Abstract
Proteorhodopsins are an extensive family of photoactive membrane proteins found in proteobacteria distributed throughout the world’s oceans which are often classified as green- or blue-absorbing (GPR and BPR, respectively) on the basis of their visible absorption maxima. GPR and BPR have significantly different properties including photocycle lifetimes and wavelength dependence on pH. Previous studies revealed that these different properties are correlated with a single residue, Leu105 in GPR and Gln105 in BPR, although the molecular basis for the different properties of GPR and BPR has not yet been elucidated. We have studied the unexcited states of GPR and BPR using resonance Raman spectroscopy which enhances almost exclusively chromophore vibrations. We find that both spectra are remarkably similar, indicating that the retinylidene structure of GPR and BPR are almost identical. However, the frequency of a band assigned to the retinal C13-methyl-rock vibration is shifted from 1006 cm−1 in GPR to 1012 cm−1 in BPR. A similar shift is observed in the GPR mutant L105Q indicating Leu and Gln residues interact differently with the retinal C13-methyl group. The environment of the Schiff base of GPR and BPR differ as indicated by differences in the H/D induced down-shift of the Schiff base vibration. Residues located in transmembrane helices (D–G) do not contribute to the observed differences in the protein–chromophore interaction between BPR and GPR based on the Raman spectra of chimeras. These results support a model whereby the substitution of the hydrophilic Gln105 in BPR with the smaller hydrophobic Leu105 in GPR directly alters the environment of both the retinal C13 group and the Schiff base.
1. Introduction
Proteorhodopsins are a recently discovered class of photoactive proteins found primarily in proteobacteria which reside in all the world’s oceans.1-5 Harvesting sunlight, they use the absorbed energy to pump protons across the cell membrane or transduce a photon signal.1,6 Due to their ubiquity, proteor-hodopsins are thought to play a major role in solar energy transduction in the biosphere.2 Since the initial discovery in Monterey Bay, shotgun genetic techniques have identified over 4000 variants spread across every ocean and also in freshwater.3-5,7 Most variants can be divided into two main classes depending on their properties: green-absorbing proteorhodopsin (GPRs),1,8,9 and blue-absorbing proteorhodopsin (BPRs).2,10
GPR, the earliest discovered proteorhodopsin in SAR-86 γ-proteobacteria, has been the most extensively studied. GPR exists in shallow ocean depths, pumps protons with a fast photocycle (~20 ms) and has been shown to provide power for the cell under anaerobic conditions.11 BPR which is typically found near the edge of the photic zone (75 m) and in the open ocean away from the shore,5 has a blue-shifted visible absorption near 490 nm and slower photocycle (~200 ms) relative to GPR. These differences have been attributed to natural Darwinian selection given the lower intensity and blue shift of available sunlight at 75 m below the surface.2-12
It has been found that the substitution of only one residue in GPR (L105Q) switches the visible absorption and photocycle kinetics to the BPR phenotype.10,13 Likewise the reverse mutation in BPR (Q105L) produces a red-shifted absorption maximum similar to GPR, but does not accelerate the photocycle.
In order to better understand the molecular basis for the above effects, we used resonance Raman spectroscopy (RRS) to compare chromophore structure and protein–chromophore interactions of GPR and BPR. RRS has been used previously to study other rhodopsins including mammalian visual rhodopsin,14 bacteriorhodopsin,15-20 sensory rhodopsin I and II,21-23 halorhodopsin,24 and Anabaena sensory rhodopsin.25,26 Due to resonant enhancement of vibrations of the retinal chromophore when the excitation wavelength is near its visible absorption wavelength, retinal bands dominate the Raman spectra of rhodopsins, even when the excitation wavelength is far from the chromophore absorption maximum wavelength, such as in the case of BR. With a 1060 nm excitation and 570 nm visible absorption maximum, preresonance conditions still lead to significant enhancement of vibrational bands of the chromophore relative to protein bands.27 Thus, the use of near-IR 785 nm excitation allows us to probe the vibrational spectrum of the retinal chromophore without significant interference from protein bands or photocycle intermediates that appear due to driving the photocycle with visible light.
In this study, Raman spectroscopy with 785 nm excitation was used to investigate the chromophore structure and protein-chromophore interaction of residue 105 in GPR and BPR. It is found that the chromophore structures of both variants of proteorhodopsin are remarkably similar. However, differences are detected which are attributable to changes in the interaction of residue 105 near the C13-methyl and Schiff base groups of retinal which may account for most of the differences in the properties of BPR and GPR.
2. Methods and Materials
Protein Expression and Purification
Escherichia coli cells containing the GPR (Monterey Bay strain EBAC 31A8) or BPR (Hawaii Ocean Time strain HOT75M4) were cultured on standard Luria Broth Media (Sigma Aldrich) and supplied with either all-trans retinal or all-trans retinal containing a deuterium on carbon 15 (15C–2 H). All procedures for the site-directed mutagenesis, plasmid construction and expression in the E. coli UT5600 strain were identical to those described previously.8 Two chimeric constructs were made by PCR using the megaprimer method: B3G4 encodes the N-terminal portion of BPR up to Leu128 followed by the C-terminal portion of GPR. Conversely, G3B4 encodes the N-terminal portion of GPR up to Leu128 followed by the C-terminal portion of BPR. The nomenclature indicates the numbers of transmembrane helices from BPR and GPR that comprise the proteins encoded.
