Abstract
In the nonobese diabetic (NOD) mouse model of type 1 diabetes (T1D), insulin-dependent diabetes (Idd) loci control the development of insulitis and diabetes. Independently, protective alleles of Idd3/Il2 or Idd5 are able to partially protect congenic NOD mice from insulitis and diabetes, and to partially tolerize islet-specific CD8+ T cells. However, when the two regions are combined, mice are almost completely protected, strongly suggesting the existence of genetic interactions between the two loci. Idd5 contains at least three protective sub-regions/causative gene candidates, Idd5.1/Ctla4, Idd5.2/Slc11a1 and Idd5.3/Acadl, yet it is unknown which of them interacts with Idd3/Il2. Through the use of a series of novel congenic strains containing the Idd3/Il2 region and different combinations of Idd5 sub-region(s), we defined these genetic interactions. The combination of Idd3/Il2 and Idd5.3/Acadl was able to provide nearly complete protection from T1D, but all three Idd5 sub-regions were required to protect from insulitis and fully restore self-tolerance. By backcrossing a Slc11a1 KO allele onto the NOD genetic background, we have demonstrated that Slc11a1 is responsible for the diabetes protection resulting from Idd5.2. We also used Slc11a1 KO-SCID and Idd5.2-SCID mice to show that both loss-of-function alleles provide protection from insulitis when expressed on the SCID host alone. These results lend further support to the hypothesis that Slc11a1 is Idd5.2.
Introduction
T1D is a complex genetic disease caused by genetic interactions amongst numerous genes and influenced by various environmental factors (1–4). In humans, over 50 T1D-associated genes or gene regions have been identified (5–9). A number of genes/gene pathways have been found in common between human T1D and T1D susceptible NOD mice including those encoding HLA/MHC, insulin, IL-2/IL-2R, CTLA-4, PTPN22 and SLC11A1 (3, 4, 10–13). In the human population, the effect size of most T1D-associated genes are small (odds ratio <1.5) and genetic interactions between T1D-associated genes are near impossible to detect (14). However, similar gene variants are found to have large effects on disease frequency in NOD mice, where the background genome is held constant except for the region under investigation (15–18). These regions are named insulin-dependent diabetes (Idd) loci, amongst which the Idd3 and the Idd5 regions, when combined, provide nearly complete protection from T1D, insulitis, and the development of autoantibodies (16–19).
Introduction of B6 derived alleles at the Idd3 region on chromosome 3 onto the NOD parental strain results in significant protection from T1D (20). Using positional cloning, the Idd3 congenic interval has been narrowed to a 650 kb region that includes the genes encoding IL-2, IL-21, and several other genes (21–25). Yamanouchi and colleagues provided evidence that the IL-2 gene is Idd3 by correlating disease incidence with the amount of expression of IL-2 (25). Among the six Idd3 haplotypes tested in this study, no association between Il21 production and diabetes protection was observed (25), although two other studies reported that the expression of Il21 correlated with Idd3 haplotype (26, 27). Nonetheless, it is still under debate whether the expression of Il2 influences the expression of Il21, which in turn signals APCs that promote Th17 cell development. Both the IL-2 and IL-2 receptor alpha chain genes are associated with human T1D (9, 28) and the IL2 susceptibility allele is associated with a younger age of disease onset (14). A decrease in IL-2 availability in NOD mice is thought to result in impaired regulatory T cell (Treg) function (25). Interestingly, Idd3 has also been shown to function outside of the lymphocyte compartment most likely in dendritic cells (DCs) (29, 30).
When B10 derived alleles of the Idd5 region on chromosome 1 are introduced onto the NOD strain, significant protection from T1D results (16). It is now known that the Idd5 region consists of at least three Idd loci, namely, Idd5.1, Idd5.2, and Idd5.3 (16, 18). The Idd5.1 congenic interval contains two notable candidate genes, Ctla4 and Icos. The Idd5.2 congenic interval contains solute carrier family 11 member A1 (Slc11a1, previously Nramp1) (17). Idd5.3 contains 11 genes including the candidate gene Acadl (18, 31).
Evidence that Ctla4 is Idd5.1 includes that human T1D was found to be most strongly associated with a region immediately 3’ of the CTLA4 structural gene, (32, 33). Functional studies demonstrated that the resistant allele of Ctla4 produces more ligand-independent splice isoform of CTLA-4 (liCTLA-4) than does the disease-associated alleles (32), and that liCTLA-4 mediates negative signaling in T cells (34). liCTLA-4 transgenic NOD mice were found to be protected from T1D to the same extent as Idd5.1 congenic mice (35). Lastly, haplotype analysis and the development of Idd5.1 congenic strains that differ at the disease-susceptible Ctla4 exon 2 single nucleotide polymorphism (SNP) found that increased expression of liCTLA-4 correlated with reduced T1D frequency (35).
Slc11a1 is the most compelling T1D candidate gene located in the Idd5.2 region, since a functional missense polymorphism distinguishes the NOD diabetes-susceptible wild-type (WT) allele and the B10 diabetes-resistant loss-of-function mutant allele (17). In humans, SLC11A1 is associated with T1D and rheumatoid arthritis (11, 36, 37). SLC11A1 is a divalent cation transporter expressed on the membrane of late endosomes and lysosomes in macrophages and DCs, and expression of the WT SLC11A1 confers resistance to intracellular pathogens by promoting phagocytosis and rapid acidification of phago/lysosomes (38–44). By lentiviral transgenesis of congenic NOD mice, in vivo knock-down of Slc11a1 provided protection from T1D as well as susceptibility to Salmonella infection, supporting the hypothesis that Slc11a1 is Idd5.2 (45). Subsequent studies showed that WT Slc11a1 expressed in the DCs enhanced antigen processing and presentation (44, 46).
Microarray analysis of activated CD4+ T cells of NOD mice and Idd5.3 congenic mice, found Acadl to be the most differentially expressed gene, establishing Acadl as the strongest gene candidate for Idd5.3 (31). Acadl encodes acyl-coenzyme A dehydrogenase, long chain, an enzyme that catalyzes the first step of fatty-acid β-oxidation (47), but has no known direct immune function. However, considering that activated T cells require enormous expenditures of energy to divide and/or perform their functions and fatty acid metabolism occurs preferentially in naïve and memory but not effector T cells (48–50), it was suggested that Acadl may affect the T cell function and survival by altering the efficiency of energy utilization (31).
Previously, studies have found evidence of potent genetic interactions between the Idd3 and Idd5 protective alleles (16, 18, 51). Since the Idd5 protective region consists of three protective sub-regions, in the current study, we set out to identify which Idd5 sub-regions were required for this interaction. In a prior study, we showed that Idd3 and Idd5.1 interactions did not result in increased protection from diabetes (18). We have now developed a series of novel congenic mice isolating the Idd5.2 and Idd5.3 regions and recombining the isolated regions back together to verify that an additional Idd gene was not evident in between these two regions. These strains were crossed with Idd3 mice to create various combinations, and the frequency of diabetes, the degree of insulitis and islet-specific T cell tolerance were investigated. Furthermore, to substantiate the candidacy of Slc11a1 being Idd5.2, we backcrossed the 129-derived Slc11a1 KO allele onto the NOD genetic background.
Materials and Methods
Congenic mouse strains
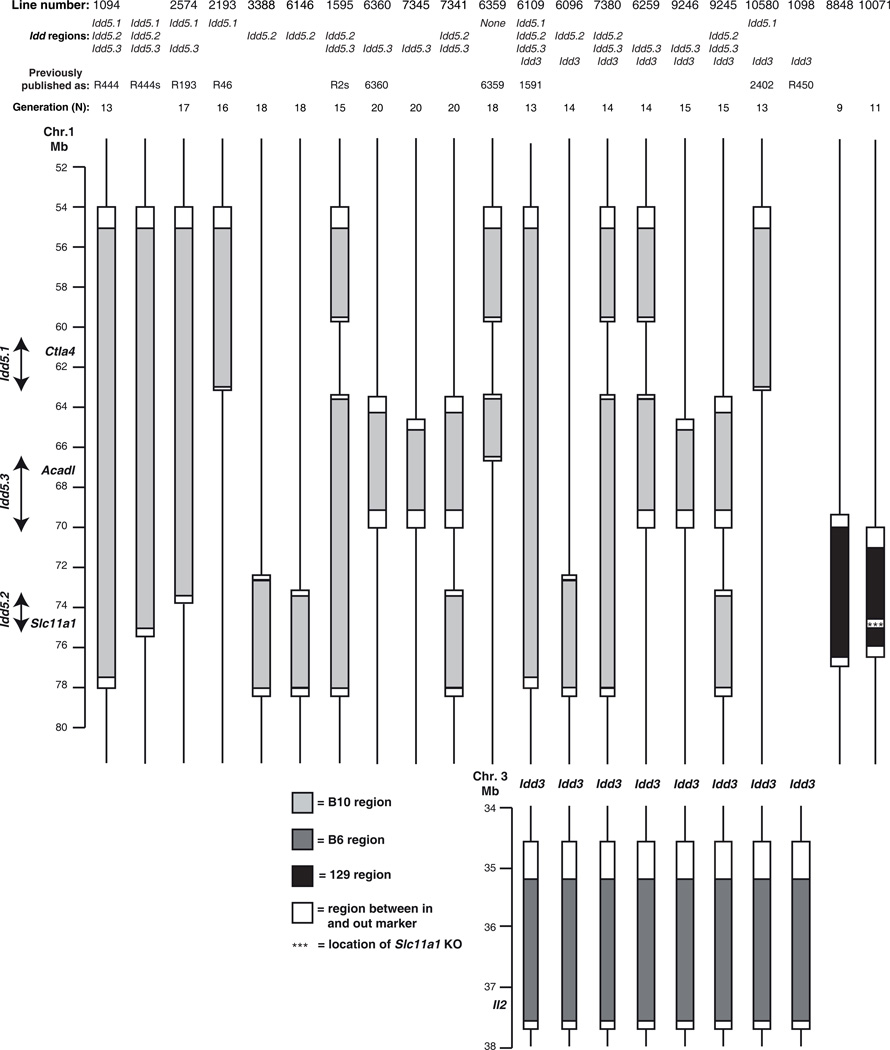
All experimental procedures were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (IACUC #09-0074). The breeding and genotyping strategies to develop the congenic murine strains with various Idd protective alleles have been described (22). For clarity, all the murine strains pertinent to the current study are summarized in Fig. 1, and all the novel markers developed for genotyping are summarized in Supplemental Table I. NOD and NOD-SCID mice were purchased from Taconic. Lines 1094, 1098, 1595, 2193, 3700, 6109, 6359, 6360, 7341 and 10580 were previously described (16–18, 23). Nine novel congenic strains have been developed specifically for this study. Two Idd5.2 congenic Lines 3388 and 6146 were generated from Line 1595. Idd5.3 congenic Line 7345 was generated from Line 6360. Idd5.2+5.3 congenic Line 7341, in which the isolated Idd5.2 and Idd5.3 congenic intervals were combined, was generated from crossing Line 6146 and Line 6360. Idd3+5.2 congenic Line 6096 was generated by crossing Line 1098 with Line 3388. Two Idd3+5.3 congenic Lines 6259 and 9246 were generated by crossing Line 1098 with Lines 3700 that has the Idd5 segment shown for Line 6259 in Fig. 1 (18) and 7345. Two Idd3+5.2+5.3 congenic Lines 7380 and 9245 were generated by crossing Line 1098 with Lines 1595 and 7341. Lines 6109, 2193, 7341, 6146, 7345, 1094, 2574 and 1098 have been cryopreserved at The Jackson Laboratory (www.jaxmice.jax.org) and are available as lines 012393, 012391, 012396, 012394, 012395, 004344, 012392 and 007934 respectively.
