Abstract
Nanodiscs are self-assembled discoidal phospholipid bilayers surrounded and stabilized by membrane scaffold proteins (MSP), that have become a powerful and promising tool for the study of membrane proteins. Even though their reconstitution is highly regulated by the type of MSP and phospholipid input, a biophysical characterization leading to the determination of the stoichiometry of MSP, lipid and membrane protein is essential. This is important for biological studies, as the oligomeric state of membrane proteins often correlates with their functional activity. Typically combinations of several methods are applied using, for example, modified samples that incorporate fluorescent labels, along with procedures that result in nanodisc disassembly and lipid dissolution. To obtain a comprehensive understanding of the native properties of nanodiscs, modification-free analysis methods are required. In this work we provide a strategy, using a combination of dynamic light scattering and analytical ultracentrifugation, for the biophysical characterization of unmodified nanodiscs. In this manner we characterize the nanodisc preparation in terms of its overall polydispersity and characterize the hydrodynamically resolved nanodisc of interest in terms of its sedimentation coefficient, Stokes’ radius and overall protein and lipid stoichiometry. Functional and biological applications are also discussed for the study of the membrane protein embedded in nanodiscs under defined experimental conditions.
Keywords: Membrane protein, nanodisc, lipid bilayer, dynamic light scattering, analytical ultracentrifugation, lipid charge
1. Introduction
In a typical cell, up to 30% of all open reading frames are predicted to encode membrane proteins [1]. They play key roles in many biological pathways [2] and represent important drug targets. In particular, G protein-coupled receptors (GPCRs) have intensively been targeted for therapeutic purposes [3, 4]. Despite the tremendous progress in understanding the function of membrane proteins, limitations still exist, for example, due to the difficulties associated with obtaining sufficient quantities of the membrane protein of interest and incorporating them into a lipid environment in functional form. To fully understand the biology of a target membrane protein, biochemical information relating to the functional role of the membrane protein (e.g. ligand binding) needs to be complemented by structural studies (e.g. nuclear magnetic resonance or x-ray experiments).
Cell membranes are permeable barriers that maintain the internal environment of cells. The lipids do not only provide the physiological environment for membrane proteins but also control their function [5], in part through their composition that is known to change with organism [6], age [7] and disease [8]. A number of membrane proteins require unique lipids to modulate their mechanism of action [6, 9, 10], and for this reason, in vitro studies should preferably be carried out with membrane proteins embedded in a native lipid bilayer environment. Therefore, the following terms need to be satisfied to conduct a thorough biological characterization of the membrane protein: (i) precise control of the lipid environment surrounding the target membrane protein; (ii) control and definition of the membrane protein’s oligomeric state; and (iii) access of the interacting partner protein or ligand to the membrane protein independent of the topology of insertion. Even though detergents, artificial lipid bilayers (e.g. liposomes) or bicelles have been used to provide native-like environments for membrane proteins [11, 12], none of these satisfy all the criteria described above.
The nanodisc technology developed by Sligar and co-workers, on the other hand, does satisfy all of the above requirements for the study of membrane proteins in vitro [13, 14]. A nanodisc is a discoidal phospholipid bilayer surrounded and stabilized by two molecules of a membrane scaffold protein (MSP). Unlike detergent micelles, bicelles, or liposomes, nanodisc reconstitution is highly regulated by the MSP to lipid ratio and the length of the MSP; at the proper lipid to MSP ratio this generates homogeneous and monodisperse entities. A membrane protein reconstituted within a nanodisc does not only gain the stability and functional activity provided by native environments, it can also be treated much like any soluble protein. To date, a number of membrane proteins have been successfully introduced into nanodiscs reflecting a variety of protein types, number of the transmembrane helices and oligomeric states. These include the rhodopsin monomer and dimer [15], the bacteriorhodopsin monomer and trimer [16], the SecYEG translocon complex [17], and cytochrome P450 [13] among others. In this manner various functional studies, such as protein-protein interaction studies in solution [18–20], ligand binding studies by immobilized surface plasmon resonance [21], and fusion pore assays [22] have been carried out demonstrating the power and versatility that the nanodisc technology provides for the study of membrane proteins.
In this review, we focus on the biophysical characterization of membrane proteins inserted into nanodiscs. We provide a brief overview of the various techniques using modified membrane proteins and focus on novel methodological applications to the analysis of unmodified membrane protein targets using the class A GPCR neurotensin receptor 1 (NTS1) as a model system. We describe the purification of the protein components needed for generating nanodiscs, along with a detailed reconstitution procedure, followed by a description of their characterization by dynamic light scattering (DLS) and analytical ultracentrifugation (AUC). The last section includes examples of the functional and biological analyses of membrane proteins inserted into nanodiscs, emphasizing the versatility of this technology.
2. Reconstitution of the target membrane protein into nanodiscs
A variety of membrane proteins have been reconstituted into nanodiscs (summarized in Table 2.1). The basic protocol for reconstitution requires mixing of the purified membrane protein, detergent solubilized phospholipids and MSP in a previously determined stoichiometric ratio, followed by detergent removal to initiate the self-assembly and formation of the membrane protein-nanodisc. Typically this is followed by further purification on an affinity column or by size exclusion chromatography. In this section, we describe the reconstitution of NTS1 into nanodiscs and provide particular considerations for each step of the protocol.
Table 2.1. Biological studies with membrane proteins inserted into nanodiscs.
The list is not exhaustive and the reader is referred to excellent recent reviews [14, 49, 103, 104] for additional details. Representative references for each example are given.
| Membrane proteins | Number of transmembrane domains in monomer | Inserted into nanodisc as | Measurements | Ref. |
|---|---|---|---|---|
| Bacterial chemoreceptor (Tar) | 1 | Dimer/trimer of dimers | Methylation and deamidation; activation of histidine kinase | [53] |
| Cytochrome P450 (3A4) | 1 | Monomer/co-incorporation with cytochrome P450 reductase | Redox potential upon ligand binding; cooperativity of ligand binding; role of lipids | [13, 64] |
| Bacteriorhodopsin | 7 | Monomer/trimer | Spectroscopic properties | [14, 68] |
| GPCRs | 7 | |||
| μ-opioid receptor | Monomer | Agonist and antagonist binding; G protein activation; role of lipids | [18, 19, 59] | |
| β2-adrenergic receptor | ||||
| Neurotensin receptor 1 | ||||
| Rhodopsin | Monomer/dimer | G protein activation; role of lipids | [15, 61, 69] | |
| Monomer | Phosphorylation by kinases; arrestin binding; role of lipids | [20, 101] | ||
| Translocon complex SecYEG | 15 | Monomer | Interaction with SecA; role of lipids | [17] |
Neurotensin (NT) is a 13 amino acid residue peptide [23, 24] with diverse biological activities. It plays key roles in antinociception [25], hypothermia [26], modulation of dopamine neurotransmission [27], cancer growth [28], Parkinson’s disease [29] and the pathogenesis of schizophrenia [30]. Most of the known effects of NT are mediated through NTS1, a class A GPCR transmembrane protein [31], and the targeting of NTS1 with synthetic agonists and antagonists for therapeutic purposes has been discussed [32]. NTS1 binds NT at its extracellular surface and couples preferentially to a Gq-type G protein at its intracellular surface. These properties distinguish NTS1 from the well-studied rhodopsin and beta-adrenergic receptors, which bind small ligands within their transmembrane cores and interact with Gi- and Gs-type proteins, respectively. The importance of the phospholipid environment for rhodopsin function has been well documented [9, 33–39]; however, these observations may not be extrapolated to other GPCRs because of rhodopsin’s specific properties and the unique lipid environment for visual signal transduction. We were therefore interested in studying the effect of lipid head group charges on the signaling properties of NTS1 and utilized the defined experimental conditions afforded by the nanodisc technology.
2.1 The target membrane protein
Nanodisc reconstitution requires relatively large amounts of the membrane protein both for the optimization of the nanodisc reconstitution procedure, as well as for the subsequent series of experiments we describe in this work. As approximately 0.5 mg of purified NTS1 is needed for the reconstitution of NTS1-nanodiscs at a 1 ml scale, direct purification from natural sources is not feasible, because NTS1, like many membrane proteins, is expressed in vivo at very low levels. Note that rhodopsin is among the very few membrane proteins, which are found in high quantities in natural tissue. For most targets under consideration, the establishment of a recombinant production system is essential and to date several heterologous overexpression systems have been developed and successfully utilized to express membrane proteins [40–45], such as E. coli for the production of prokaryotic membrane proteins [46], and yeast or insect cells for the expression of eukaryotic membrane proteins [40]. In the case of NTS1, which is bacterially expressed, 100 gram (wet weight) of E. coli cells are required to purify approximately 10 mg of the protein. Solubilization and purification are carried out in the presence of detergents, chosen for their suitability to maintain the receptor in a functional state.
2.2 The nanodisc size
The MSP, based on the human apolipoprotein A-I [47], functions as a belt that wraps around the phospholipid bilayer and stabilizes the nanodisc. Several MSP constructs of various lengths have been developed in the Sligar laboratory [48], allowing for control of the nanodisc size through choice of the appropriate MSP and an optimized amount of phospholipid [13, 14, 49, 50]. Currently, the size of an empty-nanodisc (nanodisc devoid of the membrane protein, just containing lipid and MSP) can be tuned from ~9.5 nm in diameter using MSP1D1 to ~17 nm using MSP2N3 [14]. Two aspects govern the choice of the appropriate nanodisc size for experiments: (i) the size of the transmembrane portion of the target protein that needs to be accommodated within the nanodisc, and (ii) the number of target membrane proteins that need to be incorporated. When the membrane protein is reconstituted in a nanodisc, a number of phospholipid molecules equivalent in volume to the transmembrane portion of the membrane protein need to be displaced and, for this reason, the receptor nanodisc contains less phospholipid when compared to the corresponding empty-nanodisc using a particular MSP. Furthermore, in order to provide the native-like environment, the MSP of choice needs to be large enough such that space is available for both the membrane protein and sufficient phospholipids within the nanodisc.
2.3 Choice of phospholipid
The lipid composition surrounding of membrane proteins depends on the native cell type. For instance, the E. coli membrane contains by 70–80% phosphatidyl ethanolamine, 15–20% phosphatidyl glycerol and 5% cardiolipin, whereas rat hepatocyte membranes are composed of 14–20% phosphatidyl ethanolamine, 32–47% phosphatidyl choline, 8% phosphatidyl inositol, 4–8% phosphatidyl serine, and 13–14% sphingomyelin and small amounts of other components [6, 51]. These differences in composition characterize cell membrane properties such as fluidity and surface charge, all of which influence the functional and structural properties of membrane proteins. Hence, it is important to mimic aspects of the native-like lipid properties to probe the function of the target membrane protein in vitro.
Three considerations regarding the choice of phospholipid for nanodisc reconstitution need to be taken into account; the lipid composition, the molecular shape, and the phase transition temperature (Tm). Phospholipid molecules are classified into three major shape groups: cylinder, cone, and inverted-cone that are determined by the relative sizes of the polar head group and hydrophobic domains [52]. Cylinder shaped phospholipids such as the zwitterionic lipid 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and the negatively charged 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) adopt lamellar phases, whereas cone- and inverted cone-shaped lipids favor hexagonal and micellar lipid phases, respectively [52]. Although the number of lipid molecules in nanodiscs is small compared to that in lipid vesicles, lipid shape may well be an important determinant for the function of a given membrane protein. Most nanodisc applications use POPC, or a mixture of POPC and POPG or 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), which are all cylinder shaped lipids. In some cases, nanodisc reconstitution was carried out using E. coli lipid [53] or porcine polar brain lipid extract combined with POPC and POPG [19].
