Abstract
In many studies of HIV replication, it is useful to quantify the number of HIV proviruses in cells against a background of unintegrated forms of the HIV DNA. A popular method for doing so involves quantitative PCR using one primer complementary to the HIV long terminal repeat (LTR), and a second primer complementary to a cellular Alu repeat, so that PCR product only forms from templates where a provirus is integrated in the human genome near an Alu repeat. However, several recent studies have identified conditions that alter distributions of HIV integration sites relative to genes. Because Alu repeats are enriched in gene rich regions, this raises the question of whether altered integration site distributions might confound provirus abundance measurements using the Alu-PCR method. Here modified versions of the HIV tethering protein LEDGF/p75 were used to retarget HIV integration outside of transcription units, and show that this has a negligible effect on Alu-PCR quantitation of proviral abundance. Thus altered integration targeting, at least to the degree achieved here, is not a major concern when using the Alu-PCR assay.
Keywords: HIV, integration, Alu, quantitative PCR, LEDGF/p75, Taqman
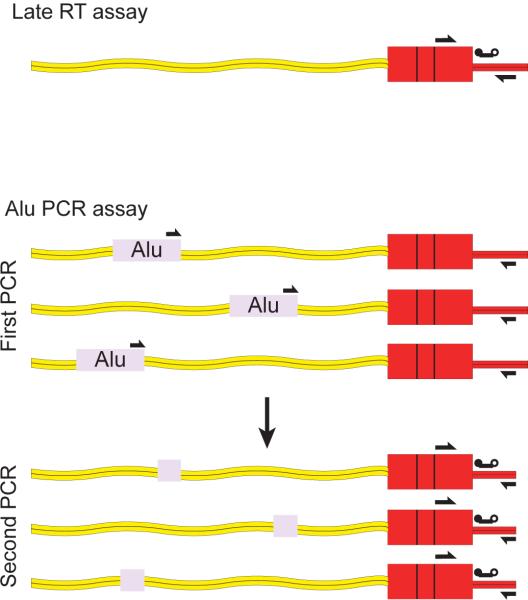
The Alu-PCR method (Butler et al., 2001; Butler et al., 2002; O'Doherty et al., 2002) allows quantitation of HIV proviruses in cells despite the presence of unintegrated forms of HIV DNA (Butler et al., 2001; Farnet and Haseltine, 1991). Amplification is carried out using primers complementary to the HIV LTR and cellular Alu repeats, so that only proviruses integrated near Alu repeats in the human genome support amplification (Figure 1). The progression of the PCR can be quantified using the Taqman PCR method or molecular beacons. For increased sensitivity, a two-step nested PCR method can be used (O'Doherty et al., 2002). Because each provirus in a cell resides a defined distance from the nearest Alu repeat, and because this distance varies for each provirus in the population, the control used for absolute quantitation must be chosen carefully. To match the heterogeneity in the analyate, standard DNA is prepared from heavily infected cells after long term culture, the long term culture being important to dilute out unintegrated DNA. The abundance of integrated proviruses can then be determined in purified genomic DNA from these cultures, and the heavily infected cell DNA diluted serially into uninfected cell DNA to make a standard curve. After analysis by Alu-PCR, comparison of the unknown to the standard allows estimation of the absolute number of proviruses in a genomic DNA sample (Butler et al., 2001; O'Doherty et al., 2002).
Figure 1.
Methods for quantifying proviral abundance using the Alu-PCR assay. Arrows indicate the positions of PCR primers, the line with two balls indicates the Taqman probe.