Figure 1.
Genetic intervals of the strains pertinent to this study. Line numbers, or strain names, as well as the Idd regions that the strains carry respectively, were labeled on top of the figure. Generation (N) indicates the number of generations of backcrossed to parental NOD strain. Filled light grey regions indicate B10-derived DNA segment on chromosome 1. Filled black regions, on which three stars denote the location of the Slc11a1 KO, indicate 129-derived DNA segment on chromosome 1. Filled dark grey regions indicate B6-derived DNA segment on chromosome 3. Open regions represent DNA segment between the in and out allelic markers at the boundaries. Lines delineate NOD-derived DNA. Vertical arrows designate the Idd5.1, Idd5.2, and Idd5.3 regions. The diagram is to scale.
To further investigate the strongest candidate gene for Idd5.2 allele, namely, Slc11a1, two novel NOD.129 murine strains were developed (Fig. 1). Line 8848 NOD.129 Slc11a1 WT (Slc11a1 WT) was developed by backcrossing the 129S6 strain (Taconic) 9 times with the NOD/MrkTac (Taconic) strain, selectively breeding progeny with proximal and distal recombination events close to the Slc11a1 gene: the 129-derived region of interest is defined by the proximal in and out markers rs30495565 and rs32083833, respectively, and the distal in and out markers rs30150991 and rs32807660, respectively. Line 10071 NOD.129 Slc11a1 knockout (Slc11a1 KO) was developed in a similar manner by backcrossing Slc11a-null 129S4 mice (46, 52) 11 times with the NOD strain; the 129-derived region of interest is defined by the proximal in and out markers rs32912547 and rs30495565, respectively, and the distal in and out markers D1mit46 and rs30150991, respectively. A 1449 SNP marker panel across 19 autosomes and the X chromosome, averaging a genetic interval of 5 Mbps (1449 marker panel, Taconic) was used to identify and remove by selective breeding any remaining 129-derived regions other than the desired congenic interval.
Idd5-SCID congenic strains have been described previously (30). Idd5.2-SCID mice were generated by inter-crossing Line 6146 and NOD-SCID mice and selecting mice homozygous for both alleles. Slc11a1 KO-SCID was generated by inter-crossing Slc11a1 KO and NOD-SCID mice and selecting progeny homozygous for both alleles.
Variant identification in Idd5.2
The NOD and 129 genomes have been sequenced using next generation sequencing technology (http://www.sanger.ac.uk/resources/mouse/genomes/). Single nucleotide polymorphisms (SNPs) from the B6, NOD and 129 Idd5.2 region were entered into T1DBase (53, 54) and displayed graphically using Gbrowse (55).
Reconstitution of SCID mice
As previously described (30), female SCID mice were reconstituted with total spleen and lymph node cells prepared from female donor mice 3 to 5 weeks of age. DCs were depleted with Pan-DC microbeads (Miltenyi Biotec) according to the manufacturer’s instructions before transfer of 2–3 × 107 cells intravenously. Mice were rested for 10 weeks before harvesting pancreata.
Assessment of diabetes and insulitis
Female mice were monitored on a bi-weekly basis, and mice having elevated urinary glucose >500 mg/dl (detected using Diastix (Myles, Elkhart, IN)) for two consecutive daily tests were classified as diabetic and euthanized thereafter. During the diabetes frequency study, a few non-diabetic mice became sick due to infection or a tumor, or were found dead with unknown cause and these were censored from the analysis. The diabetes frequency studies represented in Figures 2A and 2D were performed at Merck Research Laboratories whilst the studies in Figures 2B, 2C, 2E5B and supplemental Fig. 1 were conducted at Taconic. The diabetes frequency of the control NOD strain was notably reduced in the study reported in Fig. 5B due to unknown environmental influences since the genetic integrity of the NOD strain used at Taconic for the study depicted in Fig. 5B was confirmed.
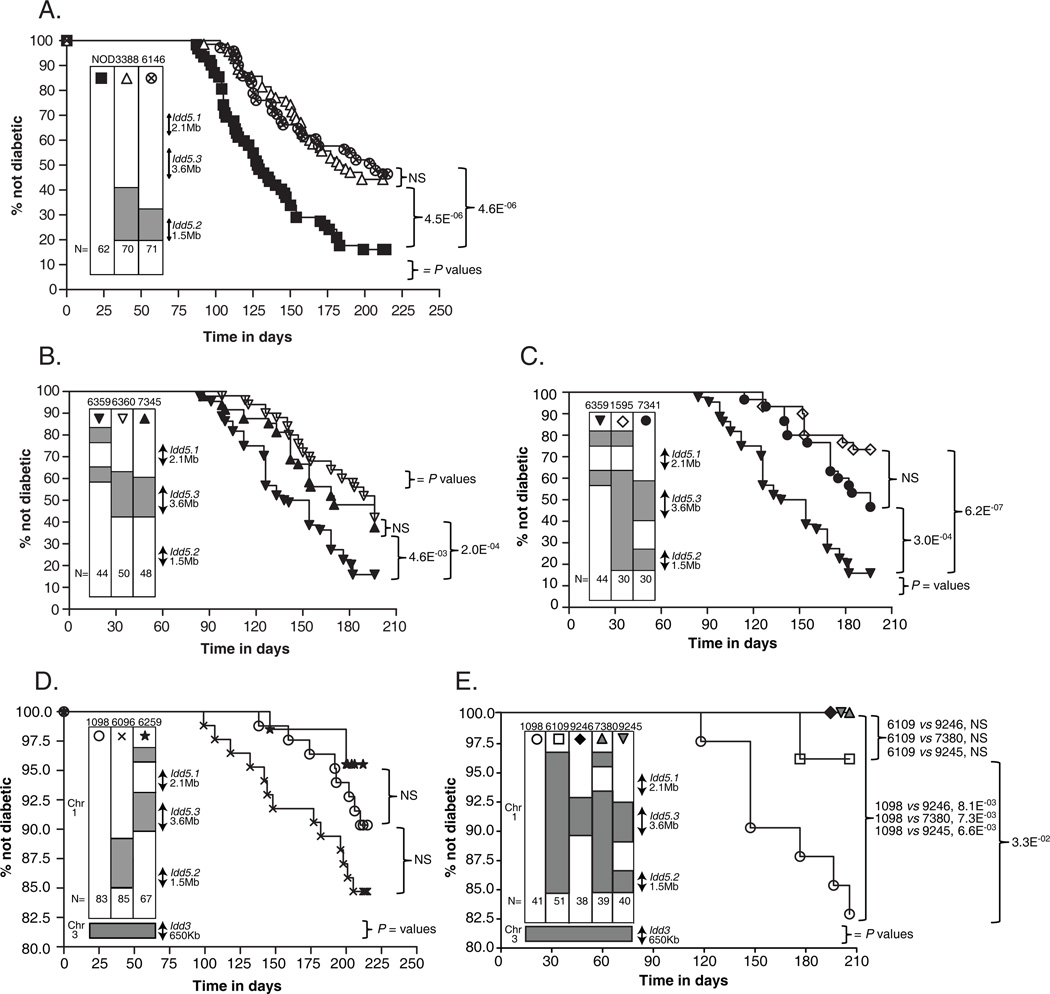
Figure 2.
Diabetes frequencies of the novel congenic strains developed specifically for this study. The diabetes frequency studies represented in Figures 2A and 2D were performed at Merck Research Laboratories whilst the studies in Figures 2B, 2C, and 2E were conducted at Taconic. Group size N is indicated at the bottom of the genetic map of each strain. The p-values were calculated by comparing the Kaplan-Meier survival curves of the indicated strains using the log-rank test. NS, not significant, p-value ≥0.05. (A) Diabetes frequencies of the Idd5.2 congenic Lines 3388 and 6146 females were compared with NOD females. (B) Diabetes frequencies of the Idd5.3 congenic Lines 6360 and 7345 females were compared with NOD females. (C) Diabetes frequencies of the Idd5.2+5.3 congenic Lines 1595 and 7341 females were compared with NOD-like Line 6359 females. (D) Diabetes frequencies of the Idd3 congenic Line 1098, the Idd3+5.2 congenic Line 6096, and the Idd3+5.3 congenic Line 6259 females were compared. Note that the y-axis is truncated due to the low frequency of T1D in the strains examined. (E) Diabetes frequencies of the Idd3 congenic Line 1098, the Idd3+5.3 congenic Line 9246, two Idd3+5.2+5.3 congenic Lines 7380 and 9245, the Idd3/5 congenic Line 6109 females were compared. Note that the y-axis is truncated due to the low frequency of T1D in the strains examined.
