Abstract
Histone acetylation regulates activation and repression of multiple inflammatory genes known to play critical roles in chronic inflammatory diseases. However, proteins responsible for translating the histone acetylation code into an orchestrated pro-inflammatory cytokine response remain poorly characterized. Bromodomain extra terminal (BET) proteins are “readers” of histone acetylation marks with demonstrated roles in gene transcription, but the ability of BET proteins to coordinate the response of inflammatory cytokine genes through translation of histone marks is unknown. We hypothesize that members of the BET family of dual bromodomain-containing transcriptional regulators directly control inflammatory genes. We examined the genetic model of brd2 lo mice, a BET protein hypomorph, to show that Brd2 is essential for pro-inflammatory cytokine production in macrophages. Studies that utilize siRNA knockdown and a small molecule inhibitor of BET protein binding, JQ1, independently demonstrate BET proteins are critical for macrophage inflammatory responses. Furthermore, we show that Brd2 and Brd4 physically associate with the promoters of inflammatory cytokine genes in macrophages. This association is absent in the presence of BET inhibition by JQ1. Finally, we demonstrate that JQ1 ablates cytokine production in vitro and blunts the “cytokine storm” in endotoxemic mice by reducing levels of IL-6 and TNF-α while rescuing mice from LPS-induced death. We propose that targeting BET proteins with small molecule inhibitors will benefit hyper-inflammatory conditions associated with high levels of cytokine production.
Introduction
Regulation of inflammatory gene expression is tightly controlled through chromatin “readers” that specifically bind histone post-translational modifications and provide a scaffold which, in addition to sequence-specific transcription factors, is an integral component of the transcriptional activation complex (1). The biological effects of chromatin-dependent, multi-protein complexes include both transcriptional co-activation and co-repression of inflammatory genes in differentiated adult cells (2), thus chromatin readers play critical roles in exquisitely tuned inflammatory responses to a variety of immune system stimuli.
Pro-inflammatory stimuli such as bacterial endotoxin (LPS) arouse extensive transcriptional reprogramming through their ability to activate acetylation of ε-amino groups of nucleosomal histone lysines, a general mark of gene activation (3–5). The acetylated lysines residues are recognized by chromatin readers, many of which contain a conserved structure designated the bromodomain. Bromodomains are highly conserved, left-twisted bundles of four-α-helices, with a hydrophobic cleft between two conserved loops that connect the helices (6). The motif uses hydrogen bonding, often at asparagine residues, to bind to acetylated histones (7). In humans, there are at least forty bromodomain proteins (8, 9), which include histone acetyltransferases (HATs), helicases, scaffolding proteins and other co-factors that control gene transcription. These findings raise the possibility that bromodomain proteins regulate acetylated, histone-packaged inflammatory genes through multiple downstream mechanisms to significantly contribute to outcomes from pro-inflammatory stimuli.
The bromodomain and extra-terminal (BET) family is distinct group of bromodomain proteins that in mammals includes Brd2, Brd3, Brd4, all of which are ubiquitously expressed in mammalian tissues (10–13). Brd2 and Brd4 have been extensively studied in the context of cell cycle control (14–18) and transcription elongation (19–21), but potential roles in inflammatory responses have been explored poorly. Establishing links between BET proteins and inflammation has become clinically critical due in part to recent drug development efforts, which have shown that drugs able to interrupt interactions between Brd4 and thienodiazepines (22) have efficacy in BET-protein related cancers (23, 24). JQ1 was the first drug developed that specifically interacts with the hydrophobic pocket of the BET bromodomain to block interaction between multiple BET proteins (Brd2/3/4) and acetylated histones (9). JQ1 effectively prompted squamous differentiation and reduced tumor volume of Brd4-dependent, human NUT midline carcinoma (NMC) xenografts in mice (9), and was proven efficient to block growth of various leukemic cells (25–27). However, other possible applications of JQ1, including those targeting BET protein functions in non-malignant cells, are untested (Reviewed in 28). Establishing the importance of BET proteins in inflammation is a first critical step toward evaluating the possibility that JQ1 may be exploited as a next-generation anti-inflammatory treatment.
Considering the involvement of BET proteins in control of the most fundamental cell growth and proliferation processes, it is not surprising that multiple attempts to create mouse strains of BET genes knockouts have not succeeded. Both Brd2 and Brd4 knockouts have early lethality phenotypes (13, 29–31). We have generated a mouse model with gene disruption of Brd2 that exhibits a hypomorph phenotype with expression of about half the wild-type level of Brd2 in all tissues tested. These “brd2 lo” hypomorphs develop a complex whole-body phenotype, the key feature of which is severe obesity without insulin resistance (IR) (13). Given that inflammation from the macrophage compartment is critical for obesity-associated IR (32, 33), these data predicted that appropriate levels of BET protein expression in macrophages may be a required component of inflammation in obesity, amongst other inflammatory diseases (Reviewed in 33).
To establish rigorously the link between BET protein function and inflammation, we investigated the inflammatory response of macrophages derived from brd2 lo mice in detail and showed that low Brd2 levels severely blunt pro-inflammatory cytokine production. Complementary studies that test BET protein knockdown and the BET inhibitor JQ1 in vivo demonstrate that BET proteins play important roles in acute inflammatory responses. Finally, we show that the functions of BET proteins in inflammation are regulated by direct contact with the promoter chromatin of a select subset of cytokine genes. Taken together, these studies establish a role for BET proteins in mouse macrophage stimulation and justify further testing of BET protein-targeting drugs in chronic inflammatory diseases.
Materials and Methods
Mice
C57BL/6J (The Jackson Laboratory) and BALB/cJ (Charles River) males, 6–8 wk of age, were maintained in our specific pathogen-free animal facility according to institutional guidelines, with protocols approved by the Institutional Animal Care and Use Committee of Boston University School of Medicine. brd2 lo mice were generated as previously reported (13). NF-κBEGFP mice (34) were obtained from Christian Jobin, University of North Carolina, Chapel Hill, North Carolina, USA. In these mice, a single copy of eGFP reporter was placed under control of cis-NF-κBEGFP and was inserted 5′ of the X-linked hypoxanthine phosphoribosyltransferase locus. Male hemizygous mice were used for the experiments. Sacrifice was by isoflurane narcosis followed by cervical dislocation in accordance with recommendations of the American Veterinary Association.
Reagents
Dulbecco’s Minimum Essential Medium (DMEM) plus glutamine and 25mM glucose was from Mediatech. Penicillin, streptomycin, and fungizone were purchased from Gibco. LPS from E. coli clone 0111:B4 was purchased from Sigma-Aldrich. JQ1 (+) and JQ1(−) were a generous gift from James Bradner (Dana Farber Cancer Institute). For in vitro assays, we prepared 10 mM stock solutions of JQ1 in DMSO that were diluted in PBS before addition to cell culture medium. For in vivo studies, 50 mg/ml JQ1 stock in DMSO was diluted 1:10 with 10% (w:v) solution of 2-hydroxypropyl-β-cyclodextrin (Sigma-Aldrich) immediately prior to administration.
