Abstract
In mice, two T-box transcription factors, T-bet and Eomes, drive the differentiation of CD8 T-cell lineages; however, little is known regarding their role in human CD8 T-cell differentiation. Here, we characterized T-bet and Eomes expression and localization within human CD8 memory T-cell populations. We find T-bet and Eomes are broadly expressed in human memory CD8 T cells, with increasing levels of T-bet and Eomes strongly correlating with differentiation from central memory to effector memory and effector subpopulations. In resting T-cells, T-bet levels directly correlate to subcellular localization, with a higher propensity for nuclear expression of T-bet within T-bethi cells and predominately cytoplasmic expression in T-betlo cells. Additionally, Eomes is also localized to either the nucleus or cytoplasm. Upon T-cell receptor stimulation, the percentage of T-cells that express T-bet dramatically increases, while the percentage of cells expressing Eomes remains largely unchanged across all memory populations. Interestingly, T-bet, but not Eomes, relocalizes to the nucleus in the majority of cells across all populations within 24 hours post-stimulation. These data indicate that T-bet and Eomes are likely regulated at the level of subcellular localization, potentially via different mechanisms. Together, these findings suggest a novel model for CD8 T-cell differentiation in humans based on the localization of T-bet and Eomes.
INTRODUCTION
The T-box transcription factors, T-bet (T-box expressed in T-cells) and Eomes (Eomesodermin), have been implicated as master regulators of CD8 T-cell differentiation and function (1, 2). During the early stages of CD8 T-cell activation, T-bet and Eomes cooperate to promote cytotoxic lymphocyte formation by inducing the expression of the cytolytic molecules perforin and granzyme B (1, 3–6). In the context of CD8 memory T-cells, both T-bet and Eomes sustain memory phenotypes by stabilizing the expression of IL2Rβ, thus promoting IL15 signaling and continued proliferation of memory cells (1, 4–6).
While T-bet and Eomes have cooperative and redundant roles in CD8 T-cells, they also have unique influences on CD8 T-cell function. In murine models, early effector CD8 T-cells are characterized by high levels of T-bet which gradually decline as cells progress towards a memory phenotype (7). In contrast, while Eomes is upregulated in early effectors, its expression increases as cells progress from effector to a memory cell (5–7). Single knockout murine studies, as well as T-bet overexpression studies, suggest that consistently high levels of T-bet drive terminal effector differentiation (3). In contrast, Eomes knockouts are deficient in long-term memory formation and homeostatic renewal (1, 6, 8). Taken together, these data suggest that the levels of T-bet and Eomes are important factors in determining the function and fate of a given T-cell.
A key feature of eukaryotic cells is the physical compartmentalization of genetic material in the nucleus from translational machinery in the cytoplasm. Transport between the nucleus and the cytoplasm is a highly regulated process and, in the case of many transcription factors, is key to downstream gene regulation within a particular cell type (9). The activity of Tbx5, another member of the T-box transcription factor family, is modulated by its localization within the nucleus or cytoplasm, depending on cell signaling and differentiation stimuli (10). Similarly, many other transcription factors are tightly regulated by their subcellular localization, such as NF-κB (11–13) and the nuclear factor of activated T-cells (NFAT) subfamily (14–16). To date, localization studies of T-bet and Eomes within human CD8 T-cells have been relatively limited (17) and, thus, it is unclear if T-bet and Eomes might be regulated at the level of cellular localization and, in turn, how, and in what cellular context, they drive T-cell differentiation and function.
In this study we sought to investigate the expression and localization patterns of T-bet and Eomes within human CD8 memory T-cells to begin to dissect their possible functions in driving specific human CD8 T-cell subsets. Here, we present a characterization of T-bet and Eomes expression and localization in carefully delineated CD8 naïve, central memory, effector memory, and effector T-cell subsets from healthy human donors. We show that T-bet and Eomes expression correlate with effector and effector memory CD8 T-cell populations, respectfully. Interestingly, we found that both T-bet and Eomes can be differentially localized to the nucleus or cytoplasm (or both), and the localization of both factors correlates with their expression level. Upon T-cell receptor (TCR) stimulation, we show that T-bet expression dramatically increases in blasting cells while Eomes expression does not change considerably in any CD8+ T-cell population. T-bet, but not Eomes, are relocalized to the nucleus depending on cellular context suggesting that TCR activation might be sufficient to signal shuttling of T-bet, but not Eomes, across the nuclear envelope. Taken together, our data support a novel model of CD8 T-cell function regulated, in part, by the modulation of T-bet and Eomes subcellular localization.
MATERIALS AND METHODS
Human Cells
Donor peripheral blood mononuclear cells (PBMC) were collected after written, informed consent from the University of Pennsylvania's Center for AIDS Research Human Immunology Core (IRB #705906) in compliance with IRB guidelines. PBMCs were cryopreserved in fetal bovine serum (FBS; ICS Hyclone, Logan, UT) containing 10% dimethyl sulfoxide (DMSO; Fisher Scientific, Pittsburgh, Pennsylvania) and stored at −140°C until further use.
