-induced DNA methylation changes. To address this question, we infected elp2 and wild-type leaves with Pst DC3000/avrRpt2 and analyzed DNA methylation in NPR1 and PAD4 at 0, 4, 8, 16, and 24 hpi. Surprisingly, during the first 24 h after Pst DC3000/avrRpt2 infection, DNA methylation levels changed dramatically in the wild type but remained relatively high and stable in elp2 (Figure 6; see Supplemental Figure 7 online). The most dramatic changes in DNA methylation levels occurred at the CG sites in the PAD4 coding region (Figure 6D; see Supplemental Figures 7J to 7L online). The overall average methylation level of the eleven CG sites in the sequenced PAD4 coding region increased from ∼56.7% at 0 hpi to ∼69.2% at 4 hpi, then dropped to ∼60.1, ∼48.9, and ∼47.7% at 8, 16, and 24 hpi, respectively (Figure 6D). At the PAD4 C2612G site, the methylation level increased from ∼50.2% at 0 hpi to ∼75.9% at 4 hpi, then dropped to ∼64.7, ∼24.6, and ∼14.4% at 8, 16, and 24 hpi, respectively (see Supplemental Figure 7K online). In the NPR1 promoter region, methylation levels at CG and CHG sites increased and reached the highest levels at 16 hpi, then decreased slightly at 24 hpi (Figures 6A and 6B; see Supplemental Figures 7A to 7F online), whereas methylation levels at CHH sites were low and did not display any reproducible dynamic patterns (Figure 6C; see Supplemental Figures 7G to 7I online). Taken together, these results indicate that ELP2 contributes to pathogen-induced dynamic changes in DNA methylation levels of two major defense genes.
Figure 6.
Pathogen-Induced Dynamic DNA Methylation Changes in NPR1 and PAD4.
(A) to (C) Average methylation levels of methylcytosines at CG sites (A), CHG sites (B), and CHH sites (C) of the NPR1 promoter region in elp2 and the wild type (WT).
(D) Average methylation levels of methylcytosines at CG sites of the PAD4 coding region in elp2 and the wild type.
DNA samples were extracted from three biological replicates of each genotype/time point. After bisulfite conversion and PCR amplification, the PCR products were cloned into pGEM-T easy vector. A total of 45 independent clones were sequenced for each genotype/time point (15 for each DNA sample). The 15 clones from the same DNA sample were used to calculate methylation levels, which were then used for statistical analysis. Data represent the mean of three independent samples with sd. Different letters above the bars indicate significant differences (P < 0.05, t test). Note that the comparison was made separately among time points for each genotype.
DISCUSSION
Elongator has been implicated in diverse biological processes, including exocytosis, embryogenesis, cell migration, cell proliferation, and responses to abiotic stresses (Nelissen et al., 2005; Rahl et al., 2005; Chen et al., 2006; Close et al., 2006; Creppe et al., 2009; Okada et al., 2010; Bauer et al., 2012). Our previous characterization of the Arabidopsis elp2 mutant uncovered a role for Elongator in plant immune responses (DeFraia et al., 2010). We showed that elp2 mutations delay or/and decrease the induction of several defense genes, but the underlying mechanism remains unclear. Here, we provide evidence that ELP2 functions in DNA demethylation/methylation and histone acetylation and is involved in plant immunity by directly or indirectly affecting the kinetics of pathogen-induced transcriptome reprogramming.
Induction of many defense genes is delayed in the elp2 mutant (Figure 1E). Similar expression patterns were observed for several stress-inducible genes in yeast elpΔ cells (Otero et al., 1999). Expression of GAL1-10, PHO5, and ENA1 is delayed in elpΔ cells following transfer to media containing Gal, low phosphate, and high salt, respectively. Although these results clearly showed that Elongator is involved in regulating the expression kinetics of individual stress-inducible genes, whether Elongator governs the kinetics of stress-induced transcriptome changes is not yet known. Our results suggest that Elongator plays a critical role in shaping the kinetics of pathogen-induced transcriptome changes. Compared with the npr1 mutation, elp2 has a much stronger impact on the kinetics of pathogen-induced transcriptional reprogramming (Figure 1C). The npr1 mutation does not dramatically change the numbers of genes that are up- or downregulated after Pst DC3000/avrRpt2 infection, whereas elp2 significantly reduces the gene numbers at early time points. The numbers of genes that are up- or downregulated in elp2 reach higher levels at a later time point compared with the wild type and npr1, suggesting that elp2 delays genome-wide transcriptional responses to pathogen infection. These results, together with the pathogen susceptibility phenotype of elp2, indicate that ELP2 is required for the Arabidopsis genome to rapidly and efficiently reprogram its transcriptome to fend off pathogen attack.