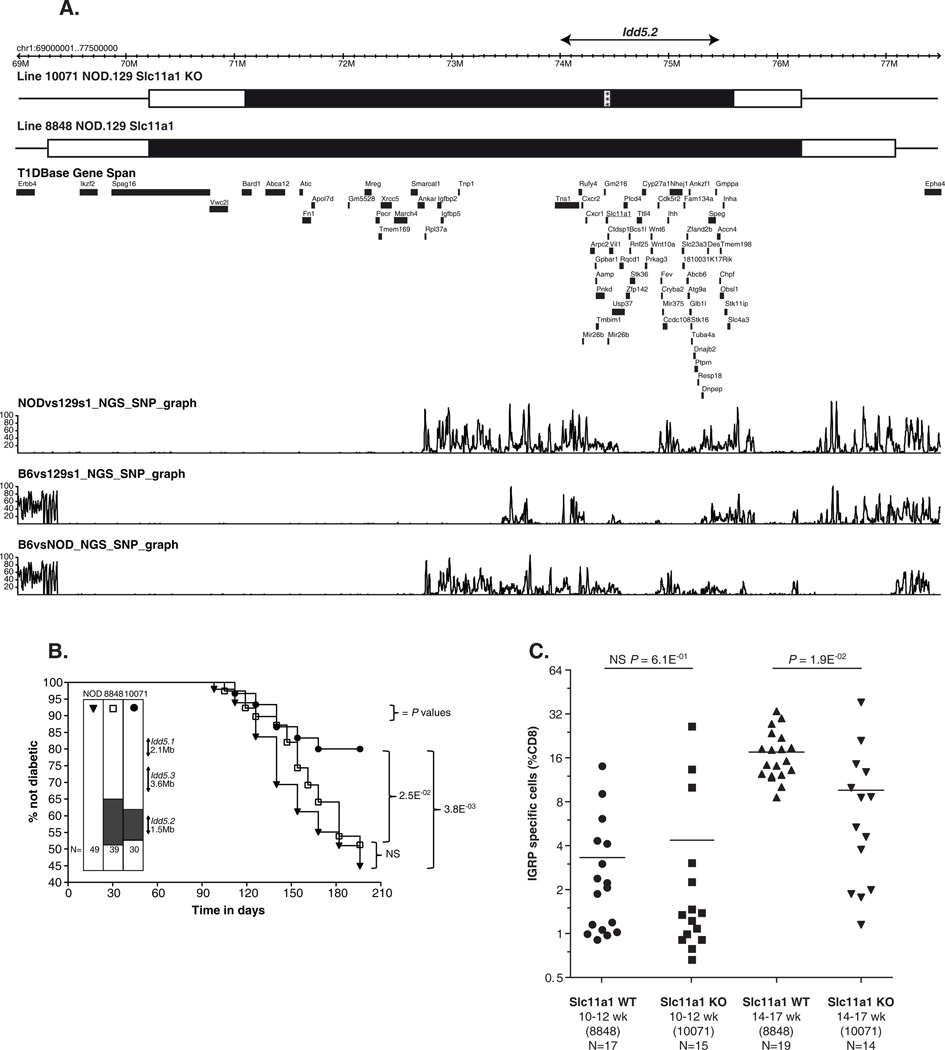
Figure 5.
Slc11a1 KO strain is protected from T1D and shows partially restored CD8+ T cell tolerance to endogenous islet antigen IGRP. (A) Genetic intervals of the Slc11a1 WT and Slc11a1 KO strains. Filled black region indicates the 129-dervied DNA and white bars indicate the regions between the ‘in’ and ‘out’ markers. The gene content is displayed in the T1Dbase Gene Span tract (based on GRCm37 genome build), with genes shown in black and Slc11a1 underlined. The SNP density tracks are based on the next generation sequencing (NGS) data and displays SNPs per 10 kb. (B) Diabetes frequencies of the NOD, the Slc11a1 WT and the Slc11a1 KO females were compared. The diabetes frequency studies were conducted at Taconic. (C) Slc11a1 KO alleles contribute to the reduced percentage of self-reactive IGRP specific CD8+ T cells. Results were compiled from 6 independent experiments, each of which analyzed 6–8 female mice per strain, at either 10–12 weeks of age, or 14–17 weeks of age. Female Slc11a1 WT and Slc11a1 KO mice were infected with Vac-KdIGRP. One week later, spleens were harvested, and analyzed by FACS for CD8+ IGRP-tetramer+ cells. Horizontal lines depict means. Pairs of strains were compared using unpaired t-test with Welch’s correction. NS, not significant, p-value ≥0.05.
Pancreata were fixed in 10% neutral buffered formalin, processed for paraffin embedding, stained with hematoxylin and eosin, and cut to obtain two non-contiguous tissue sections. Blinded tissue sections were evaluated for the presence of mononuclear cell infiltrates in the pancreatic islets. At least 20 islets were scored as either 0: no infiltration, 1: peri-insulitis, 2: mild-invasive insulitis, or 3: severe invasive insulitis. The average score of each pancreas was calculated and used for statistical analysis.
Virus
Recombinant vaccinia virus (Vac-KdIGRP) expressing the H-2Kd restricted epitope VYLKTNVFL, amino acid residues 206–214 of murine Islet-specific glucose-6-phosphatase catalytic subunit related protein (IGRP) was previously described (56). Mice were infected intraperitoneally with 1×107 p.f.u. of virus and CD8+ T cell responses measured in the spleen 7 days later by tetramer staining.
Flow cytometry
CD8+ T cells were stained with H-2Kd-IGRP206–214 –PE tetramers (NIAID MHC Tetramer core facility) for 15 minutes RT followed by staining with anti-CD8-FITC at 4°C for 15 minutes. All mAbs were obtained from either eBioscience (San Diego, CA), BioLegend (San Diego, CA) or BD Pharmingen (San Diego, CA). Cells were acquired with either a FACS Calibur or LSRII (Becton Dickinson, Mountain View, CA.) and analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Statistical analysis
Kaplan Meier survival analysis of diabetes was performed for each group of mice and T1D frequencies were compared using the log-rank test (Graphpad Prism software). Differences in the degrees of insulitis were compared between groups via the Mann-Whitney test (Graphpad Prism software). Differences in the proportion or number of IGRP tetramer binding cells were compared between groups via the unpaired student t-test with Welch’s correction (Graphpad Prism software).
Results
Recombining isolated Idd5.2 and Idd5.3 congenic intervals recapitulates the T1D protection observed with contiguous protective alleles
The previous observation of approximately 2% diabetes frequency amongst Idd3/5 females strongly suggests that potent interlocus genetic interactions between Idd3 and Idd5 exist (16, 18, 51). In our previous study (18), we combined the isolated protective Idd5.1 and Idd3 regions in a double congenic strain, and showed that the Idd3+5.1 strain and Idd3 strain were equally protected from T1D, and that both strains were significantly less protected from T1D than Idd3/5 strain, suggesting that either Idd5.2, or Idd5.3, or both, are essential for the interlocus genetic interactions with Idd3. However, concurrent with the development of such double and triple congenic strains combining Idd5.2, or Idd5.3, or both, with Idd3 (18), we wanted to include a comparison of the diabetes protection provided by the Idd5.2 and Idd5.3 regions together on a single congenic segment (Line 1595, Fig. 1) to that of the isolated congenic regions recombined back together (Line 7341) in the presence or absence of protective Idd3 alleles. This strategy would enable us to determine if another Idd subregion is present between the Idd5.2 and Idd5.3 regions as did a similar experiment of recombining the isolated Idd5.1 and Idd5.2 regions facilitated the discovery of the Idd5.3 region (18). In pursuing this strategy, we developed novel strains having the isolated Idd5.2 and Idd5.3 congenic intervals. These novel strains enabled us to confirm the T1D protection using independently-derived congenic segments and to reduce the overall size of the introgressed congenic interval as compared to the analogous strains previously published.
Two novel Idd5.2 congenic strains were developed, Lines 3388 and 6146 (Fig. 1). We observed that both strains were indistinguishable from each other in their respective T1D frequency and significantly more protected than control NOD mice (Fig. 2A). This confirms our previous findings (18, 45) that the presence of a B10-derived congenic region encompassing the 1.5 Mb Idd5.2 interval including the candidate gene Slc11a1 (17) was sufficient to confer highly significant, albeit moderate, protection from T1D, compared with NOD strain (Fig. 2A).
We also developed a novel Idd5.3 congenic Line 7345, and observed that its T1D frequency was indistinguishable from that of Line 6360 (Fig. 2B), an Idd5.3 congenic strain that we developed in a previous study (18). This demonstrated that the presence of the B10-derived resistance allele at Idd5.3 locus alone was sufficient to confer highly significant, albeit moderate, protection from T1D, compared with NOD strain (Fig. 2B). The protective effect of Line 7345 corroborates previous work mapping Idd5.3 to a 3.5 Mb interval including the candidate gene Acadl (18, 31).
We developed a novel Idd5.2+5.3 congenic Line 7341 in which the isolated Idd5.2 (from Line 6146) and Idd5.3 (from Line 6360) congenic intervals were combined by selective breeding of an inter-subregion recombinant (Fig. 1). We observed that the T1D frequency of Line 7341 was indistinguishable from that of Line 1595 (Fig. 2C), an Idd5.2+5.3 congenic strain that we developed in a previous study where the Idd5.2 and Idd5.3 regions are both in the same congenic segment (18), indicating that it is unlikely there are additional Idd genes located between the Idd5.2 and Idd5.3. The two Idd5.2+5.3 congenic strains were compared to Line 6359, which contains the proximal end of Line 1595, and was previously shown to have a diabetes incidence similar to NOD mice (18). The high incidence of Line 6359 found here (Fig 2C) replicates the finding that Line 6359 does not contain a protective gene and Idd5.3 is located in the distal half of the congenic segments present in Lines 6360 and 7345.
Interlocus genetic interaction between Idd3 and Idd5 sub-regions in the protection from T1D
We now combined the Idd5.2, Idd5.3 and Idd5.2+5.3 congenic intervals with Idd3 in order to test which region(s) are required for the interlocus genetic interaction and prevention of diabetes. The Idd5.2 congenic Line 3388 was crossed with Idd3 mice to generate a novel Idd3+5.2 congenic Line 6096 (Fig. 1). Line 3700, an Idd5.3 congenic strain that we developed in a previous study (18) that has the Idd5 segment shown for Line 6259 in Fig. 1, was crossed with Idd3 mice to generate a novel Idd3+5.3 congenic Line 6259 (Fig. 1). The Idd5.3 congenic Line 7345 was crossed with Idd3 mice to generate another novel Idd3+5.3 congenic Line 9246 (Fig. 1). Both Idd5.2+5.3 congenic Lines 1595 and 7341 were crossed with Idd3 mice to generate two novel Idd3+5.2+5.3 congenic Lines 7380 and 9245 (Fig. 1).