It is now clear that various treatments can alter HIV integration target site selection, raising the question of how this influences the Alu-PCR assay. HIV integration in human cells occurs most commonly in active transcription units (Schröder et al., 2002). HIV integration is promoted by binding of the cellular LEDGF/p75 protein (product of the PSIP1 gene) to HIV integrase protein (Cherepanov et al., 2003; Emiliani et al., 2005; Llano et al., 2006; Maertens et al., 2003; Turlure et al., 2004) and depletion of LEDGF/p75 reduces the targeting of integration to transcription units (Ciuffi et al., 2005; Marshall et al., 2007; Shun et al., 2007). LEDGF/p75 appears to act via a simple tethering mechanism, in which LEDGF/p75 binds to integrase with its C-terminal domain and to chromatin in transcription units with its N-terminal domains. Three reports have shown that re-engineering the LEDGF/p75 tether to contain a CBX/HP1β heterochromatin binding domain promotes integration outside of transcription units (Ferris et al., 2010; Gijsbers et al., 2010; Silvers et al., 2010). Further studies suggest that modulating the levels of additional cellular genes or cell growth status can also have detectable effects on integration frequency in transcription units (Barr et al., 2006; Ciuffi et al., 2006; Ocwieja et al., 2011; Schaller et al., 2011). Because Alu repeats are predominantly located in gene-rich regions (Lander, 2001; Venter, 2001), not randomly distributed in chromosomes, it is thus possible that factors altering integration targeting would confound Alu-PCR quantitation of proviral abundance.
In this study, infections were carried out in the presence of LEDGF/p75 knockdowns, CBX1-LEDGF fusions, or controls, to generate genomic DNA with different distributions of HIV integration sites (Gijsbers et al., 2010) and quantified the effects on the Alu-PCR assay. LEDGF/p75 was knocked down using optimized siRNAs, then genes encoding three altered tethering proteins introduced. These included the LEDGF/p75 C-terminal region fused to CBX1, a version of this chimera defective for integrase binding (the D366N mutation (Cherepanov et al., 2004; Llano et al., 2006)), and an intact version of LEDGF/p75 expressed from a modified gene engineered to be insensitive to the siRNAs used for knock down. Cells were challenged with a GFP-bearing NL4-3 based HIV vector (Lu et al., 2004b), then vector supernatants were removed, and cells were allowed to grow for 14 days to dilute out unintegrated forms of the HIV DNA.
To verify altered integration targeting, genomic DNA was isolated from cells, and DNA containing integration sites amplified using ligation-mediated PCR. PCR products were then sequenced using the 454/Roche pyrosequencing method and human DNA flanking integrated proviruses mapped to the human genome (Ciuffi et al., 2009). The resulting integration site data sets are listed in Supplementary Table 1.
To assess the differences in proviral distribution between samples, the relationship of integration sites to nearby genomic features was analyzed (Figure 2) and found to parallel results from previous studies with these reagents (Gijsbers et al., 2010). Briefly, wild type HeLaP4-CCR5 cells infected with HIV-1 showed favored integration in transcription units, whereas reduction in LEDGF/p75 expression diminished the proportion of integration sites in transcription units (Figure 2A). Back-complementing the depleted cells with an LEDGF/p75 allele insensitive to the siRNA restored integration in transcription units to WT levels. In the cells depleted for LEDGF/p75 but containing the CBX1-LEDGF fusion, integration in transcription units was no longer favored. In contrast, control cells encoding a version of the CBX1-LEDGF fusion containing an amino-acid substitution at the integrase binding site (D366N) showed no difference from the LEDGF/p75 knockdown cells.
Figure 2.