ELP2 regulates the expression of a group of major defense genes, including NPR1, EDS1, PAD4, EDS5, ICS1, NDR1, ALD1, and FMO1 (Table 1, Figure 1E) (Cao et al., 1997; Century et al., 1997; Falk et al., 1999; Jirage et al., 1999; Wildermuth et al., 2001; Nawrath et al., 2002; Song et al., 2004; Mishina and Zeier, 2006), which suggests that ELP2 may regulate plant immune responses through these major defense genes. ELP2 may also function independently of these genes, as seen for NPR1. In the NPR1 transcriptional cascade, ELP2 not only regulates NPR1 itself, but also regulates almost all of the NPR1 target genes (Table 1) (Wang et al., 2005). Overexpression of the previously characterized transgene NPR1-GFP in elp2 does not restore the induction pattern of most of the NPR1 target genes tested in our experiment (Figure 2A) (Kinkema et al., 2000), indicating that ELP2 also functions independently of NPR1 and is involved in the transcription activation of NPR1 target genes. Consistently, increasing basal immunity by overexpression of NPR1-GFP requires ELP2 (Figure 2B) (Cao et al., 1998). Thus, in NPR1-mediated signal transduction, ELP2 functions in a transcriptional feed-forward loop, in which it regulates both NPR1 and its target genes.
Although ELP2 regulates the NPR1 transcriptional cascade, ELP2 and NPR1 appear to function largely independently of each other in effector-triggered immunity (ETI) (DeFraia et al., 2010). This paradox can be reconciled by the fact that other ELP2-regulated major defense genes, such as PAD4, EDS1, and NDR1, influence subsets of NPR1-independent genes (Wang et al., 2008). Mutations in ELP2 delay the induction of both NPR1-dependent and -independent defense genes, whereas mutations in NPR1 block the transcription of only NPR1-dependent defense genes. Our previous work has shown that the elp2 and npr1 single mutants are moderately susceptible to Pst DC3000/avrRpt2, whereas the elp2 npr1 double mutant is significantly (>130-fold) more susceptible than either elp2 or npr1 (DeFraia et al., 2010). Therefore, both ELP2-regulated kinetics and the NPR1-dependent magnitude of defense gene induction are crucial for ETI. Intriguingly, although ELP2 regulates the induction kinetics of a group of major defense genes that encode important regulators of systemic acquired resistance (SAR) (Table 1), ELP2 itself does not play a significant role in SAR (DeFraia et al., 2010). It is possible that the basal expression levels of the major defense genes, which are not affected by the elp2 mutation (see Supplemental Figure 2 online), are sufficient for the establishment of SAR, as shown for the NPR1 gene (van Wees et al., 2000). It is also possible that establishment of SAR does not depend on the kinetics of defense gene induction, but rather on the magnitude of the induction. In any case, elp2, as a unique mutant in which both basal immunity and ETI are compromised but SAR is not, is invaluable for dissecting the mechanistic differences between basal immunity or ETI and SAR (DeFraia et al., 2010).
It has been well documented that Elongator possesses HAT activity and is required for maintaining normal histone acetylation levels (Winkler et al., 2002; Close et al., 2006; Nelissen et al., 2010). Consistent with this, loss of ELP2 reduces basal histone acetylation levels in the coding regions of several defense genes (Figure 3). Elongator might also bear DNA demethylase activity, affecting paternal genome demethylation in mouse zygotes (Okada et al., 2010). However, it is unknown whether Elongator influences DNA demethylation in somatic cells and whether this function is conserved in other organisms. In this study, we found that elp2 causes dramatic genome-wide DNA methylation changes, including increased total number of methylcytosines (Figure 5; see Supplemental Figures 4 and 5 online). These changes appear to be more profound than those observed in the DNA demethylase triple mutant rdd, in which the total number of methylcytosines identified is similar to the wild type (Lister et al., 2008). Many regions in elp2 exhibit increased DNA methylation levels (Figures 5E and 5G to 5I), but some regions are hypomethylated (Figures 5F and 5J to 5L). This result suggests that, similarly to the human DNA methyltransferases Dnmt3a and 3b, Elongator may act in both DNA demethylation and methylation (Métivier et al., 2008). However, presently, no evidence is available indicating that Elongator acts upon DNA as a DNA demethylase or a DNA methyltransferase (Okada et al., 2010). This aspect of Elongator requires further investigation.
Methylation of genomic DNA in Arabidopsis is thought to be dynamically regulated by both active DNA demethylation and DNA methylation mechanisms (He et al., 2011). Consistent with this idea, we found that Pst DC3000/avrRpt2 infection induces biphasic changes in DNA methylation levels in the PAD4 coding region and, to a lesser extent, the NPR1 promoter region in mature mesophyll cells in the wild type (Figure 6; see Supplemental Figure 7 online). However, DNA methylation levels of these regions in elp2 are high (higher than the highest levels reached in the wild type) and do not change upon Pst DC3000/avrRpt2 infection. Since mature mesophyll (nondividing somatic) cells cannot lose methylation through the loss of maintenance, this result indicates that the elp2 mutation blocks pathogen infection-induced active DNA demethylation. Therefore, Elongator regulates genomic DNA methylation landscape likely through its DNA demethylation function in Arabidopsis.