The diabetes frequency of the Idd3+5.2 congenic Line 6096 was indistinguishable from that of its contemporaneous control Idd3 mice (Fig. 2D), suggesting that Idd5.2 is not sufficient for the interlocus genetic interactions when diabetes phenotype was assessed. The diabetes frequency of the Idd3+5.3 congenic Line 6259 was indistinguishable from that of its contemporaneous control Idd3 mice, although we observed a trend of Idd3+5.3 strain having less incidence of disease than Idd3 strain (Fig. 2D). The diabetes frequency of the more recently developed Idd3+5.3 congenic Line 9246 was significantly lower than that of its contemporaneous control Idd3 mice, and was indistinguishable from that of its contemporaneous control Idd3/5 mice (Fig. 2E). Considering the stochastic nature of the onset and development of T1D, we assessed both studies together in a combined analysis (Supplemental Fig. 1). The combined Idd3+5.3 mice were significantly more protected from T1D than the control Idd3 mice (Supplemental Fig. 1), suggesting that that Idd5.3 is essential for the interlocus genetic interactions when diabetes phenotype was assessed.
The disease frequencies of Idd3+5.2+5.3 congenic Lines 7380 and 9245, as well as their contemporaneous control Idd3/5 mice, were next assessed and found to be indistinguishable from each other (Fig. 2E). All three strains were significantly more protected from T1D than the contemporaneous control Idd3 mice (Fig. 2E), providing strong support to our above mentioned hypothesis that Idd5.3 is essential for the interlocus genetic interactions when diabetes phenotype was assessed.
Genetic interactions between Idd3 and Idd5 sub-regions that lend protection from insulitis
Our previous observation that Idd3+5.1+5.3 females were protected from disease to a degree indistinguishable from that of Idd3/5 females (18) appears to suggest that Idd5.2 is not essential for the interlocus genetic interactions with Idd3. However, when the occurrence and severity of insulitis of the two strains were compared, it was discovered that Idd3+5.1+5.3 mice had significantly more insulitis than Idd3/5 mice (18), indicating that Idd5.2 is required for the interlocus genetic interactions with Idd3 to reduce insulitis. Hence, in regard to the NOD genetic background, although both diabetes and insulitis have a certain degree of stochastic nature, the insulitis readout tends to provide a larger window and more power than the diabetes readout, in distinguishing a relatively smaller yet clearly existing difference between two different strains.
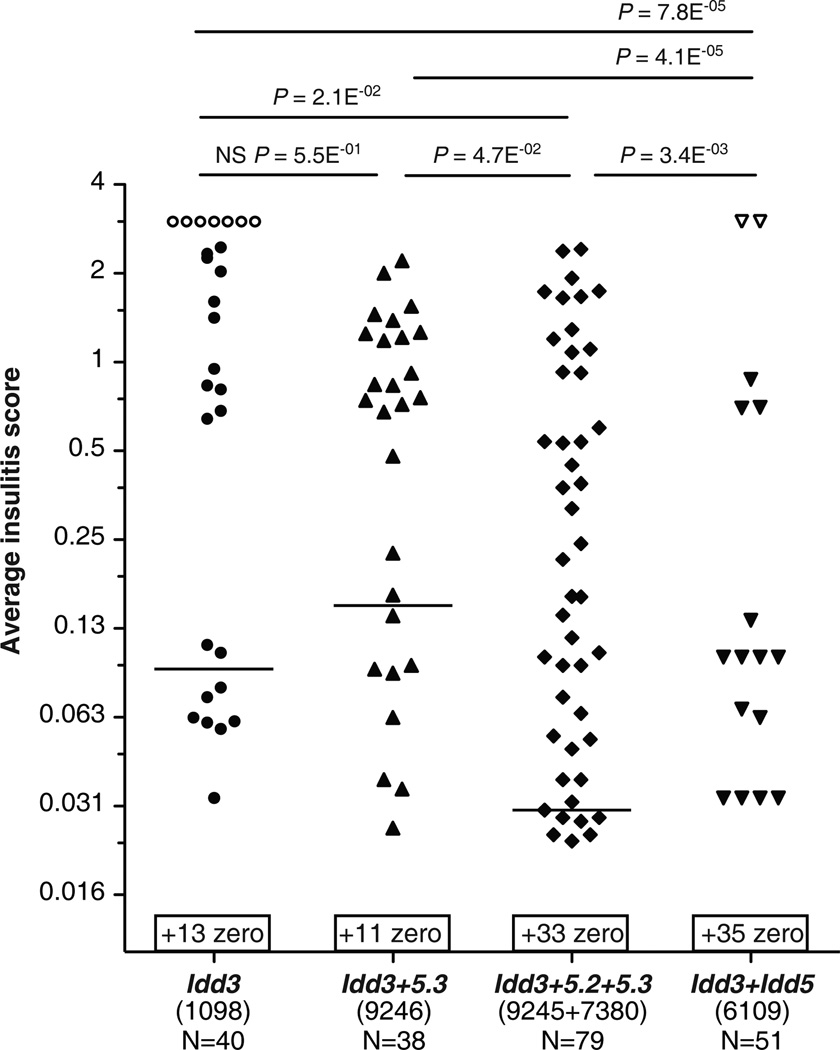
In order to further investigate the genetic interactions between Idd3 and Idd5 sub-regions that provide protection from insulitis, five strains: Idd3 mice, Idd3+5.3 congenic Line 9246, Idd3+5.2+5.3 congenic Lines 7380 and 9245, and Idd3/5 mice, were examined for the occurrence and severity of insulitis (Fig. 3). The latter four strains were specifically selected because among all the newly developed strains, they were the only four that were significantly more protected from diabetes than their contemporaneous Idd3 controls (Fig. 2E). The occurrence and degrees of insulitis of both Idd3+5.2+5.3 congenic Lines 7380 and 9245 were indistinguishable from each other (Supplemental Fig. 2); therefore, we combined these two strains in subsequent analyses (Fig. 3). Reminiscent of our previous study (18), a hierarchy in the protection from insulitis was discovered: first, Idd3/5 strain was more protected than Idd3+5.2+5.3 strains (Fig. 3), suggesting that Idd5.1 contributes to reduced insulitis in the presence of Idd3, Idd5.2 and Idd5.3 protective alleles; second, Idd3+5.2+5.3 mice were more protected than the Idd3+5.3 and Idd3 mice (Fig. 3), suggesting that Idd5.2 contributes to reduced insulitis in the presence of Idd3 and Idd5.3 protective alleles in the NOD genome; third, Idd3+5.3 mice and Idd3 mice had equivalent levels of insulitis (Fig. 3), suggesting that although Idd5.3 contributes to reduced diabetes in the presence of Idd3 (Fig. 2E and Supplemental Fig. 1), it does not contribute to insulitis reduction. It is important to note that Idd3+5.3 mice, the two lines of Idd3+5.2+5.3 mice and Idd3/5 mice had indistinguishable T1D frequency (Fig. 2E), yet the first three strains all had significantly more occurrence and greater severity of insulitis than Idd3/5 strain (Fig. 3). Taking into account both diabetes incidence and insulitis, we conclude that all three Idd5 sub-loci on chromosome 1 are required to interact with Idd3 locus on chromosome 3 to provide protection from diabetes and insulitis to the extent observed in Idd3/5 mice.
Figure 3.
A hierarchy in the insulitis protection in Idd3+Idd5-sub-region(s) congenic strains. Non-diabetic females of the congenic Idd3 (Line 1098), Idd3+5.3 (Line 9246), Idd3+5.2+5.3 (Lines 9245 and 7380), and Idd3/5 (Line 6109) at 196 days of age were sacrificed and insulitis scored. Each diabetic mouse was assigned an insulitis score of ‘3’ indicated by an open symbol. For each group, the number (X) of pancreases that have an average insulitis score of zero was indicated on top of the x-axis and written as ‘+(X) zero’, because of the Log2 scale on the y-axis. Horizontal lines on the graph depict median values. Note that Idd3/5 strain had a median of zero. Each pair of strains is compared using Mann-Whitney U test. NS, not significant, p-value ≥0.05.
Idd5.2 alone is able to partially restore self-tolerance to endogenous islet antigen IGRP
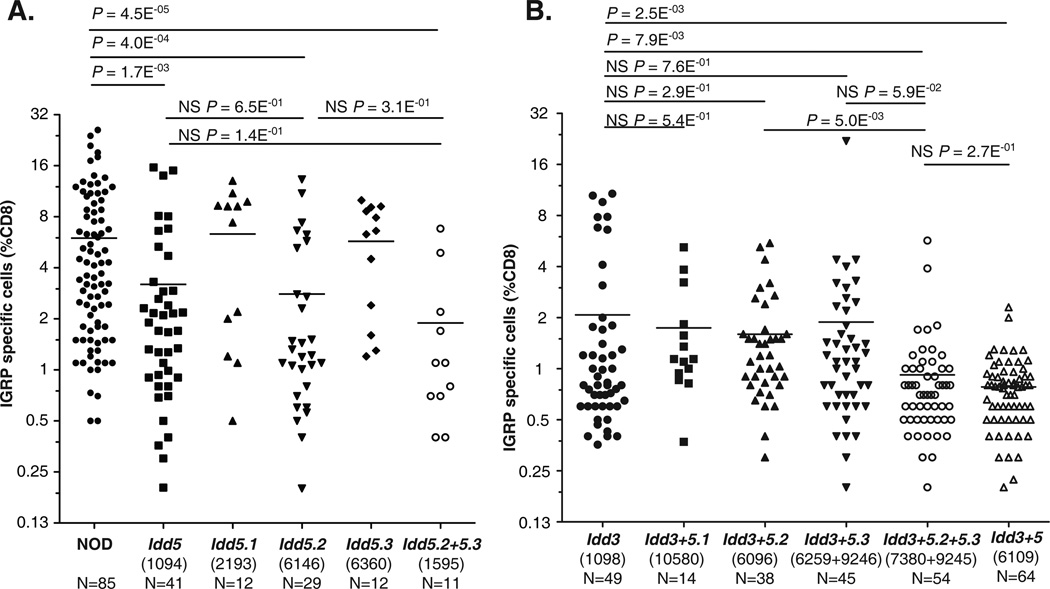
IGRP is an endogenous pancreatic islet antigen. The number of IGRP specific CD8+ T cells is an indication of auto-reactivity and loss of self-tolerance, and is predictive of T1D progression in individual NOD mouse (57). Compared with the NOD strain, Idd3/5 mice exhibited noticeably restored CD8+ T cell tolerance (29, 58, 59). Previously, we found that both the Idd3 and Idd5 regions independently contributed to the partially restored CD8+ T cell tolerance, and that their effects were additive (30).