After the induction period, the cells expressing His-tagged wild-type or mutant PRs were centrifuged at 1000g, resuspended in 5 mM MgCl2, 150 mM Tris-HCl, pH 7.0, and disrupted by sonication. Unbroken cells were removed by low speed centrifugation. The membranes containing pigment were collected by centrifugation (39000g, 30 min) and solubilized in a wash buffer (50 mM KPi, 300 mM NaCl, 5 mM imidazole and 1.5% octylglucoside (OG), pH 7.0) for at least 1 h at 4 °C. Unsolubilized membranes were removed by centrifugation at 28,000g for 30 min. The supernatant was incubated with a Hisbinding resin on a shaker at 4 °C for at least 1 h. The bound resin was applied to a 10 cm chromatography column and washed with 3× volume of wash buffer followed by elution buffer (50 mM KPi, 300 mM NaCl, 250 mM imidazole and 1.0% OG, pH 7.0). The sample purity was assessed by the UV–visible spectroscopy and SDS-PAGE analysis.1
Proteoliposome Reconstitution
Purified His-tagged PR was reconstituted in E. coli polar lipids (Avanti, Alabaster AL) at a 1:10 protein-to-lipid (w/w) ratio. Lipids initially dissolved in chloroform were dried under argon and resuspended in the dialysis buffer (50 mM K phosphate, 300 mM NaCl, pH 7.0) to which OG was added to a final concentration of 1%. The lipid solution was incubated with the OG-solubilized protein for 1 h on ice and dialyzed against the dialysis buffer with 3 buffer changes every 24 h. The reconstituted proteoliposomes were pelleted by centrifugation and resuspended in the sample buffer (50 mM CHES, 150 mM NaCl, pH 9.5).
Resonance Raman Spectroscopy
Approximately 50 μ g of protein was deposited on a quartz window and dried under a stream of Argon. The films were then rehydrated via the vapor phase and sealed using a second window. Raman spectra were obtained on a Bruker Senterra confocal Raman microscope using 785 nm laser excitation at room temperature. A laser power of 100 mW was used with 300 s of integration time. The Raman scattered light was dispersed onto a CCD (Andor, iDus, South Windsor CT) at a 3–5 cm−1 resolution. A background of buffer and CaF2 was subtracted from each spectrum measured.
Samples were left for 24 h in a darkened room to test for evidence of dark adaptation. A 300 s dark spectrum was acquired, followed by illumination with the halogen lamp for >5 min. A second 300 s spectrum was acquired immediately afterward and subtracted.
3. Results
GPR and BPR Have Very Similar Chromophore Structures
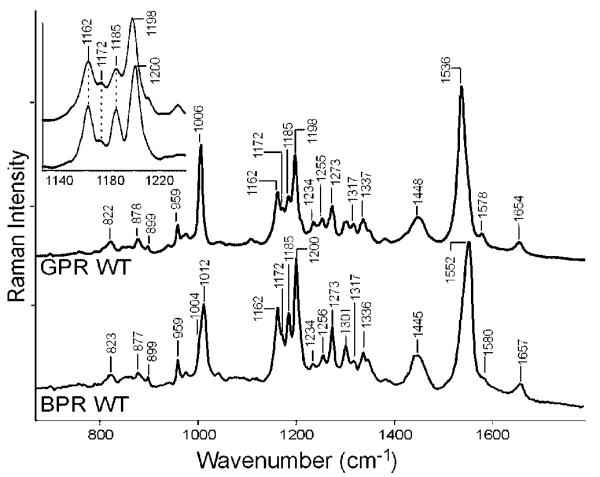
Figure 1 compares GPR WT and BPR WT spectra at pH 9.5. The resonance Raman spectrum (532 nm excitation)28 and FT Raman spectrum (1064 nm excitation)8 have previously been reported only for GPR and at lower resolution. The GPR spectrum in Figure 1 agrees with the earlier reported spectra, although as expected the intensity of some bands are different due to the dependence of Raman intensity on exciting wavelength. The most intense band is the ethylenic stretch vibration at 1536 cm−1 in GPR and 1552 cm−1 in BPR. The upshift of 16 cm−1 of the major ethylenic mode is expected on the basis of an empirical linear correlation between the visible absorption wavelengths and the ethylenic stretch frequency observed for other rhodopsins.29
Figure 1.
RRS of the green (GPR) and blue (BPR) blue absorbing proteorhodopsins. Data was taken using a 785 nm probe laser for 300 s. A background spectrum of the quartz coverslip and buffer was subtracted from the sample data. The spectra were scaled using the intensity of the ethylenic peak at 1536/1552 cm−1.
Bands between 1150 and 1300 cm−1 which include C–C single bond stretching modes of the retinal are sensitive to the isomeric configurations of the retinal chromophore.30 The BPR spectrum has bands at 1162, 1172, 1185, and 1200 cm−1 which correspond almost exactly in frequency and intensity to bands in GPR at 1162, 1172, 1185, and 1198 cm−1 indicating that the retinal conformation of the two species is almost identical. Earlier studies have identified these bands as characteristic of the all-trans configuration of retinal.30,31 For example, in bacteriorhodopsin, bands appear in the all-trans state in this region at 1169, 1184, and 1201 cm−1.30 In BR, a partial switch occurs thermally to a 13-cis configuration in the dark which is evident by changes in the fingerprint region and also the appearance of bands at 800 and 1348 cm−1.30 However in the case of GPR and BPR, no evidence was found of dark adaptation after leaving the sample in the dark for 24 h.