Cell isolation from spleen and bone marrow for flow cytometry
Splenocytes were isolated from freshly harvested, minced spleens of brd2 lo mice or littermate controls by gently pressing the spleens through a 70 µm cell strainer (BD Biosciences), followed by treatment with 1X red blood cell lysis buffer (eBioscience). Bone marrow (BM) cells were isolated as described below. Cells were counted and stained with directly conjugated monoclonal antibodies against CD3, CD4, CD8, B220, CD11b and Gr-1 in the presence of anti-mouse CD16/CD32 from eBioscience. All stains were performed along with isotype controls. Flow cytometry was performed on an LSRII cytometer (BD) and data were analyzed from 50,000 – 100,000 gated events using FlowJo 8.7 (TreeStar).
Isolation, culture and stimulation of bone marrow-derived macrophages
Femurs, tibiae and sterna were isolated under sterile conditions. Bones were crushed in a sterile glass Dounce homogenizer to liberate BM cells into a suspension of RPMI1640 medium buffered with 20 mM HEPES pH 7.4 and supplemented with 10% fetal bovine serum, penicillin, streptomycin, fungizone and 50 µM 2-mercaptoethanol. Bone fragments were removed by sterile filtration with 70 µm cell strainers (BD) and erythrocytes were lysed as above. Nucleated BM cells were recovered and plated overnight in 75 ml flasks to allow resident macrophages to attach, and then suspension cells were plated in DMEM medium supplemented as above. To initiate differentiation, the medium was supplemented with 50 ng/ml of recombinant M-CSF (eBioscience) for 5–7 days. Equal numbers of macrophages were seeded into 6-, 12-, or 24-well plates prior to stimulation with E. coli LPS 0111:B4 (50 ng/ml; Sigma). IFNγ priming was performed with 100 u/ml IFNγ (eBioscience). At indicated time points, cell-free supernatants were removed for multiplex protein/cytokine analysis, and/or RNA was isolated from adherent cells.
NF-κB luciferase reporter assay
RAW264.7 cells were transfected using Lipofectamine 2000 (Invitrogen) and components of the Cignal NF-κB Luc Reporter kit (SA Biosciences) plus either pSiBrd2 (13) or control vectors. Cells were harvested 48 h after transfection and luciferase activity was measured using a Dual Luciferase Reporter Assay System (Promega) on a TD-20/20 luminometer (Turner Biosystems).
Protein assays
The supernatants from bone marrow-derived macrophage (BMDM) or RAW264.7 cultures, or serum, were collected and frozen in aliquots. Cytokines were determined using Singleplex reagent kits (Invitrogen/Life Sciences) according to the manufacturer's instructions and a Bio-Plex 200 (Bio-Rad) reader. Serum was diluted with Singleplex Assay diluent buffer prior to analysis or was analyzed for serum amyloid A (SAA) by ELISA (Immunology Consultants Laboratory). For TNF-α intracellular staining, we used a fixation/permeabilization buffer kit (eBioscience). RAW264.7 cells were collected from the plates in trypsin-free ice-cold PBS supplemented with 2 mM EDTA. Cells were washed, fixed and permeabilized according to the manufacturer’s protocol, then stained with allophycocyanin (APC)-anti-TNF-α (eBioscience). Labeled cells were analyzed by flow cytometry as described above.
siRNA knockdown in BMDMs
Twenty nM Dharmacon ON-TARGETplus SMARTpool siRNA with DharmaFECT formulation #4 reagent (Dharmacon) was used for Brd2, Brd3 and Brd4 knockdown. Cells were used for experiments 48 hrs post-transfection. Knockdown was validated by RT-qPCR.
RT-qPCR
RNA from macrophages and RAW264.7 cells was isolated and quantified as published using 7500 Fast Real-Time PCR System (Applied Biosystems), Power SYBR Green PCR Master Mix (Applied Biosystems) and Quantitech primers for mouse Brd2, Brd3, Brd4, TNF-α, IL-6, MCP-1, eGFP and GAPDH (Qiagen). For whole blood, 50–100 µl of mouse blood was obtain from the tail vein and RNA was purified with Mouse RiboPure Blood RNA kit (Ambion). For 28S rRNA amplification, the following primers were used: 5’-GCGAAATACCGGCACGAGACCGATAG-3’ and 5’-GGTTTCACGCCCTCTTGAACTCTCTC-3’. Melt curve analysis indicated formation of a single product in all cases. Relative mRNA expression levels were determined using ΔCt values and were normalized to GAPDH or 28S rRNA levels as indicated.
Chromatin immunoprecipitation
One hour after LPS stimulation and/or JQ1 treatment, BMDMs were fixed in 1% formaldehyde at 37 °C for 10 min, then subjected to chromatin immunoprecipitation (ChIP) as previously published (35). Chromatin was precipitated with 2 µg of α-acetylated histone H3 (Upstate), α-Brd2 (Bethyl Labs), α-Brd4 (Bethyl Labs) or α-GST (Upstate). Two nanograms of each sample were then analyzed in duplicate or triplicate by qPCR. Fold difference was calculated as 2^(Ct(input)-Ct(ChIP)), then fold enrichment over an unrelated antibody (α-GST) was assessed. Oligonucleotides were: IL-6: 5′-TGTGGGATTTTCCCATGAGT-3′ and 5′-TGCCTTCACTTACTTGCAGAGA-3′; TNF-α: 5′-AGCGAGGACAGCAAGGGA-3′ and 5′-TCTTTTCTGGAGGGAGTGTGG-3′.
LPS-induced shock in mice
Weight-matched 8-week-old male mice were injected i.p. with 50 mg/kg of JQ1 (+) or JQ1(−). Two hours later, 20 mg/kg of E. coli LPS was injected into the contralateral side of the abdomen. For survival experiments, mice were monitored for mortality. Serum cytokines were measured in tail vein sera just prior to LPS injection, then 20, 40, 60, 90, 120 and 240 min after LPS injection.
Statistical Analysis
For comparison of treatment group differences, we used the unpaired, two-tailed Student t test or one-factor ANOVA in conjunction with the Dunnett’s or Tukey-Kramer multiple comparisons tests. Error bars indicate standard errors of the mean (SEM), with significant differences (P <0.05) indicated. Analyses were performed in Graphpad Prism 5 or, for survival curves, Kaplan-Meier analysis was performed in SigmaStat 3.1 software.