Flow Cytometry Analysis
Antibodies used in our flow cytometry studies are listed in Table I. Cryopreserved PBMCs were thawed and rested overnight at 37°C, 5% CO2 in complete medium (RPMI (Mediatech Inc; Manassas, Virginia) supplemented with 10% FBS, 1% L-glutamine (Mediatech Inc), and 1% penicillin-streptomycin (Lonza; Walkersville, Maryland), sterile filtered) at a concentration of 2×106 cells/mL. Cells were washed with 1×PBS and was added to each sample and incubated at 37°C, 5% CO2 for 15 min. Samples were then stained with aqua amine-reactive viability dye for 10 min at room temperature (RT) in the dark. Surface stain antibodies against various cell markers were then added to the cells and incubated for an additional 20 min at RT in the dark. Cells were then washed with PBS containing 1% bovine serum albumin (BSA, Fisher Scientific; Pittsburgh, Pennsylvania) and 0.1% sodium azide (Fisher Scientific). Cells were permeabilized for 20 min at RT using the Cytofix/Cytoperm kit (BD Biosciences) and then washed twice with Perm Wash buffer (BD Biosciences). ICS antibodies were then added to the cells and incubated for one hour at RT in the dark. Cells were then washed again with Perm Wash buffer and fixed in PBS containing 1% paraformaldehyde (Sigma-Aldrich; St. Louis, Missouri). For each sample, 1,000,000 total events were acquired on a modified flow cytometer (LSRII; BD Immunocytometry Systems; San Jose, CA) equipped for the detection of 18 fluorescent parameters. Antibody capture beads (BD Biosciences) were used as individual compensation tubes for each fluorophore in the experiment and data analysis was performed using FlowJo version 9.0.1 (TreeStar, Ashland, Oregon). Statistical analyses were performed using GraphPad Prism software (Version 5.0a). Where applicable, non-parametric Wilcoxon matched paired T-tests were used where Gaussian distribution is not assumed because we analyzed less than 25 subjects. To define positive and negative populations, we employed Fluorescence Minus One (FMO) controls for each fluorophore used in this study when initially developing staining protocols. Additionally, we further optimized gating by examining known negative cell populations for background level expression.
Table I.
Antibodies used in this study.
| Technique | Protein Target | Clone | Fluorophore | Company |
|---|---|---|---|---|
|
| ||||
| Flow Cytometry | ||||
|
| ||||
| Surface Molecules | CD3 | OKT3 | Qdot585 | Custom |
|
| ||||
| CD8 | 3B5 | Qdot605 | Invitrogen | |
|
| ||||
| CCR7 | 3D12 | APC eFluor780 | eBioscience | |
|
| ||||
| CD45RO | UCHL1 | ECD | Beckman Coulter | |
|
| ||||
| CD14 | 61D3 | PE Cy5 | Invitrogen | |
|
| ||||
| CD16 | 3G8 | PE Cy5 | Biolegend | |
|
| ||||
| CD19 | HIB19 | PE Cy5 | Invitrogen | |
|
| ||||
| Intracellular Molecules | T-bet | 4B10 | PE | eBioscience |
|
| ||||
| Eomes | WD1928 | Alexa 647 | eBioscience | |
|
| ||||
| Confocal Microscopy | ||||
|
| ||||
| Intracellular Molecules | T-bet | O4–46 | Alexa 488 | BD Pharmagen |
|
| ||||
| Eomes | WD1928 | Alexa 647 | eBioscience | |
|
| ||||
| Lamin A | 133A2 | Unconjugated | Abcam | |
|
| ||||
| ImagestreamX Analysis | ||||
|
| ||||
| Surface Molecules | CCR7 | 3D12 | APC eFluor780 | eBioscience |
|
| ||||
| CD45RO | UCHL1 | ECD | Beckman Coulter | |
|
| ||||
| Intracellular Molecules | T-bet | 4B10 | PE | eBioscience |
|
| ||||
| Eomes | WD1928 | Alexa 647 | eBioscience | |
Confocal Microscopy
CD8 T-cells were prepared for fixed cell immunoflourescent confocal microscopy as previously described (18). Antibodies used in our confocal microscopy studies are listed in Table I. Human α-T-bet, α-Eomes, and α-Lamin A were used in the range of 0.5–2 μg/ml. A secondary goat α-rabbit IgG (H+L conjugated to Alexa Fluor 568 (Molecular Probes, Grand Island, NY) was used to detect Lamin A. Slides were then covered with 0.15mm coverslips (VWR Scientific, Philadelphia, PA) using DAPI (Invitrogen) containing mounting media (Vectashield, Burlingame, CA) to visualize chromatin. 2D micrographs were obtained using a multi-laser based spinning disk confocal microscope (Zeiss).
Polychromatic Imaging Flow Cytometry of Resting CD8 T-cells
Antibodies used in the polychromatic imaging analysis are listed in Table I. Purified CD8 T-cells from five normal donors were isolated from whole PBMCs using a MACS negative selection CD8 T-cell isolation kit (Miltenyi Biotec; Boston, MA). Cells were stained with DAPI for 5 min at RT. Fixed cells were then analyzed on a polychromatic imaging flow cytometer. For each sample, 30,000 events were collected on an ImageStream (Amnis Corp.; Seattle, WA). Antibody capture beads (BD Biosciences) were used as individual compensation tubes for each fluorophore. Images were captured using a 60× lens with the extended depth of field upgrade through Inspire software (Amnis Corp.) and data analysis was performed using IDEAS 4.0 software (Amnis Corp.). Our gating strategy for delineating CD8 memory populations is shown in Supplemental Figure 1. Nuclear and cytoplasmic T-bet and Eomes were defined using masking functions within IDEAS 4.0.
Cell Fractionation and Immunoblot Analysis
Purified CD8 T-cells were fractionated into nuclear and cytoplasmic compartments using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Rockford, IL). Lysate concentrations for the nuclear and cytoplasmic compartments were determined using a Biocinchonic Acid (BCA) Protein Assay Reagent kit (Thermo Scientific). 20 μg of total lysate from each compartment was analyzed for the presence of T-bet or Eomes. Standard methods for immunoblot analysis were used (19). T-bet was detected using a 1:200 dilution of a monoclonal α-T-bet antibody (Santa Cruz Biotechnology). Eomes was detected using a 1:500 dilution of a polyclonal α-Eomes antibody (Abcam, Cambridge, MA). As controls for efficient fractionation, a polyclonal α-HDAC1 antibody (1:1000, Abcam) was used to detect the nuclear fraction while a polyclonal α-Hsp90 antibody (Thermo Scientific) was used to detect the cytoplasmic fraction.