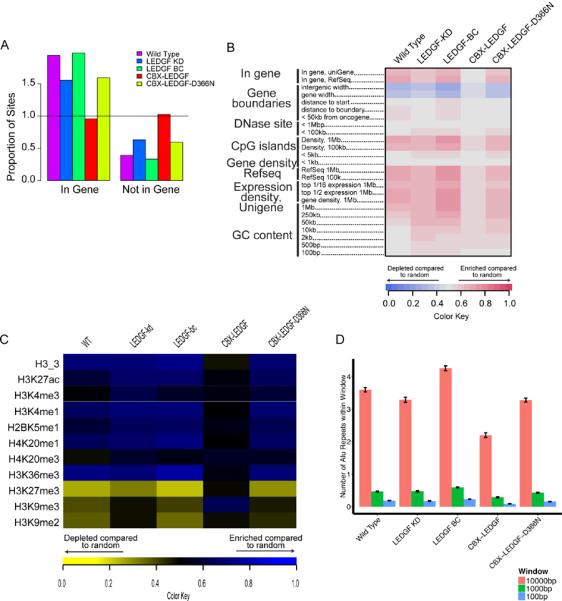
Retargeting HIV DNA integration in cells using fusions of LEDGF/p75 to CBX. (A) Integration frequency in genes. The proportion of integration sites falling within a RefSeq gene is shown with the frequency of a random distribution represented by the horizontal line at one. (B) Genomic heatmap of integration site associations with genomic features. Each column represents a dataset, each row a genomic feature. Some genomic features were tested at several interval sizes surrounding the integration site. Increasing shades of red show favored association with a feature, increasing shades of blue show disfavored association compared with a random distribution. For a description of the ROC area method used to construct the heat map see (Berry et al., 2006). (C) Epigenetic heatmap of integration site associations with epigenetic marks. Heatmap structure is similar to the Genomic heatmap but with epigenetic marks indicated in the rows and shades of blue showing favoring and yellow showing disfavoring. Chip-Seq data on epigenetic marks in HeLa cells used to generate the heatmap was taken from (Meylan et al., 2011). (D) Average number of Alu repeats in a specified window surrounding integration sites. The sizes of the genomic intervals surrounding each integration site (“Window”) is shown at the right.
Detailed comparisons of integration site distributions over genomic features and epigenetic marks (Figure 2B and C) also showed patterns paralleling those reported previously (Gijsbers et al., 2010; Marshall et al., 2007; Shun et al., 2007). For associations with genomic features such as gene density, transcription units, CpG islands, and DNase sensitive sites, integration in CBX1-LEDGF cells appeared globally shifted toward a more random distribution compared to other conditions (Figure 2B). For associations with epigenetic features (Figure 2C), the CBX1-LEDGF sites showed many opposite associations compared to the other datasets. For example, all cell types except CBX1-LEDGF favored integration near marks associated with transcription, including H4K20me3 and H3K79me3. CBX1-LEDGF expressing cells, however, did not favor integration near these marks. Similarly, HIV integration near regions with H3K9me3, a mark associated with centromeric heterochromatin, transcriptionally silent regions and regions of CBX1 binding, was favored only in the presence of CBX1-LEDGF fusions and disfavored for all other conditions. Together, these changes confirm a significant shift in the HIV integration profile in CBX1-LEDGF cells compared to the other cell types.
Figure 2D presents a summary of the mean number of Alu repeats in each cell line, as assessed in a range of genomic window sizes (100bp to 10kb) around integration sites in each data set. In a 1kb window, the wild type control cells had a mean of 0.47 annotated Alu repeats surrounding each integration site. The mean number of Alu repeats in the LEDGF/p75 knockdown cells was not significantly different from wild type in the 1kb window, but the cells knocked down for LEDGF/p75 and containing the CBX1-LEDGF fusion were significantly different (mean 0.29 Alu repeats per 1kb window). Whether there were significant differences in the number of Alu repeats around integration sites was assessed using a Kruskal-Wallis test followed by pairwise Wilcoxon rank sum tests. The CBX1-LEDGF cell line differed significantly from wild type cells in all windows tested, but the LEDGF KD line differed significantly from wild type cells only in the 10kb window (Bonferroni-corrected p<0.05 for all tests). The differences in numbers of Alu repeats were consistently less than two-fold over all windows.
To investigate how differences in proximity of proviruses to Alu repeats affected the Alu-PCR assay in samples containing many proviruses, results were compared for two Taqman PCR assays that quantify the copy number of integrated proviruses per cell Figure 3). For Alu-independent quantitation, a one-step assay was used with primers and probe binding to internal sequences within the HIV vector DNA, specifically in the R-U5 and PBS regions of HIV (the “late RT” amplicon) (Butler et al., 2001). Results were compared to those with the two-step version of the Alu-PCR protocol (Agosto et al., 2007; O'Doherty et al., 2002). In the first step, PCR was carried out using primers binding to an Alu repeat and HIV gag. In the second step, the late RT Taqman amplicon was used to quantify the amount of Alu-PCR product formed. Importantly, the same procedure was applied to control DNA comprised of a dilution series of long-term infected cell DNA, so that the differential abundance due to the preceding Alu-PCR step could be quantified.
Figure 3.