Results
Decreased Brd2 levels correspond to decreased macrophage inflammatory responses
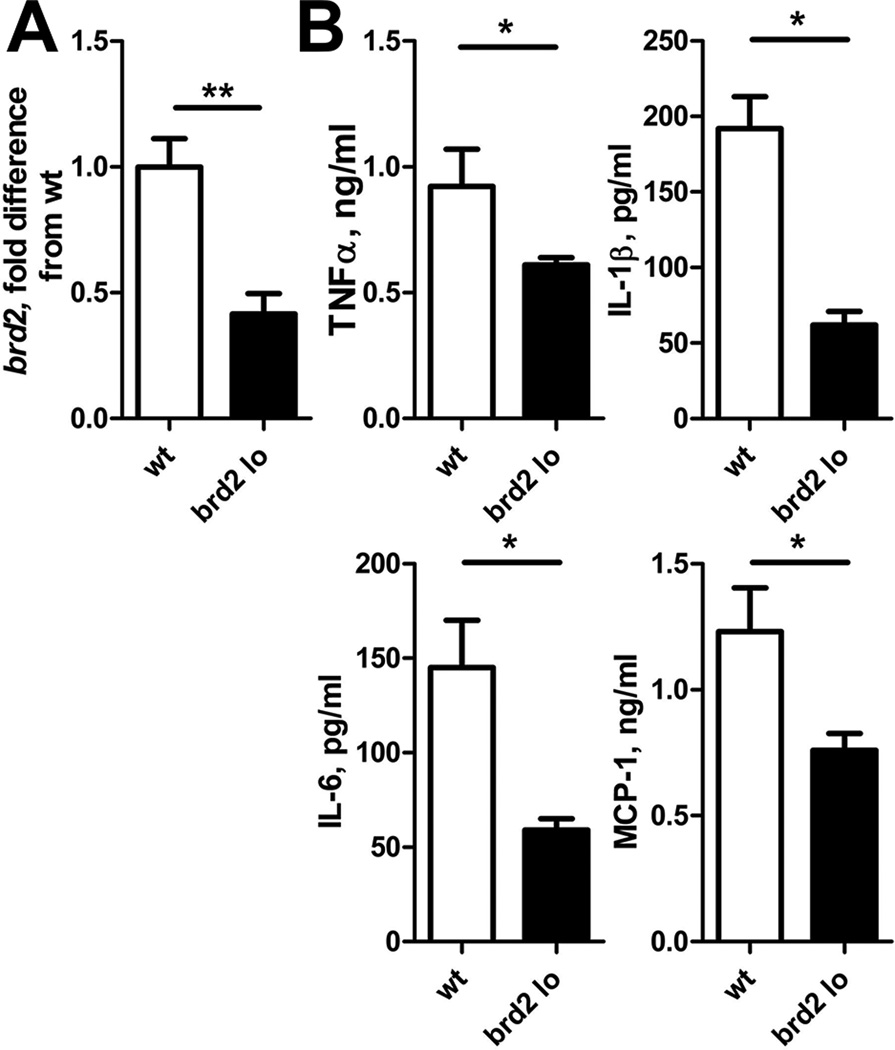
Our previous demonstration that brd2 lo mice are protected from obesity-induced inflammation (13) predicted that Brd2 regulates inflammation in response to a broad array of pro-inflammatory stimuli. To begin testing this prediction, we first asked whether Brd2 hypomorphs (confirmed in Fig. 1A) resist obesity-associated inflammation due to constitutive changes in numbers and/or ratios of immune cells, a possibility consistent with our demonstration that Brd2 shRNA expression in hematopoietic stem cells decreases their contribution to total bone marrow (Belkina et al, in preparation). Table I shows similar percentages of B cells, T cells and macrophages in spleen and BM of brd2 lo and WT mice. Absolute numbers of splenocytes and BM cells from brd2 lo were also similar compared to WT (not shown). To test the alternative possibility that Brd2 regulates pro-inflammatory responses rather than in vivo cell distribution, we stimulated bone marrow derived macrophages (BMDMs) from brd2 lo mice with LPS and quantified production of inflammatory cytokines. brd2 lo BMDMs produced lower levels of pro-inflammatory TNF-α, IL-1β, IL-6 and MCP-1/CCL2, compared to WT macrophages (Fig. 1B). Importantly, the number of CD11b+ BMDMs differentiated from BM of brd2 lo mice was similar to BM of WT mice, consistent with lower cytokine production by brd2 lo macrophages on a per cell basis (data not shown), and in agreement with a lack of developmental defects as suggested by the fresh ex vivo immunophenotyping data (Table I). We conclude that brd2 lo macrophages have a cell intrinsic impairment in pro-inflammatory responses.
Figure 1. brd2 lo macrophages demonstrate a diminished inflammatory response.
A. Brd2 mRNA levels in leukocytes from WT (white bar) compared to brd2 lo (black bar) mice as measured by RT-qPCR. Relative mRNA expression was normalized to GAPDH. N = 4; **, P < 0.01, as calculated by Student's t test. B. Production of pro-inflammatory cytokines by brd2 lo and WT BMDMs stimulated for 24 hours with 50 ng/ml E. coli LPS. All bars in all figures show mean values and SEM. N = 4; *, P < 0.05, as calculated by Student's t test.
Table I.
Major immune cells subsets in brd2 lo mice spleen and BM.*
| wt | brd2 lo | ||
|---|---|---|---|
| Spleen | |||
| B220+ | 52.7 ± 2.1 | 50.4 ± 1.1 | |
| CD3+ | 31.1 ± 1.9 | 29.9 ± 0.9 | |
| CD3+CD4+CD8 (of CD3+ gate) | 44.7 ± 2.2 | 40.8 ± 3.3 | |
| CD3+CD4-CD8+ (of CD3+ gate) | 26.5 ± 1.4 | 28.7 ± 2.4 | |
| CD11b+ | 9.3 ± 0.9 | 8.8 ± 1.4 | |
| CD11b+ low (of CD11b+ gate) | 77.6 ± 2.5 | 80.6 ± 3.1 | |
| CD11b+ high (of CD11b+ gate) | 6.7 ± 3.6 | 4.7 ± 0.6 | |
| Bone marrow | |||
| B220+ | 23.6 ± 4.9 | 22.0 ± 2.4 | |
| CD11b+ | 29.1 ± 2.5 | 29.9 ± 2.8 | |
| Gr-1+ | 33.3 ± 3.6 | 33.7 ± 1.5 | |
Major immune cells subsets in spleen and BM of brd2 lo mice, compared to WT. Shown are mean percentages of the live cell gate or parent gate ± SEM. N = 4.
BET knockdown ablates cytokine expression in BMDMs
To verify independently a role for Brd2 in primary macrophage inflammation absent any remaining concern about differentiation defects in brd2 lo cells, we tested the effect of Brd2 knockdown on cytokine responses of WT BMDMs. We transfected WT BMDMs with Brd2-specific siRNA and confirmed gene knockdown by qPCR 48 hrs post-transfection. Importantly, down regulation of any single BET protein did not alter the levels of expression of the other two members of the BET family (Fig. S1). We then stimulated knockdown BMDMs with LPS and quantified inflammatory cytokine mRNA by RT-qPCR. TNF-α, IL-6 and MCP-1 mRNA levels were significantly lower in cells transfected with the siBrd2 pool compared to the non-targeting siRNA pool control (Fig. 2A). Taken together with decreased inflammatory cytokine production by brd2 lo macrophages, these data indicate that Brd2 promotes pro-inflammatory cytokine production in macrophages. Functional significance of BET protein knockdown was evidenced by decreased TNF-α, IL-6 and MCP-1 protein levels in Brd2, Brd3 and Brd4 knockdown macrophages (Fig. 2B). Interestingly, the effects of BET protein ablation were somewhat selective, as evidenced by failure of BET knockdown to affect protein levels of keratinocyte chemoattractant (KC), a mouse ortholog of IL-8 and a potent pro-inflammatory chemokine (36) (Fig. 2B-you need to mention the result here). We conclude that Brd2, Brd3 and Brd4 have non-redundant and selective roles in the macrophage inflammatory response through their ability to regulate cytokine production.