Activation and analysis of CD8 T-cell memory populations
To study the effect of T-cell receptor activation on CD8 T-cell populations, CD8 T-cells from 5 donors were sorted using a FACS Aria (BD Immunocytometry Systems; San Jose, CA) into naïve (CCR7+ CD45RO−), central memory (CCR7+ CD45RO+), effector memory (CCR7− CD45RO+) and effector (CCR7− CD45RO−) to obtain purified memory populations. Cell populations were rested overnight at 37°C, 5% CO2 in complete medium (RPMI (Mediatech Inc; Manassas, Virginia) supplemented with 10% FBS, 1% L-glutamine (Mediatech Inc), and 1% penicillin-streptomycin (Lonza; Walkersville, Maryland), sterile filtered). Cells were then cultured in 5 μg/mL CD3, 3 μg/mL, CD28/CD49d, and 1 μg/mL BSA. Cells were collected at 0, 24, and 72 hours post-stimulation and stained for T-bet and Eomes and analyzed on the ImageStreamX. To determine if changes in blasting cells were different from changes in non-blasting cells we performed statistical analysis on the area under each curve followed by a paired student T-test. Paired T-tests were also performed on localization analysis (Figures 7C and 8C).
Figure 7. Activation-induced changes of T-bet expression and localization in CD8 T-cell memory populations.

CD8+ T-cells from three normal human donors were sorted into memory populations as follows: naïve (CD45RO−CCR7+), central memory (TCM, CD45RO+CCR7+), effector memory (TEM, CD45RO+CCR7−), and effector (CD45RO−CCR7−) and stimulated with αCD3, αCD28, and αCD49d for 72 hours. At 24-hour intervals, cells were collected and data analyzed using IDEAS software. Blasting (solid line) vs. non-blasting (dashed line) cells were delineated based on size. (A) Longitudinal assessment of the average percentage of T-bet+ cells (B) and the median fluorescence intensity within each CD8+ memory population is plotted. (C) Changes in the percentage of cells expressing exclusively nuclear (black bars), nuclear and cytoplasmic (grey bars), and exclusively cytoplasmic (white bars) T-bet is plotted for central memory, effector memory, and effector CD8+ T-cells. (D) Longitudinal analysis of T-bet MFI within the nucleus of central memory, effector memory, and effector CD8+ T-cells is plotted. Statistically significant differences are delineated by *. * p<0.05, ** p<0.001, *** p<0.0001.
Figure 8. Activation-induced changes of Eomes expression and localization in CD8 T-cell memory populations.
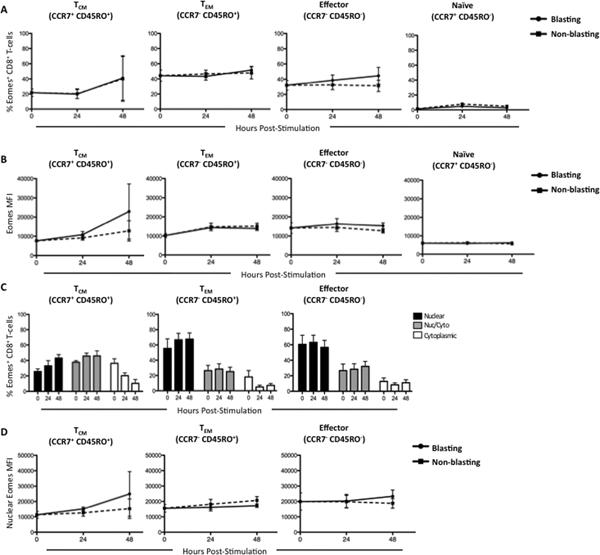
CD8+ T-cells from three normal human donors were sorted into memory populations as follows: naïve (CD45RO−CCR7+), central memory (TCM, CD45RO+CCR7+), effector memory (TEM, CD45RO+CCR7−), and effector (CD45RO−CCR7−) and stimulated with αCD3, αCD28, and αCD49d for 72 hours. At 24-hour intervals, cells were collected and data analyzed using IDEAS software. Blasting (solid line) vs. non-blasting (dashed line) cells were delineated based on size. (A) Longitudinal assessment of the average percentage of Eomes+ cells (B) and the median fluorescence intensity within each CD8+ memory population is plotted. (C) Changes in the percentage of cells expressing exclusively nuclear (black bars), nuclear and cytoplasmic (grey bars), and exclusively cytoplasmic (white bars) Eomes is plotted for central memory, effector memory, and effector CD8+ T-cells. (D) Longitudinal analysis of Eomes MFI within the nucleus of central memory, effector memory, and effector CD8+ T-cells is plotted. Statistically significant differences are delineated by *. * p<0.05, ** p<0.001, *** p<0.0001.
RESULTS
T-bet and Eomes expression in CD8 T-cells populations
Extensive studies in mice have suggested that T-bet and Eomes play key roles in effector function and long-term memory formation, respectively (1–3, 20, 21); however, the roles of T-bet and Eomes in human CD8 T-cells remain unclear. As shown in Figure 1A, human CD8 T-cells display two T-bet+ populations, T-bethi and T-betlo, whereas Eomes is bimodally distributed. While the percentage of T-bet+ or Eomes+ cells can vary greatly between individuals, these distinct populations for each transcription factor were observed in all donors we analyzed in our studies (data not shown).
Figure 1. T-bet and Eomes expression correlates with effector and TEM human CD8 subsets.