Consistent with the notion that acetylation of histones H3 is generally associated with active transcription (Li et al., 2007), reduced histone H3 acetylation levels in the coding regions of several defense genes, including NPR1 and PAD4, are correlated with delayed or/and decreased induction in elp2 (Figures 1E and 3). Although basal expression levels of these defense genes are not significantly changed in the elp2 mutant (see Supplemental Figure 2 online), a correlation between reduced histone H3 acetylation and decreased basal expression of several auxin-related genes has been seen in the Arabidopsis Elongator mutant elo3-6 (Nelissen et al., 2010). Therefore, Elongator may help establish a transcriptionally active chromatin state at specific defense loci. A role for histone modification in establishment of active chromatin structures at plant defense loci is not without precedent. The Arabidopsis histone methyltransferase SET DOMAIN GROUP8 maintains H3K36me3 levels and regulates both basal and induced expression of particular R genes (Palma et al., 2010). ARABIDOPSIS HOMOLOG OF TRITHORAX1 establishes H3K4me3 patterns and regulates both basal and induced expression of WRK70, which encodes a key transcription factor of plant immunity (Li et al., 2006; Alvarez-Venegas et al., 2007). Histone modification may even be a molecular basis for defense priming, a phenomenon resulting in enhanced defense gene transcription upon a subsequent stress (Conrath, 2011; Berr et al., 2012). Indeed, priming of WRKY6, WRKY29, and WRKY53 is associated with an increase in histone H3 acetylation and H3K4me3 at their promoters (Jaskiewicz et al., 2011). Although ELP2 does not contribute significantly to the establishment of SAR (DeFraia et al., 2010), whether Elongator is involved in priming-mediated histone acetylation deserves further investigation.
Interestingly, besides histone acetylation, DNA methylation levels are elevated in the NPR1 promoter region and the PAD4 coding region in elp2 plants (Figure 4), which may also contribute to the delayed or/and decreased induction. Furthermore, we found that pathogen-induced dynamic changes in DNA methylation levels in the PAD4 coding region and, to a lesser extent, the NPR1 promoter region are correlated with the impulse response of the genes to pathogen infection (Figures 1E and 6; see Supplemental Figure 7 online). Dynamic DNA methylation changes have previously been implicated in regulation of gene transcription. During mouse muscle cell line differentiation, the dynamics of DNA demethylation in the 5′-flanking region and exon 1 of the myogenin gene is strongly correlated with its expression (Lucarelli et al., 2001). In human MDA-MB231 cells, transcriptional regulation of the human pS2 gene involves cyclical variation in CG methylation of the pS2 promoter (Métivier et al., 2008). The elp2 mutation blocks pathogen-induced DNA methylation changes in NPR1 and PAD4 and delays the induction of the defense genes, suggesting that, in Arabidopsis, pathogen-induced Elongator-dependent dynamic DNA methylation changes may play a role in regulating defense gene transcription.
Compared with other epigenetic regulators, Elongator is unique in that it regulates both global histone acetylation levels and genome-wide DNA methylation profiles (Figure 5) (Winkler et al., 2002; Nugent et al., 2010; Xu et al., 2012). The delayed genome-wide transcriptional response of elp2 to pathogen infection likely results from altered genome chromatin structure caused by both reduced histone acetylation levels and altered DNA methylation profiles. Proteins regulating global histone acetylation or/and genome-wide DNA methylation have been implicated in plant innate immunity. For instance, the Arabidopsis HISTONE DEACETYLASE19 (HDA19) appears to be involved in basal defense against the bacterial pathogen Pst DC3000 but results from examining the effect of hda19 mutations on disease resistance in several reports are contradictory (Tian et al., 2005; Kim et al., 2008; Choi et al., 2012). Knockout of SIRTUIN2, a homolog of yeast Silent information regulator2, which encodes an NAD+-dependent HDA, enhances resistance to Pst DC3000 (Wang et al., 2010). Mutations in several components of the RNA-directed DNA methylation pathway have been shown to alter immune responses to Pst DC3000. While mutations in AGRONAUTE4 (AGO4) compromise resistance to Pst DC3000 (Agorio and Vera, 2007), mutations in NRPE1 and NRPD2, which encode the largest subunit of the RNAP V complex and second largest subunit of the RNAP IV and V complexes, respectively (Law and Jacobsen, 2010), enhance resistance to this pathogen (López et al., 2011). The DNA methylation mutants met1-3 and ddc (drm1-2 drm2-2 cmt3-11), which are deficient in CG maintenance methylation and non-CG maintenance/de novo methylation, respectively (Saze et al., 2003; Chan et al., 2006; Penterman et al., 2007a), are highly resistant to Pst DC3000 (Dowen et al., 2012; Luna et al., 2012). As AGO4 might play some unknown functions independent of the RNA-directed DNA methylation pathway (López et al., 2011), it has been proposed that DNA methylation represses immune responses to Pst DC3000 in the absence of the pathogen (Dowen et al., 2012). However, we found that the DNA demethylase triple mutant rdd (ros1-3 dml2-1 dml3-1), which contains genome-wide DNA hypermethylation (Penterman et al., 2007b; Lister et al., 2008), also exhibits constitutively elevated resistance to bacterial pathogens (see Supplemental Figure 8 online). Therefore, changing global histone acetylation levels or/and genomic DNA methylation profiles exerts great influence on plant immune responses. Interestingly, although both elp2 and rdd are DNA demethylation mutants, their defense phenotypes are opposite (see Supplemental Figure 8 online) (DeFraia et al., 2010). The enhanced disease susceptibility of elp2 might be attributed to impaired histone acetylation. Although the intrinsic relationship between histone acetylation and DNA demethylation/methylation in elp2 is unknown, it would be interesting to test whether histone acetylation is epistatic to DNA methylation in plant immune responses. Furthermore, recent studies have shown that HDA6 and MET1 interact directly and function together in locus-directed heterochromatin silencing (To et al., 2011; Liu et al., 2012). Whether Elongator counteracts HDA/DNA methyltransferase complexes in epigenetic regulation of plant immunity merits further investigation.