Bands from 1000–800 cm−1 include the hydrogen-out-of-plane (HOOP) modes of the chromophore which are also sensitive to chromophore conformation. HOOP modes are expected to have little intensity for planar polyenes,29,32 but twists around single and double bonds which create a nonplanar configurations produce intensification of these modes in the RRS.32-34 In contrast to previous studies8,28 which only noted a band at 959 cm−1, several bands were observed in this region which are candidates for HOOP modes based on their similarity with the BR spectrum.30 The largest peaks in this region appear at 959, 899, 878, and 822 cm−1 in both GPR and BPR with similar intensity indicating that the out-of-plane structure of retinal polyene due to torsions around single and double bounds is very similar in the unphotolyzed states of GPR and BPR, as well as BR (959, 898, 882, and 830 cm−1).30
Environment of the Schiff base in GPR and BPR is different
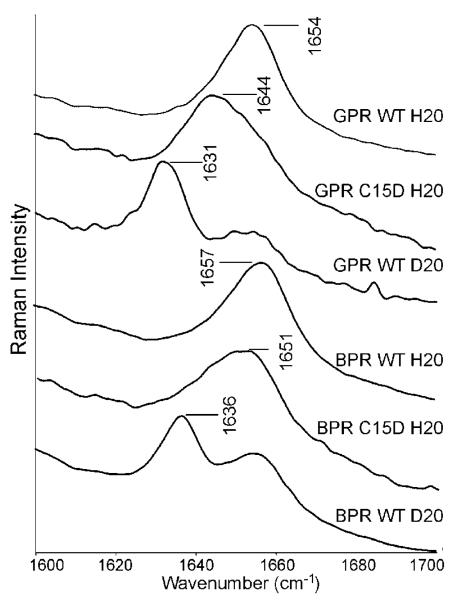
A band near 1640 cm−1 in the RRS of BR was assigned to the C=N stretching vibration of the protonated Schiff base (SB).15 In the case of GPR purified on a phenylsepharose column and solubilized in octylglucoside (OG), a doublet at 1643 and 1656 cm−1 was previously reported28 and attributed to the presence of two different forms of GPR. In the current study where GPR is reconstituted in E. coli polar lipids (see Materials and Methods), only a single band is observed at 1654 cm−1. A single band at a similar, but not identical frequency (1657 cm−1) is found in BPR (see Figure 2). The difference of 3 cm−1 was highly reproducible based on more than 4 independently prepared samples of GPR and BPR.
Figure 2.
The RRS of the region showing the C=N stretch of the Schiff base. The assignment is made on the basis of C15D and D2 O exchange.
In order to confirm that these bands are due to the C=N SB vibration, GPR and BPR were regenerated using a C15-2 H labeled retinal (see Materials and Methods), which is expected to produce a downshift in the C=N stretch frequency. For example in BR this causes a downshift from 1640 to 1629 cm−1 31 and in HR from 1633 to 1627 cm−1.35 As seen in Figure 2, the band in GPR undergoes a similar C15-2H induced shift of 11 cm−1 whereas BPR undergoes a smaller shift of 6 cm−1. There is also an indication that a shoulder still remains in the isotope labeled samples near 1655 cm−1 likely due to a small contribution from the amide I vibration of the protein backbone. This is not unexpected due to the preresonance 785 nm excitation and the fact that the amide I mode of the protein should be relatively intense relative to other protein vibrations.
Hydrogen/deuterium (H/D) exchange was used to probe the hydrogen bonding strength of the SB in GPR and BPR. H/D exchange induces a downshift in C=N frequency which has previously been related to the strength of the SB-counterion hydrogen-bond.15,36 The differences in GPR (23 cm−1) and BPR (21 cm−1) are significantly larger than in BR (16 cm−1) indicating a stronger hydrogen bond strength in both proteor-hodopsins. However, even after repeated exposure of the sample to D2 O a small band remains near 1656 cm−1, again indicating the presence of a contribution at this frequency from the amide I vibration. The differences in both the frequency of the C=N stretching vibration and the H/D induced downshift while small were highly reproducible. Furthermore, as discussed below the differences depend on the identity of the residue 105, thus indicating that this residue largely controls the differences in environment of the retinal Schiff base in GPR and BPR.
Difference Detected in Interaction of Residue 105 with the Retinylidene C13-Methyl Group in BPR and GPR
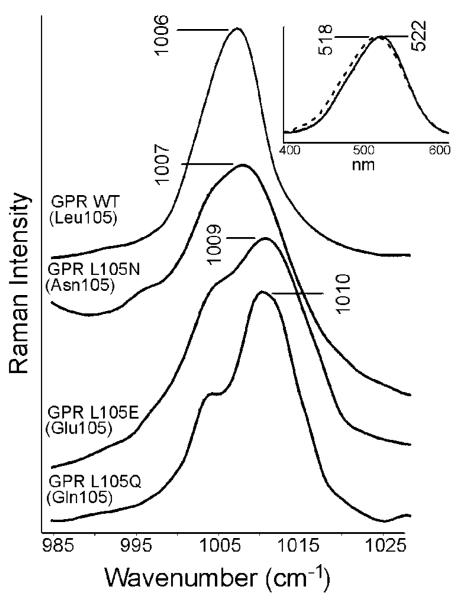
In addition to the ethylenic stretch frequency between GPR and BPR, the most striking difference occurs in the methyl rock region of the spectrum (1000–1020 cm−1). This band in principle could be assigned to either methyl group in the retinylidene chromophore (C13, C9), however based on the evidence presented below, it is most likely assigned predominantly to the C13 methyl rock vibration. GPR has a single band similar in frequency to BR at 1006 cm−1 (Figure 3) in agreement with earlier studies.8,28,37 BPR has a more intense peak which is shifted to 1012 cm−1 with a shoulder near 1004 cm−1 (Figure 4). This frequency is higher compared to the many other microbial rhodopsins studied including BR, SRI, SRII, and HR.17,21,23,24 In the case of BR, the band frequency has been found to shift when Leu93, the homologous residue for Leu105 in GPR, is substituted with an Ala.38 This is not surprising since Leu93 is located close to the C13-methyl group in BR as determined by high resolution crystallographic structures.39 This residue has also been shown to influence the 13-cis to all-trans isomerization in the late N to O transition of the photocycle of BR.38
Figure 3.