The phospholipid phase behavior and temperature also need to be considered when working with nanodiscs. At low temperatures, lipids organize into an ordered gel phase, where the hydrophobic lipid tails are closely packed, whereas at higher temperatures a liquid crystalline phase with fluid tails is noted. The transition temperature, Tm, that characterizes this change is a reflection of the length of fatty acid chains, the nature of the head group and charge. For instance, the Tm for the pure zwitterionic POPC and the negatively charged POPG are −2.6°C [54] and −4.0°C [55], respectively, whereas POPS, which is the major negatively charged phospholipid in eukaryotic membranes, has a Tm of 14°C [56]. To maintain a liquid crystalline phase, both the nanodisc reconstitution and downstream experiments need to be performed at temperatures above the Tm of the respective lipid(s) used. Studies on the Tm behavior of phospholipids in nanodiscs resulted in broader ordered gel to liquid crystalline transitions with a shift of the transition midpoint to higher temperatures when compared to vesicles, reflecting a more ordered state of the lipids in close contact with the MSP compared to core lipids [57, 58]. For example, the transition midpoint temperatures of dipalmitoyl phosphatidylcholine and dimyristoyl phosphatidylcholine increase by 3–4°C when incorporated into nanodiscs [58]. Hence Tm values of pure lipids provide guidance, but explicit studies with nanodiscs would be required to quantitate the behavior of the respective lipids in close contact with the MSP and the membrane protein. We conducted our experiments with POPC and POPG, but not with POPS, which allowed us to work at 0–4°C in order to maintain the integrity of NTS1 in nanodiscs [59].
A variety of detergents such as cholate [13, 16, 18], Triton X-100 [36], and n-dodecyl-β-D-maltoside (DDM) [60] have been used for dissolving phospholipids to be used in nanodisc reconstitution. The detergents used to dissolve lipids need to be compatible with the membrane protein; hence a detergent screen may need to be carried out with careful attention to both the solubility of the phospholipids and the activity of the target membrane protein.
2.4 Detergent removal
Nanodisc formation is initiated by detergent removal, which can be carried out in several ways. These include dialysis [61], the use of detergent binding columns (e.g. Extracti-Gel D columns) [61], or detergent binding beads such as polystyrene-based Bio-beads SM-2 (BioRad) [13, 14] or Amberlite XAD2 [62]. Here we describe the use of Bio-beads and note that the kinetics and efficiency of detergent removal by Bio-beads depends on the type of detergent, the amount of beads, and temperature [63]. In the specific case of the NTS1-nanodisc preparation, we found that prolonged incubation times or addition of an excess amount of Bio-beads resulted in the loss of the NTS1 function (data not shown), possibly because NTS1 was gradually adsorbed to the Bio-beads and thus denatured. To avoid this, we accurately controlled both the amount of Bio-beads and the incubation time, based on the quantity of detergent present in the reconstitution mix, accounting for cholate used to dissolve the lipid, and for DDM, 3-[(3-cholamidopyropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and cholesteryl hemisuccinate Tris salt (CHS) used for the purification of NTS1. Reconstitution with MSP1D1 required a 35-fold weight excess of Bio-beads and an incubation time of 2 hours at 4°C for optimal detergent removal and nanodisc formation [59]. Nanodiscs reconstituted with MSP1E3D1 required the addition of a 50-fold weight excess of Bio-beads and an incubation time of 3 hours at 4°C (data not shown), reflecting the larger amount of lipid and hence cholate present in the reconstitution mix.
2.5 Determination of phospholipid/MSP molar ratio
The phospholipid to MSP molar ratio is absolutely critical for nanodisc reconstitution. This ratio is determined by the length of the respective MSP and thus the amount of lipid accommodated within the nanodisc, the type of phospholipid, and the presence or absence of the target membrane protein [13, 14, 49]. Nanodiscs of a homogeneous size are produced when the proper phospholipid to MSP ratio is used. However, in the case of an incorrect ratio, aggregation of MSP and/or the formation of large lipid-MSP-membrane protein complexes is observed instead [59]. Compared to an empty-nanodisc, the presence of the membrane protein requires a reduction in the number of phospholipid molecules, corresponding to the size of the transmembrane region of the membrane protein [14] as the nanodisc diameter is determined by the MSP utilized [48]. This step is specific for each membrane protein under investigation as each target displaces a different number of phospholipid molecules.
A successful strategy for the incorporation of monomeric membrane proteins suggests the use of an excess of nanodisc components (i.e. lipid and MSP) compared to the target receptor in the reconstitution mixture. A subsequent purification step is therefore required to separate membrane protein-nanodiscs from empty-nanodiscs for their characterization (see below). This can be achieved by exploiting an affinity tag of the membrane protein such as a histidine tag for immobilized metal affinity chromatography (IMAC) [59], a FLAG epitope for Flag M1 anti-FLAG immunoaffinity chromatography [19], or the 1D4 tag using 1D4 resin [61]. Alternatively, size-exclusion chromatography has been utilized for the enrichment of membrane protein-nanodiscs [17]. In the case of NTS1f-nanodiscs, we found a two-step titration experiment helpful for determining the optimal conditions for NTS1f-nanodisc formation in the presence of excess nanodisc components. The first titration is carried out to establish the correct phospholipid to MSP ratio for empty-nanodisc formation. The prepared samples were analyzed by size exclusion chromatography [Superdex 200 HR 10/30 (GE Healthcare)] to assess the nanodisc quality. In the case of empty-nanodiscs prepared with MSP1D1 and lipid (POPC, a mixture of POPC and POPG, or POPG), a 55:1 molar ratio of lipid to MSP1D1 resulted in well-defined nanodiscs displaying a symmetrical size exclusion chromatography peak profile (data not shown). Based on these results, a second phospholipid/MSP titration experiment is then carried out using a fixed amount of NTS1f (20% of the total reconstitution volume). Due to the excess of phospholipid and MSP compared to the receptor, a large excess of empty-nanodiscs is found in the product. A subsequent purification step was therefore employed to separate the NTS1f-nanodiscs from empty-nanodiscs exploiting a histidine tag at the receptor C-terminus for IMAC [59]. We found that IMAC-purified NTS1f-nanodiscs were not amenable to analysis by size exclusion chromatography, unlike nanodiscs containing other membrane proteins [15, 18, 19, 64]. We therefore used DLS to directly assess for NTS1f-nanodisc formation, and propose its use as a universal method for the analysis of nanodiscs independent of their behavior in size exclusion chromatography (Section 3). This approach allowed for the identification of optimal reconstitution conditions using lipid:MSP1D1 and MSP1D1:NTS1f molar ratios of 30:1 and 100:~4, respectively, in the initial reaction setup [59].
2.6 Nanodisc reconstitution with NTS1
2.6.1 Expression and purification of NTS1
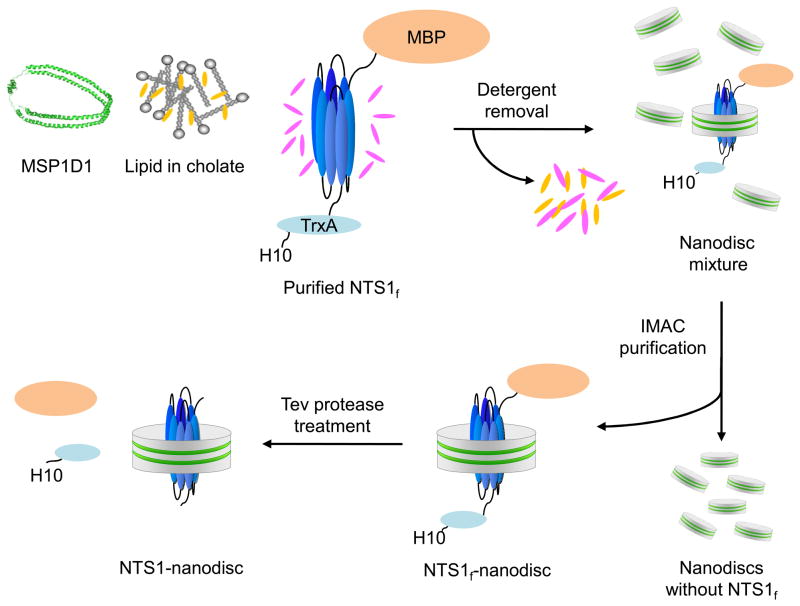
NTS1 was expressed in E. coli as a fusion protein (NTS1f) containing the E. coli maltose-binding protein (MBP), followed by a tobacco etch virus (Tev) protease recognition site, the rat NTS1, a second Tev protease recognition site, the E. coli thioredoxin (TrxA), and a decahistidine (H10) tag. The expression and purification of NTS1f was performed as described [65] with modifications. Briefly, one hundred gram of cells were homogenized in 200 ml of 2x solubilization buffer (100 mM Tris pH 7.4, 60% (v/v) glycerol, 400 mM NaCl, 10 mM MgCl2) in a chilled Waring blender for 2 minutes. All subsequent steps were carried out at 4°C or on ice. Following the addition of 200 μl of a DNase I stock (Sigma P/N D-4527, 10 mg/ml), 400 μl of 1.4 mg/ml Pepstatin A, 400 μl of 1 mg/ml Leupeptin, and 400 μl of 0.1 M AEBSF, 40 ml of each detergent stock solution {6% (w/v) CHAPS, 1.2% (w/v) CHS and 10% DDM} were added dropwise with gentle stirring. The mixture was sonicated for 13 minutes (Misonix sonicator 3000, 1/2 inch flat tip, level 4, 1 s on, 2 s off). After a subsequent addition of 400 μl of each protease inhibitor, the volume was adjusted to 400 ml with cold water. Cell debris was then removed by centrifugation (Beckman 45Ti rotor, 45,000 rpm, 1 h, 4ºC). Imidazole (2 M stock solution, adjusted to pH 7.4) was added to the supernatant to a final concentration of 50 mM. The sample was passed through a 0.2 μm filter (Stericup, Millipore) and loaded onto a Ni-NTA column (5 ml bed volume, Qiagen) equilibrated with buffer A [50 mM Tris pH 7.4, 30% (v/v) glycerol, 200 mM NaCl, 50 mM imidazole, 0.5% (w/v) CHAPS, 0.1% (w/v) CHS, 0.1% (w/v) DDM]. The resin was washed with 10 column volumes of buffer A and the NTS1f protein was eluted with 10 column volumes of buffer B [50 mM Tris pH 7.4, 30% (v/v) glycerol, 200 mM NaCl, 200 mM imidazole, 0.5% (w/v) CHAPS, 0.1% (w/v) CHS, 0.1% (w/v) DDM]. The protein content was measured as described [65] typically resulting in 9.5 mg of purified NTS1f.
2.6.2 Preparation of MSP
The expression and purification of MSP1D1 is hereby described as an example for the preparation of an MSP. The expression plasmid pMSP1D1 (#20061, Addgene), a derivative of pET28a, codes for the deletion mutant (Δ1–54) of human apolipoprotein A-I preceded by an N-terminal hepta-histidine (H7) tag, a spacer sequence and Tev protease recognition site (H7-MSP1D1). Expression was carried out as described in [14] with modifications. BL21Gold(DE3) cells (Stratagene) harboring pMSP1D1 were grown at 37°C to an OD600 of 4 in double strength YT (2xYT) medium containing 50 μg/ml kanamycin. Following induction with 0.3 mM isopropyl-β-D-thiogalactopyranoside, the temperature was decreased to 28°C. The cells were harvested 4 hours later, frozen in liquid nitrogen and stored at −80°C until further use. The weight of the wet cell pellet was usually 6.5–7.0 g per 1 L culture.