The development of isolated Idd5 sub-region strains allowed us to investigate which of the three Idd5 sub-regions contribute to restored CD8+ T cell tolerance observed in Idd5 congenic strain. Hence, we applied the same method to characterize the CD8+ T cell tolerance to IGRP in these strains (Fig. 4A). Consistent with our previous report (30), Idd5 mice exhibited a significantly reduced percentage of IGRP-specific CD8+ T cells (Fig. 4A), suggesting that the entire Idd5 genetic segment contributes to the restoration of CD8+T cell tolerance. Among the three Idd5 sub-regions, only the Idd5.2 region significantly reduced the percentage of IGRP-specific CD8+ T cells, to the same degree as that of the Idd5 strain (Fig. 4A). The effect of the Idd5.2 region was also observed in the Idd5.2+5.3 strain, which showed significantly reduced percentage of IGRP-specific CD8+ T cells, to the same degree as that of the Idd5 strain (Fig. 4A).
Figure 4.
The contribution of Idd3 and Idd5 sub-regions to the restoration of CD8+ T cell tolerance to endogenous islet antigen IGRP. Results were compiled from 13 independent experiments, each of which analyzed 6–12 female mice per strain, at 10–14 weeks of age. Female NOD, Idd5, Idd5.1, Idd5.2, Idd5.3, Idd5.2+5.3, Idd3, Idd3+5.1, Idd3+5.2, Idd3+5.3, Idd3+5.2+5.3, and Idd3/5 mice were infected with Vac-KdIGRP. One week later, spleens were harvested, and analyzed by FACS for CD8+ IGRP-tetramer+ cells. 30 out of 91 NOD females, 18 out of 46 Idd5 females, 29 out of 58 Idd3 females, and 30 out of 71 Idd3/5 females shown on the graph were published previously (30). These mice were included in the analysis because they were contemporaneous controls of other congenic strains that were examined at the same time. Horizontal lines depict the means. Pairs of strains were compared using unpaired t-test with Welch’s correction. NS, not significant, p-value ≥0.05. (A) Idd5.2 alone is sufficient to partially restore CD8+ T cell tolerance to self-antigen IGRP. (B) More than one Idd5 sub-regions need to be combined with Idd3 to further restore CD8+ T cell tolerance to self-antigen IGRP.
When combined with Idd3, more than one Idd5 sub-regions is needed to restore self-tolerance to the same degree observed in Idd3/5 mice
The development of Idd3+5.1, Idd3+5.2, Idd3+5.3 and Idd3+5.2+5.3 strains presented an opportunity to investigate the potential interactions between each of three Idd5 sub-regions and Idd3 for restoration of CD8+ T cell tolerance. Consistent with our previous report (30), Idd3/5 mice exhibited a significantly reduced percentage of IGRP-specific CD8+ T cells than that of Idd3 mice (Fig. 4B), suggesting that at least one gene within Idd5 contributes to the genetic interactions with Idd3. Since both Idd3+5.3 congenic Lines 6259 and 9246 were indistinguishable from each other (Supplemental Fig. 3), we combined them in the subsequent analyses. Both Idd3+5.2+5.3 congenic Lines 7380 and 9245 were also indistinguishable from each other (Supplemental Fig. 3), and were combined in the subsequent analyses. None of the three Idd5 sub-regions, when individually combined with Idd3, was able to significantly reduce the percentage of IGRP-specific CD8+ T cells (Fig. 4B). However, the combination of Idd5.2 and Idd5.3 sub-regions was able to interact with Idd3 to provide additional restoration of CD8+ T cell tolerance, to a degree that was indistinguishable from that of Idd3/5 mice (Fig. 4B). These results suggest that more than one Idd5 sub-region is required to interact with Idd3 to further restore CD8+ T cell tolerance.
NOD.129 Slc11a1 KO strain is protected from T1D and shows partially restored self-tolerance to endogenous islet antigen IGRP
As the protective B10 allele at Slc11a1 is a loss of function mutation (17), we reasoned that if Slc11a1 was the causative gene in the Idd5.2 locus, Slc11a1 KO NOD mice should be protected from T1D. Hence, we crossed a Slc11a1-null allele from the 129S4 genetic background (46, 52) onto the NOD genetic background and generated a novel Slc11a1 KO NOD strain. As a control, we crossed the Slc11a1-WT allele from 129S6 mice onto the NOD genetic background. The control strain expresses the functional Slc11a1 allele in the context of the 129 genome at the Idd5.2 region matching closely to the 129-derived congenic interval present in the NOD.129 Slc11a1-null strain (Fig. 1 and Fig. 5A). The two congenic 129 intervals are not identical in size as it is nearly impossible by chance to achieve identical recombination sites between two strains. However, the congenic regions in both strains encompass the previously defined Idd5.2 region shown as a horizontal arrow in Fig. 5A. The gene content and SNP density of the congenic intervals are also shown in Fig. 5A.
After the development of these two strains, we first tested the diabetes incidence, in comparison to contemporaneous control NOD (Fig. 5B). Diabetes frequency of Slc11a1 WT was indistinguishable from NOD (Fig. 5B), even though NOD and 129 have different haplotypes across this region (Fig. 5A). This observation strongly suggests that the DNA segment derived from the 129 genetic background carrying Slc11a1 WT alleles does not provide significant protection from diabetes, and SNPs in other 129-derived genes in the congenic region also do not influence disease susceptibility. In contrast, diabetes frequency of Slc11a1 KO was significantly lower than that of Slc11a1 WT and NOD controls (Fig. 5B). Since the Slc11a1 KO 129 congenic segment is contained entirely within the WT 129 congenic segment, the T1D frequencies observed indicate that the Slc11a1 WT alleles in the context of NOD genome are T1D susceptibility alleles and Slc11a1 KO alleles confer protection from diabetes. It is also notable that where the two congenic strains differ, the NOD and 129 strains either share the same sequence (proximal boundary region) or no coding genes are present (distal boundary region); this is consistent with the hypothesis that the Slc11a1 disruption is the genetic event causing reduced diabetes susceptibility rather than a complex interaction amongst two or more 129-derived genes that are variably present in the two NOD.129 congenic strains and that Idd5.2 is Slc11a1.
As the Idd5.2 strain provides partial restoration of CD8+ T cell tolerance to IGRP, we reasoned that the Slc11a1 KO allele on the NOD genetic background should also mediate this effect. However, the examination of young female mice 10–12 weeks of age revealed no difference between the percentages of IGRP specific CD8+ T cells of the Slc11a1 WT and KO strains (Fig. 5C). This was a surprising result, considering that the diabetes frequencies of the two strains clearly differ from each other (Fig. 5B). Therefore, we decided to assess older female mice, 14–17 weeks of age, as unpublished data (E.H.W. personal observations) strongly indicates that IGRP specific CD8+ T cells accumulate in various NOD congenic mice as they age. We found that comparing the mice at 14–17 weeks of age, Slc11a1 KO has significantly reduced percentage of IGRP specific CD8+ T cells than Slc11a1 WT (Fig. 5C), suggesting the Slc11a1 KO alleles contribute to a partial restoration of CD8+ T cell tolerance.
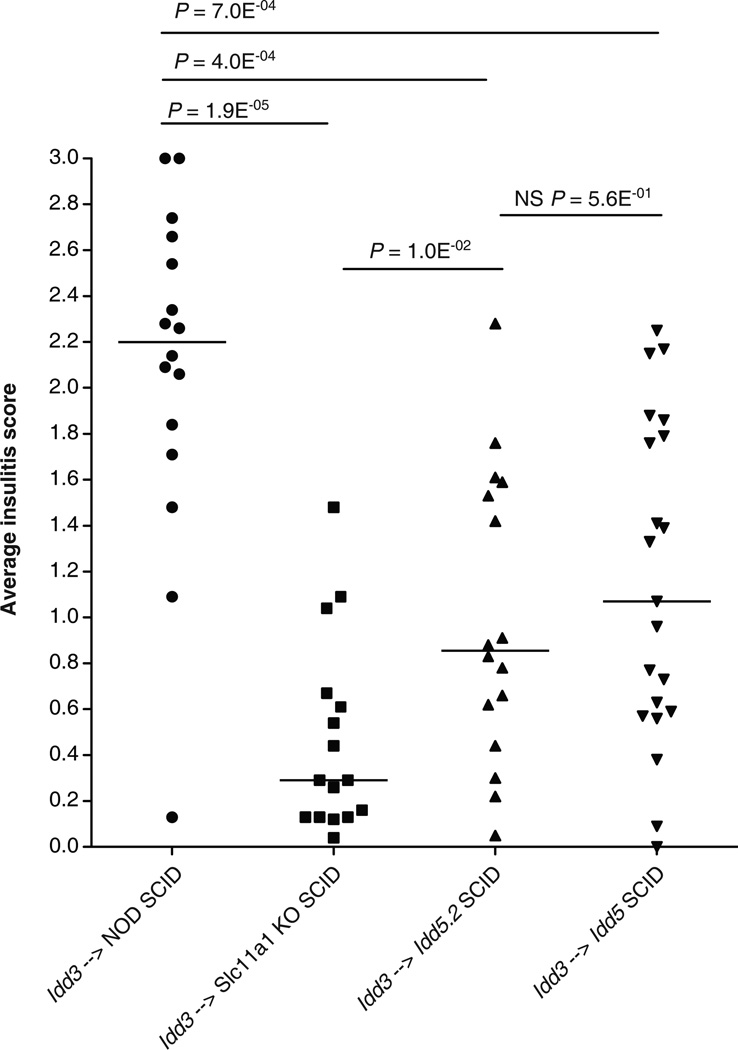
Slc11a1 loss of function alleles expressed in the SCID hosts provides insulitis protection
SLC11A1 is a divalent cation transporter that is exclusively expressed in macrophages and DCs (29, 46). Therefore, we tested if the expression of either the B10-derived Idd5.2 alleles encompassing the gene encoding the loss of function mutant SLC11A1 protein, or the Slc11a1 KO alleles in non-lymphocytes would provide protection against insulitis, by reconstituting NOD-SCID, Idd5-SCID, Idd5.2-SCID, and Slc11a1 KO-SCID with DC-depleted lymphocytes purified from young Idd3 congenic donor mice. The rationale of using Idd3 donor lymphocytes was that the incomplete protection from insulitis provided by the genes in the donor cells allows the detection of insulitis protection conferred by the genes in the host (30). As shown in Fig. 6, the presence of either Idd5 or Idd5.2 in non-lymphocytes conferred significant protection from insulitis, and the degree of insulitis protection provided by Idd5 or Idd5.2 was indistinguishable, suggesting that the insulitis protection conferred by Idd5 should be attributed to Idd5.2, and that Idd5.1 and Idd5.3 alleles were not required. Notably, Slc11a1 KO-SCID mice were also strongly protected from insulitis, compared to NOD-SCID mice (Fig. 6), providing further evidence that Slc11a1 is the causative gene in the Idd5.2 region.