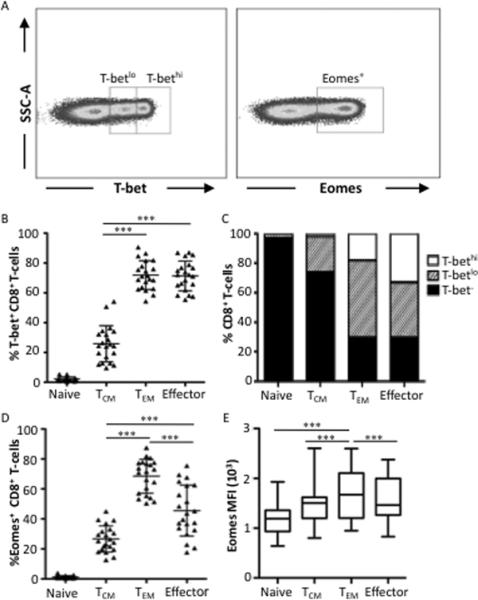
(A) Representative flow cytometry plots showing T-bet and Eomes. CD8 T-cells exhibit two distinct T-bet+ and one distinct Eomes+ populations. A representative donor is shown. (B) T-bet expression was characterized in CD8 T-cells with the following phenotypes: naïve (CD45RO−CCR7+), Central memory (TCM, CD45RO+CCR7+), effector memory (TEM, CD45RO+CCR7−), and effector (CD45RO−CCR7−). The percentage of CD8+ T-bet+ T-cells is plotted for each memory population for each of 29 donors. Each symbol represents an individual donor. (C) Graphical representation of mean percentages of T-bethi, T-betlo or T-bet− cells within each CD8 T-cell population. Filled bar sections represent T-bet− cells, hatched sections represent T-betlo cells, and open sections represent T-bethi cells. (D) Eomes expression by CD8 T-cells is plotted in each memory phenotype for each donor. (E) Eomes median fluorescence intensity (MFI) values in CD8 subpopulations is shown. The box and whisker graph displays the 25–75% (box) and 10–90% (whisker). The line in the box represents the median value. Specific statistical differences of interest, as measured by non-parametric Wilcoxon matched paired T-tests are described in the text. All flow cytometry data shown were gated as follows: singlets, AquaBlue− (live cells), CD14− CD16− CD19−, small lymphocytes, CD3+, CD4−, CD8+. *** p<0.0001.
To further characterize T-bet and Eomes expression, we assessed the expression of these transcription factors within naive (CCR7+CD45RO−), central memory (CCR7+CD45RO+), effector memory (CCR7−CD45RO+), and effector (CCR7−CD45RO−) CD8 T-cells from normal donors. T-bet is undetectable in the majority of naïve CD8 T-cells (Figure 1B). ~25% of central memory express T-bet and these cells are mainly T-betlo (Figure 1B and 1C). There is a significant increase (p<0.001) in the number of T-bet+ effector memory and effector cells compared to central memory cells; however there is a higher proportion of T-bethi cells in the effector population. Similar to T-bet, Eomes is also undetectable in the majority of naïve CD8 T-cells (Figure 1D). While a small percentage of central memory cells are Eomes+, there is a significant increase (p<0.001) in the proportion of Eomes+ effector memory and effector T-cells compared to central memory. Interestingly, there is a statistically significant decrease (p<0.001) in the proportion of Eomes+ effector cells compared to Eomes+ effector memory. Additionally, Eomes was most highly expressed within effector memory cells compared to all other memory subsets (p<0.001) (Figure 1E). Taken together, these data reveal that effector CD8 T-cells have the highest expression levels of T-bet whereas Eomes is expressed in the highest frequency and expression levels in effector memory cells.
Localization of T-bet and Eomes in total resting human CD8 T-cells
While much characterization of T-bet and Eomes has focused on the presence or absence of these factors in various cell types, little work has investigated the localization, and thus the potential activity, of these factors. Because the function of many transcription factors is regulated by subcellular localization (9), we directly assessed the localization of T-bet and Eomes in human CD8 T-cells. Using a high resolution spinning disc confocal microscope, we visualized T-bet and Eomes in cells costained with DAPI and Lamin A, which localizes to the inner nuclear envelope. We found that T-bet can be localized almost exclusively in the nucleus as indicated by DAPI stain (Figures 2A, top row); however, we also observed cells where T-bet is localized throughout the cell (nuclear and cytoplasmic) and, in some cases, is almost exclusively localized to the cytoplasm (Figures 2A, middle and bottom rows, respectively). Similarly, CD8 T-cells also contain exclusively nuclear Eomes (Figures 2B top row), however, Eomes was also observed occasionally in both the nucleus and the cytoplasm or in the cytoplasm alone (middle and bottom rows).
Figure 2. Confocal microscopy of T-bet localization in resting human peripheral blood CD8 T-cells.

Representative images of resting CD8 T-cells displaying transmitted light (TL, left) images and maximal projections of confocal fluorescence microscopy for DAPI (blue), Lamin A (peach), T-bet (green), and Eomes (red). (A) T-bet is detected in the nucleus (top row), the nucleus and cytoplasm (middle row), and in the cytoplasm (bottom row) as defined by Lamin A and DAPI staining. Overlays of fluorescent channels of DAPI, T-bet, and Lamin A are shown. (B) Eomes is detected in the nucleus (top row), nucleus and cytoplasm (middle row), and in the cytoplasm (bottom row) as defined by Lamin A and DAPI staining. Overlays of fluorescent channels of DAPI, Eomes, and Lamin A are shown. Scale bar = 5 microns. TL = transmitted light.