The resonance Raman spectra of GPR WT and mutants in the methyl rock region. Inset shows the visible absorption of GPR WT (solid) and GPR L105N (dashed). Even with a polar asparagine at residue 105, it still shows the GPR WT phenotype.
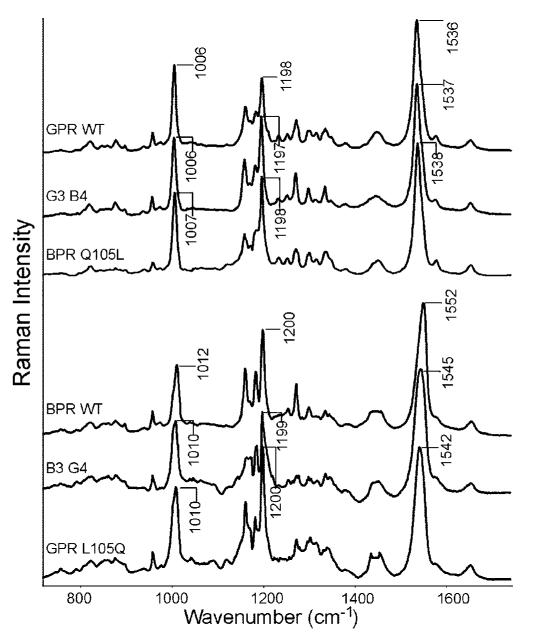
Figure 4.
Top three plots: RRS spectra of proteorhodopsins with leucine at position 105, GPR WT, G3B4 chimera, and BPR Q105L. Bottom three plots: RRS spectra of proteorhodopsins with glutamine at residue 105; BPR WT, B3G4 chimera, GPR Q105L. All data were scaled to the ethylenic peaks.
In order to determine if residue 105 in GPR and BPR also interact with the C13-methyl group in retinal the RRS of several mutants were measured. The GPR L105Q and L105E mutants shift the methyl rock band to 1010 cm−1, very similar to BPR (Figure 3). The reciprocal mutation in BPR (Q105L) shows the ethylenic C=C stretch and the SB C=N stretch frequencies of GPR and BPR are switched in the mutants GPR L105Q and BPR Q105L, respectively (Figure 4). For example, the bands at 1655, 1538, and 1006 cm−1 in BPR Q105L match almost exactly the bands in GPR WT. Note that in the GPR L105Q mutant the ethylenic band at 1542 cm−1 is not fully restored to the frequency in BPR in agreement with the partial shift of the visible maximum from 523 to 498 nm (not the full blue-shifted value observed in BPR of 488 nm).13
A mutant replacing leucine 105 with a polar asparagine exhibits a methyl rock band at 1007 cm−1, very close to the GPR WT frequency of 1006 cm−1. This residue is smaller than glutamine by a single carbon, yet it has almost no effect on color tuning (see inset to Figure 3). This shows C13 methyl interaction with values resembling GPR WT. This shows that the distance of residue 105 to the retinal is of crucial importance in determining the interaction with the SB and C13 methyl group. In agreement, when a glutamic acid residue replaces Gln 105, the methyl stretch bands appear at 1009 and 1004 cm−1, close to WT BPR. Due to the similarity in frequency, the carboxyl group is most likely neutral, even at pH 9.5. The side chain is the same length as a glutamine and therefore in a position to interact with both the SB and the C13 methyl group.
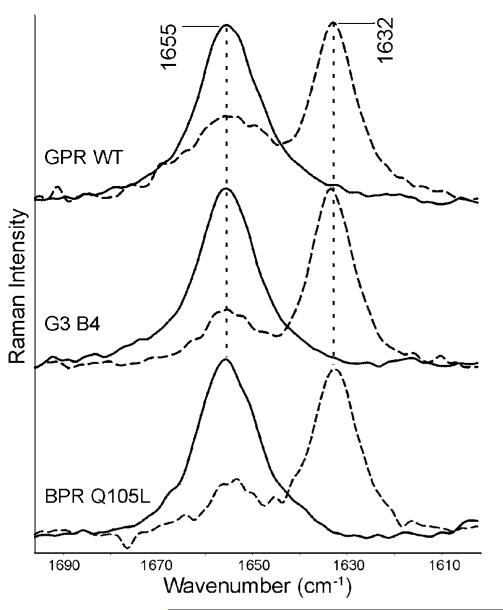
To test the hypothesis that the residue at position 105 is substantially responsible for the differences in the hydrogen bonding strength of the Schiff base in GPR and BPR, H/D exchange was measured for the mutant BPR Q105L. This caused a downshift of the 1655 cm−1 band by 23 cm−1 to 1632 cm−1 (Figure 5), exactly the same as the H/D induced downshift of the Schiff base in GPR WT. The L105Q mutant of GPR showed a reduced shift of 20 cm−1 from 1655 to 1635 cm−1 similar to the H/D induced downshift in BPR WT (Figure 6).