As the MSP may aggregate within the cell when expressed at very high levels, the standard purification method incorporates Triton X-100 and cholate to help improve its solubility [14]. Because Triton X-100 denatures NTS1 [44], we purified MSP in the absence of any detergent [59]. Briefly, cells (10 g) were resuspended in 60 ml of buffer C (50 mM Tris pH 7.4, 200 mM NaCl), and then passed twice through a French Press. Cell debris was removed by centrifugation (Beckman 70Ti rotor, 55,000 rpm, 45 minutes, 4ºC) and imidazole was added to the supernatant to a final concentration of 25 mM. The sample was passed through a 0.2 μm filter (Stericup, Millipore) and loaded onto a Ni-NTA column (8 ml bed volume, Qiagen), equilibrated with buffer D (buffer C containing 25 mM imidazole). The resin was washed with buffer D and H7-MSP1D1 was eluted with buffer E (buffer C containing 280 mM imidazole). Fractions containing H7-MSP1D1 were identified by SDS-PAGE (NuPAGE 4–12% Bis-Tris gel, 1×MES running buffer, Invitrogen) and a NanoDrop 1000 spectrophotometer (version 3.6.0, Thermo Scientific). Pooled fractions containing H7-MSP1D1 were concentrated to 9 mg/ml using a Centriprep YM-10 device (Millipore) and the imidazole removed by passage over a PD10 column (GE Healthcare) equilibrated with buffer C. The concentration of purified H7-MSP1D1 was ~6.5 mg/ml. This was frozen in liquid nitrogen and stored at −80°C until further use. The protein concentration was determined by measuring the absorbance at 280 nm using a calculated extinction coefficient of 21,430 M−1cm−1 and a calculated molar mass of 24,793 Da (http://web.expasy.org/protparam/). The protein content was also determined by the method of Schaffner and Weissmann [66] using bovine serum albumin as the standard.
Prior to reconstitution, the H7-tag was removed by incubation with Tev protease (150:1 M/M H7-MSP1D1/Tev protease, in the absence of DTT) at room temperature for 1.5 hours and subsequent incubation with Talon resin (1 ml resin per 5 mg of H7-MSP1D1, equilibrated in buffer C, Clontech Laboratories) for 1 hour at 4ºC. MSP1D1 was recovered from the flow-through and the subsequent wash with buffer F (buffer C containing 20 mM imidazole), concentrated in the presence of 2.5 mM cholate to 10–15 mg/ml using an Amicon Ultra-0.5 Ultracel-30 concentrator (Millipore) and used for reconstitution. The protein concentration was determined by measuring the absorbance at 280 nm using a calculated extinction coefficient of 18,450 M−1cm−1 and a calculated molar mass of 22,044 Da.
2.6.3 Reconstitution and purification of NTS1-nanodiscs
To observe the effects of lipid head group charges on the signaling properties of NTS1, nanodiscs were prepared using zwitterionic POPC, negatively charged POPG, or a mixture of POPC/POPG at a 1:1 molar ratio. All reconstitution experiments were conducted at 4°C or on ice to preserve NTS1 activity, requiring the use of POPC and POPG, both of which have phase transition temperatures below 0°C. Although POPG is a minor phospholipid in eukaryotic membranes, we did not utilize the more abundant POPS because of its higher transition temperature of 14°C. A detergent solubilized lipid solution was prepared as follows: chloroform dissolved phospholipid was dried in a vacuum desiccator overnight protected from light. Dried phospholipids were solubilized in buffer C containing 50 mM cholate (the molar ratio of phospholipid to cholate was 3:1), sonicated for 20–30 minutes at 4°C and then stored on ice. The phospholipid/cholate solution was freshly prepared for each reconstitution experiment.
Purified NTS1f was concentrated to a volume of ~200 μl on an Amicon Ultra-0.5 Ultracel-30 concentrator (14,000 rpm, 30 min, 4°C, Eppendorf centrifuge). The phospholipid/cholate solution, ice-cold buffer C and concentrated NTS1f were combined, and concentrated MSP1D1 was then added such that the final concentrations of the components were 3 mM lipids, 3–4 μM NTS1 fusion protein, and 100 μM MSP1D1 in a total volume of 1 ml (Figure 2.1). The mixture was incubated for 1 hour at 4°C and the detergent removed by addition of 35-fold the weight in detergent of Bio-Beads. After mixing gently for 2 hours at 4°C, the Bio-Beads were removed by centrifugation (10,000 rpm, 3 min, 4°C, Eppendorf centrifuge). This procedure generated a mixture of NTS1f-nanodiscs and nanodiscs devoid of receptor. To enrich for the former, the mixture was incubated in batch for 1 hour at 4°C with 0.3 ml Talon resin (Clontech Laboratories), equilibrated with buffer F, transferred into a small column and washed with 4 bed volumes of buffer F. NTS1f-nanodiscs were eluted from the resin in 0.3 ml steps using buffer G (buffer C with 200 mM imidazole). The fractions were analyzed for protein by measuring the absorbance at 280 nm with a NanoDrop 1000 spectrophotometer using a calculated extinction coefficient of 176,185 M−1cm−1 and a calculated combined molar mass of 144.3 kDa, assuming the presence of 2 MSP1D1 and 1 NTS1f per nanodisc. The protein content was also determined by the method of Schaffner and Weissmann [66] with bovine serum albumin as the standard. The fraction with the highest NTS1f-nanodisc concentration was used for further experiments. These NTS1f-nanodiscs were characterized by DLS and AUC as is. Prior to their pharmacological characterization, the NTS1f-nanodiscs were treated with Tev protease to remove the N-terminal MBP and C-terminal TrxA-H10 tail, thus generating NTS1 with near authentic N- and C-termini. For this, purified NTS1f-nanodiscs were incubated with a stoichiometric amount of Tev protease (in the absence of DTT) for 2 hours on ice; the completeness of the digest was confirmed by SDS-PAGE.
Figure 2.1.
Preparation of NTS1-nanodiscs. The purified NTS1f, MSP and detergent-solubilized phospholipid are mixed, and then detergent is removed by the addition of Bio-beads. During detergent removal, nanodisc self-assembly is initiated. After completion of the detergent removal, nanodiscs without NTS1f were separated from NTS1f-nanodiscs by immobilized metal affinity chromatography exploiting the H10 tail of NTS1f. Purified NTS1f-nanodiscs were treated with Tev protease prior to pharmacological studies. Figure taken from [59].
For the preparation of empty-nanodiscs (nanodiscs devoid of receptor), the respective lipid/cholate solution, concentrated MSP1D1, and buffer C were mixed to a final concentration of 5.5 mM lipid and 100 μM MSP1D1 in a volume of 250 μl. The mixture was incubated for 1 hour at 4°C and Bio-Beads were added for detergent removal to 35-fold of the weight of cholate present in the mixture. After mixing for 2 hours at 4°C, the Bio-Beads were removed by centrifugation (10,000 rpm, 3 min, 4°C, Eppendorf centrifuge).
3. Biophysical characterization
The study of membrane proteins embedded in a nanodisc requires the characterization of the nanodisc size and component stoichiometry, with particular emphasis on the number of target membrane proteins per nanodisc. For example, the functional activity of NTS1 is dramatically different for its monomeric and dimeric forms when analyzed in detergent solution [67], and therefore a similar behavior is expected for NTS1 inserted into nanodiscs. Several methods have been described in the literature to determine the dimensions of nanodiscs. These include size exclusion chromatography, small angle x-ray scattering, scanning probe microscopy, and electron microscopy [15, 47, 68]. Lipid stoichiometries have been determined by scintillation counting of nanodiscs prepared using tritiated phospholipids [47], 1H-NMR [59], and HPLC combined with tandem quadrupole mass spectrometry (HPLC/MS/MS) [59]. Density gradient centrifugation monitoring the absorbance at 500 nm [15] and circular dichroism [16] have also been used to analyze the oligomeric states of rhodopsin and bacteriorhodopsin, respectively. The membrane protein stoichiometry in nanodiscs has also been determined by the use of FRET or single-molecule imaging techniques using Cy3- and Cy5-modified membrane proteins [18, 19]. Although such techniques provide useful information, methods that involve membrane protein labeling may lead to membrane protein properties that differ from that of an unlabeled protein. Rhodopsin [69] and bacteriorhodopsin [68] are notable exceptions in this respect as the spectroscopic properties of their natural retinal chromophores provide a convenient measure for their quantification.
Here, we present approaches for membrane proteins without chromophores that do not require any modification or labeling. These methods, namely DLS and AUC, report on the total nanodisc preparation and enable for a characterization of the nanodisc of interest. Even though DLS is ideally suited for the characterization of monodisperse preparations, we show that it is also useful for the analysis of polydisperse systems. As DLS measurements are rapid and sensitive to the presence of larger aggregates, we have found this technique to be extremely helpful in determining the appropriate protein to lipid stoichiometry in the preparation of empty- and receptor-nanodiscs. Sedimentation velocity (SV) affords a much better hydrodynamic resolution, even distinguishing between nanodiscs based on their lipid composition. SV does not just result in an improved characterization of the typically paucidisperse nanodisc preparation, the method also allows for an estimate of the shape and molar mass of the resolved species. Furthermore, by collecting both absorbance and interference sedimentation data, we show the analysis further allows for a determination of the nanodisc protein to lipid ratio. Combined with sedimentation equilibrium, these techniques provide non-destructive solution methods for the characterization of nanodisc preparations. In this manner, we were able to characterize receptor nanodisc preparations by DLS and use the recovered sample for downstream functional studies described in Section 4. Due to the practical advantages afforded by these hydrodynamic methods for the characterization of nanodisc preparations, we first present a brief overview of the methods, prior to discussing their application to the nanodiscs studied.
3.1 Dynamic light scattering
DLS analyzes the fluctuations in the intensity of scattered light arising from the Brownian motion and diffusion of the particles of interest [70, 71]. It is a fairly straightforward technique allowing for the rapid characterization of both receptor nanodiscs and nanodiscs devoid of receptors [59, 72, 73]. The rapid characterization has been particularly useful in helping us optimize the lipid, MSP and receptor ratios required for nanodisc preparation. A successful implementation of DLS requires the preparation of dust-free samples, proper sample temperature equilibration and an understanding of the appropriate mathematical to model the experimental autocorrelation function. Even though the data collection software implements a dust filter by monitoring the photon count rate, dust-free samples should be prepared. This is readily done by briefly spinning the sample at 14,000 × g in a table top centrifuge or filtration, followed by transfer and handling in a dust-free hood. As the nanodisc samples have been spun to remove the Bio-Beads or filtered through an affinity Talon resin, we found the samples to be dust-free. To minimize further contamination, all handling was carried out using plastic pipette tips that have been rinsed with the appropriate filtered dust-free buffer.
Interpretation of the experimental autocorrelation function, which reports on all of the macromolecular species present in solution, requires implementation of the appropriate analysis method. Analysis is usually carried out in real time during data collection, thus providing important feedback on the quality of the autocorrelation function. Typically, data analysis tests all possible models, ranging from the simplest single exponential function describing a monodisperse species (equation 3.3); to stretched exponential functions describing a monomodal, paucidisperse and non-interacting distribution of species (equation 3.8) and a multimodal polydisperse distribution (equation 3.6).