To confirm that T-bet and Eomes can be present in both the nucleus and cytoplasm, CD8 T-cells were fractionated into nuclear and cytoplasmic compartments and analyzed for the presence of T-bet or Eomes. Both T-bet and Eomes were detectable in the nuclear and cytoplasmic fractions of CD8 T-cells (Figure 3). As controls for efficient fractionation, the exclusively nuclear protein histone deacetylase 1 (HDAC1) was detected only in the nuclear fraction whereas heat shock protein 90 (Hsp90), a cytoplasmic protein, was observed only in the cytoplasmic fraction. Taken together, these data show that T-bet and Eomes can be found in both the nuclear and cytoplasmic compartments within CD8 T-cells and suggest that their functions might be regulated at the level of nuclear localization.
Figure 3. Fractionation and immunoblot analysis of CD8 T-cells.
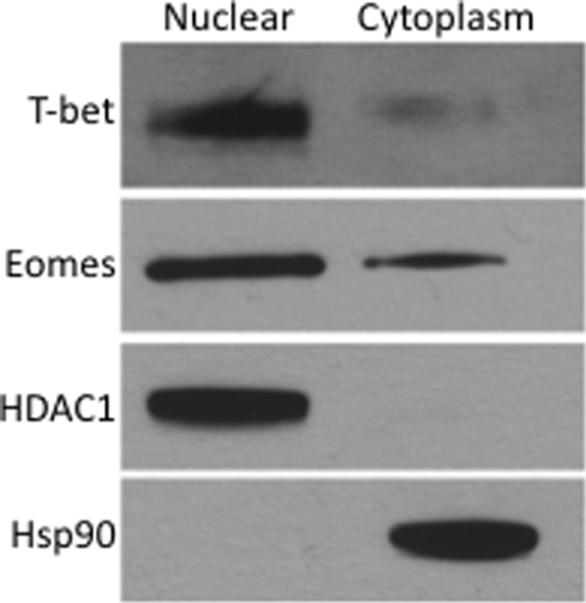
Immunoblot analysis of the nuclear and cytoplasmic fraction of purified CD8 T-cells from one healthy human donor. T-bet or Eomes was detected in both fractions. The control for exclusive nuclear localization was histone deacetylase 1 (HDAC1). The control for exclusive cytoplasmic localization was heat shock protein 90 (HSP90). Each control served as negative controls for the opposite cellular compartment.
Higher levels of nuclear T-bet and Eomes correlate with CD8 T-cell progression from memory to effector phenotypes
To better understand how the localization of T-bet and Eomes factors might contribute to CD8 memory subsets, we took advantage of Amnis ImageStream technology, which allows for simultaneous population-based flow cytometry and fluorescence microscopy. Purified, resting CD8 T-cells from five donors were stained for memory markers, CCR7 and CD45RO, as well as T-bet and Eomes. Cells were gated through memory markers and T-bet and Eomes localization was investigated. Representative effector memory cells are shown in Figure 4. T-bethi cells contain either exclusively nuclear or nuclear and cytoplasmic T-bet (Figure 4A) while T-betlo cells contain exclusively nuclear, nuclear and cytoplasmic, or exclusively cytoplasmic T-bet (Figure 4B). Similar to the confocal microscopy data, Eomes is detectable in the nucleus, nucleus and cytoplasm, or cytoplasm in CD8 T-cells.
Figure 4. Imagestream analysis of T-bet and Eomes localization in resting effector memory human CD8 T-cells.
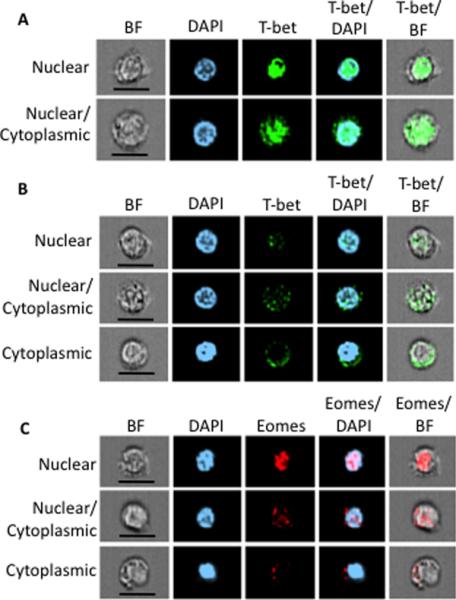
Representative images of resting CD8 effector memory T-cells are displayed. (A) T-bet (green) from T-bethi cells is detected in the nucleus (top row) or in the nucleus and cytoplasm (bottom row) as defined by DAPI (blue) staining. Overlays of fluorescent channels of DAPI and T-bet or brightfield (BF) and T-bet are shown. (B) T-bet from T-betlo cells is detected in the nucleus (top row), the nucleus and cytoplasm (middle row), or exclusively in the cytoplasm (bottom row) as defined by DAPI staining. Overlays of fluorescent channels of DAPI and T-bet or BF and T-bet are shown. (C) Eomes (red) is detected in the nucleus (top row), the nucleus and cytoplasm (middle row), or exclusively in the cytoplasm (bottom row) as defined by DAPI staining. Overlays of fluorescent channels of DAPI and Eomes or BF and Eomes are shown. Scale bar = 7 microns. Cells shown are gated as follows: singlets, focused, DAPI+, CCR7−CD45RO+.