Figure 2. In vitro knockdown of BET protein genes reduces the inflammatory response.
A. WT BMDMs were transfected with ON-TARGET Plus siRNA pools specific for knockdown of Brd2, Brd3 or Brd4, or non-targeting control pools, then stimulated with 50 ng/ml E. coli LPS for 2 hrs before quantitation of the indicated cytokine mRNAs by RT-qPCR. Relative mRNA expression was normalized to GAPDH. N = 5; **, P < 0.01, ***, P < 0.001 as calculated by ANOVA with Dunnett’s multiple comparison. B. Cytokine protein concentrations in supernatants from LPS-stimulated (24 hours, 50 ng/ml) BMDMs after individual BET knockdown. N = 5; *. P < 0.05; **, P < 0.01; ***, P < 0.001 as calculated by ANOVA with Dunnett’s multiple comparison.
Brd2 regulates NF-κB activity
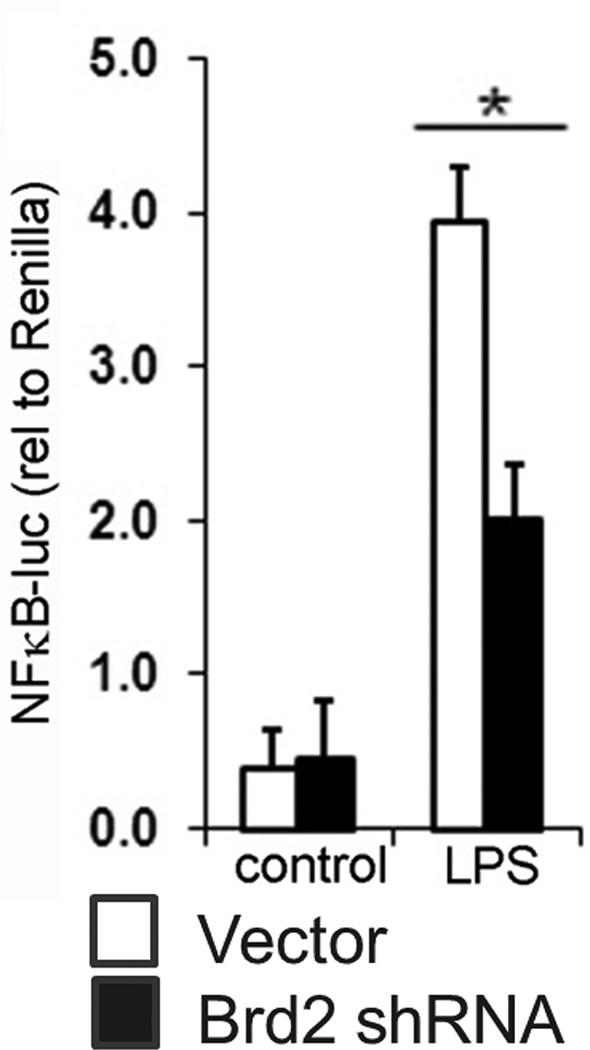
Given that all of the cytokines affected by decreased Brd2 levels are targets of NF-κB, a master pro-inflammatory transcription factor, we tested the possibility that Brd2 regulates cytokine production by altering NF-κB activity. We modeled brd2 lo macrophages in vitro with siRNA-mediated Brd2 knockdown in a RAW264.7 macrophage cell line. We then transiently transfected Brd2 knockdown macrophages with an NF-κB luciferase reporter gene, and stimulated cells with LPS. NF-κB reporter activity was significantly decreased in Brd2 knockdown cells (Fig. 3), supporting the conclusion that Brd2 regulates pro-inflammatory cytokines through its ability to support NF-κB-regulated transcription.
Figure 3. In vitro knockdown of Brd2 decreases NF-κB-luciferase activity.
RAW264.7 cells were transfected with Brd2-specific shRNA (black bars) or empty vector (white bars) plus NF-κB luciferase reporter plasmid. Forty-eight hours after transfection, cells were stimulated with E. coli LPS for 2 hrs. NF-κB-driven luciferase activity is shown (mean and SEM). N = 3. *, P < 0.05 as calculated by Student's t test comparing empty vector and Brd2 shRNA-containing plasmids.
BET inhibitor JQ1 ablates inflammatory responses in LPS-stimulated BMDMs
Given that down regulation of multiple individual BET proteins is anti-inflammatory (Fig. 2), we tested the efficacy of JQ1, a broad-spectrum BET protein inhibitor, as an anti-inflammatory therapeutic drug that could counter macrophage-mediated inflammation. We exposed RAW264.7 macrophage-like cells to LPS and concomitantly treated them with either JQ1(+), the biologically active enantiomer, or JQ1(−), the inactive enantiomer. Levels of secreted TNF-α and MCP-1 were dramatically lower in JQ1(+) treated RAW 264.7 macrophages compared to JQ1(−) treated macrophages (Fig. 4A; >50% and >80% decrease, respectively). Furthermore, IL-6 production was 90% lower in the presence of JQ1(+) (Fig. 4A). Protein data were consistent with mRNA analyses. For example, TNF-α mRNA was reduced to less than 25% of the levels produced by cells treated with JQ1(−) (Fig. S2A). Intracellular TNF-α stain independently confirmed lower protein levels in JQ1(+)-treated cells (Fig. S2B). Quantitatively similar dose-dependent decreases in TNF-α, IL-6 and MCP-1 were produced by JQ1(+) treated primary BMDMs (Fig. 4B), independently confirming the BET protein inhibitor JQ1 had relatively broad anti-inflammatory effects on macrophages. KC was produced at similar concentrations by LPS- stimulated BMDMs in the presence of JQ1(+) and JQ1(−) (Fig. 4B). This observation is in concordance with the demonstration that KC levels are unaffected by knockdown of BET proteins with siRNA (Fig. 2B), and may be due to the fact that LPS-induced elevation of KC transcription depends on the TLR4-AP1 axis rather than NF-κB (37).
Figure 4. The BET family bromodomain inhibitor JQ1 reduces cytokine production in vitro.
A. RAW 264.7 cells were exposed simultaneously to 50 ng/ml E. coli LPS plus 400 nM JQ1(+) or JQ1(−) for 24 hrs. Mean value and SEM for 3 determinations are shown. JQ1 enantiomers are abbreviated by (+) or (−), respectively. *, P < 0.05, ***, P < 0.001 as calculated by Student's t test comparing JQ1(+) and JQ1(−) treated samples. The chemokine KC was not detectable in RAW 264.1 macrophages under these conditions. B. BMDMs were exposed simultaneously to 50 ng/ml E. coli LPS or PBS and different concentrations of JQ1(+) or JQ1(−) for 24 hrs. Supernatant cytokines were quantified in multiplex protein assays. N = 6; ***, P < 0.001 as calculated by ANOVA with Dunnett’s multiple comparison. ND – not detected.