In DLS the scattered laser light from a small volume of solution is collected and correlated with itself over time scales τ ranging from 100 ns to 100 ms, so as to include the time-scale at which Brownian motion occurs. The temporal fluctuations in intensity I(t) determined experimentally are used to generate a normalized second order or intensity (photon count) autocorrelation function g(2)(τ):
| Equation 3.1 |
This is related to the first order or electric field E(t) electric field autocorrelation function g(1)(τ) by the Siegert equation:
| Equation 3.2 |
where β is an instrumental correction factor and B a baseline correction. In the case of a monodisperse solution of non-interacting nanodiscs, g(1)(τ) is related to the decay constant Γ and nanodisc translational diffusion coefficient D:
| Equation 3.3 |
where q represents the scattering vector:
| Equation 3.4 |
and n0 the buffer refractive index, θ the scattering angle and λ0 the laser wavelength. The Stokes-Einstein equation relates the diffusion coefficient to the hydrodynamic radius Rh, thus linking the experimental autocorrelation function to the effective particle size:
| Equation 3.5 |
where k is the Boltzmann constant, T the absolute temperature and η the buffer viscosity. In the case of polydisperse systems, typically observed for most of our nanodisc preparations, the autocorrelation function becomes an intensity-weighted G(Γ) combination of contributions from movements of the individual particles:
| Equation 3.6 |
The determination of Γ or its distribution, more frequently in terms of a distribution of hydrodynamic radii, represents the commonest challenge in DLS as very few systems behave as the monodisperse system described by equation 3.3. Typically, the simplest model that fits the autocorrelation function for the receptor nanodiscs is a monomodal, cumulant expansion [74–76] that includes a second moment, polydispersity term. Rearranging equations 3.2 and 3.3, it can be shown that the intensity and field autocorrelation functions for a monodisperse solution of non-interacting nanodiscs are related as follows:
| Equation 3.7 |
In the method of cumulants, the integral of exponential functions defining the field autocorrelation function (equation 3.6) is rigorously expanded in terms of a power series such that [75]:
| Equation 3.8 |
in which Γ̄ = q2 Dz now represents the average decay rate and z-averaged translational diffusion coefficient Dz, in the case of Rayleigh scatterers. The second moment μ2 is the intensity-weighted variance of the diffusion coefficient distribution and this is better understood in terms of the relative variance or polydispersity term, μ2/Γ̄2 as it provides a measure of the spread in the size distribution. Samples are narrowly distributed or essentially monodisperse when the polydispersity is less than 0.05. We have observed good data fits with polydispersity values in the range of 0.08 to 0.3, noting that these became worse as the polydispersity parameter increased beyond 0.3. Even though previous preparations of nanodiscs devoid of receptor were found to be essentially paucidisperse by sedimentation velocity (SV), the light scattering autocorrelation functions clearly indicated the presence of two distributions of species [59] representing the paucidisperse nanodiscs observed by SV and a significantly larger lipid-protein aggregate. Much like SV, DLS reports on all of the species present in solution – even though the larger lipid-protein aggregate is expected to have a significantly larger sedimentation coefficient and perhaps sediment before the maximal rotor speed is attained, a more likely explanation is that this aggregate is present at very low concentrations. This follows as the intensity of scattered light I(θ) is proportional to McP(θ), where M is the molar mass, c the mass concentration and P(θ) the scattering form factor, and thus highly biased towards contributions from larger species. Using the Rayleigh approximation it can be shown that the intensity of scattered light scales as the sixth power of the particle diameter. Experimental data for these nanodiscs devoid of receptor were best modeled in terms of contributions from two noninteracting species [59]:
| Equation 3.9 |
The autocorrelation functions can also be analyzed in terms of a distribution of particles, not necessarily centered on a mean value as in the case of the cumulant expansion (equation 3.8). We have utilized both a regularized CONTIN [77–79] implemented as part of the Brookhaven Instruments Corporation 9KDLSW 2.12 data collection and analysis package, as well as a regularized intensity based continuous I(Rh) distribution implemented in SEDFIT 13.0d [80] based on an inverse Laplace transform of equation 3.6. As discussed below, these methods do not provide the same hydrodynamic resolution afforded by SV. Furthermore, it is important to note that in all cases the data analysis and interpretation assumes a non-interacting distribution of particles.
Experimental data collection was performed at room temperature using a Brookhaven Instruments Corporation BI-200 goniometer and light scattering system coupled to a BI-9000 AT autocorrelator and Lexel Model 95 argon ion laser operating in the TEM00 mode at 514.5 nm. In a typical experiment we accumulate a series of autocorrelation functions for 2 – 4 minutes at a laser intensity (100 – 200 mW) and photomultiplier tube aperture (100 or 200 μm) such that the experimental count rate ranges from 100 to 400 kHz. The stability of the count rate is monitored during the experiment and the use of a software dust filter is implemented. To ensure that high quality data are analyzed, data having small differences between the calculated (obtained from the autocorrelation function) and measured (obtained from four time points beyond the autocorrelation time range) baselines are chosen. Ideally the difference should be much less than 0.1%. Autocorrelation functions are collected at 90.0° using sampling times of 1.0 μs to 5 ms with receptor nanodisc concentrations in the range of 4 – 9 μM; higher concentrations in the range of 30 – 50 μM are used for nanodiscs devoid of receptors. Data analysis is carried out in real time using the Brookhaven Instruments 9KDLSW 2.12 software package and subsequently in SEDFIT 13.0d.
3.2 Analytical ultracentrifugation
We have also utilized AUC to characterize the various nanodisc preparations. SV has proven to be particularly useful as it provides a greatly improved hydrodynamic resolution when compared to DLS, even though the method is not as rapid. In addition, by simultaneously collecting absorbance and interference data, SV allows for an estimate of the protein to lipid ratio for each of the resolved sedimenting species. A description of the method, experimental setup and strategies for data analysis has been the subject of many reviews and published works, and is not the primary goal of this work (see [81–84] and references therein). As most nanodisc preparations consist of a pauci- or polydisperse mixture of non-interacting species, we found the regularized continuous c(s) distribution analysis implemented in SEDFIT to be the most appropriate model for the description of SV data [85]. In this model, data are analyzed in terms of a discretized set of Lamm equation solutions spanning a defined range of sedimentation coefficients, s and a diffusion coefficient, D(s) that scales with the best-fit weight-averaged frictional ratio f/f0. A few important features of the c(s) distribution are highlighted – one is that the distribution is not very sensitive to f/f0, another is the fact that a defined integral of the distribution returns the loading concentration of material spanning the sedimentation coefficients of interest [85, 86]. Furthermore, discrete species observed in the distribution can be characterized by their weight-averaged sedimentation coefficient sw, loading concentration c and molar mass M(s), obtained by use of the Svedberg equation:
| Equation 3.10 |
where R is the gas constant, T the absolute temperature, ν̄ the nanodisc partial specific volume and ρ the buffer density. As the nanodisc of interest was usually the predominant species, we also analyzed the sedimentation data in terms of a hybrid model incorporating Lamm equation solutions for the discrete species of interest and c(s) distributions describing larger and smaller, such that the diffusion envelope for the discrete species does not overlap with the c(s) distribution. This model, implemented in SEDPHAT 10.41, provides a weight average f/f0 independent estimate of the molar mass [87].
SV experiments were conducted at 10.0°C on a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge. Nanodisc preparations, typically at concentrations that provide a measured absorbance at 280 nm between 0.5 and 1.5, are loaded into prechilled 2-channel centerpiece cells. Following temperature equilibration at 0 krpm under vacuum, SV data are collected at 40 krpm using both the absorbance (280 nm) and interference optical systems. Data analysis requires both the solution densities ρ and solution viscosities η and these were measured experimentally [59]. Another important parameter required for the proper estimate of the molar mass is the nanodisc partial specific volume. We initially calculated this based on estimated partial specific volumes ν̄ for the protein and lipid components. Protein partial specific volumes were calculated at 10.0°C in SEDNTERP 1.09 [88], whereas the partial specific volume for POPC was based on experimentally published values for ν̄ [89] and dν̄/dT [90, 91]. In the absence of published data for POPG, we initially assumed that the partial specific volume was identical to that of POPC. In this manner we estimated the partial specific volume for the nanodisc ν̄nanodisc:
| Equation 3.11 |
where M and ν̄ represent the molar masses and partial specific volumes of the various components, and n, x and y represent the expected NTS1f, POPC and POPG stoichiometries, respectively.
In the course of SV experiments we observed that the sedimentation coefficients for the POPC, 50% POPC/50% POPG and POPG empty-nanodiscs were significantly different [59]. We attributed this to differences in the lipid molar mass and possibly their partial specific volume. As the nanodiscs devoid of receptor were reasonably monodisperse, we were able to investigate this matter further by sedimentation equilibrium (SE). Unlike SV, SE is carried out at lower rotor speeds such that the transport forces of sedimentation and diffusion balance each other (see [81, 82, 92] and references therein). At equilibrium a steady concentration gradient c(r) is formed across the centrifuge cell such that:
| Equation 3.12 |
where r is the radial position, r0 a reference radial position and ω the angular rotor speed. In this manner SE allows for a measure of the buoyant molar mass M(1 − ν̄ρ) and thus a measure of ν̄ in cases where the molar mass is known or assumed. Whereas SV experiments are complete within 18 hours, SE requires at least two days for equilibrium. Furthermore, to improve the quality of the data analysis as well as report on all of the possible species present in solution, we routinely collect multi-speed equilibrium data further lengthening the time required for the experiment. The long-term stability of the nanodiscs was a matter of concern; however we found that preparations of the MSP1D1 stabilized nanodiscs devoid of receptor were stable in SE experiments. In addition, based on essentially identical SV c(s) profiles, we founds that empty MSP1D1 nanodiscs prepared using POPC were stable for more than a month when stored at 4°C (data not shown).
SE experiments were conducted at 10.0°C on a Beckman Optima XL-A analytical ultracentrifuge. Empty-nanodisc preparations at a single loading concentration corresponding to loading A280 of 0.3 – 0.4 were loaded into prechilled 6-channel centerpiece cells. Absorbance (280 nm) data were collected at rotor speeds 10, 15 and 20 krpm and analyzed globally in SEDPHAT 10.41 in terms of a single ideal noninteracting species, following preprocessing in SEDFIT 13.0d (sorting, splitting and assembly of experiment files) [93, 94]. Soft mass conservation was implemented and the value of the best-fit buoyant molar mass M (1 − ν̄ρ) was determined by setting the partial specific volume ν̄ to zero.
To determine the protein to lipid ratio from the SV data, we required values for the absorbance extinction coefficient ε280 and interference molar signal increment εJ. Protein extinction coefficients were calculated in SEDNTERP 1.09 and we assumed that it was only the protein that contributed to the absorbance signal. Interference signal increments were determined using refractive increment dn/dc values of 0.185 cm3g−1 for the proteins [95] and 0.164 cm3g−1 for the lipids [96]:
| Equation 3.13 |
where λ is the wavelength in cm. Both the absorbance extinction coefficients and interference signal increments were assumed to scale linearly with protein and lipid composition:
| Equation 3.14 |
| Equation 3.15 |
In this manner, the integrated c(s) absorbance signal for the nanodisc species of interest was used to determine the MSP1D1 and NTS1f concentrations. Their corresponding contribution to the interference signal was then used, by difference, to determine the extent of the lipid contributions. We have subsequently refined these calculations by estimating the values of dn/dc for MSP1D1 and NTS1f based on their amino acid composition using the calculator function in SEDFIT 13.0d [97].
3.3 Hydrodynamic characterization of nanodiscs
The primary goal for the biophysical characterization of the nanodiscs was to determine the NTS1f stoichiometry in the reconstituted receptor nanodiscs. Ultimately we developed a set of methods that allow for a more extensive characterization of the nanodisc preparations addressing the issues of sample polydispersity and receptor, MSP and lipid stoichiometry for the hydrodynamically resolved single nanodiscs. The characterization of the sample polydispersity was particularly important as we found that the NTS1f-nanodiscs, in contrast to empty-nanodiscs, were not amenable to purification by size exclusion chromatography.