To begin to investigate how T-bet and Eomes localization relates to different memory T-cell phenotype, we next quantified T-bet and Eomes localization within naïve, central memory, effector memory, and effector CD8 cells. To this end, T-bet+ cells were separated into T-bethi and T-betlo populations (Supplemental Figure 1). Using a masking function within IDEAS software (Amnis), the intensity of T-bet within the nucleus was plotted against the intensity of T-bet within the cytoplasm allowing the quantification of differentially localized T-bet within memory populations. T-bet was not detectable in naïve cells (data not shown). The percentage of T-bet+ cells, and specifically the percentage of T-bethi cells, increases as cells progress towards an effector phenotype (Figure 5, top row; Supplemental Table I). In all CD8 T-cell populations, T-bethi cells contain T-bet that is predominantly exclusively nuclear with a smaller fraction containing nuclear and cytoplasmic and exclusively cytoplasmic T-bet (Figure 5, middle row, naïve -data not shown). T-betlo cells contain mixed populations of nuclear, nuclear and cytoplasmic, or cytoplasmic T-bet across all memory populations (Figure 5, bottom row). The majority of T-bet+ central memory cells are T-betlo and, interestingly, ~75% of these cells contain cytoplasmic T-bet, suggesting T-bet is not active in the majority of central memory cells. ~70% of effector memory cells are T-bet+ with approximately a third of these being T-bethi and and two thirds being T-betlo. The majority of T-bethi effector memory cells are nuclear while the majority of T-betlo cells are cytoplasmic. In contrast, ~50% of effector cells are T-bethi and of these ~85% contain exclusively nuclear T-bet while ~30% of T-betlo cells contain some nuclear T-bet suggesting T-bet is likely active in most effector cells.
Figure 5. T-bet localization in resting CD8 T-cell memory populations.
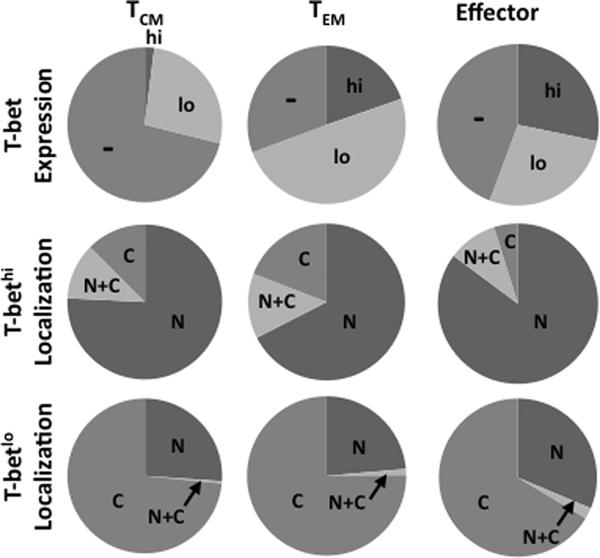
Graphical representation of the average percentage of cells expressing T-bethi or T-betlo in central memory (TCM), effector memory (TEM), or effector cells is shown (top row). The localization of T-bet in T-bethi (middle row) or T-betlo (bottom row) within each memory population is shown. N = exclusively nuclear; N+C = nuclear and cytoplasmic; C = exclusively cytoplasmic. Pie charts represent average responses of 3 normal donors.
In correlation with the data in Figure 1D, Eomes is expressed in ~15% of central memory, ~50% in effector memory, and in just less than half of effector cells (Figure 6A, top row; Supplemental Table I). Across all memory populations, between 60–75% of Eomes+ cells contain some nuclear Eomes (Figure 6A, bottom row) with central memory cells having the lowest amount of nuclear Eomes. Like T-bet, as cells become more effector-like, nuclear Eomes as well as the amount of Eomes in a given cell, increases (Figure 6B).
Figure 6. Eomes localization in resting CD8 T-cell memory populations.
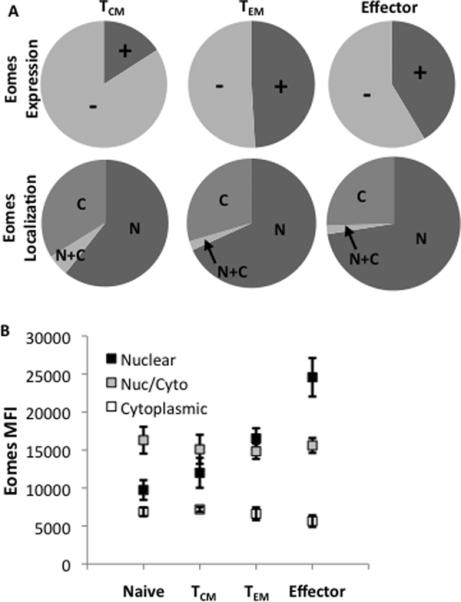
Purified CD8 T-cells analyzed using IDEAS software were gated as shown in Supplemental Figure 1. (A) Graphical representation of the average percentage of cells expressing Eomes in central memory (TCM), effector memory (TEM), or effector cells is shown (top row). The localization of Eomes within each memory population is shown (bottom row). + = Eomes+; − = Eomes−; N = exclusively nuclear; N+C = nuclear and cytoplasmic; C = exclusively cytoplasmic. Pie charts represent average responses of 3 normal donors. (B) Graphical representation of the average median fluorescence intensity (MFI) from three donors of Eomes within cells containing nuclear, nuclear and cytoplasmic, or cytoplasmic Eomes within each memory population is displayed. Diamonds = nuclear; Squares = nuclear and cytoplasmic; Triangles = cytoplasmic.
T-bet expression and localization is modulated upon T-cell receptor activation CD8 T-cells
We next investigated how generalized TCR activation affects T-bet expression and localization dynamics. To this end, purified CD8 T-cells from 5 donors were sorted into naïve, central memory, effector memory, and effector subsets based on CCR7 and CD45RO expression. Purified memory populations were activated with αCD3, αCD28, and αCD49d for up to 48 hours and cells were collected at 24-hour intervals. Blasting and non-blasting cells, as defined by size, were analyzed on the ImageStream. Within 24 hours, T-bet expression was robustly induced in greater than 80% of blasting cells following TCR stimulation in all memory phenotypes (Figure 7A, solid line). The MFI of T-bet also increases in blasting cells in all populations (Figure 7B, solid line). While non-blasting cells also can increase T-bet expression, blasting cells show a statistically significant increase in the observed overall percentage of T-bet+ cells compared to non-blasting cells (Figure 7A, dashed line; naïve p=0.002, central memory p=0.017, effector memory p=0.003, effector p=0.003). Similarly, T-bet MFI in blasting cells from naïve (p=0.016), central (p=0.013), and effector memory (p=0.030) cells is significantly higher than in non-blasting cells, where the MFI did not change over the course of 48 hours following TCR stimulation across all populations (Figure 7B, dashed line).