Figure 6.
Idd5.2 or Slc11a1 KO alleles expressed by non-lymphocytes in the host conferred insulitis protection. Female NOD-SCID, Idd5-SCID, Idd5.2-SCID and Slc11a1 KO-SCID mice were reconstituted with DC-depleted spleen and lymph node cells from Idd3 female donor mice. Ten weeks after reconstitution, pancreata were harvested and H&E stained sections were assessed for insulitis. Each diabetic mouse was given an arbitrary insulitis score of ‘3’. Data are pooled from 7 independent experiments. Horizontal lines depict median values. Pairs of strains were compared with the Mann-Whitney U test.
Discussion
The Idd3/5 mouse on the NOD genetic background contains the B6-derived Idd3 region on chromosome 3 and B10-derived Idd5 region on chromosome 1, and it was nearly completely protected from diabetes and insulitis (16, 18). Cordell and colleagues modeled the interlocus genetic interactions of protective alleles at Idd3 and Idd5, and concluded that synergistic gene-gene interactions between two loci were evident (51). The combination of Idd3 and Idd5 protective alleles has also been shown to potently resist autoimmunity in other experimental conditions. For instance, an AIRE knockout crossed onto the B6 genetic background resulted in minimal autoimmunity, on the NOD genetic background, it resulted in very severe multi-organ autoimmunity (60); however, the combination of Idd3 and Idd5 protective alleles was found to abrogate the severe autoimmunity caused by the AIRE knockout on the NOD genetic background (60).
As reviewed in Ridgway et al., a major obstacle in identifying the disease-causing gene variants and their molecular mechanisms acting in the immune system is that many of those candidate genes function in more than one cell types, some of which could have opposing effects during the course of the development of self-tolerance or autoimmunity (3). Yamanouchi et al. provided strong evidence that the reduced IL-2 production by the NOD Idd3/Il2 allele correlates with reduced suppressor function of Tregs (25), and this correlation was confirmed in another study (61). However, IL-2 may well play a pivotal role in not only Tregs, but also DCs (30). It was shown that DCs stimulated with microbial antigens are able to produce IL-2 and prime T cells (29, 62). An unorthodox mechanism by which CD25 molecules expressed on the surface of mature DCs could present IL-2 produced by the DCs to prime the T cells and to enhance the T cell activation was recently described (63).
Functional studies demonstrated that the protective allele of Idd5.1 produces more ligand-independent splice isoform of CTLA-4 (liCTLA-4) than does the disease-associated alleles (32). The liCTLA-4 molecules mediate negative signaling in T cells by dephosphorylating the TCR zeta chain (34) and increased expression of liCTLA-4 correlated with reduced T1D frequency (35). Since Ctla4 is a critical gene expressed exclusively in lymphocytes (17), Idd3 and Idd5.1 could act together in Tregs to enhance their suppressor function.
The most compelling candidate gene within Idd5.2, Slc11a1, encodes a cation transporter expressed on the phago-lyosomal membrane of DCs and macrophages (38–44). There is a known functional missense polymorphism (Gly169) > (Asp169) that distinguishes the disease-associated WT allele and the disease-protective loss-of-function mutant allele (17). Functional studies demonstrated that, compared with the loss-of-function mutant allele, WT Slc11a1 expressed in the DCs enhanced antigen processing and presentation (44, 46), induced more Th1 cytokine production by T cells (46), increased the proliferation of diabetogenic T cells (46), and that in vivo knock-down of WT Slc11a1 provided congenic mice on the NOD genetic background with significant protection from T1D (45). Considering the above discussed effects of IL-2 production by DCs, it is possible that Idd3 and Idd5.2 could act together in DCs to enhance self-tolerance.
The strongest candidate gene within Idd5.3 is Acadl, encoding acyl-coenzyme A dehydrogenase (31), an enzyme that catalyzes the first step of fatty-acid β-oxidation (47). Mice carrying Acadl KO alleles had severe defects in maintaining body temperature (64–66). It was shown that activated T cells require enormous energy to divide and/or carry out their functions, and that fatty acid metabolism occurs preferentially in naïve and memory but not effector T cells (48–50). It was proposed that Idd5.3 may affect the T cell function and survival by altering fatty acid metabolism (31). Therefore, it is possible that Idd3, Idd5.1, and Idd5.3 could interact in Tregs; it is also possible that Idd3, Idd5.2, and Idd5.3 could interact in DCs, considering that ACADL is an enzyme ubiquitously expressed in all cell types.
Of the new bi-congenic strains, only Idd3+5.3 strains were significantly more protected from T1D than the Idd3 strain, suggesting that that Idd5.3 is essential for the interlocus genetic interactions with Idd3 when diabetes phenotype was assessed. Supporting this interpretation, the Idd3+5.1+5.3 strain (18) and the two Idd3+5.2+5.3 strains that we developed in this study were all significantly more protected from T1D than the Idd3 mice. It was surprising that when present alone, among the three Idd5 sub-regions, Idd5.3 is the least protective (18) (Figs. 2A and 2B), but when combined with Idd3, Idd5.3 provides most additional protection from the disease (18) (Figs. 2D and 2E).
Although Idd3+5.1+5.3 females appeared to be protected from T1D to a degree indistinguishable from that of Idd3/5 females, they were found to have significantly more insulitis than Idd3/5 females (18). Therefore, we concluded that the insulitis readout tends to be more sensitive in distinguishing a small difference between two different strains, even when they appeared to be equally protected from T1D. Indeed, we found that each of the three Idd5 sub-regions when combined with the Idd3 strain provided an additional degree of protection from insulitis development, highlighting the requirement of the presence of multiple protective alleles for synergistic genetic interactions. Hence, by assessing both diabetes and insulitis, we concluded that all three Idd5 sub-regions were required to interact with Idd3 to provide the nearly complete protection from diabetes and insulitis as observed in the Idd3/5 strain.
We have found that the expression of Idd3 and Idd5 protective alleles in not only DCs, but also CD4+ T cells, contributes to the self-reactive CD8+ T cell deletional tolerance (29). In order to dissect the cellular basis by which one or more Idd protective alleles contribute to self tolerance, we examined the extent of restoration of CD8+ T cell tolerance in the various strains of Idd3+Idd5-sub-region congenic mice. We found that only the Idd3+5.2+5.3 strains and the Idd3/5 strain had significantly reduced IGRP-specific CD8+ T cells as compared with the Idd3 strain, whereas none of the Idd3+5.1, Idd3+5.2, and Idd3+5.3 strains had significantly less IGRP-specific CD8+ T cells than the Idd3 strain. These observations are similar to the observations that we made when we examined the insulitis of those strains, highlighting the requirement and necessity of having more than one Idd5 sub-region interacting with the Idd3 region to provide significant additional synergistic effects in both situations. Interestingly, the isolated Idd5.2 strain provided equivalent protection from loss of IGRP-specific CD8+ T cell tolerance to the full Idd5 strain. As Idd5.2/Slc11a1 functions in antigen presenting cells, this is likely due to cognate interactions between the CD8+ T cells and the antigen presenting DCs.
Although the development of congenic strains that are protected from T1D due to the presence of a genetic interval derived from a T1D-resistant strain is one of the most important tools in defining genes underlying T1D in the NOD mouse model, a limitation is that the genetic segment containing the allele of interest can not be reduced to a length less than 0.5–3 Mb, depending on the recombination frequency of the region under investigation (3). Therefore, in a gene-dense region such as the Idd5.2 locus that contains at least 45 genes (17), reducing the congenic interval by positional cloning to include only one gene is technically impossible. In order to test the hypothesis that Slc11a1 is Idd5.2, a lentiviral transgenesis and in vivo RNA interference strategy was taken to generate an NOD strain in which Slc11a1 was knocked down by shRNA (45). It was demonstrated that in vivo knock-down of Slc11a1 significantly reduced the T1D incidence (45). However, with the RNA interference experimental approach, it is nearly always impossible to completely eliminate a concern of the possibility of off-target effects (67, 68).
In order to directly test the consequences of lacking Slc11a1 in the NOD genome, in this study we backcrossed the 129-derived Slc11a1 KO allele onto the NOD genetic background, and demonstrated significant protection from T1D due to the lack of Slc11a1. However, in a complex genetic disease such as T1D, not only disease-promoting genes, but also environmental factors, influence the onset and development of the disease (1, 2). In the case of our T1D frequency study of Slc11a1 WT, Slc11a1 KO and the control NOD strains (Fig. 5B), the disease incidence was particularly low compared to our previous studies of other Idd congenic strains (18) (Figs. 2A, 2B and 2C). Noticing that even the NOD females of the control group had a much lower T1D frequency than the same NOD strain in our other T1D frequency studies carried out previously (18) (Figs. 2A, 2B and 2C), we propose that the most likely explanation of the low T1D incidence is the variation of environmental factors.
To further test the hypothesis that Slc11a1 is Idd5.2, we examined the percentage of self-reactive IGRP specific CD8+ T cells in the Slc11a1 WT and Slc11a1 KO strains, and found in young female mice (10–12 weeks of age) there was only a small statistically insignificant difference (Fig. 5C). Since our unpublished data (E.H.W. personal observations) suggest that as the mice age, IGRP specific CD8+ T cells accumulate in various strains of congenic mice on the NOD background, we chose to compare Slc11a1 WT and Slc11a1 KO females at 14–17 weeks of age, and found that Slc11a1 KO has significantly less self-reactive CD8+ T cells than Slc11a1 WT (Fig. 5C), suggesting that Slc11a1 KO allele functions similarly to Idd5.2 disease-resistant allele encoding the SLC11A1 loss-of-function mutant by contributing to a partial restoration of CD8+ T cell tolerance. As illustrated in Figure 5A, the Idd5.2 locus is a gene-dense region encoding at least 45 genes (17), and a large number of SNPs exist in that genetic interval between NOD allele and 129 allele, between NOD allele and B6 allele, between B6 allele and 129 allele. It is plausible, even probable, that one or more SNPs, in addition to the disrupted Slc11a1 gene contribute to the difference in the CD8+ T cell tolerance as measured by the percentage of self-reactive IGRP specific T cells between the Slc11a1 KO and Idd5.2 strains (Figs. 4A and 5C).