Anti-inflammatory effect of JQ1 is not strain-specific and cannot be overcome by IFNγ priming
Genetic background is known to influence the patterns of innate immune responses in mice. In particular, C57BL/6 mice are often referred to as “M1” strain, while Balb/c mice tend to “M2” type of response (38, 39). To test whether the anti-inflammatory effects of JQ1 are influence by genetic background, we exposed primary macrophages derived from C57BL/6 or Balb/c bone marrow to JQ1(+) and stimulated the cells with LPS. Overall, in Balb/c BMDMs the effect of JQ1 (+) was very similar to what we observed in C57BL/6, and included blockade of TNFα, IL-6, and MCP-1 (but not KC; Fig. 5A). Furthermore, IL-10, produced at detectable levels by macrophages from Balb/c but not C57BL/6 mice, was inhibited by JQ1(+) treatment (Fig. 5A). These data indicate that the anti-inflammatory effect of JQ1(+) is independent of IL-10 up regulation. Overall, we conclude the anti-inflammatory effect of JQ1(+) is not genotype-specific.
Figure 5. JQ1 effect on cytokine production is strain-independent and is not overcome by IFNγ priming of macrophages.
A. Open bars show C57Bl/6 and BALB/c BMDMs exposed to PBS, 100 U/ml IFNγ, 50 ng/ml E. coli LPS, 50 ng/ml E. coli LPS with 400 nM JQ1(+), or primed with 100 U/ml IFNγ and stimulated with LPS 4 hours later as indicated below graph. Black bars show BMDMs primed with IFNγ or PBS and stimulated with LPS 4 hours later, with JQ1(+) being added either simultaneously with LPS (4 hours after priming with IFNγ) or simultaneously with IFNγ. Cytokine production was measured 24 hours after LPS stimulation. Supernatant cytokines were quantified in multiplex protein assays. N = 5; ***, P < 0.001; **, P < 0.01; *, P < 0.05 as calculated by ANOVA with Dunnett’s multiple comparison. #, P < 0.05 as calculated by Student's t test; ND – not detected. B. eGFP NF-κB BMDMs were stimulated were stimulated with PBS, 50 ng/ml LPS or LPS + JQ1(+) as indicated. N = 3 **, P < 0.01; *, P < 0.05 as calculated by ANOVA with Dunnett’s multiple comparison.
IFNγ priming is a first event in the classical cascade of macrophage activation. The effect of the subsequent trigger stimulus, like LPS, bacterial DNA, or TNFα, is vastly amplified and expanded by IFNγ priming (40, 41). To test whether IFNγ priming overcomes the effect of JQ1(+) on macrophages, we exposed cells to IFNγ prior to LPS stimulation. As previously reported, the effect of IFNγ priming on cytokine production is modest in this model (40). We found that pre-treatment with IFNγ does not prevent the inhibition of cytokine production by subsequent exposure to JQ1(+) and LPS. Interestingly, addition of JQ1(+) simultaneously with IFNγ significantly potentiated inhibition of TNFα, IL-6 and MCP-1 production in BALB/c macrophages (Fig. 5A). As expected from previous results, KC production was not robustly inhibited.
JQ1 treatment disrupts association of Brd2 and Brd4 with the regulatory chromatin of pro-inflammatory cytokine promoters
We have demonstrated that Brd2 regulates NF-κB activity outside the context of naturally assembled chromatin (Fig. 3). To assess the effect of blocking BET protein function on NF-κB activity within chromosomal DNA, we generated BMDMs from male NF-κBEGFP knockin mice (34) and stimulated cells with LPS in presence of JQ1(+) or JQ1(−). Like other myeloid immune cells originating from these mice (34), the BMDMs demonstrated high background eGFP fluorescence even when not stimulated with LPS, probably mirroring the basal NF-κB activation. We therefore quantified LPS response by eGFP mRNA. eGFP mRNA increased approximately 5-fold in response to LPS stimulation, and was significantly reduced when macrophages were simultaneously exposed to LPS and JQ1(+) (Fig. 5B).
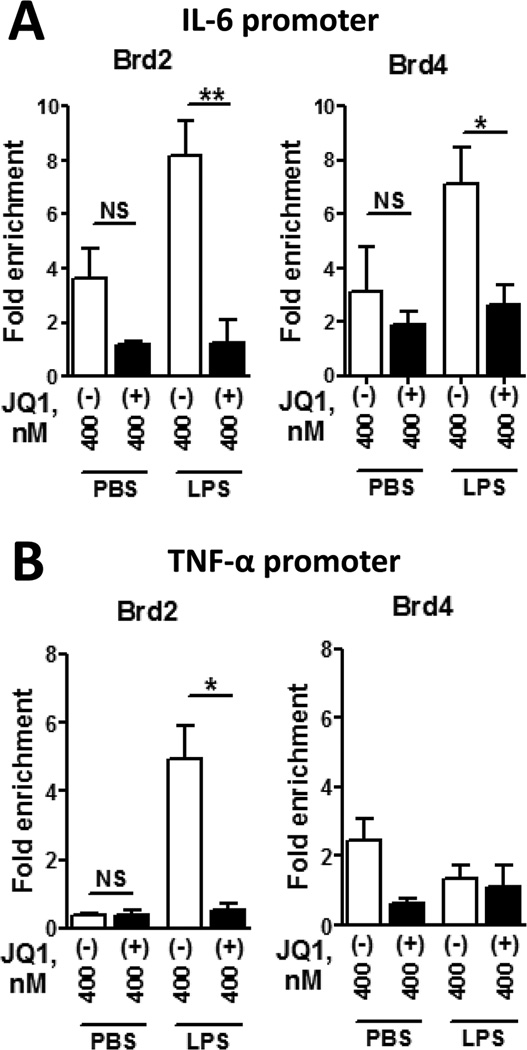
Findings thus far agree with the current understanding that BET proteins regulate acetylated histone-packaged genes through physical association with modified histones, the molecular target of JQ1(+)(9, 42). Because active cytokine promoters are packaged by acetylated histone proteins, we reasoned that JQ1(+) functions as an anti-inflammatory compound by altering BET association with acetylated histone-packaged cytokine promoters (4, 5). To test this possibility, we quantified BET protein/cytokine promoter interactions by ChIP. For these studies, we used anti-Brd2 or anti-Brd4 antibodies to precipitate sheared chromosomal DNA from BMDMs stimulated with LPS in the presence of either JQ1(+) or JQ1(−). Both Brd2 and Brd4 modestly bound the IL-6 promoter in the non-stimulated cells (Fig. 6A); however, when cells were activated with LPS, we observed significant amounts of IL-6 promoter DNA precipitated with anti-Brd2 and anti-Brd4 antibodies (Fig. 6A). Importantly, JQ1(+), but not JQ1(−), treatment prevented association of both Brd2 and Brd4 with the IL-6 promoter. An alternative, but functionally equivalent, interpretation is that JQ1(+) promoted dissociation of BET proteins from the IL-6 promoter. These data show the LPS-inducible association of BET proteins with the IL-6 promoter is significantly decreased in the presence of JQ1(+), which explains the decreased production of pro-inflammatory cytokines in macrophages.