Figure 5.
Resonance Raman spectra of the C=N vibration of the Schiff base for the GPR WT, G3B4 chimera, and BPR Q105L mutant all with a leucine at residue 105. The H2 O spectrum (solid) is compared to the D2 O spectrum (dashed) to reveal the H-bond strength of the counterion.
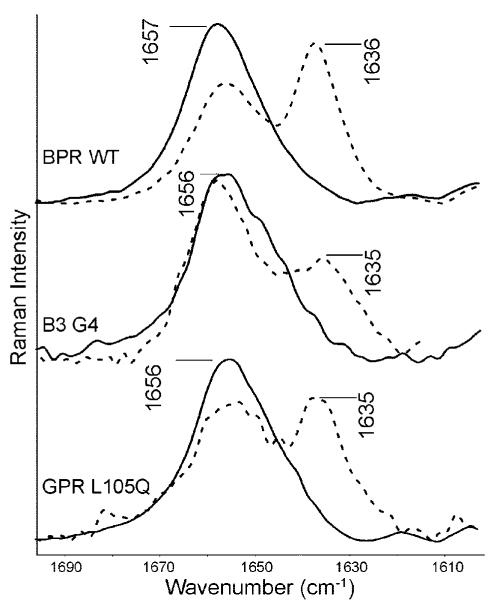
Figure 6.
Resonance Raman spectra of the C=N vibration of the Schiff base for the BPR WT, B3G4 chimera, and GPR L105Q mutant all with a glutamine at residue 105. The H2 O spectrum (solid) is compared to the D2 O spectrum (dashed).
Substitution of the BPR Sequence in GPR after Helices A–C Does Not Affect Its Chromophore Vibrational Spectrum
In order to test whether residues other than at position 105 account for the observed differences in the RRS of GPR and BPR, chimeras of GPR and BPR were expressed and measured. One chimera designated G3B4 consisted of the first three helices of GPR and the final four helices of BPR (see Materials and Methods). A second chimera designated B3G4 consisted of the first three helices and BPR and the last four helices of GPR. The G3B4 spectrum shown in Figure 4 has a leucine at residue 105 and is almost identical to the GPR WT spectrum particularly in the methyl rock, ethylenic and Schiff base vibrations. The H/D induced shift of the Schiff base frequency is exactly the same as in GPR WT and BPR Q105L mutant (Figure 5). This result demonstrates that none of the sequence differences in helices D–G between GPR and BPR have an influence on the overall chromophore structure in the unexcited state. This result provides further evidence that the differences observed are attributable to residue 105 and its inferred interaction with the C13 methyl group and Schiff base. In the case of B3G4, the spectrum resembles BPR including an upshift of the methyl-rock mode to 1010 cm−1. However the ethylenic stretch vibration at 1545 cm−1 is not at the same value as BPR indicating that other residues in helices D–G may also play a role in the chromophore environment. In agreement the wavelength of the visible absorption at 496 nm does not fully shift to that of BPR at 488 nm.13 One possibility is that residue Glu 142 on helix D which has recently been determined to exist in a protonated state in GPR but ionized state in BPR, may play a role in this difference.40
4. Discussion
This study compares the resonance Raman spectra of the two most extensively studied representatives of the GPR and BPR classes of proteorhodopsins. These two variants are typical of the two major classes in general and have visible absorption maxima near 532 and 488 nm, respectively. The visible wavelength maxima are believed to have evolved to match the spectral environment of the different marine photic zones which these variants occupy.2-12 Although both GPR and BPR are capable of pumping protons using solar energy,2 BPR is exposed to a much lower photon flux at 75 m depth compared to the more shallow waters where GPR exists. At least for the GPR and BPR variants thus far characterized BPR exhibits a much slower photocycle compared to GPR (200 ms vs 20 ms).10 This difference has led to the suggestion that the primary function of BPR may not be the utilization of solar energy to power cellular processes as in the case for GPR10,41 but rather a photoregulatory function. Although the visible absorption properties of BPR and GPR have been modeled using the high resolution structures of BR and SRII,41,42 a high resolution crystallographic structure for any PR has not yet been elucidated. Hence, an atomic structural basis for the different properties of BPR and GPR is currently unknown.
A key observation is that switching residue 105 in GPR (Leu) to BPR (Gln) reverses the visible absorption maximum and photocycle time back to the BPR phenotype.10,13 One proposed model suggests that spectral tuning may derive from the altered hydrogen bonding of the chromophore charge complex when polar side groups are nearby.13 The homologous residue in mouse cone rhodopsins causes a similar effect.43 The RRS data of the residue 105 mutants confirms that the hydrophobicity and length of the side chain will directly change the environment of the SB.