Since SLC11A1 is expressed in macrophages and DCs (38–44), we tested whether the lack of functional SLC11A1 expressed in the host would be associated with increased protection from the disease. Although we were unable to successfully discern if the lack of SLC11A1 expression in the host compartment provided protection from diabetes, we found that the 129-derived Slc11a1 KO allele, the Idd5.2/Slc11a1 mutant allele, as well as the Idd5 allele (which contains the Idd5.2 region) in the host provided significant protection from insulitis (Fig. 6), a result that strongly supports our hypothesis that Slc11a1 is Idd5.2.
Our study provides new insights into the synergistic genetic interactions among Idd3/Il2, Idd5.1/Ctla4, Idd5.2/Slc11a1, Idd5.3/Acadl, the first three of which are all associated with human T1D (10–13, 32, 33), emphasizing the common pathogenesis for human and mouse T1D and highlighting the relevance of the NOD and its derived congenic strains for modeling human variants. This study also supports the hypothesis that Slc11a1 is Idd5.2, a finding that highlights SLC11A1 as a molecular target for immune-based therapeutic interventions.
Supplementary Material
Acknowledgements
The authors thank all members of the Sherman laboratory for helpful discussion; Kristi Marquardt for technical assistance; Kathleen Cairns and Michelle Gassert for administrative assistance. We also thank the TSRI histology core facility who prepared histology sections and the TSRI Flow Cytometry core facility for sorting and help with FACS analysis. This is manuscript #22049 from The Scripps Research Institute.
Abbreviations used in this paper
- Acadl
acyl CoA dehydrogenase long chain
- DC
dendritic cell
- Idd
insulin-dependent diabetes
- IGRP
islet-specific glucose-6-phosphatase catalytic subunit related protein
- Slc11a1
solute carrier family 11 member a1
- T1D
type 1 diabetes
- Treg
T regulatory cell.
Footnotes
This work was supported by National Institute of Allergy and Infectious Disease (NIAID) grant AI 070351. X.L. is supported by a graduate student fellowship from The Scripps Research Institute (TSRI) Kellogg School of Science and Technology and by a TSRI Dean’s fellowship. E.H.W. was supported by postdoctoral fellowships from the Juvenile Diabetes Research Foundation (JDRF). L.S.W. is supported by the Wellcome Trust (096388) and the Juvenile Diabetes Research Foundation International (9-2011-253). The Cambridge Institute for Medical Research is the recipient of a Wellcome Trust Strategic Award (100140). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of National Institute of Allergy and Infectious Disease or the National Institutes of Health.
Author Contributions
XL designed the study, performed experiments, analyzed data, prepared figures, and wrote the manuscript. EHW designed the study, performed experiments, analyzed data and revised the manuscript. DBR, KMH, LSW and LBP developed congenic mouse strains, performed experiments, analyzed data, prepared figures and revised the manuscript. JC maintained the murine lines and performed experiments. YDD developed congenic mouse strains and revised the manuscript. LSW and LAS conceived and designed the study, analyzed data, and revised the manuscript.
References
- 1.Kikutani H, Makino S. The murine autoimmune diabetes model: NOD and related strains. Adv. Immunol. 1992;51:285–322. doi: 10.1016/s0065-2776(08)60490-3. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh S, Palmer SM, Rodrigues NR, Cordell HJ, Hearne CM, Cornall RJ, Prins JB, McShane P, Lathrop GM, Peterson LB, et al. Polygenic control of autoimmune diabetes in nonobese diabetic mice. Nat. Genet. 1993;4:404–409. doi: 10.1038/ng0893-404. [DOI] [PubMed] [Google Scholar]
- 3.Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv. Immunol. 2008;100:151–175. doi: 10.1016/S0065-2776(08)00806-7. [DOI] [PubMed] [Google Scholar]
- 4.Maier LM, Wicker LS. Genetic susceptibility to type 1 diabetes. Curr. Opin. Immunol. 2005;17:601–608. doi: 10.1016/j.coi.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 5.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper JD, Howson JM, Smyth D, Walker NM, Stevens H, Yang JH, She JX, Eisenbarth GS, Rewers M, Todd JA, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Pociot F, Rich SS. Confirmation of novel type 1 diabetes risk loci in families. Diabetologia. 2012;55:996–1000. doi: 10.1007/s00125-012-2450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium TWTCC. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicker LS, Clark J, Fraser HI, Garner VE, Gonzalez-Munoz A, Healy B, Howlett S, Hunter K, Rainbow D, Rosa RL, Smink LJ, Todd JA, Peterson LB. Type 1 diabetes genes and pathways shared by humans and NOD mice. J. Autoimmun. 2005;25 Suppl:29–33. doi: 10.1016/j.jaut.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Yang JH, Downes K, Howson JM, Nutland S, Stevens HE, Walker NM, Todd JA. Evidence of association with type 1 diabetes in the SLC11A1 gene region. BMC medical genetics. 2011;12:59. doi: 10.1186/1471-2350-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rainbow DB, Esposito L, Howlett SK, Hunter KM, Todd JA, Peterson LB, Wicker LS. Commonality in the genetic control of Type 1 diabetes in humans and NOD mice: variants of genes in the IL-2 pathway are associated with autoimmune diabetes in both species. Biochem. Soc. Trans. 2008;36:312–315. doi: 10.1042/BST0360312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Wicker LS, Santamaria P. IL-2 and its high-affinity receptor: genetic control of immunoregulation and autoimmunity. Semin. Immunol. 2009;21:363–371. doi: 10.1016/j.smim.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Howson JM, Cooper JD, Smyth DJ, Walker NM, Stevens H, She JX, Eisenbarth GS, Rewers M, Todd JA, Akolkar B, Concannon P, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Pociot F, Rich SS. Evidence of Gene-Gene Interaction and Age-at-Diagnosis Effects in Type 1 Diabetes. Diabetes. 2012 doi: 10.2337/db11-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podolin PL, Denny P, Armitage N, Lord CJ, Hill NJ, Levy ER, Peterson LB, Todd JA, Wicker LS, Lyons PA. Localization of two insulin-dependent diabetes (Idd) genes to the Idd10 region on mouse chromosome 3. Mamm. Genome. 1998;9:283–286. doi: 10.1007/s003359900749. [DOI] [PubMed] [Google Scholar]
- 16.Hill NJ, Lyons PA, Armitage N, Todd JA, Wicker LS, Peterson LB. NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes. 2000;49:1744–1747. doi: 10.2337/diabetes.49.10.1744. [DOI] [PubMed] [Google Scholar]
- 17.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, Howson JM, Smink LJ, Kingsnorth A, Lyons PA, Gregory S, Rogers J, Todd JA, Peterson LB. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J. Immunol. 2004;173:164–173. doi: 10.4049/jimmunol.173.1.164. [DOI] [PubMed] [Google Scholar]
- 18.Hunter K, Rainbow D, Plagnol V, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J. Immunol. 2007;179:8341–8349. doi: 10.4049/jimmunol.179.12.8341. [DOI] [PubMed] [Google Scholar]
- 19.Robles DT, Eisenbarth GS, Dailey NJ, Peterson LB, Wicker LS. Insulin autoantibodies are associated with islet inflammation but not always related to diabetes progression in NOD congenic mice. Diabetes. 2003;52:882–886. doi: 10.2337/diabetes.52.3.882. [DOI] [PubMed] [Google Scholar]
- 20.Wicker LS, Todd JA, Prins JB, Podolin PL, Renjilian RJ, Peterson LB. Resistance alleles at two non-major histocompatibility complex-linked insulin-dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect nonobese diabetic mice from diabetes. J. Exp. Med. 1994;180:1705–1713. doi: 10.1084/jem.180.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny P, Lord CJ, Hill NJ, Goy JV, Levy ER, Podolin PL, Peterson LB, Wicker LS, Todd JA, Lyons PA. Mapping of the IDDM locus Idd3 to a 0.35-cM interval containing the interleukin-2 gene. Diabetes. 1997;46:695–700. doi: 10.2337/diab.46.4.695. [DOI] [PubMed] [Google Scholar]
- 22.Lyons PA, Wicker LS. Localising quantitative trait loci in the NOD mouse model of type 1 diabetes. Curr Dir Autoimmun. 1999;1:208–225. doi: 10.1159/000060488. [DOI] [PubMed] [Google Scholar]
- 23.Lyons PA, Armitage N, Argentina F, Denny P, Hill NJ, Lord CJ, Wilusz MB, Peterson LB, Wicker LS, Todd JA. Congenic mapping of the type 1 diabetes locus, Idd3, to a 780-kb region of mouse chromosome 3: identification of a candidate segment of ancestral DNA by haplotype mapping. Genome Res. 2000;10:446–453. doi: 10.1101/gr.10.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podolin PL, Wilusz MB, Cubbon RM, Pajvani U, Lord CJ, Todd JA, Peterson LB, Wicker LS, Lyons PA. Differential glycosylation of interleukin 2, the molecular basis for the NOD Idd3 type 1 diabetes gene? Cytokine. 2000;12:477–482. doi: 10.1006/cyto.1999.0609. [DOI] [PubMed] [Google Scholar]