Figure 6. JQ1(+) displaces BET proteins Brd2 and Brd4 from inflammatory cytokine promoters.
Mouse BMDMs were treated with PBS or 50 ng/ml E. coli LPS in presence of either 400 mM JQ1(−) or 400 nM JQ1(+) as indicated, then harvested for ChIP 1 hour later. A. Brd2 or Brd4 association with the IL-6 promoter. B. Brd2 or Brd4 association with the TNF-α promoter. Mean values and SEM are shown; N=3 for all datapoints except for Brd4-precipitated chromatin treated with PBS+JQ1(−), where N=2. *, P < 0.05; **, P < 0.01 as calculated by Student's t test comparing JQ1(+) and JQ1(−) results.
To measure the effect of JQ1(+) on BET protein/promoter interactions more universally, we re-amplified ChIP samples from Fig. 6A to quantify Brd2 and Brd4 association with the TNF-α promoter. As expected, LPS induced Brd2 association with the TNF-α promoter in the absence of JQ1(+) (Fig. 6B, left). Furthermore, Brd2 was not associated with the TNF-α promoter in LPS/JQ1(+) treated cells. Interestingly, we did not find significant Brd4/TNF-α promoter association in BMDMs under any of the conditions tested (Fig. 6B, right). This negative result may reflect bona fide differences between the molecular mechanisms that regulate IL-6 and TNF-α. Alternatively, the antibody-binding Brd4 epitope may be masked by other transcriptional proteins present only on the TNF-α promoter.
BET protein inhibition with JQ1 protects mice from an LPS-induced cytokine storm and death
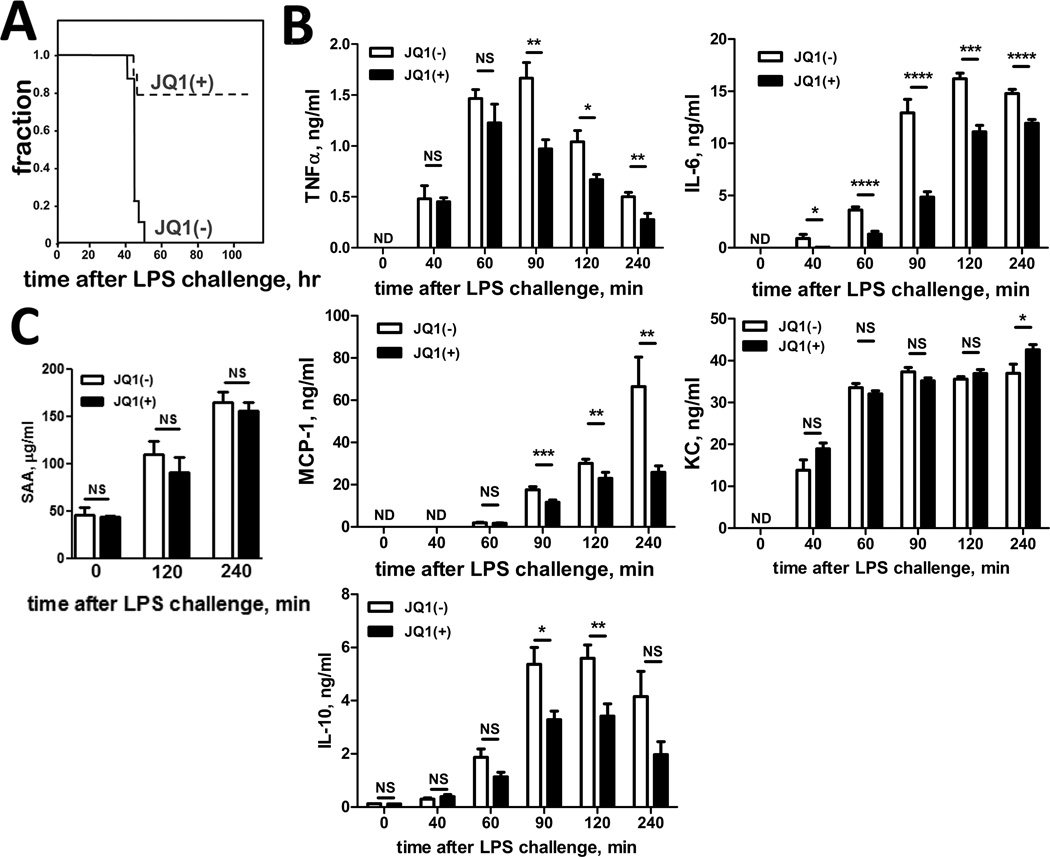
To test the efficacy of JQ1 to ablate macrophage cytokine production in vivo, we asked whether JQ1 administration could ameliorate the LPS-induced ‘cytokine storm’, a severe, systemic production of inflammatory cytokines dominated by macrophages (and their monocyte precursors) in response to endotoxemic shock (43). We administered 50 mg/kg of JQ1(+) or JQ1(−) (a dose well-tolerated in daily chemotherapy regimens (9)) 2 hours before challenging the mice with a lethal intraperitoneal dose of E. coli LPS (20 mg/kg). Although both JQ1(+)- and JQ1(−)-pretreated mice demonstrated clinical signs of endotoxemia (reduced movement, hunched posture, diarrhea), JQ1(+) treatment dramatically improved survival: by 48 hours post-LPS, 100% of JQ1(−) mice were dead, while more than 80% of JQ1(+) mice survived with almost complete resolution of clinical symptoms (Fig. 7A). Furthermore, all JQ1 mice that survived for 48 hrs post-LPS recovered completely, as documented ≥ 14 days following LPS challenge,.
Figure 7. JQ1 protects mice from LPS-induced death.
Mice were injected with a lethal dose of E. coli LPS (20 mg/kg, i.p.). At 2 h before and 24 h after LPS injection, mice also received JQ1 (50 mg/kg, i.p.) of one or the other enantiomer, as indicated. A. Survival curves, Kaplan-Meier analysis. N=9 per arm, P=0.001. B. Serum levels of TNF-α, IL-6, MCP-1, KC and IL-10 in LPS challenged mice. Cytokines in serum were determined 40, 60, 90, 120, and 240 min after challenge. N = 7–9. *, P < 0.05; **, P < 0.01, ***, P < 0.001, ****, P < 0.0001 as calculated by Student's t test and comparison of JQ1(+) and JQ1(−). C. Serum SAA levels in mice challenged with lethal dose of LPS 2 hours and 4 hours after challenge. N = 6. NS indicates insignificant difference (P >0.05) as calculated by Student’s t test.