It is noted that the RRS measurements were performed at pH 9.5. The generally accepted typical value for the pH of seawater is 8.3. The pKa of the Schiff base proton acceptor in proteorhodopsins (Asp 97) is approximately 7.5,10,40 and an ionized acceptor Asp is required for their proton transport activity. The presence of the nonfunctional acid form (e.g., which is substantial at pH 8.3) complicates molecular spectroscopic analysis by creating a subpopulation of molecules with an altered nonproductive photocycle with different photointer-mediates.44-46 Therefore, to have a homogeneous population of functional pumping forms of BPR, it is necessary to make measurements at pH well above the Asp pKa. Studies of proteorhodopsins are generally made at alkaline pH values; we choose 2 pH units above the Asp pKa to ensure that >99% of the molecules are functionally active in proton transport. This pH is consistent with many other published papers on proteor-hodopsin including studies using resonance Raman spectroscopy (pH 9.5),28 FTIR (pH 9.5),47 (pH 9.2–10)8 and (pH 8.5),9 ultrafast visible and IR (pH 9.5)48,49 and NMR and EM (pH 10).50
Our measurements reveal that the chromophore conformation of GPR and BPR are very similar. For example, differences in the retinal structure in the polyene chain would be expected to result in significant changes in frequencies and intensities of the C–C stretching and HOOP modes of the chromophore are not observed. On the other hand, the observed changes in the ethylenic stretching frequency between BPR and GPR can occur without a significant change in chromophore structure such as predicted by the point charge model of retinal chromophore color tuning.51 In this model, polar and charged groups near the SB cause alterations in the bond conjugation along the polyene chain thereby leading to changes in the absorption wavelengths.
Despite the strong similarity of the GPR and BPR RRS, one major difference which cannot be accounted for as a direct consequence of alteration in the polyene bond conjugation is the shift in the methyl-rock frequency from 1006 cm−1 in GPR to 1012 cm−1 in BPR. In particular, normal mode calculations reflect only pure methyl rock vibrations in this region.30,52 Instead, the changes observed most likely reflect real differences in the interaction of this C13-methyl group with the residue at position 105. In particular, our evidence shows that the methyl-rock shift is almost completely reversible depending on the presence of a leucine or glutamine residue in position 105. The wavelength of visible absorption (and correlated ethylenic stretch) is also sensitive to other substitutions. However, replacement of an asparagine for a leucine cause nearly no shift as compared to the GPR WT even though asparagine is nearly as hydrophilic as glutamine.
A second finding is the detection of a shift in the hydrogen bonding strength of the Schiff base depending on the identity of residue 105. Although the difference of the H/D induced downshift is relatively small (~2–3 cm−1) it is consistently reproduced in not only GPR/BPR but mutants of GPR or BPR that switch the visible absorption, e.g. a small H/D induced shift is associated with the BPR blue-shifted visible absorption. This pattern is also preserved in the chimeras G3B4 and B3G4 as discussed above and hence appears to be highly correlated with the change in visible absorption and ethylenic stretch frequency.
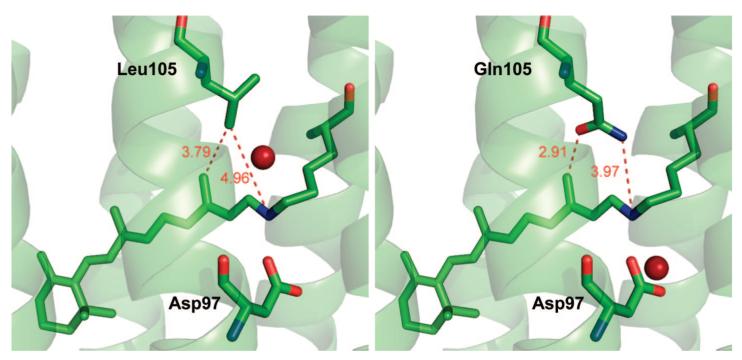
A model that accounts for all of these findings is shown in Figure 7. BPR and GPR are modeled using a high resolution structure of BR39 with the homologous residue Leu93 in BR replaced by Gln or Leu, respectively. In the case of BPR (see Figure 7, right panel), the Gln105 Cγ–Cδ bond has been rotated so that the NH group of the Gln residue interacts directly with the Schiff base with a distance of 3.97 Å while the oxygen atom on the Gln105 carbonyl group is 2.91 Å away from the carbon of the C13 methyl. This model fits the RRS data indicating an interaction of Gln105 with both the methyl group and the SB. In the case of GPR, a water molecule is hypothesized to be present near Leu105 which interacts directly with the Schiff base (see Figure 7, left panel) instead of Gln105. Further evidence for the existence of this model is found from low-temperature FTIR difference measurements on BPR.53
Figure 7.
Models of GPR (left) and BPR (right) based on the BR structure (pdb file 1C3W39). The replacement of leucine with glutamine in the case of BPR shows that Gln105 is in a position to interact both with the Schiff base as well as the C13 methyl group. In the case of GPR, a water molecule has been inserted which is located 2.91 Å away from the Schiff base nitrogen. Distances shown in Å.
Note that in this model, a hydrogen bond is formed with the Schiff base either from the Gln residue (BPR) or from a water molecule in the case of GPR. Hence the environment near the Schiff base is expected to be similar but not exactly the same as deduced by RRS. In addition the interaction with the C13-methyl retinal group is expected to be considerably different since in one case a nonpolar Leu residue is located nearby whereas in the case of BPR the polar C=O group of Gln is present.