Elongator employs distinct molecular mechanisms to play diverse functions in different biological processes (Svejstrup, 2007; Versées et al., 2010). Our data revealed a new role for Elongator in regulating DNA demethylation/methylation in nondividing somatic cells and demonstrated an important function for Elongator-mediated chromatin regulation in plant immune responses, though this function could be direct or indirect. We propose that Elongator plays a key role in genome-wide transcriptomic responses to diverse stresses, likely by regulating the histone acetylation and DNA methylation status of stress-responsive genes. The NPR1 transcriptional cascade exemplifies a signaling cascade where Elongator modulates the chromatin structure of both the key transcription regulator and its target genes, forming a transcriptional feed-forward loop and determining the kinetics of the transcription. Further investigation on the relationship between NPR1 and Elongator in regulating gene transcription during immune responses will shed light on the cooperative interaction between specific transcription regulators and chromatin structure.
METHODS
Plant Materials and Pathogen Infection
The wild type used was the Arabidopsis thaliana Columbia-0 (Col-0) ecotype, and the mutant alleles used were npr1-3 (Glazebrook et al., 1996), elp2-5 (DeFraia et al., 2010), met1-3 (Saze et al., 2003), drm1-2 drm2-2 cmt3-11 (Chan et al., 2006), and ros1-3 dml2-1 dml3-1 (Penterman et al., 2007b). The 35S:NPR1-GFP transgenic line has been described previously (Kinkema et al., 2000). Plant growth, pathogen infection, and determination of in planta pathogen growth were performed as previously described (Cao et al., 1997).
RNA Analysis
RNA extraction, reverse transcription, and real-time qPCR analysis were performed as described by DeFraia et al. (2010). The primers used for real-time qPCR in this study are shown in Supplemental Table 3 online.
Microarray Analysis
Four-week-old soil-grown plants were inoculated with the bacterial pathogen Pst DC3000/avrRpt2. Total RNA samples extracted from the inoculated leaves were subjected to microarray analysis. Briefly, RNA concentration was determined on a NanoDrop Spectrophotometer (Thermofisher Scientific), and sample quality was assessed using the 2100 Bioanalyzer (Agilent Technologies). cDNA was synthesized from 200 ng of total RNA and used as a template for in vitro transcription in the presence of T7 RNA polymerase and cyanine-labeled CTPs using the Quick Amp Labeling kit (Agilent Technologies) according the manufacturer’s protocol. The amplified, labeled complementary RNA was purified using the RNeasy mini kit (Qiagen). For each array, 1650 ng of Cy 3–labeled complementary RNA was fragmented and hybridized with rotation at 65°C for 17 h. Samples were hybridized to Arabidopsis 4 × 44k arrays (Agilent Technologies). The arrays were washed according to the manufacturer’s protocol and then scanned on a G2505B scanner (Agilent Technologies). Data were extracted using Feature Extraction 10.1.1.1 software (Agilent Technologies).