3.3.1 Nanodiscs devoid of receptor (empty-nanodiscs)
We combined SV and DLS to characterize empty-nanodiscs reconstituted using MSP1D1 and various lipids. SV experiments indicated the presence of a major species at 2.9 – 3.7 S, depending on the lipid composition [59] (Table 3.1). In all cases this species represented ~90% of the loading absorbance. These observations are in broad agreement with DLS studies indicating the presence of a major species having a hydrodynamic radius of ~5 nm (Table 3.1), along with contaminating quantities of a larger aggregate having a hydrodynamic radius larger than 100 nm [59]. The sedimentation coefficient of the empty-nanodiscs appears to increase with increasing POPG content, reflecting in part the increased molecular mass of this lipid or, possibly a smaller partial specific volume for POPG, although we initially assumed that POPC and POPG have identical partial specific volumes. Even though the best-fit frictional ratios f/fo of 1.2 – 1.3 are consistent with the globular shape of the empty-nanodiscs, we found that the best-fit molar masses are larger than expected. Much like the sedimentation coefficient, the overestimate in the molar mass appeared to correlate with the POPG content [59]. To address this issue, we have reconstituted a new set of empty-nanodiscs utilizing a H7-MSP1D1 and characterized each preparation by DLS, SV and SE. Unlike earlier reconstitutes, DLS data for all preparations were best-fit using a cumulant analysis resulting in z-averaged hydrodynamic radii of 5.3 – 5.4 nm and polydispersity indices of 0.19 – 0.22 (Figure 3.1, Table 3.1). As in the case of the MSP1D1 nanodiscs, a SV analysis indicates the presence of a major species at 3.3 – 4.1 S representing ~90% of the loading absorbance (Figure 3.1, Table 3.1). The remainder of the absorbing material consisted of ~0.4, 0.6, 0.8 MDa and larger aggregates, contributing to the polydispersity of 0.2 observed by DLS. As above, the sedimentation coefficients (Table 3.1) and molar mass estimates (data not shown), based on the best-fit frictional ratios f/fo of 1.1 – 1.2, increase with increasing POPG content. We therefore we analyzed these preparations by SE to determine the buoyant molar masses of the monomeric empty-nanodiscs.
Table 3.1.
Hydrodynamic properties of empty- and NTS1f-nanodiscs.
| Nanodisc | DLS | SV | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Rh,1 (nm)a | s20,w (S), f/fob | M (kDa)b | % load, c (μM)c | Lipidd (εJ/ε280) | Rh (nm)e | |
| Empty-MSP1D1 | ||||||
| POPC | 4.7 | 2.88 ± 0.01 (1.25) | 158 ± 8 (127.696) | 91 (20) | 114 | 4.2 |
| POPC/POPG | 5.2 | 3.21± 0.01 (1.23) | 169 ± 8 (128.297) | 88 (17) | 139 | 3.9 |
| POPG | 5.1 ± 0.3 | 3.64 ± 0.02 (1.20) | 185 ± 12 (128.897) | 91 (20) | 102 | 3.6 |
| Empty H7-MSP1D1 | ||||||
| POPC | 5.4 | 3.29 ± 0.01 (1.20) | 160 ± 11 (133.194) | 87 (16) | 113 | 4.1 |
| POPC/POPG | 5.3 | 3.60 ± 0.01 (1.16) | 166 ± 12 (133.794) | 89 (20) | 107 | 3.9 |
| POPG | 5.4 | 4.05 ± 0.01 (1.08) | 169 ± 14 (134.394) | 87 (19) | 95 | 3.6 |
|
|
||||||
| Average | 112 ± 15 | |||||
|
|
||||||
| NTS1f MSP1D1 | ||||||
| POPC | 6.2 ± 0.5 | 6.50 ± 0.02 (1.40) | 205 ± 8 (189.893) | 81 (3.7) | 63 | 5.4 |
| POPC/POPG | 6.4 ± 0.5 | 6.66 ± 0.02 (1.40) | 210 ± 8 (190.221) | 79 (3.6) | 105 | 5.3 |
| POPG | 6.6 ± 0.8 | 6.84 ± 0.02 (1.34) | 205 ± 6 (190.548) | 64 (2.7) | 52 | 5.2 |
|
|
||||||
| Average | 73 ± 23 | |||||
|
|
||||||
Data were analyzed in terms of two discrete species (empty-MSP1D1 nanodiscs) or a second moment cumulant (empty-H7-MSP1D1 nanodiscs, NTS1f-nanodiscs). The hydrodynamic radii (Rh,1) values were calculated from the averages of independent experiments (± SD) (empty-nanodiscs with MSP1D1: POPC, n=1; POPC/POPG, n=2; POPG, n=2; empty-nanodiscs with H7-MSP1D1: POPC, n=1; POPC/POPG, n=1; POPG, n=1; NTS1f-nanodiscs: POPC, n=3; POPC/POPG, n=2; POPG, n=2). In addition to monodisperse species, aggregates (Rh,2 > 100 nm) were found in empty-nanodisc MSP1D1 preparations.
Sedimentation coefficients (s20,w) and experimental molar masses (M) represent average values for the major species observed in the continuous c(s) distribution in a single experiment for each nanodisc preparation. The frictional ratios f/fo indicated in brackets represent an average of the best-fit values obtained from the absorbance and interference continuous c(s) distributions. Averages in s20,w and M were obtained from both the absorbance and interference data analyzed using a continuous c(s) distribution in SEDFIT, as well as various hybrid local continuous distributions and global discrete species models in SEDPHAT. Sedimentation coefficient corrections to s20,w are carried out using revised values of the partial specific volumes at 10.0 and 20.0°C, as are estimates of the molar mass. Molar masses in brackets are the calculated values based on the expected stoichiometries of 110:2 lipid:MSP1D1 and 60:2:1 lipid:MSP1D1:NTS1f for the empty and receptor nanodiscs, respectively.
Percent of the loading absorbance that represents the major species of interest, based on the best-fit continuous c(s) distribution observed with SEDFIT. The observed concentration c, based on the absorbance data, of the major nanodisc species is indicated in brackets.
Lipid stoichiometries per single nanodisc based on the presence of 2 MSP1D1 molecules, and in the case of the receptor-nanodiscs, 1 NTS1f. Stoichiometries are based on signal contributions of the major species to the absorbance (protein alone) and interference (protein and lipid) data.
Hydrodynamic radii based on the observed sedimentation coefficient, calculated molar masses and experimental partial specific volumes obtained from the sedimentation equilibrium data.
Figure 3.1.
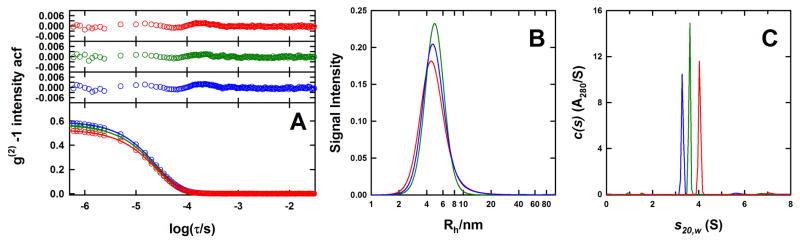
Hydrodynamic properties of empty-nanodiscs. (A) DLS autocorrelation functions for empty-H7-MSP1D1 nanodiscs reconstituted with POPC (blue), POPC/POPG (green) and POPG (red) modeled in terms of the presence of a paucidisperse distribution of species using a cumulant analysis shown together with the corresponding residuals. (B) Regularized intensity based continuous I(Rh) distributions obtained using SEDFIT for the same samples described in (A). Light scattering data were collected at 20°C. (C) SV c(s) distributions obtained in SEDFIT for the same samples analyzed in (A) and (B). Sedimentation data were collected at 40 krpm and 10.0°C.
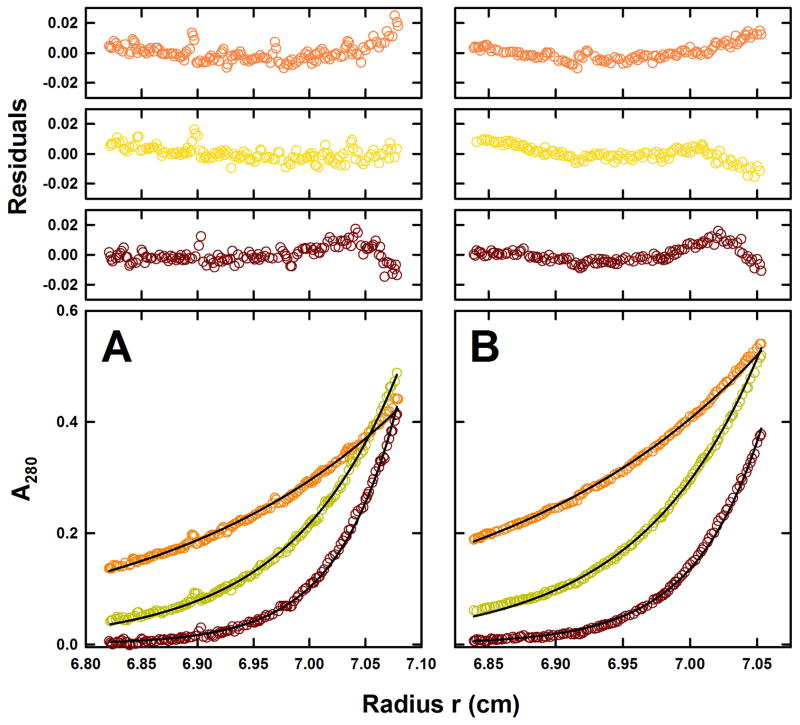
SE experiments on POPC and POPG containing nanodiscs were consistent with the presence of a single species at the rotor speeds studied, returning buoyant molar masses of 13.9 ± 0.4 and 15.1 ± 0.5 kDa, respectively (Table 3.2, Figure 3.2). Whereas the derived partial specific volume for the POPC nanodiscs is, within the error of the method, identical to that calculated, this is not the case for the POPG nanodiscs (Table 3.2). Based on these observations we determine partial specific volumes of 0.981 and 0.968 cm3g−1 for the POPC and POPG lipids, respectively. The smaller partial specific volume observed for POPG is in agreement with the observations made by SV, and differences in the buoyant molar mass account for most of the differences in the sedimentation coefficient. As these lipids have different surface charge properties, differences in the partial specific volume are currently attributed to possible variation in their degree of surface hydration and counterion binding which contribute to the effective partial specific volume [98]. Counterion binding effects are expected to be more pronounced for POPG which is negatively charged. We have therefore re-evaluated previous [59] and current SV data for both empty and receptor nanodiscs utilizing these experimental partial specific volumes (Table 3.1). In this manner we found that molar mass estimates, which depend critically on use of the correct partial specific volume [85], are now slightly closer to the calculated values (see [59] and Table 3.1). These studies illustrate the usefulness of SV in which the hydrodynamic resolution can not only distinguish between monomeric nanodiscs and small amounts of aggregate, but also distinguish between the POPC, POPC/POPG and POPG containing nanodiscs. These observations also confirm the mixed lipid composition for the nanodiscs assembled in the presence of equimolar amounts of POPC and POPG.
Table 3.2.
Sedimentation properties of empty-nanodiscs.
| Empty-nanodisc | SE | |||
|---|---|---|---|---|
|
| ||||
| M (kDa)a | M (1 − ν̄ρ) (kDa)b | Calculated ν̄ (cm3/g)c | Experimental ν̄ (cm3/g) | |
| POPC | 133.194 | 13.9 ± 0.4 | 0.8899 | 0.886 ± 0.003 |
| POPC/POPG | 133.794 | --d | 0.8904 | 0.883d |
| POPG | 134.394 | 15.1 ± 0.5 | 0.8908 | 0.879 ± 0.004 |
Expected molar mass for H7-MSP1D1 reconstituted empty-nanodiscs containing 110 lipid molecules.