5. Conclusions
We have compared the resonance Raman spectra of two forms of proteorhodopsin and found that despite their different absorption and photochemical characteristics, their RR spectra are remarkably close. The spectral similarity shows that the retinal chromophore is in a nearly identical environment in both proteins. However, two differences between GPR and BPR are evident, namely the environments of the C13 methyl group and of the Schiff base. These differences can be explained by a molecular model involving the replacement of Leu105 (GPR) with Gln105 (BPR). This simple substitution has profound effects on spectral tuning and photocycle rate and is possibly related to different functions in the native organisms.
Acknowledgment
We thank Dr. Weiwu Wang for his assistance with plasmid constructions and Dr. Jason Amsden for his helpful comments. This work was supported by National Institutes of Health Grants R01GM069969 (to K.J.R.) and R37GM27750 (to J.L.S.), Department of Energy Grant DE-FG02-07ER15867, and the Robert A. Welch foundation (to J.L.S.).
References and Notes
- (1).Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Science. 2000;289:1902. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- (2).Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Nature. 2001;411:786. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- (3).de la Torre JR, Christianson LM, Beja O, Suzuki MT, Karl DM, Heidelberg J, DeLong EF. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12830. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Science. 2004;304:66. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- (5).Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Spudich JL. Trends Microbiol. 2006;14:480. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- (7).Sabehi G, Beja O, Suzuki MT, Preston CM, DeLong EF. Environ. Microbiol. 2004;6:903. doi: 10.1111/j.1462-2920.2004.00676.x. [DOI] [PubMed] [Google Scholar]
- (8).Dioumaev AK, Brown LS, Shih J, Spudich EN, Spudich JL, Lanyi JK. Biochemistry. 2002;41:5348. doi: 10.1021/bi025563x. [DOI] [PubMed] [Google Scholar]
- (9).Friedrich T, Geibel S, Kalmbach R, Chizhov I, Ataka K, Heberle J, Engelhard M, Bamberg E. J. Mol. Biol. 2002;321:821. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- (10).Wang WW, Sineshchekov OA, Spudich EN, Spudich JL. J. Biol. Chem. 2003;278:33985. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- (11).Gomez-Consarnau L, Gonzalez JM, Coll-Llado M, Gourdon P, Pascher T, Neutze R, Pedros-Alio C, Pinhassi J. Nature. 2007;445:210. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- (12).Bielawski JP, Dunn KA, Sabehi G, Beja O. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14824. doi: 10.1073/pnas.0403999101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Beja O. Embo J. 2003;22:1725. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Mathies R, Freedman TB, Stryer L. J. Mol. Biol. 1977;109:367. doi: 10.1016/s0022-2836(77)80040-5. [DOI] [PubMed] [Google Scholar]
- (15).Lewis A, Spoonhower J, Bogomolni RA, Lozier RH, Stoeckenius W. Proc. Natl. Acad. Sci. U. S. A. 1974;71:4462. doi: 10.1073/pnas.71.11.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mendelsohn R, Verma AL, Bernstein HJ, Kates M. Can. J. Biochem. 1974;52(9):774. doi: 10.1139/o74-110. [DOI] [PubMed] [Google Scholar]
- (17).Campion A, El-Sayed MA, Terner J. Biophys. J. 1977;20(3):369. doi: 10.1016/S0006-3495(77)85555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Argade PVR, Kenneth J. Biochemistry. 1983;22:3460. [Google Scholar]
- (19).Argade PV, Rothschild KJ, Kawamoto AH, Herzfeld J, Herlihy WC. Proc. Natl. Acad. Sci. U.S.A. 1981;78:1643. doi: 10.1073/pnas.78.3.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Smith SO, Lugtenburg J, Mathies RA. J. Membr. Biol. 1985;85:95. doi: 10.1007/BF01871263. [DOI] [PubMed] [Google Scholar]
- (21).Fodor SP, Gebhard R, Lugtenburg J, Bogomolni RA, Mathies RA. J. Biol. Chem. 1989;264:18280. [PubMed] [Google Scholar]
- (22).Dratz EA, Furstenau JE, Lambert CG, Thireault DL, Rarick H, Schepers T, Pakhlevaniants S, Hamm HE. Nature. 1993;363:276. doi: 10.1038/363276a0. [DOI] [PubMed] [Google Scholar]
- (23).Gellini C, Luttenberg B, Sydor J, Engelhard M, Hildebrandt P. FEBS Lett. 2000;472:263. doi: 10.1016/s0014-5793(00)01472-1. [DOI] [PubMed] [Google Scholar]
- (24).Alshuth T, Stockburger M, Hegemann P, Oesterhelt D. FEBS Lett. 1985;179(1) [Google Scholar]