Data (individual signal intensity values) obtained from the microarray probes were background corrected using a normexp+offset method, in which a small positive offset (k = 50) was added to move the corrected intensities away from zero (Ritchie et al., 2007). The resulting data were log transformed (using 2 as the base) and normalized between individual samples by scaling the individual log-transformed signal intensities so that all data sets had comparable lower quartile, median, and upper quartile values (Smyth, 2005). After normalization, the Student’s t test was performed considering a probe-by-probe comparison between different genotypes at the same time point using the wild type (Col-0) as the reference sample and between different time points of the same genotype using the 0-h sample as the reference. In each comparison, a P value and fold change were computed for each gene locus. The gene expression fold changes were computed based on the normalized log-transformed signal intensity data. The comparison results were further explored to obtain numbers of overlapped genes between/among different comparisons.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described by Saleh et al. (2008). Briefly, ∼3 g of 4-week-old soil-grown plants were submerged in 50 mL of cross-linking buffer (10 mM Tris-HCl, pH 8, 0.4 M Suc, 1 mM PMSF, 1 mM EDTA, and 1% formaldehyde) and vacuum infiltrated three times for 3 to 4 min each at room temperature. The cross-linking reaction was stopped by adding 2.5 mL of 2 M Gly to a final concentration of 100 mM and vacuum infiltration for 5 min. Plant tissues were washed three times with cold sterile deionized water. After removing water, plants tissues were submerged in liquid nitrogen, ground to a fine powder, and resuspended in 20 to 25 mL cold nuclei isolation buffer (15 mM PIPES, pH 6.8, 0.25 M Suc, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 0.9% Triton X-100, 20 mM sodium butyrate, 1 mM PMSF, 2 μg/mL pepstatin A, and 2 μg/mL aprotinin). After brief vortex and incubation, the homogenized slurry was filtered through one layer of Miracloth. After centrifugation at 3220g for 20 min, the pellet (nuclei) was resuspended in 1.5 mL of cold nuclei lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 20 mM sodium butyrate, 1 μg/mL pepstatin A, and 1 μg/mL aprotinin). DNA was sheared into ∼500-bp (200 to 1000 bp) fragments by 6 to 10 min of 3-s pause sonication at 40 to 43% amplitude using a TekMar TM-100 sonic disruptor (TekMar). After centrifugation at 13,800g for 10 min, the supernatant (200 μL) was diluted fivefold with nuclei lysis buffer and precleared by adding 50 μL salmon sperm DNA/protein A agarose beads. After removing the agarose beads, 5 μL of Ac-Histone H3 (Lys-9/14) antibody (sc-8655-R; Santa Cruz Biotechnology) was added and the mixture was incubated at 4°C for 5 h to overnight with gentle rotation, and then 60 to 75 μL salmon sperm DNA/protein A agarose beads was added and the incubation was continued for 2 to 3 h. After centrifugation at 3800g for 2 min, the agarose beads were sequentially washed with low salt wash buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 0.2% SDS, 0.5% Triton X-100, and 2 mM EDTA), high salt wash buffer (20 mM Tris-HCl, pH 8, 500 mM NaCl, 0.2% SDS, 0.5% Triton X-100, and 2 mM EDTA), LiCl wash buffer (10 mM Tris-HCl, pH 8, 0.25 M LiCl, 1% sodium deoxycholate, 1% Nonidet P-40, and 1 mM EDTA), and TE buffer (twice; 1 mM EDTA and 10 mM Tris-HCl, pH 8). The immunocomplexes were eluted with freshly prepared elution buffer (0.1 M NaHCO3 and 0.5% SDS) and incubation at 65°C for 15 min with gentle rotation. Twenty microliters of 5 M NaCl was added to 500 μL of the immunocomplex solution and the mixture was incubated at 65°C for 4 h to overnight to reverse cross-linking. Then, 20 μL of 1 M Tris-HCl, pH 6.5, 10 μL of 0.5 M EDTA, and 2 μL proteinase K (10 mg/mL) was added, and the mixture was incubated at 45°C for 1.5 h to digest the proteins. Immunoprecipitated DNA was purified using a mixture of phenol:chloroform:isoamyl alcohol (25:24:1), and the resulting DNA was used for real-time qPCR with the primers in Supplemental Table 4 online. The amount of precipitated DNA corresponding to a specific gene region was determined by real-time qPCR and normalized to both input DNA and a constitutively expressed gene (ACTIN2/7) as described (Mosher et al., 2006). The resulting values were used as measures for the levels of histone H3K9/14ac in specific gene regions.
Bisulfite Sequencing
For locus-specific DNA methylation analysis, strand-specific and bisulfite-specific primers (see Supplemental Table 1 online) were used to amplify the target regions from bisulfite-converted genomic DNA. The PCR products were cloned into pGEM-T easy vector (Promega). Individual clones were sequenced, and the sequence data were analyzed using the Web-based tool Kismeth (http://katahdin.mssm.edu/kismeth) (Gruntman et al., 2008). Genome-wide bisulfite sequencing and data analysis were performed as described by Lister et al. (2008). Briefly, genomic DNA was extracted from ∼1 g of fresh rosette leaves of 4-week-old soil-grown plants using a hexadecyltrimethylammonium bromide (CTAB) DNA extraction protocol (Aldrich and Cullis, 1993). Approximately 2 µg of high molecular weight genomic DNA was dissolved in 125 μL TE (10 mM Tris-HCl, pH 8.0, and 0.5 mM EDTA) and transferred to 6 × 16-mm glass microtubes with AFA fiber and snap-caps (Covaris). DNA samples were sheared into fragments with an average size of 300 bp in a Covaris S2 ultrasonic disruptor following the manufacturer’s recommended settings. AMPure magnetic beads (Beckman Coulter) with a bead-to-sample ratio of 73:100 were used to clean up the samples prior to sequencing library construction. Sequencing libraries were made using the TruSeq DNA Sample Preparation kits (Illumina) following the manufacturer’s protocol with a few modifications. Briefly, DNA fragments were end-repaired, adenylated, adaptor-ligated, and size-selected (250 to 500 bp) in a 2% agarose gel. The gel was stained with Invitrogen SYBR safe (Life Technologies) and viewed on a blue light transilluminator (Life Technologies) in order to avoid UV damage. Libraries were quantitated in a Qubit fluorometer (Life Technologies). The final yield was ∼160 ng (in 25 μL Tris-HCl, pH 8.0), which was subjected to sodium bisulfite treatment using the EpiTect Bisulfite kit (Qiagen) following the manufacturer’s instructions. The resulting library was amplified using uracil-insensitive PfuTurbo Cx Hotstart DNA polymerase (Agilent Technologies) under the following conditions: denaturation at 98°C for 30 s, 18 cycles (98°C for 15 s, 60°C for 30 s, and 72°C for 1 min), and final extension at 72°C for 5 min. The PCR amplification products were then cleaned twice using AMPure magnetic beads with a bead-to-sample ratio of 85:100, and the resulting DNA was quantitated by the Qubit fluorometer and qPCR with the Kapa SYBR Fast qPCR reagents (Kapa Biosystems) on an ABI7900HT real-time PCR system (Life Technologies). The average insert size of the libraries was ∼360 bp. Libraries were then diluted to 9 pM for cluster generation on the cBOT (Illumina), and a 101 cycle multiplex single-end sequencing run for pooled barcoded libraries and a 2 × 101 cycle multiplex pair-end sequencing run for each library was performed on an Illumina Genetic Analyzer IIx (running SCS2.9) using an eight-lane flow cell.