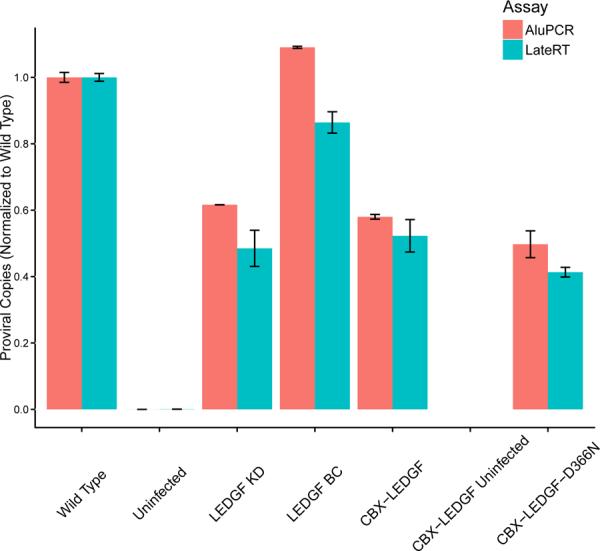
Effects of different integration site distributions on detection by the Alu-PCR assay. The graph compares the relative provirus copy number determined using the one-step late RT assay (blue) versus the Alu-PCR assay (red). Engineered HeLa cells were infected with an HIV-based vector and DNA harvested 14 days after infection. The long time was used to allow unintegrated DNA to be diluted out by growth of cells. The x-axis shows the cell type studied, the y-axis shows relative provirus copies/genome normalized to wild-type, with standard error. Data represent four technical replicates. The sample with the largest difference between assays (22%) was LEDGF BC. Statistical tests comparing all Alu-PCR versus all Late-RT samples (Kruskal-Wallis) showed no significant differences. Follow up tests of individual pairs of samples (Wilcoxon rank-sum test) also showed no significant differences.
Note that in many experimental infections the proportion of unintegrated DNA would be unknown, so that quantifying only the early reverse-transcription product would not provide a satisfactory measurement of the integrated fraction. Here cells were grown for two weeks after infection and prior to harvest, which results in loss of unintegrated DNA due to dilution during growth. Because of the prolonged growth phase, it was possible to compare results from the one-step late RT and Alu-PCR assays directly.
The results of the two assays are compared in Figure 3. Values were normalized to the wild type within each assay, then compared between assays. The largest difference between assays for any sample was no greater than 22%, indicating good general concordance. Pairwise Wilcoxon rank-sum tests, comparing the Alu-PCR assay to the one-step Late RT assay in each cell line, failed to detect any significant differences between the two assays under any condition.
Thus redistributing integration sites away from transcription units did not strongly affect provirus quantitation using the Alu-PCR assay. It is possible that even more extreme redistribution of HIV integration sites might disrupt this assay more significantly, but given current technology, use of the CBX1-LEDGF fusion provides the most extreme redistribution that is experimentally accessible at present.
Methods
The production of cell lines depleted for LEDGF/p75 was performed by knockdown with murine leukemia virus (MLV)-based retroviral vectors encoding two miRNA-based short-hairpin RNAs and a zeocin-resistance cassette. After transduction of HeLaP4-CCR5 with this vector, monoclonal cell lines were established, with LEDGF/p75 mRNA levels reduced to <4% that of wild-type. LEDGF/p75 hybrids were produced by fusion of CBX1 in place of the DNA-binding region of LEDGF/p75 and re-introduction into LEDGF/p75 depleted cell lines via MLV-based viral vectors. Control cell lines with RNAi-resistant LEDGF/p75 (LEDGF BC) were generated in the same manner (Gijsbers et al., 2010).
To generate cells with different integration site distributions, cells were infected for 48 hours in a 96-well plate using 20,000 cells/well and media containing 10ug/ml DEAE-Dextran. Media was then changed and cells allowed to expand for two weeks before harvest to dilute out unintegrated forms of HIV DNA. Infection ranged from 50–70% as measured by FACS of GFP+ cells. Infection was performed with an HIV NL4-3 derivative carrying a GFP expression cassette and capable of single round infection (VRX494(VSV-G)). The vector was provided courtesy of VIRxSYS (Humeau et al., 2004; Lu et al., 2004a).
