Abstract
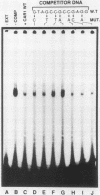
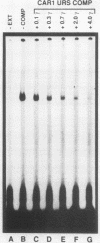
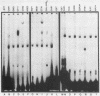
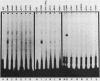
Induction of the arginase (CAR1) gene expression in Saccharomyces cerevisiae has previously been shown to require participation of a cis-dominantly regulated upstream repression sequence (URS). Deletion of this element results in high-level expression of the CAR1 gene without inducer. To determine the structure of the CAR1 URS element, we performed a saturation mutagenesis. Results of the mutagenic analysis indicated that the CAR1 URS was a 9-base-pair palindromic sequence, 5'-AGCCGCCGA-3'. A DNA fragment containing this sequence was shown to bind one or more proteins by a gel shift assay. DNA fragments containing point mutations that completely eliminated URS function were not effective competitors in this assay, whereas those which supported URS function were effective competitors. Sequences in the 5'-flanking regions of 14 other genes were found to be homologous to the CAR1 URS. These sequences were shown to support varying degrees of URS function in the expression vector assay, to bind protein as demonstrated by the gel shift assay, and to compete with a DNA fragment containing the CAR1 URS for protein binding. These results indicate that the CAR1 URS element possesses the characteristics of a repressor binding site. Further, they are consistent with the suggestion that sites homologous to the CAR1 URS may be situated in the 5'-flanking regions of multiple unrelated yeast genes. The widespread occurrence of this element raises the possibility that it is the target site for one or more negatively acting general transcription factors.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cohen R., Yokoi T., Holland J. P., Pepper A. E., Holland M. J. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol. 1987 Aug;7(8):2753–2761. doi: 10.1128/mcb.7.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- MIDDELHOVEN W. J. THE PATHWAY OF ARGININE BREAKDOWN IN SACCHAROMYCES CEREVISIAE. Biochim Biophys Acta. 1964 Dec 9;93:650–652. doi: 10.1016/0304-4165(64)90349-6. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Enzyme repression in the arginine pathway of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1969;35(2):215–226. doi: 10.1007/BF02219132. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. Induction and repression of arginase and ornithine transaminase in baker's yeast. Antonie Van Leeuwenhoek. 1970;36(1):1–19. doi: 10.1007/BF02069003. [DOI] [PubMed] [Google Scholar]
- Middelhoven W. J. The derepression of arginase and of ornithine transaminase in nitrogen-starved baker's yeast. Biochim Biophys Acta. 1968 Mar 11;156(2):440–443. doi: 10.1016/0304-4165(68)90284-5. [DOI] [PubMed] [Google Scholar]
- Rai R., Genbauffe F. S., Sumrada R. A., Cooper T. G. Identification of sequences responsible for transcriptional activation of the allantoate permease gene in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):602–608. doi: 10.1128/mcb.9.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak W., Fessl F., Rytka J., Traczyk A., Skoneczny M., Ruis H. Isolation of the catalase T structural gene of Saccharomyces cerevisiae by functional complementation. Mol Cell Biol. 1983 Sep;3(9):1545–1551. doi: 10.1128/mcb.3.9.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Nucleotide sequence of the Saccharomyces cerevisiae arginase gene (CAR1) and its transcription under various physiological conditions. J Bacteriol. 1984 Dec;160(3):1078–1087. doi: 10.1128/jb.160.3.1078-1087.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Point mutation generates constitutive expression of an inducible eukaryotic gene. Proc Natl Acad Sci U S A. 1985 Feb;82(3):643–647. doi: 10.1073/pnas.82.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Ubiquitous upstream repression sequences control activation of the inducible arginase gene in yeast. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3997–4001. doi: 10.1073/pnas.84.12.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney P. A., Magasanik B. The induction of arginase in Saccharomyces cerevisiae. J Biol Chem. 1973 Sep 10;248(17):6197–6202. [PubMed] [Google Scholar]
- Yoo H. S., Cooper T. G. The DAL7 promoter consists of multiple elements that cooperatively mediate regulation of the gene's expression. Mol Cell Biol. 1989 Aug;9(8):3231–3243. doi: 10.1128/mcb.9.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]