The cleanup module of the Paracel Transcript Assembler version 3.0.0 was applied for raw reads cleanup. Consecutive ambiguous characters (Ns) were removed from both ends of a read, and all reads were checked and masked for adaptors. Low-quality sequences were trimmed from ends of individual reads, and reads with length shorter than 40 nucleotides were excluded from further analysis (see Supplemental Table 5 online). Reads were aligned against in silico bisulfite converted sense and antisense references of the Arabidopsis Col-0 genome and the nonconverted normal reference using the Novoalign software (Novocraft Technologies, V2.07.15b). A read was considered to be “mapped” to the sense reference if the number of mismatches with the antisense reference is at least twice that with the sense reference and vice versa. Mapped reads were filtered out as follows: reads either with three consecutive cytosines in CHH context (possible nonconversion in bisulfite conversion) or with mismatches more than 10% of the total nucleotides were removed (Bormann Chung et al., 2010; Otto et al., 2012); clonal reads potentially produced during PCR amplification from the same template molecule (based on a common start position) were removed. The uniquely mapped nonclonal reads were used for methylcytosine identification. A binomial probability distribution was used to calculate the minimum sequence depth at a cytosine position at which a methylcytosine could be called while maintaining a false positive rate below 5%. The false methylcytosine discovery rate was estimated by the sum of the rates of nonconversion and thymidine to cytosine sequencing errors at cytosine positions in the chloroplast reference genome.
Statistical Methods
Except those used in microarray analysis, all statistical analyses were performed with the data analysis tools (t test: two samples assuming unequal variances) in Microsoft Excel of Microsoft Office 2004 for Macintosh.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ELP2 (At1g49540), NPR1 (At1g64280), EDS1 (At3g48090), PAD4 (At3g52430), EDS5 (At4g39030), ICS1 (At1g74710), NDR1 (At3g20600), FMO1 (At1g19250), ALD1 (At2g13810), PR1 (At2g14610), PR2 (At3g57260), PR5 (At1g75040), WRKY18 (At4g31800), WRKY33 (At2g38470), WRKY38 (At5g22570), WRKY54 (At2g40750), WRKY58 (At3g01080), and VSR6 (At1g30900); NCBI Gene Expression Omnibus Series number GSE38986 (microarray data); and NCBI Short Read Archive accession number SRA055073 (BS-Seq data).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Pst DC3000/avrRpt2-Induced Transcriptome Changes in elp2.
Supplemental Figure 2. Basal Expression Levels of Several Defense Genes in elp2 and the Wild Type.
Supplemental Figure 3. DNA Methylation Levels in NPR1 and PR2.
Supplemental Figure 4. Numbers of Methylcytosines Identified in elp2 and the Wild Type.
Supplemental Figure 5. The Density of Methylcytosines and Ratio of the Number of Methylcytosines Identified in elp2 versus the Wild Type.
Supplemental Figure 6. DNA Methylation Status at the PAD4 Locus Revealed by the Genome-Wide Bisulfite Sequencing.
Supplemental Figure 7. Pathogen-Induced Dynamic Methylation Changes at Specific Methylcytosines in NPR1 and PAD4.
Supplemental Figure 8. Immune Responses in Arabidopsis Mutants Deficient in DNA Methylation/Demethylation.
Supplemental Table 1. Primers Used for Bisulfite-Sequencing PCR.
Supplemental Table 2. Rates of Nonconversion and T-to-C Sequencing Errors in the Chloroplast Reference Genome.
Supplemental Table 3. Primers Used for Real-Time qPCR Analysis.
Supplemental Table 4. Primers Used for ChIP Real-Time qPCR Analysis.
Supplemental Table 5. Results of the Sequence Clean Process.