In addition to changes in T-bet expression, there was a significant increase in the percentage of naïve (data not shown) and effector memory cells containing nuclear T-bet that correlated with a decrease in the percentage of cells containing cytoplasmic T-bet (Figure 7C). In naïve cells, ~80% of T-bet+ cells contain nuclear T-bet following 24 hours of stimulation (data not shown). Interestingly, there was no significant increase in the percentage of central memory and effector cells containing nuclear T-bet (Figure 7C). We also observed varying amounts of nuclear and cytoplasmic T-bet over 48 hours in each population, likely due to new T-bet translation within the cytoplasm. There was no obvious difference in T-bet localization between blasting and non-blasting cells (data not shown).
To understand the dynamics of T-bet within the nuclear compartment, we next measured the MFI of T-bet within the nucleus. We found that the MFI of T-bet within the nucleus of blasting cells increases in naïve, central memory, and effector CD8 T-cells (Figure 7D, solid line; naïve - data not shown) suggesting that T-bet is being actively shuttled into the nucleus, though at different levels depending on memory type. These changes are significantly greater in blasting naïve (p=0.006), central memory (p=0.011), and effector (p=0.022) cells compared to non-blasting cells where the MFI of nuclear T-bet does not change (Figure 7D, dashed line) suggesting that the net overall amount of T-bet within the nucleus of these cells is not changing.
Taken together, these data reveal that TCR stimulation induces a robust increase in T-bet expression in all blasting memory populations. Additionally, these data provide evidence to suggest that TCR signaling elicits cell-type specific changes to T-bet expression and localization.
Eomes localization, but not Eomes expression, is modulated by T-cell receptor stimulation in purified CD8 T-cell populations
We next examined Eomes expression and localization in purified CD8 memory populations following TCR stimulation. In contrast to T-bet, the percentage of Eomes+ CD8 T-cells, as well as the MFI of Eomes, does not significantly increase in either blasting or non-blasting cells following TCR activation in naïve and effector cells within 48 hours (Figure 8A and 8B). We also analyzed the localization of Eomes and the MFI of nuclear Eomes within CD8 T-cell memory populations and found there were no significant changes in localization induced by TCR stimulation in purified CD8 T-cells. Taken together, these data reveal that, in contrast to T-bet, there is neither a net overall increase in Eomes expression, nor are there changes in Eomes localization following TCR stimulation.
DISCUSSION
In recent years, many studies have contributed to defining the mechanisms underlying control of CD8 T-cell effector function in mice (21–24), however little is known about CD8 effector differentiation in humans. In this study, we characterized the expression and localization patterns of the T-box transcription factors, T-bet and Eomes, in human CD8 memory T-cell populations to begin to dissect their functions within these T-cell subsets. In agreement with murine data (3), T-bet was observed in a graded expression pattern where high levels of T-bet are associated with effector and some TEM cells and low levels of T-bet correlate with TCM and some TEM cells. While Eomes is present in the majority of effector and TEM cells, it is more predominant and more highly expressed in TEM subsets.
A key and novel finding from our studies is that T-bet and Eomes can be localized outside of the nucleus in CD8 T-cells. To date, T-bet has been observed in rare instances in the cytoplasm in dividing cells (17), however our data provide some of the first evidence that T-bet and Eomes can be differentially localized within resting CD8 T-cells. Additionally, we show that high levels of T-bet and Eomes are more associated with nuclear localization than cells that express low levels of T-bet or Eomes. Nuclear T-bet and nuclear Eomes are associated with effector and effector memory cells suggesting T-bet and Eomes are active in these populations, while cytoplasmic T-bet and Eomes is associated predominantly with central memory phenotypes, suggesting these factors are more likely to be inactive in this subset.
The compartmentalization of T-bet and Eomes to the nucleus or cytoplasm in specific cellular contexts is of particular consequence to T-cell function. There is evidence to suggest that T-bet and Runx3, another transcription factor important to CD8 T-cell differentiation (25), cooperatively bind the same genomic region to upregulate IFNγ expression and also repress IL4 expression in CD4 Th1 T-cells (26). It is possible that T-bet and Eomes could also cooperatively bind the same gene promoters in CD8 T-cells under specific conditions, depending on their cellular localization. Interestingly, recent studies suggest that there is a concentration-dependent binding of T-bet to specific sites within the genome (27). Exhausted cells are T-betlo and this decrease in T-bet expression correlates with a loss in the ability of T-bet to repress the inhibitory receptor, PD-1 (27). Additionally, this hypothesis could also explain the contradiction that T-bet, which has been shown to repress IL-2, is expressed in IL-2 producing cells (28). The localization of T-bet in IL2 or PD1-expressing cells remains to be determined; however, based upon the low expression level of T-bet in these cells, it is likely that T-bet localization is key to this regulatory network.