Experimental buoyant molar mass for the monomeric empty-nanodisc. SE data collected at 10, 15 and 20 krpm for a single loading concentration were analyzed globally, using soft mass conservation, in terms of a single ideal species. The analysis and error determination were carried out without the implementation of TI noise.
Calculated assuming that all lipids have a partial specific volume of 0.987 cm3g−1 based on published data for POPC as described in the text [59, 89–91].
No data were collected for the POPC/POPG nanodiscs. The experimental partial specific volume was calculated based on data presented for the POPC only and POPG only containing empty-nanodiscs.
Figure 3.2.
Sedimentation equilibrium studies of empty-nanodiscs. SE profiles for (A) H7-MSP1D1/POPC and (B) H7-MSP1D1/POPG empty-nanodiscs at 10.0°C plotted as a distribution of the absorbance at 280 nm versus radius at equilibrium. Data were collected at 10 (orange), 15 (yellow) and 20 (brown) krpm and loading concentrations of (A) 5.8 and (B) 8.2 μM. The solid lines show the best-fit analysis in terms of a single non-interacting species, returning a buoyant molar mass consistent with the presence of single nanodiscs. The corresponding residuals for this best-fit are show in the plots above.
Analogous to methods developed for the study of membrane proteins in the presence of solubilizing detergents [99, 100], we have combined the absorbance and interference signal intensities of the major species to determine the lipid to protein content (Table 3.1). Briefly, the absorbance signal for the species of interest, obtained from an integration of the c(s) distribution, was used to determine MSP concentration. The corresponding contribution to the interference signal is then used to determine the lipid concentration. Stoichiometries of 91 to 139 lipid molecules per empty-nanodisc are noted (Table 3.1). The variation in these values more than likely reflects the variability in sample preparation and error in the method, as the average of 112 ± 15 lipid molecules per empty-nanodisc corresponds to the expected lipid stoichiometry [49]. We note that this method provides an indirect confirmation of the nanodisc molar mass that is independent of the partial specific volume. SV experiments also indicate that no free detergent is present in the empty-nanodisc preparations, confirming that the optimized amount of Bio-beads SM2 lead to essentially complete detergent removal.
3.3.2 Nanodisc with receptor (NTS1f- nanodiscs)
We have also characterized the reconstituted receptor-nanodisc complexes utilizing both SV and DLS. SV experiments showed the presence of a major species at 6.2 – 6.9 S, representing 50 – 80% of the loading absorbance (Table 3.1), along with aggregates presumed to represent dimeric NTS1f-nanodisc complexes, as reported for SecYEG-nanodisc preparations [17]. DLS experiments indicate that the amount of aggregate present appears to be preparation dependent – for example, three separate preparations of NTS1f-MSP1D1-POPC nanodiscs lead to best-fit (z-averaged) hydrodynamic radii of 5.7, 6.3 and 6.7 nm. Importantly, the larger radii correlate with an increased polydispersity, reflecting the increasing contributions from dimers and higher order species. Even though, DLS does not resolve the contributions from the various species, SV does, allowing us to characterize the monomeric receptor-nanodisc complex. As in the case of the empty-nanodiscs, we find that the sedimentation coefficient for the major species increases with POPG content, indicating the correct incorporation of the expected lipids. Furthermore, the now revised best-fit molar masses are still slightly larger than expected, however the differences are not as large as observed for the empty-nanodiscs, due in part to the decreased lipid contribution. The best-fit frictional ratios f/fo of 1.3 – 1.4 are identical to those reported for the SecYEG receptor nanodisc complex [17] and also consistent with the overall globular shape of the receptor nanodisc. An analysis of the absorbance and interference signal intensities for the major species leads to the expected lipid stoichiometry of 50 to 70 molecules per receptor nanodisc. It is important to note that both the observed sedimentation coefficient and signal intensity based lipid stoichiometry confirm the presence of one NTS1f molecule and two MSP1D1 molecules per receptor nanodisc. As described in [59], simple volume and shape calculations return an expected sedimentation coefficient of approximately 6.45 S for a receptor nanodisc having a stoichiometry of 60:2:1 lipd:MSP1D1:NTS1f. Furthermore, if we assume the presence of two NTS1f per nanodisc, the absorbance and interference signal intensities result in an unlikely lipid content that far exceeds that of the empty-nanodisc [59].
3.3.3 Discussion
We have combined SV and DLS to characterize preparations of empty- and NTS1f-nanodiscs. In addition to providing parameters such as the sedimentation coefficient and Stokes’ radius, these methods have allowed us to determine the lipid stoichiometry and establish the presence of a single NTS1f per nanodisc. We have thus shown that the empty-nanodisc preparations are essentially monodisperse with the expected stoichiometry of ~110 lipid molecules and 2 MSP1D1 molecules. Even though molar mass estimates obtained from the Lamm equation modeling of the sedimentation data are slightly larger than expected, we note that small errors in the partial specific volume will lead to larger mass changes as the lipids have very small buoyant molecular mass contributions with (1 − ν̄ρ) ~ 0.01. Although our biophysical studies show that the NTS1f-nanodisc preparations are polydisperse, with a polydispersity that appears to be batch specific, SV experiments allowed us to resolve the monomer species and show that it has the expected stoichiometry of approximately 60 molecules per 2 MSP1D1 and 1 NTS1f. Furthermore, based on the hydrodynamic resolution of SV experiments, we can confirm that the approximately 60 lipid molecules in the POPC/POPG receptor nanodisc correspond to approximately 30 POPC and 30 POPG species.
3.4. Determination of the lipid stoichiometry
The lipid composition for the NTS1f-nanodiscs analyzed by SV was in broad agreement with lipid analyses by 1H-NMR and HPLC/MS/MS [59]. Unlike SV, both these methods require nanodisc disassembly and lipid dissolution, i.e. these methods do not analyze the native nanodisc. In our hands, 1H-NMR underestimated the lipid compositions due to lipid losses during the last step of sample preparation: nanodiscs were freeze-dried, dissolved in deuterated chloroform, and then filtered through a glass wool filter to remove chloroform-insoluble salt precipitations, which resulted in sample losses. In terms of the characterization of the native nanodisc, the results derived from SV are a better reflection of the lipid content of the native nanodiscs compared to 1H-NMR and HPLC/MS/MS procedures because no additional manipulation of the nanodisc was required.
4. Functional studies of the membrane protein in nanodisc
Nanodiscs are ideal tools to investigate the biological and signaling properties of integral membrane proteins in a lipid environment under strictly defined conditions. A particularly powerful aspect of the nanodisc technology is that it can be used to study the protein of interest in a known monomeric or oligomeric state, which is more difficult to achieve in liposomes or detergents [11]. In addition, ligands binding from the extracellular side to the membrane protein, and molecules, interacting with the intracellular receptor face, have simultaneous access and can thus be studied in conjunction. Lastly, the influence of lipid on the biological properties can be interrogated because the lipid environment surrounding the membrane protein can be manipulated with high precision.
Membranes are the physiological environment for membrane proteins containing many different types of lipids in relative proportions that depend on the cell type and cellular compartments. Lipid components directly impact the function of membrane proteins, and changes in membrane lipid composition have been associated with age-related diseases. Hence nanodiscs provide the unique opportunity to probe the effect of particular lipid molecules on the properties of membrane proteins.
In the following, we discuss the effect of lipid head group charges as an example of how lipids modulate the signal transduction properties of NTS1 [59]. NTS1 belongs to the rhodopsin family of GPCRs and is predominantly coupled to the Gq heterotrimeric protein. As described earlier, NTS1 with near-authentic N- and C-termini was embedded into nanodiscs containing the zwitterionic lipid POPC, or the negatively charged lipid POPG, or 50% POPC/50% POPG, with ~65 lipid molecules per nanodisc, likely providing one layer around NTS1. This experimental setup thus allowed studying the relevance of lipids for signaling in the immediate vicinity of NTS1. Binding of the agonist neurotensin to NTS1 occurred with similar affinities and was essentially unaffected by the phospholipid composition, as shown by [3H]NT saturation binding and homologous competition experiments in the absence of G protein, and by dose-response experiments in the presence of the G protein (Figure 4.1, Table 4.1). In stark contrast, Gq protein coupling to NTS1 in various lipid nanodiscs was significantly different, and the apparent affinity of Gαq and Gβ1γ1 to activated NTS1 increased with increasing POPG content (Figure 4.1, Table 4.1). Variation of lipids in the immediate vicinity of NTS1 may therefore provide a mechanism to regulate cell-specific NTS1 signaling by specific interactions between lipid and receptor residues, and/or local net charges at the NTS1/lipid interface. Furthermore, these studies demonstrated that nanodiscs provide an environment in which receptors can adopt conformations that appear identical to signaling-competent receptors located in plasma membranes, as demonstrated for NTS1 by ligand binding and G protein activation. It has also been documented that the interaction of arrestin with rhodopsin, reconstituted into nanodiscs, was enhanced in the presence of phospholipids with acidic head groups [101]. Interestingly, phosphorylation of rhodopsin by GRK1 was not dependent on lipid charge [20].
Figure 4.1.
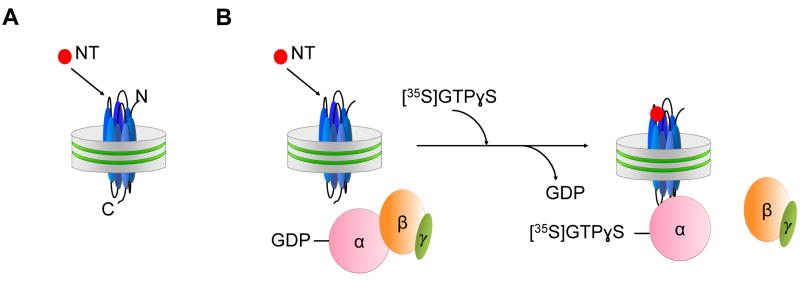
Assays for assessing the effect of lipid charges on NTS1 signaling. NTS1 with near-authentic N- and C-termini was embedded into nanodiscs containing the zwitterionic lipid POPC, or the negatively charged lipid POPG, or 50% POPC/50% POPG. (A) The agonist binding was investigated by radio-ligand binding assays using the agonist [3H]NT in the absence of G protein (saturation binding and homologous competition assays). (B) NTS1 catalyzed nucleotide exchange was analyzed by GDP/[35S]GTPγS exchange reactions in the presence of Gαq-Gβ1γ1. The cartoon does not imply mechanistic details of the G protein activation process.
Table 4.1.
Pharmacological properties of NTS1 in nanodiscs. (see also Figure 4.1)
| POPC | POPC/POPG | POPG | |
|---|---|---|---|
| Neurotensin binding | |||
| Kd (nM) | 1.04 ± 0.46 (n=3) | 0.24 ± 0.03 (n=2) | 0.31 ± 0.11 (n=3) |
| IC50 (nM) | 3.46 ± 0.07 (n=4) | 2.34 ± 0.36 (n=4) | 2.66 ± 0.24 (n=3) |
| EC50 (nM) | 4.92 ± 1.33 (n=3) | 4.54 ± 0.94 (n=3) | 3.05 ± 0.41 (n=3) |
| G protein activation | |||
| Km Gαq (μM) | n.d. (n=2) | > 5 (n=3) | 0.55 ± 0.07 (n=3) |
| Km Gβ1γ1 (μM) | n.d. (n=2) | 8.2 ± 4.3 (n=4) | 2.40 ± 0.12 (n=4) |
| Normalized data {catalyzed [35S]GTPγS bound (cpm) (nM NTS1 protein)−1 min−1} | |||
| Gαq saturation | 21 ± 6 (n=2) | 656 ± 166 (n=3) | 3496 ± 50 (n=3) |
| Gβ1γ1 saturation | 10 ± 4 (n=2) | 280 ± 174 (n=4) | 2784 ± 170 (n=4) |
| NT saturation | 45 ± 30 (n=3) | 2177 ± 548 (n=3) | 7392 ± 1288 (n=3) |
Values are given ± SD. Kd, equilibrium dissociation constant (saturation binding); IC50, half maximal inhibitory concentration (homologous competition); EC50, half maximal effective concentration (dose-response experiment, NT saturation of Gαq activation); Km, binding constants for Gαq and Gβ1γ1 (Gαq and Gβ1γ1 saturation); normalized data from Gαq, Gβ1γ1 and NT saturation experiments given as catalyzed [35S]GTPγS bound (cpm) (nM NTS1 protein)−1 min−1; n, number of independent experiments; n.d., not determined. For details see [59]. Data taken from [59].