Acknowledgments
We thank Robert Fischer (University of California, Berkeley, CA) for rdd seeds and the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH) for met1-3 and ddc seeds. This work was supported by a grant from the National Science Foundation (IOS-0842716) awarded to Z.M. We thank Jin Yao for help with microarray data analysis and Christopher DeFraia for critical comments on the article.
AUTHOR CONTRIBUTIONS
Z.M. conceived and designed the experiments. Y.W., C.A., X.Z., Y.Z., and D.M.A. performed the experiments. J.Y., Y.S., F.Y., and Z.M. analyzed the data. Z.M. wrote the article.
Glossary
- HAT
histone acetyltranferase
- SAM
S-adenosylmethionine
- Pst
Pseudomonas syringae pv tomato
- NCBI
National Center for Biotechnology Information
- qPCR
quantitative PCR
- hpi
hours after inoculation
- BS-Seq
bisulfite deep sequencing
- ETI
effector-triggered immunity
- SAR
systemic acquired resistance
- Col-0
Columbia-0
References
- Agorio A., Vera P. (2007). ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19: 3778–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrich J., Cullis C.A. (1993). CTAB DNA extraction from plant tissues. Plant Mol. Biol. Rep. 11: 128–141 [Google Scholar]
- Alvarez-Venegas R., Abdallat A.A., Guo M., Alfano J.R., Avramova Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2: 106–113 [DOI] [PubMed] [Google Scholar]
- Anderson S.L., Coli R., Daly I.W., Kichula E.A., Rork M.J., Volpi S.A., Ekstein J., Rubin B.Y. (2001). Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68: 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., Hermand D. (2012). Translational control of cell division by elongator. Cell Rep. 1: 424–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berr A., Ménard R., Heitz T., Shen W.H. (2012). Chromatin modification and remodelling: A regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell. Microbiol. 14: 829–839 [DOI] [PubMed] [Google Scholar]
- Bormann Chung C.A., Boyd V.L., McKernan K.J., Fu Y., Monighetti C., Peckham H.E., Barker M. (2010). Whole methylome analysis by ultra-deep sequencing using two-base encoding. PLoS ONE 5: e9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cao H., Li X., Dong X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95: 6531–6536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century K.S., Shapiro A.D., Repetti P.P., Dahlbeck D., Holub E., Staskawicz B.J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chan S.W., Henderson I.R., Zhang X., Shah G., Chien J.S., Jacobsen S.E. (2006). RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in Arabidopsis. PLoS Genet. 2: e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Zhang H., Jablonowski D., Zhou X., Ren X., Hong X., Schaffrath R., Zhu J.K., Gong Z. (2006). Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 26: 6902–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. (2002). A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem. Sci. 27: 115–117 [DOI] [PubMed] [Google Scholar]
- Choi S.M., Song H.R., Han S.K., Han M., Kim C.Y., Park J., Lee Y.H., Jeon J.S., Noh Y.S., Noh B. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 71: 135–146 [DOI] [PubMed] [Google Scholar]
- Close P., Hawkes N., Cornez I., Creppe C., Lambert C.A., Rogister B., Siebenlist U., Merville M.P., Slaugenhaupt S.A., Bours V., Svejstrup J.Q., Chariot A. (2006). Transcription impairment and cell migration defects in elongator-depleted cells: Implication for familial dysautonomia. Mol. Cell 22: 521–531 [DOI] [PubMed] [Google Scholar]
- Conrath U. (2011). Molecular aspects of defence priming. Trends Plant Sci. 16: 524–531 [DOI] [PubMed] [Google Scholar]
- Creppe C., et al. (2009). Elongator controls the migration and differentiation of cortical neurons through acetylation of α-tubulin. Cell 136: 551–564 [DOI] [PubMed] [Google Scholar]
- DeFraia C.T., Zhang X., Mou Z. (2010). Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 64: 511–523 [DOI] [PubMed] [Google Scholar]
- Dowen R.H., Pelizzola M., Schmitz R.J., Lister R., Dowen J.M., Nery J.R., Dixon J.E., Ecker J.R. (2012). Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A., Feys B.J., Frost L.N., Jones J.D.G., Daniels M.J., Parker J.E. (1999). EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc. Natl. Acad. Sci. USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J., Rogers E.E., Ausubel F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes N.A., Otero G., Winkler G.S., Marshall N., Dahmus M.E., Krappmann D., Scheidereit C., Thomas C.L., Schiavo G., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. (2002). Purification and characterization of the human elongator complex. J. Biol. Chem. 277: 3047–3052 [DOI] [PubMed] [Google Scholar]
- He X.J., Chen T., Zhu J.K. (2011). Regulation and function of DNA methylation in plants and animals. Cell Res. 21: 442–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Johansson M.J., Byström A.S. (2005). An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11: 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D., Frohloff F., Fichtner L., Stark M.J., Schaffrath R. (2001). Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol. Microbiol. 42: 1095–1105 [DOI] [PubMed] [Google Scholar]
- Jaskiewicz M., Conrath U., Peterhänsel C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12: 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim J.H., Lane W.S., Reinberg D. (2002). Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20: 2357–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Greenblatt J.F. (2001). Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 8203–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. (2007). The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li J., Brader G., Kariola T., Palva E.T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46: 477–491 [DOI] [PubMed] [Google Scholar]