Our studies also revealed key differences in the dynamics of T-bet and Eomes expression and localization. First, we show that generalized TCR stimulation robustly induces T-bet expression to greater than 80% in blasting CD8 T-cells within 24 hours of TCR activation, suggesting that CD8 T-cells alone have the ability to quickly respond to TCR stimulation to modulate T-bet. Whether TCR activation directly signals for T-bet upregulation or if it signals for upregulation of a secondary signal that then induces T-bet expression remains to be determined. Additionally, T-bet localization changes following TCR activation across all memory populations except central memory, resulting in an increase in cells expressing nuclear T-bet at 24 hours. Based on our localization in central memory cells, it is likely that upon TCR stimulation, pre-existing cytoplasmic T-bet is initially shuttled into the nucleus (Figure 7C- % Cyto T-bet), and new T-bet is subsequently produced and also shuttled into the nucleus (% Nucl/Cyto T-bet). In this regard, the lack of an overall increase in the percentage of central memory cells containing exclusively nuclear T-bet is counter-balanced by the increase in cells expressing both nuclear and cytoplasmic T-bet. In other words, T-bet is being shuttled into the nucleus, as evident by the increase in the MFI of nuclear T-bet, but these cells contain both nuclear and cytoplasmic T-bet, as evident by the increase in the percentage of these cells. If this is the case, the localization data in other memory groups likely have different mechanisms or dynamics for T-bet relocalization and upregulation. In contrast to T-bet, there are no significant changes to Eomes expression or localization in purified CD8 T-cell populations following TCR stimulation. These data suggest that TCR stimulation alone is not sufficient to affect Eomes in purified CD8 T-cells.
Overall, our data provide a baseline for future studies of T-bet and Eomes within the context of CD8 T-cells and suggest a model wherein T-bet and Eomes localization can be modulated by TCR activation in specific cellular contexts. Such a mechanism ultimately has direct impacts on the transcriptome and, consequently, overall phenotype and function of a given cell. Additionally, our localization studies represent a novel way to investigate factors involved in immune cell functions that will undoubtedly contribute to the way future studies are developed when addressing the roles of lineage defining transcription factors within the context of CD8 T-cells. Most importantly, these studies indicate the presence of previously unknown mechanisms involved in the regulation of T-bet and Eomes function. Further studies of the mechanisms that sequester T-bet and Eomes in the nucleus or cytoplasm, and the signal(s) that trigger transport of T-bet and Eomes into or out of the nucleus will provide a better understanding of T-cell development and function. Such findings will undoubtedly be beneficial towards future anti-viral therapies by potentially unlocking the mechanism(s) that could be exploited to manipulate CD8 lineage commitment and function.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI 076066 (MRB) and F32 #AI 094855 (LMM).
REFERENCES
- 1.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and Eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 2.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 3.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Intlekofer AM, Takemoto N, Kao C, Banerjee A, Schambach F, Northrop JK, Shen H, Wherry EJ, Reiner SL. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J Exp Med. 2007;204:2015–2021. doi: 10.1084/jem.20070841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi NS, Cui W, Dominguez CX, Chen JH, Hand TW, Kaech SM. Increased numbers of preexisting memory CD8 T cells and decreased T-bet expression can restrain terminal differentiation of secondary effector and memory CD8 T cells. J Immunol. 2011;187:4068–4076. doi: 10.4049/jimmunol.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 9.McLane LM, Corbett AH. Nuclear localization signals and human disease. IUBMB Life. 2009;61:697–706. doi: 10.1002/iub.194. [DOI] [PubMed] [Google Scholar]
- 10.Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Mol Cell Biol. 2008;28:1553–1564. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zabel U, Henkel T, Silva MS, Baeuerle PA. Nuclear uptake control of NF-kappa B by MAD-3, an I kappa B protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganchi PA, Sun SC, Greene WC, Ballard DW. I kappa B/MAD-3 masks the nuclear localization signal of NF-kappa B p65 and requires the transactivation domain to inhibit NF-kappa B p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beg AA, Ruben SM, Scheinman RI, Haskill S, Rosen CA, Baldwin AS., Jr. I kappa B interacts with the nuclear localization sequences of the subunits of NF-kappa B: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 14.Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 16.Zhu J, McKeon F. Nucleocytoplasmic shuttling and the control of NF-AT signaling. Cell Mol Life Sci. 2000;57:411–420. doi: 10.1007/PL00000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang JT, Ciocca ML, Kinjyo I, Palanivel VR, McClurkin CE, Dejong CS, Mooney EC, Kim JS, Steinel NC, Oliaro J, Yin CC, Florea BI, Overkleeft HS, Berg LJ, Russell SM, Koretzky GA, Jordan MS, Reiner SL. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 34:492–504. doi: 10.1016/j.immuni.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makedonas G, Banerjee PP, Pandey R, Hersperger AR, Sanborn KB, Hardy GA, Orange JS, Betts MR. Rapid up-regulation and granule-independent transport of perforin to the immunological synapse define a novel mechanism of antigen-specific CD8+ T cell cytotoxic activity. J Immunol. 2009;182:5560–5569. doi: 10.4049/jimmunol.0803945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 21.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat Rev Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- 22.Rutishauser RL, Kaech SM. Generating diversity: transcriptional regulation of effector and memory CD8 T-cell differentiation. Immunol Rev. 2010;235:219–233. doi: 10.1111/j.0105-2896.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- 23.Pipkin ME, Rao A, Lichtenheld MG. The transcriptional control of the perforin locus. Immunol Rev. 235:55–72. doi: 10.1111/j.0105-2896.2010.00905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallies A. Distinct regulation of effector and memory T-cell differentiation. Immunol Cell Biol. 2008;86:325–332. doi: 10.1038/icb.2008.16. [DOI] [PubMed] [Google Scholar]
- 25.Wong WF, Kohu K, Chiba T, Sato T, Satake M. Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology. 2011;132:157–164. doi: 10.1111/j.1365-2567.2010.03381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 27.Kao C, Oestreich KJ, Paley MA, Crawford A, Angelosanto JM, Ali MA, Intlekofer AM, Boss JM, Reiner SL, Weinmann AS, Wherry EJ. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8(+) T cell responses during chronic infection. Nat Immunol. 12:663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR. Perforin Expression Directly Ex Vivo by HIV-Specific CD8 T-Cells Is a Correlate of HIV Elite Control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