The breadth and versatility of the nanodisc technology is reflected by the diverse nature of the membrane proteins studied and the multitude of biological assays performed (Table 2.1). For example, the availability of defined GPCR monomers in nanodiscs has allowed studying details of allosteric modulation of agonist binding by G protein, basal activation, and ligand efficacy, and it was shown that monomeric receptors are the minimal functional unit in regard to G protein activation [18, 19, 59, 61, 69, 102]. Furthermore, monomeric active rhodopsin was phosphorylated by rhodopsin kinase GRK1 as efficiently as rhodopsin in the native bovine rod outer segment disc membrane [20] and was sufficient for arrestin-1 binding [20, 101]. The signaling properties of GPCR dimers have so far only been studied using rhodopsin. Although the presence of two rhodopsin molecules per nanodisc was unambiguously established, conclusions about mechanistic details of G protein activation by rhodopsin dimers have been complicated because such nanodisc preparations contained a mixture of physiologically relevant parallel dimers and non-physiological antiparallel dimers [15, 61]. The translocon, a membrane-embedded protein assembly that catalyzes protein movement across membranes, has been probed for its interaction with its cytosolic partner, the SecA dimer. It was shown that the SecYEG complex triggers dissociation of the SecA dimer and associates only with the SecA monomer [17]. Trapping the bacterial chemoreceptor Tar as dimer or trimer of dimers in nanodiscs suggested that the formation of the dimer was sufficient for signal transduction across the membrane, but that trimers of dimers were required for downstream signaling [53].
5. Conclusion
Methods and techniques have been developed to study membrane proteins for understanding biological processes. Although membrane proteins have been studied in vitro using detergent and/or lipid-based systems, providing useful information, the native phospholipid bilayer is better suited to understand their physiological function and structure. We have demonstrated the production and biophysical characterization of membrane proteins embedded into nanodiscs; though we have focused on NTS1 the method is applicable to a variety of membrane proteins. For a successful nanodisc reconstitution it is crucial to consider the size of the transmembrane region of the target membrane protein, the MSP length, the type of phospholipid and detergents used for the purification of the membrane protein and the solubilization of phospholipid. Furthermore, a biophysical characterization is necessary to confirm the purity of the nanodisc sample, the stoichiometry of each component, and the phospholipid environment of the membrane protein. We have demonstrated that DLS and AUC are particularly well suited for this as they allow us to observe the native biophysical properties and provide information of the stoichiometry in addition to the sedimentation coefficient and Stokes’ radius. As the membrane protein embedded in a nanodisc gains the stability and functional activity provided by native environments, it is well suited for biochemical and physiological studies. In this manner the nanodisc technology addresses a critical gap in our understanding of properties of membrane protein in a native environment.
Highlights.
The neurotensin receptor NTS1 was reconstituted into nanodiscs.
A biophysical characterization of nanodiscs was carried out without sample modification using a combination of dynamic light scattering and analytical ultracentrifugation.
The influence of lipid charge on the signaling properties of NTS1 was investigated.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health (SI, RGr: National Institute of Neurological Disorders and Stroke; RGh: National Institute of Diabetes and Digestive and Kidney Diseases).
Abbreviations
- 2xYT
double strength YT medium
- AUC
analytical ultracentrifugation
- CHAPS
3-[(3-cholamidopyropyl)dimethylammonio]-1-propanesulfonate
- CHS
cholesteryl hemisuccinate Tris salt
- DDM
n-dodecyl-β-D-maltoside
- DLS
dynamic light scattering
- GPCR
G protein-coupled receptor
- H10
decahistidine
- H7-MSP1D1
MSP1D1 with an N-terminal heptahistidine tag
- HPLC/MS/MS
HPLC combined with tandem quadrupole mass spectrometry
- IMAC
immobilized metal affinity chromatography
- MBP
maltose-binding protein
- MSP
membrane scaffold protein
- NT
neurotensin
- NTS1
neurotensin receptor 1
- NTS1f
NTS1 expressed as a fusion protein
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- SE
sedimentation equilibrium
- SV
sedimentation velocity
- Tev
tobacco etch virus
- Tm
phase transition temperature
- TrxA
E. coli thioredoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sayaka Inagaki, Email: inagakisn@ninds.nih.gov.
Rodolfo Ghirlando, Email: rodolfo.ghirlando@nih.gov.
Reinhard Grisshammer, Email: rkgriss@helix.nih.gov.
References
- 1.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 2.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4.Insel PA, Snead A, Murray F, Zhang L, Yokouchi H, Katakia T, Kwon O, Dimucci D, Wilderman A. GPCR expression in tissues and cells: are the optimal receptors being used as drug targets? Br J Pharmacol. 2012;165:1613–1616. doi: 10.1111/j.1476-5381.2011.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee AG. Biological membranes: the importance of molecular detail. Trends Biochem Sci. 2011;36:493–500. doi: 10.1016/j.tibs.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Opekarova M, Tanner W. Specific lipid requirements of membrane proteins--a putative bottleneck in heterologous expression. Biochim Biophys Acta. 2003;1610:11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 7.Alvarez E, Ruiz-Gutiérrez V, Sobrino F, Santa-María C. Age-related changes in membrane lipid composition, fluidity and respiratory burst in rat peritoneal neutrophils. Clin Exp Immunol. 2001;124:95–102. doi: 10.1046/j.1365-2249.2001.01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabelo N, Martín V, Santpere G, Marín R, Torrent L, Ferrer I, Díaz M. Severe alterations in lipid composition of frontal cortex lipid rafts from Parkinson’s disease and incidental Parkinson’s disease. Mol Med. 2011;17:1107–1118. doi: 10.2119/molmed.2011.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim Biophys Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spector AA, Yorek MA. Membrane lipid composition and cellular function. J Lipid Res. 1985;26:1015–1035. [PubMed] [Google Scholar]
- 11.Serebryany E, Zhu GA, Yan EC. Artificial membrane-like environments for in vitro studies of purified G-protein coupled receptors. Biochim Biophys Acta. 2012;1818:225–233. doi: 10.1016/j.bbamem.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 12.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Baas BJ, Denisov IG, Sligar SG. Homotropic cooperativity of monomeric cytochrome P450 3A4 in a nanoscale native bilayer environment. Arch Biochem Biophys. 2004;430:218–228. doi: 10.1016/j.abb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayburt TH, Leitz AJ, Xie G, Oprian DD, Sligar SG. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J Biol Chem. 2007;282:14875–14881. doi: 10.1074/jbc.M701433200. [DOI] [PubMed] [Google Scholar]
- 16.Bayburt TH, Grinkova YV, Sligar SG. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuszak AJ, Pitchiaya S, Anand JP, Mosberg HI, Walter NG, Sunahara RK. Purification and functional reconstitution of monomeric mu-opioid receptors: allosteric modulation of agonist binding by Gi2. J Biol Chem. 2009;284:26732–26741. doi: 10.1074/jbc.M109.026922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, Gurevich VV. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem. 2011;286:1420–1428. doi: 10.1074/jbc.M110.151043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glück JM, Koenig BW, Willbold D. Nanodiscs allow the use of integral membrane proteins as analytes in surface plasmon resonance studies. Anal Biochem. 2011;408:46–52. doi: 10.1016/j.ab.2010.08.028. [DOI] [PubMed] [Google Scholar]
- 22.Shi L, Shen QT, Kiel A, Wang J, Wang HW, Melia TJ, Rothman JE, Pincet F. SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carraway R, Leeman SE. The isolation of a new hypotensive peptide, neurotensin, from bovine hypothalami. J Biol Chem. 1973;248:6854–6861. [PubMed] [Google Scholar]
- 24.Leeman SE, Carraway RE. Neurotensin: discovery, isolation, characterization, synthesis and possible physiological roles. Ann N Y Acad Sci. 1982;400:1–16. doi: 10.1111/j.1749-6632.1982.tb31557.x. [DOI] [PubMed] [Google Scholar]
- 25.Clineschmidt BV, McGuffin JC, Bunting PB. Neurotensin: antinocisponsive action in rodents. Eur J Pharmacol. 1979;54:129–139. doi: 10.1016/0014-2999(79)90415-1. [DOI] [PubMed] [Google Scholar]
- 26.Bissette G, Nemeroff CB, Loosen PT, Prange AJ, Jr, Lipton MA. Hypothermia and intolerance to cold induced by intracisternal administration of the hypothalamic peptide neurotensin. Nature. 1976;262:607–609. doi: 10.1038/262607a0. [DOI] [PubMed] [Google Scholar]
- 27.Binder EB, Kinkead B, Owens MJ, Nemeroff CB. Neurotensin and dopamine interactions. Pharmacol Rev. 2001;53:453–486. [PubMed] [Google Scholar]
- 28.Carraway RE, Plona AM. Involvement of neurotensin in cancer growth: evidence, mechanisms and development of diagnostic tools. Peptides. 2006;27:2445–2460. doi: 10.1016/j.peptides.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 29.Schimpff RM, Avard C, Fénelon G, Lhiaubet AM, Tennezé L, Vidailhet M, Rostène W. Increased plasma neurotensin concentrations in patients with Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2001;70:784–786. doi: 10.1136/jnnp.70.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griebel G, Holsboer F. Neuropeptide receptor ligands as drugs for psychiatric diseases: the end of the beginning? Nat Rev Drug Discov. 2012;11:462–478. doi: 10.1038/nrd3702. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka K, Masu M, Nakanishi S. Structure and functional expression of the cloned rat neurotensin receptor. Neuron. 1990;4:847–854. doi: 10.1016/0896-6273(90)90137-5. [DOI] [PubMed] [Google Scholar]
- 32.Kitabgi P. Targeting neurotensin receptors with agonists and antagonists for therapeutic purposes. Curr Opin Drug Discov Devel. 2002;5:764–776. [PubMed] [Google Scholar]
- 33.Jastrzebska B, Debinski A, Filipek S, Palczewski K. Role of membrane integrity on G protein-coupled receptors: Rhodopsin stability and function. Prog Lipid Res. 2011;50:267–277. doi: 10.1016/j.plipres.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 35.Bubis J. Effect of detergents and lipids on transducin photoactivation by rhodopsin. Biol Res. 1998;31:59–71. [PubMed] [Google Scholar]
- 36.Gawrisch K, Soubias O, Mihailescu M. Insights from biophysical studies on the role of polyunsaturated fatty acids for function of G-protein coupled membrane receptors. Prostaglandins Leukot Essent Fatty Acids. 2008;79:131–134. doi: 10.1016/j.plefa.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jastrzebska B, Goc A, Golczak M, Palczewski K. Phospholipids are needed for the proper formation, stability, and function of the photoactivated rhodopsin-transducin complex. Biochemistry. 2009;48:5159–5170. doi: 10.1021/bi900284x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaya AI, Thaker TM, Preininger AM, Iverson TM, Hamm HE. Coupling efficiency of rhodopsin and transducin in bicelles. Biochemistry. 2011;50:3193–3203. doi: 10.1021/bi200037j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Botelho AV, Martinez GV, Brown MF. Electrostatic properties of membrane lipids coupled to metarhodopsin II formation in visual transduction. J Am Chem Soc. 2002;124:7690–7701. doi: 10.1021/ja0200488. [DOI] [PubMed] [Google Scholar]