- Lister R., O’Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133: 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yu C.W., Duan J., Luo M., Wang K., Tian G., Cui Y., Wu K. (2012). HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiol. 158: 119–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López A., Ramírez V., García-Andrade J., Flors V., Vera P. (2011). The RNA silencing enzyme RNA polymerase v is required for plant immunity. PLoS Genet. 7: e1002434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarelli M., Fuso A., Strom R., Scarpa S. (2001). The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J. Biol. Chem. 276: 7500–7506 [DOI] [PubMed] [Google Scholar]
- Luna E., Bruce T.J., Roberts M.R., Flors V., Ton J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158: 844–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R., et al. (2008). Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141: 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Durrant W.E., Wang D., Song J., Dong X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18: 1750–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Métraux J.-P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., et al. (2010). Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 107: 1678–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., Fleury D., Bruno L., Robles P., De Veylder L., Traas J., Micol J.L., Van Montagu M., Inzé D., Van Lijsebettens M. (2005). The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 102: 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent R.L., Johnsson A., Fleharty B., Gogol M., Xue-Franzén Y., Seidel C., Wright A.P., Forsburg S.L. (2010). Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation. BMC Genomics 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. (2010). A role for the elongator complex in zygotic paternal genome demethylation. Nature 463: 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Otto C., Stadler P.F., Hoffmann S. (2012). Fast and sensitive mapping of bisulfite-treated sequencing data. Bioinformatics 28: 1698–1704 [DOI] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6: e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou C., Fairhurst S.A., Lowe D.J., Brick P., Onesti S. (2006). The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol. Microbiol. 59: 795–806 [DOI] [PubMed] [Google Scholar]
- Pavet V., Quintero C., Cecchini N.M., Rosa A.L., Alvarez M.E. (2006). Arabidopsis displays centromeric DNA hypomethylation and cytological alterations of heterochromatin upon attack by Pseudomonas syringae. Mol. Plant Microbe Interact. 19: 577–587 [DOI] [PubMed] [Google Scholar]
- Penterman J., Uzawa R., Fischer R.L. (2007a). Genetic interactions between DNA demethylation and methylation in Arabidopsis. Plant Physiol. 145: 1549–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J.H., Ballinger T., Henikoff S., Fischer R.L. (2007b). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P.B., Chen C.Z., Collins R.N. (2005). Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol. Cell 17: 841–853 [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G.K. (2007). A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Ryals J., Weymann K., Lawton K., Friedrich L., Ellis D., Steiner H.-Y., Johnson J., Delaney T.P., Jesse T., Vos P., Uknes S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor I κ B. Plant Cell 9: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Shah J., Tsui F., Klessig D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant Microbe Interact. 10: 69–78 [DOI] [PubMed] [Google Scholar]
- Slaugenhaupt S.A., et al. (2001). Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68: 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K. (2005). Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R. Gentleman, V. Carey, S. Dudoit, R. Irizarry, and W. Huber, eds (New York: Springer), pp. 397–420. [Google Scholar]
- Song J.T., Lu H., Greenberg J.T. (2004). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup J.Q. (2007). Elongator complex: How many roles does it play? Curr. Opin. Cell Biol. 19: 331–336 [DOI] [PubMed] [Google Scholar]
- Tao Y., Xie Z., Chen W., Glazebrook J., Chang H.S., Han B., Zhu T., Zou G., Katagiri F. (2003). Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M.P., Wang J.J., Wei N.E., Jiang H., Doerge R.W., Chen Z.J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wees S.C.M., de Swart E.A.M., van Pelt J.A., van Loon L.C., Pieterse C.M.J. (2000). Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 8711–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versées W., De Groeve S., Van Lijsebettens M. (2010). Elongator, a conserved multitasking complex? Mol. Microbiol. 76: 1065–1069 [DOI] [PubMed] [Google Scholar]
- Wang C., Gao F., Wu J., Dai J., Wei C., Li Y. (2010). Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 51: 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308: 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wang L., Mitra R.M., Hasselmann K.D., Sato M., Lenarz-Wyatt L., Cohen J.D., Katagiri F., Glazebrook J. (2008). The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: Roles of major regulators and the phytotoxin coronatine. Mol. Plant Microbe Interact. 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Winkler G.S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99: 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittschieben B.O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C.D., Tempst P., Svejstrup J.Q. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4: 123–128 [DOI] [PubMed] [Google Scholar]
- Workman J.L., Kingston R.E. (1998). Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67: 545–579 [DOI] [PubMed] [Google Scholar]
- Xu D., Huang W., Li Y., Wang H., Huang H., Cui X. (2012). Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J. 69: 792–808 [DOI] [PubMed] [Google Scholar]
- Zhou X., Hua D., Chen Z., Zhou Z., Gong Z. (2009). Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 60: 79–90 [DOI] [PubMed] [Google Scholar]