- 25.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat. Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuire HM, Vogelzang A, Hill N, Flodstrom-Tullberg M, Sprent J, King C. Loss of parity between IL-2 and IL-21 in the NOD Idd3 locus. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19438–19443. doi: 10.1073/pnas.0903561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu SM, Lee DH, Sullivan JM, Chung D, Jager A, Shum BO, Sarvetnick NE, Anderson AC, Kuchroo VK. Differential IL-21 signaling in APCs leads to disparate Th17 differentiation in diabetes-susceptible NOD and diabetes-resistant NOD Idd3 mice. J Clin. Invest. 2011;121:4303–4310. doi: 10.1172/JCI46187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, Lowe CE, Szeszko JS, Hafler JP, Zeitels L, Yang JH, Vella A, Nutland S, Stevens HE, Schuilenburg H, Coleman G, Maisuria M, Meadows W, Smink LJ, Healy B, Burren OS, Lam AA, Ovington NR, Allen J, Adlem E, Leung HT, Wallace C, Howson JM, Guja C, Ionescu-Tirgoviste C, Simmonds MJ, Heward JM, Gough SC, Dunger DB, Wicker LS, Clayton DG. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton-Williams EE, Martinez X, Clark J, Howlett S, Hunter KM, Rainbow DB, Wen L, Shlomchik MJ, Katz JD, Beilhack GF, Wicker LS, Sherman LA. Expression of diabetes-associated genes by dendritic cells and CD4 T cells drives the loss of tolerance in nonobese diabetic mice. J. Immunol. 2009;183:1533–1541. doi: 10.4049/jimmunol.0900428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamilton-Williams EE, Cheung J, Rainbow DB, Hunter KM, Wicker LS, Sherman LA. Cellular mechanisms of restored beta-cell tolerance mediated by protective alleles of Idd3 and Idd5. Diabetes. 2012;61:166–174. doi: 10.2337/db11-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irie J, Reck B, Wu Y, Wicker LS, Howlett S, Rainbow D, Feingold E, Ridgway WM. Genome-wide microarray expression analysis of CD4+ T Cells from nonobese diabetic congenic mice identifies Cd55 (Daf1) and Acadl as candidate genes for type 1 diabetes. J. Immunol. 2008;180:1071–1079. doi: 10.4049/jimmunol.180.2.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 33.Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA. A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia. 2007;50:741–746. doi: 10.1007/s00125-007-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijayakrishnan L, Slavik JM, Illes Z, Greenwald RJ, Rainbow D, Greve B, Peterson LB, Hafler DA, Freeman GJ, Sharpe AH, Wicker LS, Kuchroo VK. An autoimmune disease-associated CTLA-4 splice variant lacking the B7 binding domain signals negatively in T cells. Immunity. 2004;20:563–575. doi: 10.1016/s1074-7613(04)00110-4. [DOI] [PubMed] [Google Scholar]
- 35.Araki M, Chung D, Liu S, Rainbow DB, Chamberlain G, Garner V, Hunter KM, Vijayakrishnan L, Peterson LB, Oukka M, Sharpe AH, Sobel R, Kuchroo VK, Wicker LS. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J. Immunol. 2009;183:5146–5157. doi: 10.4049/jimmunol.0802610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Runstadler JA, Saila H, Savolainen A, Leirisalo-Repo M, Aho K, Tuomilehto-Wolf E, Tuomilehto J, Seldin MF. Association of SLC11A1 (NRAMP1) with persistent oligoarticular and polyarticular rheumatoid factor-negative juvenile idiopathic arthritis in Finnish patients: haplotype analysis in Finnish families. Arthritis Rheum. 2005;52:247–256. doi: 10.1002/art.20772. [DOI] [PubMed] [Google Scholar]
- 37.Shaw MA, Clayton D, Atkinson SE, Williams H, Miller N, Sibthorpe D, Blackwell JM. Linkage of rheumatoid arthritis to the candidate gene NRAMP1 on 2q35. J. Med. Genet. 1996;33:672–677. doi: 10.1136/jmg.33.8.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 39.Cellier M, Govoni G, Vidal S, Kwan T, Groulx N, Liu J, Sanchez F, Skamene E, Schurr E, Gros P. Human natural resistance-associated macrophage protein: cDNA cloning, chromosomal mapping, genomic organization, and tissue-specific expression. J. Exp. Med. 1994;180:1741–1752. doi: 10.1084/jem.180.5.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malo D, Vogan K, Vidal S, Hu J, Cellier M, Schurr E, Fuks A, Bumstead N, Morgan K, Gros P. Haplotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 41.Blackwell JM. Structure and function of the natural-resistance-associated macrophage protein (Nramp1), a candidate protein for infectious and autoimmune disease susceptibility. Mol. Med. Today. 1996;2:205–211. doi: 10.1016/1357-4310(96)88773-9. [DOI] [PubMed] [Google Scholar]
- 42.Gruenheid S, Pinner E, Desjardins M, Gros P. Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome. J. Exp. Med. 1997;185:717–730. doi: 10.1084/jem.185.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Searle S, Bright NA, Roach TI, Atkinson PG, Barton CH, Meloen RH, Blackwell JM. Localisation of Nramp1 in macrophages: modulation with activation and infection. J. Cell Sci. 1998;111(Pt 19):2855–2866. doi: 10.1242/jcs.111.19.2855. [DOI] [PubMed] [Google Scholar]
- 44.Stober CB, Brode S, White JK, Popoff JF, Blackwell JM. Slc11a1, formerly Nramp1, is expressed in dendritic cells and influences major histocompatibility complex class II expression and antigen-presenting cell function. Infect. Immun. 2007;75:5059–5067. doi: 10.1128/IAI.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat. Genet. 2006;38:479–483. doi: 10.1038/ng1766. [DOI] [PubMed] [Google Scholar]
- 46.Dai YD, Marrero IG, Gros P, Zaghouani H, Wicker LS, Sercarz EE. Slc11a1 enhances the autoimmune diabetogenic T-cell response by altering processing and presentation of pancreatic islet antigens. Diabetes. 2009;58:156–164. doi: 10.2337/db07-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsubara Y, Indo Y, Naito E, Ozasa H, Glassberg R, Vockley J, Ikeda Y, Kraus J, Tanaka K. Molecular cloning and nucleotide sequence of cDNAs encoding the precursors of rat long chain acyl-coenzyme A, short chain acyl-coenzyme A, and isovaleryl-coenzyme A dehydrogenases. Sequence homology of four enzymes of the acyl-CoA dehydrogenase family. J. Biol. Chem. 1989;264:16321–16331. [PubMed] [Google Scholar]
- 48.Fox CJ, Hammerman PS, Thompson CB. Fuel feeds function: energy metabolism and the T-cell response. Nat Rev Immunol. 2005;5:844–852. doi: 10.1038/nri1710. [DOI] [PubMed] [Google Scholar]
- 49.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J. Biol. Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 50.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordell HJ, Todd JA, Hill NJ, Lord CJ, Lyons PA, Peterson LB, Wicker LS, Clayton DG. Statistical modeling of interlocus interactions in a complex disease: rejection of the multiplicative model of epistasis in type 1 diabetes. Genetics. 2001;158:357–367. doi: 10.1093/genetics/158.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J. Exp. Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hulbert EM, Smink LJ, Adlem EC, Allen JE, Burdick DB, Burren OS, Cassen VM, Cavnor CC, Dolman GE, Flamez D, Friery KF, Healy BC, Killcoyne SA, Kutlu B, Schuilenburg H, Walker NM, Mychaleckyj J, Eizirik DL, Wicker LS, Todd JA, Goodman N. T1DBase: integration and presentation of complex data for type 1 diabetes research. Nucleic Acids Res. 2007;35:D742–D746. doi: 10.1093/nar/gkl933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smink LJ, Helton EM, Healy BC, Cavnor CC, Lam AC, Flamez D, Burren OS, Wang Y, Dolman GE, Burdick DB, Everett VH, Glusman G, Laneri D, Rowen L, Schuilenburg H, Walker NM, Mychaleckyj J, Wicker LS, Eizirik DL, Todd JA, Goodman N. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 2005;33:D544–D549. doi: 10.1093/nar/gki095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamilton-Williams EE, Wong SB, Martinez X, Rainbow DB, Hunter KM, Wicker LS, Sherman LA. Idd9.2 and Idd9.3 protective alleles function in CD4+ T-cells and nonlymphoid cells to prevent expansion of pathogenic islet-specific CD8+ T-cells. Diabetes. 2010;59:1478–1486. doi: 10.2337/db09-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trudeau JD, Kelly-Smith C, Verchere CB, Elliott JF, Dutz JP, Finegood DT, Santamaria P, Tan R. Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J. Clin. Invest. 2003;111:217–223. doi: 10.1172/JCI16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinez X, Kreuwel HT, Redmond WL, Trenney R, Hunter K, Rosen H, Sarvetnick N, Wicker LS, Sherman LA. CD8+ T cell tolerance in nonobese diabetic mice is restored by insulin-dependent diabetes resistance alleles. J. Immunol. 2005;175:1677–1685. doi: 10.4049/jimmunol.175.3.1677. [DOI] [PubMed] [Google Scholar]
- 59.Hamilton-Williams EE, Martinez X, Lyman M, Hunter K, Wicker LS, Sherman LA. The use of idd congenic mice to identify checkpoints of peripheral tolerance to islet antigen. Ann. N. Y. Acad. Sci. 2007;1103:118–127. doi: 10.1196/annals.1394.003. [DOI] [PubMed] [Google Scholar]
- 60.Jiang W, Anderson MS, Bronson R, Mathis D, Benoist C. Modifier loci condition autoimmunity provoked by Aire deficiency. J. Exp. Med. 2005;202:805–815. doi: 10.1084/jem.20050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goudy KS, Johnson MC, Garland A, Li C, Samulski RJ, Wang B, Tisch R. Reduced IL-2 expression in NOD mice leads to a temporal increase in CD62Llo FoxP3+ CD4+ T cells with limited suppressor activity. Eur. J. Immunol. 2011;41:1480–1490. doi: 10.1002/eji.201040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Granucci F, Vizzardelli C, Pavelka N, Feau S, Persico M, Virzi E, Rescigno M, Moro G, Ricciardi-Castagnoli P. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nature immunology. 2001;2:882–888. doi: 10.1038/ni0901-882. [DOI] [PubMed] [Google Scholar]
- 63.Wuest SC, Edwan JH, Martin JF, Han S, Perry JS, Cartagena CM, Matsuura E, Maric D, Waldmann TA, Bielekova B. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat. Med. 2011;17:604–609. doi: 10.1038/nm.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guerra C, Koza RA, Walsh K, Kurtz DM, Wood PA, Kozak LP. Abnormal nonshivering thermogenesis in mice with inherited defects of fatty acid oxidation. J. Clin. Invest. 1998;102:1724–1731. doi: 10.1172/JCI4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kurtz DM, Rinaldo P, Rhead WJ, Tian L, Millington DS, Vockley J, Hamm DA, Brix AE, Lindsey JR, Pinkert CA, O'Brien WE, Wood PA. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cox KB, Hamm DA, Millington DS, Matern D, Vockley J, Rinaldo P, Pinkert CA, Rhead WJ, Lindsey JR, Wood PA. Gestational, pathologic and biochemical differences between very long-chain acyl-CoA dehydrogenase deficiency and long-chain acyl-CoA dehydrogenase deficiency in the mouse. Hum. Mol. Genet. 2001;10:2069–2077. doi: 10.1093/hmg/10.19.2069. [DOI] [PubMed] [Google Scholar]
- 67.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Qiu S, Adema CM, Lane T. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005;33:1834–1847. doi: 10.1093/nar/gki324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.