To monitor the dynamics of the cytokine response in the endotoxemic mice, we measured concentrations of pro-inflammatory cytokines in serum every 20 min after LPS administration. Concentrations of key LPS-responsive inflammatory cytokines (IL-6, MCP-1, TNF-α) were significantly reduced in mice pre-treated with JQ1(+) (Fig. 7B) compared to JQ1(−) controls. However (and as predicted by Fig. 2B, 4B and 5A), KC levels were similar in JQ1(+) and JQ1(−) pre-treated mice (Fig. 7B). Interestingly, serum IL-10, a known anti-inflammatory cytokine, was also reduced in JQ1(+)-treated animals. The ability of JQ1(+) to decrease IL-10 in vivo agrees with ability of JQ1(+) to block IL-10 production in vitro (Fig.5A). JQ1(+) treatment did not significantly impair early production of serum amyloid A (SAA), an acute phase, liver-derived protein that is a signature of endotoxemia in both LPS-treated mice and human sepsis patients (Fig. 7C). Given that serum SAA levels largely reflect hepatocyte function, lack of a JQ1(+) effect on SAA levels supports the interpretation that anti-inflammatory effects of JQ1(+) on macrophages are cell type-limited. Taken together, these observations therefore indicate that JQ1(+) treatment specifically targets production of pro-inflammatory cytokines and protects mice from the systemic pro-inflammatory response elicited by endotoxemic shock, without altering acute phase proteins, but this protective mechanism does not work through elevated IL-10. Although the in vivo data cannot attribute effects of JQ1(+)-mediated ablation of cytokines to a specific cell type, the effects are consistent with the interpretation that JQ1(+) blunts inflammatory responses, at least in part, through its ability to block macrophage pro-inflammatory responses to bacterial stimuli.
Discussion
Our data support the conclusion that BET proteins play critical roles in pro-inflammatory cytokine responses. We used multiple approaches that include an in vitro model of brd2 lo RAW macrophages, brd2lo primary BMDMs and siBrd2-treated WT BMDMs. In the experiments with siRNA knockdown of BET family proteins, we expanded our initial focus on Brd2 and found that all three members of the BET family were essential for pro-inflammatory cytokine expression. These data raise an interesting question about the redundancy of BET proteins: despite high homology, their functions do not overlap enough for intact BET proteins to rescue brd2null or brd4null knockout mice, both of which are embryonic lethal (13, 29–31). Notwithstanding possible untested mechanistic differences, the similar effects of Brd2, Brd3 and Brd4 knockdown on inflammatory cytokine expression indicate that developmental functions of BET proteins may be highly specific and distinct from inflammation-associated functions. Taken together, the data predict that Brd3 and Brd4 deficiency may prove as promising as Brd2 for protection from inflammatory conditions beyond Brd2-regluated insulin resistance in obesity (13), if the problems of lethality associated with whole-body ablation of these proteins can be addressed.
JQ1 treatment of BMDMs qualitatively recapitulated the phenotype of BET protein knockdown through siRNA; however, the JQ1 effect is stronger than the siRNA effect. This result may be due to the fact that JQ1 simultaneously acts on the bromodomains of all three BET proteins. In contrast, each siRNA provides only 60–70% knockdown of the targeted mRNA. We attempted to expose the WT BMDMs to the combination of all three siRNAs, but this treatment appears to be highly toxic for the cells (data not shown) as opposed to the well-tolerated JQ1 treatment. These data suggest that BET proteins likely harbor functions that are crucial for cell survival but that are bromodomain-independent, and thus not targeted by JQ1.
Although the BET bromodomain is known to associate with chromatin through protein-protein interaction with acetylated histones, our data support the unexpected possibility that histone interaction is not the only mechanism underlying BET-mediated regulation of inflammatory genes. Rather, we demonstrate that Brd2 knockdown macrophages have a limited ability to activate NF-κB dependent transcription in the context of a transfected plasmid (i.e. outside the constraints of a complex chromatin structure). Chromatin-independent action of BET proteins may be explained by data showing that Brd2, like Brd4, may bind and activate the NF-κB protein RelA through association with an acetylated RelA K310 residue (44). Because this interaction would occur through the Brd2 bromodomain, the data are consistent with the interpretation that JQ1(+) may function through disrupting the RelA K310/BET protein tether. The possibility that BET proteins can be tethered to genes through NF-κB rather than through histone acetylation is consistent with the demonstration that BET proteins can regulate both transfected NF-κB reporter plasmids and naturally chromatinized promoters. Interruption of direct BET protein/NF-κB interactions may also regulate chromatinized genes independently from mechanisms involving BET/acetylated histone interactions.
Another benzodiazepine-based small molecule BET protein inhibitor, I-BET, was described recently to block IL-6 in mice (45). However, the I-BET compound, while reported to act upon both Brd2 and Brd4 bromodomains, failed to affect TNF-α and MCP-1, although both of which are major predictors of survival in murine models of systemic inflammatory response syndrome (SIRS) and sepsis shock (46, 47). The authors of that report furthermore showed that TNF-α expression was down regulated with siRNAs to BET proteins, although surprisingly, I-BET had no effect on TNF-α protein production. Our JQ1(+) results therefore demonstrate a previously unappreciated, widespread potency of BET inhibition as a more comprehensive, anti-inflammatory strategy that may have efficacy in TNF-α-, IL-6- and/or MCP-1-mediated inflammatory conditions. Our demonstration by ChIP that JQ1(+) inhibits association between Brd2 and both TNF-α and IL-6 promoters further supports this interpretation.
The potency of BET inhibition with JQ1(+) in a standard in vivo model of endotoxemia (48) specifically links the identification of BET proteins as pro-inflammatory cytokine regulators to inflammatory disease. LPS-induced endotoxemia is associated with rapid, dramatic elevation of inflammatory cytokines (46), which, as predicted by our in vitro observations, respond to in vivo JQ1(+) therapy. Given that serum IL-6 and TNF-α are major predictors of outcome in septic and LPS-induced shock (46, 49), and that both of these cytokines were successfully targeted by JQ1, the data justify clinical tests of JQ1 in this disease category, which thus far has been dismayingly resistant to clinical intervention. The vast improvement in survival, likely a direct consequence of diminished “cytokine storm” in the challenged mice, also raises the possibility of JQ1 efficacy in pathogen-mediated cytokine storms characteristic of, for example, hemorrhagic fever viruses (48, 50, 51). Success may hinge on therapeutics developed in mice for well-defined or limited patient cohorts. Our success with JQ1(+) and its reasonable safety profile may justify efficacy tests of JQ1(+) in other inflammatory diseases, including human SIRS or sepsis.
Supplementary Material
Acknowledgments
We thank F. Wang for molecular cloning of Brd2 shRNA construct and help with brd2 lo mouse colony maintenance; J. Bradner from Dana Farber Cancer Institute for JQ1 reagents; K. Bossart for help with Luminex assays; J. Carr and M. Zhu for help with ChIP assays; D. Remick for advice on the LPS shock experiments; and S. Fried and J. Schlezinger for constructive critiques of the project.