Integration sites were sequenced and analyzed as described previously (Ciuffi et al., 2009; Wang et al., 2007; Wang et al., 2008) using MseI and Tsp509I to digest purified genomic DNA, ligating on PCR adapters, and amplifying using primers specific for the viral LTR and the PCR adapter. A round of nested PCR was used to increase specificity and incorporate 454 adapter A & B sequences on the ends of amplicons. Sequencing was done using the 454 “Junior” platform and sequence reads were required to align to the human genome (hg18) within three basepairs of the start of the sequence, have a single best hit to the genome, and have ≥ 98% identity. Statistical comparisons were performed as described by Berry et al (Berry et al., 2006). Sequence reads will be deposited to SRA and accession numbers included upon acceptance of this manuscript.
Quantitative PCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR System. For the one-step assay, using the “late RT” amplicon, Q-PCR was performed with 125ng of template DNA per well, with primers specific for the R-U5 and PBS regions of HIV, as described in Supplemental Table 2. Thermal cycler conditions were 95°C for 2 minutes, then 45 cycles of 95°C 15 seconds, 50°C 30 seconds, 72°C 1 minute, with data collection at the 50°C stage. Cycle of threshold values were converted to HIV copies using an HIV-plasmid standard curve. For the two-step Alu-PCR assay, samples were initially amplified with Alu and Gag primers using the Applied Biosystems Veriti 96-well Thermal Cycler at 95°C for 2 minutes, then 20 cycles of 95°C 15 seconds, 50°C 15 seconds, 72°C 2 minutes 30 seconds. After this initial amplification, Q-PCR was performed with 125ng of initial template DNA per well, with the same primers and thermal cycler conditions as used in the one-step assay above. For the two-step Alu-PCR assay, cycle of threshold values were converted to HIV copies using a standard curve developed from a dilution series of long-term infected cells containing HIV proviral DNA. All qPCR assays were normalized by gDNA input. (Agosto et al., 2007; Butler et al., 2001; O'Doherty et al., 2002). The absolute numbers of proviruses per genome differed between the single-step late RT assay and the two-step Alu-PCR assay by a factor of ~7. For the wild type infection in Figure 3, the mean proviral copies/genome were 7.61 by one-step late RT assay and 54.0 by Alu-PCR assay. Higher values were seen consistently in the Alu-PCR assay, for unknown reasons. Possibly the PCR products that serve as amplification templates in the two-cycle Alu-PCR reaction are used more efficiently than genomic DNA, which is the template in the late-RT assay.
Supplementary Material
Highlights
-
-
Alu-PCR is a popular method for quantifying integrated proviruses.
-
-
The Alu-PCR method is potentially sensitive to the distributions of HIV integration sites in cells.
-
-
Samples with considerably altered HIV integration site distributions were designed to test the accuracy of Alu-PCR.
-
-
Altered integration site distributions exerted negligible effects on Alu-PCR quantitation of integrated proviruses.
Acknowledgements
We are grateful to members of the Bushman laboratory for help and suggestions, and to Laurent Humeau and Nikolay Kirokhow of VIRxSYS for preparation of the NL4-3 based vector VRX496. This work was supported by NIH grants AI52845, P01-AI090935-01 and AI082020, the University of Pennsylvania Center for AIDS Research, and the Penn Genome Frontiers Institute with a grant with the Pennsylvania Department of Health. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. T. B. is a Special Fellow of the Leukemia and Lymphoma Society of America.