- (25).Bergo VB, Ntefidou M, Trivedi VD, Amsden JJ, Kralj JM, Rothschild KJ, Spudich JL. J. Biol. Chem. 2006;281:15208. doi: 10.1074/jbc.M600033200. [DOI] [PubMed] [Google Scholar]
- (26).Shi L, Yoon SR, Bezerra AG, Jr., Jung KH, Brown LS. J. Mol. Biol. 2006;358:686. doi: 10.1016/j.jmb.2006.02.036. [DOI] [PubMed] [Google Scholar]
- (27).Rath P, Krebs MP, He Y-W, Khorana HG, Rothschild KJ. Biochemistry. 1993;32:2272. doi: 10.1021/bi00060a020. [DOI] [PubMed] [Google Scholar]
- (28).Krebs RA, Dunmire D, Partha R, Braiman MS. J. Phys. Chem. B. 2003;107:7877. [Google Scholar]
- (29).Aton B, Doukas AG, Callender RH, Becher B, Ebrey TG. Biochemistry. 1977;16:2995. doi: 10.1021/bi00632a029. [DOI] [PubMed] [Google Scholar]
- (30).Smith SO, Braiman MS, Myers AB, Pardoen JA, Courtin JML, Winkel C, Lugtenburg J, Mathies RA. J. Am. Chem. Soc. 1987;109:3108. [Google Scholar]
- (31).Smith SO, Myers AB, Mathies RA, Pardoen JA, Winkel C, Van den Berg EMM, Lugtenburg J. Biophys. J. 1985;47(5) doi: 10.1016/S0006-3495(85)83961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Eyring G, Curry B, Mathies R, Fransen R, Palings I, Lugtenburg J. Biochemistry. 1980;19:2410. doi: 10.1021/bi00552a020. [DOI] [PubMed] [Google Scholar]
- (33).Braiman M, Mathies R. Proc. Natl. Acad. Sci. U.S.A. 1982;79:403. doi: 10.1073/pnas.79.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rothschild KJ, Marrero H, Braiman M, Mathies R. Photochem. Photobiol. 1984;40:675. doi: 10.1111/j.1751-1097.1984.tb05359.x. [DOI] [PubMed] [Google Scholar]
- (35).Smith SO, Marvin MJ, Bogomolni RA, Mathies RA. J. Biol. Chem. 1984;259:12326. [PubMed] [Google Scholar]
- (36).Baasov T, Friedman N, Sheves M. Biochemistry. 1987;26:3210. doi: 10.1021/bi00385a041. [DOI] [PubMed] [Google Scholar]
- (37).Bergo V, Amsden JJ, Spudich EN, Spudich JL, Rothschild KJ. Biochemistry. 2004;43:9075. doi: 10.1021/bi0361968. [DOI] [PubMed] [Google Scholar]
- (38).Subramaniam S, Greenhalgh DA, Rath P, Rothschild KJ, Khorana HG. Proc. Natl. Acad. Sci. U.S.A. 1991;88:6873. doi: 10.1073/pnas.88.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. J. Mol. Biol. 1999;291:899. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- (40).Kralj JM, Bergo VB, Amsden JJ, Spudich EN, Spudich JL, Rothschild KJ. Biochemistry. 2008;47:3447. doi: 10.1021/bi7018964. [DOI] [PubMed] [Google Scholar]
- (41).Hillebrecht JR, Galan J, Rangarajan R, Ramos L, McCleary K, Ward DE, Stuart JA, Birge RR. Biochemistry. 2006;45:1579. doi: 10.1021/bi051851s. [DOI] [PubMed] [Google Scholar]
- (42).Rangarajan R, Galan JF, Whited G, Birge RR. Biochemistry. 2007;46:12679. doi: 10.1021/bi700955x. [DOI] [PubMed] [Google Scholar]
- (43).Sun H, Macke JP, Nathans J. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8860. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Huber R, Kohler T, Lenz MO, Bamberg E, Kalmbach R, Engelhard M, Wachtveitl J. Biochemistry. 2005;44:1800. doi: 10.1021/bi048318h. [DOI] [PubMed] [Google Scholar]
- (45).Imasheva ES, Balashov SP, Wang JM, Dioumaev AK, Lanyi JK. Biochemistry. 2004;43:1648. doi: 10.1021/bi0355894. [DOI] [PubMed] [Google Scholar]
- (46).Lakatos M, Lanyi JK, Szakacs J, Varo G. Biophys. J. 2003;84:3252. doi: 10.1016/S0006-3495(03)70049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Krebs RA, Alexiev U, Partha R, DeVita AM, Braiman MS. BMC Physiol. 2002;2:5. doi: 10.1186/1472-6793-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Rupenyan A, van Stokkum IH, Arents JC, van Grondelle R, Hellingwerf K, Groot ML. Biophys. J. 2008;94:4020. doi: 10.1529/biophysj.107.121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Amsden JJ, Kralj JM, Chieffo LR, Wang X, Erramilli S, Spudich EN, Spudich JL, Ziegler LD, Rothschild KJ. J. Phys. Chem. B. 2007;111:11824. doi: 10.1021/jp073490r. [DOI] [PubMed] [Google Scholar]
- (50).Shastri S, Vonck J, Pfleger N, Haase W, Kuehlbrandt W, Glaubitz C. Biochim. Biophys. Acta. 2007;1768:3012. doi: 10.1016/j.bbamem.2007.10.001. [DOI] [PubMed] [Google Scholar]
- (51).Honig B, Dinur U, Nakanishi K, Balogh-Nair V, Gawinowicz MA, Arnaboldi M, Motto MG. J. Am. Chem. Soc. 1979;101:7084. doi: 10.1111/j.1751-1097.1979.tb07745.x. [DOI] [PubMed] [Google Scholar]
- (52).Smith SO, Pardoen JA, Lugtenburg J, Mathies RA. J. Phys. Chem. 1987;91:804. [Google Scholar]
- (53).Amsden JJ, Kralj JM, Bergo VB, Spudich EN, Spudich JL, Ziegler LD, Rothschild KJ. J. Phys. Chem. B. 2007;111:11824. doi: 10.1021/jp073490r. [DOI] [PubMed] [Google Scholar]