Abbreviations used in this paper
- BET
bromodomain and extraterminal domain
- BM
bone marrow
- BMDM
bone marrow derived macrophage
- ChIP
chromatin immunoprecipitation
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- HAT
histone acetyltransferase
- KC
keratinocyte chemoattractant
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- WT
wildtype
Footnotes
This work was supported by grants from the National Institutes of Health (R56 DK090455), American Cancer Society (RSG-05-072-01), the Leukemia and Lymphoma Society (6023-09), and the Evans Center for Interdisciplinary Biomedical Research Affinity Research Collaborative on ‘Obesity, Cancer and Inflammation’ at Boston University (GVD); R21DK089270, R56 DK096525 and The Boston Nutrition Obesity Research Center DK046200 (BSN). G. V. Denis is an American Cancer Society research scholar.
BIBLIOGRAPHY
- 1.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 2.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a000224. a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 4.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor kappaB recruitment to target promoters. J. Exp. Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan KE, Reddy ABM, Dietzmann K, Suriano AR, Kocieda VP, Stewart M, Bhatia M. Epigenetic regulation of tumor necrosis factor alpha. Mol. Cell. Biol. 2007;27:5147–5160. doi: 10.1128/MCB.02429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 7.Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. The EMBO journal. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez R, Zhou M-M. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Devel. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 9.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorpe KL, Gorman P, Thomas C, Sheer D, Trowsdale J, Beck S. Chromosomal localization, gene structure and transcription pattern of the ORFX gene, a homologue of the MHC-linked RING3 gene. Gene. 1997;200:177–183. doi: 10.1016/s0378-1119(97)00415-0. [DOI] [PubMed] [Google Scholar]
- 11.Rhee K, Brunori M, Besset V, Trousdale R, Wolgemuth D. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 1998;111:3541–3550. doi: 10.1242/jcs.111.23.3541. [DOI] [PubMed] [Google Scholar]
- 12.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem. J. 2010;425:71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo N, Faller D, Denis G. Activation-induced nuclear translocation of RING3. J. Cell Sci. 2000;113:3085–3091. doi: 10.1242/jcs.113.17.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 16.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinha A, Faller DV, Denis GV. Bromodomain analysis of Brd2-dependent transcriptional activation of cyclin A. Biochem. J. 2005;387:257–269. doi: 10.1042/BJ20041793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brès V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang MK, Mochizuki K, Zhou M, Jeong H-S, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi S, Ooike S, Iwata K, Hikawa H, Sugahara K. Antitumor agent. 2009 [Google Scholar]
- 23.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, Fletcher JA, Aster JC. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–2242. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 24.French CA. Demystified molecular pathology of NUT midline carcinomas. J. Clin. Pathol. 2010;63:492–496. doi: 10.1136/jcp.2007.052902. [DOI] [PubMed] [Google Scholar]
- 25.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertz Ja, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele Da, Bergeron L, Sims RJ. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer. 2012;12:1–14. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyuris A, Donovan DJ, Seymour KA, Lovasco LA, Smilowitz NR, Halperin ALP, Klysik JE, Freiman RN. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim. Biophys. Acta. 2009;1789:413–421. doi: 10.1016/j.bbagrm.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 2009;238:908–917. doi: 10.1002/dvdy.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RSP. Growth and Early Postimplantation Defects in Mice Deficient for the Bromodomain-Containing Protein Brd4. Mol. Cell. Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J. Clin. Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magness ST, Jijon H, Van Houten Fisher N, Sharpless NE, Brenner DA, Jobin C. In vivo pattern of lipopolysaccharide and anti-CD3-induced NF-kappa B activation using a novel gene-targeted enhanced GFP reporter gene mouse. J. Immunol. 2004;173:1561–1570. doi: 10.4049/jimmunol.173.3.1561. [DOI] [PubMed] [Google Scholar]
- 35.McCarthy KM, McDevit D, Andreucci A, Reeves R, Nikolajczyk BS. HMGA1 co-activates transcription in B cells through indirect association with DNA. J. Biol. Chem. 2003;278:42106–42114. doi: 10.1074/jbc.M308586200. [DOI] [PubMed] [Google Scholar]
- 36.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil Chemokines KC and Macrophage-Inflammatory Protein-2 Are Newly Synthesized by Tissue Macrophages Using Distinct TLR Signaling Pathways. J. Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 37.Kim HY, Kim HK, Kim JR, Kim HS. Upregulation of LPS-induced chemokine KC expression by 15-deoxy-delta12,14-prostaglandin J2 in mouse peritoneal macrophages. Immunol. Cell Biol. 2005;83:286–293. doi: 10.1111/j.1440-1711.2005.01329.x. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe H, Numata K, Ito T, Takagi K, Matsukawa A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock. 2004;22:460–466. doi: 10.1097/01.shk.0000142249.08135.e9. [DOI] [PubMed] [Google Scholar]
- 39.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 Macrophages and the Th1/Th2 Paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- 40.El Chartouni C, Rehli M. Comprehensive analysis of TLR4-induced transcriptional responses in interleukin 4-primed mouse macrophages. Immunobiology. 215:780–787. doi: 10.1016/j.imbio.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 41.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, Uda-Tochio H, Hamana H, Terada T, Adachi N, Matsumoto T, Tanaka A, Horikoshi M, Ozato K, Padmanabhan B, Yokoyama S. Crystal structure of the human BRD2 bromodomain: insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- 43.López-Bojórquez LN, Dehesa AZ, Reyes-Terán G. Molecular mechanisms involved in the pathogenesis of septic shock. Arch. Med. Res. 2004;35:465–479. doi: 10.1016/j.arcmed.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 44.Huang B, Yang X-D, Zhou M-M, Ozato K, Chen L-F. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol. Cell. Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C-W, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Remick DG, Newcomb DE, Bolgos GL, Call DR. Comparison of the mortality and inflammatory response of two models of sepsis: lipopolysaccharide vs. cecal ligation and puncture. Shock. 2000;13:110–116. doi: 10.1097/00024382-200013020-00004. [DOI] [PubMed] [Google Scholar]
- 47.Juskewitch JE, Knudsen BE, Platt JL, Nath KA, Knutson KL, Brunn GJ, Grande JP. LPS-induced murine systemic inflammation is driven by parenchymal cell activation and exclusively predicted by early MCP-1 plasma levels. Am. J. Pathol. 2012;180:32–40. doi: 10.1016/j.ajpath.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rittirsch D, Hoesel LM, Ward PA. The disconnect between animal models of sepsis and human sepsis. J. Leukoc. Biol. 2007;81:137–143. doi: 10.1189/jlb.0806542. [DOI] [PubMed] [Google Scholar]
- 49.Oberholzer A, Souza SM, Tschoeke SK, Oberholzer C, Abouhamze A, Pribble JP, Moldawer LL. Plasma cytokine measurements augment prognostic scores as indicators of outcome in patients with severe sepsis. Shock. 2005;23:488–493. [PubMed] [Google Scholar]
- 50.Marshall JC. Sepsis: rethinking the approach to clinical research. J. Leukoc. Biol. 2008;83:471–482. doi: 10.1189/jlb.0607380. [DOI] [PubMed] [Google Scholar]
- 51.Ward PA, Bosmann M. A historical perspective on sepsis. Am. J. Pathol. 2012;181:2–7. doi: 10.1016/j.ajpath.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.