- 40.Midgett CR, Madden DR. Breaking the bottleneck: eukaryotic membrane protein expression for high-resolution structural studies. J Struct Biol. 2007;160:265–274. doi: 10.1016/j.jsb.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 41.McCusker EC, Bane SE, O’Malley MA, Robinson AS. Heterologous GPCR expression: a bottleneck to obtaining crystal structures. Biotechnol Prog. 2007;23:540–547. doi: 10.1021/bp060349b. [DOI] [PubMed] [Google Scholar]
- 42.Laage R, Langosch D. Strategies for prokaryotic expression of eukaryotic membrane proteins. Traffic. 2001;2:99–104. doi: 10.1034/j.1600-0854.2001.020204.x. [DOI] [PubMed] [Google Scholar]
- 43.Frommer WB, Ninnemann O. Heterologous expression of genes in bacterial, fungal, animal, and plant cells. Annu Rev Plant Physiol Plant Molec Biol. 1995;46:419–444. [Google Scholar]
- 44.Tucker J, Grisshammer R. Purification of a rat neurotensin receptor expressed in Escherichia coli. Biochem J. 1996;317:891–899. doi: 10.1042/bj3170891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grisshammer R, Tate CG. Overexpression of integral membrane proteins for structural studies. Q Rev Biophys. 1995;28:315–422. doi: 10.1017/s0033583500003504. [DOI] [PubMed] [Google Scholar]
- 46.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 47.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002;2:853–856. [Google Scholar]
- 48.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 49.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 51.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vance DE, Vance JE. Lipoproteins And Membranes. 5. Elsevier; New York, NY: 2008. Biochemistry of Lipids. [Google Scholar]
- 53.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis PJ, Fleming BD, Coolbear KP, Keough KM. Gel to liquid-crystalline transition temperatures of water dispersions of two pairs of positional isomers of unsaturated mixed-acid phosphatidylcholines. Biochemistry. 1981;20:3633–3636. doi: 10.1021/bi00515a051. [DOI] [PubMed] [Google Scholar]
- 55.Boggs JM, Tümmler B. Interdigitated gel phase bilayers formed by unsaturated synthetic and bacterial glycerolipids in the presence of polymyxin B and glycerol. Biochim Biophys Acta. 1993;1145:42–50. doi: 10.1016/0005-2736(93)90379-e. [DOI] [PubMed] [Google Scholar]
- 56.Mattai J, Hauser H, Demel RA, Shipley GG. Interactions of metal ions with phosphatidylserine bilayer membranes: effect of hydrocarbon chain unsaturation. Biochemistry. 1989;28:2322–2330. doi: 10.1021/bi00431a051. [DOI] [PubMed] [Google Scholar]
- 57.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J Phys Chem B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 59.Inagaki S, Ghirlando R, White JF, Gvozdenovic-Jeremic J, Northup JK, Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J Mol Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalal K, Duong F. Reconstitution of the SecY translocon in nanodiscs. Methods Mol Biol. 2010;619:145–156. doi: 10.1007/978-1-60327-412-8_9. [DOI] [PubMed] [Google Scholar]
- 61.Banerjee S, Huber T, Sakmar TP. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 62.Nath A, Grinkova YV, Sligar SG, Atkins WM. Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs: the effect of model membranes. J Biol Chem. 2007;282:28309–28320. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- 63.Rigaud JL, Levy D, Mosser G, Lambert O. Detergent removal by non-polar polystyrene beads. Eur Biophys J. 1998;27:305–319. [Google Scholar]
- 64.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J Biol Chem. 2007;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 65.White JF, Trinh LB, Shiloach J, Grisshammer R. Automated large-scale purification of a G protein-coupled receptor for neurotensin. FEBS Lett. 2004;564:289–293. doi: 10.1016/S0014-5793(04)00195-4. [DOI] [PubMed] [Google Scholar]
- 66.Schaffner W, Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 67.White JF, Grodnitzky J, Louis JM, Trinh LB, Shiloach J, Gutierrez J, Northup JK, Grisshammer R. Dimerization of the class A G protein-coupled neurotensin receptor NTS1 alters G protein interaction. Proc Natl Acad Sci U S A. 2007;104:12199–12204. doi: 10.1073/pnas.0705312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bayburt TH, Sligar SG. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whorton MR, Jastrzebska B, Park PS, Fotiadis D, Engel A, Palczewski K, Sunahara RK. Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J Biol Chem. 2008;283:4387–4394. doi: 10.1074/jbc.M703346200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berne BJ, Pecora R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics. Wiley-Interscience; New York: 1976. [Google Scholar]
- 71.Chu B. Laser Light Scattering: Basic Principles and Practice. Academic press; New York: 1991. [Google Scholar]
- 72.Hernández-Rocamora VM, Reija B, García C, Natale P, Alfonso C, Minton AP, Zorrilla S, Rivas G, Vicente M. Dynamic Interaction of the Escherichia coli Cell Division ZipA and FtsZ Proteins Evidenced in Nanodiscs. J Biol Chem. 2012;287:30097–30104. doi: 10.1074/jbc.M112.388959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaw AW, Pureza VS, Sligar SG, Morrissey JH. The local phospholipid environment modulates the activation of blood clotting. J Biol Chem. 2007;282:6556–6563. doi: 10.1074/jbc.M607973200. [DOI] [PubMed] [Google Scholar]
- 74.Koppel DE. Analysis of Macromolecular Polydispersity in Intensity Correlation Spectroscopy: The Method of Cumulants. The Journal of Chemical Physics. 1972;57:4814–4820. [Google Scholar]
- 75.Pusey PN, Koppel DE, Schaefer DW, Camerini-Otero RD, Koenig SH. Intensity fluctuation spectroscopy of laser light scattered by solutions of spherical viruses: R17, Q beta, BSV, PM2, and T7. I. Light-scattering technique. Biochemistry. 1974;13:960–970. doi: 10.1021/bi00702a020. [DOI] [PubMed] [Google Scholar]
- 76.Frisken BJ. Revisiting the method of cumulants for the analysis of dynamic light-scattering data. Appl Opt. 2001;40:4087–4091. doi: 10.1364/ao.40.004087. [DOI] [PubMed] [Google Scholar]
- 77.Provencher SW. Inverse problems in polymer characterization: Direct analysis of polydispersity with photon correlation spectroscopy. Die Makromolekulare Chemie. 1979;180:201–209. [Google Scholar]
- 78.Provencher SW. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Computer Physics Communications. 1982;27:213–227. [Google Scholar]
- 79.Provencher SW. CONTIN: A general purpose constrained regularization program for inverting noisy linear algebraic and integral equations. Computer Physics Communications. 1982;27:229–242. [Google Scholar]
- 80.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. The latest version of the program can be obtained from the following address: http://www.analyticalultracentrifugation.com/sedfit.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laue T. Biophysical studies by ultracentrifugation. Curr Opin Struct Biol. 2001;11:579–583. doi: 10.1016/s0959-440x(00)00250-5. [DOI] [PubMed] [Google Scholar]
- 83.Balbo A, Schuck P. In: Protein-Protein Interactions: A Molecular Cloning Manual. 2. Golemis EA, Adams PD, editors. Chapter 14 Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2005. [Google Scholar]
- 84.Brown PH, Balbo A, Schuck P. Characterizing protein-protein interactions by sedimentation velocity analytical ultracentrifugation. Curr Protoc Immunol. 2008;Chapter 18(Unit 18):15. doi: 10.1002/0471142735.im1815s81. [DOI] [PubMed] [Google Scholar]
- 85.Dam J, Schuck P. Calculating sedimentation coefficient distributions by direct modeling of sedimentation velocity concentration profiles. Methods Enzymol. 2004;384:185–212. doi: 10.1016/S0076-6879(04)84012-6. [DOI] [PubMed] [Google Scholar]
- 86.Dam J, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys J. 2005;89:651–666. doi: 10.1529/biophysj.105.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. The latest version of the program can be obtained from the following address: http://www.analyticalultracentrifugation.com/sedphat/index.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayes DB, Laue T, Philo J. 2008 at http://www.jphilo.mailway.com.. Also see Cole JL, Lary JW, PMT, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4.
- 89.Greenwood AI, Tristram-Nagle S, Nagle JF. Partial molecular volumes of lipids and cholesterol. Chem Phys Lipids. 2006;143:1–10. doi: 10.1016/j.chemphyslip.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagle JF, Wilkinson DA. Lecithin bilayers. Density measurement and molecular interactions. Biophys J. 1978;23:159–175. doi: 10.1016/S0006-3495(78)85441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tristram-Nagle S, Petrache HI, Nagle JF. Structure and interactions of fully hydrated dioleoylphosphatidylcholine bilayers. Biophys J. 1998;75:917–925. doi: 10.1016/S0006-3495(98)77580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Howlett GJ, Minton AP, Rivas G. Analytical ultracentrifugation for the study of protein association and assembly. Curr Opin Chem Biol. 2006;10:430–436. doi: 10.1016/j.cbpa.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 93.Balbo A, Brown PH, Braswell EH, Schuck P. Measuring protein-protein interactions by equilibrium sedimentation. Curr Protoc Immunol. 2007;Chapter 18(Unit 18.18) doi: 10.1002/0471142735.im1808s79. [DOI] [PubMed] [Google Scholar]
- 94.Ghirlando R. The analysis of macromolecular interactions by sedimentation equilibrium. Methods. 2011;54:145–156. doi: 10.1016/j.ymeth.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Padrick SB, Deka RK, Chuang JL, Wynn RM, Chuang DT, Norgard MV, Rosen MK, Brautigam CA. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal Biochem. 2010;407:89–8103. doi: 10.1016/j.ab.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stora T, Dienes Z, Vogel H, Duschl C. Histidine-Tagged Amphiphiles for the Reversible Formation of Lipid Bilayer Aggregates on Chelator-Functionalized Gold Surfaces. Langmuir. 2000;16:5471–5478. [Google Scholar]
- 97.Zhao H, Brown PH, Schuck P. On the distribution of protein refractive index increments. Biophys J. 2011;100:2309–2317. doi: 10.1016/j.bpj.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Casassa EF, Eisenberg H. Thermodynamic analysis of multicomponent solutions. Adv Protein Chem. 1964;19:287–395. doi: 10.1016/s0065-3233(08)60191-6. [DOI] [PubMed] [Google Scholar]
- 99.Nury H, Manon F, Arnou B, le Maire M, Pebay-Peyroula E, Ebel C. Mitochondrial bovine ADP/ATP carrier in detergent is predominantly monomeric but also forms multimeric species. Biochemistry. 2008;47:12319–12331. doi: 10.1021/bi801053m. [DOI] [PubMed] [Google Scholar]
- 100.le Maire M, Arnou B, Olesen C, Georgin D, Ebel C, Moller JV. Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat Protoc. 2008;3:1782–1795. doi: 10.1038/nprot.2008.177. [DOI] [PubMed] [Google Scholar]
- 101.Tsukamoto H, Sinha A, DeWitt M, Farrens DL. Monomeric rhodopsin is the minimal functional unit required for arrestin binding. J Mol Biol. 2010;399:501–511. doi: 10.1016/j.jmb.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yao XJ, Vélez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, Deupi X, Sunahara RK, Kobilka B. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:9501–9506. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borch J, Hamann T. The nanodisc: a novel tool for membrane protein studies. Biol Chem. 2009;390:805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- 104.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]