Abbreviations
- HIV
human immunodeficiency virus
- LEDGF/p75
human lens epithelium-derived growth factor
- LTR
long terminal repeat
- PBS
primer-binding site
- RT
reverse transcriptase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O'Doherty U. HIV-1 integrates into resting CD4+ T cells even at low inoculums as demonstrated with an improved assay for HIV-1 integration. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr SD, Ciuffi A, Leipzig J, Shinn P, Ecker JR, Bushman FD. HIV integration site selection: targeting in macrophages and the effects of different routes of viral entry. Molecular Therapy: The Journal of the American Society of Gene Therapy. 2006;14:218–225. doi: 10.1016/j.ymthe.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of Target Sites for Mobile DNA Integration in the Human Genome. PLoS Comput Biol. 2006;2:e157–e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nature Medicine. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- Butler SL, Johnson EP, Bushman FD. HIV cDNA Metabolism Studied by Fluorescence-Monitored PCR: Notable Stability of Two-LTR Circles. J. Virol. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. The Journal of Biological Chemistry. 2004;279:48883–48892. doi: 10.1074/jbc.M406307200. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. The Journal of Biological Chemistry. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Diamond TL, Hwang Y, Marshall HM, Bushman FD. Modulating target site selection during human immunodeficiency virus DNA integration in vitro with an engineered tethering factor. Human Gene Therapy. 2006;17:960–967. doi: 10.1089/hum.2006.17.960. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nature Medicine. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- Ciuffi A, Ronen K, Brady T, Malani N, Wang G, Berry CC, Bushman FD. Methods for integration site distribution analyses in animal cell genomes. Methods (San Diego, Calif.) 2009;47:261–268. doi: 10.1016/j.ymeth.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempe D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, Christ F, Rain JC, Dargemont C, Debyser Z, Benarous R. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem. 2005;280:25517–23. doi: 10.1074/jbc.M501378200. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Haseltine WA. Circularization of human immunodeficiency virus type 1 DNA in vitro. Journal of Virology. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun M-C, Allis CD, Engelman A, Hughes SH. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Molecular Therapy: The Journal of the American Society of Gene Therapy. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, Pereira M, Slepushkina T, Barnett S, Dropulic LK, Carroll R, Levine BL, June CH, Dropulic B. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Molecular Therapy: The Journal of the American Society of Gene Therapy. 2004;9:902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Lander E. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. An essential role for LEDGF/p75 in HIV integration. Science (New York, N.Y.) 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- Lu X, Humeau L, Slepushkin V, Binder G, Yu Q, Slepushkina T, Chen Z, Merling R, Davis B, Chang Y-N, Dropulic B. Safe two-plasmid production for the first clinical lentivirus vector that achieves >99% transduction in primary cells using a one-step protocol. The Journal of Gene Medicine. 2004a;6:963–973. doi: 10.1002/jgm.593. [DOI] [PubMed] [Google Scholar]
- Lu X, Yu Q, Binder GK, Chen Z, Slepushkina T, Rossi J, Dropulic B. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. Journal of virology. 2004b;78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. The Journal of Biological Chemistry. 2003;278:33528–33539. doi: 10.1074/jbc.M303594200. [DOI] [PubMed] [Google Scholar]
- Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PloS One. 2007;2:e1340–e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan S, Groner AC, Ambrosini G, Malani N, Quenneville S, Zangger N, Kapopoulou A, Kauzlaric A, Rougemont J, Ciuffi A, Bushman FD, Bucher P, Trono D. A gene-rich, transcriptionally active environment and the pre-deposition of repressive marks are predictive of susceptibility to KRAB/KAP1-mediated silencing. BMC Genomics. 2011;12:378. doi: 10.1186/1471-2164-12-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. Journal of Virology. 2002;76:10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocwieja KE, Brady TL, Ronen K, Huegel A, Roth SL, Schaller T, James LC, Towers GJ, Young JAT, Chanda SK, König R, Malani N, Berry CC, Bushman FD. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathogens. 2011;7:e1001313–e1001313. doi: 10.1371/journal.ppat.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T, Ocwieja KE, Rasaiyaah J, Price AJ, Brady TL, Roth SL, Hue S, Fletcher AJ, Lee K, KewalRamani VN, Noursadeghi M, Jenner RG, James LC, Bushman FD, Towers GJ. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder ARW, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Shun M-C, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes & Development. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Human Gene Therapy. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlure F, Devroe E, Silver PA, Engelman A. Human cell proteins and human immunodeficiency virus DNA integration. Front Biosci. 2004;9:3187–208. doi: 10.2741/1472. [DOI] [PubMed] [Google Scholar]
- Venter JC. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Research. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.